Abstract
Vibrio vulnificus expresses a number of potential virulence determinants that may contribute to its ability to cause a severe and rapidly disseminating septicemia in susceptible hosts. We have cloned and characterized two genes encoding products related to components of the type IV pilus biogenesis and general secretory (type II) pathways by complementation of a type IV peptidase/N-methyltransferase (PilD) mutant of Pseudomonas aeruginosa with a V. vulnificus genomic library. One of the genes (vvpD) encodes a protein homologous to PilD and other members of the type IV peptidase family that completely restores this activity in a P. aeruginosa mutant deficient in the expression of PilD. The other gene (vvpC) encodes a homolog of PilC from P. aeruginosa, where it is essential for assembly of type IV pili. Phenotypic characterization of a V. vulnificus vvpD mutant, constructed by allelic exchange, showed that VvpD is required for the expression of surface pili, suggesting that the pili observed on V. vulnificus are of the type IV class. This mutant was also unable to secrete at least three extracellular degradative enzymes, and the localization of one of these (the cytolysin/hemolysin) to the periplasmic space indicates that these proteins are normally exported via the type II secretion pathway. Loss of VvpD resulted in significant decreases in CHO cell cytotoxicity, adherence to HEp-2 cells, and virulence in a mouse model. Capsule formation and serum resistance were not affected in the vvpD mutant, indicating that in addition to capsule, virulence of V. vulnificus requires type IV pili and/or extracellular secretion of several exoenzymes.
Vibrio vulnificus biotype 1 is an estuarine bacterium that can cause primary septicemia as well as serious wound infections (34, 35). While septicemia occurs primarily in immunocompromised individuals or those that suffer from cirrhosis or hemochromatosis, healthy people can become infected through wounds. Septicemia can develop after ingestion of shellfish carrying the organism, with the greatest risk coming from the consumption of raw oysters (5, 21). Mortality in these cases exceeds 50%, increasing to greater than 90% in people who go into shock or become hypotensive shortly after admission to a hospital (22). As many as 50% of all vibrio-related illnesses in the United States are caused by V. vulnificus (7). Recently, a second biotype of V. vulnificus, biotype 2, has been implicated in septicemic infections of cultured eels (63). Animal studies have shown that biotype 2 is also capable of causing septicemia in mammals, including opportunistic infections of humans (3, 4).
Among the many factors implicated as possible virulence determinants for V. vulnificus are extracellular toxins and enzymes (e.g., cytolysin and elastolytic protease) (25, 33), a polysaccharide capsule (67), resistance to phagocytosis (19, 70), resistance to the bactericidal effects of human sera (19, 67, 70), and the ability to acquire iron from transferrin (51). V. vulnificus undergoes a phase variation between virulent and avirulent forms, with the former being encapsulated and serum resistant and the latter having lost these traits (49, 67). In animal models, encapsulation is clearly an important determinant of virulence (67, 70), most likely because the capsule confers serum resistance and is antiphagocytic (49). The role of the cytolysin is less clear, as cytolysin-negative strains have the same 50% lethal dose (LD50) for mice as wild-type strains (64).
Type IV pili have been shown to be important adherence factors in many gram-negative bacteria (57). Biogenesis of these pili is in part controlled by the type IV leader peptidase, a bifunctional enzyme that proteolytically cleaves the specialized leader sequence of type IV pilin precursors followed by N-methylation of the newly exposed N-terminal amino acid before assembly into the pilus structure (38, 60). Additional proteins required for type IV pilus biogenesis also have this specialized leader sequence (1, 31), and though it has yet to be demonstrated directly, it is presumed that these pilin-like proteins are substrates of the type IV peptidase as well. In addition to pilus biogenesis, the type IV peptidase is required for extracellular secretion of proteins via the general secretory (type II secretion) pathway (GSP) (43, 45). At least four GSP-associated proteins with the type IV leader sequence are processed by this peptidase (37, 44, 60), and as demonstrated directly in Pseudomonas aeruginosa (58), Aeromonas hydrophila (40), and more recently Legionella pneumophila (9a, 27) type IV leader peptidase mutants are unable to secrete proteins via the GSP.
We have cloned and sequenced two genes from V. vulnificus that encode homologs of components of the type IV pilus biogenesis and type II secretion pathways. One of these, vvpC, encodes a polypeptide that is highly homologous to PilC, a protein of unknown function that is required for assembly of type IV pili in P. aeruginosa (36). The other, vvpD, encodes a homolog of the bifunctional type IV leader peptidase/N-methyltransferases found in many bacterial genera (28). We constructed a mutant unable to express VvpD and examined effects of the mutation with respect to expression of pili, extracellular protein secretion, capsule expression, tissue culture cytotoxicity and adherence, and virulence. We show that in the absence of VvpD, the mutant is significantly reduced in all of these functions except capsule formation. This is the first demonstration of a V. vulnificus mutation affecting expression of factors other than capsule that results in decreased virulence of the organism.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Unless noted, V. vulnificus strains were grown at 30°C in brain heart infusion (BHI) broth, while Escherichia coli and P. aeruginosa strains were grown at 37°C in Luria-Bertani (LB) broth. Antibiotic concentrations (micrograms per milliliter) used were as follows: chloramphenicol, 30; streptomycin, 25; spectinomycin, 25; and tetracycline, 20. Polymyxin B was used at 50 U/ml, and isopropylthio-β-d-galactoside (IPTG) was used at a final concentration of 1 mM.
TABLE 1.
Bacterial strains, plasmids and phage used
| Strain, plasmid, or phage | Relevant characteristics | Source or reference(s) |
|---|---|---|
| V. vulnificus | ||
| MO6-24/OP | Wild type (clinical isolate) | J. D. Oliver |
| C7184/OP | Wild type (clinical isolate) | J. D. Oliver |
| PAC1 | Wild type (environmental isolate) | M. Coyle |
| C7184-D12Ω | C7184 with disrupted vvpD | This study |
| C7184-402K | C7184 with disrupted vvhA | This laboratory |
| E. coli | ||
| DH5α | supE44 lacU169(φlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| S17-1λpir | supE44 hsdR endA1 recA thi pro RP4-2-Tc::Mu-kan::Tn7 (λpir) | 32, 50 |
| P. aeruginosa | ||
| PAK | Wild type | S. Lory |
| PAK 2B18 | pilD::Tn5 | S. Lory (36) |
| PAK-CΩ | PAK mutant with the Ω interposon inserted in the pilC gene | 23 |
| Cloning vectors | ||
| pLAFR3 | Tcr broad-host-range plasmid vector | 52 |
| pBluescript II SK+ | Apr phagemid cloning vector | Stratagene |
| pMMB67HE.cam | Cmr Apr broad-host-range cloning vector, lacIq/tac promoter | 17 |
| pEP185.2 | Cmr suicide vector | 20 |
| pZErO-2 | Kanr Zero background cloning vector | Invitrogen |
| Recombinant plasmids | ||
| pJCL12B | 25-kb Sau3AI fragment from V. vulnificus MO6-24 in pLAFR3 | This study |
| pMS450 | 4.8-kb PstI fragment from pJCL12B cloned into pLAFR3 (vvpBCD) | This study |
| pMS451 | 2.8-kb HindIII-PstI fragment cloned into pLAFR3 (vvpCD) | This study |
| pMS4514 | 2.8-kb HindIII-PstI fragment cloned into pBluescript II SK+ | This study |
| pMS451Ω | 2.8-kb HindIII-PstI fragment with Ω fragment inserted in the NarI site of vvpD | This study |
| pMS451Ω-2 | 4.8-kb KpnI-SmaI fragment from pMS451Ω in pEP185.2 | This study |
| pRPD1 | 1.0-kb SalI fragment from C7184 cloned into pMMB67HE.cam (vvpD) | This study |
| Bacteriophage PO4 | P. aeruginosa pilus-specific phage | S. Lory |
DNA manipulations.
V. vulnificus chromosomal DNA was extracted following procedures described for P. aeruginosa (56). Routine plasmid and cosmid DNA extractions were performed by the alkaline lysis method (6), while Qiagen midiprep columns (Qiagen, Santa Clarita, Calif.) were used for large-scale plasmid preparations. Standard techniques were used for enzymatic manipulations, ligations, transformations, and DNA electrophoresis (47). Probes used for Southern blot analyses were prepared by the random priming method incorporating digoxigenin-labeled dUTP, followed by chemiluminescence detection with anti-digoxigenin–alkaline phosphatase conjugate and disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5-chloro)tricyclo[3,3.1.13.7]decan-4}-yl) phenyl phosphate (Genius system nonradioactive detection kit, version 2.0; Boehringer Mannheim, Indianapolis, Ind.).
Construction of a V. vulnificus genomic library.
A V. vulnificus genomic library was constructed by ligation of chromosomal DNA partially digested with Sau3AI from strain MO6-24 into the cosmid vector pLAFR3 (52) as described by Pepe et al. (40).
Nucleotide sequence determination.
Double-stranded DNA sequencing of pMS450, containing a 4.8-kb PstI fragment from V. vulnificus MO6-24, was performed by the dideoxy-chain termination method (48). Primers were synthesized with a Gene Assembler Special (Pharmacia, Piscataway, N.J.) or purchased commercially. Subsequent homology searches were performed by using BLAST (basic local alignment search tool) (2).
The vvpC and vvpD genes of strain C7184 were amplified by PCR using V. vulnificus C7184 chromosomal DNA as the template and the following primers: 5′-CTATCAATGGAAAGGGATTAACGG-3′ and 5′-AGCCAAACAGGGAAGCT-3′ to generate a 1,308-bp fragment containing vvpC; and 5′-GCCGGGATGCCAATGTA-3′ and 5′-GACGATATCGATGGCAA-3′ to generate an overlapping 1,319-bp fragment encompassing vvpD. The fragments were purified on Nucleospin columns (Machery-Nagel GmbH & Co., Düren, Germany), and both strands were sequenced by the ABI Prism 310 Dye Terminator Cycle sequencing protocol (Applied Biosystems Inc., Perkin-Elmer, Foster City, Calif.).
Construction of a VvpD overexpression plasmid.
A plasmid construct in which vvpD is located downstream of an inducible tac promoter was constructed by the following method. A primer pair was designed to amplify the V. vulnificus vvpD gene with a HindIII site (underlined) and a ribosome binding site (boldface) immediately upstream of the initiating methionine codon (italicized) of vvpD (5′-CACCACAAAGCTTAAGGAGATTATATATATGGAC-3′) and a SalI site (underlined) (3′-TGTCTGCGTCGACGATATCGATGGCAA-5′) 92 bp downstream of the termination codon. Using these primers, a 1-kb fragment containing the entire vvpD gene was amplified from V. vulnificus C7184 by PCR, digested with HindIII and SalI, subcloned into pBluescript II SK+, and subsequently subcloned into pMMB67HE.cam immediately downstream of the tac promoter to generate pRPD1. pRPD1 was then conjugated into C7184D12Ω by triparental matings (10) with E. coli DH5α(pRK2013) for complementation experiments.
Prepilin peptidase assay.
Strains C7184 and C7184D12Ω carrying the vector pMMB67HE.cam and strain C7184D12Ω carrying pRPD1 were grown overnight at 30°C in BHI broth with chloramphenicol (2.5 μg/ml) and IPTG added to a final concentration of 1 mM. Preparation of membrane fractions and subsequent prepilin leader peptidase assays using purified prepilin from P. aeruginosa as the substrate were performed as previously described (59). Samples were run on a sodium dodecyl sulfate–Tricine–15% polyacrylamide gel and stained with Coomassie blue R-250.
Cell fractionation and cytolysin assay.
Overnight cultures of strains to be tested were incubated overnight at 37°C at 200 rpm in BHI broth with or without chloramphenicol (2.5 μg/ml), subcultured into BHI broth or BHI broth plus chloramphenicol, grown to late log phase (optical density at 600 nm of 2.0), and then centrifuged to separate the cells from the supernatant fraction. The pelleted cells were fractionated to recover the periplasmic contents by osmotic shock (69), and the cytoplasmic fractions were isolated as described by Manoil and Beckwith (30). Cytolysin activity was measured in the supernatant, periplasmic, and cytoplasmic fractions in 96-well microtiter plates. Serial twofold dilutions of 0.1-ml samples in phosphate-buffered saline, pH 7.4 (PBS), were incubated with an equal volume of a 0.8% suspension of sheep erythrocytes for 2 h at 37°C (15). Hemolytic indices were determined as the reciprocal of the last dilution in which complete lysis of erythrocytes was observed.
Other enzyme assays.
Chitinase activity was determined by the conventional plate method (39). Plates were spot inoculated on nutrient agar containing 0.5% NaCl and 5 μg of chloramphenicol per ml, overlaid with 10 ml of the same agar containing a 1% (wt/vol) colloidal chitin suspension, and incubated at 25°C for 7 days. A narrow (1- to 2-mm) zone of clearing around bacterial growth was indicative of chitinase secretion. Protease secretion was similarly determined on BHI agar plates containing 1% skim milk incubated at 30°C overnight (26). Secretion was verified by a 2- to 3-mm zone of clearing around the area of growth of the streaked culture.
Phage PO4 sensitivity assay.
P. aeruginosa strains to be tested for the presence of pili were grown overnight at 37°C on LB agar plates with the appropriate antibiotics. Cells were suspended in LB broth and then streaked onto LB agar plates. One microliter of the pilus-specific phase PO4 (∼1010 PFU) was spotted onto the center of each streak, and plates were incubated at 37°C. Sensitivity was defined as the absence of growth in the area of phage inoculation.
Serum sensitivity assay.
A 0.5-ml aliquot of saline-washed bacterial cells containing 107 to 108 CFU ml−1 was mixed with 0.5 ml of fresh human serum or heat-inactivated serum (56°C, 30 min) and incubated at 37°C for 1 h. The number of bacteria in serial 10-fold dilutions before and after incubation was counted on LB agar after overnight incubation at 37°C. Serum resistance was measured as the log of the ratio of the initial concentration of bacteria to the number recovered after 1 h.
TEM.
To visualize pili by transmission electron microscopy (TEM), bacterial cells were negatively stained with 2% phosphotungstic acid (pH 7.2) on Parlodion-coated grids and examined with a JEOL 100-B transmission electron microscope operated at 60 kV. For each strain tested, at least 100 cells on each of three separate grids were examined for the presence of pili. For observation of capsules, TEM of ultrathin slices stained with ruthenium red was performed as previously described (29, 70).
Cytotoxicity assay.
All tissue culture cells were grown in Eagle’s minimal essential medium (MEM) supplemented with 10% fetal calf serum and 200 mM l-glutamine at 37°C in 5% CO2. Cytotoxicity assays were carried out on Chinese hamster ovary cells (CHO) cells, using the procedures outlined by Guerrant et al. (13) that were previously used to measure the cytolysin activity present in supernatants of V. vulnificus cultures (65). Briefly, cultures were grown overnight in BHI broth (with chloramphenicol and IPTG where appropriate) at 30°C. Optical densities were measured at 600 nm to verify comparable growth of the cultures. Supernatants were centrifuged twice at 12,000 × g and filtered through 0.2-μm-pore-size cellulose acetate syringe filters to remove bacteria. Serial twofold dilutions were prepared in PBS, and 20 μl of each dilution was added to CHO cell monolayers covered with 200 μl of MEM in 96-well tissue culture plates, each seeded with 4 × 104 CHO cells 24 h prior to the experiment. The cells were then incubated at 37°C in 5% CO2. Cytotoxicity was determined by changes in CHO cell morphology (cell rounding and lysis; detachment of monolayer from the plastic surface) after 6 h of exposure to the bacterial supernatants. The results are reported as the reciprocal of the lowest dilution causing greater than 75% of the cells to show the altered morphologies.
Tissue culture adherence assay.
Quantitative adherence assays were performed on human epidermoid carcinoma cell (HEp-2) monolayers grown in 24-well tissue culture dishes in which each well was seeded with 2 × 105 cells and grown overnight at 37°C in 5% CO2. The HEp-2 cells were prepared for the assay by removing the medium and washing twice with Hanks’ balanced salt solution, followed by the addition of 1 ml of serum-free MEM with Earle’s salts. V. vulnificus strains were grown overnight in BHI broth (with 5 μg of chloramphenicol per ml and 1 mM IPTG if containing plasmid pMMB67EH.cam or pRPD1) at 30°C with gentle agitation. After measurement of the optical density at 600 nm to estimate bacterial cell concentration from previous growth curves, the cultures were pelleted in a microcentrifuge (6,000 × g), resuspended in PBS to their original concentration, and then diluted in PBS to 4 × 107 CFU ml−1. Cell monolayers were inoculated in triplicate with 50 μl of the diluted bacteria to give a multiplicity of infection (MOI) of ca. 10 and then centrifuged at 670 × g for 10 min at 10°C in an SH3000 rotor. The tissue culture plates were incubated at 37°C in 5% CO2 for 1 h, and then the monolayers were washed six times with PBS to remove nonadherent bacteria. Following the last wash, the cells were covered with 1 ml PBS and mechanically agitated by vigorous pipetting to suspend epithelial cells and bacteria, followed by serial 10-fold dilutions and quantitation by plating on LB agar. The assay results are presented as the percent cell-associated bacteria, which equals (CFU recovered/CFU inoculated) × 100.
Animal studies.
Female, 6- to 8-week old Swiss Webster mice (DNK Universal Inc., Kent, Wash.) were used in all experiments. Intramuscular injections of 5 mg of iron dextran (Sigma, St. Louis, Mo.) in 100 μl of water were administered 2 h prior to challenge as previously described (66). Overnight cultures of bacteria grown in BHI broth with chloramphenicol and IPTG, where appropriate, were pelleted and washed once in PBS prior to resuspension in PBS to appropriate concentrations. The mice (n = 5 for each dilution) were inoculated intraperitoneally with 200 μl of the bacteria, and the experiment was terminated after 26 h. The LD50 was calculated by the method of Reed and Muench (46).
Nucleotide sequence accession numbers.
The nucleotide sequences of vvpC and vvpD from both MO6-24 and C7184 have been submitted to the GenBank/EMBL/DDBJ data libraries under accession no. U48808 and AF070934, respectively.
RESULTS
Isolation of a V. vulnificus vvpD clone that complements a P. aeruginosa pilD mutation.
A V. vulnificus genomic library from strain MO6-24 was conjugated into P. aeruginosa PAK 2B18, which contains a Tn5 insertion in the pilD gene (36). Clones complementing the P. aeruginosa pilD defect were identified by the reappearance of twitching motility (8), which is caused by the polymerization and retraction of type IV pili and is observed in P. aeruginosa as characteristic rough and irregular spreading colonies on solid surfaces. Mutants that lack PilD produce small round colonies with smooth edges because the type IV pilin subunits are not processed and assembled into functional pili (36).
Three clones with a twitching motility phenotype, observed as a characteristic rough and irregular spreading morphology, were tested for susceptibility to the P. aeruginosa pilus-specific bacteriophage PO4. All PO4-sensitive clones contained similar cosmids, as evidenced by restriction endonuclease digests. One clone, which carried approximately 25 kb of the V. vulnificus chromosome, was chosen for further study and designated pJCL12B.
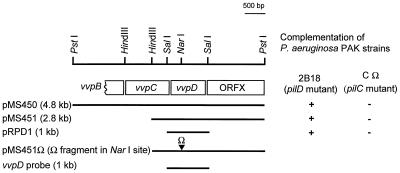
Various restriction fragments of plasmid pJCL12B were subcloned and religated into pLAFR3 in order to delimit the region containing the V. vulnificus pilD homolog (Fig. 1). Digestion with PstI produced a 4.8-kb fragment that complemented the P. aeruginosa pilD mutation (pMS450). The 4.8-kb PstI fragment contained two internal HindIII sites that permitted its subcloning as three separate fragments in pLAFR3: pMS451 carrying a 2.8-kb HindIII-PstI fragment, pMS452 carrying a 1.2-kb PstI-HindIII fragment, and pMS453 carrying a 0.8-kb HindIII fragment. Only pMS451 complemented the pilD mutation in P. aeruginosa PAK 2B18, as confirmed by phage PO4 sensitivity.
FIG. 1.
Genetic organization and partial restriction map of the 4.8-kb PstI fragment showing restriction sites used for cloning various constructs. Genes encoded on the fragment are shown as boxes. The ▾ symbol indicates the insertion site for the Ω fragment. Complementation of P. aeruginosa pilD and pilC mutants, indicated by restoration of PO4 phage sensitivity, is shown on the right. The vvpD probe derived from the 1-kb SalI fragment used in Southern blots is shown as a bar at the bottom.
Sequence analysis of V. vulnificus vvpC and vvpD.
Nucleotide sequencing of the PstI fragment from pMS450 (strain MO6-24) revealed the presence of one partial and three complete open reading frames (ORFs) (Fig. 1). Comparison of the ORFs to the GenBank database by using BLAST showed that three of the ORFs encoded proteins homologous to type IV pilus biogenesis proteins previously identified as PilBCD from P. aeruginosa (36) and TapBCD from A. hydrophila (40); the ORF products and their corresponding genes were designated VvpBCD and vvpBCD (V. vulnificus pili). The first partial ORF product had strong homology to the ABC transporter-like PilB/TapB. A second clone has been isolated since the initial isolation of pJCL12B that contains a complete vvpB gene; its characterization will be reported elsewhere. There was a complete ORF immediately downstream of the partial vvpB gene, designated vvpC, encoding a protein of 409 amino acids with strong homology to PilC/TapC, a protein of unknown function but absolutely required for pilus assembly (36). VvpC shows 55% similarity and 41% identity to PilC and 58% similarity and 45% identity to TapC (Fig. 2).
FIG. 2.
Comparison of amino acid sequences of V. vulnificus VvpC (Vv VvpC) with homologs from P. aeruginosa (Pa PilC) and A. hydrophila (Ah TapC). Differences in the C7184 and MO6-24 VvpC sequences are shaded. The alignment was generated by using the Pileup program of the Genetics Computer Group (Madison, Wis.).
Downstream and adjacent to vvpC is vvpD, which encodes a 289-amino-acid protein that shows 59% similarity and 52% identity to PilD and 62% similarity and 55% identity to TapD (Fig. 3). The sequence also reveals the presence of two pairs of cysteine residues, at positions 70 and 73 and positions 95 and 98, and an invariant glycine at position 92, which in P. aeruginosa (PilD) have been shown to be important in leader peptide cleavage and methyltransferase activities of the enzyme (41, 55). Immediately downstream of the vvpD gene is a third ORF encoding a 22.5-kDa protein highly homologous to products of several similarly sized ORFs of unknown function, located downstream of pilD homologs in A. hydrophila, Pseudomonas putida, and Xanthomonas campestris (40).
FIG. 3.
Comparison of amino acid sequences of V. vulnificus VvpD (Vv VvpD) with homologs from P. aeruginosa (Pa PilD) and A. hydrophila (Ah TapD). The conserved cysteine and glycine residues shown to be involved in leader peptidase and N-methyltransferase activities are shaded. The alignment was generated by using the Pileup program of the Genetics Computer Group.
During this work we found that allelic exchange was more efficient in V. vulnificus C7184 than in MO6-24. Therefore, a DNA fragment containing the genes encoding vvpC and vvpD from strain C7184 was generated by PCR using primers specific for the flanking regions of MO6-24 vvpC and vvpD, as described in Materials and Methods. Sequencing of the PCR-generated C7184 vvpD showed it to be identical to vvpD from MO6-24. There were some differences in the nucleotide sequences of vvpC from MO6-24 and C7184 that result in a Leu-to-Pro substitution at amino acid position 119, and a Ser to Pro substitution at position 176, in the deduced amino acid sequences (Fig. 2).
vvpD is conserved in V. vulnificus strains.
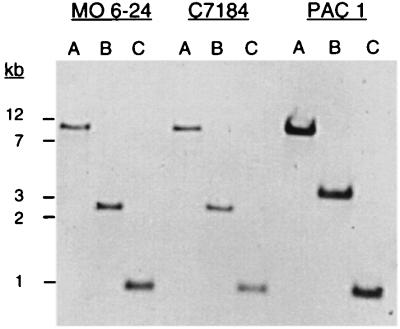
An environmental isolate, PAC1, was compared with the two clinical isolates of V. vulnificus, MO6-24 and C7184, for the presence of the type IV pilus biogenesis gene cluster. Chromosomal DNA from all of three strains was digested with the restriction enzymes PstI, PstI-HindIII, and SalI and probed under high stringency with the vvpD gene (a 1-kb SalI fragment). The vvpD sequence was present on a 1-kb SalI fragment as well as an 11-kb PstI fragment in all three strains (Fig. 4). Probing a PstI-HindIII double digest confirmed the presence of vvpD on a 2.8-kb fragment in strains MO6-24 and C7184. However, probing PAC1 DNA digested with the same enzymes showed hybridization to a 3.6-kb fragment demonstrating a minor restriction site polymorphism and a missing downstream HindIII site (Fig. 1).
FIG. 4.
Southern blot comparison of various restriction digests of V. vulnificus chromosomal DNA, using vvpD as a probe. Lanes: A, PstI; B, PstI-HindIII; C, SalI. Positions of molecular weight markers are indicated on the left.
V. vulnificus VvpC does not complement P. aeruginosa PilC.
Both TapC and TapD of A. hydrophila can complement their homologous counterparts in P. aeruginosa (40), and as shown here, VvpD can functionally replace PilD. To determine whether V. vulnificus VvpC can complement PilC, plasmids carrying vvpC and vvpD from strain MO6-24(pMS450), vvpD from strain C7184(pRPD1), and the vector alone (pMMB67HE.cam) were conjugated into P. aeruginosa PAK 2B18 and P. aeruginosa PAK-CΩ, a pilC null mutant (23). Testing for restoration of pilus expression by sensitivity to phage PO4 showed that pMS450 and pRPD1 complemented PAK 2B18 but not PAK-CΩ (Fig. 1). Therefore, while VvpD is expressed and complements its homolog in P. aeruginosa, we were unable to demonstrate complementation by VvpC. However, it is unknown whether this is due to any differences in specificity of the proteins or due to lack of expression or incorrect cytoplasmic membrane localization of VvpC in P. aeruginosa.
Construction of a V. vulnificus vvpD mutant.
To construct a V. vulnificus vvpD mutant, the 2.0-kb Ω interposon encoding spectinomycin and streptomycin resistance (42) was cloned into a unique NarI site within vvpD (137 bp downstream of the vvpD ATG start codon) in plasmid pMS4514, creating pMS451Ω (Fig. 1). The 4.8-kb DNA fragment containing vvpD::Ω was then subcloned into the suicide vector pEP185.2 (20) as a SmaI-KpnI fragment and maintained in E. coli S17-1λpir (32, 50). This construct (pMS451Ω-2) was subsequently transferred into V. vulnificus C7184 by conjugation. Double recombinants were selected for spectinomycin resistance and screened for chloramphenicol sensitivity, and then the presence of only the insertionally inactivated copy of vvpD was confirmed by Southern blot analysis (data not shown). One strain, C7184D12Ω was selected for further study.
VvpD is a type IV leader peptidase.
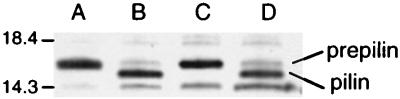
Complementation of the P. aeruginosa pilD mutation by vvpD demonstrates that VvpD can replace the type IV peptidase activity of PilD. To confirm that VvpD acts to cleave the leader sequence from a type IV prepilin, an in vitro assay was performed. Plasmids pRPD1 and pMMB67HE.cam were conjugated into C7184 and C7184D12Ω. Membrane fractions prepared from these strains were incubated with P. aeruginosa purified prepilin as the substrate (59). The prepilin incubated with membranes from C7184(pMMB67HE.cam) and C7184D12Ω(pRPD1) show an increased mobility, suggesting cleavage of the leader peptide from the prepilin. There was no shift in the mobility of the prepilin band treated with membranes derived from C7184D12Ω(pMMB67HE.cam) compared to prepilin alone, indicating the absence of a functional type IV peptidase (Fig. 5). These mobility shifts were identical to those seen with PilD-containing membranes as a source of enzyme activity (data not shown). Although this experiment does not unequivocally show that VvpD cleaves the leader peptide precisely at the site cleaved by PilD (MKAQKG▾FTLIE, where ▾ is the cleavage site), sufficient evidence has accumulated through N-terminal amino acid sequencing that expression of pili in vivo requires that the leader sequence of the P. aeruginosa prepilin (and pilin-like proteins of the type II secretion system) be cleaved between these glycine and phenylalanine residues before polymerization of the pilin monomers into pili can proceed (37, 60).
FIG. 5.
Sodium dodecyl sulfate–Tricine–15% polyacrylamide gel showing in vitro activity of VvpD on P. aeruginosa prepilin substrate. Membranes were prepared from V. vulnificus strains carrying the plasmids indicated. Lanes: A, prepilin substrate only; B, C7184(pMMB67HE.cam); C, C7184D12Ω(pMMB67HE.cam); D, C7184D12Ω(pRPD1). Positions of molecular weight markers are shown on the left in kilodaltons.
The vvpD mutation defines a type II secretion system in V. vulnificus.
In addition to pilus biogenesis, P. aeruginosa PilD has been shown to be required for extracellular protein secretion via the GSP or type II pathway (43, 58). To determine the role of VvpD in protein secretion, we compared the activities and localization of three enzymes, the cytolysin/hemolysin, protease, and chitinase. Cells from C7184(pMMB67HE.cam), C7184D12Ω(pMMB67HE.cam), and C7184D12Ω(pRPD1) were fractionated to compare the activities of the cytolysin in the cytoplasm, periplasmic space, and extracellular milieu. Activity of the cytolysin was measured by its hemolytic titer in culture filtrates of cell fractions. In C7184(pMMB67HE.cam), all activity was in the supernatant fraction; none was detected in either the periplasmic or cytoplasmic fraction (Table 2). The majority of the hemolytic activity was concentrated in the periplasmic fraction in C7184D12Ω(pMMB67HE.cam), with some residual activity in both the supernatant and cytoplasmic fractions, indicating a defect in secretion of the cytolysin from the periplasm across the outer membrane. The defect in secretion was restored in the complemented strain, C7184D12Ω(pRPD1), where the majority of the cytolysin was detected in the supernatant fraction. Cytolysin was not detected in any of the cell fractions of a cytolysin (vvhA) mutant, C7184-402K, which was included as a control.
TABLE 2.
Secretion of cytolysin, protease, and chitinase in V. vulnificus
| Strain | Hemolytic indexa
|
Extracellular secretionb
|
|||
|---|---|---|---|---|---|
| Supernatant | Periplasm | Cytoplasm | Protease | Chitinase | |
| C7184(pMMB67HE.cam) | 2,048 | 0 | 0 | + | + |
| C7184D12Ω(pMMB67HE.cam) | 8 | 272 | 4 | − | − |
| C7184D12Ω(pRPD1) | 2,048 | 16 | 4 | + | + |
| C7184-402K | 0 | 0 | 0 | ND | ND |
Reciprocal of the highest dilution of culture supernatant or cell fraction at which hemolysis was observed (average of two assays).
Detected by a zone of clearing around the bacterial colonies (see Materials and Methods). ND, not done.
The same strains were compared for extracellular secretion of protease and chitinase. C7184(pMMB67HE.cam) and C7184D12Ω(pRPD1) secreted protease extracellularly, but secretion of protease was not observed in the mutant strain, C7184D12Ω(pMMB67HE.cam) (Table 2). Extracellular secretion of chitinase by C7184(pMMB67HE.cam) and C7184D12Ω(pRPD1) was confirmed by a zone of clearing around the area of inoculation on plates containing chitin. The mutant strain C7184D12Ω(pMMB67HE.cam) showed no secretion of chitinase (Table 2).
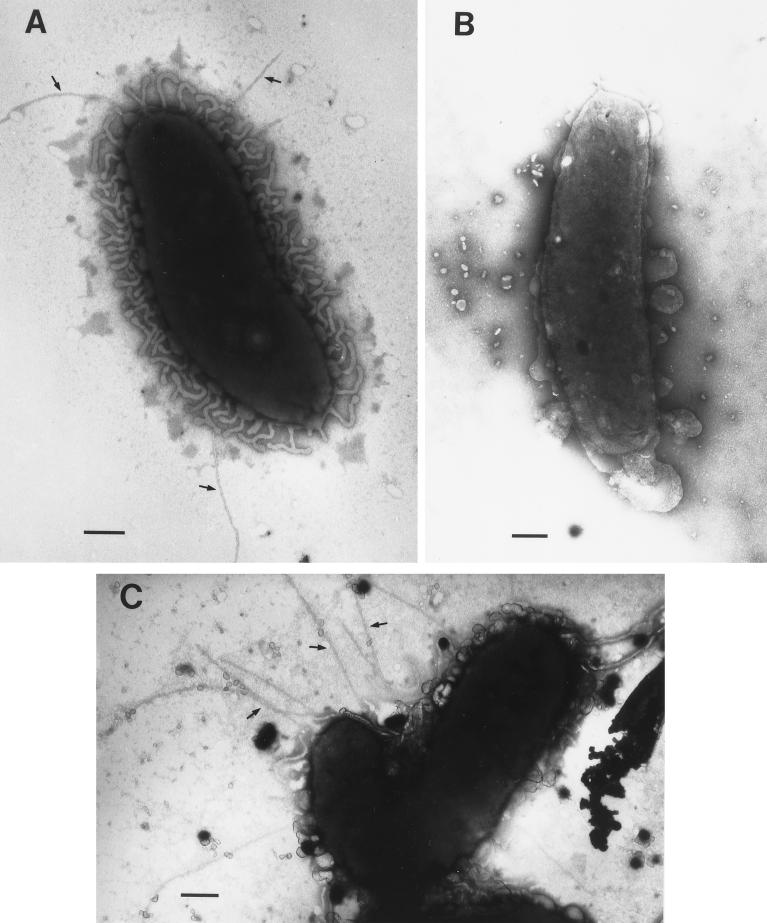
Inactivation of VvpD results in loss of surface pili.
Transmission electron micrographs of wild-type C7184 show the presence of fine hairlike structures of typical size and length for type IV pili (57) extending from the surface of the cell (Fig. 6A). Pili were not detected on the surface of C7184D12Ω (Fig. 6B), indicating that a mutation in vvpD results in loss of expression of pili on the surface of the cells. When VvpD function was restored in the complemented strain C7184D12Ω(pRPD1), pili were again observed on the surface of the cells (Fig. 6C). Greater than 50% of the wild-type and complemented mutant cells were piliated as visualized by TEM, while no surface or background pili were observed on the mutant alone.
FIG. 6.
Electron micrographs of V. vulnificus C7184 strains showing surface pili (indicated by arrows). (A) C7184; (B) C7184D12Ω; (C) C7184D12Ω(pRPD1). Bar = 200 nm.
Loss of VvpD does not affect serum resistance and encapsulation of V. vulnificus.
The specific virulence functions attributed to the V. vulnificus capsule include resistance to the bactericidal effects of serum as well as inhibition of phagocytosis by macrophages. To determine whether V. vulnificus C7184D12Ω is equally or less serum resistant than wild-type C7184, a serum sensitivity assay was performed. As shown in Table 3, the wild-type and vvpD mutant strains of C7184 were equally resistant to the bactericidal effect of human serum at levels significantly higher than the serum-sensitive E. coli S17-1λpir.
TABLE 3.
Serum resistance
| Strain | Serum resistancea
|
|
|---|---|---|
| Normal | Heat inactivated | |
| V. vulnificus | ||
| C7184 | 0.46 | 0.44 |
| C7184-D12Ω | 0.93 | 0.65 |
| E. coli S17-1λpir | >6.4 | 0.71 |
Log10 reduction of bacterial numbers before and after incubation for 1 h at 37°C in normal or heat-inactivated serum.
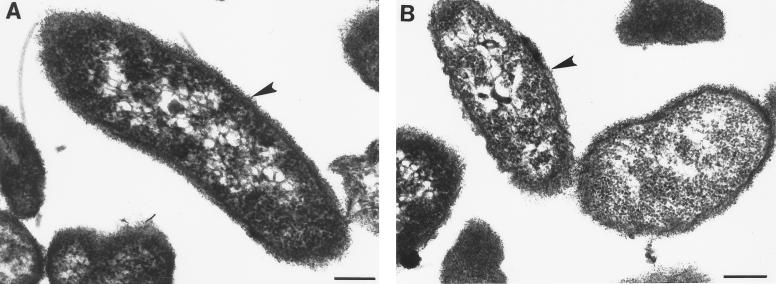
The presence of an intact polysaccharide capsule was then confirmed. Whereas unencapsulated variants of V. vulnificus are translucent in visible light (67), colonies of V. vulnificus C7184D12Ω are opaque and indistinguishable from wild-type colonies, suggesting no alteration of capsule expression. This was experimentally confirmed by demonstrating there was no difference in hydrophobicity in ammonium sulfate (67) between C7184 and C7184D12Ω (data not shown). Finally, electron micrographs (Fig. 7) of ruthenium red-stained ultra thin sections show that both the wild-type and vvpD mutant strains have an electron-dense layer outside their outer membranes typical of encapsulated V. vulnificus (67).
FIG. 7.
Electron micrographs of V. vulnificus C7184 (A) and C7184D12Ω (B) stained with ruthenium red. Bar = 200 nm.
VvpD is required for culture supernatant cytotoxicity on CHO cells.
Culture supernatants from V. vulnificus are cytotoxic to cultured epithelial cells, an activity attributed to the cytolysin, a 51-kDa protein that is hemolytic to mammalian erythrocytes. Purification and subsequent cloning and sequencing indicated that this cytotoxic and hemolytic activity was caused by the protein, designated VvhA or cytolysin, which has some homology to the V. cholerae El Tor hemolysin (12, 65, 68). To determine the effect of the vvpD mutation on cytoxicity, serial twofold dilutions of filtered supernatants from overnight cultures were added to monolayers of CHO cells, incubated for 6 h at 37°C, and monitored for cell rounding and destruction of the monolayer (65). As expected from the results obtained in the cytolysin localization assays, the supernatant from C7184D12Ω was able to cause a cytopathic effect only at the highest concentration, while the supernatant from wild-type C7184 had to be diluted more than 64-fold before no effect on the CHO cells was observed (Table 4). Complete cytotoxicity of C7184D12Ω was restored by complementation with pRPD1. Surprisingly, the supernatant from strain C7184-402K, which contains a kanamycin resistance cassette in vvhA and does not express the cytolysin, is equally cytotoxic to CHO cells (Table 4), contrary to results reported previously (64). This suggests that cytotoxicity of culture supernatants from V. vulnificus is due to a combination of several exported degradative and cytotoxic enzymes, perhaps acting in concert. However, the role of VvhA is unclear.
TABLE 4.
Cytotoxicity, adherence, and virulence
| V. vulnificus strain | CHO cell cytotoxicitya | HEp-2 cell adherence (%)b | Mouse LD50 |
|---|---|---|---|
| C7184(pMMB67HE.cam) | 64 | 9.2 ± 2.1 | 9.4 × 103 |
| C7184-D12Ω(pMMB67EH.cam) | <2 | 0.3 ± 0.1c | 1.1 × 106 |
| C7184-D12Ω(pRPD1) | 64 | 4.5 ± 2.0c | 2.1 × 105 |
| C7184-402K | 64 | 9.0 ± 3.6 | NDd |
Reciprocal of the highest dilution of filtered bacterial supernatants to cause typical CHO cell cytotoxicity (cell lysis, rounding, and detachment of monolayer) after 6 h.
Presented as (number of CFU recovered/number of input CFU) × 100 ± standard deviation.
Significantly different when compared by Tukey-Kramer analysis at P ≤0.5.
ND, not done.
VvpD is required for HEp-2 cell adherence.
To determine if pili or other type II secretion products play a role in virulence of V. vulnificus by aiding adherence to epithelial cells, we developed an adherence assay that is more quantitative and reproducible than the method previously used (11). The assay involves adding washed V. vulnificus cells to monolayers of HEp-2 cells at numbers no greater than 10 CFU per epithelial cell. The low MOI prevents the rapid cytotoxic effect that destroys the integrity of the tissue culture monolayer when higher MOIs are used. Adherent bacteria are resuspended by mechanical agitation instead of lysing the epithelial cells with detergents such as Triton X-100 because V. vulnificus is extremely sensitive to rapid lysis by these agents as well. Following the procedure outlined in the Materials and Methods, ca. 9% of wild-type strain C7184(pMMB67HE.cam) input bacteria remained adherent to the HEp-2 cell monolayer after extensive washing (Table 4). C7184D12Ω(pMMB67HE.cam) was consistently and significantly less adherent than the wild-type parent strain, with less than 0.5% of input bacteria remaining associated with the monolayers. Complementation of C7184D12Ω with pRPD1 significantly increased adherence to a value about half of that for the wild-type strain, suggesting that adherence of V. vulnificus to HEp-2 cells requires pili, a type II secretion pathway product (or products), or both. Adherence was not dependent on the presence of VvhA, as strain C7184-402K was as adherent to HEp-2 cells as the wild-type C7184 strain.
Virulence is dependent on VvpD.
The LD50s in iron-overloaded mice after intraperitoneal injections of V. vulnificus strains are shown in Table 4. Strain C7184D12Ω showed more than a 2-log increase in LD50 over the wild-type strain. This difference is not attributed to lack of or slower growth of the mutant strain in vivo. In limited analyses, 15- to 30-fold more CFU than initial challenge inocula were recovered 24 h postinfection from homogenized spleens of mice whether they were infected with the wild-type, the mutant, or the complemented mutant strain. Complementation of C7184D12Ω with vvpD in trans on plasmid pRPD1 partially restored virulence to a value 1 log less than that for the uncomplemented mutant. Also, at inoculum levels that eventually resulted in the death (within 26 h) of all five mice injected with C7184D12Ω(pMMB67HE.cam) or C7184D12Ω(pRPD1), the time to death for the mice infected with the latter was consistently 2 to 4 h less than for the uncomplemented mutant (data not shown). Again, equal numbers of bacteria were recovered from spleens of infected animals regardless of the strain or plasmid construct it carried. However, it was found during these analyses that the plasmid containing vvpD was not maintained in these strains in the absence of antibiotic selection and was rapidly lost within 24 h postinfection by approximately 99% of the bacteria during in vivo cell division. This may account for the partial complementation in C7184D12Ω(pRPD1) to an LD50 lower than that of the wild type.
DISCUSSION
With the exception of the polysaccharide capsule, it has been difficult to identify specific factors that contribute to the pathogenicity of V. vulnificus. The data presented in this report show that a mutation in a single gene encoding a type IV peptidase/N-methyltransferase in V. vulnificus causes pleiotropic defects in expression of pili and in the type II extracellular protein secretion pathway. These defects significantly decrease tissue culture adherence and cytotoxic activity, as well as virulence in the iron-overloaded mouse model. The in vitro results obtained with tissue culture experiments suggest that specific colonization factors such as type IV pili are required for adherence of V. vulnificus to epithelial cells. In addition, this mutant is significantly less cytotoxic to cultured epithelial cells than the wild-type or the cytolysin mutant, suggesting that multiple secreted products and possibly close contact mediated by the pili are necessary for cytotoxicity. Since the vvpD mutation has no measurable effect on capsule expression, these results indicate that a combination of pili and/or secreted protein(s) may play a significant role in V. vulnificus pathogenesis, as indicated by the reduced virulence in mice. Clarification of these possibilities will be aided by the construction of a mutant unable to express the type IV pilin subunit, the gene for which has recently been localized upstream of vvpC and vvpD (unpublished data).
V. vulnificus produces a number of degradative and cytotoxic enzymes that are important for survival in its normal environmental niche, namely, estuarine waters and molluscan shellfish. A number of these, including the cytolysin, an elastolytic protease (24), and an enterotoxin (54), have been suggested to be important for the rapid invasiveness, dissemination, and tissue damage caused by the organism after ingestion of contaminated shellfish or infections through wounds. Adherence to cells mediated by pili has also been suggested to be an important virulence determinant (11).
The purified cytolysin protein is cytotoxic to CHO cells, is hemolytic for mammalian erythrocytes, causes vascular permeability in guinea pig skin, and is lethal for mice (12, 25). Detection of specific anticytolysin antibody in convalescent sera suggests that it is expressed during clinical infection. However, its contribution to virulence is suspect in that a cytolysin mutant strain of V. vulnificus is just as virulent in the mouse model as isogenic wild-type strains (64). It is unclear why the mutant strain that we constructed by allelic exchange of wild-type vvhA with a copy interrupted with a kanamycin resistance cassette is still cytotoxic to CHO cells, even though the cytolysin (hemolysin) assays clearly show that VvhA is not expressed (Table 2). The vvhA mutant strain previously reported to lack CHO cell cytotoxicity (64) contains a transposon Tn10 insertion in vvhA. It is possible the Tn10 insertion had a polar effect on the expression of an adjacent gene(s) required for tissue culture cytotoxicity in addition to cytolysin. Interestingly, cytotoxicity of a nonhemolytic vvhA mutant (constructed by allelic exchange of vvhA with a copy insertionally inactivated by a non-transposon antibiotic resistance cassette) similar to our C7184-402K was not reported in this same study (64).
To our knowledge, there has been only one other report describing adherence of V. vulnificus to cultured epithelial cells (11). In that study, Gander and LaRocco described the presence of surface pili on a variety of V. vulnificus strains and reported that clinical isolates from blood or wounds of infected individuals averaged higher numbers of individual pili per cell than environmental isolates. In assays where they directly enumerated adherent bacteria on monolayers of HEp-2 cells, they correlated adherence with this increased piliation. Environmental isolates, on average, showed fewer bacteria adhering to individual epithelial cells. We are interested in determining whether the quantitative assay that we developed to measure bacterial adherence will show similar differences between environmental (i.e., from oysters) and clinical (blood or wound) isolates.
In addition to the role of pili, other virulence factors have been examined in an attempt to determine why various V. vulnificus strains differ in pathogenicity. Oysters are colonized with up to hundreds of individual V. vulnificus strains when differentiated by clamped field electrophoresis, pulsed-field gel electrophoresis, or ribotyping (9, 61), and mouse bioassays demonstrate that there are virulent and avirulent strains (53, 62). However, evidence has accumulated to suggest that human infections are caused by only a few of the strains present in the heterogeneous populations found in shellfish (16). To account for these differences in virulence, studies have focused on the cytolysin (18, 62), serum resistance (19, 53), and agglutinating property of the capsule (14). To date a definitive phenotypic difference has not been demonstrated between virulent and less virulent strains, with the exception that loss of capsule correlates with loss of virulence (49, 67, 70).
We have demonstrated the role of the type IV peptidase/N-methyltransferase VvpD from V. vulnificus in the expression of pili and extracellular secretion by the GSP or type II pathway. Pili and/or one or more of the secreted factors appear to be required for tissue culture cytotoxicity, cell adherence, and virulence. These results indicate that in addition to the capsule, V. vulnificus expresses additional virulence determinants that possibly could be targeted for therapeutic intervention.
ACKNOWLEDGMENTS
We thank Mark Peterson and Frank Poysky for assistance with the virulence studies, and we thank Jay Bell and Rachel Johnson for assistance with the adherence and cytotoxicity assays. We also thank Stephen Lory and Walt Dickhoff for critically reading the manuscript.
C.M.P. is supported by a NOAA Cooperative Education and Research Program grant (NA67FE0396) to the University of Washington School of Fisheries through a collaboration with Faye Dong. We also thank Faye Dong for serving as the U.W. School of Fisheries graduate advisor for R.N.P.
REFERENCES
- 1.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amaro C, Biosca E G. Vibrio vulnificus biotype 2, pathogenic for eels, is also an opportunistic pathogen for humans. Appl Environ Microbiol. 1996;62:1454–1457. doi: 10.1128/aem.62.4.1454-1457.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amaro C, Biosca E G, Fouz B, Toranzo A E, Garay E. Role of iron, capsule, and toxins in the pathogenicity of Vibrio vulnificus biotype 2 for mice. Infect Immun. 1994;62:759–763. doi: 10.1128/iai.62.2.759-763.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anonymous. Vibrio vulnificus infections associated with raw oyster consumption—Florida, 1981–1992. Morbid Mortal Weekly Rep. 1993;42:405–407. [PubMed] [Google Scholar]
- 6.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonner J R, Coker A S, Berryman C R, Pollock H M. Spectrum of Vibrio infections in a Gulf Coast community. Ann Intern Med. 1983;99:464–469. doi: 10.7326/0003-4819-99-4-464. [DOI] [PubMed] [Google Scholar]
- 8.Bradley D E. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 9.Buchrieser C, Gangar V V, Murphree R L, Tamplin M L, Kaspar C W. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl Environ Microbiol. 1995;61:1163–1168. doi: 10.1128/aem.61.3.1163-1168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Cianciotto, N. Personal communication.
- 10.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;79:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gander R M, LaRocco M T. Detection of piluslike structures on clinical and environmental isolates of Vibrio vulnificus. J Clin Microbiol. 1989;27:1015–1021. doi: 10.1128/jcm.27.5.1015-1021.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray L D, Kreger A S. Purification and characterization of an extracellular cytolysin produced by Vibrio vulnificus. Infect Immun. 1985;48:62–72. doi: 10.1128/iai.48.1.62-72.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrant R L, Brunton L L, Schnaitman T C, Rebhun L I, Gilman A G. Cyclic adenosine monophosphate and alteration of Chinese hamster ovary cell morphology: a rapid, sensitive in vitro assay for the enterotoxins of Vibrio cholerae and Escherichia coli. Infect Immun. 1974;10:320–327. doi: 10.1128/iai.10.2.320-327.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayat U, Reddy G P, Bush C A, Johnson J A, Wright A C, Morris J G., Jr Capsular types of Vibrio vulnificus: an analysis of strains from clinical and environmental sources. J Infect Dis. 1993;168:758–762. doi: 10.1093/infdis/168.3.758. [DOI] [PubMed] [Google Scholar]
- 15.Howard S P, Buckley J T. Activation of the hole-forming toxin aerolysin by extracellular processing. J Bacteriol. 1985;163:336–340. doi: 10.1128/jb.163.1.336-340.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson J K, Murphree R L, Tamplin M L. Evidence that mortality from Vibrio vulnificus infection results from single strains among heterogeneous populations in shellfish. J Clin Microbiol. 1997;35:2098–2101. doi: 10.1128/jcm.35.8.2098-2101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang B, Howard S P. The Aeromonas hydrophila exeE gene, required both for protein secretion and normal outer membrane biogenesis, is a member of a general secretion pathway. Mol Microbiol. 1992;6:1351–1361. doi: 10.1111/j.1365-2958.1992.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 18.Johnson D E, Calia F M. Hemolytic reaction of clinical and environmental strains of Vibrio vulnificus. J Clin Microbiol. 1981;14:457–459. doi: 10.1128/jcm.14.4.457-459.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson D E, Calia F M, Musher D M, Goree A. Resistance of Vibrio vulnificus to serum bactericidal and opsonizing factors: relation to virulence in suckling mice and humans. J Infect Dis. 1984;150:413–418. doi: 10.1093/infdis/150.3.413. [DOI] [PubMed] [Google Scholar]
- 20.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the Yen I restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 21.Klontz K C, Lieb S, Schreiber M, Janowski H T, Baldy L M, Gunn R A. Syndromes of Vibrio vulnificus infections. Clinical and epidemiologic features in Florida cases, 1981–1987. Ann Intern Med. 1988;109:318–323. doi: 10.7326/0003-4819-109-4-318. [DOI] [PubMed] [Google Scholar]
- 22.Koenig K L, Mueller J, Rose T. Vibrio vulnificus. Hazard on the half shell. West J Med. 1991;155:400–403. [PMC free article] [PubMed] [Google Scholar]
- 23.Koga T, Ishimoto K, Lory S. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosa pili. Infect Immun. 1993;61:1371–1377. doi: 10.1128/iai.61.4.1371-1377.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kothary M H, Kreger A S. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J Gen Microbiol. 1987;133:1783–1791. doi: 10.1099/00221287-133-7-1783. [DOI] [PubMed] [Google Scholar]
- 25.Kreger A, Lockwood D. Detection of extracellular toxin(s) produced by Vibrio vulnificus. Infect Immun. 1981;33:583–590. doi: 10.1128/iai.33.2.583-590.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leung K Y, Stevenson R M. Tn5-induced protease-deficient strains of Aeromonas hydrophila with reduced virulence for fish. Infect Immun. 1988;56:2639–2644. doi: 10.1128/iai.56.10.2639-2644.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lory S, Strom M S. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa—a review. Gene. 1997;192:117–121. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- 29.Luft J H. Ruthenium red and violet. I. Chemistry, purification, methods of use for electron microscopy and mechanism of action. Anat Rec. 1971;171:347–368. doi: 10.1002/ar.1091710302. [DOI] [PubMed] [Google Scholar]
- 30.Manoil C, Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986;233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 31.Mattick J S, Whitchurch C B, Alm R A. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene. 1996;179:147–155. doi: 10.1016/s0378-1119(96)00441-6. [DOI] [PubMed] [Google Scholar]
- 32.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi N, Shimizu C, Miyoshi S, Shinoda S. Purification and characterization of Vibrio vulnificus protease. Microbiol Immunol. 1987;31:13–25. doi: 10.1111/j.1348-0421.1987.tb03064.x. [DOI] [PubMed] [Google Scholar]
- 34.Morris J G., Jr Vibrio vulnificus—a new monster of the deep? Ann Intern Med. 1988;109:261–263. doi: 10.7326/0003-4819-109-4-261. [DOI] [PubMed] [Google Scholar]
- 35.Morris J G, Jr, Black R E. Cholera and other vibrioses in the United States. N Engl J Med. 1985;312:343–350. doi: 10.1056/NEJM198502073120604. [DOI] [PubMed] [Google Scholar]
- 36.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunn D N, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Brien M, Colwell R R. A rapid test for chitinase activity that uses 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide. Appl Environ Microbiol. 1987;53:1718–1720. doi: 10.1128/aem.53.7.1718-1720.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pepe C M, Eklund M W, Strom M S. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol Microbiol. 1996;19:857–869. doi: 10.1046/j.1365-2958.1996.431958.x. [DOI] [PubMed] [Google Scholar]
- 41.Pepe J C, Lory S. Amino acid substitutions in PilD, a bifunctional enzyme of Pseudomonas aeruginosa: effect on leader peptidase and N-methyltransferase activities in vitro and in vivo. J Biol Chem. 1998;273:19120–19129. doi: 10.1074/jbc.273.30.19120. [DOI] [PubMed] [Google Scholar]
- 42.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 43.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugsley A P. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 45.Pugsley A P, Francetic O, Possot O M, Sauvonnet N, Hardie K R. Recent progress and future directions in studies of the main terminal branch of the general secretory pathway in Gram-negative bacteria—a review. Gene. 1997;192:13–19. doi: 10.1016/s0378-1119(96)00803-7. [DOI] [PubMed] [Google Scholar]
- 46.Reed L J, Muench H. A simple method of estimating the 50% endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinoda S, Kobayashi M, Yamada H, Yoshida S, Ogawa M, Mizuguchi Y. Inhibitory effect of capsular antigen of Vibrio vulnificus on bactericidal activity of human serum. Microbiol Immunol. 1987;31:393–401. doi: 10.1111/j.1348-0421.1987.tb03102.x. [DOI] [PubMed] [Google Scholar]
- 50.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 51.Simpson L M, Oliver J D. Ability of Vibrio vulnificus to obtain iron from transferrin and other iron-binding compounds. Curr Microbiol. 1987;15:155–157. [Google Scholar]
- 52.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stelma G N, Jr, Reyes A L, Peeler J T, Johnson C H, Spaulding P L. Virulence characteristics of clinical and environmental isolates of Vibrio vulnificus. Appl Environ Microbiol. 1992;58:2776–2782. doi: 10.1128/aem.58.9.2776-2782.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stelma G N, Spaulding P L, Reyes A L, Johnson C H. Production of enterotoxin by Vibrio vulnificus. J Food Prot. 1988;51:192–196. doi: 10.4315/0362-028X-51.3.192. [DOI] [PubMed] [Google Scholar]
- 55.Strom M S, Bergman P, Lory S. Identification of active-site cysteines in the conserved domain of PilD, the bifunctional type IV pilin leader peptidase/N-methyltransferase of Pseudomonas aeruginosa. J Biol Chem. 1993;268:15788–15794. [PubMed] [Google Scholar]
- 56.Strom M S, Lory S. Cloning and expression of the pilin gene of Pseudomonas aeruginosa PAK in Escherichia coli. J Bacteriol. 1986;165:367–372. doi: 10.1128/jb.165.2.367-372.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 58.Strom M S, Nunn D, Lory S. Multiple roles of the pilus biogenesis protein PilD: involvement of PilD in excretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991;173:1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strom M S, Nunn D N, Lory S. Posttranslational processing of type IV prepilin and homologs by PilD of Pseudomonas aeruginosa. Methods Enzymol. 1994;235:527–540. doi: 10.1016/0076-6879(94)35168-6. [DOI] [PubMed] [Google Scholar]
- 60.Strom M S, Nunn D N, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamplin M L, Jackson J K, Buchrieser C, Murphree R L, Portier K M, Gangar V, Miller L G, Kaspar C W. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl Environ Microbiol. 1996;62:3572–3580. doi: 10.1128/aem.62.10.3572-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tison D L, Kelly M T. Virulence of Vibrio vulnificus strains from marine environments. Appl Environ Microbiol. 1986;51:1004–1006. doi: 10.1128/aem.51.5.1004-1006.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tison D L, Nishibuchi M, Greenwood J D, Seidler R J. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl Environ Microbiol. 1982;44:640–646. doi: 10.1128/aem.44.3.640-646.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright A C, Morris J G., Jr The extracellular cytolysin of Vibrio vulnificus: inactivation and relationship to virulence in mice. Infect Immun. 1991;59:192–197. doi: 10.1128/iai.59.1.192-197.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wright A C, Morris J G, Jr, Maneval D R, Jr, Richardson K, Kaper J B. Cloning of the cytotoxin-hemolysin gene of Vibrio vulnificus. Infect Immun. 1985;50:922–924. doi: 10.1128/iai.50.3.922-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wright A C, Simpson L M, Oliver J D. Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect Immun. 1981;34:503–507. doi: 10.1128/iai.34.2.503-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wright A C, Simpson L M, Oliver J D, Morris J G., Jr Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect Immun. 1990;58:1769–1773. doi: 10.1128/iai.58.6.1769-1773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto K, Wright A C, Kaper J B, Morris J G., Jr The cytolysin gene of Vibrio vulnificus: sequence and relationship to the Vibrio cholerae El Tor hemolysin gene. Infect Immun. 1990;58:2706–2709. doi: 10.1128/iai.58.8.2706-2709.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yim H H, Villarejo M. osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol. 1992;174:3637–3644. doi: 10.1128/jb.174.11.3637-3644.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoshida S, Ogawa M, Mizuguchi Y. Relation of capsular materials and colony opacity to virulence of Vibrio vulnificus. Infect Immun. 1985;47:446–451. doi: 10.1128/iai.47.2.446-451.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]