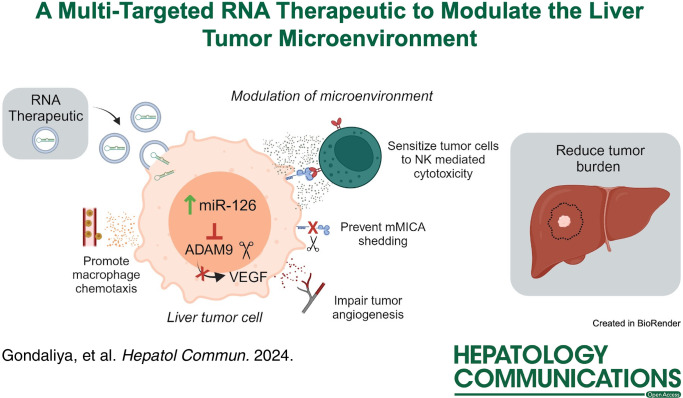
Abstract
Background:
Impaired natural killer (NK) cell-mediated antitumor responses contribute to the growth of liver tumors. Expression of a disintegrin and metalloprotease 9 (ADAM9) increases shedding of membrane-bound major histocompatibility complex class I chain-related protein A and results in evasion from NK cell-mediated cytolysis. ADAM9 is also involved in angiogenesis and tumor progression and is a target of miR-126-3p, a tumor suppressor that is downregulated and alters tumor cell behavior in the liver and other cancers. We evaluated the restoration of miR-126-3p and modulation of the miR-126-3p/ADAM9 axis as a therapeutic approach to simultaneously enhance NK cell-mediated cytolysis while targeting both tumor cells and their microenvironment.
Methods:
Precursor miRNAs were loaded into milk-derived nanovesicles to generate therapeutic vesicles (therapeutic milk-derived nanovesicles) for the restoration of functional miR-126-3p in recipient cancer cells.
Results:
Administration of therapeutic milk-derived nanovesicles increased miR-126-3p expression and reduced ADAM9 expression in target cells and was associated with an increase in membrane-bound major histocompatibility complex class I chain-related protein A. This enhanced NK cell cytolysis in adherent tumor cells and in multicellular tumor spheroids while also impairing angiogenesis and modulating macrophage chemotaxis. Moreover, IV administration of therapeutic milk-derived nanovesicles with adoptive transfer of NK cells reduced tumor burden in orthotopic hepatocellular cancer xenografts in mice.
Conclusion:
A directed RNA therapeutic approach can mitigate NK cell immune evasion, reduce angiogenesis, and alter the tumor cell phenotype through the restoration of miR-126-3p in liver tumor cells. The pleiotropic effects elicited by this multi-targeted approach to modulate the local tumor microenvironment support its use for the treatment of liver cancer.
INTRODUCTION
Primary liver cancers, such as HCC, are a serious complication of chronic liver disease and a leading cause of cancer-related mortality worldwide. The rising global prevalence of liver cancer and the dearth of effective curative therapies for advanced-stage disease highlight the critical need for more effective therapeutic approaches. Natural killer (NK) cells account for a significant population of lymphocytes within the human liver.1,2 These hepatic NK cells play a critical role as effectors of the innate immune response and in mediating tumor cell immune surveillance within the liver. Moreover, intrahepatic or circulating NK cell levels are positively related to HCC patient survival. However, NK cell dysfunction can occur due to the evolution of mechanisms that enable tumor cells to evade NK cell-mediated cytolysis, ecological changes within the tumor microenvironment (TME), and several biotic and abiotic factors. Collectively, these impair the NK cell-mediated antitumor immune response.3 Consequently, restoration of immune surveillance mechanisms, either by restoring the functional activity of NK cells or by preventing liver tumor cell immune escape, could be effective for the treatment of liver cancers such as HCC.4
An approach to modulating NK cell-mediated immune surveillance and tumor control involves targeting receptor/ligand interactions that lead to the activation of NK cells. Liver cancer cells express ligands such as major histocompatibility complex class I chain-related proteins A and B (MICA/B), B7 protein homolog 6, and CD155, which can bind to activating NK receptors such as NK receptor group 2 member D (NKG2D), Natural Killer cell p30, and DNAX Accessory Molecule-1, respectively. NK cell-mediated antitumor responses can be activated following the downregulation of major histocompatibility complex class I molecules and/or the expression of ligands, such as MICA/B.1 Targeting the NKG2D receptor-MICA/B axis provides an opportunity to therapeutically ameliorate tumor cell immune evasion and resistance to NK cell-mediated cytolysis. Expression of MICA/B and their recognition by the NKG2D receptor render tumor cells sensitive to NK cell-mediated cytolysis. The expression of these ligands correlates with disease outcomes.5,6 A reduction in MICA expression is associated with decreased disease-free and overall survival in HCC.7 To target the NKG2D receptor-MICA/B axis, we focused on A disintegrin and metalloprotease 9 (ADAM9). ADAM9 is overexpressed in advanced-stage HCC, and serum levels of ADAM9 are elevated in patients with HCC.8–10 ADAM9 can enable tumor cells to avoid NK cell-mediated cytolysis through proteolytic cleavage of membrane-bound MICA from the tumor cells and the production of soluble MICA/B (sMICA), which can bind NKG2D. By blocking ligand binding to MICA/B on tumor cells, sMICA can function as an immune decoy to reduce NK cell-mediated tumor cell cytotoxicity.11 Modulating MICA/B shedding from liver cancer cells thus provides a means to target NK cell activation and immune escape.
We have identified that ADAM9 is a downstream target of miR-126-3p and observed dysregulation in the miR-126-3p/ADAM9 axis within the liver TME.4 miR-126-3p expression is markedly reduced in HCC as well as other solid tumors, and this noncoding RNA acts as a tumor suppressor. By repressing the expression of ADAM9, miR-126-3p could modulate migration and invasion in liver cancer cells.4 Moreover, mir-126-3p can control the expression of several other genes involved in angiogenesis and metastasis and contribute to a malignant cell phenotype. Thus, we sought to restore miR-126-3p in tumor cells as a multi-targeted strategy aimed at both immune escape mechanisms enabled by ADAM9 mediated shedding of membrane-bound MICA in liver cancer cells and directly targeting the tumor cell phenotype. Restoration of miR-126-3p in tumor cells was accomplished by the cellular delivery of miR-126-3p using a biological nanoparticle delivery platform comprised of milk-derived nanovesicles (MNVs). MNVs are effective carriers for therapeutic RNA and have liver tropic distribution after systemic administration with a favorable safety profile and can be produced at high yield.12,13 A therapeutic MNV (tMNV) agent was generated by loading MNV with miR-126-3p, and its efficacy was evaluated by assessing both NK cell activation and tumor cell-induced angiogenesis in vitro in liver tumor cells and in vivo using an orthotopic HCC model. Our studies demonstrate the effective use of an RNA therapeutic approach to modulate both tumor cell behavior and the immunosuppressive liver TME by concomitantly mitigating immune escape and the malignant cell phenotype.
METHODS
TME simulation
Multicellular tumor spheroids (MTS) were generated as described.14 Autoclaved methylcellulose (Sigma-Aldrich, #M0512-100G) was used to generate a 3% solution in DMEM medium containing 10% Fetal Bovine Serum and stored at 4°C. HepG2 and LX215 were mixed in a 4:1 ratio, and the cell suspension was adjusted to 5 × 106 cells/ml. 104 cells in 2 microliters of cell suspension in 3% methyl cellulose were plated in 6-well plates coated with poly-2-hydroxyethyl methacrylate (Sigma-Aldrich, #P3932) and incubated at 37°C and 5% CO2 for 3 days. Compact spheroids were then removed, washed with PBS, and transferred to DMEM complete media for further studies.
Generation of an RNA nanotherapeutic
MNV were prepared as described.16 Nonfat milk was diluted in PBS and centrifuged for 30 minutes at 12,000 g at 4°C. The supernatant was treated with Ca2+-free EDTA and adjusted to a pH of 4.2 using 3.0 M hydrochloric acid, followed by precipitation at 4°C for 10 minutes. The supernatant was centrifuged at 3000 g for 10 minutes at 4°C, then at 10,000 g for 30 minutes at 4°C, followed by filtration through a 0.45 µm membrane, adjusted to a pH of 7.0, filtration through a 0.22 µm membrane and ultracentrifugation at 34,100 rpm for 70 minutes at 4°C. The resulting pellets were washed with PBS and ultracentrifuged again, as listed above. The pellets were resuspended in 150 µL of PBS and stored at 4°C. Nanoparticle tracking analysis (Nanosight LM10, UK) was used to assess the size distribution profile of MNVs. MNVs were loaded with pre-miR-126-3p (tMNVs) or nontargeting RNA as negative controls (control MNVs) by combining an equivalent volume of pre-miR-126-3p or negative control miRNA (5 µM) with 4 µL of lipofectamine 2000 (Fisher, #11668019) and 25 µL of OptiMEM media for 10 minutes at RT. Lipo-miR complexes (50 µL) were mixed with MNVs (2 × 1012 particles) in equal volume and incubated for 30 minutes at RT before ultracentrifugation-based isolation. The MNVs were diluted in PBS to a final volume of 4 mL and then isolated by ultracentrifugation (Beckman Coulter, SW 60Ti) at 27,200 rpm for 70 min at 4°C using Ultra-Clear centrifuge tubes (Beckman Coulter, #344062, Brea, CA). The resulting control MNV or tMNV pellets were resuspended in 1 mL of PBS.
NK cell-mediated cell cytotoxicity
Liver tumor cells (HepG2 and SNU449) were pretreated with 2 × 1011 control MNVs or tMNVs for 48 hours. To visually evaluate the NK cell-mediated cytotoxicity in liver cancer cells, MNV pretreated cells were seeded in 48 well plates at effector-to-target (E:T, NK:tumor cell) ratios of 5:1 and 10:1 for 6 hours at 37°C, 5% CO2. For cytotoxicity studies in MTS, a single spheroid was placed per well in a 96-well plate. Thereafter, the tumor spheroids were treated with 2 × 1011 control MNVs or tMNVs for 48 hours. Following MNV pretreatment, NK cells were coculture with MTS in 10:1 E:T ratio (100,000:10,000) for 48 hours. Imaging was performed at 24 and 48 hours using a Zeiss LSM880 confocal microscope. To assess the effect of IL-15 on NK cell-mediated cytotoxicity, tumor cells were incubated with 2 µM calcein AM (#C1430, Life Technologies, Carlsbad, CA) and cocultured with NK cells stained using 5 µM Cell Tracker deep red (Life Technologies, #C34565) in the presence of 50 ng/ml IL-15 for 6 hours. Images were captured using an EVOS M5000 microscope. To quantify NK cell-mediated cytotoxicity, the coculture experiments were repeated using unstained cells. After 6 hours of coculture, the conditioned media (CM) was collected and used for a lactate dehydrogenase activity assay.
Orthotopic HCC in vivo model
Eight-week-old athymic nude male mice were purchased from Jackson Laboratory and housed in individually vented cages (Allentown Inc., Allentown, NJ) with bed-o’-cobbs bedding (The Andersons, Maumee, Ohio). The mice were fed food (Pico Diet 5053, LabDiet, St Louis, MO) and water ad libitum and maintained in a 12-hour/12-hour light/dark cycle. All animals received human care, and the study was completed in compliance with the Institutional Animal Care and Use Committee guidelines (protocol A00005435-20) and the Guide for the Care and Use of Laboratory Animals. To generate an orthotopic HCC tumor model, 5 × 106 stably expressing luciferase SNU449 cells were mixed with 50 µL of phenol-red free matrigel (Fisher Scientific, #356237) and maintained on ice. Mice were anesthetized using isoflurane (1%–2%); a small incision was made, and the cell-matrigel was slowly implanted into the left lobe of the liver. Subsequently, the abdominal muscles were sutured, and the skin was closed with a staple. Animals were maintained under a heat lamp post-op until they recovered from anesthesia. In Vivo Imaging System imaging was performed weekly to monitor tumor growth. Starting 1 week after orthotopic injection, the mice were treated with 2 × 1011 MNVs (tMNVs or control MNVs) by tail vein injection every 2 days for a total of 5 doses. Two million human NK cells were adoptively transferred by tail vein injection on days 3 and 7 post-MNV treatment initiation. At the treatment end point (day 11 post-MNV treatment), the mice were sacrificed by CO2 asphyxiation followed by cervical dislocation, and serum and tissue were collected.
Statistical analysis
All analyses were performed using GraphPad Prism version 8.0.2 (GraphPad Software, San Diego, CA). For statistical comparison of tMNVs and control MNVs, a Student t-test was performed (two-tailed, unpaired). For the analysis of 3 or more treatment groups, a one-way ANOVA test was performed. A two-way ANOVA was used to analyze the differences between NCAM-positive cell infiltration in tumor and nontumor regions of MNV-treated mice. Data are expressed as the mean and SD of at least 3 replicates.
Other methods
For all other methods and supporting information, see the accompanying Supplemental Digital Content, http://links.lww.com/HC9/A808.
RESULTS
Restoration of miR-126-3p modulates MICA shedding in tumor cells
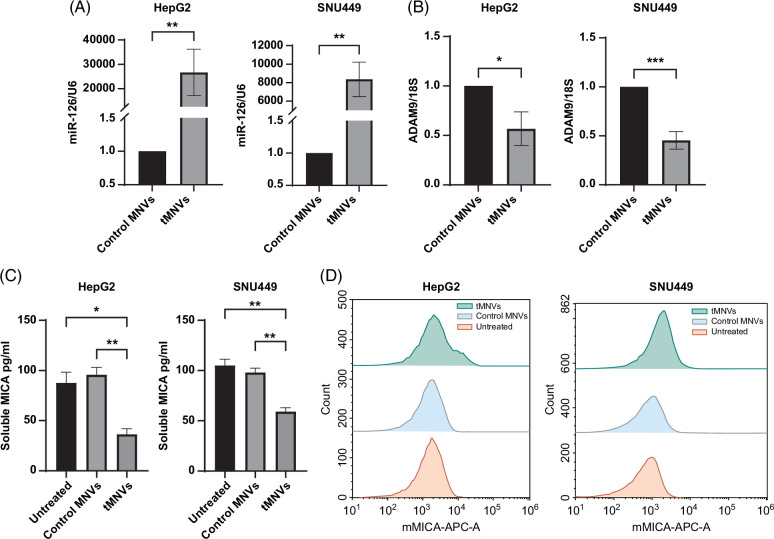
To evaluate the therapeutic potential for miR-126-3p replacement, we used MNVs as a carrier to deliver miRNA precursor molecules to liver cancer cells. tMNVs were generated by loading MNVs with pre-miR-126-3p, while control MNVs were generated by loading MNVs with a nontargeting RNA control (control MNVs). Unloaded (plain) MNVs and tMNVs exhibited a similar size distribution profile (Supplemental Figure S1, http://links.lww.com/HC9/A808). An increase in the expression of endogenous miR-126-3p was observed in both HepG2 and SNU449 cells following exposure to tMNVs (Figure 1A). Moreover, decreased expression of ADAM9, a miR-126-3p downstream target, was observed in tMNVs but not in control MNVs (Figure 1B). These results indicate that tMNVs can effectively deliver biologically active miR-126-3p to recipient tumor cells in vitro and, in doing so, can regulate ADAM9 expression. Reducing the expression of ADAM9 provides a cell-intrinsic mechanism for modulating MICA release from tumor cells because ADAM9 promotes the shedding of membrane-bound MICA from the surface of tumor cells. sMICA levels were assessed in tumor cell CM obtained after 48 hours of incubation in the absence or presence of 2 × 1011 control MNVs or tMNVs containing miR-126-3p. A decrease in sMICA levels was observed in HepG2 and SNU449 cells treated with tMNVs compared to control MNVs or untreated controls (Figure 1C). In addition, tMNVs increased the percentage of membrane-bound MICA-positive HepG2 cells and SNU449 cells when compared with control MNVs (Supplemental Figure S1B, http://links.lww.com/HC9/A808).
FIGURE 1.
tMNVs restore miR-126-3p expression in liver tumor cells. (A, B) HepG2 and SNU449 cells were incubated with 2×1011 MNVs loaded with a nontarget control miRNA (control MNVs) or pre-miR-126-3p (tMNVs), and the relative expression of (A) miR-126 and (B) ADAM9 was assessed after 48 hours. (C, D). Media was collected from HepG2 or SNU449 cells treated with control or tMNVs for 48 h, or from untreated cells. (C) The levels of secreted soluble MICA were quantified by ELISA, and (D) mMICA expression was evaluated by flow cytometry. The values are expressed as the mean±SD from 3 replicates. *p < 0.05, **p <0.01, ***p<0.001. Abbreviations: MICA, major histocompatibility complex I-related chain A; mMICA, membrane-bound MICA; tMNVs, therapeutic MNVs.
MiR-126 replacement in tumor cells modulates NK cell cytolysis
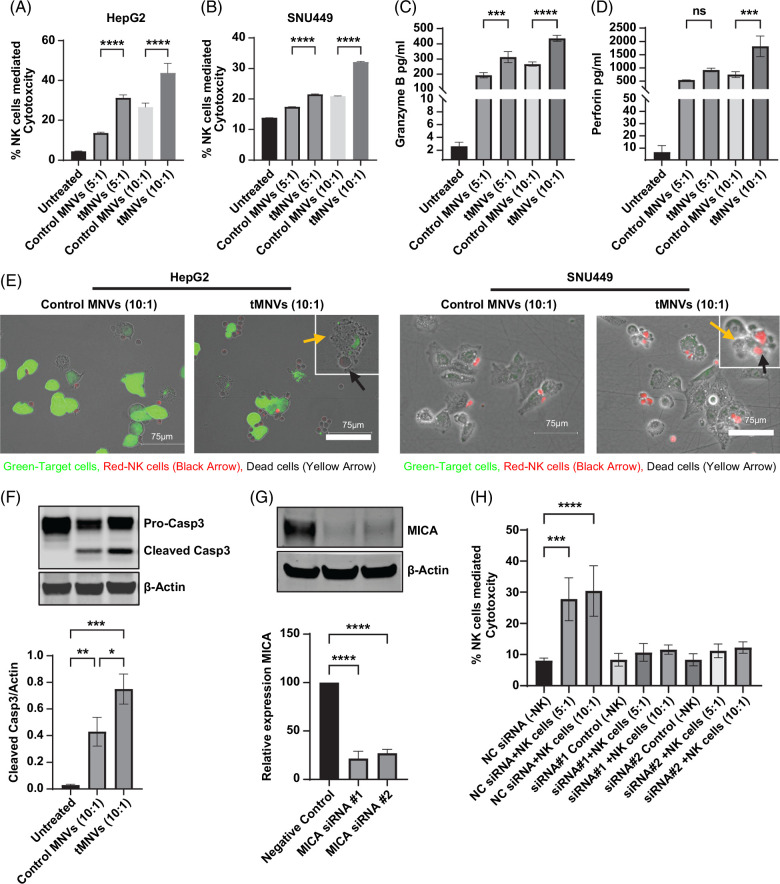
In its soluble form, MICA can attenuate NK cell-mediated antitumor responses.9 We examined the effect of targeting tumor cell ADAM9 expression with miRNA-126-3p on NK cell-mediated responses. Live cell imaging was performed on tMNV pretreated HepG2 cells cocultured with NK cells to evaluate NK cell-mediated cytolysis in real-time (Supplemental Video 1). NK cell-mediated cytolysis was evident as early as 18 minutes after coculture was initiated. Apoptotic blebbing of the target cells was observed after transient contact was established with the effector NK cells. Conversely, HepG2 cells in monoculture under identical settings in a parallel study remained viable throughout the imaging study. Treatment with plain MNVs did not affect NK cell cytotoxicity (Supplemental Figure S2 http://links.lww.com/HC9/A808). Given the retention of surface-bound MICA, we further evaluated the susceptibility of MNV-treated liver tumor cells to NK cell-mediated cytolysis. NK cells were cocultured with MNV-treated tumor cells at 5:1 and 10:1 E:T ratios. Pretreatment of tumor cells with tMNVs enhanced NK cell-mediated cytolysis of both HepG2 and SNU449 cells (Figure 2A, B). Granzyme B and perforin, characterized mediators of NK cell cytolysis, were also increased in NK cell cocultures with tumor cells pretreated with tMNVs (Figure 2C, D). Additionally, coculture of HepG2 cells with NK cells increased caspase-3 cleavage, with the greatest change observed in cocultured cells pretreated with tMNVs (Figure 2F). Fluorescence microscopy confirmed NK cell-mediated cytolysis of tMNVs-treated HepG2 and SNU449 cells (Figure 2E). Conversely, MNV did not have a direct effect on NK cell viability (Supplemental Figure S2A, http://links.lww.com/HC9/A808) and did not modulate NK cell-mediated cytotoxicity (Supplemental Figure S2B, http://links.lww.com/HC9/A808). Furthermore, NK cell-mediated cytotoxicity was reduced during coculture of NK cells with basal or ADAM9 overexpressing HepG2 cells pretreated with control or tMNVs (Supplemental Figure S2C, http://links.lww.com/HC9/A808). Moreover, minimal cytotoxicity was observed in control MNV-treated cocultured tumor cells (Supplemental Figure S3, http://links.lww.com/HC9/A808). Thus, therapeutic MNVs can restore endogenous miR-126 expression in malignant hepatocytes and sensitize them to NK cell-mediated cytolysis. To confirm the functional role of MICA, HepG2 cells were transfected with a siRNA against MICA (Figure 2G). When NK cells were cocultured with MICA-knockdown HepG2 cells, NK cell-mediated cytotoxicity was drastically diminished (Figure 2H, Supplemental Figure S4, http://links.lww.com/HC9/A808). These findings indicate that therapeutic modulation on the miR-126/ADAM9 axis can regulate MICA shedding and could thereby modulate tumor cell immune evasion.
FIGURE 2.
Tumor cell delivery of miR-126-3p increases their sensitivity to NK cell-mediated cytolysis. (A, B) HepG2 and SNU449 cells pretreated with control MNV or tMNV, or untreated controls, were cocultured with NK cells at 5:1 or 10:1 effector-to-target (NK: tumor cell) ratio. NK cell-mediated cytolysis was evaluated after 6 hours by LDH release. (C) Granzyme B and (D) perforin levels were evaluated in control MNV or tMNV pretreated HepG2 cells cocultured with NK cells. (E) Still images from live cell imaging of NK cells cocultured with HepG2 cells or SNU449 cells at 10:1 effector-to-target ratio identify NK cell-mediated cytotoxicity. (F) HepG2 cells were cocultured with NK cells. After 48 hours, adherent cells were harvested, and the expression of pro- and cleaved casp3 was evaluated by western blot. Representative immunoblot and quantitative data from 3 separate experiments are shown. (G) HepG2 cells were transfected with 2 siRNA to MICA, and an immunoblot for MICA was performed on lysates from transfected cells, showing robust silencing. (H) NK cell-mediated cytotoxicity was evaluated by LDH release in HepG2 cells transfected with NC or one of 2 siRNA to MICA and cocultured with or without NK cells at 5:1 and 10:1 effector-to-target ratios. *p < 0.05, **p <0.01, ***p<0.001, ****p<0.0001. Abbreviations: casp3, caspase-3; LDH, lactate dehydrogenase; MICA, major histocompatibility complex I-related chain A; MNVs, milk-derived nanovesicles; NC, nontargeting control; NK cells, natural killer cells; tMNVs, therapeutic MNVs.
Delivery of miR-126-3p increases NK cell sensitivity in tumor spheroids
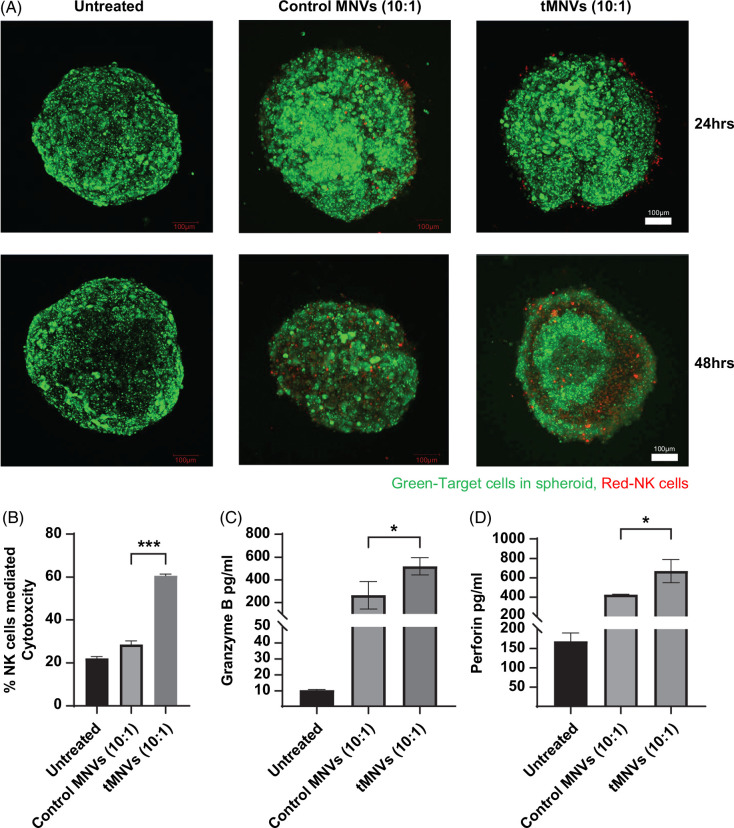
The interplay between tumor cells and nonparenchymal cells within the TME contributes to tumor cell immune evasion. To simulate the in situ environment, we generated multicellular spheroids composed of tumor cells and hepatic stellate cells. In prior studies, we had observed decreased expression of miR-126-3p within MTS compared with monocellular spheroids.4 Therefore, we assessed the therapeutic potential of tMNVs in sensitizing tumor cells within MTS to NK cell cytolysis. Confocal microscopy of tumor spheroids that were pretreated with tMNVs revealed a loss of cells around the periphery (Figure 3A, Supplemental Figure S5, http://links.lww.com/HC9/A808). Next, to evaluate NK cell-mediated cytotoxicity, MTS were pretreated with control MNVs or tMNVs and then cocultured with NK cells at an E:T ratio of 10:1. Compared to control MNVs, tMNVs increased NK cell-mediated cytolysis of tumor cells within spheroids (Figure 3B). Concomitantly, pretreatment with tMNVs increased the secretion of granzyme B (Figure 3C) and perforin (Figure 3D) from cocultures of MTS with NK cells. Thus, incubation with tMNVs can sensitize tumor cells within spheroids to NK cell-mediated cytotoxicity.
FIGURE 3.
Restoration of miR-126-3p sensitizes MTS to NK-mediated cytotoxicity. 3D MTS were prepared with HepG2 cells and LX2 cells in a 4:1 tumor cell to stellate cell ratio, using a methylcellulose-based protocol. The MTS were pretreated with control MNVs or tMNVs, stained with Calcein AM (green), and subsequently cocultured with NK cells labeled with CT Deep Red. (A) Representative confocal microscopy images of treated MCS were captured at 24 and 48 hours post-coculture. Scale bars represent 100 µm. NK cell-mediated cytotoxicity within MTS was evaluated using (B) lactate dehydrogenase activity assay, (C) granzyme B ELISA, and (D) perforin ELISA. The values are expressed as the mean ± SD from 3 replicates. Statistically significant data were represented as follows: *p < 0.05, ***p<0.001. Abbreviations: CT, CellTracker; MNVs, milk-derived nanovesicles; MTS, multicellular tumor spheroids; tMNVs, therapeutic MNVs.
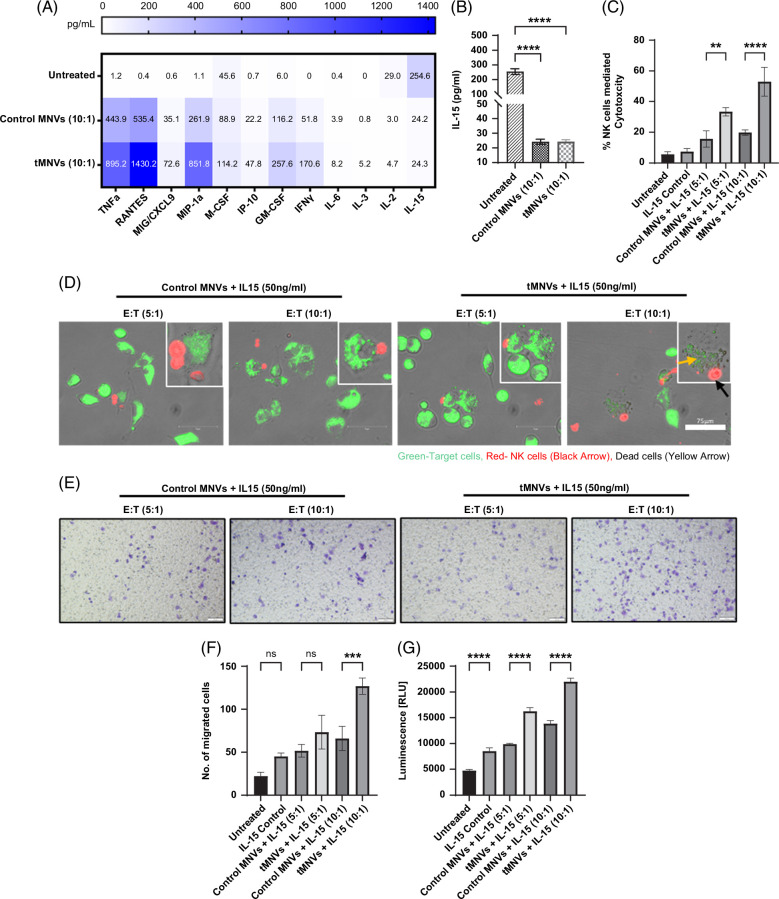
Therapeutic MNVs alter the tumor cell secretome and modulate immune cell behavior
Since NK cell function can be mediated through paracrine signaling, we evaluated the effects of tMNVs on the secretome of NK cells and tumor cells in coculture. To analyze the secretome, the concentrations of several bioactive proteins were measured in CM obtained from tumor cells in monoculture, during coculture with NK cells, or after treatment with MNVs. Twelve proteins were significantly altered in CM obtained from tMNV-treated cells compared to control MNV-treated cells (Figure 4A). Compared to monocultured cells, cocultured cells secreted less IL-15 when incubated with control MNVs or tMNVs (Figure 4B). Priming the NK-HepG2 cocultured cells with IL-15, combined with tMNV pretreated HepG2 cells, enhanced tumor cell sensitivity to NK cell-mediated cytolysis. An increase in cytotoxicity was observed in the 5:1 and 10:1 tMNV-treated coculture cells in comparison to the control MNV treatment groups (Figure 4C, D and Supplemental Figure S6, http://links.lww.com/HC9/A808).
FIGURE 4.
tMNVs alter the tumor cell-NK cell secretome and modulate macrophage migration. HepG2 cells, pretreated with MNVs loaded with nontarget control miRNA (control MNVs) or pre-miR-126 (tMNVs), were cocultured with NK cells at a 10:1 effector:target (E:T, NK cell: HepG2 cell) ratio for 24 hours. Untreated cells in monoculture were used as controls. After 24 hours, the culture supernatant was collected, and the concentrations of 48 cytokines and chemokines were measured. (A) Quantitation of the 12 most abundant proteins in the secretome. (B) IL-15 concentrations were determined by ELISA. (C). HepG2 cells were either untreated or pretreated with control MNVs or tMNVs and cocultured with NK cells in combination with IL-15 treatment. After 6 hours, NK cell-mediated cytotoxicity was evaluated using a lactate dehydrogenase activity assay. (D) Calcein AM-labeled HepG2 cells were cocultured with CT Deep Red stained NK cells in the presence of IL-15. Representative images of NK cell-mediated cytotoxicity were captured by microscopy. Scale bars represent 75 µm. (E-G) Conditioned media was collected from HepG2-NK (10:1) cocultured cells, and the chemotactic potential was evaluated using THP1-differentiated macrophages. (E) Migration of cells was visualized by crystal violet staining. (F). The number of migrating cells on the membrane was counted. (G). Migrated non-adherent cells within the bottom chamber were quantitated using Cell Titer Glo. Scale bars represent 100 µm. The values are expressed as the mean ± SD (n=3 replicates). **p <0.01, ***p<0.001, ****p<0.0001. Abbreviations: CT, CellTracker; NK cells, natural killer cells; tMNVs, therapeutic MNVs.
Several of the immune mediators that were altered by tMNVs in HepG2-NK cell cocultured cells have been implicated in the modulation of macrophage homing and function. Therefore, we evaluated the effect of the secretome on macrophage migration. Studies were performed using a transwell culture system with THP1-differentiated macrophages in the upper chamber and CM from NK-HepG2 cells primed with IL-15 and treated with either control MNVs or tMNVs in the lower chamber. The number of migrated macrophages was quantified by crystal violet staining and using a Cell Titer Glo assay. Exposure to CM from the 10:1 NK cell-HepG2 cell cocultures primed with IL-15 and treated with tMNVs enhanced macrophage migration compared to media harvested from the IL-15-primed, control MNV-treated, cocultured cells (Figure 4E-G and Supplemental Figure S7, http://links.lww.com/HC9/A808). These findings indicate that the secretome of HepG2-NK cells treated with tMNVs can stimulate macrophage chemotaxis.
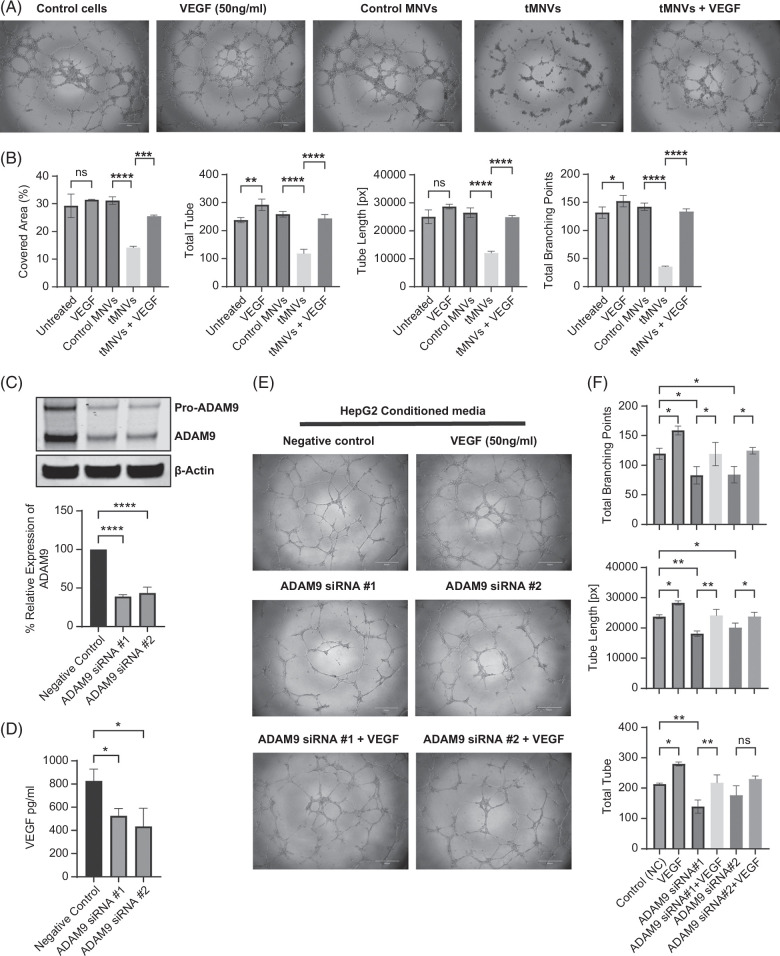
Therapeutic MNVs alter angiogenesis in vitro
In addition to modulating MICA release, the miR-126-3p/ADAM9 axis can regulate angiogenesis in solid tumors. Indeed, multitargeting different effectors of tumor growth formed the basis for the selection of miR-126-3p as a therapeutic. We assessed the effect of tMNVs on the tube formation capabilities of endothelial cells. Administration of tMNVs inhibited human umbilical vascular endothelial cell (HUVEC) cell tube formation, but this was effectively restored with concomitant administration of VEGF (Figure 5A, B). Next, we evaluated the role of endogenous ADAM9 in the anti-angiogenic effects of tMNVs. Transfection of HepG2 cells with either one of 2 ADAM9 siRNA constructs suppressed ADAM9 expression and was accompanied by a decrease in the secretion of VEGF (Figure 5C, D). As VEGF can stimulate angiogenesis, we evaluated the angiogenic potential of HUVEC cells treated with CM from ADAM9-silenced HepG2 cells. Incubation of HUVEC cells with CM from tumor cells transfected with siRNA-1 or siRNA-2 to ADAM9 significantly inhibited tube formation, with reductions in the number of tubes, tube length, and the number of tube branching points compared to negative control-treated HUVEC cells (Figure 5F). Secretion of VEGF was also markedly reduced in ADAM9-silenced HepG2 cells (Figure 5D). The anti-angiogenic effects of ADAM9-silenced tumor cell CM were reversed with co-incubation with VEGF.
FIGURE 5.
The miR-126/ADAM9 axis regulates the angiogenic potential of endothelial cells. (A) HUVEC endothelial cells were pretreated with nothing (untreated controls), 50 ng/mL VEGF, 2 × 1011 MNVs loaded with nontarget control (control MNVs), or miR-126 (tMNVs) with or without VEGF. Cells were then seeded onto a matrigel-coated 96-well plate, and tube formation was assessed after 4 hours. (B) The percentage of area covered, total number of tubes, tube length in pixels, and total number of branching points were quantitated using Wimasis software. (C) HepG2 cells were transfected with one of 2 different siRNA constructs to silence endogenous ADAM9 expression, and a western blot was performed to evaluate knockdown efficiency. Representative immunoblot and quantitative data from 3 studies are shown. (D) CM was obtained from HepG2 cells transfected with control or siRNA to ADAM9, and the VEGF concentration was measured by an ELISA. (E) HUVEC cells with or without siRNA-mediated ADAM9 knockdown were incubated with HepG2-CM or VEGF (positive control) for 24 hours, and tube formation was assessed. (F) The total branching points, tube length, and number of tubes were quantitated using the Wimasis software. The values are expressed as the mean ± SD from 3 replicates. *p< 0.05, **p<0.01, ***p<0.001, ****p<0.0001. Scale bars represent 500 µm. Abbreviations: CM, conditioned media; HUVEC, human umbilical vascular endothelial cell; MNVs, milk-derived nanovesicles; tMNVs, therapeutic MNVs.
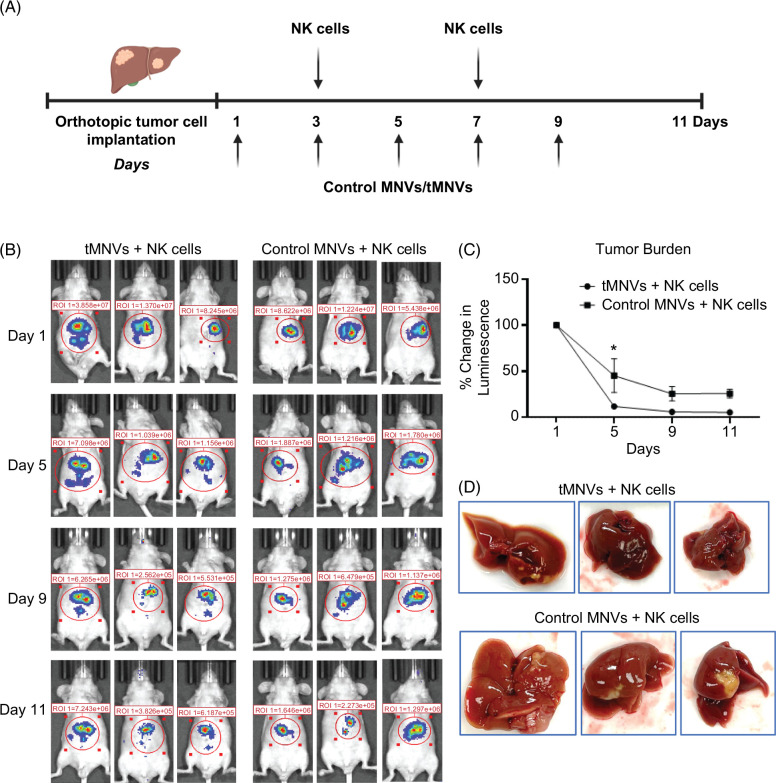
Systemic administration of therapeutic MNVs decreases tumor burden in vivo
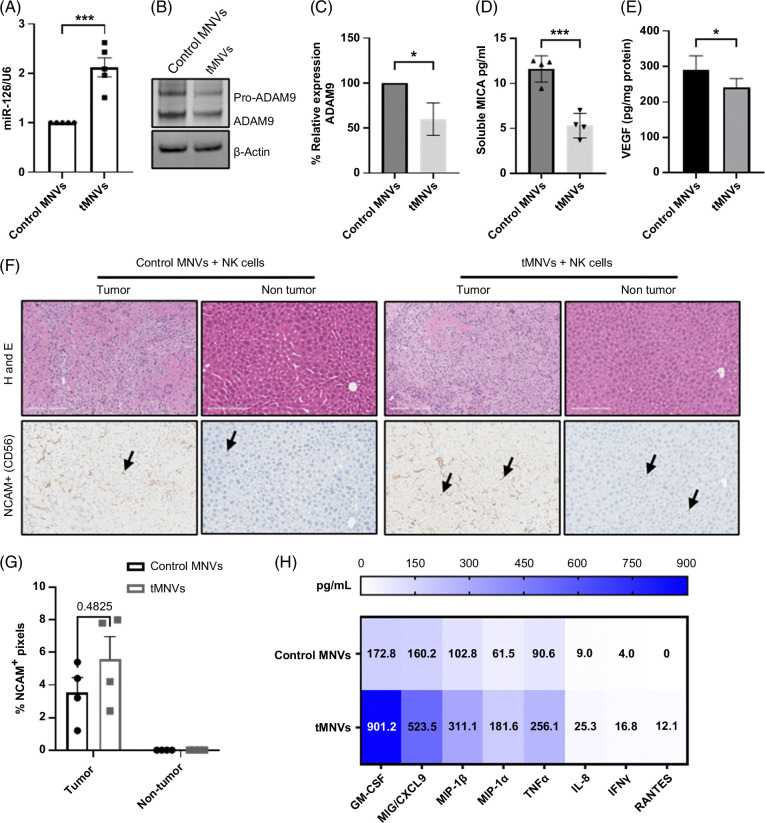
To determine if MNV could directly modulate NK cells in vivo, we performed immune profiling by mass cytometry of splenocytes after iv administration of MNVs or a control vehicle in mice. Overall, NK cells comprised only a small proportion of total hematopoietic cells, and the abundance of NK cells was comparable in MNV-treated or vehicle-treated mice. On Flt-t-distributed stochastic neighbor embedding plots, the size and location of NK cell clusters were comparable across samples (Supplemental Figure S9, http://links.lww.com/HC9/A808). This suggests that systemic administration of MNV does not significantly alter the host NK cell population. Next, to evaluate the effectiveness of therapeutic MNVs to augment NK cell antitumor immune responses in vivo, we established an orthotopic HCC mouse model with luciferase-expressing SNU449 cells. Nine days post-orthotopic injection of SNU449 cells, either control MNVs or tMNVs, were administered by tail vein injection every 2 days for a total of 5 doses. Adoptive transfer of NK cells was performed 3 and 7 days after the initiation of MNV therapy (Figure 6). The tumor burden was markedly reduced in mice treated with tMNVs in combination with adoptively transferred NK cells, compared to mice receiving control MNVs and NK cells. Reduction in tumor burden was associated with a significant increase in the hepatic expression of miR-126-3p in mice with tMNV treatment in combination with adoptively transferred NK cells, compared to mice treated with control MNVs (Figure 7A). As expected, treatment with tMNV in conjunction with NK cell adoptive transfer reduced the hepatic expression of ADAM9 compared with control MNVs/NK cells (Figure 7B and C). Consequently, the serum levels of sMICA were significantly decreased after administration of tMNVs compared with control MNVs (Figure 7D). VEGF expression was reduced in liver tissues from tMNV-treated mice compared with control MNV-treated mice (Figure 7E). Immunohistochemistry revealed an increase in NCAM-positive cell infiltration in the tumor region of livers from HCC-bearing mice treated with tMNVs in combination with NK cells in comparison to those receiving control MNVs (Figure 7F and G). Systemic treatment with tMNVs and adoptive transfer of NK cells altered the immunomodulatory profile, with global increases in the levels of several cytokines and chemokines (Figure 7H). In summary, therapeutic MNVs can effectively restore the hepatic expression of miR-126-3p and modulate the tumor microenvironment and immune escape for a robust anticancer effect.
FIGURE 6.
Systemic delivery of therapeutic MNVs reduce tumor burden in vivo. (A) An orthotopic HCC model was established by intrahepatic injection of SNU449 cells stably transfected to express luciferase. Nine days after implantation, 2 × 1011 nontarget control miRNA loaded MNVs (control MNVs) or miR-126-3p loaded MNVs (tMNVs) were administered by tail vein injection every 2 days for a total of 5 doses. NK cells (2 × 106) were adoptively transferred by tail vein injection on days 3 and 7 postinitiation of treatment with MNV. (B-C) Tumor burden was monitored by IVIS imaging. (D) The gross appearance of tumors harvested at the study endpoint showed a reduction in tumor formation. The values are expressed as the mean and SD (n = 5 mice per group). *p < 0.05. Abbreviations: IVIS, In Vivo Imaging System; MNVs, milk-derived nanovesicles; NK cells, natural killer cells; tMNVs, therapeutic MNVs.
FIGURE 7.
Therapeutic MNVs alter the tumor microenvironment. Livers were harvested, and serum samples were collected from orthotopic HCC tumor-bearing mice treated with control MNVs or tMNVs and adoptively transferred NK cells. (A) RNA was extracted, and miR-126-3p and U6 expression was assessed by RT-PCR. (B, C). Expression of ADAM9 in protein lysates. Representative immunoblots (B) and quantitative data (C) from 4 samples are shown. (D) Soluble MICA levels were quantitated in serum samples by ELISA. (E) VEGF level in tumor tissues was quantified by ELISA. (F) H&E and immunohistochemistry for CD56 were performed in sections from tumor and nontumor regions of the liver. The arrows indicate cells that express CD56. (G) Quantitation of CD56 positivity within cells in tumoral and non-tumoral regions. (H) Serum cytokine and chemokine levels. The values are expressed as the mean andSD (n = 4 mice per group). p < 0.05, ***p<0.001. Scale bars represent 200 µm. Abbreviations: H&E, Hematoxylin and Eosin; MICA, major histocompatibility complex I-related chain A; MNVs, milk-derived nanovesicles; NK cells, natural killer cells; tMNVs, therapeutic MNVs.
DISCUSSION
The combination of anti-angiogenic therapy with immunotherapy represents a first-line therapy for unresectable HCC. Both immune evasion and angiogenesis occur during tumor development and are mechanistically involved in tumor progression. However, therapeutic approaches that specifically target immune evasion by hepatic NK cells are lacking. Herein, we used miRNA restoration as a therapeutic approach to modulate ADAM9 expression within tumor cells and thereby tumor cell immune escape from NK cells by preventing MICA shedding while concomitantly impairing angiogenesis and modulating tumor and macrophage behavior within the TME.
Although NK cells are an essential effector of anticancer responses to malignant cells, their effectiveness can be muted by tumor cells, resulting in immune escape. The use of miRNA for therapeutic modulation of immune escape is appealing, as some miRNAs can target and alter the expression of surface receptors on NK cells or their ligands on tumor cells.17 Efforts have been made to develop miRNA-based therapeutics to restore NK cell functions in some human cancers.18 However, ensuring the direct targeting of NK cells is challenging. In contrast, as demonstrated herein, reducing ADAM9 expression and shedding of membrane-bound MICA using miRNA replacement in tumor cells is feasible and effective in modulating NK cell cytolysis and immune evasion. Interestingly, similar effects have also been observed as off-target effects during treatment with sorafenib.9 In addition to the inhibition of ligand-activated NKG2D signaling by sMICA, impaired NK cell-mediated immune surveillance can also result from a decrease in the number of NK cells or an alteration in the expression of NK receptors such as NKG2D and other receptors.11,19,20 While more targeted approaches either aimed at these or toward increasing NK activation, proliferation, and infiltration could potentially be used to boost immune clearance of tumor cells within the TME, they are less amenable to therapeutic intervention.21
The pleiotropic effects of microRNA on multiple mRNA targets opened up the possibility of targeting other nonimmune processes within the TME. miR-126-3p is a tumor suppressor miRNA that is significantly reduced in liver and other cancers.22–25 Aberrant expression of miR-126-3p is linked to the progression and recurrence of HCC. Restoration of miR-126-3p can be accomplished through direct replacement, as demonstrated in the current study, or through epigenetic modulation, as miR-126 is upregulated following DNA demethylation with 5-aza-2’-deoxycytidine.26,27 . Therapeutic restoration of miR-126-3p in HCC cells sensitized them to NK cell-mediated cytolysis in isolated cells as well as in multicellular spheroids in vitro. Consistent with our findings, other studies have reported an increase in NK cell-mediated cytotoxicity in response to drug-induced repression of ADAM9 expression,9,28 as well as MICA as a target of ADAM9-mediated proteolysis.9 Notably, other miRNAs such as miR-20, can directly target MICA and could provide additional opportunities for RNA therapeutic approaches to modulate tumor cell sensitivity to NK cells.29
The orthotopic tumor studies reported demonstrate the impact on NK cells in vivo but cannot fully recapitulate the human liver cancer immune microenvironment due to the lack of T cells and intra-species differences. The TME in advanced liver cancers is dominated by immunomodulatory cells such as regulatory T cells, myeloid-derived suppressor cells, and M2 polarized macrophages. tMNVs could alter macrophage migration, and further investigation into the effects of miR-126-3p restoration on tumor-associated macrophages is appropriate. These and other immune cells could also contribute to NK cell dysfunction through paracrine and juxtacrine signaling, in part by altering the balance of activating and inhibitory surface receptors expressed on NK cells.30 As this strategy targets tumor cells and does not directly involve or target NK cells, the use of agonists to boost host NK cell function or NK cell adoptive therapy, further augmented with immune adjuvants, may offer complementary approaches to enhance hepatic NK cell-based immunotherapy.31
Restoration of tumor cell miR-126-3p can inhibit tumor angiogenesis in addition to stimulating antitumor immune responses.32 miR-126-5p is encoded within intron 7 of the EGF-like domain 7 gene.33 miR-126 can inhibit tumor angiogenesis in liver cancer cells by negatively regulating EGF-like domain 7 and other targets, as well as directly targeting VEGF. VEGF secretion was reduced in ADAM9 KD liver cancer cells, and the impaired angiogenic activity on HUVEC cells elicited by CM from these cells could be rescued by VEGF. Similar effects have been observed with ADAM9 knockdown in lung cancer cells.34 Treatment with tMNVs inhibited HUVEC cell tube formation in vitro. Furthermore, VEGF expression and angiogenesis were decreased in subcutaneous xenografts of HCC cells, stably overexpressing miR-126-3p compared to wild-type control cells.12 As a tumor suppressor miRNA, miR-126 has been implicated in diverse processes such as inflammation, invasion, migration, and metastases.23,35–37 While not specifically evaluated in the present study, we have previously reported alterations in liver tumor cell migration and the growth of TS in response to miR-126 restoration in vitro.4 miR-126 has several additional targets, such as PI3K, KRAS, CRK, and HOXA9, that can contribute to tumor cell behavior.38 These effects on tumor cell gene expression and phenotype provide additional benefits that further enhance the utility of miR-126 replacement therapies for HCC.
The efficacy of MNVs as biological carriers for the delivery of RNA therapeutics such as ASO or siRNA has been reported.10,18 In this study, we extended these to demonstrate the feasibility of using this platform to deliver intact full-length miRNA to liver cancer cells. The therapeutic efficacy of MNV-based miRNA replacement therapy was shown by a reduction in serum levels of sMICA, decreased tumor burden, and increased tumoral infiltration of NCAM+ cells in vivo. While angiogenesis was not directly examined, alterations in VEGF expression were observed and are consistent with the involvement of the miR-126-ADAM9 axis that has been implicated in angiogenesis. There are several advantages to this approach for miRNA-based therapeutic delivery. The MNVs encapsulate and can thereby protect the therapeutic miRNA cargo from serum nucleases, improve cell penetration, reduce the likelihood of off-target side effects, and, when decorated with tissue-targeting aptamers, achieve selective uptake to enhance targeted delivery of a therapeutic miRNA cargo.12 Our prior studies on the MNVs as cargo shuttles have demonstrated low toxicity in vivo13,39,40 and high accumulation within the liver after iv administration, which makes them appealing for the treatment of liver cancer. However, there are some specific needs prior to translating MNV-based therapies into the clinic. First, there is a need for a high-quality control marker for bovine-derived vesicles. While milk-associated biomarkers such as miR-148a have been proposed as biomarkers of donor bovine health, their use requires further validation prior to adoption.41 Next, endogenous cargo within MNV could have undesirable effects. In vivo toxicological analysis did not reveal any significant immunological, histological, or biochemical effects following systemic administration of MNV, but it remains uncertain if endogenous cargo is abundant enough to elicit a biologically meaningful effect within tumor cells.13,42 Finally, the tMNV platform could be improved by ensuring targeted delivery to liver cancer recipient cells through engineering the MNV surface with tumor cell-targeting moieties.
In conclusion, this study indicates the feasibility and therapeutic impact of concomitantly targeting immune-mediated and nonimmune cellular processes within the liver TME. Restoration of miR-126-3p in tumor cells is an attractive multi-targeted strategy that enhances NK cell-mediated cytolysis and impairs neo-angiogenesis and tumor spread. Given the pleiotropic benefits of miRNA replacement utilizing tMNV, further research is warranted to demonstrate their efficacy in human studies and to explore the therapeutic potential of targeted RNA therapies for liver cancers such as HCC.
Supplementary Material
AUTHOR CONTRIBUTIONS
Tushar Patel: Conceptualization, resources, supervision, and funding acquisition; Tushar Patel, Piyush Gondaliya, Julia Driscoll, Irene K. Yan, and Adil Ali Sayyed: Methodology and investigation; Piyush Gondaliya: Formal analysis, data curation, and visualization; Tushar Patel, Piyush Gondaliya, and Julia Driscoll: Writing—Original Draft; Piyush Gondaliya, Julia Driscoll, Irene K. Yan, and Tushar Patel: Writing—Review and Editing. All authors have read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
Assistance with the study: The authors thank the members of the Patel Lab for their helpful feedback and Brandi Edenfield for the assistance provided with tissue processing for Hematoxylin and Eosin and immunohistochemistry. The authors also thank Drs. Al Copland, Ravi Durvasula, and Gregory J. Gores for providing cells used in these studies.
Presentation: Portions of this work were presented at the Mayo Clinic Symposium on Immuno-Oncology and Tumor Microenvironment Crosstalk at Amelia Island, FL, on December 10, 2022.
FUNDING INFORMATION
This work was supported by the National Cancer Institute [NIH grant CA217833], by the James C and Sarah K Kennedy Deanship, and by Alfred D and Audrey M Petersen Professorship at Mayo Clinic to Dr Tushar Patel.
CONFLICTS OF INTEREST
The authors have no conflicts to report.
Footnotes
Abbreviations: ADAM9, a disintegrin and metalloprotease 9; cc, cocultured; CM, conditioned media; E:T, effector-to-target; HUVEC, human umbilical vascular endothelial cells; MTS, multicellular tumor spheroids; MICA, major histocompatibility complex I-related chain A; MNVs, milk-derived nanovesicles; NK cells, natural killer cells; NKG2D, NK receptor group 2 member D; sMICA, soluble MICA; TME, tumor microenvironment; tMNVs, therapeutic MNVs; tSNE, t-distributed stochastic neighbor embedding.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's website, www.hepcommjournal.com.
Contributor Information
Piyush Gondaliya, Email: gondaliya.piyushkumar@mayo.edu.
Julia Driscoll, Email: driscoll.julia@mayo.edu.
Irene K. Yan, Email: yan.irene@mayo.edu.
Adil Ali Sayyed, Email: ali.adil@mayo.edu.
Tushar Patel, Email: patel.tushar@mayo.edu.
REFERENCES
- 1. Paul S, Lal G. The molecular mechanism of natural killer cells function and its importance in cancer immunotherapy. Front Immunol. 2017;8:1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tian Z, Chen Y, Gao B. Natural killer cells in liver disease. Hepatology. 2013;57:1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pineiro Fernandez J, Luddy KA, Harmon C, O’Farrelly C. Hepatic tumor microenvironments and effects on NK cell phenotype and function. Int J Mol Sci. 2019;20:4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moirangthem A, Gondaliya P, Yan IK, Sayyed AA, Driscoll J, Patel T. Extracellular vesicle-mediated miR-126-3p transfer contributes to inter-cellular communication in the liver tumor microenvironment. Int J Oncol. 2023;62:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gong J, Fang L, Liu R, Wang Y, Xing J, Chen Y, et al. UPR decreases CD226 ligand CD155 expression and sensitivity to NK cell-mediated cytotoxicity in hepatoma cells. Eur J Immunol. 2014;44:3758–3767. [DOI] [PubMed] [Google Scholar]
- 6. Mantovani S, Oliviero B, Lombardi A, Varchetta S, Mele D, Sangiovanni A, et al. Deficient natural killer cell NKp30-mediated function and altered NCR3 splice variants in hepatocellular carcinoma. Hepatology. 2019;69:1165–1179. [DOI] [PubMed] [Google Scholar]
- 7. Zhang J, Xu Z, Zhou X, Zhang H, Yang N, Wu Y, et al. Loss of expression of MHC class I-related chain A (MICA) is a frequent event and predicts poor survival in patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:3123–3131. [PMC free article] [PubMed] [Google Scholar]
- 8. Tao K, Qian N, Tang Y, Ti Z, Song W, Cao D, et al. Increased expression of a disintegrin and metalloprotease-9 in hepatocellular carcinoma: Implications for tumor progression and prognosis. Jpn J Clin Oncol. 2010;40:645–651. [DOI] [PubMed] [Google Scholar]
- 9. Kohga K, Takehara T, Tatsumi T, Ishida H, Miyagi T, Hosui A, et al. Sorafenib inhibits the shedding of major histocompatibility complex class I-related chain A on hepatocellular carcinoma cells by down-regulating a disintegrin and metalloproteinase 9. Hepatology. 2010;51:1264–1273. [DOI] [PubMed] [Google Scholar]
- 10. Oh S, Park Y, Lee HJ, Lee J, Lee SH, Baek YS, et al. A disintegrin and metalloproteinase 9 (ADAM9) in advanced hepatocellular carcinoma and their role as a biomarker during hepatocellular carcinoma immunotherapy. Cancers (Basel). 2020;12:745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jinushi M, Takehara T, Tatsumi T, Hiramatsu N, Sakamori R, Yamaguchi S, et al. Impairment of natural killer cell and dendritic cell functions by the soluble form of MHC class I-related chain A in advanced human hepatocellular carcinomas. J Hepatol. 2005;43:1013–1020. [DOI] [PubMed] [Google Scholar]
- 12. Ishiguro K, Yan IK, Lewis-Tuffin L, Patel T. Targeting liver cancer stem cells using engineered biological nanoparticles for the treatment of hepatocellular cancer. Hepatol Commun. 2020;4:298–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsuda A, Moirangthem A, Angom RS, Ishiguro K, Driscoll J, Yan IK, et al. Safety of bovine milk derived extracellular vesicles used for delivery of RNA therapeutics in zebrafish and mice. J Appl Toxicol. 2020;40:706–718. [DOI] [PubMed] [Google Scholar]
- 14. Sayyed AA, Gondaliya P, Mali M, Pawar A, Bhat P, Khairnar A, et al. MiR-155 inhibitor-laden exosomes reverse resistance to cisplatin in a 3D tumor spheroid and xenograft model of oral cancer. Mol Pharm. 2021;18:3010–3025. [DOI] [PubMed] [Google Scholar]
- 15. Xu L, Hui AY, Albanis E, Arthur MJ, O’Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX-1 and LX-2: New tools for analysis of hepatic fibrosis. Gut. 2005;54:142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matsuda A, Ishiguro K, Yan IK, Patel T. Extracellular vesicle-based therapeutic targeting of beta-catenin to modulate anticancer immune responses in hepatocellular cancer. Hepatol Commun. 2019;3:525–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pesce S, Greppi M, Ferretti E, Obino V, Carlomagno S, Rutigliani M, et al. miRNAs in NK cell-based immune responses and cancer immunotherapy. Front Cell Dev Biol. 2020;8:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neviani P, Wise PM, Murtadha M, Liu CW, Wu CH, Jong AY, et al. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Cancer Res. 2019;79:1151–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cai L, Zhang Z, Zhou L, Wang H, Fu J, Zhang S, et al. Functional impairment in circulating and intrahepatic NK cells and relative mechanism in hepatocellular carcinoma patients. Clin Immunol. 2008;129:428–437. [DOI] [PubMed] [Google Scholar]
- 20. Chu PS, Nakamoto N, Taniki N, Ojiro K, Amiya T, Makita Y, et al. On-treatment decrease of NKG2D correlates to early emergence of clinically evident hepatocellular carcinoma after interferon-free therapy for chronic hepatitis C. PLoS One. 2017;12:e0179096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murugan D, Murugesan V, Panchapakesan B, Rangasamy L. Nanoparticle enhancement of natural killer (NK) cell-based immunotherapy. Cancers (Basel). 2022;14:5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu MH, Ma CY, Wang XM, Ye CD, Zhang GX, Chen L, et al. MicroRNA-126 inhibits tumor proliferation and angiogenesis of hepatocellular carcinoma by down-regulating EGFL7 expression. Oncotarget. 2016;7:66922–66934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du C, Lv Z, Cao L, Ding C, Gyabaah OA, Xie H, et al. MiR-126-3p suppresses tumor metastasis and angiogenesis of hepatocellular carcinoma by targeting LRP6 and PIK3R2. J Transl Med. 2014;12:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gong C, Fang J, Li G, Liu HH, Liu ZS. Effects of microRNA-126 on cell proliferation, apoptosis and tumor angiogenesis via the down-regulating ERK signaling pathway by targeting EGFL7 in hepatocellular carcinoma. Oncotarget. 2017;8:52527–52542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xie QY, Almudevar A, Whitney-Miller CL, Barry CT, McCall MN. A microRNA biomarker of hepatocellular carcinoma recurrence following liver transplantation accounting for within-patient heterogeneity. BMC Med Genomics. 2016;9:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun. 2009;379:726–731. [DOI] [PubMed] [Google Scholar]
- 27. Liu W, Zhang Y, Huang F, Ma Q, Li C, Liu S, et al. The polymorphism and expression of EGFL7 and miR-126 are associated with NSCLC susceptibility. Front Oncol. 2022;12:772405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arai J, Goto K, Stephanou A, Tanoue Y, Ito S, Muroyama R, et al. Predominance of regorafenib over sorafenib: Restoration of membrane-bound MICA in hepatocellular carcinoma cells. J Gastroenterol Hepatol. 2018;33:1075–1081. [DOI] [PubMed] [Google Scholar]
- 29. Tang S, Fu H, Xu Q, Zhou Y. miR-20a regulates sensitivity of colorectal cancer cells to NK cells by targeting MICA. Biosci Rep. 2019;39:BSR20180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun C, Sun H, Zhang C, Tian Z. NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell Mol Immunol. 2015;12:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hu W, Wang G, Huang D, Sui M, Xu Y. Cancer immunotherapy based on natural killer cells: Current progress and new opportunities. Front Immunol. 2019;10:1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kong R, Ma Y, Feng J, Li S, Zhang W, Jiang J, et al. The crucial role of miR-126 on suppressing progression of esophageal cancer by targeting VEGF-A. Cell Mol Biol Lett. 2016;21:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J Mol Med (Berl). 2008;86:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin CY, Cho CF, Bai ST, Liu JP, Kuo TT, Wang LJ, et al. ADAM9 promotes lung cancer progression through vascular remodeling by VEGFA, ANGPT2, and PLAT. Sci Rep. 2017;7:15108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, et al. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10:3–10. [DOI] [PubMed] [Google Scholar]
- 36. Bao J, Yu Y, Chen J, He Y, Chen X, Ren Z, et al. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018;9:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meister J, Schmidt MHH. miR-126 and miR-126*: New players in cancer. ScientificWorldJournal. 2010;10:2090–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rouigari M, Dehbashi M, Ghaedi K, Pourhossein M. Targetome analysis revealed involvement of MiR-126 in neurotrophin signaling pathway: A possible role in prevention of glioma development. Cell J. 2018;20:150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. George J, Yan IK, Patel T. Nanovesicle-mediated delivery of anticancer agents effectively induced cell death and regressed intrahepatic tumors in athymic mice. Lab Invest. 2018;98:895–910. [DOI] [PubMed] [Google Scholar]
- 40. Li S, Tang Y, Dou Y. The potential of milk-derived exosomes for drug delivery. Curr Drug Deliv. 2021;18:688–699. [DOI] [PubMed] [Google Scholar]
- 41. Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010;20:1128–1137. [DOI] [PubMed] [Google Scholar]
- 42. Feng X, Chen X, Zheng X, Zhu H, Qi Q, Liu S, et al. Latest trend of milk derived exosomes: Cargos, functions, and applications. Front Nutr. 2021;8:747294. [DOI] [PMC free article] [PubMed] [Google Scholar]