Abstract
Introduction
Little is known regarding genetic factors associated with treatment outcome of psychotic depression. We explored genomic associations of remission and relapse of psychotic depression treated with pharmacotherapy.
Methods
Genomic analyses were performed in 171 men and women aged 18–85 years with an episode of psychotic depression who participated in the Study of the Pharmacotherapy of Psychotic Depression II (STOP-PD II). Participants were treated with open-label sertraline plus olanzapine for up to 12 weeks; those who achieved remission or near-remission and maintained it following 8 weeks of stabilization were eligible to participate in a 36-week randomized controlled trial that compared sertraline plus olanzapine with sertraline plus placebo in preventing relapse.
Results
There were no genome-wide significant associations with either remission or relapse. However, at a suggestive threshold, SNP rs1026501 (31 kb from SYNPO2) in the whole sample and rs6844137 (within the intronic region of SYNPO2) in the European ancestry subsample were associated with a decreased likelihood of remission. In polygenic risk analyses, participants who had greater improvement after antidepressant treatments showed a higher likelihood of reaching remission. Those who achieved remission and had a higher polygenic risk for Alzheimer’s disease had a significantly decreased likelihood of relapse.
Conclusion
Our analyses provide preliminary insights into the genetic architecture of remission and relapse in a well-characterized group of patients with psychotic depression.
Keywords: Genome-wide association study, Psychotic depression, Polygenic risk score
Introduction
Major depressive disorder with psychotic features (MDDPsy) is a severe, disabling disorder with high morbidity that presents challenges to treatment [1, 2]. Compared with major depressive disorder (MDD) without psychotic features, MDDPsy has greater severity, longer episodes, increased incapacity, and a higher risk of relapse [3]. MDDPsy affects approximately four of 1,000 individuals and accounts for up to 20% of individuals with MDD [4, 5]. Clinical studies have shown that the combination pharmacotherapy of antidepressant and antipsychotic is more efficacious in MDDPsy treatment than antidepressant or antipsychotic alone [5, 6]. Moreover, relapse should be considered in MDDPsy treatment following remission since early discontinuation of medication might have a higher risk of relapse. For example, in patients who have experienced remission of MDDPsy, sertraline alone was associated with a higher relapse rate compared with the combination pharmacotherapy of sertraline and olanzapine [7]. Therefore, exploring biomarkers for treatment outcomes in MDDPsy would allow for some precision in deciding which patients can benefit from the combination pharmacotherapy and which patients can be safely withdrawn from antipsychotic treatment after MDDPsy remission.
There is strong evidence that genetic factors contribute to response to depression pharmacotherapy. Common genetic variants with small additive effects may account for up to 40% of the inter-individual variation in response to antidepressant medication [8]. While research into the pharmacogenetics of antidepressant response is impeded by the small sample size of treatment studies, there is now an emphasis on publishing data that can be pooled to increase statistical power to detect effects [9]. As such, pharmacogenetic analyses offer an opportunity to identify key biomarkers of treatment response and to understand response mechanisms.
There has been very little research on genetic factors associated with the treatment outcome of MDDPsy. In an earlier candidate gene study in 102 individuals diagnosed with MDDPsy, the homozygous short variant of the serotonin transporter long polymorphic region (LPR; SLC6A4 5 LPR) was associated with a poor response of MDDPsy to fluvoxamine [10]. However, a subsequent investigation from the same group showed no difference across genotypes between MDDPsy (N = 52) and nonpsychotic (N = 103) depression between SLC6A4 short variant carriers [11]. In another earlier study, Arias et al. [12] reported that the A-allele of tryptophan hydroxylase 1 (TPH1) rs1800532 was associated with a worse response of MDDPsy to citalopram. Methodological factors and a small sample size limit the initial findings of these studies: psychotic features were diagnosed on the basis of an unstructured clinical interview and the treatment regimen did not include an antipsychotic, although a combination of an antidepressant and an antipsychotic is recommended for the acute treatment of MDDPsy [13].
To our knowledge, there have been no genome-wide association studies (GWASs) of treatment outcomes of MDDPsy. In addition, we are not aware of any studies that examined genetic associations of relapse of remitted MDD, either with or without psychotic features. In this exploratory study, we present the findings of GWAS analyses of remission and relapse of a well-characterized group of younger and older adults with MDDPsy treated in the Study of the Pharmacotherapy of Psychotic Depression II (STOP-PD II) [7] clinical trial. Furthermore, we also examined whether remission and relapse were associated with polygenic risks for disorders of potential relevance to MDDPsy.
Materials and Methods
Participants
The design and methods of STOP-PD II (ClinicalTrials.gov Identifier: NCT01427608) have been previously described [14]. The study was conducted at four sites (University Health Network, Toronto; University of Massachusetts Medical School; University of Pittsburgh School of Medicine; and Weill Cornell Medicine). Using procedures approved by the Local Institutional Review Boards, written informed consent was obtained from all participants or their substitute decision-maker before initiating any research procedures. Additional separate written informed consent was obtained for participation in the genetic component of STOP-PD II which is the focus of this article.
The STOP-PD II study comprised three phases: an acute phase lasting up to 12 weeks, followed by an 8-week stabilization phase, and finally a 36-week randomized controlled trial (RCT) phase. At the time of enrollment in the acute phase of the study, participants were between 18 and 85 years old, met the Diagnostic and Statistical Manual of Mental Disorders (Fourth Edition, Text Revision) (DSM-IV-TR) criteria rated by the Structured Clinical Interview for DSM-IV (SCID) for a current major depressive episode with at least one associated delusion (with or without hallucinations), and had a 17-item Hamilton Depression Rating Scale (Ham-D17) total score ≥21 [15, 16]. Exclusion criteria included DSM-IV-TR criteria for lifetime bipolar disorder, any other psychotic disorder, or intellectual disability; current body dysmorphic disorder or obsessive-compulsive disorder; substance abuse or dependence within the preceding 3 months; dementia preceding the index episode of depression or a 26-item IQCODE mean score ≥4 at acute phase baseline [17]; type I diabetes mellitus; a neurologic disease that might affect neuromuscular function; and unstable physical illness, although many of the study participants had stable chronic physical problems.
In the acute phase, participants received a combination of open-label sertraline (target dosage: 150–200 mg/day) plus open-label olanzapine (target dosage: 15–20 mg/day). Participants entered the stabilization phase as soon as they met the study’s criteria for remission or, failing that, met the criteria for near-remission at week 12 of the acute phase (see these criteria below). At the end of the stabilization phase, participants who still met remission or near-remission criteria following treatment with open-label sertraline plus olanzapine, and had a Mini-Mental State Examination (MMSE) score ≥24, were eligible for the RCT phase [18]. All participants continued to take open-label sertraline during the RCT phase; they were randomized under double-blind conditions to either continue olanzapine or switch from olanzapine to identically appearing placebo pills during a protocolized 4-week taper of olanzapine. During the RCT phase, participants were assessed weekly for the first 8 weeks and once every 4 weeks after that until they reached one of the study endpoints: study completion at RCT week 36, relapse, or early termination.
Outcomes
Remission Status
Remission was defined as the absence of delusions and hallucinations and a Ham-D17 total score ≤10 for two consecutive weeks. Near-remission at week 12 of the acute phase was defined as the absence of delusions and hallucinations, a Ham-D17 total score of 11–15 with ≥50% reduction from baseline, and being rated as very much or much improved on the Clinical Global Impression (CGI) scale [19]. Near-remission was included as an outcome to allow participants who experienced substantial improvement in depressive symptoms plus remission of psychosis to enter the RCT, even though the symptoms may not have reached the conventional Ham-D17 cut-point used in studies of nonpsychotic depression; this approach reflected “real-world” clinical practice. For our analysis, remission status included the combined remission and near-remission outcomes.
Relapse Status
Relapse was the primary outcome in the RCT phase. Relapse was defined as at least one of the following: (1) sufficient SCID-rated symptoms to meet criteria for a DSM-IV major depressive episode; (2) Ham-D17 total score ≥18; (3) SCID-rated psychosis (delusions or hallucinations); or (4) other significant clinical worsening (i.e., suicidal ideation or attempt, development of mania or hypomania, or psychiatric hospitalization). Although individuals with a history of bipolar disorder were not eligible for the study, mania and hypomania were included as an outcome because the onset of psychotic depression in younger adults may predict the subsequent development of bipolar disorder [20].
Processing of DNA
Whole blood of participants who consented to the collection of genetic material was sent directly to the NIMH Center for Collaborative Genetic Studies (“the Center”) at Rutgers University for the generation of cell lines and extraction of DNA. Blood samples were collected at the time of enrollment in STOP-PD II or as close as possible thereafter and sent to the center using the center’s established protocol designed to minimize DNA denaturation. Extracted DNA was stored at the center until completion of enrollment in STOP-PD II and then shipped to the Neurogenetics Laboratory at the Center for Addiction and Mental Health (CAMH), Toronto, for genotyping.
Genotyping, Quality Control, and Imputation
Blood samples for genetic analyses were collected from 179 STOP-PD II participants. However, the quality of two samples was too poor for analysis, and 177 participants were genotyped. We used the Illumina PsychArray BeadChip endorsed by the Psychiatric Genomics Consortium for genotyping. Using standard protocols, quality control was conducted in PLINK v.1.9[21–23]. Based on this quality control, 5 participants were excluded: one due to discordant self-reported and genetic sex and four due to excessive heterozygosity and missingness. With respect to the variants, we excluded 2,095 due to missingness, 108 due to violation of Hardy-Weinberg equilibrium, and 255,150 due to minor allele frequency less than 1%.
Subsequently, 172 individuals and 307,881 SNPs entered genome-wide imputation. Imputation was conducted using the genipe pipeline, which uses IMPUTE2 v2.2 in 5-Mb segments per chromosome after pre-phasing with SHAPEIT2 and the 1000 Genomes reference panel (phase 3) [24–27]. In total, the final imputed genome-wide data, including 8,300,065 variants, were imputed at an information threshold of 0.7 and a probability threshold of 90%, with an average completion rate of 98.8%. In the final sample, 4,261,750 (51.3%) had a minor allele frequency of ≥5% and were used for subsequent analyses (see online suppl. Fig. 1–7; for all online suppl. material, see www.karger.com/doi/10.1159/000529637).
We separately genotyped the serotonin transporter gene (SLC6A4) since it was not part of the PsychArray BeadChip. For the SLC6A4 LPR variant, we combined genomic DNA (40 ng) with MBI Fermentas PCR buffer (1X) containing (NH4)2SO4, MgCl2 (1.5 mM) (MBI Fermentas), forward primer labeled with 5’ HEX fluorescent tag (0.0325 µg) [28], dNTP (0.16 mM) (MBI Fermentas), and Taq polymerase (1 U) (MBI Fermentas), all totaling 25 μL. The PCR reactions began with a 3-min denaturation at 95°C, then 40 amplification cycles in an AB 2720 thermal cycler (Thermo Fisher Scientific, Burlington, ON), including denaturation at 95°C (30 s), annealing at 61°C (30 s), extension at 71°C (1 min), and final extension at 72°C (10 min). Subsequently, New England Biolabs Buffer 2 (1X) and MspI restriction enzyme (10 U) were added to the resulting PCR product (5 µL). The final mixture (30 µL) was digested overnight at 37°C and electrophoresed by an AB 3130-Avant Genetic Analyzer. Product sizes were calculated using GeneMapper 4.0 compared to GeneScan 500 ROX standards. For quality control, we duplicated 10% of samples from each run batch.
Data Analyses
Genome-Wide and Targeted Associations
We conducted genome-wide association analyses in PLINK v.1.9 for i) remission status and ii) relapse using logistic regressions [23]. Associations for remission status were adjusted for age, sex, acute phase baseline Ham-D17 total score and Schedule for Affective Disorder and Schizophrenia (SADS) delusion severity score, and the first three principal components from standard ancestry analysis. For relapse in the remitted/near-remitted group, we included sex, RCT phase baseline Ham-D17 total score, randomized treatment, and the first three principal components from ancestry analysis. We conducted associations across the whole mixed ancestry and the European ancestry subsample (N = 143, 83.6%). Given the small sample size of the African and other ancestries (N = 28, 16.4%), separate analyses were not performed.
For remission status, we also conducted gene-based analyses using MAGMA implanted in FUMA v1.3.5e with default parameters (SNP-wide mean model) [29, 30]. SNPs were assigned to genes within a 10 kb window based on a combined reference panel including 1000 Genomes phase 3 reference panels (2,504 individuals, approximately 84.8 million SNPs) and a subset of the UK Biobank data (10,000 individuals, approximately 17 million SNPs) [26]. The genome-wide significance was defined as 0.05/number of genes analyzed, which was 0.05/18,954 = 2.64 × 10−6.
Given the reported association of SLC6A4 with treatment outcome of nonpsychotic depression [31], we also conducted a targeted exploratory analysis of the association between SLC6A4 variants (LPR, rs25531) and (i) remission status and (ii) relapse. Similar to the genome-wide analysis, we explored the variants independently adjusting for the same covariates and the interaction between the LPR and rs25531.
Polygenic Risk Scoring
We conducted polygenic risk scoring (PRS) to evaluate the shared genetic contribution between i) remission of MDDPsy and ii) relapse of remitted MDDPsy and disorders of potential relevance to the outcome of MDDPsy. On our samples, we constructed 14 PRSs using PRSice-2 v.2.1 [32] for depression [33–35], schizophrenia [36], bipolar disorder [37], Alzheimer’s disease [38, 39], and stroke (as a proxy for cerebrovascular disease) [40], utilizing eight large genome-wide studies. The constructed scores were then evaluated for their association with (1) remission status using logistic regression analysis adjusted for age, sex, acute phase baseline Ham-D17 total score, SADS delusion severity score, and the first three principal components from standard ancestry analysis and (2) relapse status, where we included sex, RCT phase baseline Ham-D17 total score, randomized treatment, and the first three principal components from ancestry analysis as covariates. To adjust for potential overfitting, we conducted permutation testing (number of permutations = 10,000) to obtain empirical p values. Considering potential issues of population stratification, we restricted PRS analyses to the European ancestry subsample and used discovery cohorts with predominantly European ancestry contributions. The results were corrected for multiple testing with adjusted ɑ = 0.05/28 = 0.0018 (we accounted for each PRS as one study and accounted for both remission and relapse).
Results
Genome-Wide Associations
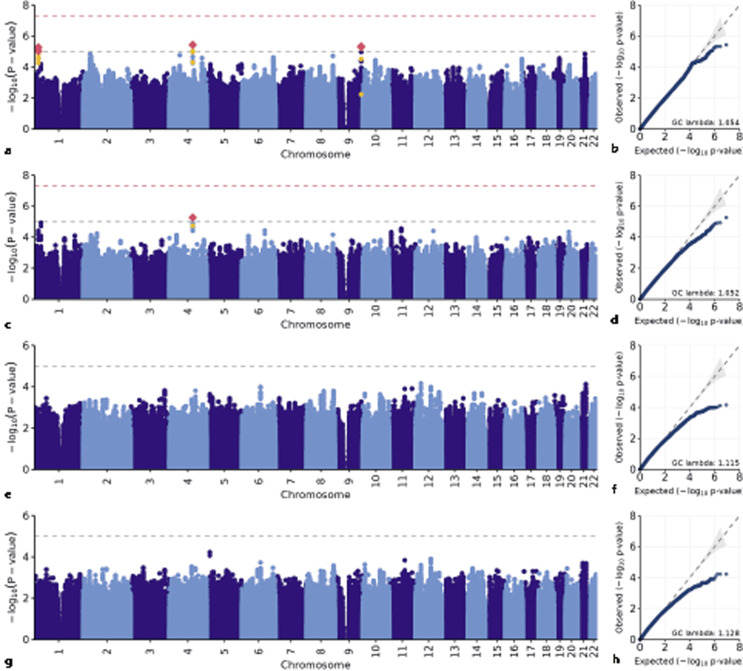
Of the 172 participants who passed quality control, one had incomplete covariate data for remission analyses and one had incomplete covariate data for relapse; therefore, statistical analyses were based on N = 171 for remission status and N = 87 for relapse status (shown in Table 1; online suppl. Table 1). After adjusting for the aforementioned covariates, there were no statistically significant genome-wide associations with remission status or relapse (shown in Fig. 1). With respect to remission status, the most strongly associated SNP in the whole sample was rs1026501 (30.9 kb from the 3’ of the SYNPO2 [synaptopodin 2] gene, OR = 0.21 [0.11, 0.40], p = 3.72 × 10−6, N = 171) and in the European ancestry subsample was rs6844137 (within the intronic region of the SYNPO2 gene, OR = 0.15 [0.07, 0.34], p = 5.36 × 10−6, N = 143) (shown in Table 2; online suppl. Fig. 8). With respect to relapse in the whole sample (N = 87), our top association was on chromosome 12 near the PTHLH (parathyroid hormone-like hormone) gene, where the genotyped variant rs7137085 was associated with a lower likelihood of relapse in the whole sample (OR = 0.08 [0.02, 0.27], p = 6.99 × 10−5, N = 87). In the European ancestry subsample, the top association rs154288 was located on chromosome 5, associated with a higher likelihood of relapse (within the intronic region of the MYO10 gene, OR = 73.74 [1.07, 603.28], p = 6.07 × 10−5, N = 73) (shown in Table 2). However, these two variants did not reach the suggestive association threshold of p = 10−5.
Table 1.
Characteristics of participants who entered the acute and RCT phases of the STOP-PD II clinical trial
| Acute phase | RCT phase | |||
|---|---|---|---|---|
| remission (N = 115) | non-remission (N = 56) | relapse (N = 34) | non-relapse (N = 53) | |
| Ethnic ancestry, N (%) | ||||
| European | 94 (81.7) | 49 (87.5) | 27 (79.4) | 46 (86.8) |
| African | 14 (12.2) | 3 (5.4) | 6 (17.6) | 2 (3.8) |
| Other | 7 (6.1) | 4 (7.1) | 1 (2.9) | 5 (9.4) |
| Sex, N (%) female | 76 (66.1) | 34 (60.7) | 25 (73.5) | 31 (58.5) |
| Age (mean [SD]) | 56.7 (14.8) | 55.8 (17.4) | 53.6 (15.5) | 57.2 (13.4) |
| Randomization, N (%) placebo | – | – | 23 (67.6) | 18 (34.0) |
| Acute baseline Ham-D17 total score (mean [SD]) | 28.7 (4.4) | 29.5 (5.3) | 29.8 (3.9) | 28.3 (4.6) |
| Acute baseline SADS delusion score (median [IQR]) | 5 [5, 6] | 6 [5, 6] | 5 [5, 6.75] | 5 [5, 6] |
| RCT baseline Ham-D17 total score (median [IQR]) | – | – | 7 [4, 8] | 4 [2, 6] |
| RCT baseline SADS delusion score (median [IQR]) | – | – | 1 [1, 1] | 1 [1, 1] |
Ham-D17, 17-item Hamilton Depression Rating Scale; SADS delusion, Schedule for Affective Disorders and Schizophrenia delusion severity score.
Fig. 1.
SNP-based genome-wide association results. SNP-based GWAS results for remission analyses in the whole sample (a, b), remission analyses in the European ancestry subsample (c, d), relapse analyses in the whole sample (e, f), and relapse analyses in the European ancestry subsample (g, h). In Manhattan plots, red diamond represents SNPs that reached suggestive significant threshold of p = 10−5, and yellow dot represents secondary clumped SNPs of the linkage disequilibrium threshold of 0.5.
Table 2.
Top associated variants for remission status and relapse within the whole sample and European ancestry subsample
| SNP | CHR | A1 | Gene | Type | Whole sample | |
|---|---|---|---|---|---|---|
| OR [95% CI] | p value | |||||
| Remission | Whole sample (N = 171) | |||||
| rs1026501a | 4 | G | SYNPO2 | ITR (30 kb) | 0.21 [0.11, 0.40] | 3.72 × 10−6 |
| rs12347401a | 9 | C | – | – | 0.11 [0.04, 0.28] | 4.62 × 10−6 |
| rs11243867a | 9 | T | – | – | 0.11 [0.04, 0.28] | 4.71 × 10−6 |
| rs12336064a | 9 | A | – | – | 0.11 [0.04, 0.28] | 5.07 × 10−6 |
| rs2745253a | 1 | A | TMCO4 | INT | 0.27 [0.15, 0.47] | 5.33 × 10−6 |
| Remission | European ancestry (N = 143) | |||||
| rs6844137 | 4 | T | SYNPO2 | INT | 0.15 [0.07, 0.34] | 5.36 × 10−6 |
| rs10493052 | 1 | A | ZSCAN20 | INT | 0.13 [0.05, 0.32] | 1.15 × 10−5 |
| rs16835507 | 1 | A | ZSCAN20 | SYN | 0.12 [0.05, 0.32] | 1.19 × 10−5 |
| rs62326864 | 4 | C | SYNPO2 | ITR (21 kb) | 0.18 [0.08, 0.39] | 1.25 × 10−5 |
| rs1026501a | 4 | G | SYNPO2 | ITR (30 kb) | 0.18 [0.09, 0.39] | 1.43 × 10−5 |
| Relapse | Whole sample (N = 87) | |||||
| rs7137085 | 12 | A | PTHLH | ITR (26 kb) | 0.08 [0.02, 0.27] | 6.99 × 10−5 |
| rs11910354 | 21 | G | PCP4 | ITR (12 kb) | 230.19 [15.55, 3,407.69] | 7.63 × 10−5 |
| rs7309492 | 12 | T | PTHLH | ITR (38 kb) | 0.07 [0.02, 0.26] | 9.65 × 10−5 |
| rs9452695 | 6 | T | TSG1 | ITR (653 kb) | 25.43 [4.97, 130.07] | 1.02 × 10−4 |
| rs1479028 | 12 | A | NAV3 | INT | 8.36 [2.86, 24.44] | 1.05 × 10−4 |
| Relapse | European ancestry (N = 73) | |||||
| rs154288 | 5 | A | MYO10 | INT | 73.74 [9.01, 603.28] | 6.07 × 10−5 |
| rs42566 | 5 | G | MYO10 | INT | 73.74 [9.01, 603.28] | 6.07 × 10−5 |
| rs153200 | 5 | C | MYO10 | INT | 67.66 [8.62, 530.99] | 6.09 × 10−5 |
| rs31313 | 5 | T | MYO10 | INT | 39.53 [6.31, 247.58] | 8.57 × 10−5 |
| rs1479028 | 12 | A | NAV3 | INT | 15.12 [3.77, 60.64] | 1.27 × 10−4 |
A1, effect allele; CHR, chromosome; kb, kilobase pairs; SNP, single nucleotide polymorphism; MYO10, myosin X; NAV3, neuron navigator 3; PCP4, Purkinje cell protein 4; PTHLH, parathyroid hormone-like hormone; SYNPO2, synaptopodin 2; TMCO4, transmembrane and coiled-coil domains 4; TSG1, tumor suppressor TSG1; ZSCAN20, zinc finger and SCAN domain containing 20.
aImputed SNPs.
For the gene-based analysis of remission, we mapped variants to 18,954 genes, of which 1,086 genes showed a nominal association (p < 0.05). However, no genes reached genome-wide significance of p = 2.64 × 10−6. The top associated genes included SLC22A3 (solute carrier family 22 member 3; Z = 4.03, p = 2.83 × 10−5), NRBF2 (nuclear receptor binding factor 2; Z = 3.61, p = 1.52 × 10−4), and ZNF317 (zinc finger protein 317; Z = 3.53, p = 2.05 × 10−4; shown in online suppl. Table 2).
Targeted Analysis
Our targeted analysis of the serotonin transporter gene (SLC6A4) included the LPR and rs25531, as well as their interaction. Neither the LPR nor rs25531 nor their interaction showed a significant association with remission status or relapse in the whole sample or the European ancestry subsample prior to correction (shown in online suppl. Table 4, 5).
Polygenic Risk Scores
The PRS for greater antidepressant treatment symptom improvement derived from the PGC-MDD cohort was associated with remission status in our STOP-PD II cohort (OR = 1.95 [1.20, 3.17]; p = 0.007; permutation test, p = 0.04). A higher PRS for Alzheimer’s disease from the IGAP cohort was associated with a lower likelihood of relapse (in IGAP 1, OR = 0.006 [0.0002, 0.14]; p = 0.002; permutation test, p = 0.01; in IGAP 1–2 meta-analysis, OR = 0.38 [0.18, 0.80]; p = 0.01; permutation test, p = 0.03). None of these associations survived Bonferroni correction before and after permutation (shown in Table 3 and online suppl. Table 3).
Table 3.
Polygenic risk score results for remission status and relapse in the European ancestry subsample
| Polygenic risk scores | Remission status (N = 143) | Relapse status (N = 73) | ||||
|---|---|---|---|---|---|---|
| OR [95% CI] | p value | Per-P | OR [95% CI] | p value | Per-P | |
| ALZ (IGAP 1) [38] | 1.34 [0.90, 1.99] | 0.15 | 0.45 | 0.006 [0.0002, 0.14] | 0.002 | 0.01* |
| ALZ (IGAP 1–2 meta-analysis) [38] | 0.91 [0.63, 1.32] | 0.63 | 0.91 | 0.38 [0.18, 0.80] | 0.01 | 0.03* |
| ALZ [39] | 1.53 [0.91, 2.57] | 0.11 | 0.39 | 0.73 [0.40, 1.36] | 0.32 | 0.81 |
| BPD [37] | 2.39 [0.16, 36.14] | 0.53 | 0.96 | – | – | –a |
| MDD (PGC) [34] | 1.10 [0.68, 1.79] | 0.70 | 0.99 | 0.64 [0.34, 1.20] | 0.16 | 0.40 |
| MDD (PGC) [33] | 0.85 [0.56, 1.28] | 0.43 | 0.66 | 0.72 [0.38, 1.37] | 0.32 | 0.51 |
| AD non-remission (PGC) [35] | 0.70 [0.41, 1.20] | 0.19 | 0.61 | 1.81 [0.93, 3.51] | 0.08 | 0.33 |
| AD improvement (PGC) [35] | 1.95 [1.20, 3.17] | 0.007 | 0.04* | 1.92 [1.05, 3.53] | 0.03 | 0.17 |
| SCZ [36] | 1.56 [1.04, 2.33] | 0.03 | 0.09 | 0.82 [0.36, 1.84] | 0.62 | 0.96 |
| All stroke [40] | 1.46 [0.97, 2.21] | 0.07 | 0.26 | 0.19 [0.04, 0.88] | 0.03 | 0.15 |
| Ischemic stroke | 0.81 [0.41, 1.58] | 0.54 | 0.96 | 0.27 [0.06, 1.28] | 0.10 | 0.37 |
| Cardioembolic stroke | 1.80 [0.88, 3.71] | 0.11 | 0.39 | 0.15 [0.01, 1.58] | 0.11 | 0.35 |
| Large vessel stroke | 2.15 [0.35, 13.22] | 0.41 | 0.90 | 1.86 [0.64, 5.41] | 0.25 | 0.70 |
| Small vessel stroke | 1.33 [0.73, 2.41] | 0.36 | 0.85 | – | – | –a |
AD, antidepressant; ALZ, Alzheimer’s disease; BPD, bipolar disorder; CI, confidence interval; IGAP, the International Genomics of Alzheimer’s Project; MDD, major depressive disorder; OR, odds ratio; p, p value; Per-P, permutation p value; PGC, Psychiatric Genomics Consortium; SCZ, schizophrenia.
*p < 0.05 (number of permutation testing = 10,000).
aModel did not converge.
Discussion
To our knowledge, this is the first study to examine genome-wide SNP, gene, and PRS associations of remission and relapse of treated MDDPsy. Although our exploratory analysis did not find genome-wide significant associations with either remission or relapse, SNPs associated with these outcomes at suggestive thresholds provide leads for future investigation.
Regarding remission in the whole sample, the variant most strongly associated with lower odds of remission was rs1026501, located 30.9 kb from the 3’ UTR of SYNPO2. Within the chromosome 4 locus, rs1026501 has a PHRED of 6.7, which suggests that it is deleterious. Furthermore, the adjacent rs6847629 is in high LD (r2 = 0.96) and is among the 1% of most deleterious variants in the genome (PHRED = 12). Rs6844137, which is located in the intronic region of SYNPO2, was associated with lower likelihood of remission in European sample. SYNPO2 encodes synaptopodin 2, which plays a role in postsynaptic autophagocytosis, a key component in maintaining cellular integrity by eliminating dysfunctional proteins and cytotoxic compounds. While aberrated proteostasis has been implicated in age-related neurodegenerative processes [41], there is also evidence that autophagy induction, such as physical exercise and short-term restriction of calories, could reduce depressive symptoms in humans [42]. Moreover, rs1026501 showed nominal significance in the PGC-MDD cohort on depression symptom improvement after antidepressant treatment (p = 0.04). As such, the SYNPO2 polymorphism may affect the clearance of cytotoxic substances and, as a result, antidepressant action.
With respect to relapse, rs7137085 near PTHLH, which encodes parathyroid hormone-like hormone, was characterized as the top locus associated with a lower probability of relapse. One of the functions of this neuroendocrine peptide is to regulate circulating calcium and epithelial calcium ion transport. Higher PTHLH expression has been associated with elevated serum calcium levels. As no study has investigated whether rs7137085 is associated with differential PTHLH expression, this variant is associated with several altered binding motifs [43]. Therefore, additional investigation is needed to understand the regulatory function of the associated variant. In the European ancestry subsample, the top SNP associated with relapse was intronic variant rs154288, located within the MYO10 gene. MYO10 encodes myosin X, which is involved in the intracellular movement of molecules [44]. Across 27 human tissues examined, MYO10 has one of the highest expressions in the brain [45]. The trend of higher MYO10 expression across brain tissues is also observed in GTEx expression data across 54 other tissues [46]. MYO10 may play an important role in neuronal development, as its mRNA expression is elevated during nerve regeneration, and it binds to important molecules in neuronal development [47]. A meta-analysis of 2,256 European individuals diagnosed with MDD (GENDEP, MARS, and STAR*D) reported a genome-wide association between depressive symptom improvement after antidepressant treatment and a rare intronic MYO10 SNP (rs17651119) [48]. In our study, although the association between rs154288 and MDDPsy relapse did not reach the suggestive threshold, taken together with previous findings, our finding suggests a possible role of MYO10 in MDDPsy treatment outcomes.
For gene-based analyses using GWAS statistics, the SLC22A3 (solute carrier family 22 member 3) gene showed the strongest association. Based on the GTEx portal V8, the expression for the SLC22A3 gene is lower in the brain compared to other tissues [46]. According to the Human Protein Atlas website, organic cation transporters encoded by SLC22A3 might be responsible for clearance of neurotoxins and transport ofneurotransmitters in the brain [49]. To our knowledge, no association between SLC22A3 and neuropsychiatric disorders, or their response to pharmacotherapy, has been reported. Our exploratory results may serve as stimulus for future replication studies investigating the SLC22A3 gene in MDDPsy remission.
We conducted PRS analyses to explore whether disorders of potential relevance to the outcome of MDDPsy had shared genetic contributions with remission and relapse in STOP-PD II. The PRS for greater symptom improvement in the PGC-MDD cohort was associated with a higher likelihood of remission in the STOP-PD II cohort. The PRS we used was derived from a meta-analysis of studies of various antidepressants, including SSRIs (e.g., citalopram and sertraline) and norepinephrine reuptake inhibitor (NRI, e.g., reboxetine). In addition, genetic predisposition for Alzheimer’s disease was associated with a lower likelihood of relapse. Of note, 44% of the genotyped STOP-PD II participants were in the 60–85 year age range. Although dementia was an exclusion for STOP-PD II, the older age of many participants in the study and their potential vulnerability to neurodegenerative changes in the absence of clinical manifestations of dementia may have contributed to these PRS findings. Antidepressants have not been shown to have superior efficacy than placebo in the treatment of depression in Alzheimer’s disease [50]. The PRS finding regarding genetic predisposition to Alzheimer’s and lower odds of relapse of MDDPsy may suggest that some STOP-PD II participants were in the pre-dementia stages, explaining both their depressive and psychotic symptoms and their lower likelihood of relapse once they remitted. Contrary to our initial assumption, our study showed that individuals with higher PRS for Alzheimer’s disease were as likely to reach remission as individuals with lower PRS. However, in those achieving remission, a higher PRS for AD appeared to be a protective factor against relapse, while a lower PRS was associated with higher risk for relapse. While this relationship is counterintuitive, it might point out that other factors contribute to relapse, while our reported finding might also be a false positive one. This shared genetic architecture may aid in illuminating the mechanism of antipsychotic action in those with MDD and psychotic features in the future.
Considerable literature exists on the relationship of the serotonin transporter to response of depression to SSRIs, including two studies in MDDPsy [10, 11]. Compared with SLC6A4 S allele, L allele results in higher expression of SLC6A4 [51]. A systematic review identified that L/L genotype was associated with better treatment outcomes after SSRI treatment [51]. Similarly, another meta-analysis of 46 studies also found that L/L and LS genotypes were more likely than S/S genotypes to be associated with response of MDD to SSRIs (but not other antidepressants) in individuals of European ancestry treated (OR = 1.55 [1.20, 2.00], p = 0.001) but not in individuals of East Asian ancestries [52]. For this reason, we investigated the association of SLC6A4 variants, including the LPR and rs25531, with remission and relapse of MDDPsy. However, we did not find statistically or clinically significant associations. There was no significant association of the combined 5-HTTLPR/rs25531 genotypes with SSRI response [52]. Nonetheless, the small sample size of our study had inadequate power to detect the relationship between SLC6A4 variants with remission or relapse of MDDPsy.
This study has several limitations. First, the study had a small sample size for a GWAS analysis, which reduced its power to detect significant associations, increasing the risk of type II error. In both the whole mixed ancestry sample and the European ancestry subsample, we observed genomic inflation with lambdas of 1.054 and 1.052, respectively. Thus, there may have been loci of relevance that were undetected by this present analysis. Second, 83.6% of genotyped participants were of European ancestry, which precluded analyses specific to other ancestries. Third, the number of genotyped participants in the RCT was too small to allow for separate analyses of each randomized group, for example, those switched from olanzapine to placebo compared to those continued on olanzapine. Given the small sample size of the relapse analyses, little can be said about the relationship of genetic factor with relapse of MDDPsy. Finally, the findings of this study pertain to treatment with sertraline plus olanzapine and may not generalize to other medications.
Strengths of this study include the well-characterized sample, the robust study design, and the comprehensive approach to genomic analysis. We identified SNPs potentially associated with remission and relapse that can be evaluated in future investigations, including those with a goal of understanding mechanisms of treatment outcome of psychotic depression. The genetic material donated by STOP-PD II participants is available to the wider scientific community through the NIMH Center for Collaborative Genetic Studies, so that it will contribute to larger samples with greater statistical power.
In summary, we found no genome-wide significant SNP associated with remission or relapse after the pharmacotherapy of sertraline and olanzapine. However, our exploratory analyses revealed some interesting observations with genetic markers which warrant further investigation. Moreover, our preliminary findings related to our PRS analysis will be of use when determining the role of PRS in antidepressant response and relapse in future studies.
Acknowledgments
We thank the members of the STOP-PD II Study Group for their contributions. Members of the STOP-PD II Study Group were as follows: Peter Giacobbe, M.D., and Brenda Swampillai, B.Sc., at the University Health Network, Toronto; James Kennedy, M.D., and Bruce Pollock M.D., Ph.D., at the Center for Addiction and Mental Health, Toronto; Kristina Deligiannidis, M.D., Chelsea Kosma, M.A., and Wendy Marsh, M.D., at the University of Massachusetts Medical School and UMass Memorial Health Care, Worcester, MA; Ariel Gildengers M.D., Joelle Kincman Ph.D., Meryl Butters Ph.D., and Michelle Zmuda, M.A., at the University of Pittsburgh School of Medicine, Pittsburgh, PA; and Judith English, M.A., James Kocsis, M.D., Barbara Ladenheim, Ph.D., Vassilios Latoussakis, M.D., and Nikhil Palekar, M.D., at Weill Cornell Medicine of Cornell University and New York Presbyterian Hospital, NY. We also thank Lara Murphy and Farhana Islam for assistance with preparation of the manuscript.
Statement of Ethics
This study protocol was reviewed and approved by the Institutional Review Boards of University Health Network, Toronto (approval number: 11-0137-A), University of Massachusetts Medical School (approval number: 14207), University of Pittsburgh (approval numbers: PRO 11020043 and 11050138), and Weill Cornell Medicine (approval number: 1103011563). Using procedures approved by the Local Institutional Review Boards, written informed consent was obtained from all participants or their substitute decision-maker prior to the initiation of any research procedures.
Conflict of Interest Statement
G. S. Alexopoulos has received NIMH grants and has served in advisory boards of Janssen and Eisai and in the speakers’ bureau of Takeda, Lundbeck, Otsuka, Allergan, AstraZeneca, and Sunovion. B. S. Meyers received research support from the NIMH at the time this work was done. A. J. Flint has received grant support from the U.S. National Institutes of Health, Patient-Centered Outcomes Research Institute, Canadian Institutes of Health Research, Brain Canada, Ontario Brain Institute, Alzheimer’s Association, AGE-WELL, and the Canadian Foundation for Healthcare Improvement. J. L. Kennedy has received funding from the Canadian Institutes of Health Research, CAMH Foundation, and the National Institutes of Health. He is an unpaid member of the Myriad Neuroscience Scientific Advisory Board and is an author on pharmacogenetics patents. D. J. Mueller has received funding from the Canadian Institutes of Health Research, CAMH Foundation, and the University of Toronto. B. H. Mulsant holds and receives support from the Labatt Family Chair in Biology of Depression in Late-Life Adults at the University of Toronto. He currently receives or has received research support within the past 3 years from Brain Canada, the Canadian Institutes of Health Research, the CAMH Foundation, the Patient-Centered Outcomes Research Institute (PCORI), the US National Institutes of Health (NIH), Capital Solution Design LLC (software used in a study funded by CAMH Foundation), and HAPPYneuron (software used in a study funded by Brain Canada). Within the past 3 years, he has been an unpaid consultant to Myriad Neuroscience. A. J. Rothschild has received grant or research support from Janssen, Otsuka, Praxis, and the Irving S. and Betty Brudnick Endowed Chair in Psychiatry; is a consultant to Alkermes, Janssen, Sage Therapeutics, Xenon Pharmaceuticals, and several generic medication companies; has received royalties for the Rothschild Scale for Antidepressant Tachyphylaxis (RSAT)®; Clinical Manual for the Diagnosis and Treatment of Psychotic Depression, American Psychiatric Press, 2009; The Evidence-Based Guide to Antipsychotic Medications, American Psychiatric Press, 2010; The Evidence-Based Guide to Antidepressant Medications, American Psychiatric Press, 2012; and, UpToDate®; and has received honorarium from Wolters Kluwer (Journal Editor). A. N. Voineskos has received funding from the NIMH, Canadian Institutes of Health Research, Canadian Foundation for Innovation, CAMH Foundation, and the University of Toronto. E. M. Whyte receives grant support from the NIMH and HRSA. S.S. Elsheikh,P. Marino, V.S. Marshe, and X. Men have nothing to disclose.
Funding Sources
STOP-PD II was funded by USPHS grants MH 62446, MH 62518, MH 62565, and MH 62624 from the National Institute of Mental Health (NIMH); Eli Lilly provided olanzapine and matching placebo pills and Pfizer provided sertraline; neither company provided funding. The National Institute of Mental Health (NIMH) participated in the implementation of STOP-PD II through the U01 mechanism. NIMH did not participate in the design of the study or the collection, management, or analysis of data. A data safety monitoring board at NIMH provided data and safety monitoring. Neither Eli Lilly nor Pfizer participated in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Author Contributions
Substantial contributions to the conception or design of the work and/or the acquisition, analysis, or interpretation of data for the work, drafting of the manuscript and/or revising it critically for important intellectual content, final approval of the version to be published, and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved: X. Men, V.S. Marshe, S S. Elsheikh, G. S. Alexopoulos, P. Marino, B. S. Meyers, B. H. Mulsant, A. J. Rothschild, A. N. Voineskos, E. M. Whyte, J. L. Kennedy, A. J. Flint, and D. J. Müller. Conduction of genomic analyses in the article: X. Men and V.S. Marshe.
Funding Statement
STOP-PD II was funded by USPHS grants MH 62446, MH 62518, MH 62565, and MH 62624 from the National Institute of Mental Health (NIMH); Eli Lilly provided olanzapine and matching placebo pills and Pfizer provided sertraline; neither company provided funding. The National Institute of Mental Health (NIMH) participated in the implementation of STOP-PD II through the U01 mechanism. NIMH did not participate in the design of the study or the collection, management, or analysis of data. A data safety monitoring board at NIMH provided data and safety monitoring. Neither Eli Lilly nor Pfizer participated in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Data Availability Statement
The data that support the findings of this study are not publicly available due to containing information that could compromise the privacy of research participants but are available from the corresponding authors [D.J.M. Daniel.mueller@camh.ca and A.J.F. alastair.flint@uhn.ca].
Supplementary Material
References
- 1. Rothschild AJ. Challenges in the treatment of major depressive disorder with psychotic features. Schizophr Bull. 2013 Jul;39(4):787–96. 10.1093/schbul/sbt046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vythilingam M, Chen J, Bremner JD, Mazure CM, Maciejewski PK, Nelson JC. Psychotic depression and mortality. Am J Psychiatry. 2003 Mar;160(3):574–6. 10.1176/appi.ajp.160.3.574. [DOI] [PubMed] [Google Scholar]
- 3. Coryell W. The treatment of psychotic depression. J Clin Psychiatry. 1998;59:22–7. [PubMed] [Google Scholar]
- 4. Ohayon MM, Schatzberg AF. Prevalence of depressive episodes with psychotic features in the general population. Am J Psychiatry. 2002 Nov;159(11):1855–61. 10.1176/appi.ajp.159.11.1855. [DOI] [PubMed] [Google Scholar]
- 5. Wijkstra J, Burger H, van den Broek WW, Birkenhäger TK, Janzing JGE, Boks MPM, et al. Treatment of unipolar psychotic depression: a randomized, double-blind study comparing imipramine, venlafaxine, and venlafaxine plus quetiapine. Acta Psychiatr Scand. 2010 Mar;121(3):190–200. 10.1111/j.1600-0447.2009.01464.x. [DOI] [PubMed] [Google Scholar]
- 6. Meyers BS, Flint AJ, Rothschild AJ, Mulsant BH, Whyte EM, Peasley-Miklus C, et al. A double-blind randomized controlled trial of olanzapine plus sertraline vs olanzapine plus placebo for psychotic depression: the study of pharmacotherapy of psychotic depression (STOP-PD). Arch Gen Psychiatry. 2009;66(8):838–47. 10.1001/archgenpsychiatry.2009.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flint AJ, Meyers BS, Rothschild AJ, Whyte EM, Alexopoulos GS, Rudorfer MV, et al. Effect of continuing olanzapine vs placebo on relapse among patients with psychotic depression in remission: the STOP-PD II randomized clinical trial. JAMA. 2019 Aug;322(7):622–31. 10.1001/jama.2019.10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A, et al. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013 Apr;73(7):679–82. 10.1016/j.biopsych.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 9. Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Børglum AD, Breen G, et al. Psychiatric genomics: an update and an agenda. Am J Psychiatry. 2018 Jan;175(1):15–27. 10.1176/appi.ajp.2017.17030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smeraldi E, Zanardi R, Benedetti F, Di Bella D, Perez J, Catalano M. Polymorphism within the promoter of the serotonin transporter gene and antidepressant efficacy of fluvoxamine. Mol Psychiatry. 1998 Nov;3(6):508–11. 10.1038/sj.mp.4000425. [DOI] [PubMed] [Google Scholar]
- 11. Zanardi R, Serretti A, Rossini D, Franchini L, Cusin C, Lattuada E, et al. Factors affecting fluvoxamine antidepressant activity: influence of pindolol and 5-HTTLPR in delusional and nondelusional depression. Biol Psychiatry. 2001 Sep;50(5):323–30. 10.1016/s0006-3223(01)01118-0. [DOI] [PubMed] [Google Scholar]
- 12. Arias B, Fabbri C, Gressier F, Serretti A, Mitjans M, Gastó C, et al. TPH1, MAOA, serotonin receptor 2A and 2C genes in citalopram response: possible effect in melancholic and psychotic depression. Neuropsychobiology. 2013;67(1):41–7. 10.1159/000343388. [DOI] [PubMed] [Google Scholar]
- 13. Farahani A, Correll CU. Are antipsychotics or antidepressants needed for psychotic depression? A systematic review and meta-analysis of trials comparing antidepressant or antipsychotic monotherapy with combination treatment. J Clin Psychiatry. 2012 Apr;73(4):486–96. 10.4088/JCP.11r07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Flint AJ, Meyers BS, Rothschild AJ, Whyte EM, Mulsant BH, Rudorfer MV, et al. Sustaining remission of psychotic depression: rationale, design and methodology of STOP-PD II. BMC Psychiatry. 2013 Jan;13:38. 10.1186/1471-244X-13-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. First MB, Spitzer RL, Gibbon M, Williams JBW. Structured clinical interview for DSM-IV-TR Axis I disorders: patient edition (SCID-I/P 2/2001 revision). New York: Biometrics Research Department; 2001. [Google Scholar]
- 16. Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960 Feb;23(1):56–62. 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jorm AF. The Informant Questionnaire on cognitive decline in the elderly (IQCODE): a review. Int Psychogeriatr. 2004 Sep;16(3):275–93. 10.1017/s1041610204000390. [DOI] [PubMed] [Google Scholar]
- 18. Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: a practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–98. 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 19. Guy W. ECDEU assessment manual for psychopharmacology. US department of Health, education, and welfare: Public Health service, alcohol Drug abuse, and mental Health administration. Psychopharmacology Research Branch, Division of Extramural Research Programs: National Institute of Mental Health; 1976:217–22. [Google Scholar]
- 20. Salvatore P, Baldessarini RJ, Khalsa H-MK, Amore M, Di Vittorio C, Ferraro G, et al. Predicting diagnostic change among patients diagnosed with first-episode DSM-IV-TR major depressive disorder with psychotic features. J Clin Psychiatry. 2013 Jul;74(7):723–31; quiz 731. 10.4088/JCP.12m08328. [DOI] [PubMed] [Google Scholar]
- 21. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010 Sep;5(9):1564–73. 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clarke GM, Anderson CA, Pettersson FH, Cardon LR, Morris AP, Zondervan KT. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011 Feb;6(2):121–33. 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–75. 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lemieux Perreault L-P, Legault M-A, Asselin G, Dubé M-P. genipe: an automated genome-wide imputation pipeline with automatic reporting and statistical tools. Bioinformatics. 2016 Dec;32(23):3661–3. 10.1093/bioinformatics/btw487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Leeuwen EM, Kanterakis A, Deelen P, Kattenberg MV; Genome of the Netherlands Consortium, Slagboom PE, et al. Population-specific genotype imputations using minimac or IMPUTE2. Nat Protoc. 2015 Sep;10(9):1285–96. 10.1038/nprot.2015.077. [DOI] [PubMed] [Google Scholar]
- 26. 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loh PR, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, Reference-based phasing using the haplotype reference Consortium panel. Nat Genet. 2016;48(11):1443–8. https://idp.nature.com/authorize/casa?redirect_uri=https://www.nature.com/articles/ng.3679.pdf%3Forigin%3Dppub&casa_token=K0H1LVsq2I4AAAAA:5T4wwiAwm5ddV2nDaJVIMertO95YFBxYJv6-0WE-kmhZOAZWgDXQWo9d6J1NT1HqCmXqeP7Pbt78pCFPEXg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cook EH, Jr, Courchesne R, Lord C, Cox NJ, Yan S, Lincoln A, et al. Evidence of linkage between the serotonin transporter and autistic disorder. Mol Psychiatry. 1997 May;2(3):247–50. 10.1038/sj.mp.4000266. [DOI] [PubMed] [Google Scholar]
- 29. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015 Apr;11(4):e1004219. 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017 Nov;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fabbri C, Corponi F, Souery D, Kasper S, Montgomery S, Zohar J, et al. The genetics of treatment-resistant depression: a critical review and future perspectives. Int J Neuropsychopharmacol. 2019 Feb;22(2):93–104. 10.1093/ijnp/pyy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Choi SW, O’Reilly PF. PRSice-2: polygenic Risk Score software for biobank-scale data. Gigascience. 2019 Jul;8(7):giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019 Mar;22(3):343–52. 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018 May;50(5):668–81. 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pain O, Hodgson K, Trubetskoy V, Ripke S, Marshe VS, Adams MJ, et al. Identifying the common genetic basis of antidepressant response. Biol Psychiatry Glob Open Sci. 2022;2(2):115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022 Apr;604(7906):502–8. 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019 May;51(5):793–803. 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013 Dec;45(12):1452–8. 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019 Mar;51(3):404–13. 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Malik R, Rannikmäe K, Traylor M, Georgakis MK, Sargurupremraj M, Markus HS, et al. Genome-wide meta-analysis identifies 3 novel loci associated with stroke. Ann Neurol. 2018 Dec;84(6):934–9. 10.1002/ana.25369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stürner E, Behl C. The role of the multifunctional BAG3 protein in cellular protein quality control and in disease. Front Mol Neurosci. 2017;10:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gassen NC, Rein T. Is there a role of autophagy in depression and antidepressant action? Front Psychiatry. 2019;10:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. HaploReg v4.1 variant detail view [Internet] . [cited 2022 Apr 21]. Available from: https://pubs.broadinstitute.org/mammals/haploreg/detail_v4.1.php?query=&id=rs7137085.
- 44.MYO10 myosin X [Homo sapiens (human)] - Gene - NCBI [Internet]. [cited 2023 Jan 18]. Available from: https://www.ncbi.nlm.nih.gov/gene/4651.
- 45. Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014 Feb;13(2):397–406. 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The genotype-tissue expression (GTEx) project. Nat Genet. 2013 May;45(6):580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sousa AD, Berg JS, Robertson BW, Meeker RB, Cheney RE. Myo10 in brain: developmental regulation, identification of a headless isoform and dynamics in neurons. J Cell Sci. 2006 Jan;119(Pt 1):184–94. 10.1242/jcs.02726. [DOI] [PubMed] [Google Scholar]
- 48. GENDEP Investigators, MARS Investigators, STAR*D Investigators, Wolfgang M, Mors O, Hauser J. Common genetic variation and antidepressant efficacy in major depressive disorder: a meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013 Feb;170(2):207–17. 10.1176/appi.ajp.2012.12020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pontén F, Jirström K, Uhlen M. The human protein atlas–a tool for pathology. J Pathol. 2008 Dec;216(4):387–93. 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 50. Marshe VS, Maciukiewicz M, Hauschild A-C, Islam F, Qin L, Tiwari AK, et al. Genome-wide analysis suggests the importance of vascular processes and neuroinflammation in late-life antidepressant response. Transl Psychiatry. 2021 Feb;11(1):127. 10.1038/s41398-021-01248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marshe VS, Islam F, Maciukiewicz M, Bousman C, Eyre HA, Lavretsky H, et al. Pharmacogenetic implications for antidepressant pharmacotherapy in late-life depression: a systematic review of the literature for response, pharmacokinetics and adverse drug reactions. Am J Geriatr Psychiatry. 2020 Jun;28(6):609–29. 10.1016/j.jagp.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 52. Ren F, Ma Y, Zhu X, Guo R, Wang J, He L. Pharmacogenetic association of bi- and triallelic polymorphisms of SLC6A4 with antidepressant response in major depressive disorder. J Affect Disord. 2020 Aug;273:254–64. 10.1016/j.jad.2020.04.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are not publicly available due to containing information that could compromise the privacy of research participants but are available from the corresponding authors [D.J.M. Daniel.mueller@camh.ca and A.J.F. alastair.flint@uhn.ca].