Figure 6. Fate mapping of serum antibodies reveals elicitation of cross-reactive recall responses and strain-specific de novo responses elicited by mosaic-8b nanoparticles in previously vaccinated animals, related to Figure S6.
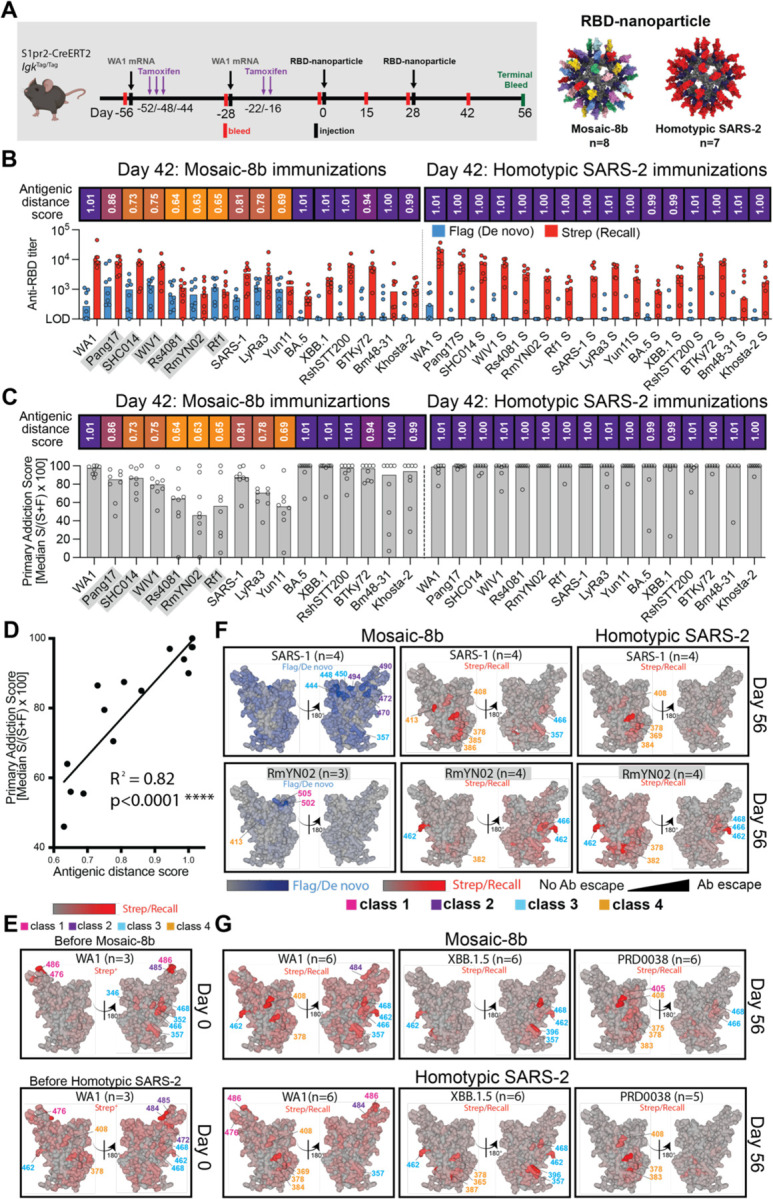
Antigenic distance score is defined as the ratio of % amino acid sequence identity of a strain to WA1 RBD divided by % sequence identity of that strain to the closest non-self RBD relative on the nanoparticle. The primary addiction index21 is defined as Strep/(Flag + Strep) x 100. See Figure 1B for RBD epitope classifications.
(A) (Left) Schematic of vaccine regimen in S1pr2-IgkTag/Tag immunized mice. Mice were vaccinated with an mRNA-LNP vaccine encoding WA1 spike at the indicated days prior to RBD-nanoparticle immunizations at day 0 and day 28. After each dose of mRNA-LNP vaccine, mice were treated with tamoxifen to switch epitope tags on the Abs produced by B cells activated by the mRNA-LNP vaccinations from Flag to Strep, so that Ab responses elicited by RBD-nanoparticle boosting can be separated into recall (Strep+) and de novo (Flag+) responses. (Right) Models of RBD-nanoparticles that were used for immunizations at days 0 and 28.
(B) Comparison of the Flag+/de novo (blue) and Strep+/recall (red) anti-RBD titers shown at day 42, two weeks after a second immunization with mosaic-8b or homotypic SARS-2 nanoparticles. For each strain, the antigenic distance score is shown above the plots and colored with a gradient from purple (score=1) to yellow (score=0). Gray shaded strains are matched to mosaic-8b.
(C) The primary addiction index21 for each RBD is plotted for day 42 serum after two doses of mosaic-8b or homotypic SARS-2. The antigenic distance score is shown above the plots and colored with a gradient from purple (score=1) to yellow (score=0). Gray shaded strains are matched to mosaic-8b.
(D) Correlation of the primary addiction index21 with the antigenic distance score. The best-fit line, R2, and p value were calculated by linear regression using Prism.
(E) DMS analysis of Strep+ Abs from day 0 (prior to RBD-nanoparticle immunization) serum from n=3 pre-vaccinated mice using an RBD mutant library derived from WA1. Ab binding sites are shaded red according to degree of Strep+ Ab escape on the surface of the WA1 RBD (PDB 6M0J). Locations of residues with high escape scores are indicated on RBD surfaces with gray indicating no escape and shades of red indicating sites with most escape. Residue numbers are color coded according to their RBD epitope classification.
(F) DMS analysis using SARS-1 (antigenic distance score = 0.81) and RmYN02 (antigenic distance score = 0.63) RBD mutant libraries of day 56 serum from n=4 or n=3 RBD-nanoparticle immunized mice. Ab binding sites are shaded according to degree of Ab escape, with blue for Flag/de novo responses and red for Strep/recall responses, on the surface of the WA1 RBD (PDB 6M0J). Comparisons are made for Flag/de novo and Strep/recall elicited by mosaic-8b and for Strep/recall elicited by homotypic SARS-2 (Flag/de novo responses after homotypic SARS-2 immunization were weak to undetectable). Gray shaded virus names represent strains that are matched to mosaic-8b. Locations of residues with high escape scores are indicated on RBD surfaces with gray indicating no escape and darker shades indicating sites with most escape. Residue numbers are color coded according to their RBD epitope classification.
(G) DMS analysis of the Strep/recall compartment of day 56 serum from n=5 or n=6 mosaic-8b or homotypic SARS-2 immunized mice using RBD mutant libraries derived from strains with antigenic distance scores of 1.01 (WA1), 1.01 (XBB.1.5), and 0.94 (PRD008). Ab binding sites are shaded red according to degree of Strep/Recall Ab escape on the surface of the WA1 RBD (PDB 6M0J). Locations of residues with high escape scores are indicated on RBD surfaces with gray indicating no escape and darker shades indicating sites with most escape. Residue numbers are color coded according to their RBD epitope classification.
