Abstract
Rationale
Obesity affects 40% of U.S. adults, is associated with a proinflammatory state, and presents a significant risk factor for the development of severe coronavirus disease (COVID-19). To date, there is limited information on how obesity might affect immune cell responses in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.
Objectives
To determine the impact of obesity on respiratory tract immunity in COVID-19 across the human lifespan.
Methods
We analyzed single-cell transcriptomes from BAL in three ventilated adult cohorts with (n = 24) or without (n = 9) COVID-19 from nasal immune cells in children with (n = 14) or without (n = 19) COVID-19, and from peripheral blood mononuclear cells in an independent adult COVID-19 cohort (n = 42), comparing obese and nonobese subjects.
Measurements and Main Results
Surprisingly, we found that obese adult subjects had attenuated lung immune or inflammatory responses in SARS-CoV-2 infection, with decreased expression of IFN-α, IFN-γ, and TNF-α (tumor necrosis factor α) response gene signatures in almost all lung epithelial and immune cell subsets, and lower expression of IFNG and TNF in specific lung immune cells. Peripheral blood immune cells in an independent adult cohort showed a similar but less marked reduction in type-I IFN and IFNγ response genes, as well as decreased serum IFNα, in obese patients with SARS-CoV-2. Nasal immune cells from obese children with COVID-19 also showed reduced enrichment of IFN-α and IFN-γ response genes.
Conclusions
These findings show blunted tissue immune responses in obese patients with COVID-19, with implications for treatment stratification, supporting the specific application of inhaled recombinant type-I IFNs in this vulnerable subset.
Keywords: COVID-19, bronchoalveolar lavage, obesity, single-cell RNA sequencing, type-I interferon
At a Glance Commentary
Scientific Knowledge on the Subject
It is known that obese people are at greater risk of developing severe coronavirus disease (COVID-19), with consequent risks of needing mechanical ventilation and subsequent death. However, the mechanisms that underpin these increased risks are not well described.
What This Study Adds to the Field
This study identified attenuated immune responses in immune cells from the respiratory tract and peripheral blood of obese patients with COVID-19 compared with nonobese patients. Reduced type-I and type-2 IFN responses were identified in all compartments and across multiple immune cell types in which reduced TNF-α (tumor necrosis factor α) signaling was identified in alveolar but not peripheral blood cells. These data provide important insights into mechanisms of severe COVID-19 disease in obesity and suggest potential therapeutic approaches.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus responsible for the current global pandemic, with more than 551 million cases and 6.3 million deaths confirmed worldwide (World Health Organization, July 8, 2022; https://covid19.who.int/). The clinical course of SARS-CoV-2 is variable, ranging from asymptomatic disease to acute respiratory distress syndrome requiring ventilatory support (1). Those at risk of a more severe clinical course after infection include the elderly, immunosuppressed, and those with comorbidities, including obesity (1–4). Despite the expedited delivery of clinically validated SARS-CoV-2 vaccines, some “vulnerable” groups mount poor vaccine responses, and emerging viral variants are poorly neutralized by vaccine-induced antibodies (5). Thus, there is an ongoing need to better understand viral immune responses in those at the highest risk of severe disease, such as obese individuals.
Obesity, defined as a body mass index (BMI) greater than 30 kg/m2, is common and affects more than 40% of adults in the United States (6). Although an increased BMI generates mechanical factors that may compromise ventilation (7), obesity is also associated with an inflammatory state characterized by elevated circulating cytokines such as TNF (tumor necrosis factor) and IL-6 (8), chemokines, including the neutrophil recruiting chemokine CXCL8 (9), and the monocyte chemoattractant CCL2 (MCP1) that mediates the accumulation of inflammatory adipose tissue macrophages (10). Lymphocyte abnormalities have also been noted in obesity, and an increased frequency of circulating IFN-γ–secreting CD4 T cells was noted, the latter likely related to the known effects of leptin in promoting T helper type 1 (Th1) cell polarization (11). Indeed, the appetite-regulating hormone leptin, produced by adipocytes and increased in obesity, has several direct immune stimulatory effects, promoting natural killer (NK) cell cytotoxicity, antigen presentation by dendritic cells (DCs), and monocyte and B-cell secretion of TNF and IL-6 by engagement of the leptin receptor, which is expressed on many immune cells, and principally acts by triggering JAK2/STAT3 signaling (12). Leptin concentrations increase in lean animals challenged with proinflammatory cytokines, infectious agents, or pathogen-derived molecules (13), and in coronavirus disease (COVID-19), elevated serum leptin concentrations have been described in ventilated patients with SARS-CoV-2 compared with control subjects (14), with a positive correlation with BMI (15).
There has been an intense focus on delineating the nature of the immune response in SARS-CoV-2 infection; type-I IFN responses play a critical role in protective immunity, with genetic deficiency or neutralizing autoantibodies affecting this axis mediating increased susceptibility to severe infection (16). Transcriptomic studies have identified both absent/low and increased IFN response genes in patients with severe or lethal disease, and longitudinal studies revealed a diminished and/or delayed induction of type-I IFNs in patients with COVID-19 compared with patients with influenza, with an exuberant early TNF/IL-6 response (16). To date, the effect of obesity on immune responses to SARS-CoV-2, particularly tissue responses, has not been considered, although it has been proposed that the elevated leptin associated with obesity might promote an excessive inflammatory response, contributing to the worse outcomes observed in obese patients with COVID-19, with calls for antiinflammatory therapeutic strategies to be employed in this patient group (17). Here we sought to determine whether and how respiratory tract immunity differed in obese compared with nonobese patients with SARS-CoV-2 infection. To address this question, we assessed single-cell transcriptomes in BAL from adult subjects ventilated in an ICU context, pediatric nasal brushings, and peripheral blood mononuclear cells in adult patients with mild to moderate disease.
Methods
For detailed methods, see the online supplement.
Ethical Approvals
The BAL samples from UCAM (University of Cambridge) were collected under our discard lavage protocol. The use of discard samples surplus to that required for clinical testing and anonymized data review were conducted under the consent waiver granted by Leeds West NHS (National Health Service) Research Ethics Committee (ref: 20/YH/0152). For blood samples, ethical approval was obtained from the East of England—Cambridge Central Research Ethics Committee (“NIHR (National Institute for Health and Care Research) BioResource” REC ref 17/EE/0025, and GANDALF [Genetic variation AND Altered Leukocyte Function in health and disease] REC ref 08/H0308/176). All participants provided written informed consent or assent from a personal consultee in which the participant lacked capacity.
BAL
We profiled paired blood and BAL samples from four patients with severe COVID-19 requiring mechanical ventilation and intensive care treatment and four control ventilated patients without COVID-19 with bacterial pneumonia using flow cytometry and single-cell RNA sequencing (scRNAseq). To address the issue of whether patients with a high BMI have abnormal tissue immune responses to SARS-CoV-2, we integrated our scRNAseq data from UCAM with two previous COVID-19 BAL scRNAseq datasets from SZH (Shenzhen Third Hospital) and NU (Northwestern University) (18, 19) and obtained the BMI metadata associated with these samples, enabling a comparison of BAL immune cells in 14 obese (Ob) patients (BMI greater than 30) and 19 nonobese (N-Ob) (BMI less than 30) patients with COVID-19 and ventilated control subjects without COVID-19 (Figures 1A and E1A in the online supplement).
Figure 1.
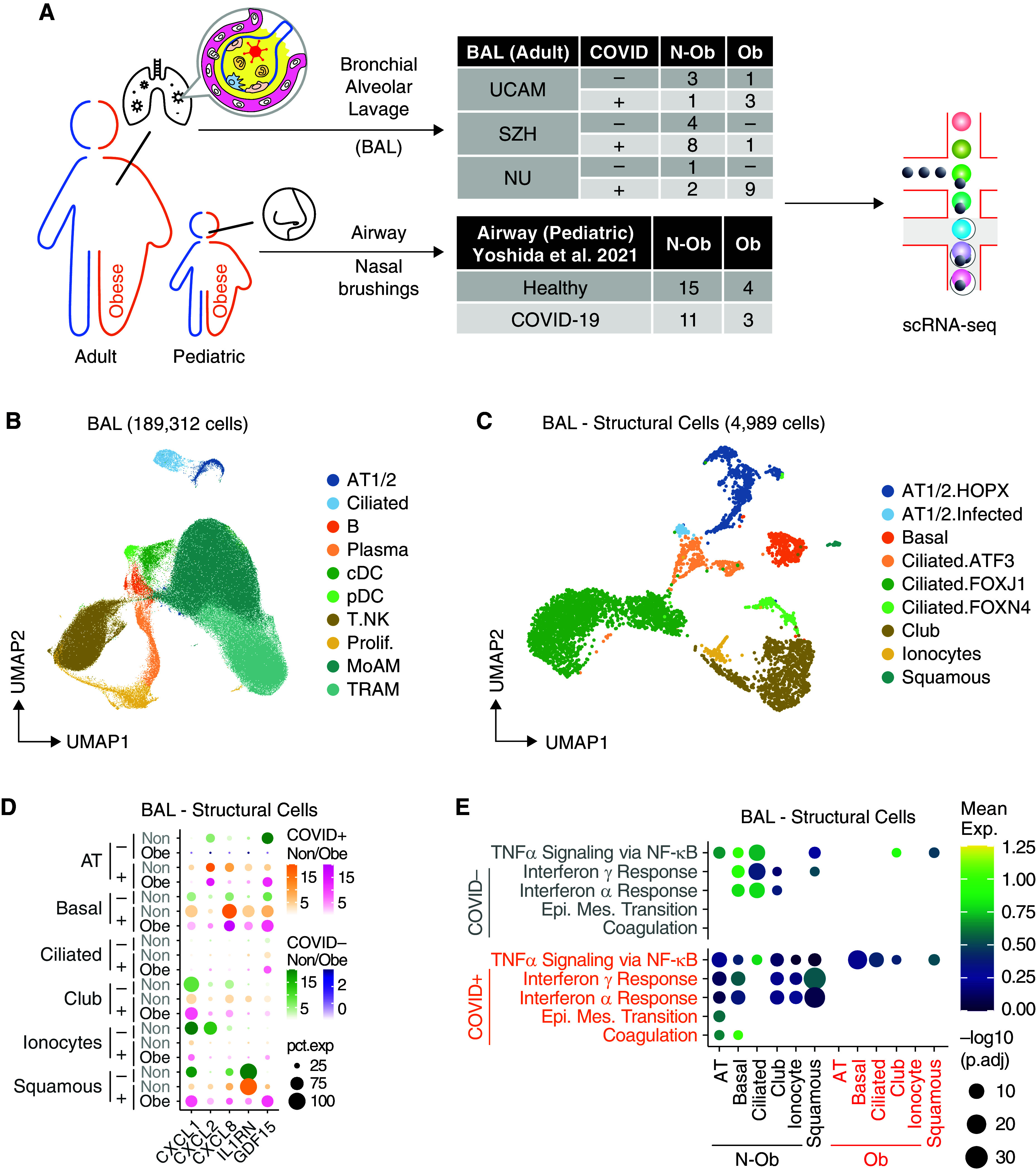
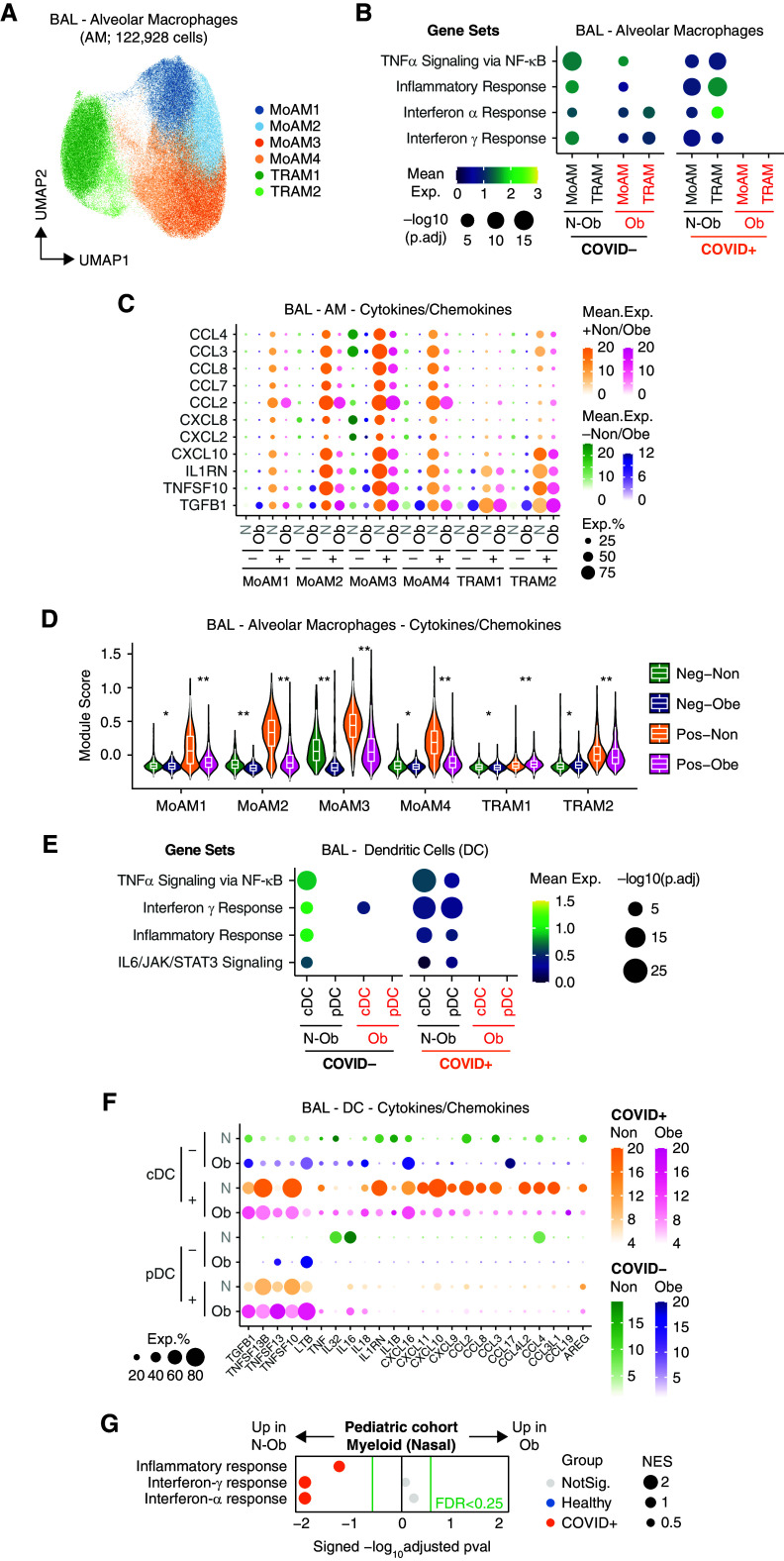
Single-cell analysis of BAL fluid samples from patients with or without coronavirus disease (COVID-19) reveals differences in gene set enrichment in structural cells in nonobese compared with obese subjects. (A) Overview of the workflow. In this study, we included BAL samples from 33 patients from three cohorts with BMI information, namely UCAM (University of Cambridge) (n = 8; ob = 3; this study), SZH (Shenzhen Third Hospital) (n = 13; ob = 1; Liao and colleagues [18]), and NU (Northwestern University) (n = 12; ob = 9; Grant and colleagues [19]). BMI data on pediatric subjects with airway sampling was obtained from Yoshida and colleagues (20). (B) Uniform manifold approximation and projection (UMAP) embedding of 189,312 cells after integration of the three datasets. Cells are colored according to harmonized broad cell type annotations. (C) UMAP embedding of 4,989 epithelial and structural cells after integration colored according to harmonized fine cell type annotations. (D) Mean expression dot plot of the top differentially expressed cytokines and chemokines in each epithelial subpopulation in the COVID+ BAL samples. Expression amounts in each case are indicated by distinct color gradients (green: nonobese without COVID-19; yellow: nonobese with COVID-19; purple: obese without COVID-19; and magenta: obese with COVID-19). Expression percentages are indicated by dot sizes. Basal, ciliated, and club cells not identified in obese without COVID-19 samples are not shown. (E) Mean expression dot plot of five most enriched immune pathways within Hallmark gene sets for epithelial/structural cells. Mean expression of genes contained in each gene set within each cell type, separated into nonobese versus obese groups, are indicated by color gradients. P values are indicated by dot sizes. AT = alveolar type; BMI = body mass index; Epi. Mes. = epithelial mesenchymal; exp = expression; N-ob = nonobese; ob = obese; pct.exp = expression percentage; Prolif. = proliferating; scRNAseq = single-cell RNA sequencing.
All patients were sampled before June 2020, and therefore were likely infected with the Wuhan SARS-CoV-2 variant.
Pediatric Nasal Tissue
For pediatric nasal brushings, BMI data were obtained (when possible) on patients included in a United Kingdom COVID-19 study (20). Details of consent and methodology can be found in the online supplement.
Isolation of the Cells from BAL and Peripheral Blood Mononuclear Cells (PBMCs)
Samples of 5–20 ml BAL were collected, filtered through a 100 μm cell strainer, topped up with phosphate-buffered saline (PBS) to 50 ml, and centrifuged (400 × g for 5 min). The cell pellet was resuspended in 100 μl of PBS. Twenty μl of Human FcR block was added (Miltenyi), followed by 10 μl of a custom TotalSeq-C Human cocktail. Cells were stained for 30 minutes, topped up with PBS, and washed as before. PBMC and granulocytes were isolated by density gradient centrifugation in Ficoll at 800 × g for 15 minutes, after which cells were washed in PBS at 300 × g for 10 minutes. Cells were counted, and 1–3 × 106 cells from each of the PBMC and granulocyte layers were stained for flow cytometry.
Single-cell RNA Sequencing
Cells were resuspended in 50 μl, counted, and loaded on Chromium Chip A (10x Genomics, 5′v3) for cell encapsulation. Complementary DNA libraries were prepared per the manufacturer’s recommendation. After quality checks with Bioanalyzer (Agilent, 2100), the libraries were pooled and sequenced with NovaSeq 6000.
Data Analysis
Flow cytometry samples were processed on a Fortessa flow cytometer (Becton Dickinson), and data were analyzed using FlowJo version 10. Single-cell RNAseq data were processed using the Cell Ranger 3.1.0 pipeline (10x Genomics). The resulting matrix files were then filtered, normalized, and integrated using SoupX (1.5.2), Seurat (4.0.4), and Harmony (1.0) in R (4.0), respectively. Data was visualized by uniform manifold approximation and projection (0.5.2). Public data were acquired from the Gene Expression Omnibus database (BAL; GSE145926, GSE155249, and GSE128033) or from the human cell atlas COVID-19 data portal (PBMC). Differentially expressed genes were identified using FindAllMarkers or FindMarkers function in Seurat with logfc.threshold = 0.5, min.pct = 0.25, and P < 0.05. Pathway analysis was performed using clusterProfiler (3.18.1) and Hallmark gene sets from msigdbr (7.4.1). The cytokine module score was calculated using the AddModuleScore function in Seurat with the cytokine and chemokine gene set from KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway. Ligand–receptor analysis was performed with CellPhoneDB 37 and was visualized with ktplots v1.1.14 using the plot_cpdb function. Plotting was performed using ggplot2 (3.3.5). Heatmap was generated using pheatmap (1.0.12). Figure layouts were edited in Affinity Designer (1.10.0). Unless otherwise specified, statistical analyses were generally performed using base R (4.0) with tidyverse (1.3.0).
Results
Demographic and clinical data, including diagnoses for patients without COVID-19, are set out in Tables E1 and E2.
Attenuated Lung Cell Responses in Obese Patients with COVID-19
For adult BAL, after quality check and integration of our scRNAseq data (UCAM) with BAL scRNAseq datasets from SZH and NU (18, 19), data were available on 189,312 BAL cells from 13 obese (Ob) patients (BMI greater than 30) and 20 nonobese (N-Ob) (BMI less than 30) patients with COVID-19 and ventilated control subjects without COVID-19 (Figures 1A, E1A, and E1B). Overall, Ob patients were sampled at an earlier time point in their ICU stay compared with N-Ob (Figure E1B) and had a lower peripheral white blood cell count (Figure E1C). Clusters were broadly annotated using canonical marker expression and comparison to previously published BAL single-cell datasets (18, 19) to identify alveolar type 1 and 2 pneumocytes (AT1/2; EPCAM and HOPX), ciliated cells (EPCAM and FOXJ1), B (MS4A1 and CD79A) and plasma (IGHM and IGHG1) cells, classical and plasmacytoid dendritic cells (cDCs and pDCs; XBP1 and CLEC9A), a broad T-cell/innate lymphocyte cluster (CD3G and GNLY), and alveolar macrophages, including monocyte-derived (MoAM; CD14 and CD163) and tissue-resident (TRAM; MRC1 and ADGRE1) clusters, with a reasonable representation of cells from Ob and N-Ob patients in all clusters (Figures 1B, E1D, and E1E).
Analysis of the nonimmune cells in isolation enabled the identification of several ciliated epithelial cell subsets, as well as basal cells, club cells, squamous cells, and alveolar type 1 and AT1/2 (Figures 1C and E2A). Organ structural cells may contribute to tissue immune responses, and indeed, several immune-related transcripts were highly expressed in some of these subsets; SARS-CoV-2–infected AT1/2 cells expressed neutrophil recruiting chemokine transcripts (CXCL1, CXCL2, and CXCL8), but this was attenuated in Ob compared with N-Ob subjects (Figure 1D). Consistent with this, flow cytometric analysis of the UCAM cohort BAL confirmed a reduction in neutrophils in Ob BAL compared with N-Ob BAL (Figure E2B). IL1RN (encoding IL1RA, a protein that binds to IL-1R, inhibiting the proinflammatory effects of IL-1β, including arterial inflammation [21]) was highly expressed in squamous epithelial cells in N-Ob, whereas barely detectable in Ob subjects (Figure 1D). Conversely, GDF15, the procachectic cytokine (22), was more highly expressed in Ob subjects across a range of airway cells (Figure 1D). Surprisingly, given the association of obesity with inflammation, there was significantly reduced expression of IFN-α and/or IFN-γ response genes in all nonimmune cell clusters in Ob compared with N-Ob COVID-19+ BAL (p.adj < 10−10), with more variable differences in these genesets in patients who test negative for COVID-19 (Figures 1E and E2F).
Obese Alveolar Macrophages Show Reduced Cytokine and Chemokine Transcripts
Considering alveolar macrophages in isolation, four subsets of MoAMs and two subsets of TRAM1/2 macrophages were evident, as noted previously (18, 19) (Figures 2A and E2C). In Ob subjects with SARS-CoV-2 infection, all alveolar macrophage subsets showed reduced expression of IFN-α and/or IFN-γ response genes, Tnfa via NFkB, inflammatory response, and complement pathway genes, compared with N-Ob COVID-19 (Figures 2B and E2D). Given the differences in the timing of BAL sampling between groups, we plotted individual patient gene pathway enrichment scores against time (Figure E2E). This confirmed that IFN-α and/or IFN-γ response genes were reduced in Ob compared with N-Ob subjects, across time points, and in all AM subsets except TRAM2, which showed only an early attenuation of responses in Ob (Figure E2E). Overall, alveolar macrophages in Ob subjects showed lower expression of several chemokine and cytokine genes compared with N-Ob subjects (Figures 2C and 2D). Similarly, in BAL DCs, there was reduced expression of IFN-α and/or IFN-γ response genes in Ob subjects with COVID-19 (Figure 2E). Genes attenuated in Ob alveolar macrophages and cDCs included CXCL10, a classical IFN-γ response gene previously proposed to form a proinflammatory circuit with T cells (19), and several monocyte- and lymphocyte-recruiting chemokines (Figures 2C and 2F). A notable exception to the muted cytokine and chemokine expression in Ob macrophages, cDCs, and pDCs was TGFB1, a tissue repair factor, which in excess can contribute to fibrosis, which was broadly more highly expressed in Ob cells in COVID-19 (Figures 2C and 2F). To validate these findings and confirm their relevance across the human lifespan, we examined the single-cell transcriptomes of tissue-immune cells isolated from nasal brushings in children with SARS-CoV-2 (20) (Figure E1A). Again, pathway analysis using all transcripts revealed reduced enrichment of IFN-α and/or IFN-γ response genes in myeloid cells in Ob children compared with N-Ob (Figure 2G).
Figure 2.
Single-cell analysis of BAL fluid samples from patients with or without coronavirus disease (COVID-19) reveals differences in gene set enrichment in myeloid cells in nonobese compared with obese subjects. (A) Uniform manifold approximation and projection (UMAP) embedding of 122,928 myeloid cells colored according to fine cell type annotation, as per Grant and colleagues (19). (B) Mean expression dot plot of most enriched immune and metabolic pathways within Hallmark gene sets for each macrophage subpopulation. Mean expression of genes contained in each gene set within each cell type, separated into nonobese versus obese and COVID− versus COVID+ groups, are indicated by color gradients. P values are indicated by dot sizes. (C) Mean expression dot plots of transcripts for the top differentially expressed cytokines and chemokines in each macrophage subpopulation in the BAL samples. Expression amounts in each case are indicated by distinct color gradients (green: nonobese without COVID-19; yellow: nonobese with COVID-19; purple: obese without COVID-19; and magenta: obese with COVID-19). Expression percentages are indicated by dot sizes. (D) Violin plot depicting mean expression amounts of transcripts for cytokines and chemokines in each macrophage subpopulation. Differences between nonobese versus obese with or without COVID-19 infection remain significant by Wilcoxon rank-sum test (*p.adj < 1 × 10−5 and **p.adj < 1 × 10−15). (E) Mean expression dot plot of most enriched immune and metabolic pathways within Hallmark gene sets for classical dendritic cells (cDCs) and plasmacytoid dendritic cells (pDCs). Mean expression of genes contained in each gene set within each cell type, separated into nonobese versus obese and COVID− versus COVID+ groups, are indicated by color gradients. P values are indicated by dot sizes. (F) Mean expression dot plots of transcripts for the top differentially expressed cytokines and chemokines in each DC subpopulation in the BAL samples. Expression amounts in each case are indicated by distinct color gradients (green: nonobese without COVID-19; yellow: nonobese with COVID-19; purple: obese without COVID-19; and magenta: obese with COVID-19). Expression percentages are indicated by dot sizes. (G) Dot plot of gene set enrichment analysis of Hallmark gene sets in myeloid cells from pediatric airway samples (Yoshida and colleagues [20]) between nonobese versus obese children. NES of each pathway is indicated by dot size. The color of the circles indicates which comparison was significantly enriched (healthy or COVID-19); gray circles are not significant. BH FDR 0.25 was used as the cut-off, as indicated in the figure in green text. BH FDR = Benjamini-Hochberg adjusted false discovery rate; exp = expression; MoAM = monocyte-derived macrophage; N-ob = nonobese; Neg = negative; NES = normalized enrichment score; ob = obese; Pos = positive; TNF = tumor necrosis factor; TRAM = tissue-resident macrophage.
Attenuated Tissue Lymphocyte IFN Responses in Obese Patients with COVID-19
We next assessed T cells and innate lymphocytes in adult BAL, annotating naive and effector memory CD4 and CD8 subsets, regulatory T cells, mucosal-associated invariant T (MAIT) cells, NKT cells, and two subsets of NK cells, CD56high and CD56low (Figures 3A and E3A). Again, we found reduced expression of IFN-α and/or IFN-γ response genes and Tnfa signaling via NFkB pathway genes across every subset present in Ob BAL (Figures 3B, E3B, and E3C). In Ob children with SARS-CoV-2, nasal T cells and innate lymphocyte cells also showed reduced enrichment of IFN-α and/or IFN-γ response genes compared with N-Ob (Figures 3C and E3D).
Figure 3.
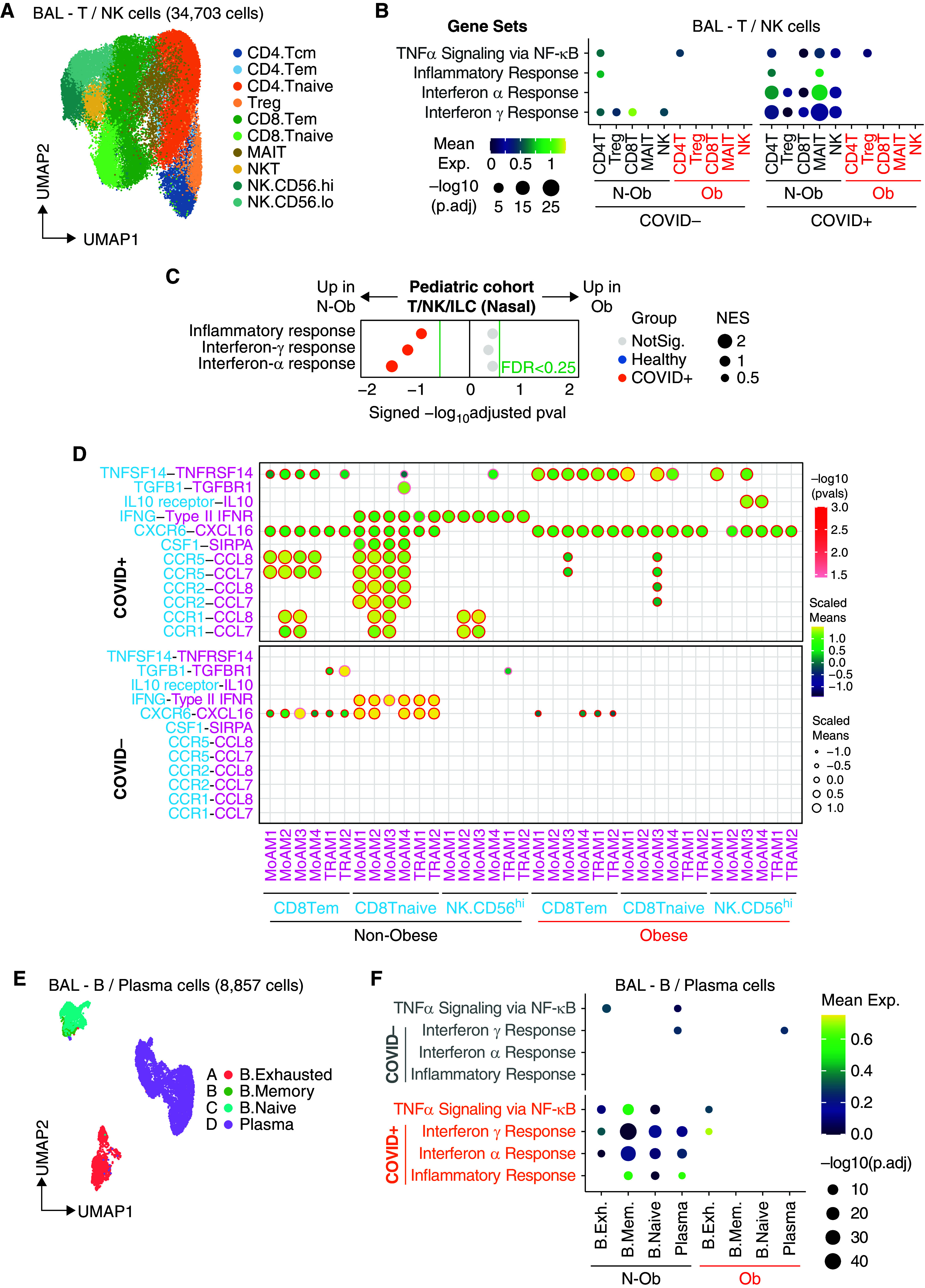
Single-cell analysis of lymphocytes shows reduced enrichment of type-I and γ IFN response genes in obese patients with coronavirus disease (COVID-19). (A) Uniform manifold approximation and projection (UMAP) embedding of 34,703 T/natural killer (NK) cells after integration. CD4 T cells (Tcm [central memory T cells], Tem [effector memory T cells], Tnaive [naive T cells], and Treg [regulatory T cells]), CD8 T cells (Tem, Tnaive, and MAIT), NKT cells, and NK cells (CD56high or CD56low). (B) Dot plot of gene set enrichment analysis of top four most enriched immune pathways within Hallmark gene sets for T/NK cells. Mean expression of genes contained in each gene set within each cell type, separated into nonobese versus obese groups, are indicated by color gradients. P values are indicated by dot sizes. (C) Dot plot of gene set enrichment analysis of Hallmark gene sets in T/NK/ILC cells from pediatric airway samples (Yoshida and colleagues [20]) between nonobese and obese children. NES of each pathway is indicated by dot size. The color of the circles indicates which comparison was significantly enriched (healthy or COVID-19); gray circles are not significant. BH FDR 0.25 was used as the cut-off, as indicated in the figure in green text. (D) Ligand–receptor analysis with CellPhoneDB infers distinct interactions between CD8.Tem/CD8.Tnaive/NK.CD56hi and alveolar macrophages. The size and color gradient of circles indicate the scaled interaction score; interaction scores are scaled row-wise. The red outline indicates P < 0.05. (E) UMAP embedding of 8,857 B/plasma cells after integration colored according to harmonized fine cell type annotations. (F) Dot plot of gene set enrichment analysis of top four most enriched immune pathways within Hallmark gene sets for B/plasma cells. Mean expression of genes contained in each gene set within each cell type, separated into nonobese versus obese groups, are indicated by color gradients. P values are indicated by dot sizes. B.Exh = B. exhausted; B.Mem = B. memory; BH FDR = Benjamini-Hochberg adjusted false discovery rate; ILC = innate lymphoid cells; MAIT = mucosal-associated invariant T cells; N-ob = nonobese; NKT = natural killer T cells; NES = normalized enrichment score; NotSig. = not significant; Ob = obese; TNF = tumor necrosis factor.
In adult BAL, cell–cell interaction analysis on the basis of receptor–ligand expression suggested reduced alveolar macrophage production of chemokines (CCL7 and CCL8) predicted to attract CD8 T and NK cell subsets in Ob subjects (Figure 3D), both important for antiviral immunity. Few predicted interactions were increased in Ob patients, notably IL-10–IL-10R (MoMAC3/4 and NKCD56hi) with the potential to suppress NK cell cytotoxicity (23), and TNFSF14–TNFRSF14 (MoMAC1/3 and naive CD8 T cells) (Figure 3D), encoding Tumor necrosis factor superfamily member 14 - Herpes Virus Entry Mediator, an axis important for stimulating lymphocytes, but high serum LIGHT concentrations have been associated with fatal COVID-19 (24).
B- and plasma cell clusters in adult BAL included naive B cells, memory B cells, exhausted B cells, and plasma cells (Figures 3E, E4A, and E4B). In COVID-19, there was again reduced enrichment of IFN-α and/or IFN-γ response and Tnfa signaling via NFkB pathway genes in all BAL B-cell subsets in Ob subjects, with more variable effects in the pediatric samples (Figures 3F, E4C, and E4D).
To determine if blunted immune responses were evident in Ob subjects beyond tissue immune cells, we obtained BMI data (when available) on an additional cohort of adult patients with COVID-19 recently included in a multiomic analysis of PBMCs (Figure E1A) (25). In this cohort, patient blood was sampled between Days 0 and 20 after symptom onset, with a similar temporal distribution of sampling in Ob and Non-Ob patients (Figure 4A). As observed in adult BAL, there was reduced expression of IFN-α and/or IFN-γ response genes in peripheral blood T cells, innate lymphocytes, and B cells in Ob compared with N-Ob patients with COVID-19 (Figures 4B and 4C). Interestingly, Tnfa signaling via NFkB pathway genes showed the opposite enrichment pattern to that observed in BAL, with an increase in Ob patients with COVID-19 (Figures 4B and 4C).
Figure 4.
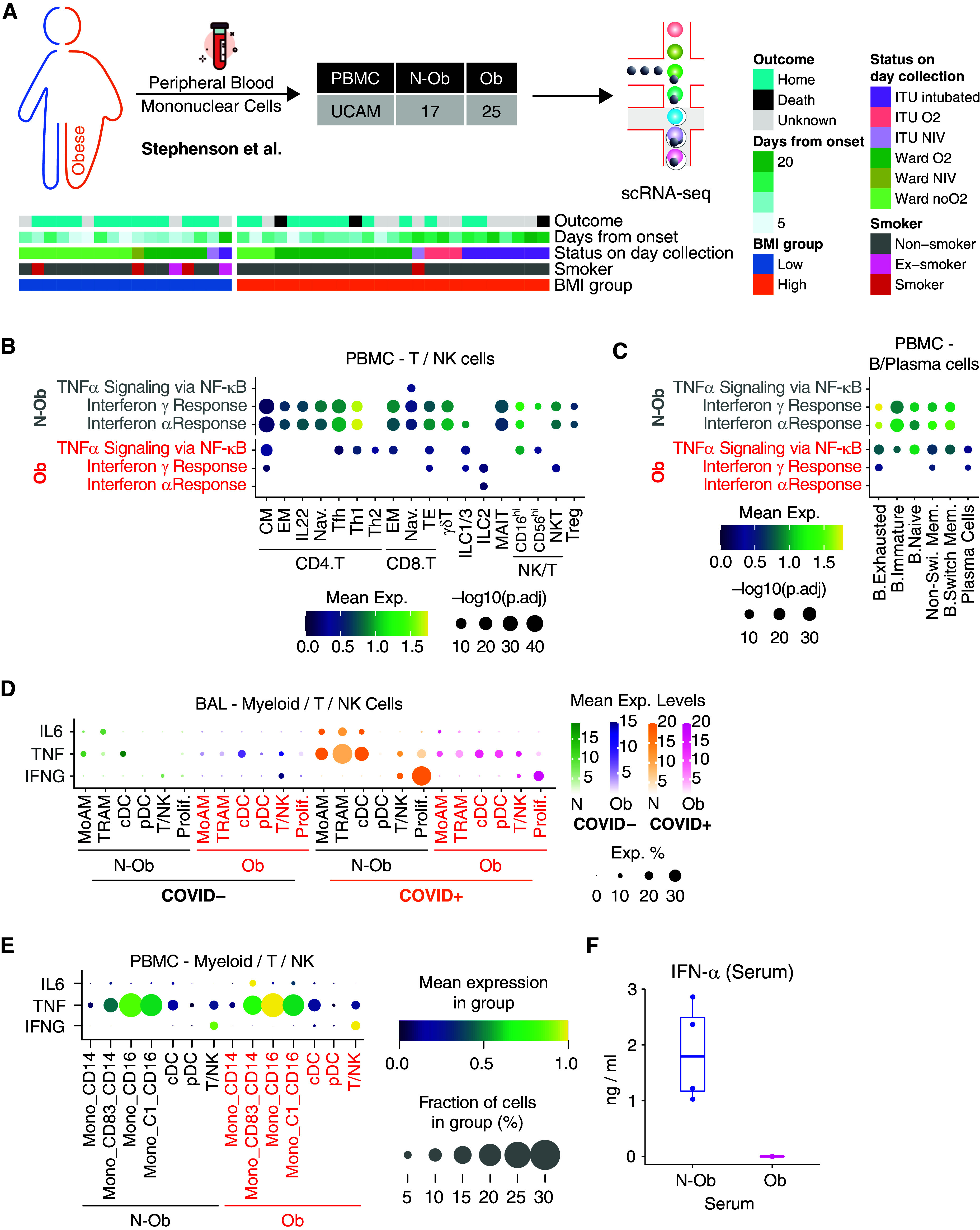
Validation of the signaling pathways in peripheral blood mononuclear cell (PBMC) samples. (A) Sample demographics of Stephenson and colleagues (25) PBMC samples used in this study. (B) Dot plot of gene set enrichment analysis of most enriched immune pathways within Hallmark gene sets for each T/natural killer (NK)/ILC subpopulation in PBMCs. Mean expression of genes contained in each gene set within each cell type, separated into nonobese versus obese groups, are indicated by color gradients. P values are indicated by dot sizes. (C) Dot plot of gene set enrichment analysis of most enriched immune pathways within Hallmark gene sets for each B/plasma subpopulation in PBMCs. Mean expression of genes contained in each gene set within each cell type, separated into nonobese versus obese groups, are indicated by color gradients. P values are indicated by dot sizes. (D) Mean expression dot plots of transcripts for IFN-γ, IL-6, and TNF-α in myeloid, T, and NK cells in the BAL samples. Expression amounts in each case are indicated by distinct color gradients (green: nonobese without COVID-19; yellow: nonobese with COVID-19; purple: obese without COVID-19; and magenta: obese with COVID-19). Expression percentages are indicated by dot sizes. (E) Mean expression dot plots for IL-6, TNF, and IFN-G in PBMC cell clusters from Stephenson and colleagues (25) grouped according to BMI status. The size of circles corresponds to the fraction of cells expressing each gene, and an increasing gradient from purple to yellow corresponds to an increasing mean expression value (standardized to 0–1 per gene). (F) Serum IFN-α measurements from n = 4 obese and n = 4 nonobese patients from the adult PBMC patient cohort (Stephenson and colleagues [25]). All four Obese samples were below detection limits. BMI = body mass index; cDC = classical dendritic cell; CM = central memory; EM = effector memory; ILC = Innate lymphoid cells; MoAM = monocyte-derived macrophage; N-ob = nonobese; Ob = obese; pDC = plasmacytoid dendritic cell; Tfh = T follicular helper; TNF = tumor necrosis factor; TRAM = tissue-resident macrophage.
Reduced Cytokine Production in Obese Patients with COVID-19
We reasoned that the reduced tissue cell responses to IFN-α, IFN-γ, and TNF-α might be because of a decreased production of these cytokines or a reduced ability to respond to them because of decreased receptor expression. To distinguish between these possibilities, we first assessed cytokine transcripts in BAL cells; IFNA1/2 and IFNB transcripts were undetectable, except in less than 0.5% of MoAM3 in N-Ob subjects with COVID-19 (Figure E5). In patients infected with SARS-CoV-2, IFNG transcripts were detectable in proliferating lymphocytes, CD4 central memory T cells, naive CD8 T cells, and NKT cells in BAL, again mainly at a higher concentration in N-Ob compared with Ob patients (Figures 4D and E5). In BAL, TNF transcripts were highest in N-Ob patients with COVID-19+ MoAM3, TRAM2, and cDC (Figures 4D and E5), but in blood, the opposite was observed, with higher TNF expression in some subsets of Ob circulating monocytes (Figures 4E and E6). There was little expression of type-I IFN transcripts in peripheral blood immune cells (Figure E7), but serum cytokine measurement showed undetectable IFN-α concentrations in Ob but elevated concentrations in N-Ob subjects (Figure 4F), with no significant differences in TNF (which was undetectable) and IFN-γ (data not shown). IFNAR1/2 expression was not decreased in Ob versus N-Ob COVID-19 BAL cells, with an increase in IFNAR1 in some alveolar macrophage subsets in Ob patients with COVID-19 (Figures E5 and E7). Altogether, this is consistent with a failure of type-I IFN production rather than a reduced ability to respond to type-I IFNs in Ob patients with COVID-19.
Discussion
Obese patients are known to have basal immune activation and inflammation, in part because of the immunostimulatory effects of elevated leptin. It has been proposed that this contributes to the increased susceptibility to severe SARS-CoV-2 infection and the worse outcomes observed in Ob subjects, generating a heightened proinflammatory cytokine response. In fact, our analysis of Ob adult BAL and Ob pediatric nasal immune cells in COVID-19 suggests that these patients exhibit a broadly immunosuppressed state in tissues compared with nonobese subjects, with reduced type-I IFN and IFN-γ signatures across almost all immune cell subsets, as well as decreased expression of monocyte and neutrophil recruiting chemokines. Of note, our findings bear similarity to studies of obese mice challenged with influenza, which not only had increased mortality but decreased Ifna, Ifnb, and Ifng transcripts in lung tissue, as well as lower concentrations of some chemokines (Ccl2 and Ccl5), compared with lean control subjects, despite a higher viral load (13, 26). In addition, these obese animals also had impaired antigen presentation by DC, decreased IFN-γ production by memory T cells, and reduced NK cell cytotoxicity in this model (27). Interestingly, serum leptin concentrations increased in lean mice during influenza infection, in contrast to obese mice, in which leptin decreased, such that during infection, concentrations were similar to lean mice (13) but with leptin resistance in the latter contributing to attenuated immune cell responses (28). Consistent with this, in COVID-19 BAL, we observed reduced JAK-STAT3 signaling pathway genes in a number of lung immune cell subsets in Ob subjects, particularly in monocyte-derived alveolar macrophages (Figure E2D). Thus, we propose that the attenuated immune responses we observed arise from a relative resistance to the proinflammatory effects of leptin in the context of infection because of chronically elevated concentrations in obesity. The distribution of obesity varied between the BAL cohorts examined (Figure E1B), reflecting the prevalence of this condition from the underlying populations they were drawn. Given the size of the study, we cannot exclude the effect of ethnicity on the effects noted in our study
Notably, although IFN response genes were reduced in Ob subjects in peripheral blood, this phenomenon was much less marked outside of tissues, and there was a disconnect between BAL and blood in terms of TNF response genes, suggesting attenuated tissue responses but a more exuberant, potentially pathogenic systemic proinflammatory landscape, and emphasizing the importance of studies assessing tissue immunity, despite the practical challenges associated.
Conclusions
Overall, our study has important translational implications; current and proposed treatments for severe COVID-19 include antiinflammatory agents such as IL6R blocking antibodies and the application of recombinant IFN-α and IFN-β to promote early antiviral responses, with the latter showing limited efficacy in an early trial (29). However, this study was small and included only patients with mild disease, and there is increasing recognition of the need to tailor the different treatment strategies available to the correct patient group at the correct time (16). The muted responses to type-I IFN and IFN-γ in tissue immune cells in the respiratory tract in Ob patients with COVID-19 revealed by our study supports the application of inhaled recombinant type-I IFNs in this vulnerable subset.
Acknowledgments
Acknowledgment
The authors are grateful to the Evelyn Trust (20/75), Addenbrooke’s Charitable Trust, Cambridge University Hospitals (12/20A), the NIHR Cambridge Biomedical Research Centre, Rosetrees Trust (M944), Action Medical Research (GN2911), and the UKRI/NIHR through the UK Coronavirus Immunology Consortium (UK-CIC) for their financial support.
Footnotes
Supported by the National Institute of Allergy and Infectious Diseases (U19AI35964 [R.G.W. and A.V.M.]); Wellcome Trust (WT211276/Z/18/Z and Sanger core grant WT206194 [K.B.M]); the Chan Zuckerberg Foundation (2017-174169 and 2019-202654 [K.B.M.]); the Rutherford Fund Fellowship allocated by the Medical Research Council and the UK Regenerative Medicine Platforms 2 (MR/5005579/1 [M.Z.N.]); the CL3 for this research was partly funded by the National Institute for Health Research AMR Research Capital Funding Scheme (NIHR200640); the European Union’s Horizon 2020 Framework Program, research and innovation program, under grant agreement No 874656 (K.B.M.); collection of lavage samples in Cambridge was supported by a grant from LifeArc (900244); Wellcome Strategic Scientific award (WT211276/Z/18/Z [G.S.B.]); Medical Research Council Research Project Grant (MR/S035842/1 [Z.K.T. and M.R.C.]); the National Institute of Health Research (NIHR) Blood and Transplant Research Unit in Organ Donation (J.R.F. and M.R.C.); the NIHR Cambridge Biomedical Research Centre (M.M., G.D., and M.R.C.); a Clinician Scientist Fellowship from the Medical Research Council (MR/V006118/1 [A.C.M.]); and by an NIHR Research Professorship RP-2017-08-ST2-002 [M.R.C. and S.A.G.]). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
Author Contributions: M.R.C., A.C.M., and S.A.T. designed the project. A.C.M. and V.N. collected BAL specimens. J.R.F., M.M., G.D., and E.G. processed BAL specimens and performed single-cell RNA-seq. G.S.B. processed blood samples and performed flow cytometry and data analysis. M.L., Z.Z., R.G.W., A.V.M., and C.A.G. provided demographic information for published BAL datasets. M.Y., K.W., M.Z.N., R.G.H.L., and K.B.M. provided pediatric airway data and matched BMI data. H.G., E.S., M.H., P.A.L., and K.G.C.S. provided matched BMI data for published PBMC datasets. S.A.G. and Z.K.T. performed single-cell RNA-seq data analysis. M.R.C. performed data interpretation and wrote the original manuscript draft. S.A.G., G.S.B., and Z.K.T. wrote the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Some of the results of these studies have been previously reported in the form of a preprint (bioRxiv, 18 Jan 2,022 www.biorxiv.org/content/10.1101/2022.01.14.475727v1)
Originally Published in Press as DOI: 10.1164/rccm.202204-0751OC on September 12, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med . 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature . 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tartof SY, Qian L, Hong V, Wei R, Nadjafi RF, Fischer H, et al. Obesity and mortality among patients diagnosed with COVID-19: results from an integrated health care organization. Ann Intern Med . 2020;173:773–781. doi: 10.7326/M20-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson MR, Geleris J, Anderson DR, Zucker J, Nobel YR, Freedberg D, et al. Body mass index and risk for intubation or death in SARS-CoV-2 infection: a retrospective cohort study. Ann Intern Med . 2020;173:782–790. doi: 10.7326/M20-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collier DA, De Marco A, Ferreira IATM, Meng B, Datir RP, Walls AC, et al. CITIID-NIHR BioResource COVID-19 Collaboration COVID-19 Genomics UK (COG-UK) Consortium. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature . 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- 6. Hales CM, Carroll MD, Fryar CD, Ogden CL. Prevalence of obesity and severe obesity among adults: United States, 2017-2018. NCHS Data Brief . 2020:1–8. [PubMed] [Google Scholar]
- 7. Dixon AE, Peters U. The effect of obesity on lung function. Expert Rev Respir Med . 2018;12:755–767. doi: 10.1080/17476348.2018.1506331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab . 2001;280:E745–E751. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 9. Straczkowski M, Dzienis-Straczkowska S, Stêpieñ A, Kowalska I, Szelachowska M, Kinalska I. Plasma interleukin-8 concentrations are increased in obese subjects and related to fat mass and tumor necrosis factor-alpha system. J Clin Endocrinol Metab . 2002;87:4602–4606. doi: 10.1210/jc.2002-020135. [DOI] [PubMed] [Google Scholar]
- 10. Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa K, Kitazawa R, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest . 2006;116:1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature . 1998;394:897–901. doi: 10.1038/29795. [DOI] [PubMed] [Google Scholar]
- 12. Naylor C, Petri WA., Jr Leptin regulation of immune responses. Trends Mol Med . 2016;22:88–98. doi: 10.1016/j.molmed.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 13. Smith AG, Sheridan PA, Harp JB, Beck MA. Diet-induced obese mice have increased mortality and altered immune responses when infected with influenza virus. J Nutr . 2007;137:1236–1243. doi: 10.1093/jn/137.5.1236. [DOI] [PubMed] [Google Scholar]
- 14. van der Voort PHJ, Moser J, Zandstra DF, Muller Kobold AC, Knoester M, Calkhoven CF, et al. Leptin levels in SARS-CoV-2 infection related respiratory failure: a cross-sectional study and a pathophysiological framework on the role of fat tissue. Heliyon . 2020;6:e04696. doi: 10.1016/j.heliyon.2020.e04696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J, Xu Y, Zhang X, Wang S, Peng Z, Guo J, et al. Leptin correlates with monocytes activation and severe condition in COVID-19 patients. J Leukoc Biol . 2021;110:9–20. doi: 10.1002/JLB.5HI1020-704R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim YM, Shin EC. Type I and III interferon responses in SARS-CoV-2 infection. Exp Mol Med . 2021;53:750–760. doi: 10.1038/s12276-021-00592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andrade FB, Gualberto A, Rezende C, Percegoni N, Gameiro J, Hottz ED. The weight of obesity in immunity from influenza to COVID-19. Front Cell Infect Microbiol . 2021;11:638852. doi: 10.3389/fcimb.2021.638852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med . 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 19. Grant RA, Morales-Nebreda L, Markov NS, Swaminathan S, Querrey M, Guzman ER, et al. NU SCRIPT Study Investigators Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature . 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoshida M, et al. Local and systemic responses to SARS-CoV-2 infection in children and adults. Nature . 2022;602:321–327. doi: 10.1038/s41586-021-04345-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicklin MJ, Hughes DE, Barton JL, Ure JM, Duff GW. Arterial inflammation in mice lacking the interleukin 1 receptor antagonist gene. J Exp Med . 2000;191:303–312. doi: 10.1084/jem.191.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnen H, Lin S, Kuffner T, Brown DA, Tsai VW, Bauskin AR, et al. Tumor-induced anorexia and weight loss are mediated by the TGF-beta superfamily cytokine MIC-1. Nat Med . 2007;13:1333–1340. doi: 10.1038/nm1677. [DOI] [PubMed] [Google Scholar]
- 23. Littwitz-Salomon E, Malyshkina A, Schimmer S, Dittmer U. The cytotoxic activity of natural killer cells is suppressed by IL-10+ regulatory T cells during acute retroviral infection. Front Immunol . 2018;9:1947. doi: 10.3389/fimmu.2018.01947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perlin DS, Zafir-Lavie I, Roadcap L, Raines S, Ware CF, Neil GA. Levels of the TNF-related cytokine LIGHT increase in hospitalized COVID-19 patients with cytokine release syndrome and ARDS. MSphere . 2020;5:e00699-20. doi: 10.1128/mSphere.00699-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, et al. Cambridge Institute of Therapeutic Immunology and Infectious Disease-National Institute of Health Research (CITIID-NIHR) COVID-19 BioResource Collaboration Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med . 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Easterbrook JD, Dunfee RL, Schwartzman LM, Jagger BW, Sandouk A, Kash JC, et al. Obese mice have increased morbidity and mortality compared to non-obese mice during infection with the 2009 pandemic H1N1 influenza virus. Influenza Other Respir Viruses . 2011;5:418–425. doi: 10.1111/j.1750-2659.2011.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith AG, Sheridan PA, Tseng RJ, Sheridan JF, Beck MA. Selective impairment in dendritic cell function and altered antigen-specific CD8+ T-cell responses in diet-induced obese mice infected with influenza virus. Immunology . 2009;126:268–279. doi: 10.1111/j.1365-2567.2008.02895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karlsson EA, Sheridan PA, Beck MA. Diet-induced obesity impairs the T cell memory response to influenza virus infection. J Immunol . 2010;184:3127–3133. doi: 10.4049/jimmunol.0903220. [DOI] [PubMed] [Google Scholar]
- 29. Monk PD, Marsden RJ, Tear VJ, Brookes J, Batten TN, Mankowski M, et al. Inhaled Interferon Beta COVID-19 Study Group Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir Med . 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]