Abstract
Endogenous cannabinoids acting at CB1 receptors stimulate appetite, and CB1 antagonists show promise in the treatment of obesity. CB1–/– mice are resistant to diet-induced obesity even though their caloric intake is similar to that of wild-type mice, suggesting that endocannabinoids also regulate fat metabolism. Here, we investigated the possible role of endocannabinoids in the regulation of hepatic lipogenesis. Activation of CB1 in mice increases the hepatic gene expression of the lipogenic transcription factor SREBP-1c and its targets acetyl-CoA carboxylase-1 and fatty acid synthase (FAS). Treatment with a CB1 agonist also increases de novo fatty acid synthesis in the liver or in isolated hepatocytes, which express CB1. High-fat diet increases hepatic levels of the endocannabinoid anandamide (arachidonoyl ethanolamide), CB1 density, and basal rates of fatty acid synthesis, and the latter is reduced by CB1 blockade. In the hypothalamus, where FAS inhibitors elicit anorexia, SREBP-1c and FAS expression are similarly affected by CB1 ligands. We conclude that anandamide acting at hepatic CB1 contributes to diet-induced obesity and that the FAS pathway may be a common molecular target for central appetitive and peripheral metabolic regulation.
Introduction
Maintenance of energy homeostasis and body weight involves the coordinated regulation of appetitive behavior and peripheral energy metabolism (1), as illustrated by the ability of the appetite-reducing hormone leptin to regulate fat metabolism in the liver (2). Endocannabinoids are novel lipid mediators that modulate appetitive behavior through the activation of CB1 (3–11). Sites in the hypothalamus (4, 10, 12), limbic forebrain (11–13), and peripheral sensory nerve terminals (7) have been implicated in mediating the orexigenic effect of endocannabinoids, which is potentiated by hunger or in hyperphagia associated with obesity (4–6, 9) and antagonized by CB1 blockade. Indeed, CB1 antagonists show promise in the treatment of obesity (14). A number of recent observations suggest that reduction of food intake alone cannot fully account for the antiobesity effects of CB1 antagonists. In a mouse model of diet-induced obesity, chronic treatment with the CB1 antagonist SR141716 caused a transient reduction in food intake and a more prolonged reduction in body weight (15). Mice lacking CB1 are resistant to diet-induced obesity even though their total caloric intake is similar to that of wild-type littermates, which become obese on the same diet (16). CB1–/– mice display a moderately lean phenotype throughout adulthood but only a temporary hypophagia in the first few weeks of life (17). These observations suggest that endocannabinoids and CB1 also regulate peripheral energy metabolism. Indeed, adipocytes have been found to express CB1, stimulation of which may affect fat metabolism by regulating the level of the adipocyte-derived hormone adiponectin (18) or by increasing lipoprotein lipase activity (17). However, the role of adipose tissue in de novo lipogenesis is minor compared with that of the liver (19). Therefore, we have examined the possible role of the liver as a peripheral target of the metabolic actions of endocannabinoids and explored the underlying molecular targets. The results indicate that hepatocytes express CB1, stimulation of which induces the expression of the lipogenic transcription factor SREBP-1c (20) and its target enzymes acetyl coenzyme-A carboxylase-1 (ACC1) and fatty acid synthase (FAS), and also increases de novo fatty acid synthesis. This mechanism may contribute to the development of diet-induced obesity, which is found to be associated with an increase in the hepatic levels of the endocannabinoid anandamide (arachidonoyl ethanolamide) and CB1-mediated fatty acid synthesis in mice. CB1 activation induces the same molecular targets in the hypothalamus, where inhibitors of FAS have been reported to cause anorexia (21, 22). Thus, the fatty acid biosynthetic pathway may represent a common molecular target for the central appetitive and peripheral metabolic effects of endocannabinoids.
Results
Activation of CB1 increases lipogenic gene expression.
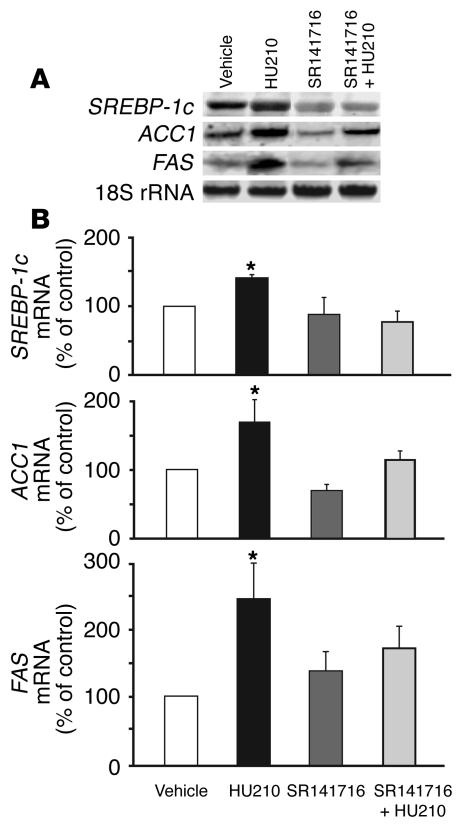
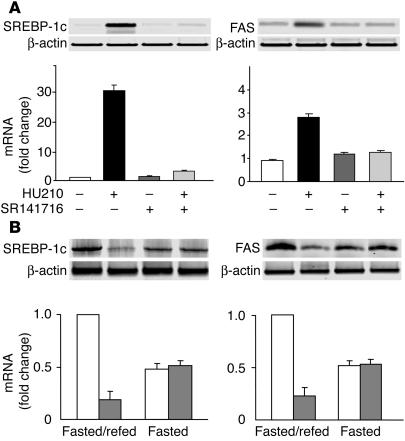
In order to identify likely peripheral molecular targets of CB1, we profiled the expression of a group of genes involved in fat metabolism in the liver and adipose tissue of wild-type and CB1–/– mice. Gene expression was analyzed by RT-PCR, using mRNA extracted from the liver and adipose tissue of CB1+/+ and CB1–/– mice maintained on regular chow. The level of expression of the lipogenic transcription factor SREBP-1c (20) was consistently lower in liver and adipose tissue of CB1–/– compared with that of CB1+/+ mice. Because SREBP-1c regulates the expression of enzymes involved in fatty acid synthesis (20), we next examined whether treatment of wild-type mice with a CB1 agonist could increase the gene expression of SREBP-1c and its target enzymes ACC1 and FAS in the liver and adipose tissue, using Northern hybridization. In vivo treatment with the potent CB1 agonist HU210 (20 ng/g i.p.) resulted in a marked increase in the hepatic levels of SREBP-1c, ACC1, and FAS mRNA, and pretreatment of the animals with 3 μg/g of the CB1 antagonist SR141716 blocked the effect of HU210 (Figure 1). Similar changes were observed in Northern blots from combined fat pads, where mRNA levels of the same 3 proteins increased by 45% ± 15%, 105% ± 32% and 90% ± 24%, respectively. Increased hepatic expression of the SREBP-1c protein in response to treatment with HU210 was also evident in DNA mobility shift assays, using nuclear extracts isolated from the livers of wild-type mice treated for 3 days with daily injections of vehicle, 20 ng/g HU210, 3 μg/g SR141716, or 20 ng/g HU210 plus 3 μg/g SR141716. Relative to vehicle-treated controls, HU210 treatment increased the radioactivity in the specific shifted bands by 30% ± 11% (P < 0.05; n = 4), whereas in mice pretreated with SR141716, HU210 treatment had no effect (13% ± 11%; n = 4).
Figure 1.
Activation of CB1 increases the gene expression of SREBP-1c, ACC1, and FAS in the liver of CB1+/+ mice. Mice were injected i.p. with vehicle, 20 ng/g HU210, 3 μg/g SR141716, or 20 ng/g HU210 plus 3 μg/g SR141716 (SR141716 + HU210) 1 hour prior to sacrifice and removal of the liver. RNA was isolated and Northern hybridization performed as described in Methods. An original blot (A) as well as the mean ± SEM from 5 replicate experiments in each group (B) are shown. Relative mRNA levels were quantified by densitometry, corrected for 18S ribosomal RNA levels used as loading control, and expressed as a percentage of the value measured in vehicle-treated controls.
Activation of CB1 stimulates de novo fatty acid synthesis in the liver.
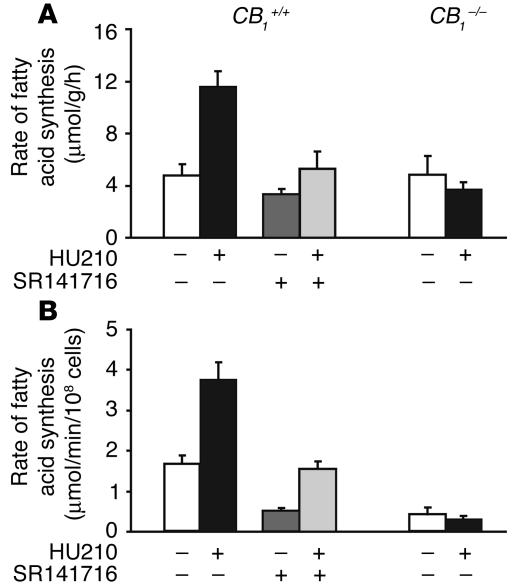
The liver is the main source of fatty acids synthesized de novo. To test the functional consequence of increased lipogenic gene expression, we analyzed de novo hepatic fatty acid synthesis in vivo, by measuring the incorporation of tritium into fatty acids in the liver following intrahepatic injection of 3H2O. As summarized in Figure 2A, pretreatment of mice with 20 ng/g HU210 caused more than a 2-fold increase in the rate of hepatic fatty acid synthesis. This increase was mediated by CB1 because no increase was observed in mice pretreated with 3 μg/g SR141716 or in CB1–/– mice. Similar effects were observed in hepatocytes isolated from wild-type mice: incubation of the cells with 100 nM HU210 for 60 minutes significantly increased the incorporation of subsequently added 3H2O into fatty acids. The effect was attenuated in the presence of 10 nM SR141716 and was absent in cells from CB1–/– mice (Figure 2B).
Figure 2.
Activation of CB1 stimulates de novo fatty acid synthesis in the liver in vivo and in vitro. (A) In vivo fatty acid synthesis in CB1+/+ and CB1–/– mice on normal diet was assayed by the conversion of 3H2O into fatty acids in the liver. Mice were treated i.p. with vehicle (white bars), 20 ng/g HU210 (black bars), 3 μg/g SR141716 (dark gray bars), or 20 ng/g HU210 plus 3 μg/g SR141716 (light gray bars), and the rate of fatty acid synthesis was expressed as micromoles of 3H20 converted into fatty acids per hour per gram of tissue weight. (B) In vitro fatty acid synthesis in isolated hepatocytes from mice. Isolated cells were treated with drugs in vitro and fatty acid synthesis quantified as described in Methods. Bars represent mean ± SEM from 4–5 separate experiments.
Presence of CB1 receptors in the mouse liver.
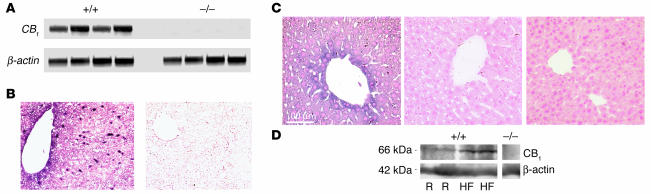
The above findings predicate the presence of CB1 in hepatocytes, which was verified using multiple methods. The results of RT-PCR indicated the presence of CB1 mRNA in the livers of control mice and its absence in livers from CB1–/– mice (Figure 3A). CB1 mRNA was also detected by in situ hybridization, with strong labeling detected in Kupffer cells and lower levels evident in endothelial cells as well as hepatocytes, particularly in perivascular areas (Figure 3B). The presence of CB1 protein in the liver was demonstrated by immunohistochemistry (Figure 3C). Using an antibody against the N terminus of CB1, the expression of immunoreactive CB1 was more prominent in hepatocytes around the central veins, a pattern similar to that seen with in situ hybridization. Staining was stronger around the cell perimeter, suggesting membrane localization. The specificity of the staining was indicated by the ability of blocking peptide to suppress it (Figure 3C, middle) and by its absence in liver sections from CB1–/– mice (Figure 3C, right). The presence of CB1 protein in purified liver plasma membranes was further documented by Western blotting using an antibody against the C terminus of rat CB1 (Figure 3D), which also indicated increased receptor levels in preparations from mice kept on a high-fat diet for 3 weeks compared with those in mice on regular mouse chow.
Figure 3.
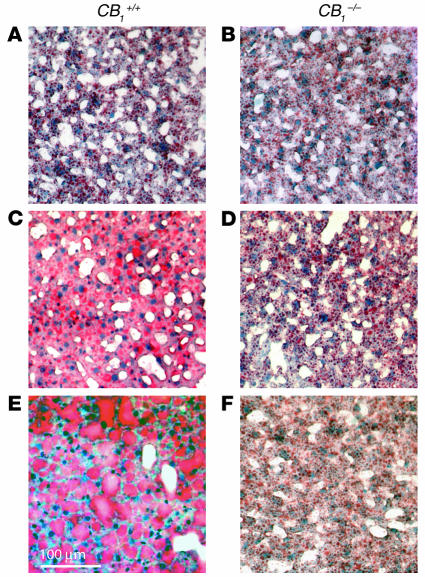
Presence of CB1 in mouse liver. (A) CB1 mRNA is present in the liver of CB1+/+ but not CB1–/– mice, as tested by RT-PCR. β-actin mRNA was amplified as internal control. (B) Localization of CB1 mRNA in normal mouse liver by in situ hybridization in the presence (left panel) and absence (right panel) of the cRNA probe. (C) Immunoreactive CB1 is present in hepatocytes in the liver of a CB1+/+ (left panel) but not a CB1–/– mouse (right panel). Center panel: the effect of preincubation of the N-terminal antibody with its blocking peptide in a section from the same liver as shown at left. Tissue structure is visualized by nuclear fast red counterstaining. (D) Immunoreactive CB1 in purified liver plasma membranes was visualized by Western blot analysis using an antibody against the C terminus of rat CB1. The specificity of the reaction is indicated by its absence in a preparation from a CB1–/– mouse. The expression of CB1 is upregulated in mice on a high-fat diet (HF) compared to regular diet (R).
CB1–/– mice are resistant to diet-induced obesity.
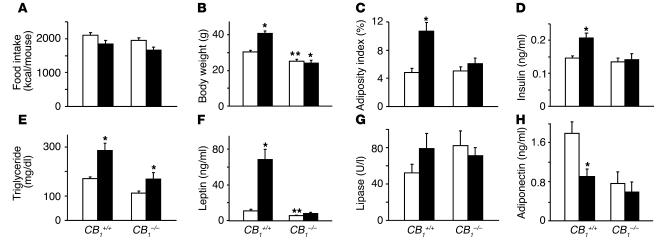
High-fat diet has been recently reported to induce obesity in wild-type but not in CB1–/– mice, despite similar caloric intake (16). To uncover the reason for this difference in phenotype, we placed CB1–/– mice and their wild-type littermates on a high-fat diet, while respective controls remained on regular mouse chow. Daily food intake increased transiently in wild-type mice during the first 10 days on the high-fat diet and then returned to the levels seen in the other 3 groups, resulting in similar overall caloric intake in the 4 groups for the 3-month diet period (Figure 4A). In contrast to wild-type mice fed normal chow, wild-type mice on the high-fat diet became obese (Figure 4B), demonstrated associated hormonal and metabolic changes (Figure 4, C–H), and developed fatty liver (Figure 5, A, C, and E). CB1–/– mice on the high-fat diet, however, remained lean (Figure 4B), their metabolic and hormonal profile remained unchanged (Figure 4, C–H), and they did not develop fatty liver (Figure 5, B, D, and F), despite having caloric intake similar to that of wild-type mice on the high-fat diet. In wild-type mice, feed efficiency increased significantly from 3.23 ± 0.19 mg/kcal on normal chow to 5.41 ± 0.53 mg/kcal on the high-fat diet (P < 0.02), whereas in CB1–/– mice, it remained unaffected by the diet (1.87 ± 0.11 mg/kcal on normal chow versus 2.40 ± 0.55 mg/kcal on the high-fat diet; P > 0.2).
Figure 4.
Effects of high-fat diet (black bars) versus regular chow (white bars) on physiological and biochemical parameters in CB1+/+ (n = 9 regular, 10 high-fat) and CB1–/– mice (n = 8 in each group). Food intake reflects cumulative intake over the diet period; the other parameters were measured at the time the mice were sacrificed. Adiposity index was calculated as the total fat pad weight ([subcutaneous + retroperitoneal + inguinal]/eviscerated body weight × 100). Body weight at the start of the diet was slightly, but significantly, lower in CB1–/– mice than in their CB1+/+ littermates, in agreement with published results (17). *P < 0.01 versus corresponding group on control diet; **P < 0.05 versus corresponding value in CB1+/+ mice.
Figure 5.
High-fat diet induces fatty liver in CB1+/+ but not CB1–/– mice. Fat deposition is visualized by Oil Red O staining in liver sections from mice on normal diet (A and B) or on high-fat diet for 3 weeks (C and D) or for 14 weeks (E and F).
Diet-induced obesity is associated with increased CB1-mediated fatty acid synthesis in the liver.
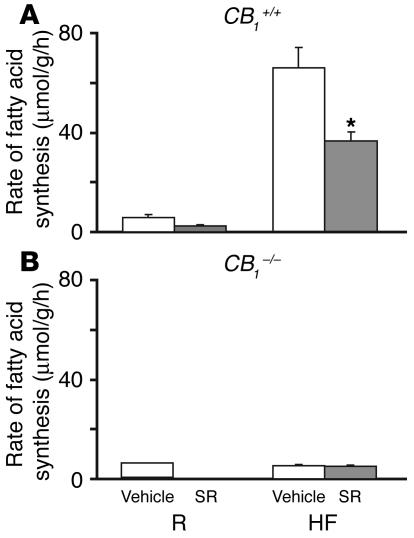
The resistance of CB1-deficient mice to diet-induced obesity in spite of similar caloric intake suggests a role for CB1 in the lipogenic response of wild-type mice to high-fat diet. In a group of mice, we tested the effect of the high-fat diet on de novo hepatic fatty acid synthesis 3 weeks following the initiation of the diet, before a significant effect on body weight could be detected. When tested at this time, the basal rates of de novo fatty acid synthesis were markedly increased compared with those of lean controls, and pretreatment of mice on the high-fat diet with 3 μg/g SR141716 significantly reduced the rate of fatty acid synthesis (Figure 6A). In CB1–/– mice, the high-fat diet did not induce any change in the basal rate of fatty acid synthesis, which remained unaffected by SR141716 (Figure 6B). Interestingly, deposition of lipid droplets was already evident at this time in wild-type, but not CB1–/–, mice (compare Figure 5C and Figure 5D).
Figure 6.
High-fat diet increases de novo hepatic fatty acid synthesis via activation of CB1 receptors. The rate of conversion of 3H2O to fatty acids in the liver was assayed in wild-type (A) and CB1–/– mice (B) 15 minutes after the i.p. injection of vehicle (white bars) or 3 mg/kg SR141716 (SR) (gray bars). Animals were on control or high-fat diets for 3 weeks prior to testing, as indicated. The marked increase in hepatic fatty acid synthesis in wild-type mice was inhibited in the presence of SR141716 and was absent in CB1–/– mice. *P < 0.01 versus corresponding vehicle-treated group. Bars indicate mean ± SEM from 5–8 animals in each group.
In mice sacrificed at the same time, hepatic levels of anandamide were greatly elevated in animals on the high-fat diet (81.5 ± 8.2 fmol/mg protein; n = 6) compared with lean controls (25.2 ± 3.5 fmol/mg protein; n = 6; P < 0.005), with no difference in the hepatic levels of 2-arachidonoylglycerol (2–AG) (630 ± 48 fmol/mg in high-fat diet versus 814 ± 94 fmol/mg in control). Hepatic anandamide levels declined through the remaining diet period but remained slightly elevated compared with controls when measured at the end of the 14-week diet period (20.1 ± 1.5 fmol/mg tissue in high-fat diet versus 14.2 ± 1.6 fmol/mg tissue in control; P < 0.05). In CB1–/– mice fed the high-fat diet for 3 weeks, hepatic anandamide levels were also increased (33.4 ± 3.2 fmol/mg protein) compared with those of controls fed normal chow (17.8 ± 6.1 fmol/mg protein, P < 0.05), although the change was less marked than in wild-type mice. Again, there was no significant difference in hepatic 2-AG levels.
Mechanism of high-fat diet–induced increase in hepatic anandamide.
To test the role of altered synthesis versus degradation in the elevation of hepatic anandamide levels, we measured the activities of N-acyltransferase (NAT), the rate-limiting step in anandamide synthesis (23), and of fatty acid amidohydrolase (FAAH), the enzyme responsible for the metabolism of anandamide (24), using purified liver plasma membrane preparations from mice on normal versus high-fat diets. NAT activity was unchanged at 570 ± 73 fmol/min/mg protein versus 543 ± 302 fmol/min/mg protein in normal versus high-fat diet groups, respectively, of N-[14C]arachidonoyl phosphatidylethanolamine (NAPE) generated from 1,2-phosphatidylcholine-L-α-[14C]diarachidonoyl. On the other hand, FAAH activity, as measured by the release of [3H]ethanolamine from [3H]anandamide, was dramatically reduced from 460 ± 15 fmol/min/mg protein in 4 plasma membrane preparations from controls to 87 ± 29 fmol/min/mg protein in 4 preparations from mice on the high-fat diet (P < 0.005). However, the level of FAAH protein, as quantified in Western blots of liver plasma membranes using a FAAH antibody, was similar in mice on normal and high-fat diets (data not shown).
CB1 regulation of fatty acid synthesis in the hypothalamus.
The enzyme FAS is widely expressed in the brain, including in neurons of the hypothalamus linked to appetite control, and inhibitors of FAS dramatically reduce food intake, most likely by an action in the hypothalamus (21, 22, 25). We therefore examined whether hypothalamic FAS is regulated by CB1. Treatment of wild-type mice with 20 ng/g HU210 caused a significant increase in hypothalamic SREBP-1c and FAS mRNA levels, as measured by RT-PCR, and this effect was abrogated in animals pretreated with 3 μg/g SR141716, although SR141716 alone did not reduce SREBP-1c and FAS mRNA levels (Figure 7A). Similar results were obtained using real-time quantitative RT-PCR, where the fold increase in mRNA levels over control, in response to HU-210, SR141716, and combined HU-210 and SR141716 treatment, respectively, was 11.2, 1.8, and 2.3 for SREBP-1c, and 3.3, 1.8, and 2.3 for FAS.
Figure 7.
Cannabinoid regulation of lipogenic gene expression in the hypothalamus. (A) Activation of CB1 by HU210 increases SREBP-1c and FAS gene expression in the hypothalamus of CB1+/+ mice (n = 4). The hypothalamus was dissected as previously described (39). RT-PCR analysis was performed as described in Methods, and its results were confirmed by real-time quantitative RT-PCR. (B) The CB1 antagonist SR141716 inhibits SREBP-1c and FAS gene expression in fasted/refed (n = 6) but not in fasted-only mice (n = 6). Mice received an i.p. injection of vehicle (white bars) or 3 μg/g SR141716 (gray bars) at the end of the 24-hour fast.
The appetite- and weight-reducing effects of CB1 antagonists are more pronounced in obese, hyperphagic animals than in their lean controls (5, 6), and in non-obese animals, the anorexigenic effect of SR141716 is more prominent when tested in animals during refeeding after temporary food restraint or deprivation (4, 7–9) than in animals with free access to food. This fasting/refeeding approach is a potent inducer of SREBP-1c (26) and FAS gene expression (27). Therefore, we tested the effect of SR141716 on hypothalamic SREBP-1c and FAS gene expression in mice fasted for 24 hours, followed by a 3-hour period of either continued fasting or refeeding with a high-carbohydrate diet. Fasting/refeeding increased SREBP-1c and FAS mRNA levels significantly more in mice treated with vehicle at the start of the refeeding period than in mice treated with 3 μg/g SR141716, whereas SR141716 treatment did not affect the much lower mRNA levels in fasted-only mice (Figure 7B).
In the fasted/refed groups, food intake during the 3-hour refeeding period was significantly reduced by SR141716 treatment compared with that in vehicle treatment (1.12 ± 0.21 g/mouse versus 1.85 ± 0.22 g/mouse; n = 6; P < 0.05). In contrast, in mice with free access to food, in which SR141716 treatment did not affect SREBP-1c and FAS mRNA levels (Figure 7A), food intake was similarly unaffected by SR141716 treatment (0.41 ± 0.12 g/mouse versus 0.45 ± 0.16 g/mouse; n = 6; P > 0.5).
Discussion
The activation of CB1 by endocannabinoids is considered a central factor in neural control of appetite (3–13). The present findings demonstrate that endocannabinoids also target the liver, where activation of CB1 results in increased de novo fatty acid synthesis through the induction of the lipogenic transcription factor SREBP-1c and its target enzymes ACC1 and FAS. The results also suggest that activation of this pathway by endogenous anandamide in the liver has a key role in the development of diet-induced obesity and fatty liver. Importantly, the same molecular targets are induced by CB1 activation in the hypothalamus, where inhibition of FAS has been previously shown to result in profound anorexia (21, 22, 25). Thus, these findings suggest that the same molecular pathway is involved in both the central appetitive and the peripheral anabolic effects of cannabinoids.
In agreement with a recent report (16), CB1-deficient mice were resistant to diet-induced obesity despite the fact that their overall caloric intake was similar to that in wild-type mice, which strongly suggests the existence of peripheral metabolic targets of endocannabinoids. We present several lines of evidence that endocannabinoids directly target the liver to stimulate fat synthesis and document, for the first time to our knowledge, the presence of CB1 in hepatocytes by using a combination of techniques including RT-PCR, in situ hybridization, immunohistochemistry, and Western blotting. The latter technique necessitated the use of purified plasma membranes owing to the low density of hepatic CB1, which may explain earlier failures to detect CB1 using crude liver homogenates (28).
In an earlier study, anandamide was reported to decrease rather than increase fatty acid synthesis in rat hepatocytes through a noncannabinoid mechanism mediated by arachidonic acid, as indicated by the ability of the nonspecific FAAH inhibitor PMSF to block this effect (29). Although in that study PMSF did not unmask a CB1-mediated stimulatory response to anandamide (29), this could be due to the weak partial agonist properties of anandamide and/or its rapid elimination from the medium. When suspended at high cell-to-medium ratios, hepatocytes have been shown to rapidly deplete drugs from the medium by uptake and metabolism, an effect that can be minimized by using low cell-to-medium ratios similar to those used in the current study (30).
The lipogenic response to CB1 activation in isolated hepatocytes argues strongly for a direct hepatic effect under in vivo conditions, although it does not rule out an additional, centrally mediated effect through neuronal or hormonal pathways. Inhibition of this lipogenic response by SR141716 and its absence in CB1–/– mice further confirms the lipogenic role of hepatic CB1. SR141716 alone reduced basal levels of fatty acid synthesis, which were similar in CB1+/+ and CB1–/– mice. This could suggest the presence of an endocannabinoid tone, the absence of which may be compensated when CB1 is absent from birth. We also demonstrate that activation of CB1 induces the expression of the lipogenic transcription factor SREBP-1c and its target enzymes ACC1 and FAS, which likely account for the CB1-mediated increase in fatty acid synthesis. CB1 is negatively coupled to adenylate cyclase via Gi/Go (31), and SREBP-1c and FAS gene expression are negatively regulated by cAMP (32). This suggests that CB1 may regulate lipogenic gene expression via inhibition of adenylate cyclase.
The ability of exogenous cannabinoids to stimulate hepatic fatty acid synthesis does not, in itself, prove that CB1 signaling contributes to diet-induced obesity. This is supported by the present findings that the increase in de novo lipogenesis in mice on a high-fat diet can be inhibited by a CB1 antagonist. Obesity induced by high-fat diets is usually associated with a decrease in de novo fatty acid synthesis. However, in the short term, high-fat diets can induce lipogenic gene expression and fatty acid synthesis (33). Even after prolonged exposure to a high-fat diet, increased de novo fatty acid synthesis can be maintained through the hyperinsulinemia present in the insulin-resistant obese animals (ref. 34, see also Figure 4), as insulin is a known inducer of SREBP-1c and lipogenic genes (35). Furthermore, such a diet-induced increase in de novo fatty acid synthesis appears to be selective for the liver (36, 37). In the current study we show that exposure of wild-type mice to a high-fat diet for 3 weeks, which is not sufficient to induce obesity, results in a marked increase in the basal rate of hepatic fatty acid synthesis as well as in the development of hepatic steatosis. The findings that the increased rate of hepatic fatty acid synthesis is blunted by SR141716 (Figure 6A) and is absent in CB1–/– mice (Figure 6B) indicate tonic activation of hepatic CB1 in wild-type mice, most likely mediated through the parallel increase in the hepatic levels of both anandamide (Figure 6B) and CB1 (Figure 3D). This mechanism likely contributes to the development of not only obesity but also fatty liver in control mice, as indicated by the absence of both changes in CB1–/– mice on a high-fat diet (Figure 4B and Figure 5). More importantly, these findings strongly suggest that a critical component in diet-induced obesity is an early, endocannabinoid-mediated increase in de novo lipogenesis in the liver, and raise the possibility that CB1 antagonists may be effective not only as antiobesity agents, but also in preventing/reversing the development of fatty liver.
With caloric intake being similar in wild-type and CB1–/– mice on the high-fat diet, there must be a difference in their energy expenditure to account for the difference in total body fat. Feeding a high-fat diet to C57Bl6 mice has been shown to decrease energy expenditure, as documented by indirect calorimetry (38), and the increase in feed efficiency we observed in such animals is in agreement with such a change. Although the underlying mechanism has not been analyzed in detail, the parallel decline in the plasma levels of adiponectin (Figure 4), a hormone that promotes fatty acid β-oxidation, is likely to be involved. In CB1–/– mice, feed efficiency as well as plasma adiponectin levels remained unaffected by the high-fat diet, suggesting that energy expenditure was not similarly reduced. Furthermore, ongoing experiments indicate that exposure to the high-fat diet for 14 weeks results in a significant increase in the expression of carnitine palmitoyltransferase-1, the rate-limiting enzyme in fatty acid β-oxidation, in CB1–/– but not in wild-type mice, as detected by Western blotting.
The present findings indicate that a high-fat diet increases hepatic anandamide owing to a major reduction in its degradation by FAAH, whereas anandamide synthesis appears to be unchanged. Because the membrane levels of the FAAH protein are not significantly altered, a high-fat diet may inhibit the activity rather than the expression of FAAH by a mechanism that remains to be determined. The upregulation of hepatic CB1 observed in mice on the high-fat diet is reminiscent of the reported upregulation of CB1 expression in adipose tissue from genetically obese versus lean rats (18) and could involve similar underlying mechanisms.
Lipid metabolism in selective hypothalamic neurons acts as a primary sensor of nutrient availability (39), and its modulation by enzyme inhibitors has been found to affect appetitive behavior (21, 22, 25). Here we demonstrate that the fatty acid synthetic pathway in the hypothalamus is regulated by endocannabinoids via CB1, which is known to be involved in the control of appetite. Fasting/refeeding unmasks the orexigenic effects of endocannabinoids (4, 9), which is in agreement with the present findings that SR141716 inhibited food intake in fasted/refed mice but not in mice with free access to food. Similarly, SR141716 treatment inhibited the hypothalamic expression of SREBP-1c and FAS in fasted/refed mice (Figure 7B) but not in free-feeding mice (Figure 7A). This suggests that lipogenic gene expression in the hypothalamus is linked to the control of food intake, although the nature of this relationship and its possible presence in other brain regions implicated in the appetitive effects of endocannabinoids (11–13) remain to be explored. Nevertheless, the present findings provide evidence for a common molecular pathway involved in central appetitive and peripheral metabolic regulation by an endogenous signaling system, thus providing a molecular basis for the coordinated regulation of nutrient intake and energy metabolism.
Methods
Mice.
All animal experiments conformed to NIH guidelines and were approved by the Institutional Animal Care and Use Committee of the National Institute on Alcohol Abuse and Alcoholism. CB1–/– mice and their wild-type littermates were developed and backcrossed to a C57Bl/6J background as previously described (40). For experiments not involving CB1–/– mice, wild-type C57Bl/6J mice were obtained from The Jackson Laboratory. To induce obesity, 8- to 10-week-old mice of both sexes were fed a diet containing 33.5% fat (60% of calories), 26.5% carbohydrate, and 27.4% protein (TD97070; Harlan Teklad) for 14 weeks. Controls received regular chow (NIH-31 rodent diet). CB1–/– mice had slightly lower body weights than their wild-type littermates (Figure 1B). The mice had free access to food and water and were maintained on a 12-hour light/12-hour dark cycle. Food intake and body weight were measured daily. Feed efficiency was calculated as milligrams of body weight gain per kilocalorie of food eaten over the 14-week diet period. For fasting/refeeding experiments, mice were placed on a low-carbohydrate diet (5789C; Purina Mills) for 3 days, followed by 24 hours of fasting and re-exposure to a high-carbohydrate diet (88122; Harlan Teklad) or continued fasting for 3 hours. Mice were sacrificed by cervical dislocation. The liver and combined adipose tissue (inguinal, retroperitoneal, and subcutaneous fat pads) were removed, weighed, and snap-frozen, and blood was collected for determining endocrine and biochemical parameters.
Blood chemistry.
Blood was collected at the time the mice were sacrificed. Serum triglycerides and lipoprotein lipase activity were determined using a clinical chemistry analyzer system (PROCHEM-V; Drew Scientific). Serum leptin (Quantikine M; R&D Systems) and adiponectin levels were determined using commercial sandwich ELISA assays (B-Bridge International) in accordance with the manufacturer’s instructions. Serum insulin was determined using the Ultrasensitive Mouse Insulin EIA kit (ALPCO Diagnostics).
Endocannabinoids.
For measuring endocannabinoid levels, mice were sacrificed by decapitation and their livers were removed and extracted (41). Anandamide and 2-AG levels were determined by liquid chromatography/mass spectrometry as previously described (41).
Anandamide synthesis.
The activity of NAT, the rate-limiting step in anandamide synthesis, was quantified in liver homogenates from mice on normal and high-fat diets by measuring the conversion of 1,2-phosphatidylcholine-L-α-[1-14C]diarachidonoyl to the anandamide precursor NAPE. Details of the assay, including separation of the lipids by thin-layer chromatography, have been described previously (42).
Anandamide degradation.
FAAH activity in liver homogenates was quantified by the amount of [3H]ethanolamine released from [3H]anandamide labeled on the ethanolamine moiety as previously described (42).
Immunohistochemistry and histology.
Affinity-purified CB1 primary polyclonal antibody, raised in rabbits against a 14–amino acid peptide near the N terminus of the human CB1 (CB11-A), was obtained from Alpha Diagnostic International. Formalin-fixed, paraffin-embedded 10-μm-thick liver sections were immunostained with primary and biotinylated secondary antibody and visualized using the VECTASTAIN Elite ABC kit as described by the manufacturer (Vector Laboratories). Specificity of the reaction was controlled by preabsorption of the antibodies with 1–10 μg/ml of the immunizing peptide and by doing parallel immunostaining in liver sections from CB1–/– mice. For analysis of fat accumulation in liver, the tissue was fixed in 10% formalin, and 10-μm-thick frozen sections prepared using a cryostat were stained with Oil Red O (Vector Laboratories), counterstained with Mayer’s haematoxylin, and analyzed by light microscopy.
In situ hybridization histochemistry.
A CB1 cDNA clone ligated into the pcDNA3 vector (43) was used to produce DIG-11-dUTP-labeled cRNA probe (Roche Diagnostics Corp.). In situ hybridization was performed on paraffin-embedded tissue sections as described previously (44). CB1 mRNA transcript was detected by reaction with anti-DIG antibody coupled to alkaline phosphatase using NBT/BCIP as substrate (Roche Diagnostics Corp.). Methods for creating negative controls included: (a) replacement of the specific antisense probe with a sense probe, (b) omission of either the RNA probe or the anti-DIG antibody, and (c) pretreatment of tissue sections with RNase.
Western hybridization.
Purified plasma membranes were isolated from mouse liver by sucrose density gradient centrifugation (45). This preparation is enriched 10-fold in membrane proteins, as indicated by an approximately 10-fold increase in the number of α1-adrenergic receptor binding sites per milligram of protein, measured by [3H]prazosin binding (G. Kunos, unpublished observations). Two hundred micrograms of membrane protein was solubilized in lysis buffer, size-fractionated by 10% SDS-PAGE, and transblotted to a nitrocellulose membrane, as previously described (46). Western blotting using rabbit antibody raised against the C terminus of the rat CB1 receptor or a rabbit antibody against human FAAH (Alpha Diagnostics) was done as previously described (46).
RT-PCR analyses.
Total RNA was isolated from liver and adipose tissue using TRIZOL and reverse-transcribed using the SuperScript First-Strand Synthesis System, in accordance with the manufacturer’s instructions (Invitrogen Corp.). The resulting single-stranded cDNA (5 μl) was denatured at 94°C for 5 minutes and, after the addition of the polymerase, subjected to 35 cycles of amplification, each consisting of 15 seconds at 94°C, 30 seconds at 58°C, and 1 minute at 68°C, with a 30-second final extension at 68°C during the last cycle. Each PCR reaction mixture (100 μl) contained the cDNA template, 1 μM of the primers, 200 μM of dNTPs, 1.5 mM MgCl2, 10 mM Tris/HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, and 2.5 μM Taq polymerase (Invitrogen Corp.). The following forward and reverse primers were used, with the size of the amplicon in brackets (the same primers were used to generate the double-stranded oligonucleotides used as probes in Northern blots): SREBP-1c, 5′-CTCCAAGGTTTCGTCTGACG-3′, 5′-TCCAGTGGCAAAGAAACACC-3′ (447 nt); FAS, 5′-TACCAGTGCCACAGGAGTCTCA-3′, 5′-TAAACACCTCGTCGATTTCGTTC-3′ (158 nt); ACC1, 5′-GAAGTGAAACGGAAACCCTC-3′, 5′-ATTTGGTCGTGAGGGCTAAG-3′ (426 nt). CB1: 5′ GTACCATCACCACAGACCTCCTC 3′, 5′ GGATTCAGAATCATGAAGCACTCCA 3′ (300). The mouse β-actin gene was amplified as a control. The PCR products were separated by electrophoresis on a 1% agarose gel. RNA without reverse transcriptions did not yield any amplicons, indicating the absence of genomic DNA contamination.
Real-time quantitative RT-PCR.
Estimates of cannabinoid-induced changes in SREBP-1c and FAS mRNA levels in the hypothalamus obtained by standard RT-PCR (see above) were confirmed by real-time quantitative PCR, using a model 7700 Sequence Detection System (Applied Biosystems Inc.). Details of the method and calculations of the fold increases over control were as previously described (47).
Northern hybridization.
Total RNA was prepared from individual mouse livers as described above. Northern analysis was performed in accordance with an established protocol (48). The signal was detected using the CDPstar chemiluminescence assay (Amersham Biosciences). Oligonucleotide probes for the respective genes were labeled using the AlkPhos-Direct kit in accordance with the manufacturer’s instructions (Amersham Biosciences). mRNA levels were quantified by densitometry, using the 18S ribosomal RNA band for loading control. Values represent the percentage of the levels expressed by vehicle-treated controls.
Hepatic fatty acid synthesis in vivo and in vitro.
Rates of in vivo hepatic fatty acid synthesis were measured as previously described (49). Briefly, following an overnight fast, mice were injected i.p. with vehicle, 20 ng/g HU210, 3 μg/g SR141716, or 20 ng/g HU210 plus 3 μg/g SR141716. One hour later, each animal received an intrahepatic injection of 50 mCi of 3H2O in saline and, following an additional hour, the animals were sacrificed, the liver removed and homogenized, and fatty acids extracted with petroleum ether and quantified by liquid scintillation spectrometry. To analyze the effects of cannabinoids on fatty acid synthesis in vitro, hepatocytes were isolated from wild-type mice as previously described (50). Cells (5–7 × 106 cells/25 ml flask) were suspended in DMEM at 37°C under 5% CO2 in O2. 3H2O (0.5 mCi/ml) was added to the cell suspension and incubated for a further 60 minutes. The incubation was terminated by the addition of chloroform/methanol (1:1 vol/vol), and lipids were extracted as previously described (51). Total lipids were saponified at 75°C for 2 hours with 0.3 M NaOH in 90% (vol/vol) methanol. The saponifiable fractions were extracted with petroleum ether. Aliquots were placed onto Whatman filter paper discs and evaporated to dryness. Radioactivity on the filters was quantified by liquid scintillation spectrometry. Lipogenesis was expressed as micromoles of 3H2O incorporated into fatty acids per 108 cells (52).
Acknowledgments
We thank A. Zimmer for providing breeding pairs of CB1-deficient mice and R.L. Veech for helpful comments.
Footnotes
See the related Commentary beginning on page 1130.
Nonstandard abbreviations used: 2-AG, 2-arachidonoylglycerol; ACC1, acetyl coenzyme-A carboxylase-1; FAAH, fatty acid amidohydrolase; FAS, fatty acid synthase; NAPE, N-[14C]arachidonoyl phosphatidylethanolamine; NAT, N-acyltransferase.Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Seeley RJ, Woods SC. Monitoring of stored and available fuel by the CNS: implications for obesity. Nat. Neurosci. 2003;4:901–909. doi: 10.1038/nrn1245. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P et al. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science. 2002;297:240–243. doi: 10.1126/science.1071527. [DOI] [PubMed] [Google Scholar]
- 3.Williams CM, Kirkham TC. Anandamide induces overeating: mediation by central cannabinoid (CB1) receptors. Psychopharmacology (Berl). 1999;143:315–317. doi: 10.1007/s002130050953. [DOI] [PubMed] [Google Scholar]
- 4.Di Marzo V, et al. Leptin-regulated endocannabinoids are involved in maintaining food intake. Nature. 2001;410:822–825. doi: 10.1038/35071088. [DOI] [PubMed] [Google Scholar]
- 5.Hildebrandt AL, Kelly-Sullivan DM, Black SC. Antiobesity effects of chronic cannabinoid CB1 receptor anntagonist treatment in diet-induced obese mice. Eur. J. Pharmacol. 2003;462:125–132. doi: 10.1016/s0014-2999(03)01343-8. [DOI] [PubMed] [Google Scholar]
- 6.Vickers SP, Webster LJ, Wyatt A, Dourish CT, Kennett GA. Preferential effects of the cannabinoid CB1 receptor antagonist, SR141716, on food intake and body weight gain of obese (fa/fa) compared to lean Zucker rats. Psychopharmacology (Berl). 2003;167:103–111. doi: 10.1007/s00213-002-1384-8. [DOI] [PubMed] [Google Scholar]
- 7.Gómez R, et al. A peripheral mechanism for CB1 cannabinoid receptor-dependent modulation of feeding. J. Neurosci. 2002;22:9612–9617. doi: 10.1523/JNEUROSCI.22-21-09612.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedland CS, Poston JS, Porrino LJ. Effects of SR141716A, a central cannabinoid receptor antagonist, on food-maintained responding. Pharmacol. Biochem. Behav. 2000;67:265–270. doi: 10.1016/s0091-3057(00)00359-2. [DOI] [PubMed] [Google Scholar]
- 9.Kunos G, Osei-Hyiaman D, Wang L, Liu J, Bátkai S. Endocannabinoids: novel lipid mediators in the neural control of appetite. In Progress in obesity research. G Medeiros-Neto, A Halpern, and C Bouchard, editors John Libbery Eurotext Surrey, United Kingdom. 2002;9:826–828. [Google Scholar]
- 10.Jamshidi N, Taylor DA. Anandamide administration into the ventromedial hypothalamus stimulates appetite in rats. Br. J. Pharmacol. 2001;134:1151–1154. doi: 10.1038/sj.bjp.0704379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkham TC, Williams CM, Fezza F, Di Marzo V. Endocannabinoid levels in rat limbic forebrain and hypothalamus in relation to fasting, feeding and satiation: stimulation of eating by 2-arachidonoyl glycerol. Br. J. Pharmacol. 2002;136:550–557. doi: 10.1038/sj.bjp.0704767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freedland CS, Whitlow CT, Smith HR, Porrino LJ. Functional consequences of the acute administration of the cannabinoid receptor antagonist, SR141716A, in cannabinoid-naïve and –tolerant animals: a quantitative 2-[14C]deoxyglucose study. Brain Res. 2003;962:169–179. doi: 10.1016/s0006-8993(02)03999-9. [DOI] [PubMed] [Google Scholar]
- 13.Harrold JA, Elliott JC, King PJ, Widdowson PS, Williams G. Down-regulation of cannabinoid CB1 receptors in specific extrahypothalamic regions of rats with dietary obesity: a role for endogenous cannabinoids in driving appetite for palatable food? Brain Res. 2002;952:232–238. doi: 10.1016/s0006-8993(02)03245-6. [DOI] [PubMed] [Google Scholar]
- 14.Vastag B. Experimental drugs take aim at obesity. JAMA. 2003;289:1763–1764. doi: 10.1001/jama.289.14.1763. [DOI] [PubMed] [Google Scholar]
- 15.Ravinet Trillou C, et al. Anti-obesity effect of SR141716, a CB1 receptor antagonist, in diet-induced obese mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;284:R345–R353. doi: 10.1152/ajpregu.00545.2002. [DOI] [PubMed] [Google Scholar]
- 16.Ravinet Trillou C, Delgorge C, Menet C, Arnone M, Soubrie P. CB1 cannabinoid receptor knockout in mice leads to leanness, resistance to diet-induced obesity and enhanced leptin sensitivity. Int. J. Obes. 2004;28:640–648. doi: 10.1038/sj.ijo.0802583. [DOI] [PubMed] [Google Scholar]
- 17.Cota D, et al. The endogenous cannabinoid system affects energy balance via central orexigenic drive and peripheral lipogenesis. J. Clin. Invest. 2003;112:423–431. doi:10.1172/JCI200317725. doi: 10.1172/JCI17725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bensaid M, et al. The cannabinoid CB1 receptor antagonist SR141716 increases Acrp30 mrNA expression in adipose tissue of obese fa/fa rats and in cultured adipocyte cells. Mol. Pharmacol. 2003;63:908–914. doi: 10.1124/mol.63.4.908. [DOI] [PubMed] [Google Scholar]
- 19.Diraison F, et al. Differences in the regulation of adipose tissue and liver lipogenesis by carbohydrates in humans. J. Lipid Res. 2003;44:846–853. doi: 10.1194/jlr.M200461-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Brown MS, Goldstein JL. Sterol regulatory element binding proteins (SREBPs): controllers of lipid synthesis and cellular uptake. Nutr. Rev. 1998;56:S1–S3. doi: 10.1111/j.1753-4887.1998.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 21.Loftus TM, et al. Reduced food intake and body weight in mice treated with fatty acid synthase inhibitors. Science. 2000;288:2379–2381. doi: 10.1126/science.288.5475.2379. [DOI] [PubMed] [Google Scholar]
- 22.Kim E-K, et al. Expression of FAS within hypothalamic neurons: a model for decreased food intake after C75 treatment. Am. J. Physiol. Endocrinol. Metab. 2002;283:E867–E879. doi: 10.1152/ajpendo.00178.2002. [DOI] [PubMed] [Google Scholar]
- 23.Di Marzo V, De Petrocellis L, Bisogno T, Melck D. Metabolism of anandamide and 2-arachidonoylglycerol: an historic overview and some recent developments. Lipids. 1999;34:S319–S325. doi: 10.1007/BF02562332. [DOI] [PubMed] [Google Scholar]
- 24.Cravatt BF, et al. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9371–9376. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z, Cha SH, Chohnan S, Lane MD. Hypothalamic malonyl-CoA as a mediator of feeding behavior. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12624–12629. doi: 10.1073/pnas.1834402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang G, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 27.Paulauskis JD, Sul HS. Cloning and expression of mouse fatty acid synthase and other specific mRNA. Developmental and hormonal regulation in 3T3-L1 cells. J. Biol. Chem. 1988;263:7049–7054. [PubMed] [Google Scholar]
- 28.Porcella A, et al. Evidence for functional CB1 cannabinoid receptor expressed in the rat thyroid. Eur. J. Endocrinol. 2002;147:255–261. doi: 10.1530/eje.0.1470255. [DOI] [PubMed] [Google Scholar]
- 29.Guzman M, Fernandez-Ruiz JJ, Sanchez C, Velasco G, Ramos JA. Effects of anandamide on hepatic fatty acid metabolism. Biochem. Pharmacol. 1999;50:885–888. doi: 10.1016/0006-2952(95)00198-9. [DOI] [PubMed] [Google Scholar]
- 30.Kunos G. The hepatic α1-adrenoceptor. Trends Pharmacol. Sci. 1984;5:380–383. [Google Scholar]
- 31.Howlett AC. Cannabinoid inhibition of adenylate cycalse. Biochemistry of the response in neuroblastoma cell membranes. Mol. Pharmacol. 1985;27:429–436. [PubMed] [Google Scholar]
- 32.Paulauskis JD, Sul HS. Hormonal regulation of mouse fatty acid synthase gene transcription in liver. J. Biol. Chem. 1989;264:574–577. [PubMed] [Google Scholar]
- 33.Gregoire FM, et al. Diet-induced obesity and hepatic gene expression alterations in C57Bl/6J and ICAM-1-deficient mice. Am. J. Physiol. Endocrinol. Metab. 2002;282:E703–E713. doi: 10.1152/ajpendo.00072.2001. [DOI] [PubMed] [Google Scholar]
- 34.Becker W, et al. Differential hepatic gene expression in a polygenic mouse model with insulin resistance and hyperglycemia: evidence for a combined transcriptional dysregulation of gluconeogenesis and fatty acid synthesis. J. Mol. Endocrinol. 2004;32:195–208. doi: 10.1677/jme.0.0320195. [DOI] [PubMed] [Google Scholar]
- 35.Shimomura I, et al. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillgartner FB, Charron T, Chesnut KA. Alterations in nutritional status regulate acetyl-CaA carboxylase expression in avian liver by a transcriptional mechanism. Biochem. J. 1996;319:263–268. doi: 10.1042/bj3190263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diraison F, Dusserre E, Vidal H, Sothier M, Beylot M. Increased hepatic lipogenesis but decreased expression of lipogenic gene in adipose tissue in human obesity. Am. J. Physiol. Endocrinol. Metab. 2002;282:E46–E51. doi: 10.1152/ajpendo.2002.282.1.E46. [DOI] [PubMed] [Google Scholar]
- 38.Hu CC, Qing K, Chen Y. Diet-induced changes in stearoyl-CoA desaturase 1 expression in obesity-prone and –resistant mice. Obes. Res. 2004;12:1264–1270. doi: 10.1038/oby.2004.160. [DOI] [PubMed] [Google Scholar]
- 39.Obici S, Feng Z, Arduini A, Conti R, Rossetti L. Inhibition of hypothalamic carnitine pelmitoyltransferase-1 decreases food intake and glucose production. Nat. Med. 2003;9:756–761. doi: 10.1038/nm873. [DOI] [PubMed] [Google Scholar]
- 40.Zimmer A, Zimmer AM, Hohmann AG, Herkenham M, Bonner TI. Increased mortality, hypoactivity, and hypoalgesia in cannabinoid CB1 receptor knockout mice. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5780–5785. doi: 10.1073/pnas.96.10.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Liu J, Harvey-White J, Zimmer A, Kunos G. Endocannabinoid signaling via cannabinoid receptor 1 is involved in ethanol preference and its age-dependent decline in mice. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1393–1398. doi: 10.1073/pnas.0336351100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu J, et al. Lipopolysaccharide induces anandamide synthesis in macrophages via CD14/MAPK/phosphoinositide 3-kinase/NF-κB independently of platelet-activating factor. J. Biol. Chem. 2003;278:45034–45039. doi: 10.1074/jbc.M306062200. [DOI] [PubMed] [Google Scholar]
- 43.Abood ME, Ditto KE, Noel MA, Showalter VM, Tao Q. Isolation and expression of a mouse CB1 cannabinoid receptor gene. Comparison of binding properties with those of native CB1 receptors in mouse brain and N18TG2 neuroblastoma cells. Biochem. Pharmacol. 1997;53:207–214. doi: 10.1016/s0006-2952(96)00727-7. [DOI] [PubMed] [Google Scholar]
- 44.Radaeva S, Ferreira-Gonzalez A, Sirica AE. Overeexpression of c-neu and c-met during rat liver cholangiocarcinogenesis: a link between biliary intestinal metaplasia and mucin-producing cholangiocarcinoma. Hepatology. 1999;29:1453–1462. doi: 10.1002/hep.510290524. [DOI] [PubMed] [Google Scholar]
- 45.Neville DM., Jr Isolation of an organ specific protein antigen from cell-surface membrane of rat liver. Biochim. Biophys. Acta. 1968;154:540–552. doi: 10.1016/0005-2795(68)90014-7. [DOI] [PubMed] [Google Scholar]
- 46.Batkai S, et al. Endocannabinoids acting at cannabinoid-1 receptors regulate cardiovascular function in hypertension. Circulation. 2004;110:1996–2002. doi: 10.1161/01.CIR.0000143230.23252.D2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Lay S, Lefrere I, Trautwein C, Dugail I, Krief S. Insulin and sterol-regulatory element-binding protein-1c (SREBP-1c) regulation of gene expression in 3T3-L1 adipocytes. J. Biol. Chem. 2002;277:35625–35634. doi: 10.1074/jbc.M203913200. [DOI] [PubMed] [Google Scholar]
- 48.Shimizu T, Yaguchi H, Ohtani K, Banu S, Hayashi H. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol. Microbiol. 2002;43:257–265. doi: 10.1046/j.1365-2958.2002.02743.x. [DOI] [PubMed] [Google Scholar]
- 49.Shimano H, et al. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorenzo M, Roncero C, Benito M. The role of prolactin and progesterone in the regulation of lipogenesis in maternal and foetal rat liver in vivo and in isolated hepatocytes during the last day of gestation. Biochem. J. 1986;239:135–139. doi: 10.1042/bj2390135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundler R, Akesson B. Regulation of phospholipid biosynthesis in isolated rat hepatocytes. Effect of different substrates. J. Biol. Chem. 1975;250:3359–3367. [PubMed] [Google Scholar]
- 52.Harris RA. Studies on the inhibition of hepatic lipogenesis by N-6,O-2’-dibutyryl adenosine 3′,5′-monophosphate. Arch. Biochem. Biophys. 1975;169:168–180. doi: 10.1016/0003-9861(75)90330-6. [DOI] [PubMed] [Google Scholar]