Abstract
HOXB13 is a key lineage homeobox transcription factor that plays a critical role in the differentiation of the prostate gland. Several studies have suggested that HOXB13 alterations may be involved in prostate cancer development and progression. Despite its potential biological relevance, little is known about the expression of HOXB13 across the disease spectrum of prostate cancer. To this end, we validated a HOXB13 antibody using genetic controls and investigated HOXB13 protein expression in murine and human developing prostates, localized prostate cancer, and metastatic castration-resistant prostate cancers.
We observed that HOXB13 expression increases during later stages of murine prostate development. All localized prostate cancers showed HOXB13 protein expression. Interestingly, lower HOXB13 expression levels were observed in higher grade tumors, although no significant association between HOXB13 expression and recurrence or disease-specific survival was found. In advanced metastatic prostate cancers, HOXB13 expression was retained in the majority of tumors. While we observed lower levels of HOXB13 protein and mRNA levels was observed in tumors with evidence of lineage plasticity, 84% of androgen receptor-negative castration resistant prostate cancers and neuroendocrine prostate cancers (NEPC) retained detectable levels of HOXB13. Notably, the reduced expression observed in NEPC was associated with a gain of HOXB13 gene body CpG methylation.
In comparison to the commonly used prostate lineage marker NKX3.1, HOXB13 showed greater sensitivity in detecting advanced metastatic prostate cancers. Additionally, in a cohort of 837 patients, 383 with prostatic and 454 with non-prostatic tumors, we found that HOXB13 immunohistochemistry had a 97% sensitivity and 99% specificity for prostatic origin. Taken together, our studies provide valuable insight into the expression pattern of HOXB13 during prostate development and cancer progression. Furthermore, our findings support the utility of HOXB13 as a diagnostic biomarker for prostate cancer, particularly to confirm the prostatic origin of advanced metastatic castration resistant tumors.
INTRODUCTION
Cell lineage is characterized by the expression of cell type-specific transcription factors, which are crucial for orchestrating core developmental programs that define lineage-defining expression and differentiation patterns [1]. Among these, HOX transcription factors play an important role in determining cell fate during development. Each of the 39 HOX genes shows distinct expression patterns and biological functions [1–3]. During development, HOXB13, the most distal of the HOXB cluster, is expressed in the caudal aspect of the embryo, including the hindgut, tail, urogenital sinus and the developing prostate [4–6].
In adult tissues, the highest expression levels of HOXB13 are observed in the prostate, indicating that HOXB13 could be a lineage-restricted transcription factor [7,8]. The discovery of recurrent germline variants of HOXB13, which account for approximately 5% of hereditary prostate cancer in men of European ancestry has amplified the interest in determining the role of HOXB13 in prostate cancer over the past years [9–12].
Although the biological function of HOXB13 is likely complex and currently poorly understood, several studies have shown that HOXB13 is one of the core dependency genes in prostate cancer models, and its depletion results in significant growth suppression [9–11]. Furthermore, earlier mechanistic studies suggested that HOXB13 cooperates with the androgen receptor (AR) and is necessary for establishing canonical AR signaling [11–13]. In addition to the well described AR associated effects, HOXB13 has AR-independent functions in regulating key cellular processes in prostate cancer [14,15].
Despite its functional association with AR signaling, the expression of HOXB13 itself has been suggested to be mostly AR independent, setting it apart from other commonly used prostatic lineage markers (e.g., PSA, NKX3.1) [16–19]. These unique characteristics make HOXB13 a potential valuable diagnostic biomarker for establishing prostatic origin. This is particularly relevant in the context of advanced castration resistant prostate cancers where a subset of tumors (~30%) have been shown to lose AR expression [20,21] and lineage markers that are regulated by AR signaling are no longer contributory [20,21]. Therefore, from a diagnostic perspective, considering the increasing number of metastatic biopsies that are being performed on men that have received AR targeting therapies, there is a need for robust AR-independent prostatic lineage markers.
More broadly, given the crucial role of HOXB13 in prostate cancer, it is essential to better understand its expression during development and tumor progression. Detailed insights in the expression pattern of HOXB13 would be relevant for prostate cancer biomarker development and would also allow to contextualize results from mechanistic studies.
Unfortunately, many of the previous reports that have assessed HOXB13 expression have been constrained by their limited scope, small sample size or utilization of antibodies lacking analytic validation. Importantly, though numerous commercial HOXB13 antibodies are available, many are polyclonal and subject to batch variations making them unsuitable for widespread application. Furthermore, the majority of previously published antibodies have not been formally validated for use in formalin fixed paraffin embedded (FFPE) tissues. It is therefore not surprising that the rate of reported HOXB13 expression in prostate cancer varies greatly [18,22,23].
To systematically assess HOXB13 expression patterns, we validate a novel HOXB13 antibody with high sensitivity and specificity and investigate the expression of HOXB13 across > 2000 tissue specimens spanning early organ development to advanced metastatic cancer. Our data provide a comprehensive compendium of HOXB13 expression across developmental and tumor progression stages, revealing important insights into HOXB13 biology.
MATERIALS AND METHODS
Cell lines
Human cell lines (LNCaP, NCI-H660, and DU145) were obtained from the American Type Culture Collection (ATCC) and were grown in the respective recommended media. All cell lines were maintained at 37°C with 5% CO2. Short tandem repeat genotyping was used to authenticate the lines, and cells were confirmed to be mycoplasma free using the MycoAlert Detection Kit (Lonza, LT07-418). Cells were cultured no longer than 10 passages after thawing and before experimental use.
HOXB13 expression constructs
Lentiviral HOXB13 and the corresponding control expression vectors were purchased from Addgene (Addgene ID: 70089) [11]. For lentiviral packaging, 293T cells were transfected with either pLX302 or pLX302_HOXB13 vectors along with packaging plasmids using calcium phosphate as described previously [24]. Lentiviral particles were concentrated by ultracentrifugation and cells were transduced with a multiplicity of infection (MOI) of 2.
Human samples
Specimens from fetal prostate tissues were obtained from the Birth Defects Research Laboratory at the University of Washington with ethics board approval and maternal written consent. This study was performed in accordance with ethical and legal guidelines of the Fred Hutchinson Cancer Center institutional review board. All other FFPE patient tissue samples were analyzed in tissue microarray (TMA) format. Primary prostate cancer samples came from radical prostatectomy specimens (N=331) and were described previously [24]. Metastatic cancer samples from 52 patients with mCRPC were collected as part of the Prostate Cancer Donor Program at the University of Washington [24,25]. For each patient, tissues specimens from different metastatic sites (median number of sites per patient 7, range 1 to 21) were included [24,25]. In addition, a total of 454 non-prostatic tumor samples representative of urothelial carcinoma of the bladder (N=93), colonic adenocarcinoma (N=100), gastric adenocarcinoma (N=32) [26], a broad spectrum of renal cell carcinomas (N=217), germ cell tumors (N=7), and melanomas (N=5) were included.
Murine tissues and patient-derived xenografts
Timed breeding studies to obtain urogenital sinus (UGS) or prostate tissues from defined post-conception and post-partum timepoints were performed as described previously [27]. Genetically modified mouse models used in this study (PbCre:Rosa26mT/mG Ptenfl/fl [single knock out, SKO], PbCre:Rosa26mT/mG Trp53fl/flRbfl/fl [double knock out, DKO], PbCre:Rosa26mT/mG Ptenfl/flTrp53fl/flRbfl/fl [triple knock out, TKO]) were described previously [28,29]. Prostate tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and used for in situ IHC labeling. Prostate cancer patient-derived xenografts (LuCaP) were established using previously published protocols, and tumor tissues were collected at necropsy for tissue-based studies [30].
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Quantitative real-time PCR was performed as described previously [31]. Total RNA was purified with the RNeasy Minikit (Qiagen). First-strand cDNA was synthesized using random hexamer primers (Applied Biosystems) and Ready-To-Go You-Prime First-Strand Beads (GE Healthcare). Quantitative PCR was performed using TaqMan Universal PCR Master Mix (ThermoFisher) and TaqMan probes specific to mouse Hoxb13 (Mm00433968; ThermoFisher), mouse Ar (Mm00442688; Applied Biosystems), and the reference gene mouse Hprt (Mm03024075; ThermoFisher).
In silico expression analyses
Differential expression analyses of publicly available RNA sequencing (RNA-Seq) data was carried out as described previously [24]. Tumors were classified into AR/NE phenotypic groups and ARG10 AR signaling scores were derived as described previously [20,21,32]. Boxplots of HOXB13 gene expression were created with ggplot2 v3.3.6 and statistical comparisons between groups were assessed by fitting linear regression models with random effects for LuCaP lines or patients to account for repeated sampling. scRNA-seq data were generated and analyzed as described previously [33]
DNA methylation analyses
For whole-genome bisulfite sequencing analyses, DNA was extracted from LuCaP patient-derived xenograft (PDX) lines (77, 78, 93 and 173.1), subjected to bisulfite conversion, and sequenced on an Illumina HiSeq 2500 instrument (Illumina) to an average coverage of 30x. Raw whole-genome bisulfite sequencing (WGBS) reads were first trimmed using Trim Galore [0.6.6] and then aligned to the UCSC hg19 reference genome using Bismark [0.23.0] [34]. Bismark was further used to deduplicate the alignments and extract methylation call files to report the percentage of methylated cytosines and the coverage at each position. Genome scale methylation analyses of 32 LuCaP PDX DNAs were carried out using Infinium MethylationEPIC BeadChip arrays (Illumina) as described previously [35]. Raw data were analyzed in the minfi package in R, and samples were normalized using the subset-quantile within array normalization method [36,37]. Probes with a detection p-value of >0.01 in 50% or more of samples and probes that contained a SNP at the CpG interrogation site or at the single nucleotide extension were removed. DNA methylation pattern at the HOXB13 locus were inspected in the Integrative Genomics Viewer (IGV) and methylation levels of the differentially methylated regions (DMRs) were extracted [38]. Whole-genome bisulfite sequencing (WGBS) and EPIC array data are publicly available on Gene Expression Omnibus (GEO), accession number GSE227853.
Immunohistochemical and immunofluorescence staining
For immunohistochemical staining, slides were deparaffinized and steamed for either 30 min in 10 mM sodium citrate (pH=6, Vector Labs) or for 45 min in Target Retrieval Solution (Dako). Primary antibodies and dilutions used were as follows: HOXB13 (Cell Signaling Technology, 90944S, 1:50), AR (Cell Signaling Technology, 5153T, 1:100), and NKX3.1 (Thermo Fisher, 5082788, 1:50). Immunocomplexes were detected using the UltraVision Quanto Detection System (Thermo Fisher, TL060QHD) for human tissues and PowerVision Poly-HRP anti-Rabbit IHC detection system (Leica Microsystems, PV6119) for mouse tissues with DAB as the chromogen. Tissue sections were counterstained with hematoxylin and slides were digitized on a Ventana DP 200 Slide Scanner (Roche). Immunoreactivity was scored in a blinded manner by two pathologists (M.R., E.S.) whereby the optical density level (“0” for no brown color, “1” for faint and fine brown chromogen deposition, “2” for moderate chromogen deposition, and “3” for prominent chromogen deposition) was multiplied by the percentage of cells at each staining level, resulting in weighted average H-scores that range between 0 and 300 [39]. The final score for each sample was the average of two duplicated tissue cores. For multiplex-immunofluorescence staining, sequential dual IF was done for HOXB13/AR, HOXB13/SYP, and HOXB13/INSM1. Primary antibodies and dilutions used were as follows: HOXB13 (Cell Signaling Technology, 90944S, 1:50), human AR (Cell Signaling Technology, 5153T, 1:100), murine AR (Millipore Sigma, 06-680, 1:300), SYP (Santa Cruz Biotechnology, SC-17750, 1:50), and INSM1 (Santa Cruz Biotechnology, SC-271408, 1:50). Immunocomplexes were detected using the Tyramide Signal Amplification system (Thermo Fisher).
Immunoblots
Cell lysates were prepared in RIPA buffer (Sigma) supplemented with phosphatase and protease inhibitors (Roche) and subsequently separated by SDS-PAGE. Proteins were transferred onto nitrocellulose membranes and probed with the following antibodies at the indicated dilutions at 4°C for 16 hours: HOXB13 (Cell Signaling Technology, 90944) 1:1000, and β-actin (Cell Signaling Technology 4970S) 1:1000. Immunocomplexes were detected using (HRP)-conjugated anti-mouse or anti-rabbit secondary antibody and visualized using a ChemiDoc Imaging System (Bio-Rad).
Statistical analyses
Associations between mean HOXB13 H-scores and univariate clinicopathological variables were evaluated using 2-sample t-tests or linear regressions using R version 4.2.2. Repeated measures from the same patients were accommodated using random effects using R packages lme4 and lmerTest [40–42]. Associations between mean HOXB13 H-scores and time-to-event variables were evaluated using Kaplan-Meier estimation, log-rank tests, and Cox regression using R packages survival and survminer. Follow-up for time-to-event variables was estimated using reverse Kaplan-Meier estimation [43]. Discrimination to identify prostatic origin was characterized by bootstrap resampling from patients with advanced prostate cancer and evaluating robust statistics (median and interquartile range) of sensitivity and specificity across 100 replicates using the R package pROC [44]. P < 0.05 indicated statistical significance.
RESULTS
HOXB13 antibody validation
To address the need for a reliable antibody to evaluate HOXB13 tissue expression, we validated a recently developed rabbit monoclonal antibody (clone D7N8O, Cell Signaling Technologies). We first performed western blot analysis on cell lines and PDX tissues with known HOXB13 expression, which revealed a single band at the predicted molecular weight of HOXB13 (~34 kDa) in LNCaP, LuCaP 35, and LuCaP 78 (Figure 1A). In contrast, DU145 cells, which demonstrated no detectable HOXB13 mRNA expression, showed no reactivity. To further evaluate the specificity of the antibody in FFPE samples, we formalin fixed and paraffin embedded LNCaP, NCI-H660, DU145 LuCaP PDXs [45] and observed strong nuclear immunoreactivity in LNCaP and LuCaP PDX tissues, but no detectable signal in DU145 and NCI-H660 cells (Figure 1B). Importantly, we further demonstrated the specificity of the antibody by transducing HOXB13 negative NCI-H660 and DU145 cells with HOXB13 expression constructs (HOXB13OE), resulting in a specific gain of immunoreactivity (Figure 1B).
FIGURE 1. Validation of a monoclonal HOXB13 antibody.
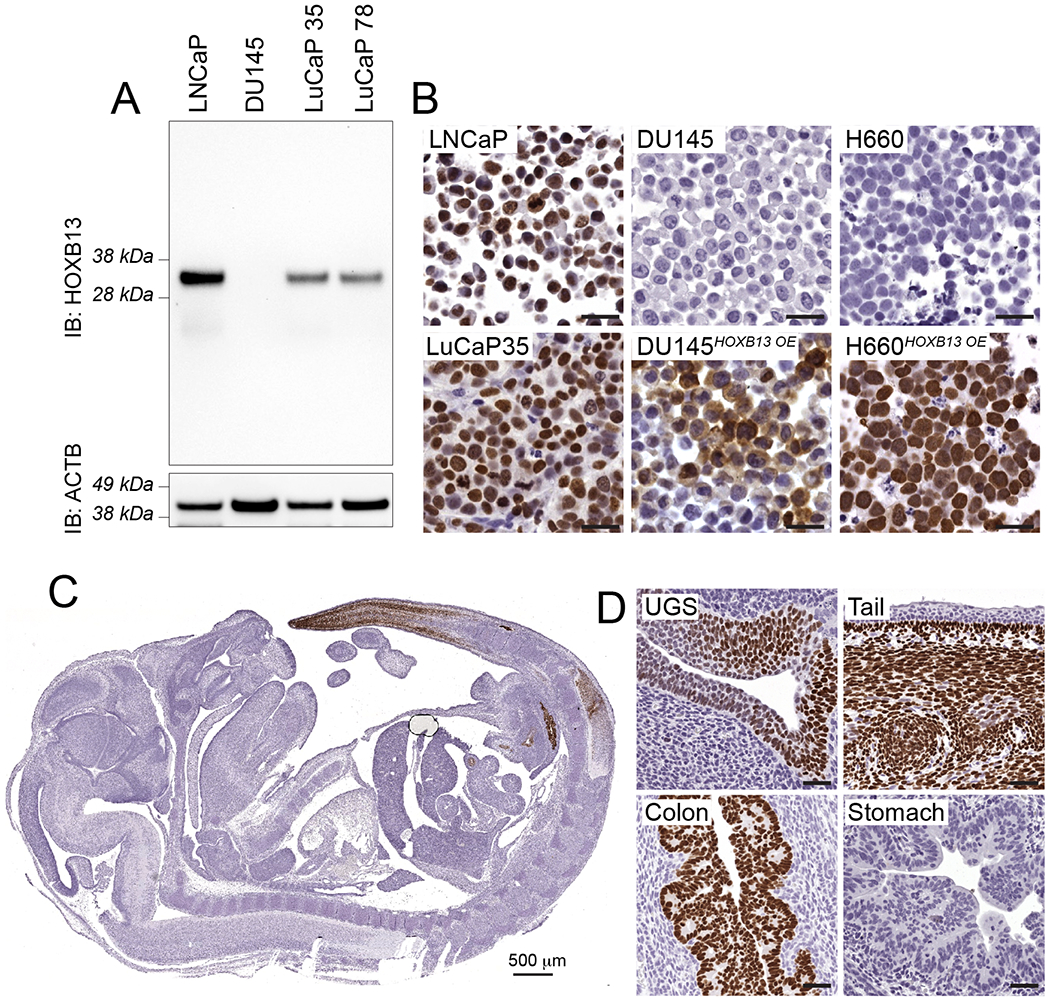
A. Western blot of lysates from prostate cancer cell lines (LNCaP, DU145) and tissues from prostate cancer patient-derived xenografts (LuCaP 35, LuCaP 78) probed with HOXB13 antibody (clone D7N8O) show a single band at the predicted molecular weight of HOXB13 (34 kDa). B. IHC on formalin fixed paraffin embedded cell lines with known HOXB13 expression define the specificity of clone D7N8O. LNCaP and LuCaP35 show strong reactivity; DU145 and H660 are negative for HOXB13. Lentiviral over-expression (HOXB13 OE) results in robust labeling of HOXB13 in DU145HOXB13OE and H660HOXB13OE cells. C. HOXB13 expression in developing mouse embryo (E17.5). HOXB13 labeling is restricted to the urogenital sinus (UGS), distal colon/rectum and tail, recapitulating the expression pattern described by reporter and RNA in situ labeling previously shown [6] . Scale bars denote 50 μm, if not otherwise specified.
Additionally, we sought to determine the concordance of HOXB13 expression based on IHC with previously published Hoxb13 genetic reporter-based expression pattern in the developing mouse embryo [6]. Our findings showed that in mouse embryos at E17.5, HOXB13 expression was restricted to the terminal segments of the tail, the distal rectum, and the urogenital sinus (Figure 1C). These finding paralleled the previously reported patterns of HOXB13 expression determined by in situ hybridization or lacZ knock-in allele reporters [5,6,46]. Notably, while HOXB13 expression was only seen in epithelial cells in the colon and urogenital sinus, in the tail, HOXB13 was predominantly expressed by stroma cells (Figure 1D). Collectively, these results indicate that the HOXB13 antibody clone D7N8O is specific for the detection of human and murine HOXB13 protein.
Assessment of HOXB13 expression during prostate development
Prior studies have established the role of HOXB13 in murine prostate development and its expression in the urogenital sinus (UGS) epithelium from which the prostate arises during organogenesis [5,6,46,47] (Figure 1). However, there is a paucity of detailed information on the temporal changes of Hoxb13 expression during murine and human prostate development.
To address this, we conducted qRT-PCR for Hoxb13 and Ar on UGS or prostate tissue collected at various stages of murine prostate development (16.5 and 17.5 days post conception; 0, 15, 22, 35 days postnatal; and 4-month-old adult) (Figure 2A). Although Hoxb13 transcripts were detected at early stages of development, a marked increase in expression (>60-fold), peaking in adult prostate tissues, was noted. Conversely, Ar transcript levels increased only modestly (maximum 4-fold) during this developmental window (Figure 2A). These findings were corroborated by in situ co-immunolabeling, which revealed patchy and low level HOXB13 expression confined to the urogenital epithelium at E17.5 (Figure 2B) but high levels of HOXB13 expression predominantly in prostatic luminal cells, with faint basal cell reactivity in the adult murine prostate. AR on the other hand, was expressed in both the UGS epithelium and the urogenital mesenchyme at E17.5. Its expression pattern in the adult murine prostate mimicked that of HOXB13, with predominant luminal epithelial reactivity (Figure 2B).
FIGURE 2. HOXB13 expression in murine and human prostate development.
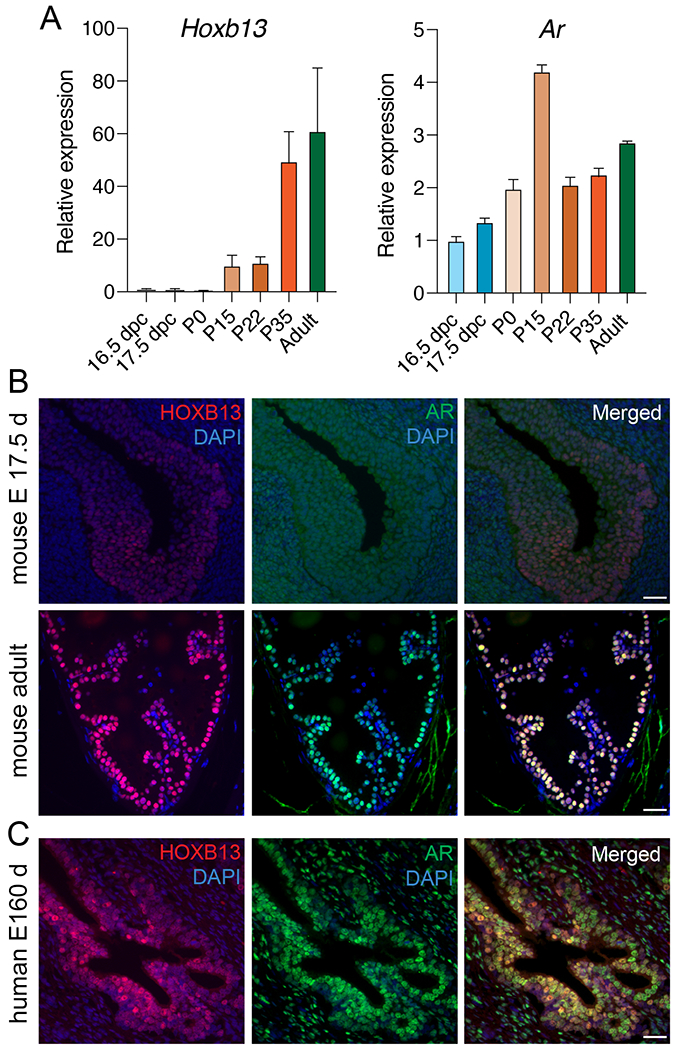
A. Relative mRNA expression of Hoxb13 and Ar in murine urogenital sinus tissue at 16.5 and 17.5 days post conception (dpc), post-partum (P) prostates at days 0, 15, 22, and 35, and 4 months (Adult). Shown are mean and standard deviation of three replicates. B. Micrographs showing HOXB13 and AR protein expression in different developmental stages of the mouse prostate. C. Micrographs of separate and merged HOXB13 and AR protein expression in fetal human prostate tissue (16 weeks of gestation). Scale bars denote 50 μm.
In human fetal prostate tissue (E160d), we detected predominantly luminal cell HOXB13 expression. Similar to human adult prostate tissues, AR was expressed in luminal epithelial cells as well as stroma cells (Figure 2C). These data show a dynamic change in HOXB13 expression during development and suggest a role of HOXB13 in terminal differentiation.
Association of HOXB13 expression in localized prostate cancer with Gleason grade and disease-specific outcomes
We next assessed HOXB13 expression in adult prostate tissues (Figure 3A–F). In benign prostate glands, HOXB13 was predominantly found in luminal cells with significantly lower expression in the basal cells (mean H-scores: 211.4 vs. 25.71, P < 0.0001) (Figure 3A,C). Increased HOXB13 expression in luminal cell compartments in both benign and cancer cells was also corroborated in previously published single cell RNA sequencing data (Supplementary Figure 1)[33]. Prostatic stroma cells were negative for HOXB13, which is consistent with prior reports [48].
FIGURE 3. HOXB13 expression in adult prostate tissue and localized prostate cancer.
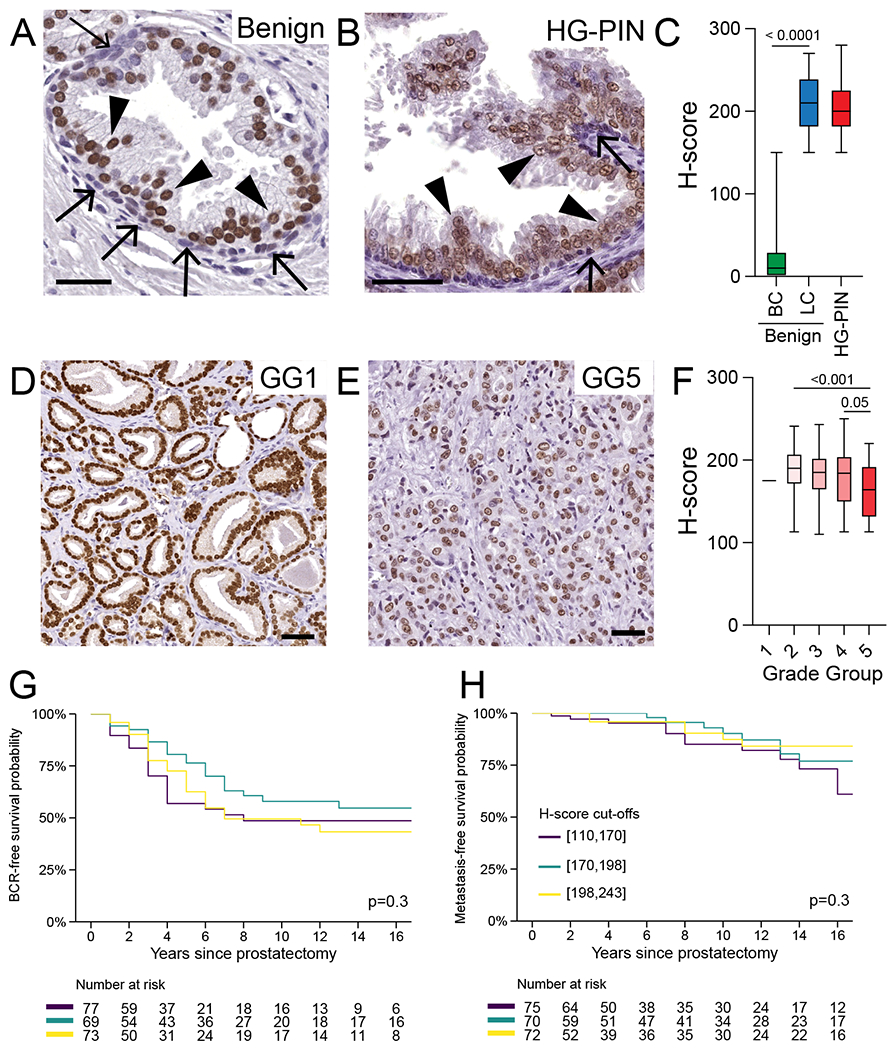
A. HOXB13 expression in benign prostate tissue. Note the higher level of HOXB13 expression luminal (arrowheads) versus basal (arrows) cells. B. Semiquantitative assessment of HOXB13 expression in benign prostatic luminal and basal cells (N=12) C. Comparison of HOXB13 expression in benign luminal cells, high-grade prostatic intraepithelial neoplasia (HG-PIN) and invasive carcinoma (N=12). D. Representative micrographs of HOXB13 expression in HG-PIN. E. Box-plots showing HOXB13 expression across grade groups (GG) and pathological stage. F. Micrographs showing HOXB13 reactivity in grade group 1 (GG1) and GG5. Kaplan-Meier plots shows G. biochemical recurrence-free (BCR) and H. metastasis-free survival. Colored lines indicate tertiles of HOXB13 H-scores. Scale bars denote 50 μm.
We next compared HOXB13 expression levels between benign prostate glands and high grade prostatic intra-epithelial neoplasia (HG-PIN) and observed no statistically significant differences in nuclear labeling intensities (mean H-scores: 211.4 vs. 206.9, P=0.6) (Figure 3B,C). Importantly, HOXB13 expression was restricted to prostatic epithelial cells and was not found in seminal vesicle/ejaculatory duct tissue (Supplementary Figure 2).
To assess the expression of HOXB13 in primary localized prostate cancers, we stained tissue micro arrays (TMAs) containing prostate cancer tissues from 297 hormone-naïve cases that were enriched for intermediate- to high-risk disease [49] (Supplementary Table 1). Applying our optimized IHC protocol, we observed HOXB13 positivity in all prostate cancers. Despite this uniform positivity, which contrasts to prior reports using other antibodies [22], we found a range of staining intensities across the cohort (median H-score 185, interquartile range 161 to 203) (Figure 3D–F). Notably, HOXB13 expression levels decreased with Gleason score/Grade Group (P<0.001) (Figure 3D–F). In particular, tumors with a Gleason Score ≥ 9/Grade Group 5 showed significantly lower HOXB13 levels (mean difference −24, 95% CI −38 to −11) compared to Gleason score 3+4/Grade Group 2 (Figure 3D,E,F, Supplementary Figure 3). We also noted significantly lower HOXB13 levels in African American men (mean difference −14, 95% CI −27 to −0.81, P=0.04). No other statistically significant associations between HOXB13 and other clinicopathological features were noted (Supplementary Table 2). In addition to the pathologist-based semiquantitative HOXB13 protein H-score, we also applied quantitative digital image analyses to determine HOXB13 expression in a subset of cases and observed a tight correlation between visual and digital expression scores (Supplementary Figure 4).
Since prior studies had suggested that higher HOXB13 expression was associated with earlier biochemical recurrence and progression to metastatic disease [22,50], we next determined the association between HOXB13 protein expression and clinical outcomes in 219 men for which follow-up data were available (median follow up 9 years, interquartile range 2 to 16 years). We observed no significant difference in biochemical recurrence-free-survival and distant-metastasis-free survival across patients grouped by tertiles of HOXB13 H-scores (Figure 3G,H) or using continuous H-scores after adjusting for Gleason score/Grade Group (Supplementary Table 2, 3).
HOXB13 expression in metastatic castration-resistant prostate cancer
To investigate the expression of HOXB13 in advanced metastatic castration-resistant prostate cancer (mCRPC), we initially conducted in silico analyses of HOXB13 transcript levels in 121 LuCaP PDX tissues (Figure 4A), 98 patients with metastatic prostate cancer (185 samples) from the University of Washington rapid autopsy program (Tissue Acquisition Necropsy, UW-TAN) (Figure 4B), and 266 mCRPC patients from the Stand Up To Cancer (SU2C) international dream team (270 samples) (Figure 4C) [21,25,30,51]. To contextualize the expression patterns, we divided each cohort into four molecular subgroups based on AR signaling and NE marker expression [20,21]. We observed high levels of HOXB13 expression in AR+/NE− (AR positive prostate cancer, ARPC) and AR+/NE+ tumors (Figure 4A–C). However, AR−/NE+ (NEPC) and AR−/NE− (double negative prostate cancer, DNPC) tumors exhibited reduced but detectable HOXB13 mRNA levels across all three sample sets. This finding is consistent with prior reports showing lower levels of HOXB13 in AR−/NE+ tumors [48].
FIGURE 4. HOXB13 expression in advanced castration resistant prostate cancer.
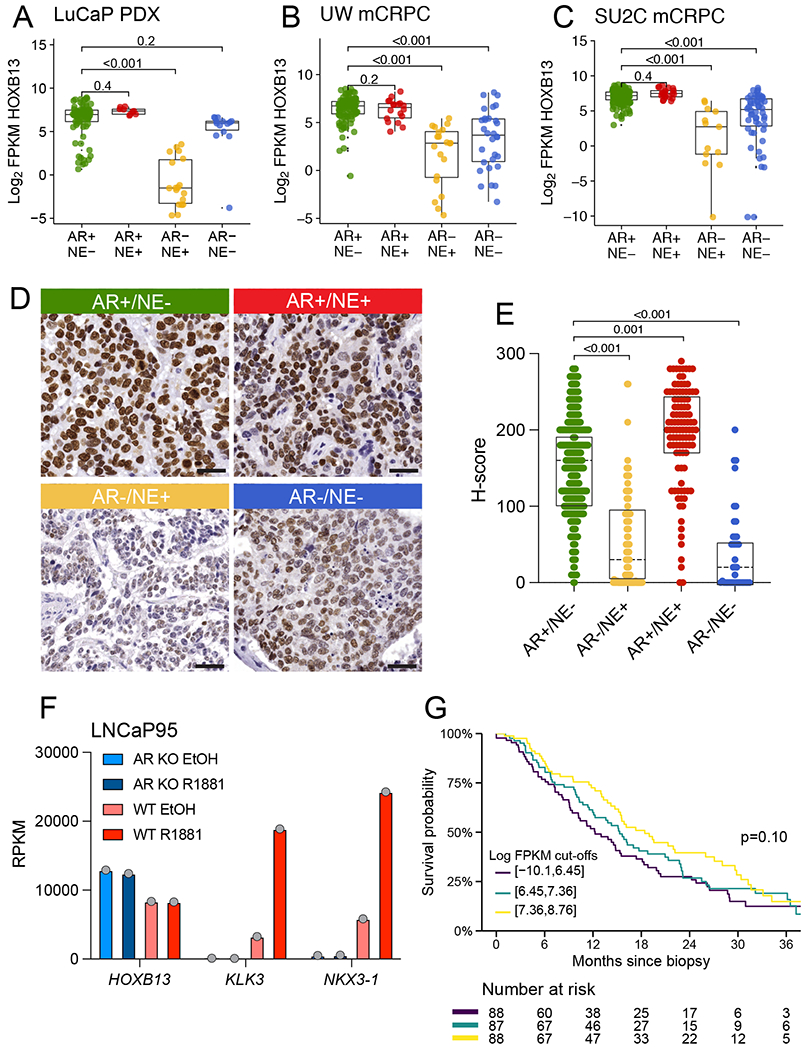
HOXB13 mRNA expression across molecular subtypes of CRPC (AR+/NE−, AR+/NE+, AR−/NE+, AR−/NE−) in A. PDX tissues (N=121 samples from 54 PDX lines), B. University of Washington rapid autopsy (TAN) cohort (N=185 samples from 98 patients) and C. the SU2C-IDT (N=270 samples from 266 patients). D. Representative micrographs showing HOXB13 protein expression in UW-TAN tissues. E. Boxplot showing HOXB13 IHC H-scores across molecular subtypes. F. HOXB13, KLK3 and NKX3.1 mRNA expression analyses in wild type (WT) and AR knock out (ARKO) LNCaP95 cells treated with vehicle (EtOH) or the synthetic androgen R1881 demonstrate the AR− and androgen-independence of HOXB13 levels G. Kaplan-Meier plots shows overall survival in the SU2C cohort stratified by HOXB13 expression. Colored lines indicate tertiles of HOXB13 log2 FPKM.
To further validate these in silico RNA expression studies on the protein level, we evaluated HOXB13 expression in 52 patients with advanced mCRPC (581 samples) from the UW-TAN prostate rapid autopsy cohort [21,24,25], which represents an extensively pretreated patients population with prior androgen deprivation and AR signaling inhibitor therapy (Supplementary Table 4) [20,21,24,25,39,52]. We found reduced HOXB13 H-scores in AR−/NE+ and AR−/NE− tumors compared to AR+/NE− (mean differences −97, 95% CI −118 to −76, and −101, 95% CI −137 to −65, both P<0.001) and increased H-scores in AR+/NE+ tumors compared to AR+/NE− (mean difference 21, 95% CI 8.4 to 33, P=0.001) (Figure 4D,E). In tumors for which matched tissues for IHC and RNA-seq analyses were available, we observed a tight correlation between HOXB13 mRNA transcript and protein levels (Supplementary Figure 5). Notably, despite the reduced expression found in AR− tumors, 11/13 (85%) of patients with AR−/NE+ tumors and 5/6 (83%) of patients with AR−/NE− tumors had HOXB13 H-scores greater than 0 in at least one tumor.
To further investigate the relationship between AR and HOXB13 expression, we utilized expression data from wild type and AR knockout LNCaP95 cell lines [19,53]. We observed that AR knock out did not result in a decrease in HOXB13 mRNA expression. However, the levels of KLK3 (encoding PSA) and NKX3.1 were significantly reduced in this context (Figure 4F). Moreover, in contrast to KLK3 and NKX3.1, treatment with the synthetic androgen R1881 did not increase HOXB13 expression levels. Additionally, HOXB13 mRNA levels showed no correlation with AR and SPOP genomic alterations and remained consistent across a wide range of AR activity scores (Supplementary Figure 6). In summary, these studies underscore that, unlike other commonly used prostate lineage markers such as PSA and NKX3.1, HOXB13 expression is AR-independent.
The unique design of our sample cohort further allowed us to determine the inter- and intra-patient heterogeneity of HOXB13 expression (Supplementary Figure 7). While the vast majority of AR+ cases showed robust HOXB13 expression, with only limited inter-tumoral heterogeneity, a subset of cases, particularly those with inter-tumoral subtype heterogeneity, exhibited differences in HOXB13 expression between different metastatic sites.
To determine the association between HOXB13 expression and clinical outcome in the metastatic setting, we evaluated RNA-seq data from 263 patients of the SU2C cohort for which follow-up data were available (median follow-up 30 months, interquartile range 19 to 62 months). Although we observed no significant differences in overall survival across patients grouped by tertiles of HOXB13 expression (Figure 4G) or using continuous HOXB13 log2-transformed FPKM values after adjusting for molecular subgroup (Supplementary Table 5), our data were consistent with higher HOXB13 expression being associated with modestly longer survival (HR=0.98, 95% CI 0.92 to 1.04).
Overall, these analyses demonstrate that HOXB13 expression differs across different molecular subtypes of mCRPC but was not found to be significantly prognostic for overall survival. Although HOXB13 levels are reduced in AR−/NE+ and AR−/NE− tumors, expression is retained at detectable levels in the majority of patients, suggesting that HOXB13 could serve as a useful marker for prostatic lineage even in advanced AR-negative mCRPC.
HOXB13 silencing in NEPC is mediated by CpG methylation
Given the observed differences in the levels of HOXB13 between AR+/NE− and AR−/NE+ tumors, we sought to further investigate the relationship between HOXB13 and neuroendocrine marker expression in biphasic tumors, which exhibit distinct AR+/NE− and AR−/NE+ cell populations (Figure 5A–C). In human mCRPC rapid autopsy cases from the UW-TAN that displayed a biphasic appearance, we found a substantial reduction of HOXB13 expression in cell populations expressing the neuroendocrine transcription factor INSM1 (Figure 5A).
FIGURE 5. Silencing of HOXB13 in NEPC is mediated by epigenetic alterations.
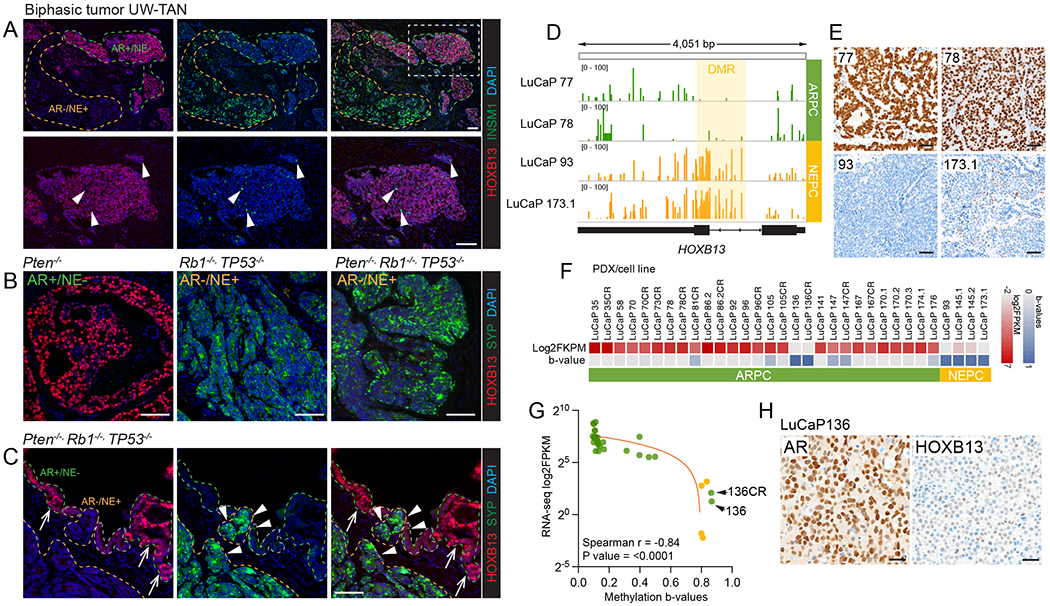
A. HOXB13 and INSM1 expression in a case with mixed/biphasic adenocarcinoma and neuroendocrine carcinoma phenotype. Note the distinct staining pattern or NE (INSM1) positive cells, which are uniformly HOXB13 low, and the HOXB13 positive cells, which are INSM1 negative. B. HOXB13 and NE (synaptophysin, SYP) expression in prostate GEM models. SKO (Pten−/−) tumors show robust HOXB13 expression in the absence of NE marker expression. TKO (Trp53−/−/Rb1−/−/Pten−/−) tumors show NE transdifferentiation characterized by the emergence of NE+ HOXB13 cell populations. C. Gradual loss of HOXB13 expression in early incipient NEPC lesions with transition from HOXB13 high NE marker negative (arrows) to HOXB13 low NE marker positive (arrowheads). Note the intermediate cell population which is characterized by reduced HOXB13 and focal NE marker expression. D. Whole genome bisulfite sequencing identifies a differentially methylated regions (DMRs) encompassing the first intron and second exon of HOXB13 that is hypermethylated in HOXB13 low/negative NEPC PDX lines (LuCaP93, LuCaP173.1) and hypomethylated in HOXB13 expressing ARPC lines (LuCaP77, LuCaP78). E. Micrographs of HOXB13 immunohistochemistry in LuCaP77, LuCaP78, LuCaP93, and LuCaP173.1. F. Heat map showing HOXB13 expression (RNA-seq, log2-transformed FPKM) and HOXB13 methylation (beta-values based on EPIC arrays) across different PDX models. G. Correlation plot shows HOXB13 DMR b-values and HOXB13 expression. Note that LuCaP136 and LuCaP136CR are the only AR positive lines with HOXB13 silencing. H. Micrographs of LuCaP136 show retained high level expression of AR but greatly reduced HOXB13 reactivity.
In murine prostate cancer models, we found that HOXB13 was highly expressed in adenocarcinomas that developed in a Pten−/− (SKO) background. However, in tumors from Rb1−/−, Trp53−/− (DKO), and Rb1−/−, Trp53−/−, Pten−/− (TKO) mice that recapitulate the progression of adenocarcinoma to neuroendocrine prostate cancer (NEPC), we observed a substantial reduction of HOXB13 in cells undergoing NE transdifferentiation (Figure 5B).
Importantly, we noted a gradual decrease in HOXB13 expression and a concurrent increase in NE marker expression in tumor regions that showed an incipient transition from adenocarcinoma to NEPC (Figure 5C).
The decreased expression of HOXB13 observed in NEPC tumors suggests a tight regulation by epigenetic mechanisms. Indeed, a prior report suggested a correlation between HOXB13 expression and CpG methylation [15]. This observation prompted us to investigate the DNA methylation changes of HOXB13 in NEPC tumors.
Using WGBS, we analyzed two well-characterized AR+/NE− (LuCaP 77, 78) and two AR−/NE+ (NEPC, LuCaP 93, 173.1) PDX models with markedly different HOXB13 protein expression. These WGBS studies revealed a DMR spanning ~2000bp from intron 1 to exon 2 of HOXB13, encompassing an intragenic CpG island that was hypermethylated in NEPC (Figure 5D). We found that HOXB13 hypermethylation was associated with significant reductions in protein expression, with focal HOXB13 reactivity by IHC in LuCaP 173.1 (Figure 5E).
To further understand the methylation pattern of the HOXB13 locus in mCRPC, we performed EPIC methylation array experiments on prostate cancer PDX lines (n=32; 4 NEPC, 28 ARPC) and compared methylation changes to previously published transcriptomic data [17]. We observed a tight correlation (Spearman r = −0.84, P=0.0001) between methylation beta-values and HOXB13 expressed based on RNA-seq (Figure 5F,G). Importantly, HOXB13 hypermethylation was predominantly observed in NEPC tumors, with only one ARPC line (LuCaP 136/136CR) displaying significant hypermethylation and associated transcriptional repression (Figure 5F).
HOXB13 is a relevant marker to determine prostatic origin
Given the relative prostate-specific expression of HOXB13, prior studies have evaluated the potential diagnostic value of HOXB13 as a marker for prostatic origin [16,18,54,55]. Since several of these studies used antibodies with less ideal specificity, we aimed to revisit this question and determine HOXB13 protein expression using a validated antibody across a larger number of tumor types (Figure 6). Prior studies have demonstrated that NKX3.1 is a specific marker for prostatic origin [17]. Despite its reported specificity, the sensitivity of NKX3.1 has not been evaluated in advanced therapy refractory prostate cancer. We therefore compared the rates of positivity of HOXB13 and NKX3.1 across different molecular subtypes of prostate cancer (Figure 6). We estimated similar probabilities of HOXB13 and NKX3.1 positivity in AR+/NE− and AR+/NE+ tumors (all between 98% and 100%) but observed lower probabilities of any positivity (H-score ≥ 1) positivity for NKX3.1 in AR−/NE+ (18% vs 77%, p=0.15) and AR−/NE− tumors (10% vs 90%, p=0.02) (Figure 6A) while HOXB13 expression was retained. Similar trends were observed when positivity was defined as H-score of ≥ 40 (Supplementary Figure 8). HOXB13 reactivity in the majority of AR-negative subtypes suggested HOXB13 is a more sensitive marker for prostatic origin in particular in cases with AR loss (Figure 6A,B). This observation was also corroborated in murine prostate cancer models where adenocarcinomas arising in the Pten−/− background, which are known to show low AR activity exhibited greatly reduced/absent of NKX3.1 reactivity but retained high HOXB13 expression (Supplementary Figure 9).
FIGURE 6. HOXB13 is a valuable prostatic linage marker for prostate cancer diagnosis.
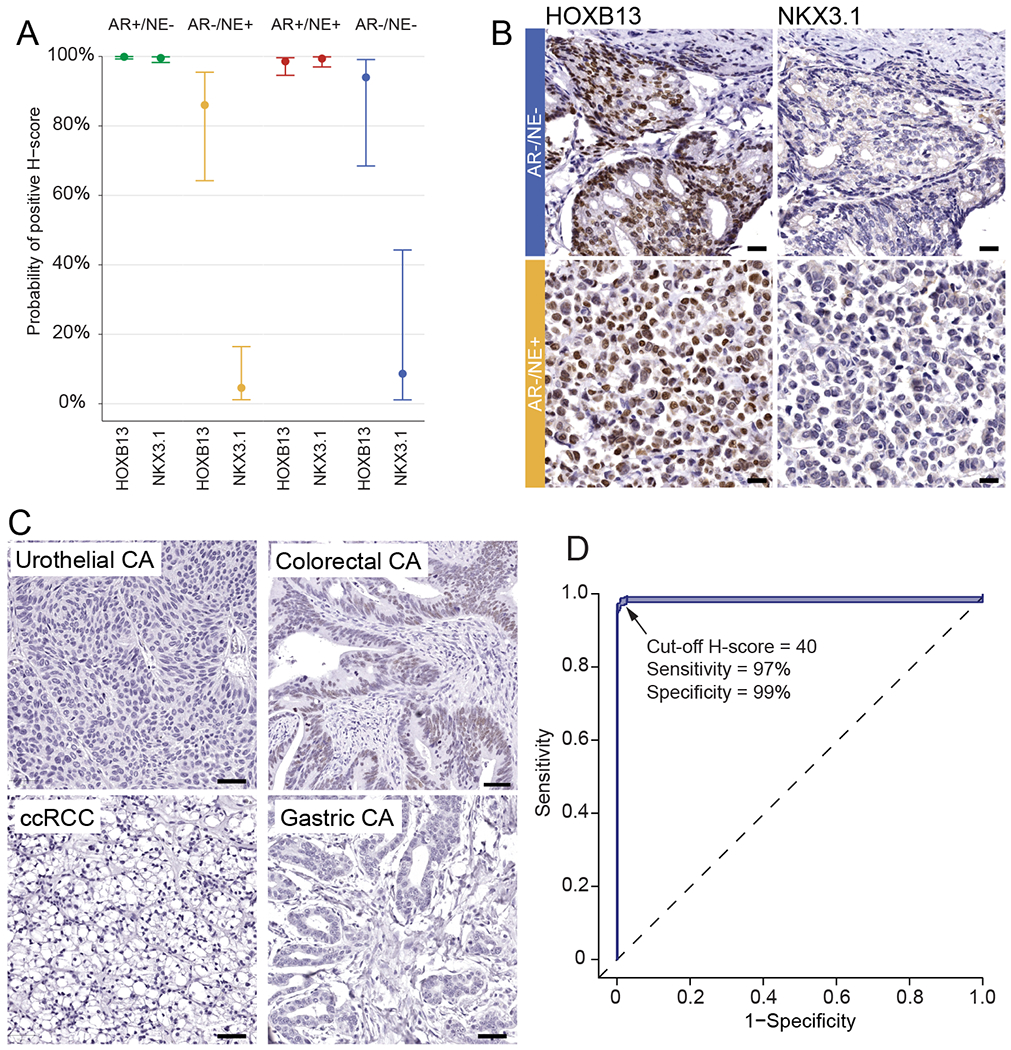
A. Probabilities of any positive immunoreactivity of HOXB13 and NKX3.1 across different molecular subtypes in 52 mCRPC cases. B. Representative micrographs of HOXB13 and NKX3.1 in AR−/NE− and AR/NE+ mCRPC tumors C. HOXB13 expression in non-prostatic primary tumors. D. Receiver operator characteristic curve for HOXB13 in 383 prostatic and 454 non-prostatic primary tumors.
We next aimed to determine the sensitivity and specificity of HOXB13 for prostatic carcinoma. To this end we studied cancer specimens from 837 patients: 454 patients with cancers of non-prostatic origin, 341 patients with localized prostate cancers, and 52 patients with advanced prostate cancer (Figure 6C, Table 1). We observed HOXB13 expression above an H-score of 40 (Supplementary Figure 8) in all localized prostate cancers. Furthermore, HOXB13 expression was only observed in 4/100 (4%) colorectal cancers, while all other tumors showed no reactivity above this threshold. Since clinically the differentiation between high-grade prostatic vs. urothelial carcinoma can be particularly challenging, we specifically assessed the frequency of HOXB13 positivity in a series of 34 high-grade prostate cancers (Grade Group 5) and 93 invasive urothelial carcinomas of the bladder. While all prostate cancers were positive for HOXB13, none of the urothelial carcinomas showed HOXB13 expression, suggesting that HOXB13 is a valuable marker for distinguishing tumors of prostatic and urothelial origin. This sensitivity compared favorably to other more commonly used markers of prostatic lineage (PSA, NKX3.1) which were previously assessed on the same sample set (Supplementary Table 5) [56]. Collectively, including all samples studied here, we calculated the sensitivity of HOXB13 for tumors of prostatic origin to be 97% (95% CI 96% to 99%) and the specificity to be 99% (95% CI 98% to 100%) (Figure 6D).
TABLE 1.
Expression (H-score ≥ 40) of HOXB13 in prostatic and non-prostatic tissues.
| No. Cases | Positive cases | Percent positive | |
|---|---|---|---|
|
| |||
| Prostatic adenocarcinoma | 331 | 331 | 100 |
| Invasive urothelial carcinoma | 93 | 0 | 0 |
| Gastric adenocarcinoma | 32 | 0 | 0 |
| Colorectal adenocarcinoma | 100 | 4 | 4 |
| Papillary renal cell carcinoma | 65 | 0 | 0 |
| Chromophobe renal cell carcinoma | 46 | 0 | 0 |
| Clear cell renal cell carcinoma | 90 | 0 | 0 |
| Oncocytoma | 16 | 0 | 0 |
| Seminoma | 2 | 0 | 0 |
| Embryonal Carcinoma | 2 | 0 | 0 |
| Mixed Germ Cell Tumor | 3 | 0 | 0 |
| Melanoma | 5 | 0 | 0 |
DISCUSSION
HOXB13 is a homeobox transcription factor with important roles in prostate biology [11,12,57]. Genetic ablation of HOXB13 in mice results in morphologic and expression changes suggesting that HOXB13 is involved in the terminal differentiation, but not initial organogenesis, of the prostate gland [4]. In our detailed analyses of HOXB13 expression across different stages of murine prostate development, we observed an increase in Hoxb13 levels at later stages of development, supporting the notion that HOXB13 is involved in establishing a terminal luminal epithelial phenotype [47].
To further elucidate the expression of HOXB13 across the disease spectrum of prostate cancer, we performed the most comprehensive tissue-based assessment to date. By utilizing a HOXB13-specific antibody, which underwent orthogonal validation with genetic controls, we found that HOXB13 was expressed in benign luminal epithelial cells, high-grade prostatic intraepithelial neoplasia (HGPIN), and 331/331 (100%) of primary treatment naïve prostatic adenocarcinomas. This differs from prior studies, which reported a significantly lower rate of HOXB13 positivity using different antibodies [18,22]. More broadly, this finding emphasizes the importance of antibody validation and assay optimization to ensure reliable labeling in IHC studies [58]
Given the putative role of HOXB13 in prostate cancer biology, previous studies have attempted to evaluate its prognostic significance in primary prostate cancers. Two reports have utilized large-scale TMA-based analyses or in silico analyses of RNA expression data to suggest that higher HOXB13 levels are associated with adverse clinical outcomes [22,50]. However, our results demonstrated no statistically significant difference in biochemical recurrence or disease-free survival when stratified by HOXB13 expression. Although we must acknowledge technical variances in the antibodies used in IHC studies and differences between bulk RNA and IHC analyses, as well as cohort differences such as a higher representation of higher-grade tumors in our cohort, these findings suggest that HOXB13 levels alone are likely not a robust prognostic marker in localized and metastatic prostate cancer.
To comprehensively study the association of HOXB13 expression in distinct molecular subclasses of mCRPC, we leveraged the unique tissue resources of the UW-TAN rapid autopsy cohort, which is representative of patients with extensive pretreatment histories involving different AR-directed therapies. Our findings revealed that the majority of tumors that retained an AR-positive luminal epithelial phenotype (ARPC) displayed high levels of HOXB13 expression. However, we observed a significant reduction of HOXB13 levels in tumors lacking prostatic luminal epithelial differentiation, with NEPC demonstrating the lowest levels of HOXB13, as suggested previously [48].
The importance of HOXB13 as a key lineage-defining factor is likely exerted at least partly through its interaction with AR [9,11,57,59]. Given this tight functional relationship, it is not surprising that AR-positive tumors show a dependency on HOXB13, which may explain its retained high expression in ARPC [9,10,13,57]. Conversely, in the context of AR loss, downregulation of HOXB13 could be an early event that sets up a more lineage plastic state [60]. Although the mechanisms involved in this lineage transition are still under active investigation [20,21,28,60–62], our results demonstrate that silencing of HOXB13 in NEPC is associated with a gain of CpG methylation across the HOXB13 gene body. Together with our mechanistic data showing a lack of direct AR regulation of HOXB13, these results suggest that epigenetic changes associated with lineage transition and not loss of AR per se are likely responsible for the reduced HOXB13 expression observed in NEPC.
Moreover, in incipient NEPC tumors that spontaneously arise in Rb1−/−, Trp53−/−, Pten−/− prostate genetically modified mouse models, we have observed a gradual decrease in HOXB13 expression, accompanied by an increase in neuroendocrine marker expression [28] . Collectively, our findings suggest that HOXB13 plays a crucial role in establishing a prostatic luminal cell identity, and reduced HOXB13 expression is associated with a more lineage-plastic cell state.
From a clinical standpoint, there is a need for robust IHC markers that allow for the assessment of prostatic origin. In mCRPC patients who have undergone androgen deprivation and androgen receptor signaling inhibitor therapies, conventional AR-regulated prostate lineage markers such as PSA and NKX3.1 are less reliable. HOXB13, on the other hand is not AR-regulated and has been proposed as a more reliable prostatic lineage marker [54]. Several smaller-scale studies have explored the clinical diagnostic utility of HOXB13 IHC [18,23,54,55] . While the sensitivity in the reported studies varies, much of these analytical differences can be attributed to the use of different antibodies and staining protocols.
Considering the clinical need for a lineage biomarker, we conducted a comprehensive assessment of HOXB13 IHC sensitivity and specificity in a large cohort of 839 tumor specimens. We show that HOXB13 has a high sensitivity (97%) and specificity (99%) for prostatic origin. A significant finding of our study is that, even in treatment-emergent NEPC and DNPC cases, a significant fraction still showed retained HOXB13 protein expression, allowing to infer prostatic origin even in high-grade lineage-plastic tumors. Given the increasing number of metastatic biopsies performed in men with suspected advanced prostate cancer, it is important to add markers (such as HOXB13) to our immunohistochemical panels to accurately diagnose these emerging and clinically relevant molecular subtypes. Additionally, worth noting is that previous studies have reported lower sensitivity of HOXB13 antibodies in decalcified bone tissue [16,23,55]. However, our study did not find any reduced sensitivity in bone specimens, indicating that clone D7N8O is less vulnerable to pre-analytic differences.
Although our data suggest that HOXB13 could serve as a valuable diagnostic marker, it’s crucial to acknowledge potential pitfalls. Other than the prostate, a limited number of tumor tissues show expression of HOXB13. In addition, while not studied in this manuscript, neuroendocrine tumors of the cauda equina and rare pediatric tumors including Ewing’s sarcoma and embryonal rhabdomyosarcoma have been suggested to express HOXB13 [63,64]. As no single marker can exhibit ideal sensitivity and specificity, it will be important to validate our experience with HOXB13 clone D7N8O in additional cohorts and clinical practice.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the patients and their families, Celestia Higano, Evan Yu, Elahe Mostaghel, Heather Cheng, Bruce Montgomery, Andrew Hsieh, Jonathan Wright, Daniel Lin, Funda Vakar-Lopez, and the rapid autopsy teams for their contributions to the University of Washington Medical Center Prostate Cancer Donor Rapid Autopsy Program and the Development of the LuCaP PDX models. We thank the members of Haffner, Lee, and Nelson laboratories for their constructive suggestions and to Yibai Zhao for helping to implement the bootstrap resampling algorithm to evaluate discrimination of prostatic origin. This work was supported by the NIH/NCI (P50CA097186, R01CA234715, R01CA266452, R01 CA234162, R50CA22183, U54CA224079, P30CA015704, P01CA163227, P50CA58236, U54CA224079-05) NIH Office of Research Infrastructure Programs (ORIP) (S10OD028685), the U.S. Department of Defense Prostate Cancer Research Program (W81XWH-20-1-0111, W81XWH-21-1-0229, W81XWH-22-1-0278, W81XWH-18-1-0347, W81XWH-18-1-0689, W81XWH-21-1-0264) Grant 2021184 from the Doris Duke Charitable Foundation, the V Foundation, the Prostate Cancer Foundation, the Safeway Foundation, the Institute for Prostate Cancer Research, the Richard M. Lucas Foundation the FredHutch/UW Cancer Consortium, the Brotman Baty Institute for Precision Medicine and the UW/FHCC Institute for Prostate Cancer Research.
Conflict of interest disclosure statement:
P.S. Nelson has served as a paid advisor for Bristol Myers Squibb, Pfizer, and Janssen. A.M.D. serve as consultant for Cepheid Inc and Merck Inc. and receives sponsored research funding from Janssen R&D. L. D. True is co-founder and has equity in Lightspeed Microcopy, LLC. No potential conflicts of interest were disclosed by the other authors. E. Corey is a consultant of DotQuant and obtained institutional SRAs from AbbVie, Gilead, Bayer Pharmaceuticals, Janssen Research, Forma Therapeutics, Foghorn, Kronos, MacroGenics, Astra Zeneca and GlaxoSmithKline.
REFERENCES
- 1.Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet 2005; 6: 893–904 [DOI] [PubMed] [Google Scholar]
- 2.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer 2010; 10: 361–371 [DOI] [PubMed] [Google Scholar]
- 3.Javed S, Langley SEM. Importance of HOX genes in normal prostate gland formation, prostate cancer development and its early detection. Bju Int 2013; 113: 535–540 [DOI] [PubMed] [Google Scholar]
- 4.Economides KD, Capecchi MR. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development 2003; 130: 2061–2069 [DOI] [PubMed] [Google Scholar]
- 5.Economides KD, Zeltser L, Capecchi MR. Hoxb13 mutations cause overgrowth of caudal spinal cordand tail vertebrae. Dev Biol 2003; 256: 317–330 [DOI] [PubMed] [Google Scholar]
- 6.McMullin RP, Mutton LN, Bieberich CJ. Hoxb13 regulatory elements mediate transgene expression during prostate organogenesis and carcinogenesis. Dev Dyn 2009; 238: 664–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brechka H, Bhanvadia RR, VanOpstall C, et al. HOXB13 mutations and binding partners in prostate development and cancer: Function, clinical significance, and future directions. Genes Dis 2017; 4: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker B, Ostrander EA. Dysregulation of the homeobox transcription factor gene HOXB13: role in prostate cancer. Pharmacogenomics Personalized Medicine 2014; 7: 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stelloo S, Nevedomskaya E, Kim Y, et al. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene 2018; 37: 313–322 [DOI] [PubMed] [Google Scholar]
- 10.Pomerantz MM, Qiu X, Zhu Y, et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat Genet 2020; 52: 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pomerantz MM, Li F, Takeda DY, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015; 47: 1346–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim EH, Cao D, Mahajan NP, et al. ACK1–AR and AR–HOXB13 signaling axes: epigenetic regulation of lethal prostate cancers. Nar Cancer 2020; 2: zcaa018- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z, Wu D, Thomas-Ahner JM, et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc National Acad Sci 2018; 115: 6810–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao J, Chen Y, Nguyen DT, et al. The Homeobox gene, HOXB13, Regulates a Mitotic Protein-Kinase Interaction Network in Metastatic Prostate Cancers. Sci Rep-uk 2019; 9: 9715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu X, Fong K, Gritsina G, et al. HOXB13 suppresses de novo lipogenesis through HDAC3-mediated epigenetic reprogramming in prostate cancer. Nat Genet 2022; 54: 670–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varinot J, Furudoï A, Drouin S, et al. HOXB13 protein expression in metastatic lesions is a promising marker for prostate origin. Virchows Arch 2016; 468: 619–622 [DOI] [PubMed] [Google Scholar]
- 17.Gurel B, Ali TZ, Montgomery EA, et al. NKX3.1 as a Marker of Prostatic Origin in Metastatic Tumors. Am J Surg Pathology 2010; 34: 1097–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristiansen I, Stephan C, Jung K, et al. Sensitivity of HOXB13 as a Diagnostic Immunohistochemical Marker of Prostatic Origin in Prostate Cancer Metastases: Comparison to PSA, Prostein, Androgen Receptor, ERG, NKX3.1, PSAP, and PSMA. Int J Mol Sci 2017; 18: 1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennen WN, Zhu Y, Coleman IM, et al. Resistance to androgen receptor signaling inhibition does not necessitate development of neuroendocrine prostate cancer. JCI Insight 2021; 6: e146827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bluemn EG, Coleman IM, Lucas JM, et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017; 32: 474–489.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labrecque MP, Coleman IM, Brown LG, et al. Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J Clin Invest 2019; 129: 4492–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zabalza CV, Adam M, Burdelski C, et al. HOXB13 overexpression is an independent predictor of early PSA recurrence in prostate cancer treated by radical prostatectomy. Oncotarget 2015; 6: 12822–12834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varinot J, Comperat E. HOXB13 expression in metastatic prostate cancer. Virchows Arch 2016; 469: 121–121 [DOI] [PubMed] [Google Scholar]
- 24.Patel RA, Coleman IM, Roudier MP, et al. Comprehensive assessment of anaplastic lymphoma kinase in localized and metastatic prostate cancer reveals targetable alterationsALK alterations in prostate cancer. Cancer Res Commun 2022; 2: 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayar E, Patel RA, Coleman IM, et al. Reversible epigenetic alterations mediate PSMA expression heterogeneity in advanced metastatic prostate cancer. Jci Insight 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Brien VP, Kang Y, Shenoy MK, et al. Single-cell profiling uncovers a Muc4-expressing metaplastic gastric cell type sustained by Helicobacter pylori-specific inflammation. Biorxiv 2022: 2022.12.20.521287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurley PJ, Marchionni L, Simons BW, et al. Secreted protein, acidic and rich in cysteine-like 1 (SPARCL1) is down regulated in aggressive prostate cancers and is prognostic for poor clinical outcome. Proc National Acad Sci 2012; 109: 14977–14982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan JM, Zaidi S, Love JR, et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science 2022; 377: 1180–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ku SY, Rosario S, Wang Y, et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017; 355: 78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen HM, Vessella RL, Morrissey C, et al. LuCaP Prostate Cancer Patient‐Derived Xenografts Reflect the Molecular Heterogeneity of Advanced Disease and Serve as Models for Evaluating Cancer Therapeutics. Prostate 2017; 77: 654–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley PJ, Hughes RM, Simons BW, et al. Androgen-Regulated SPARCL1 in the Tumor Microenvironment Inhibits Metastatic Progression. Cancer Res 2015; 75: 4322–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corella AN, Ordonio MVAC, Coleman I, et al. Identification of Therapeutic Vulnerabilities in Small-cell Neuroendocrine Prostate Cancer. Clin Cancer Res 2020; 26: 1667–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong HY, Sheng Q, Hesterberg AB, et al. Single cell analysis of cribriform prostate cancer reveals cell intrinsic and tumor microenvironmental pathways of aggressive disease. Nat Commun 2022; 13: 6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011; 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016; 17: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maksimovic J, Gordon L, Oshlack A. SWAN: Subset-quantile Within Array Normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol 2012; 13: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014; 30: 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roudier MP, Winters BR, Coleman I, et al. Characterizing the molecular features of ERG‐positive tumors in primary and castration resistant prostate cancer. Prostate 2016; 76: 810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bates D, Mächler M, Bolker B, et al. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw 2015; 67 [Google Scholar]
- 41.Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest Package: Tests in Linear Mixed Effects Models. J Stat Softw 2017; 82 [Google Scholar]
- 42.Fox J, Hong J. Effect Displays in R for Multinomial and Proportional-Odds Logit Models: Extensions to the effects Package. J Stat Softw 2009; 32 [Google Scholar]
- 43.Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials 1996; 17: 343–346 [DOI] [PubMed] [Google Scholar]
- 44.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. Bmc Bioinformatics 2011; 12: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sfanos KS, Aloia AL, Hicks JL, et al. Identification of Replication Competent Murine Gammaretroviruses in Commonly Used Prostate Cancer Cell Lines. Plos One 2011; 6: e20874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeltser L, Desplan C, Heintz N. Hoxb-13: a new Hox gene in a distant region of the HOXB cluster maintains colinearity. Development 1996; 122: 2475–2484 [DOI] [PubMed] [Google Scholar]
- 47.Huang L, Pu Y, Hepps D, et al. Posterior Hox Gene Expression and Differential Androgen Regulation in the Developing and Adult Rat Prostate Lobes. Endocrinology 2007; 148: 1235–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng S, Yang S, Shi Y, et al. Neuroendocrine prostate cancer has distinctive, non-prostatic HOX code that is represented by the loss of HOXB13 expression. Sci Rep-uk 2021; 11: 2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haffner MC, Guner G, Taheri D, et al. Comprehensive Evaluation of Programmed Death-Ligand 1 Expression in Primary and Metastatic Prostate Cancer. Am J Pathology 2018; 188: 1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weiner AB, Faisal FA, Davicioni E, et al. Somatic HOXB13 Expression Correlates with Metastatic Progression in Men with Localized Prostate Cancer Following Radical Prostatectomy. European Urology Oncol 2021; 4: 955–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLucia DC, Cardillo TM, Ang L, et al. Regulation of CEACAM5 and Therapeutic Efficacy of an Anti-CEACAM5–SN38 Antibody–drug Conjugate in Neuroendocrine Prostate Cancer. Clin Cancer Res 2021; 27: 759–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brady L, Kriner M, Coleman I, et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat Commun 2021; 12: 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y, Dalrymple SL, Coleman I, et al. Role of androgen receptor splice variant-7 (AR-V7) in prostate cancer resistance to 2nd-generation androgen receptor signaling inhibitors. Oncogene 2020; 39: 6935–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varinot J, Cussenot O, Roupret M, et al. HOXB13 is a sensitive and specific marker of prostate cells, useful in distinguishing between carcinomas of prostatic and urothelial origin. Virchows Arch 2013; 463: 803–809 [DOI] [PubMed] [Google Scholar]
- 55.Barresi V, Ieni A, Cardia R, et al. HOXB13 as an immunohistochemical marker of prostatic origin in metastatic tumors. Apmis 2016; 124: 188–193 [DOI] [PubMed] [Google Scholar]
- 56.Chuang A-Y, DeMarzo AM, Veltri RW, et al. Immunohistochemical Differentiation of High-grade Prostate Carcinoma From Urothelial Carcinoma. Am J Surg Pathol 2007; 31: 1246–1255 [DOI] [PubMed] [Google Scholar]
- 57.Norris JD, Chang C-Y, Wittmann BM, et al. The Homeodomain Protein HOXB13 Regulates the Cellular Response to Androgens. Mol Cell 2009; 36: 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sfanos KS, Yegnasubramanian S, Nelson WG, et al. If this is true, what does it imply? How end-user antibody validation facilitates insights into biology and disease. Asian J Urology 2019; 6: 10–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johng D, Torga G, Ewing CM, et al. HOXB13 interaction with MEIS1 modifies proliferation and gene expression in prostate cancer. Prostate 2019; 79: 414–424 [DOI] [PubMed] [Google Scholar]
- 60.Beltran H, Hruszkewycz A, Scher HI, et al. The Role of Lineage Plasticity in Prostate Cancer Therapy Resistance. Clin Cancer Res 2019; 25: 6916–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mu P, Zhang Z, Benelli M, et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 2017; 355: 84–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng S, Wang C, Wang Y, et al. Ectopic JAK–STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat Cancer 2022; 3: 1071–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aspuria P-J, Cheon D-J, Gozo MC, et al. HOXB13 controls cell state through super-enhancers. Exp Cell Res 2020; 393: 112039. [DOI] [PubMed] [Google Scholar]
- 64.Asa SL, Mete O, Schüller U, et al. Cauda Equina Neuroendocrine Tumors: Distinct Epithelial Neuroendocrine Neoplasms of Spinal Origin. Am J Surg Pathol 2022; Publish Ahead of Print [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


