Abstract
A central mechanism of mTOR complex 1 (mTORC1) signaling is the coordinated translation of ribosomal protein and translation factor mRNAs mediated by the 5′-terminal oligopyrimidine motif (5′TOP). Recently, La-related protein 1 (LARP1) was proposed to be the specific regulator of 5′TOP mRNA translation downstream of mTORC1, while eIF4E-binding proteins (4EBP1/2) were suggested to have a general role in translational repression of all transcripts. Here, we use single-molecule translation site imaging of 5′TOP and canonical mRNAs to study the translation of single mRNAs in living cells. Our data reveal that 4EBP1/2 has a dominant role in repression of translation of both 5′TOP and canonical mRNAs during pharmacological inhibition of mTOR. In contrast, we find that LARP1 selectively protects 5′TOP mRNAs from degradation in a transcriptome-wide analysis of mRNA half-lives. Our results clarify the roles of 4EBP1/2 and LARP1 in regulating 5′TOP mRNAs and provide a framework to further study how these factors control cell growth during development and disease.
Single-molecule imaging reveals the roles of regulatory proteins in mTORC1-dependent translation of ribosomal protein mRNAs.
INTRODUCTION
For cellular homeostasis, ribosome biogenesis needs to be tightly coupled to nutrient availability. In eukaryotic cells, mechanistic target of rapamycin complex 1 (mTORC1) is the central signaling hub that integrates nutrient cues to match cell growth by stimulating or inhibiting ribosome biogenesis (1, 2). When nutrients are available, active mTORC1 promotes translation by the phosphorylation of key substrates such as ribosomal S6 kinases (S6K1/2) and eukaryotic translation initiation factor 4E-binding proteins (4EBP1/2) that stimulate eukaryotic initiation factor 4F (eIF4F) assembly and translation. When nutrients are limited, mTORC1 substrates are dephosphorylated, allowing 4EBP1/2 to bind and sequester the cap-binding eukaryotic translation initiation factor 4E (eIF4E), thereby inhibiting mRNA translation initiation. In addition, the La-related protein 1 (LARP1) has recently been described as a direct mTORC1 substrate and translational regulator (3–7). While mTORC1-dependent translation regulation acts on all mRNAs via multiple routes, it exerts a much more rapid and pronounced effect on ribosomal protein and translation factor mRNAs (~100 mRNAs) that carry a 5′-terminal oligopyrimidine motif (5′TOP, 4 to 15 pyrimidines) directly adjacent to the 5′ cap (8).
While it has been well established that the 5′TOP motif is both essential and sufficient for rapid mTORC1-mediated translational regulation (9, 10), the underlying molecular mechanism has been challenging to resolve (11). Both 4EBP1/2 and LARP1 have been found to contribute to 5′TOP translational inhibition, as loss of either factor partially relieved 5′TOP translational repression in cells acutely treated with the mTOR inhibitor Torin1 (4, 5, 12–14). Although binding of 4EBP1/2 to eIF4E reduces cap-dependent translation of all transcripts, eIF4E may have lower affinity for 5′TOP mRNAs, which could make them more sensitive to mTORC1 inhibition (15, 16). Recently, cocrystal structures of LARP1 bound to both the 5′ cap and the first five nucleotides of a 5′TOP oligo suggested that LARP1 could specifically repress 5′TOP mRNAs upon mTORC1 inhibition, leading to a model in which dephosphorylated LARP1 specifically binds the 5′ end of 5′TOP mRNAs to prevent assembly of the eIF4F complex (17).
An additional layer of ribosome biogenesis control is the pool of 5′TOP mRNAs available for translation, which are among the most highly expressed and stable transcripts in eukaryotic cells (18, 19). LARP1 has been found to associate with poly(A)-binding protein cytoplasmic 1 (PABPC1) and inhibit deadenylation of mRNA transcripts (20–23). It is, however, unclear how LARP1 is recruited to the mRNAs it stabilizes, the importance of the 5′TOP motif for target selection, and the relevance of mTORC1 activity in this process. Crosslinking studies have found LARP1 associates with thousands of mRNAs including 5′TOP mRNAs, and a subset of these transcripts have increased binding upon mTORC1 inhibition (7, 24, 25). In contrast, polyA tail sequencing in mTOR active cells has found LARP1 to inhibit mRNA deadenylation globally but that 5′TOP transcripts were among the most strongly affected transcripts upon LARP1 depletion (26).
In this study, we sought to clarify the roles of LARP1 and 4EBP1/2 in regulating the translation and stability of 5′TOP mRNAs. Direct measurements of translation of 5′TOP and non-5′TOP (canonical) mRNAs using single-molecule SunTag imaging revealed a dominant role of 4EBP1/2 in mediating 5′TOP translational repression. In contrast, we find a highly selective role of LARP1 in protecting 5′TOP mRNAs from degradation by measuring transcriptome-wide changes in mRNA half-lives using metabolic mRNA labeling (SLAMseq). Our study provides insights into the distinct roles of LARP1 and 4EBP1/2 in mediating 5′TOP regulation and a framework for further investigations into the mechanisms by which these factors regulate cell growth under normal physiological conditions and disease.
RESULTS
Single-molecule imaging of translation during mTOR inhibition
To study the regulation of translation during mTOR inhibition, we engineered a HeLa cell line that expresses fluorescent proteins for single-molecule imaging of mRNA [nuclear localized MS2 coat protein (MCP)-Halo] and translation [single-chain-variable fragment (scFv)-GFP], together with the reverse tetracycline-controlled transactivator to enable induction of reporter mRNAs (27). Into this cell line, we integrated two different constructs into a single genomic locus under the control of a doxycycline-inducible promoter. The reporter mRNAs were identical except for their 5′ untranslated region (5′UTR), where one contains the full-length 60S ribosomal protein L32 (RPL32) 5′UTR that begins with a 5′TOP motif, and the other has a canonical 5′UTR that does not contain a 5′TOP sequence. The coding sequence encodes 24 GCN4 epitope tags for translation site imaging [SunTag; (28)] followed by Renilla luciferase for bulk measurements of translation and the FK506 binding protein 1A (FKBP1A)–derived destabilization domain to reduce the accumulation of mature proteins (29). In addition, the 3′UTR contains 24 MS2 stem loops for mRNA imaging (Fig. 1A). The 5′-end of both reporter mRNAs was sequenced to determine the transcription start sites. All 5′TOP transcripts contained a 5′TOP motif and the canonical transcripts initiated with AGA, which is similar to the most common transcription start site (fig. S1) (30). To inhibit mTOR, we used the adenosine triphosphate (ATP)–competitive inhibitor Torin1, which has been widely used to study the translation of 5′TOP mRNAs. For both the 5′TOP and canonical mRNA reporter cell lines, Torin1 treatment for 1 hour resulted in inhibition of mTORC1 as seen by 4EBP1 dephosphorylation, consistent with previous results (fig. S2) (24, 31).
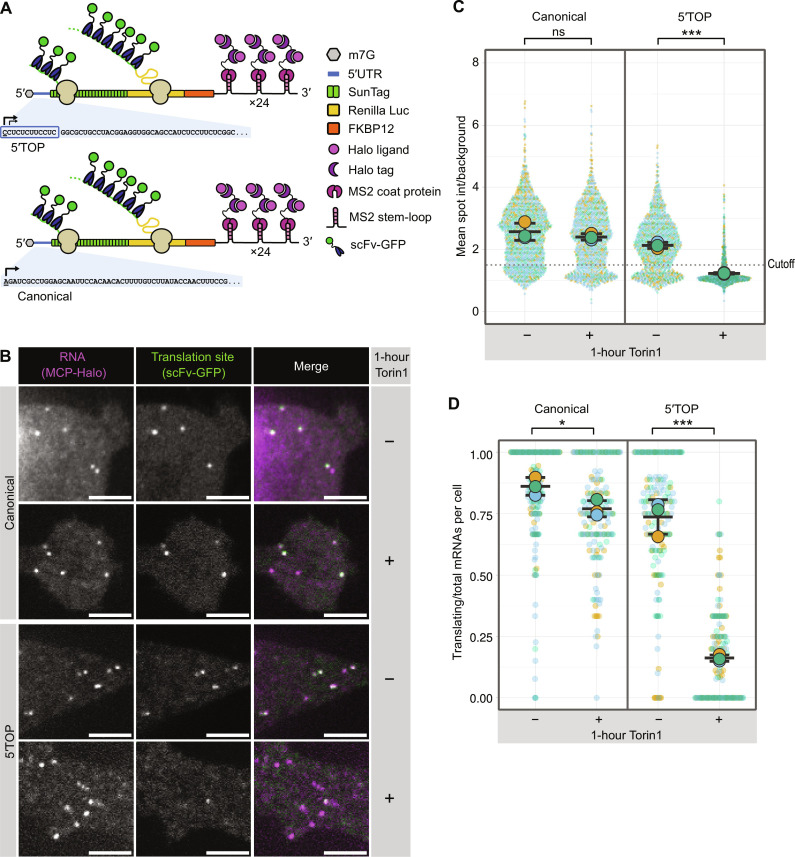
Fig. 1. Single-molecule imaging recapitulates 5′TOP translational repression.
(A) Schematic representation of reporter mRNAs for single-molecule imaging of translation. The 5′TOP reporter contains the full-length RPL32 5′UTR, whereas the canonical reporter has a control 5′UTR of similar length. Black arrows indicate transcription start sites. (B) Representative images of canonical and 5′TOP reporter mRNAs (MCP-Halo foci, magenta) undergoing translation (scFv-GFP foci, green) in the absence or presence of mTOR inhibitor Torin 1 (250 nM, 1 hour). Scale bars, 5 μm. (C) Translation site intensities of canonical and 5′TOP reporter mRNAs quantified in absence or presence of Torin 1 (250 nM, 1 hour). SunTag intensities are plotted for all mRNAs (colored circles) overlaid with the mean ± SD (≥1089 mRNAs per condition, n = 3). (D) Fraction of mRNAs undergoing translation quantified per cell for canonical and 5′TOP reporter in the absence or presence of Torin 1 (250 nM, 1 hour). Values are plotted for each cell (colored circles) overlaid with the mean ± SD (≥162 cells per condition, n = 3). For statistics, unpaired t tests were performed, with statistical significance claimed when P < 0.05 (ns, not significant; *P < 0.05; ***P < 0.001).
To observe the effect of mTORC1 inhibition on translation, we induced expression of the reporter mRNAs in both cell lines and imaged them in either the presence or absence of Torin1. Treatment with Torin1 was found to strongly repress translation of most 5′TOP transcripts as seen by the disappearance of scFv-GFP spots that colocalized with mRNA spots, whereas the canonical mRNAs were largely unaffected (Fig. 1B). For quantification of single mRNAs and their translation, we used a high-throughput image analysis pipeline that tracks individual mRNAs and measures the corresponding SunTag intensities. mRNA trajectories were determined using single-particle tracking of MCP-Halo spots, and scFv-GFP intensities at those same coordinates were quantified for each mRNA as background-corrected mean spot intensity (SunTag intensity). Using this analysis pipeline, we quantified the translation of >1000 mRNAs for both the 5′TOP and canonical mRNA cell lines (Fig. 1C), which revealed a broad distribution of SunTag intensities for both types of transcripts indicating a heterogeneity of ribosomes engaged in translation of individual transcripts (27, 28). The average SunTag intensity for the 5′TOP mRNAs was slightly lower compared to the canonical mRNAs indicating fewer ribosomes engaged in translation when mTOR is active (Fig. 1C). The mean SunTag intensity for the 5′TOP mRNAs decreased markedly upon Torin1 treatment, whereas the mean SunTag intensity of the canonical mRNAs decreased only slightly.
While changes in SunTag intensity indicate differences in ribosome number, translation site imaging can also be used to quantify the fraction of transcripts actively translating within a cell. Puromycin treatment, which inhibits translation due to premature termination, was used to measure SunTag spot intensities in the absence of translation to calibrate a threshold for identifying translating mRNAs (>1.5-fold over background; fig. S3). Quantifying translation as the fraction of translating mRNAs per cell revealed slightly fewer translating 5′TOP mRNAs (mean: 74%) compared to the canonical mRNAs (mean: 86%) when mTOR is active (Fig. 1D). Upon 1-hour Torin1 treatment, the fraction of translating 5′TOP mRNAs per cell decreased drastically (mean: 16%), although many cells retained a minor fraction of translating 5′TOP mRNAs. In contrast, the fraction of translating canonical mRNAs decreased only slightly upon Torin1 treatment (mean: 77%). To determine whether the remaining fraction of translating 5′TOP mRNAs after 1-hour Torin1 treatment represented stalled ribosomes, Torin1-treated cells were cotreated with harringtonine, which stalls ribosome at the start codon and allows elongating ribosomes to run-off. Addition of harringtonine abolished the remaining translation sites in the Torin1-treated 5′TOP cell line within 10 min (fig. S4), demonstrating that the low number of 5′TOP mRNAs that colocalize with SunTag signal are still actively translating.
To verify our findings with other mTOR inhibitors, we repeated the imaging of canonical and 5′TOP mRNAs with the allosteric mTOR inhibitor Rapamycin (32) and the ATP-competitive mTOR inhibitors PP242 and TAK228 (33, 34). While PP242 and TAK228 treatment closely mirror the response seen for Torin1 treatment, Rapamycin treatment did not significantly alter the translation of either canonical or 5′TOP mRNAs (fig. S5). Rapamycin insensitivity has been described for a number of cell lines including HeLa (35), and, in agreement with previous studies (31, 36), we find that Rapamycin selectively inhibits S6K1 phosphorylation, while levels of phosphorylated 4EBP1 remain high (fig. S6). Together, our data capture both inter- and intracellular variability in the translation of canonical and 5′TOP mRNAs in the presence and absence of mTOR inhibitors, providing direct translation measurements independent of effects arising from transcriptional regulation or mRNA stability.
LARP1 KO partially rescues translation of 5′TOP mRNAs during Torin1 treatment
Recently, LARP1 has been found to specifically bind the 5′TOP motif in an mTOR-dependent manner to regulate translation (3, 17, 24). To further investigate the role of LARP1 in translational repression of 5′TOP mRNAs during mTOR inhibition, we generated LARP1 CRISPR-Cas9 knockouts (KOs) in the 5′TOP and canonical mRNA cell lines. Genomic DNA sequencing confirmed frameshift mutations in all alleles of LARP1 exon 4 that are upstream of any domain of known function (amino acids 205 to 240) (fig. S7, A and B). Loss of LARP1 protein in the KO cell lines was confirmed by Western blot analysis using two LARP1 antibodies targeting either the N- or C-terminal regions, which did not detect alternative LARP1 isoforms (fig. S7C). Loss of LARP1 did not disrupt the regulation of other mTORC1 targets, as seen by dephosphorylation of 4EBP1, S6K1, and RPS6 upon 1-hour Torin1 treatment (fig. S7D). Consistent with earlier reports in human embryonic kidney (HEK) cells, deletion of LARP1 in HeLa cells resulted in decreased cell proliferation (4, 5).
Following the validation of the LARP1 KO cell lines, we quantified the translation of 5′TOP and canonical mRNAs (>600 mRNAs per condition) in the absence of LARP1 (fig. S8A). Analysis of SunTag intensities of the canonical and 5′TOP mRNAs revealed similar translation levels in the LARP1 KO compared to wild type (WT), indicating that LARP1 does not regulate 5′TOP mRNA translation in cells when mTOR is active. Upon 1-hour Torin1 treatment, canonical mRNAs decreased slightly in mean SunTag intensity, whereas the 5′TOP mRNAs decreased more strongly (Fig. 2A). Calculating the fraction of translating mRNAs per cell revealed that the canonical mRNAs show a mild response to Torin1 in the absence of LARP1 (mean untreated: 87%, mean 1-hour Torin1: 75%; Fig. 2B), mirroring the response observed for the canonical mRNAs in LARP1 WT cells. The 5′TOP mRNAs in LARP1 KO cells displayed a partial rescue of translation upon Torin1 treatment (mean untreated: 79%, mean 1-hour Torin1: 41%) compared to LARP1 WT cells (mean 1-hour Torin1: 16%). The incomplete rescue of 5′TOP mRNA translation in the absence of LARP1 suggested the existence of additional trans-acting factors in mediating 5′TOP translational repression.
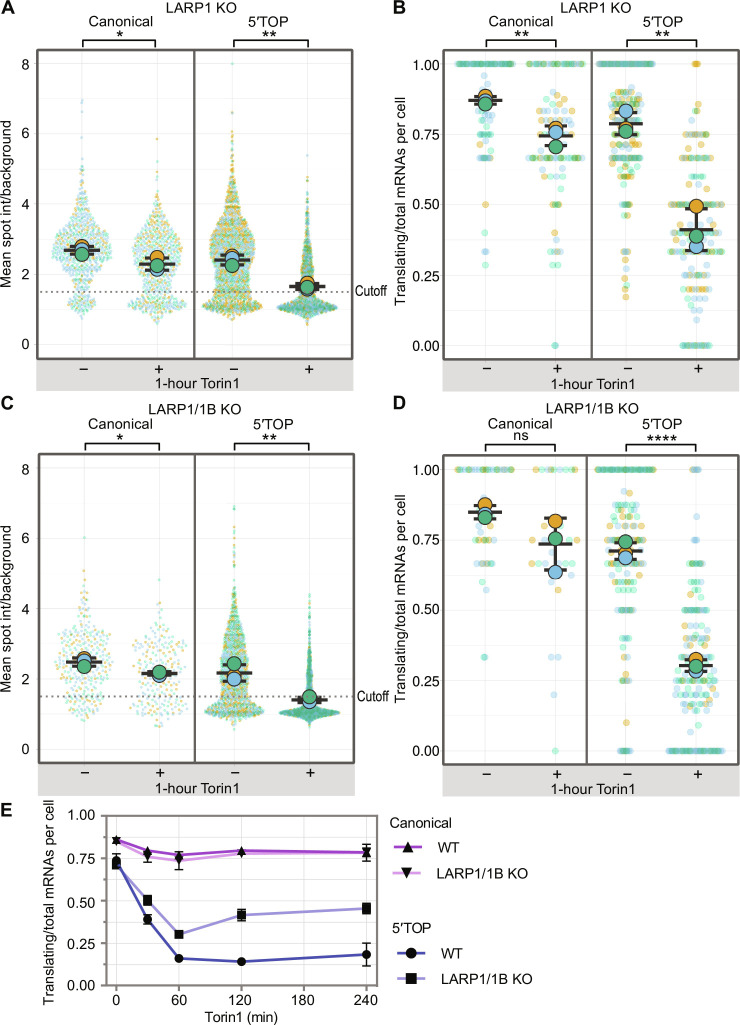
Fig. 2. Loss of LARP1 partially alleviates 5′TOP translational repression during Torin1 treatment.
(A) Quantification of translation site intensities in LARP1 KO cells ± Torin1 (250 nM, 1 hour). SunTag intensities are plotted for all mRNAs overlaid with the mean ± SD (≥652 mRNAs per condition, n = 3). (B) Fraction of mRNAs undergoing translation quantified per cell in LARP1 KO cells ± Torin1 (250 nM, 1 hour). Values are plotted for each cell (colored circles) overlaid with the mean ± SD (≥91 cells per condition, n = 3). (C) Quantification of translation site intensities in LARP1/1B KO cells ± Torin1 (250 nM, 1 hour). SunTag intensities are plotted for all mRNAs (colored circles) overlaid with the mean ± SD (≥218 mRNAs per condition, n = 3). (D) Fraction of mRNAs undergoing translation quantified per cell in LARP1/1B KO cells ± Torin1 (250 nM, 1 hour). Values are plotted for each cell (colored circles) overlaid with the mean ± SD (≥30 cells per condition, n = 3). For statistics, unpaired t tests were performed, with statistical significance claimed when P < 0.05 (*P < 0.05, **P < 0.01, ****P < 0.0001). (E) Time course of fraction of translating mRNAs per cell for canonical and 5′TOP reporter cell lines in the presence (WT) or absence of LARP1/1B (KO). Cells were treated with 0-, 30-, 60-, 120-, and 240-min Torin1. Values are plotted as the mean ± SEM (≥30 cells per condition, n = 3).
One possible trans-acting factor that could repress 5′TOP mRNAs in the absence of LARP1 is the homolog LARP1B (also called LARP2), which shares the DM15 domain that binds the 5′TOP motif, although it is lowly expressed in HeLa cells. To test this possibility, we used CRISPR-Cas9 to generate KOs of LARP1B in the LARP1 KO background (fig. S9A). Genomic DNA sequencing of the edited alleles identified frameshift mutations in all alleles of LARP1B exon 4, and polymerase chain reaction (PCR) confirmed loss of WT LARP1B mRNA (fig. S9, B and C). Western blot analysis confirmed unperturbed mTORC1 signaling in the LARP1/1B KO cells, as seen by dephosphorylation of 4EBP1, S6K1, and RPS6 upon 1-hour Torin1 treatment (fig. S9D).
Following the validation of the LARP1/1B KO cell lines, we quantified the translation of 5′TOP and canonical mRNAs (>200 mRNAs per condition) in the absence of LARP1/1B (fig. S8B). Both the distribution of SunTag intensities (Fig. 2C) and the fraction of translating canonical and 5′TOP mRNAs per cell (Fig. 2D) responded similarly to Torin1 treatment (1 hour) as obtained for LARP1 KO cells, suggesting that LARP1B does not affect 5′TOP translational regulation. These results are consistent with an earlier study in HEK cells, which also found that translational repression of endogenous 5′TOP transcripts could not be rescued further by combinatory deletion of LARP1/1B (37), arguing against functional redundancy of LARP1 and LARP1B.
While we did not observe a rescue of 5′TOP translation when cells were treated with Torin1 for 1 hour, we could not exclude the possibility that the effect of loss of LARP1/1B might be more pronounced at other time points. To characterize the kinetics of Torin1-mediated translational inhibition, we performed SunTag imaging of the canonical and 5′TOP cell lines at additional time points (30 min, 2 hours, and 4 hours; Fig. 2E). For both WT and LARP1/1B KO cells, the canonical mRNAs showed a gradual decrease in translation during the first hour of Torin1 treatment that remained low at later time points. To test whether prolonged mTOR inhibition is required to repress canonical mRNA translation, we quantified translation of canonical mRNAs in WT cells treated with Torin1 for 24 hours. The majority of canonical mRNAs remained translating at this longer time point (fig. S10). 5′TOP mRNAs in WT cells decreased in translation within 1 hour of Torin1 to a minor fraction of translating mRNAs (30-min Torin1: 39%, 1-hour Torin1: 16%) and remained at this level at the 2-hour (14%) and 4-hour (19%) time points (Fig. 2E). In LARP1/1B KO cells, translation of 5′TOP mRNAs also decreased with no change in the timing of repression but a decrease in its extent (30-min Torin1: 50%, 1-hour Torin1: 30%); however, at the 2-hour (42%) and 4-hour (46%) time points, we observed a slight increase in translation. These results suggested that while LARP1 may contribute to translational repression of 5′TOP mRNAs, it is not the dominant regulatory factor during mTOR inhibition.
4EBP1/2 knockdown rescues translation of 5′TOP mRNAs during Torin1 treatment
4EBP1/2 are thought to generally repress translation during mTORC1 inhibition but have also been previously implicated in specifically affecting 5′TOP mRNAs (12). Using lentiviral infection, stable short hairpin RNA (shRNA)–mediated knockdown (KD) cell lines were generated in the WT and LARP1/1B KD background for both 5′TOP and canonical mRNA cell lines. Western blot analysis confirmed the depletion of 4EBP1/2 levels in all four cell lines (fig. S11, A and B). Furthermore, the dephosphorylation of RPS6 and residual 4EBP1 upon 1-hour Torin1 indicated that mTORC1 signaling was unperturbed in the 4EBP1/2 KD cell lines (fig. S11, C and D).
Having validated the 4EBP1/2 KD cell lines, we measured the translation of canonical and 5′TOP mRNAs in the absence of 4EBP1/2 (fig. S12). In untreated cells, the reduction of 4EBP1/2 resulted in increased SunTag intensities for both canonical and 5′TOP mRNAs in cell lines with WT LARP1 (Fig. 3A, >1000 mRNAs per condition) and LARP1/1B KO (Fig. 3C, >700 mRNAs per condition) compared to cells with WT levels of 4EBP1/2 (Fig. 1C). This suggested that when mTOR is active, 4EBP1/2 can still weakly repress translation initiation presumably through fluctuations in mTOR signaling during cell growth. Unexpectedly, the SunTag intensities of both canonical and 5′TOP mRNAs were not reduced upon 1-hour Torin1 treatment in 4EBP1/2 KD cell lines with WT LARP1 (Fig. 3A) or LARP1/1B KO (Fig. 3C). Analyzing the fraction of translating canonical mRNAs revealed no change in translation upon Torin1 treatment for cells with WT LARP1 (Fig. 3B) and LARP1/1B KO (Fig. 3D), in contrast to the previously observed mild decrease in translation of canonical mRNAs upon 1-hour Torin1 (Fig. 1D). The fraction of translating 5′TOP mRNAs was not significantly reduced by Torin1 treatment in 4EBP1/2 KD cell lines with WT LARP1 (Fig. 3B; untreated: 77%, treated: 70%) or LARP1/1B KO (Fig. 3D; untreated: 79%, treated: 77%), indicating a full rescue of translation compared to the previous partial rescue observed in LARP1 KO cells. To exclude the possibility that translational repression is delayed in the absence of 4EBP1/2, we investigated the kinetics of mTOR inhibition in the 4EBP1/2 KD cell lines (Fig. 3E), which revealed that canonical and 5′TOP mRNAs remain similarly insensitive to Torin1 treatment at prolonged Torin1 treatment (2 and 4 hours). These experiments indicate that despite the difference in magnitude of translational repression during Torin1 treatment, 4EBP1/2 is responsible for the weak inhibition of canonical mRNAs and the stronger inhibition of 5′TOP mRNAs. Our data support a model where 5′TOP mRNAs are intrinsically more sensitive to 4EBP1/2-mediated translational regulation, which results in a minor difference in translation when mTOR is active and a pronounced difference in translation when mTOR is inhibited.
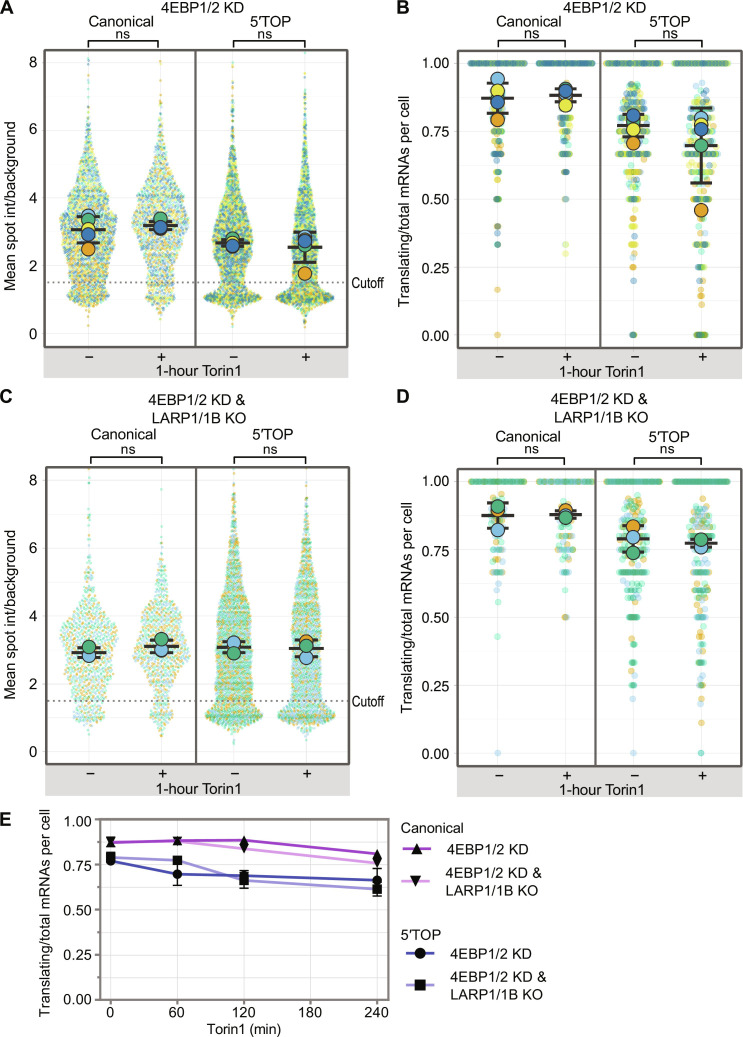
Fig. 3. Loss of 4EBP1/2 is sufficient to alleviate 5′TOP translational repression during Torin1 treatment.
(A) Quantification of translation site intensities of reporter mRNAs in 4EBP1/2 KD cells ± Torin1 (250 nM, 1 hour). SunTag intensites are plotted for all mRNAs (colored circles) overlaid with the mean ± SD (≥1344 mRNAs per condition, n = 5). (B) Fraction of mRNAs undergoing translation quantified per cell for reporter mRNAs in 4EBP1/2 KD cells ± Torin1 (250 nM, 1 hour). Values are plotted for each cell (colored circles) overlaid with the mean ± SD (≥204 cells per condition, n = 5). (C) Quantification of translation site intensities of reporter mRNAs in 4EBP1/2 KD_LARP1/1B KO cells ±1 hour Torin1 (250 nM). SunTag intensities are plotted for all mRNAs overlaid with the mean ± SD (≥783 mRNAs per condition, n = 3). (D) Fraction of mRNAs undergoing translation quantified per cell of reporter mRNAs in 4EBP1/2 KD_LARP1/1B KO cells ± Torin1 (250 nM, 1 hour). Values are plotted for each cell (colored circles) overlaid with the mean ± SD (≥97 cells per condition, n = 3). For statistics, unpaired t tests were performed, with statistical significance claimed when P < 0.05. (E) Time course of fraction of translating mRNAs per cell for canonical and 5′TOP reporters in 4EBP1/2 KD cells with WT LARP1 or with LARP1/1B KO. Cells were treated with 0-, 30-, 60-, 120-, and 240-min Torin1. Values are plotted as the mean ± SEM (≥97 cells per condition, n = 3 to 5).
Alternatively, our data could potentially be explained by the presence of additional cis-acting sequence elements within the RPL32 5′UTR of our 5′TOP mRNA reporter that were absent in the 5′UTR of the canonical mRNAs. The RPL32 5′UTR is 50 nucleotides in length and contains the 5′TOP motif (positions +1 to +11) as well as a pyrimidine-rich translational element (PRTE) at positions +38 to +47 (fig. S13A). A PRTE is found in the 5′UTRs of the majority of 5′TOP mRNAs and has been proposed to also be an alternative binding site for LARP1, although its contribution to translational regulation during mTOR inhibition remains largely unknown (7). To further dissect the contribution of the 5′TOP and PRTE motifs, we generated two additional live-cell imaging cell lines carrying a single-copy genomic integration of modified RPL32 5′UTR reporters, one where only the 5′TOP motif was mutated (Δ5′TOP) and one where the 5′TOP and the PRTE motifs were mutated (Δ5′TOP/PRTE; fig. S13A). We confirmed the sequence of the 5′UTRs in the Δ5′TOP and Δ5′TOP/PRTE mRNAs by 5′-end sequencing and imaged their translation in the absence or presence of Torin1. We found that both the Δ5′TOP and Δ5′TOP/PRTE mRNAs responded only weakly to Torin1 treatment, with a similar decrease in the fraction of translating mRNAs per cell as observed for the canonical mRNAs (fig. S13, B, C, and H), which is consistent with previous reports that the 5′TOP motif is both necessary and sufficient for the selective translational repression upon mTOR inhibition (9, 10).
Next, we generated both CRISPR-Cas9 LARP1 KO and shRNA-mediated 4EBP1/2 KD cell lines carrying the Δ5′TOP mRNAs and validated the loss of the respective protein and unperturbed mTORC1 signaling by Western blot (fig. S14, A and B). Similar to our previous experiments with canonical mRNAs (Figs. 2B and 3B), we found that loss of LARP1 did not alleviate the mild translational repression of Δ5′TOP mRNAs upon Torin1 treatment (fig. S13, D and E) and that depletion of 4EBP1/2 fully alleviated Torin1-mediated translational repression (fig. S13, F to H).
LARP1 KO results in global decreased mRNA stability of 5′TOP mRNAs
In addition to its role in 5′TOP translational repression, LARP1 has been reported to protect mRNAs from degradation (20, 21, 26, 38, 39). It is now unclear whether this protective role of LARP1 is restricted to 5′TOP mRNAs, TOP-like mRNAs, or affects all mRNAs (37). To study the effect of LARP1 and 4EBP1/2 loss on global gene expression in growing cells when mTOR is active, we extracted total RNA from our cell lines and performed RNA sequencing (RNA-seq). The canonical and 5′TOP mRNA cell lines of the same genotype were sequenced together as biological replicates since expression of different reporter mRNAs should not have a global effect on gene expression and combining the independently generated cell lines reduces potential off-target effects caused by either CRISPR KO or shRNA KD.
To determine the effect of LARP1 loss on gene expression, we compared the transcriptome of LARP1 KO and WT cells (12,403 transcripts, counts per million (CPM) > 1, pseudogenes excluded). As expected, LARP1 transcript levels were down-regulated in the KO cell lines to 30% of WT levels (table S1). Volcano plot analysis of the transcriptome changes of KO versus WT cell lines (biological replicates: n = 2 for LARP1 WT, n = 8 for LARP1 KO) revealed that the most significantly affected mRNAs are endogenous 5′TOP mRNAs, which are almost all down-regulated in the LARP1 KO cells (Fig. 4A, blue circles). Analyzing all known 5′TOP mRNAs (table S2), 70 of 94 5′TOPs are found to be significantly down-regulated [log2 fold change (FC) ≤ −0.5, −log10 P value ≥ 5], as well as 85 significantly down-regulated non-5′TOP RNAs and 40 significantly up-regulated non-5′TOP RNAs (orange circles). In contrast, previously identified TOP-like mRNAs, which were predicted to be translationally regulated by LARP1 based on sequence similarity (37), were mostly unaffected in their expression.
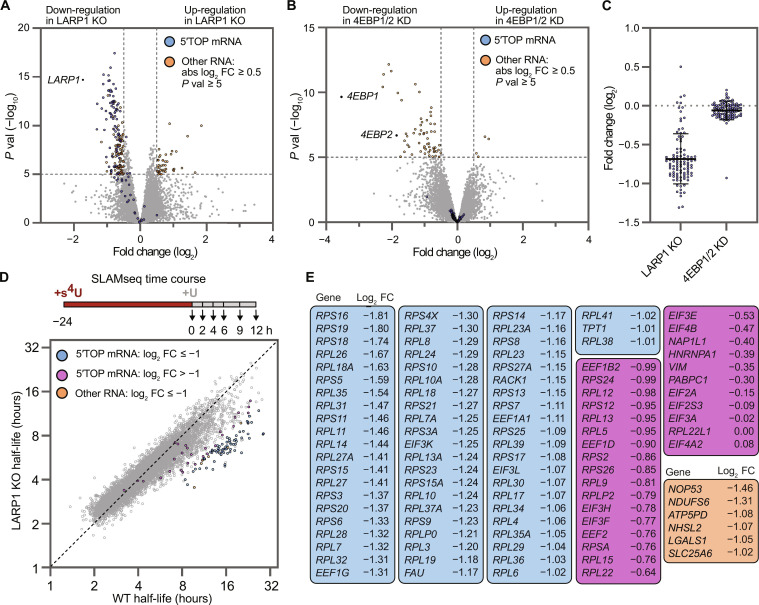
Fig. 4. Loss of LARP1 results in selective destabilization of 5′TOP mRNAs.
(A) Differential gene expression analysis of LARP1 KO cells compared to WT cell lines. Significantly down- and up-regulated mRNAs are highlighted (−log10 P value ≥5 and absolute log2 FC ≥ 0.5) indicating all classical 5′TOP mRNAs (blue circles) as well as all significant non-5′TOP mRNAs (orange circles). For WT, data are the average of n = 2 biological replicates (canonical and 5′TOP reporter cell lines, averaged). For LARP1 KO, data are the average of n = 8 biological replicates (four KO clones of each reporter cell line). (B) Differential gene expression analysis of 4EBP1/2 KD reporter cell lines compared to WT reporter cell lines. Significantly down- and up-regulated mRNAs are highlighted (orange circles, −log10 P value ≥5 and absolute log2 FC ≥ 0.5). 5′TOP mRNAs (blue) are unaffected by 4EBP1/2 KD. Data shown for n = 2 biological replicates (canonical and 5′TOP reporter cell lines, averaged). (C) FC of 5′TOP mRNAs shown in (A) and (B). Mean FC (log2) in LARP1 KO cells = −0.68, in 4EBP1/2 KD cells = −0.06. (D) Correlation plot of mRNA half-lives in LARP1 KO compared to WT reporter cells. Experimental setup of global mRNA stability analysis using s4U labeling (SLAM-seq) shown on top, with 24 hours s4U labeling, washout, and time point collection. RNAs with significantly decreased stability (log2 FC ≤ −1) are highlighted for 5′TOP mRNAs (blue circles) and non-5′TOP mRNAs (orange circles), as well as nonsignificantly down-regulated 5′TOP mRNAs (magenta circles). (E) List of all RNAs highlighted in (D), ranked by log2 FC for 5′TOP mRNAs and non-5′TOP mRNAs separately.
To determine whether depletion of 4EBP1/2 also affected the levels of 5′TOP transcripts, we compared the transcriptome of 4EBP1/2 KD cells to WT cells (13,832 transcripts, CPM > 1, pseudogenes excluded). Consistent with shRNA KD of 4EBP1/2, we found the levels of these two transcripts to be reduced by 91% (4EBP1) and 71% (4EBP2) and that LARP1 expression was unaltered in both cell lines. A small number of transcripts showed significantly altered expression; however, these do not include known 5′TOP mRNAs (65 transcripts; Fig. 4B, orange circles, absolute log2 FC ≥ 0.5, −log 10 P value ≥5, table S3). In addition, the altered mRNAs did not match mRNAs described to be sensitive to eIF4E levels in mice (40). In contrast to the dominant role of 4EBP1/2 in regulating translation during mTOR inhibition, these results indicate that LARP1 regulates levels of 5′TOP transcripts when mTOR is active (Fig. 4C). To validate our RNA-seq results with an orthogonal approach, we performed single-molecule fluorescence in situ hybridization (smFISH) on three endogenous 5′TOP transcripts (RPL5, RPL11, and RPL32) and a non-5′TOP control (MYC), which confirmed the selective decrease of 5′TOP mRNAs in the LARP1 KO cell lines, with no change of 5′TOP mRNAs in the 4EBP1/2 KD cell lines (fig. S15).
The selective down-regulation of endogenous 5′TOP mRNAs we observed in the absence of LARP1 suggested that LARP1 specifically stabilizes 5′TOP mRNAs. To confirm that the changes in steady-state expression were caused by mRNA destabilization and were not due to changes in transcription, we performed global mRNA half-life measurements using metabolic 4-thiouridine labeling [SLAM-seq (19)]. WT and LARP1 KO cells of both canonical and 5′TOP cell lines were incubated with 4-thiouridine for 24 hours, followed by washout and harvesting of cells over a 12-hour time course. Half-lives of mRNAs were calculated using a single-exponential decay fit for 9837 transcripts (R2 ≥ 0.75, pseudogenes excluded, table S4). In agreement with previous measurements of mammalian mRNA half-lives, the global median half-life for both WT and LARP1 KO HeLa cell lines was ~4 hours (fig. S16A), indicating that loss of LARP1 does not globally destabilize all mRNAs. Correlation analyses showed a high correlation in mRNA half-lives among the four cell lines (r = 0.90 to 0.94), allowing us to compare the mRNA half-lives in WT versus LARP1 KO cell lines (Fig. 4D, n = 2). In agreement with our RNA-seq results, nearly all 94 5′TOP mRNAs detected in the SLAM-seq experiment have decreased mRNA stability, with 66 5′TOP mRNAs changing by >2-fold (Fig. 4D). A few 5′TOP mRNAs seem largely unaffected by LARP1 loss (including PABPC1), suggesting the potential involvement of additional stabilizing factors for these transcripts. Furthermore, the length of the 5′TOP motif or presence of a PRTE motif within the 5′UTR does not correlate with the change in mRNA half-lives (fig. S16B). Only six non-5′TOP mRNAs were found to be destabilized >2-fold, and three of these transcripts (NOP53, LGALS1, and SLC25A6) have annotated transcription start sites that contain 5′ TOP motifs, suggesting that they could be similarly regulated in HeLa cells. Taken as a whole, our results support a model of LARP1-mediated stabilization that is highly selective for 5′TOP mRNAs.
DISCUSSION
In this study, we used single-molecule imaging to study the regulation of translation of 5′TOP mRNAs upon mTOR inhibition that allowed us to directly quantify the effect of LARP1 and 4EBP1/2. By imaging and quantifying the translation status of individual mRNAs, we find that 4EBP1/2 plays a dominant role compared to LARP1 in mediating 5′TOP translational repression during short-term (30 min to 4 hours) pharmacological inhibition of mTOR in HeLa cells. Previously, studies that used genome-wide ribosome or polysome profiling determined that LARP1 and 4EBP1/2 regulate 5′TOP translation during mTOR inhibition (3, 4, 12, 13, 37, 41, 42); however, the magnitude of their respective contributions was difficult to measure due to the inherent limitations of these approaches. We believe that this highlights the power of single-molecule imaging methods for quantifying translation in living cells to determine the specific effects of translation factors.
While our results indicate that 4EBP1/2 is the critical factor in mediating 5′TOP translational repression, the underlying molecular mechanism is not entirely clear. Although we cannot exclude the possibility of a still unknown factor acting downstream of 4EBP1/2, we favor a model where the translation of 5′TOP mRNAs is more sensitive to active eIF4E levels. In vitro experiments have determined that eIF4E binds with ~3-fold weaker affinity to m7GTP-capped oligonucleotides with a +1 cytosine than either purine, which is consistent with translation of 5′TOP mRNAs being slightly lower than a non-5′TOP mRNA when mTOR is active and then preferentially repressed when available eIF4E levels become extremely limited during mTOR inhibition (16, 43, 44). This model is also in line with previous work that found inducible overexpression of eIF4E to specifically up-regulate the translation of 5′TOP mRNAs (45), as well as recent work that showed that 5′TOP mRNAs are less sensitive to mTOR inhibition in acutely PABPC1-depleted cells where global mRNA levels are reduced (46).
In the x-ray structure of human eIF4E in complex with m7pppA, the C-terminal tail of eIF4E adopts a conformation that enables Thr205 to form a hydrogen bond with the exocyclic amine of the adenine base (47). While the position of the eIF4E C-terminal tail has not been determined when bound to longer RNA sequences, phosphorylation of Ser209 is known to enhance translation, indicating that additional residues in this region may have functional roles (48). Alternatively, other canonical translation factors (e.g., eIF4G or 4EBP1/2) may also contribute to 5′TOP specificity through additional interactions (49, 50).
While LARP1 may not be the key repressor in 5′TOP translational regulation, our data support a major role of LARP1 in mediating 5′TOP mRNA stability when mTOR is active. Previous work established a link between LARP1 and mRNA stability, with LARP1 binding both PABP and the polyA tail and inhibiting deadenylation (20–22, 24, 38, 39, 51, 52). It has been unclear whether this protective role is restricted to 5′TOP mRNAs as binding to PABP/polyA is anticipated to not be selective, and crosslinking studies have found LARP1 complexed with thousands of mRNAs (7, 24, 25). Our results show a highly selective destabilization of nearly all 5′TOP mRNAs upon loss of LARP1, with virtually all other mRNAs being largely unaffected. Similarly, a recent study found that loss of LARP1 resulted in rapid deadenylation of short polyA tails of all mRNAs, with 5′TOP mRNAs being more affected than other types of mRNAs (26). It is possible that differences in LARP1 depletion or measurement of mRNA stability or polyA-tail length may account for the differences in specificity for 5′TOP mRNAs between the studies.
Previous studies have focused on the role of LARP1 in protecting 5′TOP mRNAs in mTOR inhibited cells, as LARP1 has been shown to be recruited to 5′TOP mRNAs upon mTOR inhibition (24, 51). Our findings raise the intriguing question of how LARP1 can be specifically recruited to 5′TOP mRNAs when mTOR is active. While it has been proposed that LARP1 can interact with its La motif with both the 5′TOP motif and PABP (39), it is not clear that this interaction is compatible with eIF4F binding and translation initiation. Recent structural work of the human 48S preinitiation complex suggests that there could be a “blind spot” of ~30 nucleotides adjacent to the cap that might allow LARP1 to bind the 5′TOP sequence without blocking initiation, although this model requires biochemical and structural characterization (53). We do not observe any correlation between change in mRNA half-lives with either length of 5′TOP motif or the presence of a PRTE in the 5′UTR, suggesting that the position of the pyrimidines directly adjacent to the cap is necessary for this effect on mRNA stability. LARP1 was shown to promote the localization of ribosomal mRNAs in a PRTE-dependent manner but did not require the stricter 5′ TOP motif, suggesting that LARP1’s interaction with ribosomal mRNAs and its functional consequence could be context dependent (54).
While our translation site imaging experiments are limited to the characterization of four reporter mRNAs (canonical, 5′TOP, Δ5′TOP, and Δ5′TOP/PRTE), we have shown that the results are consistent with previous studies that characterized endogenous ribosomal protein mRNAs; however, single-molecule experiments in living cells allow more accurate quantification of the effect of loss of LARP1 and 4EBP1/2. We anticipate that similar results would be obtained with 5′TOP sequences derived from ribosomal protein mRNAs other than RPL32, although the magnitude of the difference in translational repression could be different if compared to another non-5′TOP transcript. In addition, the continued development of methodologies for imaging translation of single mRNAs for extended time periods and the interplay of translation with mRNA decay will enable the dynamics of mTOR regulation to be quantified in greater detail (55, 56).
MATERIALS AND METHODS
Materials
All antibodies used in this study are listed in Table 1. All chemicals, plasmids, viruses, cell lines, single guide RNA (sgRNA), and shRNA used in this study are listed in table S5.
Table 1. Antibodies.
IgG, immunoglobulin G.
| Primary antibodies | Provider | Catalog no. |
|---|---|---|
| LARP1 | Bethyl Labs | A302-087A |
| LARP1 | Cell Signaling Technology | 70180 |
| TUBA1B | Cell Signaling Technology | 3873 |
| MTOR | Cell Signaling Technology | 2983 |
| Pho-RPS6 (Ser235/236) | Cell Signaling Technology | 2211 |
| Pho-RPS6 (Ser235/236) | Cell Signaling Technology | 4856 |
| Pho-4EBP1 (Ser65) | Cell Signaling Technology | 9451 |
| 4EBP1 | Cell Signaling Technology | 9452 |
| 4EBP2 | Sigma-Aldrich | MABS1865 |
| Pho-S6K1 (Thr389) | Cell Signaling Technology | 9234 |
| Secondary antibodies | ||
| IRDye 680RD Goat anti-Mouse IgG | LI-COR | 926-68070 |
| IRDye 800CW Goat anti-Rabbit IgG | LI-COR | 926-32211 |
Cell culture
The HeLa-11ht cell lines expressing either RPL32 5′TOP SunTag or non-5′TOP canonical SunTag mRNAs used in this study were previously generated in the Chao lab (27), and the corresponding plasmids are available from Addgene (#119946 and #119945). The reporter cell lines were grown in 10% fetal calf serum–Dulbecco’s modified Eagle’s medium (FCS-DMEM) medium containing glucose (4.5 g/liter), penicillin and streptomycin (100 μg/ml), 4 mM l-glutamine, and 10% fetal bovine serum (FBS) at 37°C and 5% CO2. To maintain the reverse tetracycline-controlled transactivator (rtTA2-M2) for inducible expression, the medium was supplemented with G418 (0.2 mg/ml). HEK293T cells used for lentivirus production were grown in 10% FCS-DMEM medium containing glucose (4.5 g/liter), penicillin and streptomycin (100 μg/ml), 4 mM l-glutamine, and 10% FBS at 37°C and 5% CO2.
Validation of transcription start sites for 5′TOP and canonical SunTag transcripts
For mapping of transcription start sites, total RNA was converted into full-length adapter-ligated double-stranded cDNA using the TeloPrime Full-Length cDNA Amplification Kit V2 (Lexogen), which uses a cap-specific adapter selective for intact mRNAs. cDNA 5′-terminal sequences were amplified by PCR using a gene-specific primer and TeloPrime forward primer, cloned into the pCR-Blunt vector (Thermo Fisher Scientific), and sequenced.
CRISPR KO cell line generation
To generate LARP1 CRISPR KO clones, parental HeLa-11ht cell lines expressing the reporter mRNAs were transiently cotransfected with two Cas9 plasmids, each containing Cas9 and a sgRNA targeting a sequence within exon 4 of LARP1, enhancing efficiency of KO cell line generation (57, 58). Transient transfections were performed following the manufacturer’s instructions using Lipofectamine 2000 (Invitrogen) and Opti-MEM (Thermo Fisher Scientific). Two days after transient transfections, highly transfected cells were single-cell–sorted into 96-well plates for monoclonal selection (10% highest mCherry-positive cells, using Cas9-T2A-mCherry). Single clonal cell populations were screened for loss of LARP1 by immunostaining (Bethyl Labs, #A302-087A) in 96-well plates. For both 5′TOP and canonical SunTag cell lines, four KO clones each were verified by Western blot for loss of LARP1 protein expression.
To generate LARP1B CRISPR KO clones, the LARP1 KO reporter cell lines were similarly cotransfected with two Cas9 plasmids carrying two different sgRNAs targeting sequences within exon 4 of LARP1B. Following the same steps as described for generating LARP1 KO cell lines, clonal cell populations were screened by PCR for the presence of truncated LARP1B alleles and subsequently verified by genomic DNA sequencing and cDNA amplification.
shRNA stable KD cell lines
To generate 4EBP1/2 KD cells, two lentiviruses expressing different resistance genes were used that contained shRNA sequences from the RNAi Consortium public library that were previously described (12). The 4EBP1 shRNA lentivirus carrying puromycin resistance was purchased as lentiviral particles (Sigma-Aldrich). The 4EBP2 shRNA was cloned into the pLKO.1_BlastR lentiviral backbone (59). To produce 4EBP2 shRNA lentivirus, HEK293T cells were cotransfected with the 4EBP2 shRNA, the psPax2 envelope, and the vsv-G packaging plasmids using Fugene6 (Promega) according to the manufacturer’s instructions. The supernatant containing viral particles was harvested daily for the next 4 days, centrifuged at 500g for 10 min, and filtered through a 0.45-μm filter to remove cell debris. The viral particles were concentrated by precipitation using the Lenti-X concentrator (Clontech) and resuspended in cell culture medium.
For infection of HeLa cells expressing the reporter mRNAs, 10,000 cells were seeded in 12-well dishes and co-infected the next day with 4EBP1 and 4EBP2 shRNA viruses in medium supplemented with polybrene (4 μg/ml; Merck). Cells were grown until confluency and reseeded into six-well dishes before addition of puromycin (1 μg/ml; InvivoGen) and blasticidin (5 μg/ml; InvivoGen). Uninfected HeLa-11ht cells were used to determine the minimal antibiotic concentrations that resulted in lethality within 2 to 5 days. Double-resistant cell lines with dual integration of 4EBP1/2 shRNAs were validated by Western blot for efficient stable KD of the targeted proteins.
Genomic DNA extraction
For genotyping of CRISPR-edited cell lines, cells were harvested by trypsinization, and the genomic DNA was extracted using the DNeasy kit (Qiagen) according to the manufacturer’s instructions. Primers specific to the target genes were designed using the Primer-Blast tool (https://ncbi.nlm.nih.gov/tools/primer-blast/). Genomic DNA was amplified using Phusion High-Fidelity polymerase (New England Biolabs), PCR products were cleaned using a PCR purification kit (Qiagen), and purified PCR products were cloned into the pCR-Blunt vector (Thermo Fisher Scientific). For each cell line, a minimum of 10 clones were isolated and analyzed by Sanger sequencing to identify all edited alleles.
SDS-PAGE and Western blotting
For protein extraction, cells were harvested by trypsinization and lysed in radioimmunoprecipitation assay buffer (150 mM NaCl, 50 mM tris, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Triton X-100) supplemented with 1× protease inhibitor (Bimake.com) and SuperNuclease (Sino Biological). Cell lysate was centrifuged at 12,000 rpm for 10 min to remove cell debris, and the supernatant was loaded on a 4 to 15% gradient gel using loading buffer supplemented with 100 mM dithiothreitol. Following SDS–polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred onto nitrocellulose or polyvinylidene difluoride (PVDF) membranes by semi-dry transfer (Trans-Blot Turbo) and blocked in 5% bovine serum albumin–TBST buffer (tris-buffered saline supplemented with 0.1% Tween 20) for 1 hour at room temperature (RT). Primary antibodies were incubated overnight at 4°C in TBST or Intercept blocking buffer (LI-COR) supplemented with 0.1% Tween 20. The next day, the membrane was washed three to five times in TBST and incubated for 1 hour at RT with the fluorescent secondary antibodies diluted 1:10,000 in Intercept blocking buffer with 0.1% Tween 20 (supplemented with 0.01% SDS for PVDF membranes). Following three to five washes in TBST, membranes were transferred to phosphate-buffered saline (PBS), and antibody fluorescence was detected at 700 and 800 nm using an Odyssey infrared imaging system (LI-COR).
Total RNA extraction and cDNA synthesis
For total RNA extraction, cells were harvested by trypsinization and lysed in RNA lysis buffer following the RNA Miniprep kit (Agilent). Genomic DNA contamination was reduced by on-column deoxyribonuclease (DNase) digestion as described in the manual, and purified RNA was stored at −80°C. For validation of LARP1B CRISPR KO, total RNA was reverse-transcribed to cDNA, which was used as the template for cDNA amplification of edited LARP1B transcripts. LARP1B transcripts were amplified as described above for genomic DNA validation.
Live-cell imaging
For live-cell imaging, cells were seeded at low density (20 to 30,000) in 35-mm glass-bottom μ-dishes (Ibidi) and grown for 2 to 3 days. On the day of imaging, the cells were incubated with JF549 or JF646 dyes [HHMI Janelia Research Campus; (60)] to label the MCP-Halo coat protein for 30 min, unbound dye was removed (three washes, PBS), and cells were kept in culture medium until imaging. For induction of reporter mRNAs, doxycycline (1 μg/ml; Sigma-Aldrich) was added to each dish at appropriate time points before each imaging session (30 min) to ensure the same duration of doxycycline induction at the start of imaging for all dishes of an experiment.
At the start of imaging of each dish, culture medium was replaced with FluoroBrite imaging DMEM (Thermo Fisher Scientific) supplemented with 10% FCS, 2 mM glutamine, and doxycycline (1 μg/ml). To inhibit mTOR, cells were treated with 250 nM Torin1, 100 nM Rapamycin, 2.5 μM PP242, or 250 nM TAK228 (INK128) for the specified duration, and the inhibitor was maintained throughout the imaging session. To inhibit translation, cells were treated with puromycin (100 μg/ml) 5 min before the start of imaging, which was maintained throughout imaging. To allow elongating ribosomes to run-off, cells were treated with harringtonine (3 μg/ml) 10 min before the start of imaging, which was maintained throughout imaging. For all experiments, the start of the 30-min imaging window was recorded as the time point shown in the figures (e.g., imaging 60 to 90 min after Torin1 addition = 60-min time point).
Cells were kept at 37°C and 5% CO2 throughout image acquisition. All dual-color live-cell imaging was performed on an inverted Ti2-E Eclipse (Nikon) microscope equipped with a CSU-W1 scan head (Yokogawa), two back-illuminated EMCCD cameras iXon-Ultra-888 (Andor) with chroma ET525/50 m and ET575lp emission filters and an MS-2000 motorized stage (Applied Scientific Instrumentation). Cells were illuminated with 561 Cobolt Jive (Cobolt), 488 iBeam Smart, 639 iBeam Smart (Toptica Photonics) lasers, and a VS-Homogenizer (Visitron Systems GmbH). Using a CFI Plan Apochromat Lambda 100× oil/numerical aperture (NA) 1.45 objective (Nikon), images were obtained with a pixel size of 0.134 μm. To allow for simultaneous tracking of mRNA and translation sites, both channels were simultaneously acquired by both cameras at 20 Hz for 100 frames in a single Z plane (5-s movies).
Live-cell data analysis
For image analysis, the first 5 to 14 frames of each movie (500 ms) were selected for single-particle tracking. First, images were corrected for any offset between the two cameras using TetraSpeck fluorescent beads acquired on each imaging day. Using the FIJI (61, 62) descriptor-based registration plugin (63) in affine transformation mode, a transformation model was obtained to correct the bead offset and applied to all images of an imaging day using a custom macro (64). Subsequently, fine correction of remaining offsets between images were corrected for each dish individually using the FIJI translate function run in a custom macro, correcting for offsets occurring progressively throughout an imaging session.
Single-particle tracking and translation site quantification were performed as described previously (64). In short, using the KNIME analytics platform and a custom-build data processing workflow, regions of interest (ROIs) were manually annotated in the mRNA channel, selecting cytosolic regions with multiple bright spots. Annotation solely in the mRNA channel excludes any bias in selection attributable to the translational state of the cell. Next, spots in the ROIs corresponding to single mRNAs were tracked using TrackMate (65) integrated in KNIME, using the “Laplacian of Gaussian” detector with an estimated spot radius of 200-nm and subpixel localization. Detection thresholds were adjusted on the basis of the signal-to-noise ratio of images and varied between 1.25 and 2. For particle linking, the parameters linking max distance (600 nm), gap closing max distance (1200 nm), and gap closing max frame gap (2) were optimized for single-particle tracking of mRNAs. To assay whether an mRNA is translating, the mean intensity of the SunTag channel was measured at the coordinates of each mRNA spot and quantified as FC/ROI background intensity. A cutoff of <1.5 fold/background was determined to classify an mRNA as nontranslating based on calibration data using the translation inhibitor harringtonine. Excluding cells with <3 mRNAs, the fraction of translating mRNAs per cell was calculated (translating/all mRNAs per ROI). For data visualization, the fraction of translating mRNAs per cell and translation site intensities were plotted using SuperPlots (https://huygens.science.uva.nl/SuperPlotsOfData/), showing all data points together with the mean (±SD) of each biological replicate (66).
RNA sequencing
For RNA-seq, total RNA samples extracted using the RNA Miniprep kit (Agilent) were assessed for RNA integrity using the Agilent Tapestation, and library preparation was performed using the Illumina TruSeq Stranded mRNA reagents according to the manufacturer’s protocol. Libraries were sequenced on the Illumina HiSeq2500 (GEO submission GSE233183: single reads, 50 cycles) or NovaSeq6000 platforms (GEO submission GSE233182: paired-end reads, 100 cycles).
For our analysis, we used a reference list of experimentally validated 5′TOP mRNAs (37), expanded with additional experimentally verified 5′TOP mRNAs (RACK1 and EIF3K) as well as computationally predicted 5′TOP mRNAs with known roles in translation (EIF2A, EIF2S3, EIF3L, EIF4A2, and RPL22L1) (37). The presence or absence of a PRTE in the 5′UTR was taken from Hsieh et al. (14) and expanded by manual annotation for the subset of 5′TOP mRNAs not listed (table S2).
Sequenced reads were aligned against the human genome (GENCODE GRCh38 primary assembly, https://gencodegenes.org/human/release_38.html) using R version 4.1.1 with Bioconductor version 3.13 to execute the qAlign tool [QuasR package, version 1.32.0, (67)], with default parameters except for aligner = “Rhisat2,” splicedAlignment = “TRUE,” allowing only uniquely mapping reads. Raw gene counts were obtained using the qCount tool (QuasR) with a TxDb generated from gencode.v38.primary_assembly.annotation.gtf as query, with default parameters counting only alignments on the opposite strand as the query region. The count table was filtered to only keep genes which had at least 1 cpm in at least three samples.
Single-molecule fluorescence in situ hybridization
For smFISH, HeLa-11ht cell lines were seeded on high-precision glass coverslips placed in 12-well tissue culture plates, grown for 2 days to reach ~50% confluency, and fixed using 4% paraformaldehyde (10 min, RT). To detect endogenous mRNAs, atto633-conjugated smFISH probes targeting human MYC mRNA were generated by enzymatic oligonucleotide labeling (68), using Amino-11-ddUTP and Atto633-NHS. Quasar570-conjugated smFISH probes targeting RPL5, RPL11, and RPL32 were purchased ready-to-use (Biosearch Technologies). smFISH was performed as described previously (64). Briefly, fixed cells were washed twice in PBS (5 min), permeabilized overnight in 70% ethanol, washed thrice in smFISH wash buffer (2× SSC, 10% formamide, 5 min), and hybridized with smFISH probes (2× SSC, 10% formamide, 10% dextran sulfate, 125 nM Quasar570/atto633 smFISH probe) at 37°C for 4 hours. Coverslips were washed twice with smFISH wash buffer (30 min) and mounted on glass slides using Prolong Gold mounting medium containing 4′,6-diamidino-2-phenylindole. Cells were imaged using an upright spinning-disk confocal microscope equipped with a CSU-W1 scan head (Yokogawa) and sCMOS detectors. Using a Plan Apochromat 63× oil/NA 1.4 objective, Z-stacks were obtained with a pixel size of 103 nm and Z-stack spacing of 200 nm using single-camera sequential acquisition.
Analysis of smFISH data was performed using custom-build Python scripts. Nuclei were segmented in three-dimensional (3D) using the triangle threshold method, merged nuclei were split by applying a seeded watershed on the Euclidian distance transformed segmentation mask, and segmentation nuclei with an area < 200 or a solidity < 0.5 were removed. The cytoplasm was segmented on a maximum intensity projection by using the median as a threshold to obtain a semantic segmentation and then splitting this segmentation into cell instance with a seeded watershed applied to the Euclidian distance transform of the semantic segmentation. The maximum projection of the 3D nuclei labeling was taken as seeds for cell segmentation. mRNA spots were detected using a Laplacian of Gaussian filter to detect diffraction limited spots and refined by applying a h-maxima detector to remove detections below a transcript-specific threshold.
SLAM-seq
For metabolic labeling (SLAMseq), HeLa-11ht cell lines were incubated with a dilution series of 4-thiouridine (S4U) for 24 hours, exchanging S4U-containing media every 3 hours according to the manufacturer’s instructions. S4U cytotoxicity was assessed using a luminescent cell viability assay, and the half-maximal inhibitory concentration (IC50) was calculated at 209 μM (n = 2). On the basis of the IC50, a trial RNA-seq was conducted with 24-hour S4U labeling using a dilution series (0, 3, 6, 12, 24, and 48 μM) of S4U and exchanging media every 3 hours. The 12 μM S4U concentration was selected as the optimal experimental S4U concentration with minimal effects on gene expression for all cell lines.
For assessing global RNA half-lives, HeLa-11ht cell lines were labeled with 12 μM S4U for 24 hours (exchanging media every 3 hours), labeling was stopped using 100× excess uridine (1.2 mM), and cells were harvested at time points 0, 2, 4, 6, 9, and 12 hours after the uridine quench. For isolation of total RNA, RNA was extracted using an RNA miniprep kit (Agilent) with on-column DNase digestion, followed by iodoacetamide treatment and ethanol precipitation of modified total RNA. For RNA-seq, total RNA was assessed for RNA integrity using the Agilent Tapestation, and library preparation was performed using the Illumina TruSeq Stranded Total RNA Library Prep Gold kit according to the manufacturer’s protocol. Libraries were sequenced on the Illumina NextSeq500 (GEO submission GSE233186: single reads, 75 cycles). Samples were submitted as three independent technical replicates (cells harvested on separate days), with the exception of one sample with only two replicates (E6_0h).
In total, ~3.7 billion SLAMseq reads were produced corresponding to ~52 M reads per replicate. 4-Thiouridine incorporation events were analyzed using the SlamDunk software (v0.3.4) for SLAMseq analysis (69). SLAMseq reads were first reverse complemented to match the hard-coded assumed SlamDunk orientation using fastx_reverse_complement from the FASTX-toolkit (http://hannonlab.cshl.edu/fastx_toolkit) with default settings. The resulting fastq files were mapped to the reference genome (GENCODE GRCh38 primary assembly) with slamdunk map and parameters -5 0 -ss q. The mapped reads were subsequently filtered to only retain intragenic mappings according to the reference transcriptome (GRCh38, GENCODE v33) with a high identity using slamdunk filter and parameters -mi 0.9. Single-nucleotide polymorphism (SNP) variants in the samples were called with slamdunk snp with parameters -c 1 -f 0.2. The SNP variants from all samples were combined in a single master vcf file by indexing the individual vcf file using tabix of the htslib package (https://github.com/samtools/htslib, v1.9, 07/2018) and merging using vcf-merge from the VCFtools package (70). 4-Thiouridine incorporation and conversion rates were calculated separately for exonic gene segments of the reference transcriptome using slamdunk count with default parameters and the master SNP vcf file for SNP filtering. The exonic segments counts were then aggregated to obtain gene-level total mapping reads, multimapping reads, and converted reads counts. During aggregation, total and converted counts from exonic segment with multimappers were downweighted by the fraction of multimappers over the total mapped reads of the exonic segment (fm). Last, gene conversion rates were calculated as the number of gene-level aggregated converted reads over the gene-level aggregated total read counts.
Gene conversion rates in each context were fitted to an exponential time decay model to obtain gene half-life estimates. Fitting was performed by nonlinear least squares using the R stats::nls function. Example fit command: fit < - nls[rates ~ exp(a + k*time points),control = list(minFactor = 1e-7, tol = 1e-05,maxiter = 256)] where rates are the conversion rates for the gene (including all replicates) and time points are the corresponding times in hours. The half-life (in minutes) was obtained from the fitted coefficient [t1/2 = −1/k*ln(2)*60]. Half-life estimates and the fitting R2 values are listed in table S4 for all transcripts with R2 ≥ 0.75 in all conditions.
Statistical analysis
For live-cell imaging, biological replicates (n) were defined as independent days of imaging. Statistical analyses were performed using GraphPad Prism, with n numbers and statistical tests described in the figure legends. Technical replicates within biological replicates were pooled before statistical tests.
For RNA-seq analysis of LARP1 WT cells, biological replicates (n) were defined as independent clonal cell lines (canonical and 5′TOP mRNA cell lines, n = 2). For RNA-seq of LARP1 KO clones, four single-cell derived clones were sequenced for both canonical and 5′TOP cell lines and treated as independent biological replicates (n = 8 total). For each n, three independent replicates (cells harvested on separate days) were submitted for RNA-seq and averaged before statistical tests. Differential gene expression was calculated with the Bioconductor package edgeR [version 3.34.0, (71)] using the quasi-likelihood F test after applying the calcNormFactors function, obtaining the dispersion estimates and fitting the negative binomial generalized linear models.
For SLAM-seq analysis, biological replicates (n) were defined as independent clonal cell lines for both LARP1 WT and LARP1 KO cells (canonical and 5′TOP mRNA cell lines, n = 2). For analysis of changes in mRNA stability, the estimated half-lives were averaged for n1 and n2, and changes in mRNA stability were calculated between genotypes (log2 FC). Significant differences in mRNA stability were classified with abs log2 FC ≥ 1, excluding spurious transcripts overlapping in sequence with known 5′TOP mRNAs (read-through transcripts AC135178.3, AP002990.1, AC245033.1, lncRNA AL022311.1, MIR4426, and MIR3654).
Acknowledgments
We thank V. Herzog for advice on SLAMseq methodology, S. Smallwood and H.-R. Hotz for support with transcriptomics, T.-O. Buchholz, F. Voigt, L. Gelman, L. Plantard, and S. Reither for microscopy and image analysis support, H. Kohler for cell sorting, and all members of the Chao lab for input and helpful discussions. We thank E. Griesbach for MYC smFISH probe generation. A donation was made to the Henrietta Lacks Foundation to acknowledge the use of HeLa cells in our research.
Funding: This work was supported by the Novartis Research Foundation (J.A.C), a Swiss National Science Foundation Sinergia grant (CRSII5-205884), the SNF-NCCR RNA & Disease network (51NF40-205601), and a Boehringer Ingelheim Fonds PhD fellowship (T.H.).
Author contributions: Conceptualization: J.A.C. and T.H. Methodology: T.H., P.P., and E.P. Investigation: T.H. Visualization: T.H. and P.P. Supervision: J.A.C. Writing—original draft: T.H. and J.A.C. Writing—review and editing: T.H. and J.A.C.
Competing interests: The authors declare they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Sequencing data for this study has been deposited in the GEO repository (GSE233187). All reagents generated in this study are available from the lead contact, J.A.C. (jeffrey.chao@fmi.ch), with a completed material transfer agreement. All original code (KNIME workflows and ImageJ macros) and image analysis output files have been deposited at Zenodo and are publicly available (10.5281/zenodo.10057405).
Supplementary Materials
This PDF file includes:
Figs. S1 to S16
Table S2
Legends for tables S1, S3 to S6
Legends for movies S1 to S20
Other Supplementary Material for this manuscript includes the following:
Tables S1, S3 to S6
Movies S1 to S20
REFERENCES AND NOTES
- 1.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battaglioni S., Benjamin D., Wälchli M., Maier T., Hall M. N., mTOR substrate phosphorylation in growth control. Cell 185, 1814–1836 (2022). [DOI] [PubMed] [Google Scholar]
- 3.Jia J.-J., Lahr R. M., Solgaard M. T., Moraes B. J., Pointet R., Yang A. D., Celucci G., Graber T. E., Hoang H. D., Niklaus M. R., Pena I. A., Hollensen A. K., Smith E. M., Chaker-Margot M., Anton L., Dajadian C., Livingstone M., Hearnden J., Wang X. D., Yu Y., Maier T., Damgaard C. K., Berman A. J., Alain T., Fonseca B. D., mTORC1 promotes TOP mRNA translation through site-specific phosphorylation of LARP1. Nucleic Acids Res. 49, 3461–3489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca B. D., Zakaria C., Jia J. J., Graber T. E., Svitkin Y., Tahmasebi S., Healy D., Hoang H. D., Jensen J. M., Diao I. T., Lussier A., Dajadian C., Padmanabhan N., Wang W., Matta-Camacho E., Hearnden J., Smith E. M., Tsukumo Y., Yanagiya A., Morita M., Petroulakis E., González J. L., Hernández G., Alain T., Damgaard C. K., La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J. Biol. Chem. 290, 15996–16020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philippe L., Vasseur J. J., Debart F., Thoreen C. C., La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 46, 1457–1469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tcherkezian J., Cargnello M., Romeo Y., Huttlin E. L., Lavoie G., Gygi S. P., Roux P. P., Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation. Genes Dev. 28, 357–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hong S., Freeberg M. A., Han T., Kamath A., Yao Y., Fukuda T., Suzuki T., Kim J. K., Inoki K., LARP1 functions as a molecular switch for mTORC1-mediated translation of an essential class of mRNAs. eLife 6, e25237 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyuhas O., Kahan T., The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 1849, 801–811 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Biberman Y., Meyuhas O., Substitution of just five nucleotides at and around the transcription start site of rat β-actin promoter is sufficient to render the resulting transcript a subject for translational control. FEBS Lett. 405, 333–336 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Avni D., Shama S., Loreni F., Meyuhas O., Vertebrate mRNAs with a 5'-terminal pyrimidine tract are candidates for translational repression in quiescent cells: Characterization of the translational cis-regulatory element. Mol. Cell. Biol. 14, 3822–3833 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berman A. J., Thoreen C. C., Dedeic Z., Chettle J., Roux P. P., Blagden S. P., Controversies around the function of LARP1. RNA Biol. 18, 207–217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., Sabatini D. M., A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miloslavski R., Cohen E., Avraham A., Iluz Y., Hayouka Z., Kasir J., Mudhasani R., Jones S. N., Cybulski N., Rüegg M. A., Larsson O., Gandin V., Rajakumar A., Topisirovic I., Meyuhas O., Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J. Mol. Cell Biol. 6, 255–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh A. C., Liu Y., Edlind M. P., Ingolia N. T., Janes M. R., Sher A., Shi E. Y., Stumpf C. R., Christensen C., Bonham M. J., Wang S., Ren P., Martin M., Jessen K., Feldman M. E., Weissman J. S., Shokat K. M., Rommel C., Ruggero D., The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindqvist L., Imataka H., Pelletier J., Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA 14, 960–969 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamarkin-Ben-Harush A., Vasseur J. J., Debart F., Ulitsky I., Dikstein R., Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. eLife 6, e21907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahr R. M., Fonseca B. D., Ciotti G. E., al-Ashtal H. A., Jia J. J., Niklaus M. R., Blagden S. P., Alain T., Berman A. J., La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife 6, e24146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schofield J. A., Duffy E. E., Kiefer L., Sullivan M. C., Simon M. D., TimeLapse-seq: Adding a temporal dimension to RNA sequencing through nucleoside recoding. Nat. Methods 15, 221–225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herzog V. A., Reichholf B., Neumann T., Rescheneder P., Bhat P., Burkard T. R., Wlotzka W., von Haeseler A., Zuber J., Ameres S. L., Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozlov G., Mattijssen S., Jiang J., Nyandwi S., Sprules T., Iben J. R., Coon S. L., Gaidamakov S., Noronha A. M., Wilds C. J., Maraia R. J., Gehring K., Structural basis of 3′-end poly(A) RNA recognition by LARP1. Nucleic Acids Res. 50, 9534–9547 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattijssen S., Kozlov G., Gaidamakov S., Ranjan A., Fonseca B. D., Gehring K., Maraia R. J., The isolated La-module of LARP1 mediates 3′ poly(A) protection and mRNA stabilization, dependent on its intrinsic PAM2 binding to PABPC1. RNA Biol. 18, 275–289 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogami K., Oishi Y., Sakamoto K., Okumura M., Yamagishi R., Inoue T., Hibino M., Nogimori T., Yamaguchi N., Furutachi K., Hosoda N., Inagaki H., Hoshino S. I., mTOR- and LARP1-dependent regulation of TOP mRNA poly(A) tail and ribosome loading. Cell Rep. 41, 111548 (2022). [DOI] [PubMed] [Google Scholar]
- 23.K. Ogami, Y. Oishi, T. Nogimori, K. Sakamoto, S.-i. Hoshino, LARP1 facilitates translational recovery after amino acid refeeding by preserving long poly(A)-tailed TOP mRNAs. bioRxiv, 716217 (2019). 10.1101/716217. [DOI]
- 24.Smith E. M., Benbahouche N. E. H., Morris K., Wilczynska A., Gillen S., Schmidt T., Meijer H. A., Jukes-Jones R., Cain K., Jones C., Stoneley M., Waldron J. A., Bell C., Fonseca B. D., Blagden S., Willis A. E., Bushell M., The mTOR regulated RNA-binding protein LARP1 requires PABPC1 for guided mRNA interaction. Nucleic Acids Res. 49, 458–478 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mura M., Hopkins T. G., Michael T., Abd-Latip N., Weir J., Aboagye E., Mauri F., Jameson C., Sturge J., Gabra H., Bushell M., Willis A. E., Curry E., Blagden S. P., LARP1 post-transcriptionally regulates mTOR and contributes to cancer progression. Oncogene 34, 5025–5036 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park J., Kim M., Yi H., Baeg K., Choi Y., Lee Y. S., Lim J., Kim V. N., Short poly(A) tails are protected from deadenylation by the LARP1–PABP complex. Nat. Struct. Mol. Biol. 30, 330–338 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Wilbertz J. H., Voigt F., Horvathova I., Roth G., Zhan Y., Chao J. A., Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol. Cell 73, 946–958.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Tanenbaum M. E., Gilbert L. A., Qi L. S., Weissman J. S., Vale R. D., A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banaszynski L. A., Chen L.-C., Maynard-Smith L. A., Ooi A. G. L., Wandless T. J., A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C. A., Taylor M. S., Engström P. G., Frith M. C., Forrest A. R., Alkema W. B., Tan S. L., Plessy C., Kodzius R., Ravasi T., Kasukawa T., Fukuda S., Kanamori-Katayama M., Kitazume Y., Kawaji H., Kai C., Nakamura M., Konno H., Nakano K., Mottagui-Tabar S., Arner P., Chesi A., Gustincich S., Persichetti F., Suzuki H., Grimmond S. M., Wells C. A., Orlando V., Wahlestedt C., Liu E. T., Harbers M., Kawai J., Bajic V. B., Hume D. A., Hayashizaki Y., Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38, 626–635 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S., An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wullschleger S., Loewith R., Hall M. N., TOR signaling in growth and metabolism. Cell 124, 471–484 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Apsel B., Blair J. A., Gonzalez B., Nazif T. M., Feldman M. E., Aizenstein B., Hoffman R., Williams R. L., Shokat K. M., Knight Z. A., Targeted polypharmacology: Discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 4, 691–699 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jessen K., Wang S., Kessler L., Guo X., Kucharski J., Staunton J., Lan L., Elia M., Stewart J., Brown J., Li L., Chan K., Martin M., Ren P., Rommel C., Liu Y., Abstract B148: INK128 is a potent and selective TORC1/2 inhibitor with broad oral antitumor activity. Mol. Cancer Ther. 8 (Suppl. 1), B148-8 (2009). [Google Scholar]
- 35.Hassan B., Akcakanat A., Sangai T., Evans K. W., Adkins F., Eterovic A. K., Zhao H., Chen K., Chen H., do K. A., Xie S. M., Holder A. M., Naing A., Mills G. B., Meric-Bernstam F., Catalytic mTOR inhibitors can overcome intrinsic and acquired resistance to allosteric mTOR inhibitors. Oncotarget 5, 8544–8557 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huo Y., Iadevaia V., Yao Z., Kelly I., Cosulich S., Guichard S., Foster L. J., Proud C. G., Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem. J. 444, 141–151 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Philippe L., van den Elzen A. M. G., Watson M. J., Thoreen C. C., Global analysis of LARP1 translation targets reveals tunable and dynamic features of 5′ TOP motifs. Proc. Natl. Acad. Sci. 117, 5319–5328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aoki K., Adachi S., Homoto M., Kusano H., Koike K., Natsume T., LARP1 specifically recognizes the 3′ terminus of poly(A) mRNA. FEBS Lett. 587, 2173–2178 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Al-Ashtal H. A., Rubottom C. M., Leeper T. C., Berman A. J., The LARP1 La-Module recognizes both ends of TOP mRNAs. RNA Biol. 18, 248–258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Truitt M. L., Conn C. S., Shi Z., Pang X., Tokuyasu T., Coady A. M., Seo Y., Barna M., Ruggero D., Differential requirements for eIF4E dose in normal development and cancer. Cell 162, 59–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoreen C. C., The molecular basis of mTORC1-regulated translation. Biochem. Soc. Trans. 45, 213–221 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Gandin V., Masvidal L., Hulea L., Gravel S. P., Cargnello M., McLaughlan S., Cai Y., Balanathan P., Morita M., Rajakumar A., Furic L., Pollak M., Porco JA Jr, St-Pierre J., Pelletier J., Larsson O., Topisirovic I., nanoCAGE reveals 5' UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 26, 636–648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Elzen A. M. G., Watson M. J., Thoreen C. C., mRNA 5′ terminal sequences drive 200-fold differences in expression through effects on synthesis, translation and decay. PLOS Genet. 18, e1010532 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyuhas O., Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267, 6321–6330 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Mamane Y., Petroulakis E., Martineau Y., Sato T. A., Larsson O., Rajasekhar V. K., Sonenberg N., Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLOS ONE 2, e242 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kajjo S., Sharma S., Brothers R. W., Delisle V., Fabian R. M., PABPC plays a critical role in establishing mTORC-mediated translational control. Under Preparation., (2023). [Google Scholar]
- 47.Tomoo K., Shen X., Okabe K., Nozoe Y., Fukuhara S., Morino S., Ishida T., Taniguchi T., Hasegawa H., Terashima A., Sasaki M., Katsuya Y., Kitamura K., Miyoshi H., Ishikawa M., Miura K., Crystal structures of 7-methylguanosine 5'-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5',5'-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem. J. 362 (Pt. 3), 539–544 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furic L., Rong L., Larsson O., Koumakpayi I. H., Yoshida K., Brueschke A., Petroulakis E., Robichaud N., Pollak M., Gaboury L. A., Pandolfi P. P., Saad F., Sonenberg N., eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. U.S.A. 107, 14134–14139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zinshteyn B., Rojas-Duran M. F., Gilbert W. V., Translation initiation factor eIF4G1 preferentially binds yeast transcript leaders containing conserved oligo-uridine motifs. RNA 23, 1365–1375 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin H., Xu W., Rahman R., Na D., Fieldsend A., Song W., Liu S., Li C., Rosbash M., TRIBE editing reveals specific mRNA targets of eIF4E-BP in Drosophila and in mammals. Sci. Adv. 6, eabb8771 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fuentes P., Pelletier J., Martinez-Herráez C., Diez-Obrero V., Iannizzotto F., Rubio T., Garcia-Cajide M., Menoyo S., Moreno V., Salazar R., Tauler A., Gentilella A., The 40S-LARP1 complex reprograms the cellular translatome upon mTOR inhibition to preserve the protein synthetic capacity. Sci. Adv. 7, eabg9275 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gentilella A., Morón-Duran F. D., Fuentes P., Zweig-Rocha G., Riaño-Canalias F., Pelletier J., Ruiz M., Turón G., Castaño J., Tauler A., Bueno C., Menéndez P., Kozma S. C., Thomas G., Autogenous control of 5′TOP mRNA stability by 40S ribosomes. Mol. Cell 67, 55–70.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brito Querido J., Sokabe M., Kraatz S., Gordiyenko Y., Skehel J. M., Fraser C. S., Ramakrishnan V., Structure of a human 48S translational initiation complex. Science 369, 1220–1227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goering R., Arora A., Pockalny M. C., Taliaferro J. M., RNA localization mechanisms transcend cell morphology. eLife 12, e80040 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livingston N. M., Kwon J., Valera O., Saba J. A., Sinha N. K., Reddy P., Nelson B., Wolfe C., Ha T., Green R., Liu J., Wu B., Bursting translation on single mRNAs in live cells. Mol. Cell 83, 2276–2289.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dave P., Roth G., Griesbach E., Mateju D., Hochstoeger T., Chao J. A., Single-molecule imaging reveals translation-dependent destabilization of mRNAs. Mol. Cell 83, 589–606.e6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F., Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu V. T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kühn R., Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 33, 543–548 (2015). [DOI] [PubMed] [Google Scholar]
- 59.Bryant D. M., Datta A., Rodríguez-Fraticelli A. E., Peränen J., Martín-Belmonte F., Mostov K. E., A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 12, 1035–1045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grimm J. B., English B. P., Chen J., Slaughter J. P., Zhang Z., Revyakin A., Patel R., Macklin J. J., Normanno D., Singer R. H., Lionnet T., Lavis L. D., A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., Eliceiri K. W., ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Preibisch S., Saalfeld S., Schindelin J., Tomancak P., Software for bead-based registration of selective plane illumination microscopy data. Nat. Methods 7, 418–419 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Mateju D., Eichenberger B., Voigt F., Eglinger J., Roth G., Chao J. A., Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183, 1801–1812.e13 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Tinevez J.-Y., Perry N., Schindelin J., Hoopes G. M., Reynolds G. D., Laplantine E., Bednarek S. Y., Shorte S. L., Eliceiri K. W., TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Lord S. J., Velle K. B., Mullins R. D., Fritz-Laylin L. K., SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaidatzis D., Lerch A., Hahne F., Stadler M. B., QuasR: Quantification and annotation of short reads in R. Bioinformatics 31, 1130–1132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gaspar I., Wippich F., Ephrussi A., Terminal deoxynucleotidyl transferase mediated production of labeled probes for single-molecule FISH or RNA capture. Bio Protoc. 8, e2750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neumann T., Herzog V. A., Muhar M., von Haeseler A., Zuber J., Ameres S. L., Rescheneder P., Quantification of experimentally induced nucleotide conversions in high-throughput sequencing datasets. BMC Bioinformatics 20, 258 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R., 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lun A. T., Chen Y., Smyth G. K., It's DE-licious: A recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol. Biol. 1418, 391–416 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S16
Table S2
Legends for tables S1, S3 to S6
Legends for movies S1 to S20
Tables S1, S3 to S6
Movies S1 to S20