Abstract
Ecology and evolution are distinct theories, but the short lifespans and large population sizes of microbes allow evolution to unfold along contemporary ecological time scales. To document this in a natural system, we collected a two-decade, 471-metagenome time series from a single site in a freshwater lake, which we refer to as the TYMEFLIES dataset. This massive sampling and sequencing effort resulted in the reconstruction of 30,389 metagenomic-assembled genomes (MAGs) over 50% complete, which dereplicated into 2,855 distinct genomes (>96% nucleotide sequence identity). We found both ecological and evolutionary processes occurred at seasonal time scales. There were recurring annual patterns at the species level in abundances, nucleotide diversities (π), and single nucleotide variant (SNV) profiles for the majority of all taxa. During annual blooms, we observed both higher and lower nucleotide diversity, indicating that both ecological differentiation and competition drove evolutionary dynamics. Overlayed upon seasonal patterns, we observed long-term change in 20% of the species’ SNV profiles including gradual changes, step changes, and disturbances followed by resilience. Most abrupt changes occurred in a single species, suggesting evolutionary drivers are highly specific. Nevertheless, seven members of the abundant Nanopelagicaceae family experienced abrupt change in 2012, an unusually hot and dry year. This shift coincided with increased numbers of genes under selection involved in amino acid and nucleic acid metabolism, suggesting fundamental organic nitrogen compounds drive strain differentiation in the most globally abundant freshwater family. Overall, we observed seasonal and decadal trends in both interspecific ecological and intraspecific evolutionary processes. The convergence of microbial ecology and evolution on the same time scales demonstrates that understanding microbiomes requires a new unified approach that views ecology and evolution as a single continuum.
Ecology and evolution are typically considered distinct processes that unfold on different time scales. Although microbial ecology has blossomed into a rich field spanning experimental and natural systems, our understanding of microbial evolution relies primarily on phylogenetic reconstructions and laboratory experiments. Phylogenies provide insight into ancient evolutionary events1, but lack insight into contemporary evolution2 that occurs on directly observable time scales. Laboratory approaches, such as the E. coli long-term evolution experiment3, are able to directly observe evolutionary processes, but lack the context of complex communities typical of natural environments.
Here we describe a two-decade microbial time series from a freshwater lake, which allows us to directly observe the interplay between ecology and contemporary evolution in a natural ecosystem. We reconstructed tens of thousands of MAGs from 471 metagenomes spanning two decades with a sampling frequency averaging twice a month. This dataset constitutes the largest and longest metagenome dataset from a single location. We find that ecology and evolution both unfold at short, seasonal time scales as well as longer-term decadal time scales. The similar dynamics of inter- and intraspecies processes suggests that bacterial ecology and evolution unfold along a continuum that lacks a clear delineation between the two theories. We conclude that microbial ecology and evolution must be considered simultaneously and combined into a unified theory.
The TYMEFLIES dataset
We collected 471 samples over 20 years from Lake Mendota (WI, USA)4 and obtained shotgun DNA libraries (Fig. 1a, Supplementary Table 1). We refer to these Twenty Years of Metagenomes Exploring Freshwater Lake Interannual Eco/evo Shifts as the TYMEFLIES dataset. By cross-mapping reads from ~50 metagenomes to each single-sample metagenome assembly, we obtained a total of 85,684 genome bins, 30,389 of which were medium or high quality (> 50% completeness and < 10% contamination)5. We clustered these 30,389 bins at 96% average nucleotide identity (ANI) and obtained 2,855 clusters from which we chose representative metagenome-assembled genomes (MAGs)6 (Supplementary Table 2). Several previous studies have found an emergent species boundary at similar ANI cutoffs7–9, and we observed a rapid increase in the number of clusters above the 96% ANI cutoff. For the purposes of this study, we treat these 96% ANI representative MAGs as bacterial species and refer to sub-species delineations as strains.
Fig. 1. The TYMEFLIES dataset.
a, Metagenomes sample dates are indicated by black vertical lines. Microbial seasons13 are indicated by colored shading. b, Quality of the 2,855 reference genomes obtained after clustering to 96% ANI. We treat these reference genomes as species. c, Percent of metagenome reads from each sample that mapped to all reference genomes with an ANI ≥ 93%. Samples are grouped by season to highlight how well the reference genomes reflect each seasonal community. d, Rank abundance of phyla as measured by 16S rRNA gene amplicon sequencing4. The abundant Nanopelagicales order of Actinobacteria is highlighted. e, Abundance of phyla in the TYMEFLIES reference genomes, quantified as the mean relative abundance normalized by genome size and sequencing depth. The Nanopelagicales order is again highlighted.
The representative MAGs have high estimated completeness (median 86%) and low contamination (median 0.9%) (Fig. 1b), and reflect the lake’s abundant bacterial community, especially in well-sampled seasons (Fig. 1c). Using a 16S rRNA gene amplicon dataset from the same timeseries4 as a reference for the expected community composition (Fig. 1d), we found that our representative MAGs comprise most of the abundant taxa (Fig. 1e). Moreover, we obtained 168 representative MAGs from the Nanopelagicales order, which is the most abundant order in Lake Mendota and accounts for 22% of the amplicon reads and 10% of the mapped metagenomic reads. Similar to SAR11 bacteria in the oceans, this freshwater lineage is abundant in lakes globally10, difficult to culture11, and typically has highly streamlined genomes12.
Seasonal ecology and evolution
Lake Mendota has been the focus of limnological research since the late 1800s and has been routinely sampled since 1984 by the North Temperate Lakes Long-Term Ecological Research program (NTL-LTER)14. Microbial sampling began in 2000 as part of an NSF microbial observatory15. From this long history of research, we know the lake follows a consistent annual phenology, and that phenological patterns are changing in response to climate change and invasive species16–19. Rohwer et. al13 found that these phenological dynamics extend to the bacterial community. To confirm that phenological abundance patterns also exist in our more finely resolved bacterial species, we identified annual peaks in species relative abundance using periodograms (magnitude of Fourier transforms). After limiting this temporal analysis to the subset of representative MAGs that occurred at least 30 times over at least 10 years, we found that 72% of these 1,474 bacterial species have consistent seasonal abundance patterns (Fig. 2a).
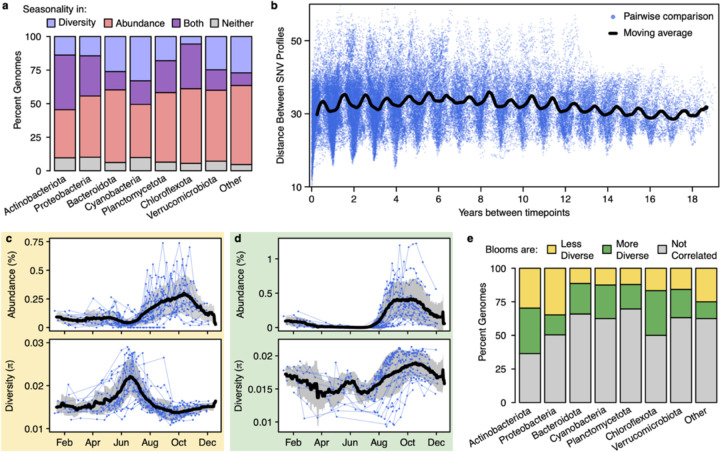
Fig. 2. Bacterial seasonality at the sub-species level.
a, The percent of species with seasonality in nucleotide diversity and/or centered log ratio-transformed relative abundance. The 1,474 reference species that occurred at least 30 times were included in this analysis. b, A time decay plot of the Euclidean distances between the SNV profiles of an abundant species in the Nanopelagicus genus (ME2017-06-13_3300043469_group7_bin14). A more similar SNV profile indicates that the strain composition is more similar. Each blue point represents a pairwise comparison between two sample dates, with the time between those dates on the x-axis. The black line is a 6-month moving average, drawn to highlight the annual periodicity of strain similarities. c, An example of a “less diverse” bloom, where nucleotide diversity decreases as relative abundance increases. Displayed is an abundant species in the Planktophila genus (ME2011-09-04_3300044729_group3_bin142). d, An example of a “more diverse” bloom, where nucleotide diversity increases as abundance increases. Displayed is an abundant species in the Nanopelagicaceae family, MAG-120802 genus (ME2012-08-31_3300044613_group4_bin150). e, The distribution of bloom diversity patterns across the 365 species that had both seasonal abundance and seasonal nucleotide diversity.
To determine whether evolutionary dynamics (i.e. changes in allele frequency within the population) also unfold seasonally, we mapped reads from each sample against each species’ reference genome to identify shifts in strain composition evidenced by shifts in nucleotide diversity (π) and the profiles of single nucleotide variants (SNVs). We found that 33% of the same 1,474 species displayed consistent seasonal nucleotide diversity patterns (Fig. 2a). For a more granular view of strain composition, we created SNV profiles for each species where each date’s profile consists of the percent reference base at each SNV position. After making our abundance cutoff more stringent by requiring a median coverage > 10x, we calculated the Euclidean distance between each sample’s SNV profile for the most abundant 263 species. We found that 80% of these species had consistent phenological patterns in their strain composition (Fig. 2b). This demonstrates that phenological patterns evident in the bacterial community extend to the finest possible taxonomic resolution. Several short-term freshwater studies have also observed changes in strain composition on seasonal timescales20,21. Phenological patterns in sub-species strains suggest that bacterial evolution, evidenced by intraspecific genomic change, also unfolds on seasonal timescales.
Given the ubiquity of seasonal patterns in both species abundance and sub-species diversity, we asked if they were correlated. We quantified whether a species’ “bloom” in abundance consisted of fewer strains or more strains than its baseline composition. Of the 365 species with seasonal patterns in both abundance and nucleotide diversity (purple bars in Fig. 2a), we found both scenarios were common; 21% of these species had less diverse blooms (Fig. 2c and yellow bars in Fig. 2e), while 19% had more diverse blooms (Fig. 2d and green bars in Fig. 2e). Further, all abundant phyla demonstrated an even mix of both bloom types (Fig. 2e). A lower-diversity bloom suggests that a subset of strains outcompeted the others, while a higher-diversity bloom suggests that micro-niches allowed rarer strains to gain abundance, resulting in higher strain diversity due to a more even strain composition. This is in agreement with a previous study that found both overlapping and distinct niches within freshwater bacterial species22. The prevalence of both bloom diversity patterns highlights the lack of a clear boundary between ecological and evolutionary processes since identical intraspecific and interspecific processes are occurring simultaneously.
Long-term ecology and evolution
Long-term changes can be masked by seasonal oscillations, lost in what is referred to as the “invisible present”23. The unprecedented length of the TYMEFLIES metagenome dataset provides a unique lens into the invisible present, enabling the identification of overlayed long-term patterns. To find long-term changes in SNV-based strain profiles, we developed a classifier trained on the distance between each date’s SNV profile and the SNV profile of that species’ first occurrence in the timeseries. We trained this classifier on 11 examples of manually identified temporal patterns, and then applied it to all 263 most abundant species. Our classifier identified gradual change (Fig. 3a), which may arise from genetic drift or in response to a slow press disturbance, as well as abrupt change (Fig 3b and c), which may arise if a disturbance reaches a tipping point threshold leading to a new stable state, or from a sudden environmental shift24,25. Among instances of abrupt change, we identified step changes (Fig. 3b), where the new strain composition persisted during the remainder of our time frame, as well as patterns of disturbance with resilience (Fig. 3c), where the strain composition recovered to baseline.
Fig. 3. Long-term changes in strain composition.
a, An example of long-term, gradual change in strain composition. Points indicate sample dates, and distance refers to the Euclidean distance between a species’ SNV profile on that sample date and that species’ first available SNV profile. A species in the Nanopelagicales order, AcAMD-5 family is shown (ME2005-06-22_3300042363_group2_bin84). b, An example of an abrupt step change in strain composition in a species in the Nanopelagicus genus (ME2011-09-21_3300043464_group3_bin69). c, An example of a disturbance/resilience pattern, where an abrupt change in strain composition is followed by recovery to the original strain composition, in a species in the Planktophila genus (ME2015-07-03_3300042555_group6_bin161). d, Long-term change patterns often overlayed seasonal patterns. Of the 263 species abundant enough to observe their strain profiles, 39 had both long-term and seasonal patterns while 16 had only long-term patterns. e, The distribution of long-term patterns across phyla. Each species that underwent long-term change is indicated by a section of the phyla’s bar, scaled by the mean abundance of that species. The sections corresponding to the examples highlighted in a-c are labelled.
We found that 21% of the most abundant species experienced one kind of long-term change in their strain profiles during our 20-year study period, and these changes overlayed both seasonal and acyclical short-term dynamics (Fig. 3d). Abrupt change was almost twice as common as gradual change (36 vs. 19 species), and resilience was only slightly more common than a lasting step change (20 vs. 16 species) (Fig. 3d). The three long-term change patterns were found in many abundant species distributed across phyla (Fig. 3e). Many abundant Actinobacteriota species experienced long-term change. These long-term changes in SNV profiles reflect shifts in intraspecific strain composition, which is typically attributed to evolutionary processes. The fact that during our observation period over a fifth of the species experienced long-term changes in their strain profiles emphasizes the importance of including contemporary evolutionary change in our understanding of microbial ecology.
Abrupt changes in Nanopelagicaceae
In general, related species did not change in unison with each other, suggesting that the drivers of evolutionary change are highly specific (Fig. 4a). One exception is an abrupt change event that impacted seven species in the Nanopelagicaceae (acI) family in 2012, specifically species in the Nanopelagicus and Planktophila genera (acI-B and acI-A). This is the most abundant family in Lake Mendota and in freshwaters globally10, and the 127 Nanopelagicaceae reference species we recovered accounted for 8% relative abundance on average. Five of these Nanopelagicaceae species displayed resilience to the abrupt change, while two experienced lasting step changes in strain composition.
Fig. 4. Abrupt changes in Nanopelagicaceae strain composition coincide with environmental extremes in 2012.
a, Dates of all abrupt changes in strain composition arranged by phyla. Most changes were isolated events, but multiple species from two abundant genera of Actinobacteriota, Planktophila and Nanopelagicus, experienced abrupt change in 2012. Point size is scaled by species abundance. b, Unusually high epilimnion water temperatures during spring and summer 2012 (relative to 1894 – 2019). c, The preceding winter had an unusually short ice duration (relative to 1853 – 2023). d, Total zooplankton biomass (excluding predatory Bythotrophes and Leptodora) was unusually high, likely enabled by warm early spring temperatures (relative to 1995 – 2018). e, Discharge from the Yahara River, the main tributary to Lake Mendota, was unusually low and lacked high runoff events typical after storms and spring snowmelt (relative to 1989 – 2021). f, Sediment transport of total phosphorus, and g, of soluble reactive phosphorus were low (relative to 1995 – 2021). h, Low phytoplankton biomass, likely resulting from both high zooplankton grazing and low nutrient availability. i, Low dissolved organic carbon (relative to 1996 – 2022), likely a result of low phytoplankton abundance30.
A myriad of possible environmental variables could have driven this event. A leading candidate is climate, which was unusually warm and dry in 2012. The lake experienced its highest epilimnion water temperatures since 189413 (Fig. 4b), the fifth shortest winter ice duration since 185626 (Fig. 4c), the 8th lowest annual discharge from its major tributary since 1976 and the second lowest peak discharge27 (Fig. 4e). These climatic conditions led to top-down and bottom-up controls on the lake’s primary productivity: the highest spring zooplankton abundance since measurements began in 199428 (Fig. 4d) was likely a result of the mild winter and spring allowing zooplankton populations, especially the prolific grazer Daphnia pulicaria to establish early, and low total phosphorus and soluble reactive phosphorus (Fig. 4f–g) was likely a result of low sediment transport associated with mild discharge events29. The resulting combination of high zooplankton grazing and low phosphorus, typically the limiting nutrient in lakes, may be responsible in turn for low phytoplankton biomass (Fig. 4h). Lake Mendota’s dissolved organic carbon (DOC) is primarily provided by phytoplankton30, and consequently DOC was also low in 2012 (Fig. 4i).
Another possible driver is the irruption of the invasive zooplankton spiny water flea (Bythorephes cedertrömii) in 2009. This major disturbance resulted in a trophic cascade that decreased water clarity19,31, increased lake anoxia28, and shifted the bacterial community composition13. Although the abrupt changes in strain composition of seven Nanopelagicaceae species were observed three years later, lag effects are common in complex ecosystems32. Ecosystem-wide drivers like the 2012 climate anomalies and the 2009 species invasion can have cascading and interacting effects on nutrient and carbon dynamics, which in turn impact the bacterial community. The observed long-term intraspecific changes suggest that such ecological drivers are also drivers of evolutionary change, further emphasizing how ecology and evolution are intertwined.
Evolutionary signals in a Nanopelagicus
To understand the dynamics of abrupt evolutionary change, we further examined one of the abundant species, a Nanopelagicus (acI-B), that experienced a step change in strain composition in August 2012 (Fig. 3b). An NMDS ordination of its SNV profiles indicated the strain composition changed abruptly at that time and settled into a new composition after a period of adjustment in 2012 and 2013 (Fig. 5a).
Fig. 5. Step change in strain composition coincides with more genes under selection.
a, An abundant Nanopelagicus species experienced a step change in strain composition in 2012 (ME2011-09-21_3300043464_group3_bin69, see also Fig. 3b). Samples with more similar SNV profiles appear closer on this NMDS plot. Years 2000–2011 cluster together and are distinct from years 2014–2019, which cluster separately. A sudden change in strain composition occurred on August 3, 2012. b, Despite the abrupt change in strain composition, the relative abundance this species remained constant over time. c, Concurrent with the shift in strain composition, nucleotide diversity increased and then remained high, indicating that the new equilibrium was comprised of a more diverse assemblage of strains. d, The absence of a spike in the number of new SNVs suggests that an increase in the evenness of existing strains occurred, rather than the introduction of new strains. e, Concurrent with the shift in strain composition, the number of genes under positive selection also increased (McDonald-Kreitman F-statistic p-value < 0.05). f, Occurrence of consistently selected genes in all the samples, in the pre-2012 period, and in the post-2012 period. X-axis indicates samples over time and Y-axis indicates genes. Shading indicates the significance level of positive selection. Amino acid-related genes and nucleic acid-related genes are indicated on the right axis. Full annotations are available in Supplementary Table 3. Note that the X-axis is evenly spaced by sample, so that years with more samples take up more space.
The relative abundance of this species was quite constant throughout our 20-year observation period (Fig. 5b), typically with higher abundances during the spring clearwater phase. The step change in strain composition (Fig. 3b) coincided with one in nucleotide diversity (Fig. 5c). These patterns could result from the spread of a new strain or with an increase in the evenness of abundance in existing strains. To distinguish between these hypotheses, we counted the number of previously unobserved SNVs in the mapped reads of every sample. We did not see large spikes in new SNVs in 2012 (Fig. 5d), suggesting that the step changes reflected a shift in the relative abundances of existing strains.
This interpretation is consistent with a dramatic increase in the number of genes under positive selection that occurred at this time (Fig. 5e). As the relative abundances of some strains increase, alleles specific to them appear to undergo partial (or “soft”) selective sweeps. As the strain composition reequilibrates, this signal dies out (Fig 5e). To identify candidate loci that may be important to adaptation that occurred during our sampling, we sought genes that consistently showed signals of selection over the entire timeseries, only during the pre-2012 period, and only during the post-2012 period. Four genes were consistently selected both pre- and post-2012, four genes were consistently selected pre-2012, and 33 genes were consistently selected post-2012. We used gene functional predictions33 to identify their potential metabolic pathways. Of the 33 consistently selected genes post-2012, ten are involved in amino acid metabolism or aminoacylation, and seven are involved with nucleic acid synthesis or degradation (Fig. 5f).
Previously, the absence of biosynthesis or auxotrophies for amino acids and nucleotides has been highlighted for microorganisms with streamlined genomes34,35. In the streamlined Nanopelagicus, auxotrophies for various amino acids12,36 coupled with an enrichment of transporters for many small organic nitrogen compounds, including amino acids12,37,38 and nucleic acid components12,36–38 are common. Moreover, the histidine pathway was found split between two different strains of Nanopelagicus growing in a mixed culture36. Our observation of consistent selection on amino acid and nucleic acid metabolism suggests that these genes differentiate the post-2012 strains, thus biosynthesis, use and reuse of small organic nitrogen compounds are key in the ecology and evolution of these globally abundant lake bacteria.
A continuum of ecology and evolution
The interface between ecology and evolution is delineated by species boundaries, but in bacteria species concepts and definitions are hotly debated39. Using a commonly chosen definition for microbial species boundaries, we found processes that unfold as interspecies ecological dynamics also occur in intraspecies evolution. Moreover, the time scales of these processes overlapped, as did likely environmental drivers. How microbes will respond to global changes in land use, invasive species, and climate40 are pressing questions that require an understanding of long-term change. Few microbial timeseries are long enough to capture such dynamics, but decadal observations are an essential approach to understand how complex ecosystems respond to global change. The two-decade TYMEFLIES dataset will serve as an invaluable community resource to continue addressing these questions and move us toward a unified approach to microbial ecology and evolution. Microbial ecology and evolution must be considered simultaneously and combined into a unified theory.
Methods
Data Availability
Metagenome sequences are available through the DOE Joint Genome Institute’s Genome Portal under Proposal 504350 (https://genome.jgi.doe.gov/portal/Exttemetagenomes/Exttemetagenomes.info.html), and through the NCBI Sequence Read Archive at accessions listed in Supplementary Table 1. Reference MAG sequences are available through the NCBI GenBank IDs listed in Supplementary Table 2. Environmental data is publicly available through the Environmental Data Initiative (https://edirepository.org/)41–49 and the U.S. Geological Survey’s Water Data for the Nation (https://waterdata.usgs.gov/nwis)27.
Code Availability
Custom scripts used for data processing are available at https://github.com/rrohwer/TYMEFLIES and on Zenodo.
Lake Mendota Samples
Lake Mendota is a eutrophic temperate lake located in Madison, Wisconsin (USA). Integrated epilimnion samples were collected from the upper 12 m at a 25 m deep location referred to as the central “deep hole” (43°05’58.2”N 89°24’16.2”W). Bacteria were collected on 0.2 μm polyethersulfone filters (Pall Corporation), stored at −80°C, and DNA was extracted after randomizing sample order by a single person in 2018–2019 using FastDNA Spin Kits (MP Biomedicals). A detailed description of the study site, sample collection, and DNA extraction procedures is provided by Rohwer and McMahon4.
Metagenome sequencing and assembly
Samples were sequenced by the US Department of Energy Joint Genome Institute (JGI) using a NovaSeq 6000 with an S4 flow cell. Sample metadata is available in Supplementary Table 1, and raw sequencing data is available through the JGI Genome Portal under Proposal 504350 (https://genome.jgi.doe.gov/portal/Exttemetagenomes/Exttemetagenomes.info.html), or from the NCBI Sequence Read Archive under accession numbers listed in Supplementary Table 1. Read filtering was performed using standard JGI protocols50, which are additionally detailed as metadata paired with each sample through the JGI IMG/M website. Briefly, BBDuk51 was used to remove adapters and quality trim reads, and BBMap51 was used to identify and remove common contaminants. In our analyses we treated the resulting filtered fastq files as the metagenome reads. Single-sample assemblies were also generated by JGI with their standard protocol50 using metaSPAdes52. These filtered fastq files and paired single-sample assemblies are available through the JGI Genome Portal under Proposal 504350.
Obtaining and characterizing genomes
Genomes were binned out of metagenomes using the Texas Advanced Computing Center’s Lonestar6 supercomputer. Metagenomic reads were mapped back to sample assemblies using BBMap (version 38.22)51, sorted BAM files were created using SAMtools (version 1.9)53, and metagenome-assembled genomes were binned using MetaBAT2 (version 2.12.1)54. Metagenomic reads from different samples were cross-mapped back to each assembly. Cross-mapping scales exponentially, so it was performed on assemblies and sample reads broken into approximately 50-sample groups of consecutive sample dates, with samples from the same year grouped together. This resulted in 85,684 genome bins. CheckM2 (version 0.1.3)5 was used to asses bin quality, including completeness and contamination estimates, and GTDB-tk (version 2.1.1)55 was used to assign GTDB taxonomy (release 207)56 to all bins. 30,389 genome bins were at least 50% complete and less than 10% contaminated, and these bins were de-replicated to 96% ANI using dRep (version 3.4.0)6. To choose 96% as our ANI cutoff, we ran dRep at ANIs ranging from 90 to 99% and examined the resulting number of de-replicated bins, as well as the number of bins from the same assembly that were combined. We chose 96% ANI because very few (one) of the 30,389 bins were combined into an ANI group with a bin created from the same assembly, and because 96% ANI was generally located right before a sudden increase in the total number of genome groups. Our goal was to separate as many species as possible, while combining strains that were so closely related they would compete for mapped reads. Applying a 96% ANI cutoff with dRep resulted in 2,855 representative genomes, which we treated as species in this study.
To quantify the relative abundance of each species in every sample, we mapped all sample reads against the concatenated 96% ANI reference genomes using bbmap (version 38.22)51, created sorted BAM files using SAMtools (version 1.9)53, and calculated relative abundance using coverM (version 0.6.1)57. With the coverM software, we required a minimum read percent identity of 93, proper pairs only, and excluded 1000 bp from each contig end from the calculation. CoverM calculates relative abundance as the mean coverage divided by the mean coverage across all genomes multiplied by the proportion of reads that mapped to the genome, thus normalizing by genome size to estimate the fraction of cells that belong to a given species in each sample. A table of representative MAGs along with taxonomy annotations, quality statistics, and abundance statistics is available as Supplementary Table 2.
To further characterize the genomes, we ran inStrain (version 1.7.1)58 using a minimum read ANI of 93%, as recommended by the inStrain documentation given our previous choice of 96% ANI to dereplicate genomes. This software called SNVs and calculated nucleotide diversity, among other metrics. To identify genes we ran prodigal (version 2.6.3)59 on each genome separately. We then used Kofamscan (version 1.3.0)60 to assign gene annotations from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (release 107.1)33. Additional custom analyses were performed using the R programming language (version 4.1.2)61, and relied extensively on the data.table R package (version 1.14.8)62, the lubridate R package (version 1.9.3)63, and GNU parallel (version ‘Chandrayaan’)64.
Classifying seasonal and long-term change
To classify each species’ abundance pattern as seasonal or not, we started with relative abundances as calculated by coverM (version 0.6.1)57 and further corrected any abundance to zero if the genome’s coverage breadth was 70% or less than its expected breadth, as calculated by inStrain (version 1.7.1)58. We then applied a centered log ratio transformation to the relative abundance values using the compositions R package (version 2.0–6)65. After taking a daily linear interpolation to obtain evenly spaced samples, we detrended the temporal profiles with a cubic fit. Finally, we performed a periodogram analysis by computing the magnitude of the fast Fourier transform. If a peak occurred within 30 days of 365 days we considered it an annual oscillation, and if any of the top five peaks corresponded to an annual period, we classified the species as having a seasonal abundance pattern. We applied this analysis only to the 1,474 species that occurred on least 30 dates over at least 10 years. To classify each species’ nucleotide diversity pattern as seasonal or not, we similarly performed a fast Fourier transform on its inStrain-calculated nucleotide diversity over time. We used the same periodogram analysis to classify it as having seasonal nucleotide diversity or not, and we applied this analysis to the same subset of 1,474 species.
To characterize blooms as more diverse or less diverse, we calculated the Pearson correlation between centered log ratio-transformed relative abundance and nucleotide diversity for the 365 species that had both seasonal abundance and seasonal nucleotide diversity annual oscillations. We considered it a positive correlation (more diverse blooms) if the Pearson correlation was at least 0.35 and a negative correlation (less diverse blooms) if the Pearson correlation was less than or equal to −0.35. We repeated this analysis with up to two weeks of lag and used the highest correlation within that window. We chose 0.35 as a reasonable cutoff after manual examination of the first 150 species’ correlations.
To calculate SNV profiles for each species, we created vectors corresponding to every SNV position in its genome, where the value of each element was the percent of mapped reads that matched the reference genome base at that position in each sample. SNV’s were called using inStrain58, and we only applied this analysis to samples where the species’ median coverage was over 10x, as at coverages less than that we observed a drop in the total SNVs called. Therefore, for both long-term and seasonal analysis of SNV profiles, we included only species that had medium coverage over 10x on at least 30 dates over at least 10 years, which resulted in a subset of 263 species. To identify changes in SNV profiles, we created a distance matrix for each species based on Euclidean distances between each sample’s SNV profile using the vegan R package (version 2.6–4)66. From this we created a table of time elapsed and Euclidean distance between each sample date.
To identify seasonal patterns in each species’ SNV profiles, we created a daily linear interpolation of pairwise distances between all samples, taking the mean when multiple sample pairs occurred 20 with the same time interval. After detrending with a cubic fit, we performed a periodogram analysis to identify annual oscillations and the presence of seasonal patterns using the same criteria as with our abundance and nucleotide diversity annual oscillation analysis.
To identify long-term change patterns, we subset our pairwise distance table to the distance of each sample from the first sample. We developed a classifier for these temporal profiles of distances between SNV profiles using 11 manually chosen species. Our classifier criteria was hierarchical: first gradual change was identified, then step change was identified, and finally disturbance/resilience patterns were identified. After training, the classifier was applied to all 263 species above the abundance cutoff. Gradual change was identified if a linear fit to the daily linearly interpolated distances, excluding dates closer than a month to the starting date, resulted in an adjusted R2 of at least 0.55. Dates closer than a month to the starting date were excluded because they tended to be highly similar, and a linear interpolation was applied to account for uneven sampling dates, particularly the high frequency of summer sampling in the latter decade of the timeseries. Possible step change locations were identified after excluding dates closer than a month to the starting date and applying an F test to the linearly interpolated distances using the strucchange R package (version 1.5–3)67. If a breakpoint was identified by the F test, the means of measured (as opposed to interpolated) before and after distances were different (Mann-Whitnes p-value < 0.01), and the step resulted in a new mean at least 33% higher than the previous mean, a step change pattern was identified. Disturbance/resilience patterns were then identified using outlier distances calculated by the default boxplot statistics in R. If a date’s distance was > 1.5 times the difference between the 3rd and 1st quartile of observed distances a date was considered an outlier, and if outlier values were maintained for at least a month the species was classified as having a disturbance event with resilience.
Analyzing abrupt change in Nanopelagicaceae
To place environmental conditions in 2012 in context, historical environmental data was collected from the North Temperate Lakes Long-Term Ecological Research program (NTL-LTER) through the Environmental Data Initiative (EDI) interface (https://edirepository.org/) and the US Geological Survey (USGS) Water Data for the Nation (https://waterdata.usgs.gov/nwis) using the USGS dataRetrieval R package (version 2.7.14)68. EDI datasets analyzed included ice duration26; nutrients, pH, and carbon41; major ions42; water temperatures combined from multiple datasets45–49 as described in Rohwer et al.13; phytoplankton43; and zooplankton44 converted to biomass as described in Rohwer, Ladwig, et al.28. River discharge measurements were obtained from the USGS for the Yahara River, the primary tributary into Lake Mendota (site ID: 05427718)27. After exploring all parameters included in these datasets, the occurrence of a hot, dry year with low primary productivity became apparent.
Relative abundance and nucleotide diversity of the Nanopelagicus MAG ME2011-09-21_3300043464_group3_bin69 were calculated as for the seasonal analysis. New SNVs were identified as SNV positions that were called by inStrain58 for the first time in a given sample. Genes under selection were identified using dN/dS and pN/pS ratios as calculated by inStrain58. A McDonald-Kreitman test69 was used to identify positively selected genes where the bias of unfixed SNVs to be nonsynonymous was lower than the bias of fixed SNVs to be nonsynonymous (pNpS/dNdS < 1), and positive selection was considered statistically significant when the Fisher p-value was less than or equal to 0.05. A gene was considered consistently selected if it appeared under significant positive selection with high frequency (in the 4th quartile). Consistently selected genes were identified for the pre-2012 and post-2012 time periods separately.
Gene annotations were analyzed in the context of the KEGG pathways33 they belonged to. For each potential pathway, all genes present in the genome were visualized with KEGG Pathway Maps (https://www.genome.jp/brite/br08901). When multiple genes that surrounded the selected gene existed in the genome, that pathway was considered a likely annotation. When likely pathways involved amino acid metabolism or aminoacylation, they were considered amino acid-related. When likely pathways involved purine or pyrimidine metabolism, they were considered nucleic acid-related.
Supplementary Material
All the JGI, GOLD, NCBI, and internal McMahon Lab identifiers that pair with each metagenome sample.
NCBI identifiers corresponding to each reference genome, as well as the quality results from CheckM25, the taxonomy results from GTDB-tk55, and average relative abundances as calculated by coverM57.
KEGG annotations of consistently selected genes. Table row order matches heatmap row order in Fig. 5f.
Acknowledgements
Long-term datasets such as TYMEFLIES rely on researchers who contribute a portion of their time and effort to future projects. This work would not be possible without the generosity of many, including Lake Mendota sampling leads Angela Kent, Tony Yannarell, Ashley Shade, Stuart Jones, Ryan Newton, Georgia Wolfe, Emily Kara Read, Lucas Beversdorf, James Mutschler, and the original Microbial Observatory lead Eric W. Triplett. We thank Peter Golightly for his advice on interpreting the genes under selection data.
Funding Statement
This project was supported by an E. Michael and Winona Foster WARF Wisconsin Idea Fellowship (R.R.R.), the U.S. National Science Foundation (NSF) (DBI-2011002) (R.R.R.), the U.S. National Institutes of Health (RO1-GM116853) (M.Kirk.), the U.S. NSF (DEB-1831730) (M. Kirk.), the U.S. Department of Agriculture (WIS01516 and WIS01789) (K.D.M.), the U.S. NSF (DEB-0702395, DEB-1344254) (K.D.M.) and a Simons Foundation Investigator in Aquatic Microbial Ecology Award (LI-SIAME-00002001) (B.J.B.). Support for sampling was provided by the U.S. NSF North Temperate Lakes Long-Term Ecological Research program (DEB-9632853, DEB-0217533, DEB-0822700, DEB-1440297, DEB-2025982) and the U.S. NSF Microbial Observatory program (MCB-9977903, DEB-0702395). The work (proposal: https://doi.org/10.46936/10.25585/60001198) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No. DE-AC02-05CH11231. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper (http://www.tacc.utexas.edu).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Hug L. A. et al. A new view of the tree of life. Nature Microbiology 1, (2016). [DOI] [PubMed] [Google Scholar]
- 2.Brennan G. L. & Logares R. Tracking contemporary microbial evolution in a changing ocean. Trends in Microbiology 31, 336–345 (2023). [DOI] [PubMed] [Google Scholar]
- 3.Lenski R. E. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J 11, 2181–2194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohwer R. R. & McMahon K. D. Lake iTag measurements over nineteen years, introducing the limony dataset. 2022.08.04.502869 Preprint at 10.1101/2022.08.04.502869 (2022). [DOI] [Google Scholar]
- 5.Chklovski A., Parks D. H., Woodcroft B. J. & Tyson G. W. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. 2022.07.11.499243 Preprint at 10.1101/2022.07.11.499243 (2022). [DOI] [PubMed] [Google Scholar]
- 6.Olm M. R., Brown C. T., Brooks B. & Banfield J. F. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J 11, 2864–2868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varghese N. J. et al. Microbial species delineation using whole genome sequences. Nucleic Acids Research 43, 6761–6771 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain C., Rodriguez-R L. M., Phillippy A. M., Konstantinidis K. T. & Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olm M. R. et al. Consistent Metagenome-Derived Metrics Verify and Delineate Bacterial Species Boundaries. mSystems 5, 10.1128/msystems.00731-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipko I. A. & Belykh O. I. Environmental Features of Freshwater Planktonic Actinobacteria. Contemp. Probl. Ecol. 14, 158–170 (2021). [Google Scholar]
- 11.Kim S., Kang I., Seo J.-H. & Cho J.-C. Culturing the ubiquitous freshwater actinobacterial acI lineage by supplying a biochemical ‘helper’ catalase. ISME J 13, 2252–2263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neuenschwander S. M., Ghai R., Pernthaler J. & Salcher M. M. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J 12, 185–198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohwer R. R., Hale R. J., Vander Zanden M. J., Miller T. R. & McMahon K. D. Species invasions shift microbial phenology in a two-decade freshwater time series. Proceedings of the National Academy of Sciences 120, e2211796120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magnuson J. J., Kratz T. K. & Benson B. J. Long-Term Dynamics of Lakes in the Landscape: Long-Term Ecological Research on North Temperate Lakes. (Oxford University Press, 2006). [Google Scholar]
- 15.Yannarell A. C., Kent A. D., Lauster G. H., Kratz T. K. & Triplett E. W. Temporal Patterns in Bacterial Communities in Three Temperate Lakes of Different Trophic Status. Microb Ecol 46, 391–405 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Magee M. R. & Wu C. H. Effects of changing climate on ice cover in three morphometrically different lakes. Hydrol. Process. 31, 308–323 (2017). [Google Scholar]
- 17.Magee M. R. & Wu C. H. Response of water temperatures and stratification to changing climate in three lakes with different morphometry. Hydrology and Earth System Sciences 21, 6253–6274 (2017). [Google Scholar]
- 18.Snortheim C. A. et al. Meteorological drivers of hypolimnetic anoxia in a eutrophic, north temperate lake. Ecological Modelling 343, 39–53 (2017). [Google Scholar]
- 19.Matsuzaki S.-I. S. et al. Climate and food web effects on the spring clear-water phase in two north-temperate eutrophic lakes. Limnol. Oceanogr. n/a, 1–17 (2020).32704188 [Google Scholar]
- 20.Okazaki Y., Nakano S., Toyoda A. & Tamaki H. Long-Read-Resolved, Ecosystem-Wide Exploration of Nucleotide and Structural Microdiversity of Lake Bacterioplankton Genomes. mSystems 7, e00433–22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meziti A. et al. Quantifying the changes in genetic diversity within sequence-discrete bacterial populations across a spatial and temporal riverine gradient. The ISME Journal 13, 767 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia S. L. et al. Contrasting patterns of genome-level diversity across distinct co-occurring bacterial populations. The ISME Journal 1 (2017) doi: 10.1038/s41396-017-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magnuson J. J. Long-Term Ecological Research and the Invisible Present. BioScience 40, 495–501 (1990). [Google Scholar]
- 24.Turner M. G. et al. Climate change, ecosystems and abrupt change: science priorities. Philosophical Transactions of the Royal Society B: Biological Sciences 375, 20190105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheffer M., Carpenter S., Foley J. A., Folke C. & Walker B. Catastrophic shifts in ecosystems. Nature 413, 591–596 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Magnuson J. J., Carpenter S. R. & Stanley E. H. North Temperate Lakes LTER: Ice Duration - Madison Lakes Area 1853 - current. Environmental Data Initiative 10.6073/PASTA/69B3391E13955392587413ECBFC7C298 (2023). [DOI] [Google Scholar]
- 27.U.S. Geological Survey National Water Information System data available on the World Wide Web. USGS 05427718 Yahara River at Windsor, WI, Daily Discharge 00060. U.S. Geological Survey National Water Information System data available on the World Wide Web (2023).
- 28.Rohwer R. R. et al. Increased anoxia following species invasion of a eutrophic lake. Limnology and Oceanography Letters n/a, (2023). [Google Scholar]
- 29.Carpenter S. R., Booth E. G. & Kucharik C. J. Extreme precipitation and phosphorus loads from two agricultural watersheds. Limnology and Oceanography 63, 1221–1233 (2018). [Google Scholar]
- 30.Berg S. M., Peterson B. D., McMahon K. D. & Remucal C. K. Spatial and Temporal Variability of Dissolved Organic Matter Molecular Composition in a Stratified Eutrophic Lake. Journal of Geophysical Research: Biogeosciences 127, e2021JG006550 (2022). [Google Scholar]
- 31.Walsh J. R., Carpenter S. R. & Zanden M. J. V. Invasive species triggers a massive loss of ecosystem services through a trophic cascade. PNAS 113, 4081–4085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rastetter E. B. et al. Time lags: insights from the U.S. Long Term Ecological Research Network. Ecosphere 12, e03431 (2021). [Google Scholar]
- 33.Kanehisa M., Furumichi M., Sato Y., Kawashima M. & Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Research 51, D587–D592 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramoneda J., Jensen T. B. N., Price M. N., Casamayor E. O. & Fierer N. Taxonomic and environmental distribution of bacterial amino acid auxotrophies. Nat Commun 14, 7608 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castelle C. J. et al. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol 16, 629–645 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Garcia S. L. et al. Auxotrophy and intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol Ecol 24, 4449–4459 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Garcia S. L. et al. Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. The ISME Journal 7, 137 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamilton J. J. et al. Metabolic Network Analysis and Metatranscriptomics Reveal Auxotrophies and Nutrient Sources of the Cosmopolitan Freshwater Microbial Lineage acI. MSystems 2, e00091–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosselló-Móra R. & Amann R. Past and future species definitions for Bacteria and Archaea. Systematic and Applied Microbiology 38, 209–216 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Tiedje J. M. et al. Microbes and Climate Change: a Research Prospectus for the Future. mBio e00800–22 (2022) doi: 10.1128/mbio.00800-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magnuson J. J., Carpenter S. R. & Stanley E. H. North Temperate Lakes LTER: Chemical Limnology of Primary Study Lakes: Nutrients, pH and Carbon 1981 - current. Environmental Data Initiative 10.6073/PASTA/325232E6E4CD1CE04025FA5674F7B782 (2023). [DOI] [Google Scholar]
- 42.Magnuson J. J., Carpenter S. R. & Stanley E. H. North Temperate Lakes LTER: Chemical Limnology of Primary Study Lakes: Major Ions 1981 - current. Environmental Data Initiative 10.6073/pasta/bb563f16c7338fdb3ddf82057ef43cc6 (2023). [DOI] [Google Scholar]
- 43.Magnuson J. J., Carpenter S. R. & Stanley E. H. North Temperate Lakes LTER: Phytoplankton - Madison Lakes Area 1995 - current. Environmental Data Initiative 10.6073/PASTA/43D3D401AF88CC05C6595962BDB1AB5C (2022). [DOI] [Google Scholar]
- 44.Magnuson J., Carpenter S. & Stanley E. North Temperate Lakes LTER: Zooplankton - Madison Lakes Area 1997 - current. Environmental Data Initiative 10.6073/PASTA/D5ABE9009D7F6AA87D1FCF49C8C7F8C8 (2022). [DOI] [Google Scholar]
- 45.Rohwer R. R. & McMahon K. D. Lake Mendota Microbial Observatory Temperature, Dissolved Oxygen, pH, and conductivity data, 2006-present. Environmental Data Initiative 10.6073/PASTA/7E533C197ED8EBD27777A89A2C8D7DFE (2022). [DOI] [Google Scholar]
- 46.Magnuson J. J., Carpenter S. R. & Stanley E. H. North Temperate Lakes LTER: Physical Limnology of Primary Study Lakes 1981 - current. Environmental Data Initiative 10.6073/PASTA/316203040EA1B8ECE89673985AB431B7 (2021). [DOI] [Google Scholar]
- 47.Magnuson J., Carpenter S. & Stanley E. North Temperate Lakes LTER: High Frequency Water Temperature Data - Lake Mendota Buoy 2006 - current. Environmental Data Initiative 10.6073/PASTA/8CEFF296AD68FA8DA6787076E0A5D992 (2020). [DOI] [Google Scholar]
- 48.Robertson D. Lake Mendota water temperature secchi depth snow depth ice thickness and meterological conditions 1894 – 2007. Environmental Data Initiative 10.6073/PASTA/F20F9A644BD12E4B80CB288F1812C935 (2016). [DOI] [Google Scholar]
- 49.Magnuson J. J., Carpenter S. R. & Stanley E. H. Lake Mendota Multiparameter Sonde Profiles: 2017 - current. Environmental Data Initiative 10.6073/PASTA/5F15BF453851987FC030B2F07A110B21 (2021). [DOI] [Google Scholar]
- 50.Clum A. et al. DOE JGI Metagenome Workflow. mSystems 6, 10.1128/msystems.00804-20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bushnell B. BBMap Short Read Aligner and Other Bioinformatic Tools. https://www.osti.gov/biblio/1241166 (2014). [Google Scholar]
- 52.Nurk S., Meleshko D., Korobeynikov A. & Pevzner P. A. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7, e7359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaumeil P.-A., Mussig A. J., Hugenholtz P. & Parks D. H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics btz848 (2019) doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parks D. H. et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Research 50, D785–D794 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aroney S. T. N. et al. CoverM: Read coverage calculator for metagenomics. Zenodo 10.5281/zenodo.10531254 (2024). [DOI] [Google Scholar]
- 58.Olm M. R. et al. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol 39, 727–736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyatt D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aramaki T. et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2022). [Google Scholar]
- 62.Barrett T. et al. Data.Table: Extension of `data.Framè. (2024). [Google Scholar]
- 63.Grolemund G. & Wickham H. Dates and Times Made Easy with lubridate. Journal of Statistical Software 40, 1–25 (2011). [Google Scholar]
- 64.Tange O. GNU Parallel 20230822 (‘Chandrayaan’). Zenodo 10.5281/zenodo.8278274 (2023). [DOI] [Google Scholar]
- 65.Boogaart K. G. van den, Tolosana-Delgado R. & Bren M. compositions: Compositional Data Analysis. (2023). [Google Scholar]
- 66.Oksanen J. et al. vegan: Community Ecology Package. (2022). [Google Scholar]
- 67.Zeileis A. et al. strucchange: Testing, Monitoring, and Dating Structural Changes. (2022). [Google Scholar]
- 68.DeCicco L., Hirsch R., Lorenz D., Watkins D. & Johnson M. dataRetrieval: R Packages for Discovering and Retrieving Water Data Available from U.S. Federal Hydrologic Web Services. (U.S. Geological Survey, Reston, VA, 2023). doi: 10.5066/P9X4L3GE. [DOI] [Google Scholar]
- 69.McDonald J. H. & Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351, 652–654 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All the JGI, GOLD, NCBI, and internal McMahon Lab identifiers that pair with each metagenome sample.
NCBI identifiers corresponding to each reference genome, as well as the quality results from CheckM25, the taxonomy results from GTDB-tk55, and average relative abundances as calculated by coverM57.
KEGG annotations of consistently selected genes. Table row order matches heatmap row order in Fig. 5f.
Data Availability Statement
Metagenome sequences are available through the DOE Joint Genome Institute’s Genome Portal under Proposal 504350 (https://genome.jgi.doe.gov/portal/Exttemetagenomes/Exttemetagenomes.info.html), and through the NCBI Sequence Read Archive at accessions listed in Supplementary Table 1. Reference MAG sequences are available through the NCBI GenBank IDs listed in Supplementary Table 2. Environmental data is publicly available through the Environmental Data Initiative (https://edirepository.org/)41–49 and the U.S. Geological Survey’s Water Data for the Nation (https://waterdata.usgs.gov/nwis)27.