Abstract
Enteropathogenic Escherichia coli (EPEC) strains are a common cause of infantile diarrhea in developing countries. EPEC strains induce a characteristic attaching and effacing (A/E) lesion on epithelial cells. A/E lesion formation requires intimin, an outer membrane adhesin protein. The cell-binding activity of intimin is localized at the C-terminal 280 amino acids of the polypeptide (Int280). So far, four distinct Int280 types (α, β, γ, and δ) have been identified. The aim of this study was to identify immunodominant regions within the Int280α and Int280β domains. Recombinant DNA was used to construct and express overlapping polypeptides spanning these domains. Rabbit anti-Int280 antisera and human colostral immunoglobulin A were reacted with these polypeptides in Western blots and enzyme-linked immunosorbent assays. The results obtained with the rabbit antisera showed the presence of two separate immunodominant regions which are common to both Int280α and Int280β. The first localized within the N-terminal region of Int280, and the second localized between amino acids 80 and 130. The results with the human colostra revealed one reactivity pattern against the Int280α fragments but two different reactivity patterns against the Int280β domain.
Diarrheal diseases are among the leading causes of early childhood mortality in the developing world. A common etiological agent of severe diarrhea in infants is enteropathogenic Escherichia coli (EPEC) (32). EPEC strains colonize the small intestinal mucosa and, by subverting intestinal epithelial cell function, produce a characteristic histopathological feature known as the attaching and effacing (A/E) lesion (36). The A/E lesion is characterized by localized destruction (effacement) of brush border microvilli, intimate bacterial adhesion to the host cell membrane, and induction of gross cytoskeletal reorganization leading to formation of a pedestal-like structure in the host cell consisting of polymerized actin, α-actinin, ezrin, talin, and myosin (13, 28, 41). A/E lesions are produced by EPEC in a variety of tissue culture cell lines (28). Similar lesions have been associated with several other human and animal bacterial mucosal pathogens, including enterohemorrhagic E. coli (EHEC) (11, 27), rabbit diarrheagenic E. coli (RDEC-1) (5), and the mouse pathogen Citrobacter rodentium (39).
The first gene to be associated with A/E activity was the eae gene (23) encoding intimin, an outer membrane protein, required for intimate bacterial attachment and full virulence in volunteers (10). Subsequently, the eae gene was shown to be part of a large pathogenicity island, the LEE region (34), which contains all of the genes required for the A/E phenotype (35). In addition, to intimin, the LEE encodes a type III secretion system (22), a translocated intimin receptor (Tir [EspE]) (8, 24), and three EPEC-secreted proteins (Esps [EspA, EspB, and EspD]) required for protein translocation (24, 29, 44), signal transduction in host cells, and A/E lesion formation (12, 25, 31).
Studies of the intimin family of proteins showed that their cell-binding activity is localized to the C-terminal 280 amino acids (Int280) (14) and that a specific cysteine residue (Cys937) in EPEC intimin is essential for binding activity (15, 17). Moreover, antisera raised to Int280 polypeptides, used to investigate the regulation of intimin expression during bacterial growth and A/E lesion formation, have shown that intimin expression is induced during the logarithmic growth phase at 37°C but is down regulated following A/E lesion formation (1, 20, 26).
Population genetic surveys with multilocus enzyme electrophoresis have shown that the classical EPEC strains have diverged into two major groups of related clones, designated EPEC clone 1 and EPEC clone 2 (37, 38, 43). In a recent study, we used immunological (anti-Int280 antisera) and genetic (PCR) approaches to study antigenic variation and classify the cell-binding domain of intimin expressed by the different EPEC clones (1). Our results revealed the presence of at least five distinct intimin subtypes: intimin α, intimin β, intimin γ, and intimin δ. Importantly, intimin α was specifically expressed by strains which belong to EPEC clone 1 (O55:H6, O127:H6, O142:H6, and O142:H34), intimin β was mainly associated with EPEC strains belonging to clone 2 (O26:H−, O111:H−, O111:H2, O114:H2, O119:H2, O119:H6, and O128:H2) and EHEC O26:H11, intimin γ was associated with EHEC O157:H7 and EPEC O55:H− and O55:H7, and intimin δ was associated with EPEC O86:H34. Significantly, a very low level of cross-reactivity was observed between antisera made with Int280α (Int280 of intimin α) and Int280β (Int280 of intimin β) as immunogens (1).
Clinical and epidemiological investigations have demonstrated that breast-feeding is protective against infantile infectious diarrhea, including EPEC infection (6, 7). Immunoglobulin A (IgA) obtained from pools of colostrum from mothers living in high-risk areas of the city São Paulo, Brazil, strongly recognized a 94-kDa EPEC-associated protein; these IgA preparations prevented localized adherence of EPEC to cultured human epithelial cells (4). In a recent study, we analyzed IgA antibodies from pools of human colostrum obtained from mothers 2 to 3 days after giving birth to healthy infants at the São Paulo University Hospital. We found that IgA reactive with all of the EPEC virulence-associated proteins tested, including intimin (33). Mucosal IgA antibodies to intimin were also detected in mice 28 days after challenge with C. rodentium (18). Therefore, these studies suggest that anti-intimin antibodies might play a key role in protection against infection with eae-positive bacterial pathogens.
The aim of the present study was to use rabbit polyclonal antisera and individual human colostrum samples to map immunodominant regions within the C-terminal domains of intimin α and intimin β. This was achieved by using various overlapping fragments of Int280 fused to maltose binding protein (MBP). We report the identification of multiple immunodominant regions within Int280α and Int280β.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Representative strains of EPEC clone 1 (ICC64, O127:H6) and EPEC clone 2 (ICC61, O114:H2) (1) were used for construction of MBP fusion proteins. E. coli TG1 cells, harboring recombinant pMal-c2 plasmids, were grown in L broth or L agar. Medium was supplemented with 100 μg of ampicillin per ml where appropriate.
Construction and purification of MBP-Int fusion proteins.
MBP fusion proteins MBP-Int280α and MBP-Int280β and derivatives of these domains containing N- and/or C-terminal deletions were constructed as previously described (15). Briefly, purified DNAs from EPEC ICC64 (O127:H6) and EPEC ICC61 (O114:H2) were used as templates for PCR amplification of the different Int280 regions. The primer pairs used for the construction of each of the MBP-Int fusions are shown in Table 1. The relevant characteristics of the MBP fusion proteins used in this study are given in Table 2. The amplified DNA products were cloned into pMal-c2 and the MBP fusion protein, purified as described previously (15).
TABLE 1.
Primers used to amplify various regions of the eae genes from O127:H6 and O114:H2
| pMal-Int construct | Primer
|
|
|---|---|---|
| Forwarda | Reverseb | |
| Int80α | CTGAATTCGCCAGCATTACTGAGATTAAG | CTTCTAGATTAGACATCGCTAACACGGGCAC |
| Int40-(1)α | CTGAATTCGCCAGCATTACTGAGATTAAG | CTTCTAGATTACGTCGTAAAGGTCACTTCCTG |
| Int40-(2)α | CTGAATTCACGACCTTAGCTAAGTTAAGTAATCC | CTTCTAGATTAGACATCGCTAACACGGGCAC |
| Int40-(3)α | CTGAATTCTACACTGTTAAAGTGATGAAGGGG | CTTCTAGATTATGTTACTTTGGCATAGCCATTCG |
| Int80β | CTGAATTCGCCAGCATTACTGAGATTAAG | CTTCTAGATTAACTCACTTTTGCACTAACAATG |
| Int130β | CTGAATTCGCCAGCATTACTGAGATTAAG | CTTCTAGATTAATTGCCCCCTGTTGTTGCCTGTAC |
| Int150β | CTGAATTCAATGCAAAATACACATGGAAATCCAGTAAAAC | CTTCTAGATTATTTTACACAARYKGCAWAAGC |
| Int100β | CTGAATTCGGTACAGAAGTTAAGGCTACTACC | CTTCTAGATTAACTACCCGGTGCATTTAATGTGT |
| Int120β | CTGAATTCACAATTACTGTAGTATCTGGTGATC | CTTCTAGATTATTTTACACAARYKGCAWAAGC |
| Int70β | CTGAATTCTCAAAAGAACTATTGGCCATTATC | CTTCTAGATTATTTTACACAARYKGCAWAAGC |
| Int40-(1)β | CTGAATTCGCCAGCATTACTGAGATTAAG | CTTCTAGATTACTTAGAAAAGGTCACTTTCTGATG |
| Int40-(2)β | CTGAATTCAAGGATTTTGGGACCCTGAATAAG | CTTCTAGATTAACTCACTTTTGCACTAACAATG |
| Int40-(3)β | CTGAATTCTATACTGTCAGAGTGATGAAGG | CTTCTAGATTATACAGTAGCATAACCATTCTCATCG |
Underlined sequences represent the EcoRI site.
Underlined sequences represent the XbaI site.
TABLE 2.
MBP fusion proteins used in immunodominant mapping experiments
| Fusion protein | Relevant features |
|---|---|
| MBP-Int280 | MBP fused to the C-terminal 280 amino acids (residues 661–939) |
| MBP-Int280(C/S) | MBP fused to the C-terminal 280 amino acids (residues 661–939) from O126:H6. Cys937 substituted for Ala |
| MBP-Int130 | MBP fused to amino acid residues 661–791 of intimin |
| MBP-Int150 | MBP fused to the C-terminal 150 amino acids (residues 791–939) |
| MBP-Int100 | MBP fused to residues 741–841 of intimin |
| MBP-Int120 | MBP fused to the C-terminal 120 amino acids (residues 821–939) |
| MBP-Int70 | MBP fused to the C-terminal 70 amino acids (residues 871–939) |
| MBP-Int80 | MBP fused to amino acid residues 661–741 |
| MBP-Int40-(1) | MBP fused to amino acid residues 661–701 |
| MBP-Int40-(2) | MBP fused to amino acid residues 701–741 |
| MBP-Int40-(3) | MBP fused to amino acid residues 681–721 |
PAGE.
Polyacrylamide gel electrophoresis (PAGE) in the presence of sodium dodecyl sulfate (SDS) was performed as described by Laemmli (30). Protein samples were diluted in an equal volume of 2× sample buffer (2% SDS [wt/vol], 2% 2-mercaptoethanol [vol/vol], 20% glycerol, and 0.01% bromophenol blue [wt/vol] in 65 mM Tris [pH 6.8]) and boiled for 5 min prior to being loaded onto 10 to 15% polyacrylamide gels. Molecular weights were estimated with Rainbow molecular markers (Amersham). Following electrophoresis, the separated proteins were visualized by staining the gel with Coomassie stain or were transferred to nitrocellulose membrane.
Western blotting (immunoblotting) with rabbit anti-Int280 antisera.
Proteins separated by SDS-PAGE were transferred electrophoretically onto nitrocellulose membranes (0.45-μm pore size; Schleicher & Schuell) and immunoblotted according to the methods of Towbin et al. (40) and Burnette (3). Proteins were blotted with a Bio-Rad Wet Blot apparatus at 80 V for 90 min in transfer buffer consisting of 250 mM Tris (pH 8.3), 192 mM glycine, and 20% (vol/vol) methanol. The membranes were blocked overnight in 3% bovine serum albumin (BSA) and washed thoroughly with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST). Membranes were probed with rabbit (diluted 1:1,000) anti-Int280α and anti-Int280β antisera, made by using Int280α and Int280β, respectively, as immunogens, as described previously (1). Briefly, following a 2-h incubation at room temperature with the intimin antiserum, diluted in PBST containing 0.1% BSA, the membranes were washed three times with PBS, and the bound antibodies were reacted with horseradish peroxidase (HRP)-conjugated swine anti-rabbit antibody (1:1,000 dilution; DAKO) for a further 2 h at room temperature. Thereafter, the membranes were washed again and developed with substrate consisting of 15 ml of 50 mM Tris (pH 7.6) 12 μl of 30% hydrogen peroxide, and 10 mg of 3′,3′-diaminobenzidine (Sigma). The reaction was stopped by washing the membrane several times in distilled water.
Colostrum samples.
Colostrum was collected from four healthy and well-nourished 18- to 35-year-old mothers (2 to 5 ml/mother) who gave birth to a normal infant in the Gynaecology and Obstetric Clinics of the São Paulo University Hospital. This institution provides medical support to a low-social-level population living in the neighborhood. Fully informed consent was obtained, and all of the mothers were seronegative for human immunodeficiency virus, hepatitis B, and syphilis.
Western blotting (immunoblotting) with the human colostrum.
A chemiluminescent substrate system was used to detect membranes that were probed with colostrum. The Supersignal CL-HRP substrate system (Pierce, Rockford, Ill.) was used according to manufacturer’s instructions. After an overnight block in 3% BSA, the membranes were washed thoroughly with PBST. Subsequently, the membranes were incubated with the colostrum samples (1:500 dilution) for 2 h, washed thoroughly with PBST, and incubated for a further 2 h with HRP-conjugated goat anti-human α-chain antibodies (Sigma) at a 1:30,000 dilution. The immunoblots were washed again and incubated with the Supersignal CL-HRP substrate working solution for 5 min. The immunoblots were then exposed to a high-performance chemiluminescence film, Hyperfilm ECL (Amersham), and the film was developed with a Fuji X-ray film developer.
ELISA.
For the enzyme-linked immunosorbent assay (ELISA), briefly, 96-well enzyme immunoassay or radioimmunoassay plates (Costar) were coated overnight at 4°C with 50 μl of 5-μg/ml MBP-Int280 derivatives in PBS per well. The wells were washed three times in PBST and blocked for 1 h at 37°C with PBST–1% BSA. The plates were washed again and incubated with fivefold serial dilutions of the primary antibody in order to determine the antiserum titer. Two-hour incubations with primary and secondary antibodies (HRP-conjugated swine anti-rabbit antibody at a 1:1,000 dilution) diluted in PBST–0.1% BSA were carried out at 37°C with PBST washes after each step. Fifty microliters of substrate (10 mg of o-phenylenediamine tablet [Sigma] in 12.125 ml of 0.1 M citric acid, 12.875 ml of 0.25 M NaHPO4, and 10 μl of 30% H2O2) was added to each well. The enzymatic reaction was terminated by the addition of 50 μl of 12.5% H2SO4. The calorimetric reactions were recorded with a Ceres 900 HDi (Bio-Tek Instruments, Inc.) microtiter plate reader. The optical density values were plotted for each sample, and the titers were determined as the reciprocal dilution giving an A490 of 0.3 above the background. All titrations and experiments were performed in duplicate. A positive reference serum was used on each plate, and the results were adjusted accordingly.
RESULTS
Construction of overlapping Int280 polypeptides.
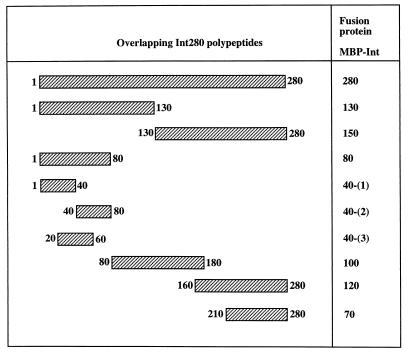
In previous reports, we have tested the cell-binding properties of MBP-Int280α and derivatives of this domain containing N- and C-terminal deletions (15). Cell-binding activity was observed only with the MBP-Int280 and MBP-Int150 fusions, localizing the cell-binding function of intimin to the C-terminal 150 amino acids. In addition, we have established that Cys937 in EPEC intimin is essential for binding activity (15–17). The aim of this study was to localize and compare the immunodominant regions within the Int280α and Int280β domains (Fig. 1). For this purpose, we have generated additional overlapping polypeptides of Int280α, expressed as MBP fusions, and a complete new set of overlapping peptides from Int280β. A schematic presentation of the overlapping fragments is shown in Fig. 2.
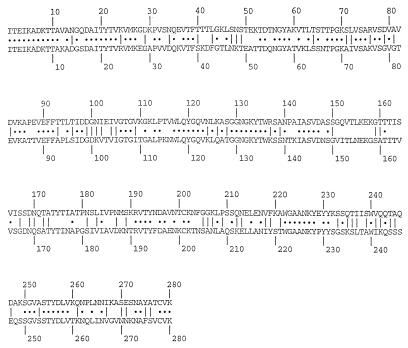
FIG. 1.
Alignment of the amino acid sequence of Int280α from the EPEC E2348/69 domain (upper row) and Int280β from the EPEC O114:H2 domain (lower row). Dots represent identical residues. Lines represent conserved substitutions.
FIG. 2.
Schematic representation of the overlapping MBP-Int280-derived polypeptides. The numbers on both sides of the fragments mark the first and last amino acids of each fragment within the Int280 domain.
Identification of immunodominant regions within Int280α by using the anti-Int280α antiserum.
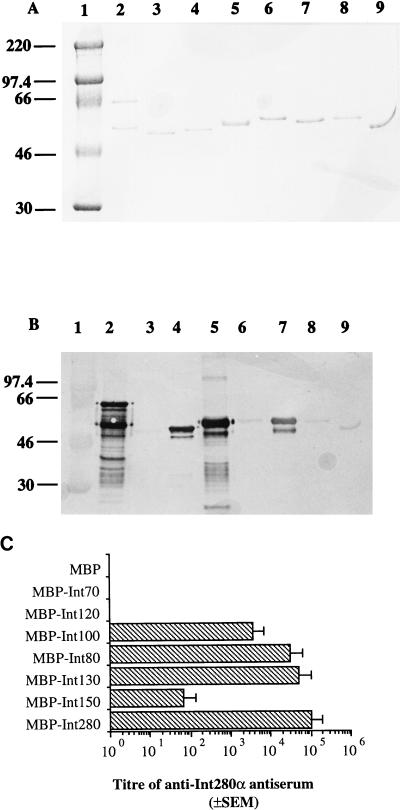
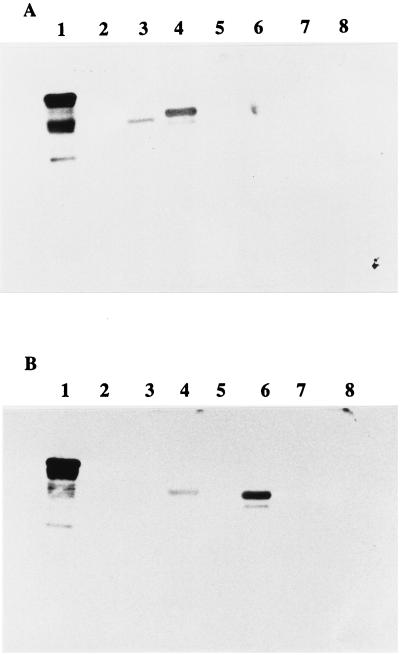
We have recently shown that the Int280 antiserum was reactive with the whole intimin polypeptide (1). To map immunodominant epitopes and regions within the C-terminal domain of intimin α, the MBP-Int fusion proteins 280, 130, 150, 80, 100, 120, and 70 (Fig. 2) were analyzed by Western blotting and ELISA with the rabbit anti-Int280α antiserum. A Coomassie blue-stained gel illustrating the different MBP-Intα fusion proteins is shown in Fig. 3A. Reaction of the rabbit Int280α antiserum with Western blots of the different fusion proteins revealed strong reactivity with four MBP-Intα fusion proteins: MBP-Int280, MBP-Int130, MBP-Int80, and MBP-Int100 (Fig. 3B). A low, but detectable reactivity was observed with MBP-Int150, MBP-Int120, and MBP-Int70 (Fig. 3B). No reactivity was observed with the MBP-negative control. Preimmune rabbit antisera showed no reactivity with any of the MBP fusion proteins (reference 1 and data not shown).
FIG. 3.
Coomassie blue staining of SDS-PAGE gel (A) and immunoblotting (B) and ELISA (C) of anti-Int280α antiserum with Int280α-derived fusion proteins. Strong reactivity on the Western blot is observed with Int280 (B, lane 2), Int80 (B, lane 4), Int130 (B, lane 5), and Int100 (B, lane 7). Low reactivity is observed with Int150 (B, lane 6), Int120 (B, lane 8), and Int70 (B, lane 9). No reactivity is seen with MBP (lane 3). The electrophoretic migration of molecular mass markers (in kilodaltons) is shown in lane 1 (A and B). The reactivity of Int280α antiserum with the different overlapping Int280α-derived fusion proteins in the ELISA produced results in agreement with those of the Western blot (C). High antibody titers were observed for Int280, -130, -80, and -100, and a low titer was observed for Int150. No reactivity was seen for Int120, Int70, or MBP.
ELISA analysis with the Int280α antiserum and MBP-Int280α as the coating antigen revealed a titer of 105 (Fig. 3C). Reaction of the antiserum in the ELISA with the other fusion proteins confirmed the observation made with the Western blots; the highest titers were seen against the MBP-Int130 (5 × 104), MBP-Int80 (3 × 104), and MBP-Int100 (3 × 103) fusion proteins (Fig. 3C). A similar level of reactivity with MBP-Int280 and MBP-Int280(C/S) was observed (data not shown). MBP-Int280(C/S) is identical to MBP-Int280, except that Cys937 was replaced with Ser (15). These results indicate the presence of two immunodominant regions within Int280α: one localized within Int80 and the second localized within Int100.
Identification of immunodominant regions within Int280β by using the anti-Int280β antiserum.
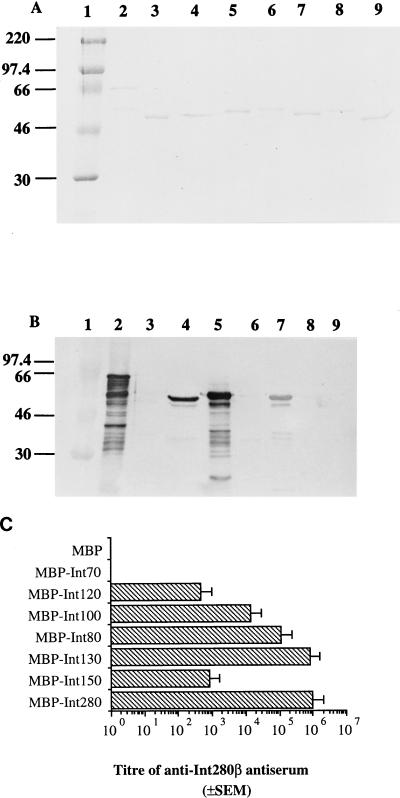
Following the approach used to map immunodominant regions in Int280α, we searched for immunodominant regions within Int280β. The overlapping MBP-Intβ fusion proteins were separated by SDS-PAGE (10% polyacrylamide); a Coomassie blue-stained gel is shown in Fig. 4A. Reaction of Int280β antiserum with the overlapping polypeptides revealed, similar to the results with Int280α, strong reactivity with MBP-Int280, MBP-Int130, MBP-Int80, and MBP-Int100 (Fig. 4B). However, no reactivity was observed with MBP-Int150, MBP-Int120, and MBP-Int70 or the MBP-negative control.
FIG. 4.
Coomassie blue staining of SDS-PAGE gel (A) and immunoblotting (B) and ELISA (C) of the reactivity of anti-Int280β antiserum (1:10,000 dilution) with the MBP-Int280β-derived fusion proteins. Strong reactivity on the Western blot is observed with Int280 (B, lane 2), Int80 (B, lane 4), Int130 (B, lane 5), and Int100 (B, lane 7). Low reactivity is seen with Int150 (B, lane 6). No reactivity is seen with Int120 (B, lane 8), Int70 (B, lane 9), or MBP (B, lane 3). MBP-Int280 preparations consistently resulted in doublet bands; this appears to represent proteolytic sensitivity introduced into the MBP part of the fusion protein (15). Molecular mass markers (A and B, lane 1) are given in kilodaltons. Analysis of the Int280β antiserum with the different overlapping Int280b-derived fusion protein ELISA (C) revealed that high antibody titers were observed for Int280, -130, -80, and -100, while lower titers were observed against Int150 and Int120. No reactivity was seen for Int70 or MBP.
ELISA analysis with the Int280β antiserum and MBP-Int280β as the coating antigen revealed a titer of 106 (Fig. 4C). Reaction of the antiserum in ELISA with the overlapping MBP-Int280β fusion proteins revealed that the strongest reactivity was with MBP-Int130 (titer of 8 × 105). The titers against MBP-Int80 and MBP-Int100 were 105 and 104, respectively. Although not reactive with the antiserum in Western blots, low reactivity (2.8 × 102 and 5 × 102) was detected against MBP-Int150 and MBP-Int120, respectively, which may reflect the presence of conformational epitopes. Overall, these results demonstrate the presence of two immunodominant epitopes within Int280β located on the same fragments as those of Int280α derivatives.
Mapping of the immunodominant regions within Int280α and Int280β.
The results presented in the previous two sections showed the presence of two immunodominant regions which are common to both Int280α and Int280β. Because strong reactivity was observed with Int130 and Int100, and no reactivity was seen with Int150, we concluded that one of the immunodominant regions is located between amino acids 80 and 130. The second immunodominant region lies along Int80 of both intimin types. In order to map the immunodominant region within Int80 more accurately, three MBP fusion proteins overlapping this region were made for both intimin α and intimin β (Fig. 2). Western blotting was performed with fusions MBP-Int40-(1)α, MBP-Int40-(2)α, and MBP-Int40-(3)α, which were probed with the Int280α antiserum, revealing reactivity only with MBP-Int40-(1)α (Fig. 5A). In contrast, Western blots with the Int280β antiserum and MBP-Int40-(1)β, MBP-Int40-(2)β, and MBP-Int40-(3)β revealed reactivity only with MBP-Int40-(3)β (Fig. 5B). These results map the immunodominant region within Int80α to amino acids 1 to 20 and within Int80β to a region within amino acids 20 to 60.
FIG. 5.
Reactivity of Int280α and Int280β antisera with Int80-derived overlapping polypeptides. Anti-Int280α antiserum reacted with Int40-(1)α (A, lane 1) but showed no reactivity against Int40-(2)α or Int40-(3)α (A, lanes 2 and 3, respectively). In contrast anti-Int280β antiserum reacted with Int40-(3)β (B, lane 3) and showed no reactivity with Int40-(1)β or Int40-(2)β (B, lanes 1 and 2, respectively).
Reactivity of human colostrum samples with MBP-Int280α and MBP-Int280β derivatives.
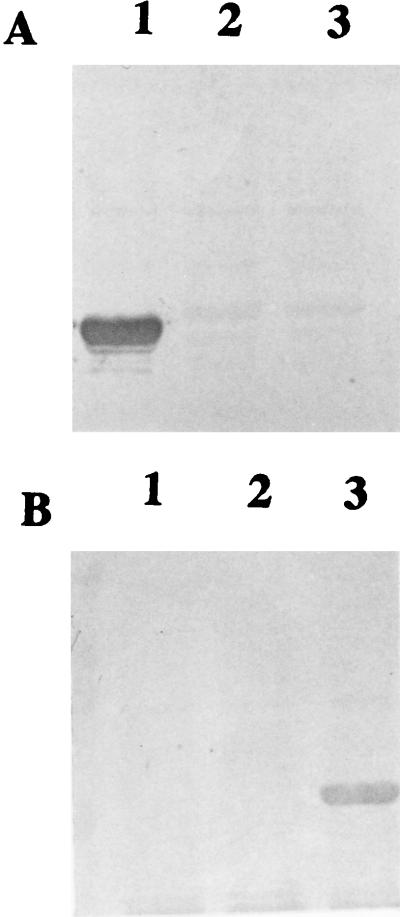
In order to study whether the immunodominant regions identified by the rabbit antisera were also recognized by humans, colostrum samples were assayed by Western blotting with the MBP-Int280α- and MBP-Int280β-derived fusion proteins. Since the colostrum samples have a low antibody titer, a chemiluminescence detection system was employed. Four individual colostrum samples were used in the analysis. Reaction of the colostrum with MBP-Int280α-derived fusion proteins 280, 130, 150, 80, 100, 120, and 70 revealed strong reactivity with MBP-Int280, moderate reactivity with MBP-Int130, and low reactivity with MBP-Int80 (Fig. 6). No reactivity was observed with the other fusion proteins or MBP. All four individual samples produced the same reactivity pattern with the Int280α derivatives.
FIG. 6.
Reactivity of colostrum samples with MBP-Int fusion proteins. Reaction of the four individual colostra with Int280α-derived MBP fusion protein revealed a single reactivity pattern (A) in which the IgA antibodies reacted strongly with Int280α (lane 1), moderately with Int130α (lane 4), and weakly with Int80α (lane 3). No reactivity was seen with Int150α, Int120α, Int70α, or MBP (A, lanes 5, 7, 8, and 2, respectively). Reaction of the same four colostrum samples with Int280β-derived fusion proteins revealed, in two samples, the same reactivity pattern as that seen with reaction against the Int280α-derived polypeptides (data not shown), while the other two samples reacted strongly with Int280β (B, lane 1), moderately with Int100β (B, lane 6), and weakly with Int130β (B, lane 4). No reactivity was seen with Int80, Int150, Int120, Int70, or MBP (B, lanes 3, 5, 7, 8, and 2, respectively).
Reaction of the same colostra with MBP-Int280β-derived fusion proteins 280, 130, 150, 80, 100, 120, and 70 revealed strong reactivity with MBP-Int280 by all four samples. However, the reactivity of the colostra with the other fusion proteins revealed two different reactivity patterns. Two of the samples showed identical reactivity patterns, as seen with Int280α, moderate reactivity with MBP-Int130β, and low reactivity with MBP-Int80β (data not shown), while the other two samples showed moderate reactivity with MBP-Int100 and low reactivity with MBP-Int130. No reactivity was observed with the other fusion proteins or MBP. These results show that although human colostra contain IgA antibodies to both intimin α and intimin β, the reactivity against the latter shows some variability among individual samples.
DISCUSSION
Our results show the presence of two immunodominant regions which are common to both Int280α and Int280β. The use in the first phase of the study of overlapping polypeptides spanning Int280α and Int280β and of rabbit polyclonal antisera in both Western blots and ELISA revealed that one of these epitopes is localized within the first 80 amino acids (MBP-Int80) and the second is mapped to a 50-amino-acid sequence between amino acids 80 and 130 of both intimin types. Accordingly, the strongest reactivity of the antisera was observed, in addition to the immunogen used in their production (Int280), against Int130, which included both immunodominant regions.
In the second phase of the study, we have attempted to map the immunodominant region within Int80α and Int80β in greater accuracy. For that purpose, overlapping polypeptides spanning this domain were made from both intimin types and tested against their corresponding antiserum. Interestingly, these assays revealed that while the Int280α antiserum reacted only with Int40-(1), the Int280β antiserum reacted only with Int40-(3). These results suggest that the immunodominant region within Int80α is localized within the first 20 amino acids of the Int280α polypeptide, while the immunodominant region of Int80β is localized between amino acids 20 and 60 of Int280β. The reason for the difference in locations of the immunodominant regions within Int80 between the two antisera is not known. However, this study clearly shows the presence of immunodominant regions within this domain which showed greater reactivity (demonstrated by ELISA) than that of the second immunodominant region localized between amino acids 80 and 130. In a previous report, we have shown little cross-reactivity between the Int280α and Int280β antisera (1). Moreover, reports by Voss et al. (42) and Agin and Wolf (2) have also shown differential reactivity of human and rabbit antisera with intimin from different EHEC and EPEC isolates, respectively. In this study, we have identified different immunodominant regions within the Int80 domains, which is consistent with these observations.
In our previous report (33), we have shown that pools of human colostra collected from mothers living in São Paulo, Brazil, where EPEC diarrhea is endemic in infants, contain IgA antibodies which are reactive to intimin, bundle-forming pili (Bfp), and EspA and EspB, bacterial virulence-associated factors that according to proposed models of infection operate at different stages of EPEC pathogenesis (9, 21). In the present study, we have shown the presence of anti-intimin α and anti-intimin β IgA antibodies in individual colostrum samples. The colostrum IgA reacted with both intimin α and intimin β. Significantly, testing of the individual colostrum samples with the overlapping Int280α polypeptides revealed a single reactivity pattern (i.e., all samples reacted strongly with Int280, moderately with Int130, and weakly with Int80). In contrast, reaction of the human colostrum samples with the overlapping Int280β polypeptides revealed two reactivity patterns, one similar to the reactivity observed with Int280α polypeptides and the other revealing high reactivity with Int280β, moderate reactivity with Int100, and low reactivity with Int130. The different reactivity patterns obtained against Int280β peptides may represent variations in immune responses elicited by individual mothers as a result of multiple immunodominant regions or may be due to exposure of individual mothers to different clonal intimin types during their lifetimes.
It was suggested that since intimin is highly immunogenic, diversity within the polypeptide cell-binding domain is driven by natural selection (1). It is important to note that despite the high variability in this region between the different intimin types, two stretches of six and seven amino acids (WLQYGQ and WGAANKY [amino acids 119 to 125 and 224 to 239, respectively]) are conserved in all intimin types, but are not found in any other sequences in the databases. It has been suggested, although not yet proven, that these amino acids may form part of the binding site and that according to the level of the immunological cross-reaction between intimin α and intimin β, these conserved amino acids sequences are not highly immunogenic (1). The results of the present study support the latter suggestion. Moreover, our data showing that the immunodominant regions within Int280 are located upstream of Int150, the smallest intimin domain that was shown to bind to host cells, are in accordance with our finding that the same reactivity was seen against Int280 and Int280(C/S).
An important aspect of the pathogenic scheme of many bacterial species is their ability to adhere to and colonize mucosal surfaces. Breast-feeding is clearly associated with mucosal protection against a number of mucosal pathogens (e.g., EPEC, enterotoxigenic E. coli, and cholera), probably through IgA inhibiting adherence to the epithelium (7, 19). The high immunogenicity of intimin in infected hosts provides a rational basis to support the concept of engineering an intimin molecule as a basis for an EPEC vaccine. However, although the relevance to human protection of the immunodominant epitopes and regions recognized by the rabbit antiserum is at present not clear, the fact that more than one immunodominant region was identified within Int280, even by the human colostra, means that additional research is needed if an intimin-based broad-spectrum vaccine against A/E-positive bacterial pathogens (which will also include EHEC O157:H7 and EPEC O55:H− strains expressing intimin γ [1]) is sought.
ACKNOWLEDGMENTS
We are grateful to Raymond Fowler, Imperial College, for advice with the chemiluminescence techniques; Roscelice Fernandez and Ivana Louriero for the colostrum samples; and Gill Douce for critical reading of the manuscript.
J. Adu-Bobie is a Ph.D. student supported by the BBSRC, WHO, and Murex diagnostics. This study was supported by a Welcome Trust program grant to G. Dougan.
REFERENCES
- 1.Adu-Bobie J, Frankel G, Bain C, Goncalves A Z, Trabulsi L R, Douce G, Knutton S, Dougan G. Detection of intimins α, β, γ, and δ, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol. 1998;36:662–668. doi: 10.1128/jcm.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agin T S, Wolf M K. Identification of a family of intimins common to Escherichia coli causing attaching-effacing lesions in rabbits, humans, and swine. Infect Immun. 1997;65:320–326. doi: 10.1128/iai.65.1.320-326.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnette W N. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulphate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 4.Camara L M, Carbonare S B, Silva M L, Carneiro Sampaio M M. Inhibition of enteropathogenic Escherichia coli (EPEC) adhesion to HeLa cells by human colostrum: detection of specific IgA related to EPEC outer-membrane proteins. Int Arch Allergy Immunol. 1994;103:307–310. doi: 10.1159/000236645. [DOI] [PubMed] [Google Scholar]
- 5.Cantey J R, Blake R K. Diarrhea due to Escherichia coli in rabbit: a novel mechanism. J Infect Dis. 1977;135:454–462. doi: 10.1093/infdis/135.3.454. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson B, Hanson L A. Immunological effects of breast-feeding on the infant. London, United Kingdom: Academic Press; 1994. [Google Scholar]
- 7.Carneiro-Sampaio, M. M. S., M. L. M. Silva, S. B. Carbonare, P. Palmeira, M. T. Delneri, A. C. Honorio, and L. R. Trabulsi. 1996. Breast feeding protection against enteropathogenic Escherichia coli. Rev. Microbiol. São Paulo 27(Suppl. 1):130–133.
- 8.Deibel C, Krämer S, Chakraborty T, Ebel F. EspE, a novel secreted protein of attaching and effacing bacteria, is directly translocated into infected host cells, where it appears as a tyrosine-phosphorylated 90 kDa protein. Mol Microbiol. 1998;28:463–474. doi: 10.1046/j.1365-2958.1998.00798.x. [DOI] [PubMed] [Google Scholar]
- 9.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 10.Donnenberg M S, Tacket C O, James S P, Losonsky G, Nataro J P, Wasserman S S, Kaper J B, Levine M M. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Investig. 1993;92:1412–1417. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg M S, Tzipori S, McKee M L, O’Brien A D, Alroy J, Kaper J B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Investig. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg M S, Yu J, Kaper J B. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finlay B B, Rosenshine I, Donnenberg M S, Kaper J B. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect Immun. 1992;60:2541–2543. doi: 10.1128/iai.60.6.2541-2543.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frankel G, Candy D C, Everest P, Dougan G. Characterization of the C-terminal domains of intimin-like proteins of enteropathogenic and enterohemorrhagic Escherichia coli, Citrobacter freundii, and Hafnia alvei. Infect Immun. 1994;62:1835–1842. doi: 10.1128/iai.62.5.1835-1842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frankel G, Candy D C A, Fabiani E, Adu-Bobie J, Gil S, Novakova M, Phillips A D, Dougan G. Molecular characterization of a carboxy-terminal eukaryotic-cell-binding domain of intimin from enteropathogenic Escherichia coli. Infect Immun. 1995;63:4323–4328. doi: 10.1128/iai.63.11.4323-4328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel, G., A. D. Phillips, J. Adu-Bobie, D. C. A. Candy, G. Douce, and G. Dougan. 1996. Intimin-mediated adherence to epithelial cells. Rev. Microbiol. São Paulo 27(Suppl. 1):99–103.
- 17.Frankel G, Phillips A D, Novakova M, Batchelor M, Hicks S, Dougan G. Generation of Escherichia coli intimin-derivatives with differing biological activities using site-directed mutagenesis of the intimin C-terminus domain. Mol Microbiol. 1998;29:559–570. doi: 10.1046/j.1365-2958.1998.00950.x. [DOI] [PubMed] [Google Scholar]
- 18.Frankel G, Phillips A D, Novakova M, Field H, Candy D C A, Schauer D B, Douce G, Dougan G. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect Immun. 1996;64:5315–5325. doi: 10.1128/iai.64.12.5315-5325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glass R E, Svennerholm A M, Stoll B J, Khan M R, Hossein K M B, Huq M I, Holmgren J. Protection against cholera in breast-fed children by antibodies in breast-milk. N Engl J Med. 1983;308:1389–1392. doi: 10.1056/NEJM198306093082304. [DOI] [PubMed] [Google Scholar]
- 20.Gómez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks S, Frankel G, Kaper J B, Dougan G, Phillips A D. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis K G, Giron J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 25.Kenny B, Lai L C, Finlay B B, Donnenberg M S. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol Microbiol. 1996;20:313–323. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 26.Knutton S, Adu-Bobie J, Bain C, Phillips A D, Dougan G, Frankel G. Down regulation of intimin expression during attaching and effacing enteropathogenic Escherichia coli adhesion. Infect Immun. 1997;65:1644–1652. doi: 10.1128/iai.65.5.1644-1652.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knutton S, Lloyd D R, McNeish A S. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect Immun. 1987;55:69–77. doi: 10.1128/iai.55.1.69-77.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knutton S, Rosenshine I, Pallen M J, Nisan I, Neves B C, Bain C, Wolff C, Dougan G, Frankel G. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 31.Lai L-C, Wainwright L A, Stone K D, Donnenberg M S. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine M M, Edelman R. Enteropathogenic Escherichia coli of classic serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol Rev. 1984;6:31–51. doi: 10.1093/oxfordjournals.epirev.a036274. [DOI] [PubMed] [Google Scholar]
- 33.Loureiro, I., G. Frankel, J. Adu-Bobie, D. Dougan, L. R. Trabulsi, and M. M. S. Carnerio-Sampaio. Human colostrum contains IgA antibodies reactive to enteropathogenic Escherichia coli-virulence-associated proteins: intimin, BfpA, EspA and EspB. J. Pediatr. Gastroenterol. Nutr. 27:166–171. [DOI] [PubMed]
- 34.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 36.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ørskov F, Whittam T S, Cravioto A, Orskov I. Clonal relationships among classic enteropathogenic Escherichia coli (EPEC) belong to different O groups. J Infect Dis. 1990;162:76–81. doi: 10.1093/infdis/162.1.76. [DOI] [PubMed] [Google Scholar]
- 38.Robins Browne R M, Yam W C, O’Gorman L E, Bettelheim K A. Examination of archetypal strains of enteropathogenic Escherichia coli for properties associated with bacterial virulence. J Med Microbiol. 1993;38:222–226. doi: 10.1099/00222615-38-3-222. [DOI] [PubMed] [Google Scholar]
- 39.Schauer D B, Falkow S. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect Immun. 1993;61:2486–2492. doi: 10.1128/iai.61.6.2486-2492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulshen M H, Rollo J L. Pathogenesis of Escherichia coli gastroenteritis in man: another mechanism. N Engl J Med. 1980;302:99–102. doi: 10.1056/NEJM198001103020207. [DOI] [PubMed] [Google Scholar]
- 42.Voss E, Paton A W, Manning P A, Paton J C. Molecular analysis of Shiga toxigenic Escherichia coli O111:H− proteins which react with sera from patients with hemolytic-uremic syndrome. Infect Immun. 1998;66:1467–1472. doi: 10.1128/iai.66.4.1467-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whittam, T. S., and E. A. McGraw. 1996. Clonal analysis of EPEC serogroups. Rev. Microbiol. São Paulo 27(Suppl. 1):7–16.
- 44.Wolff C, Nisan I, Hanski E, Frankel G, Rosenshine I. Protein translocation into HeLa cells by infecting enteropathogenic Escherichia coli. Mol Microbiol. 1998;28:143–155. doi: 10.1046/j.1365-2958.1998.00782.x. [DOI] [PubMed] [Google Scholar]