Abstract
The endoplasmic reticulum (ER) is the major intracellular membrane system. The ER is essential for protein and lipid biosynthesis, transport of proteins along the secretory pathway, and calcium storage. Here, we describe our investigations into the dynamics and regulation of the ER in the early Caenorhabditis elegans embryo. Using a GFP fusion to the ER-resident signal peptidase SP12, we observed the morphological transitions of the ER through fertilization and the early cell-cycles in living embryos. These transitions were tightly coordinated with the division cycle: upon onset of mitosis, the ER formed structured sheets that redispersed at the initiation of cleavage. Although microtubules were not required for the transition of the ER between these different states, the actin cytoskeleton facilitated the dispersal of the ER at the end of mitosis. The ER had an asymmetric distribution in the early embryo, which was dependent on the establishment of polarity by the PAR proteins. The small GTPase ARF-1 played an essential role in the ER dynamics, although this function appeared to be unrelated to the role of ARF-1 in vesicular traffic. In addition, the ER-resident heat shock protein BiP and a homologue of the AAA ATPase Cdc48/p97 were found to be crucial for the ER transitions. Both proteins have been implicated in homotypic ER membrane fusion. We provide evidence that homotypic membrane fusion is required to form the sheet structure in the early embryo.
INTRODUCTION
The endoplasmic reticulum (ER) is the site of protein and lipid synthesis. It is the first compartment of the secretory pathway and is involved in the regulation of intracellular calcium concentration. The ER has distinct functional domains: the continuity with the nuclear envelope, the rough ER and the smooth ER. Therefore, it is perhaps not surprising that the ER has a shape unlike that of the more energetically favorable spherical structures of other organelles like the endosomes, lysosomes, or peroxisomes. The nuclear ER, or nuclear envelope (NE), is distinct from the peripheral ER, which is a network of interconnected sheets and/or tubules that extends throughout the cytoplasm (Terasaki and Jaffe, 1991). The luminal space of the peripheral ER is continuous with the nuclear envelope and together they can comprise more than 10% of the total cell volume (Voeltz et al., 2002). These different levels of organization make it difficult to study the dynamics and reorganization during mitosis and development. However, from studies in different organisms, it appears as though the ER membranes exhibit distinct behavior and organization depending on the stage of development or differentiation. Neurons, for example, display an extensive ER network from the cell body to the tip in dendrites, which are likely to be involved in both Ca2+ regulation and in local translation (Terasaki, 1994; Kiebler and DesGroseillers, 2000). The ER plays an important role during fertilization by regulating the intracellular Ca2+ concentration. In some organisms, this function is attributed to a specialized subcortical layer of ER (Henson et al., 1990; Terasaki et al., 2001).
During cell division, organelles need to be distributed between the newly forming daughter cells. For the ER, it is thought that this distribution is an active process involving the cytoskeleton with microtubules and motor proteins acting to stabilize and transport ER tubules (Bannai et al., 2004). However, it is not clear how the ER is inherited. One view is that the ER vesiculates, probably together with the Golgi apparatus, during mitosis and reforms by a segregation of the ER and the Golgi vesicles and their concomitant fusion into the characteristic sheets of these structures upon completion of cytokinesis (Zaal et al., 1999). Alternatively, the Golgi would not fuse back to the ER during mitosis with the two compartments being inherited independently (Shima et al., 1998; Jokitalo et al., 2001; Shorter and Warren, 2002; Pecot and Malhotra, 2004). Another strategy might be for the ER to maintain its continuity during mitosis, as observed in tissue culture cells where ER markers retain interphase mobility during mitosis (Ellenberg et al., 1997). In contrast, in the syncytial Drosophila embryo the ER forms sheets only during mitosis (Bobinnec et al., 2003). In sea urchin eggs, the ER does not vesiculate during mitosis but accumulates at the mitotic poles (Terasaki, 2000). Thus, ER dynamics and structure appear to vary with organism and developmental state. Yet, one underlying theme for ER dynamics could be the intimate connection with the cytoskeleton. In a variety of cell types the ER tubules often coalign with microtubules, which can lead to ER tubule extension (Terasaki et al., 1986; Allan and Vale, 1991; Waterman-Storer and Salmon, 1998). However, the cytoskeleton is not necessary for the maintenance of the existing ER network because although depolymerizing microtubules with nocodazole inhibits ER tubule growth, the basic tubular-cisternal structure of the ER remains intact (Terasaki et al., 1986).
To gain insight in the ER dynamics in the C. elegans embryo, we generated a GFP marker for the ER that is expressed in the early embryo. We used the C. elegans homologue of the signal peptidase SP12, an ER-resident protein. Such a fusion protein has been successfully used to study ER dynamics in body wall muscles and neurons of the C. elegans adult worm (Rolls et al., 2002). The SP12::GFP fusion protein allowed us to observe changes in ER morphology in living embryos using time-lapse fluorescence microscopy. One of the advantages of using the early C. elegans embryo for our studies is that we could also gain insights in the influence of establishment of polarity on ER dynamics. The C. elegans zygote is highly polarized. On fertilization, which occurs at the posterior pole of the zygote, cytoplasmic streaming occurs and polarity cues are localized; the most prominent of which are the PAR proteins (Kemphues et al., 1988). While the two PDZ domain containing proteins PAR-3 and PAR-6, which together with the atypical protein kinase C (PKC) PKC-3 form a complex at the anterior cortex, the serine threonine kinase PAR-1 and the ring finger protein PAR-2 define the posterior of the embryo (Guo and Kemphues, 1995). The correct localization of these proteins represents the most prominent and earliest hallmark of polarity establishment.
Our results show that the ER in these embryos is highly dynamic. The ER cycles between a dispersed reticulate state and a highly organized sheet state. The transitions between these states are tightly synchronized with the cell-cycle. The sheet structures appear upon pronuclear rotation in the first cell-cycle or just before nuclear envelope breakdown in subsequent cycles. This extensive organization rapidly disperses at the end of mitosis, precisely at the time of initiation of the cleavage furrow. We combined this time-lapse fluorescence microscopy with knockdown of proteins by RNAi or drug treatment to study the factors that might regulate the morphology and dynamics of the ER. We found that the ER exhibits an anterior/posterior asymmetry, which is influenced by the localization of the PARs. We also determined that microtubules are not required for the transitions of the different ER forms whereas the actin cytoskeleton seems to play a role in the dispersal of the ER in late anaphase. Although the small GTPase ARF-1 is required for the cycling of the ER between the reticulate and the sheet structures, the vesicular transport machinery is not essential for this process. Furthermore, we provide evidence that these transitions between the different states require homotypic ER membrane fusion.
MATERIALS AND METHODS
Construction of a Germ-line–targeted ER Marker
The signal peptidase SP12 (C34B2.10) has been used as an ER marker in somatic C. elegans tissues (Rolls et al., 2002). N2 genomic DNA of the C34B2.10 gene coding for an ER resident protein signal peptidase (SP12) was amplified by PCR. The resulting product lacking the first methionine of the coding sequence was cloned into pENTR/D Gateway vector (Invitrogen, Carlsbad, CA). The vector pID3.01 was a generous gift from G. Seydoux. It contains the unc-119 rescuing sequence, EGFP cDNA under the germline-specific pie-1 promoter and a Gateway cassette allowing creation of N-terminal fusions with GFP. SP12 was placed into this vector by the LR Clonase (Invitrogen) reaction between pENTR/D donor and pID3.01 acceptor plasmids.
Biolistic Transformation
Transformation of worms with the vector containing the GFP::SP12 fusion sequence was performed as described by Praitis et al. (2001). Basically, the plasmid described above was coated onto 1-μm gold beads (Bio-Rad, Hercules, CA). unc-119 mutant worms were then bombarded with these coated beads using the Bio-Rad PDS-1000/He Particle Delivery System (Bio-Rad). After bombardment, worms were permitted to grow on 100-mm NGM plates for at least 2 wk. Worms that were rescued by the plasmid (phenotypically wild-type) were examined for GFP expression and those lines expressing GFP were maintained for experimental use. To assist with the timing analysis in utero, one of these lines was crossed with the H2B::GFP-expressing line, AZ212 (Praitis et al., 2001).
Immunocytochemistry
Mouse monoclonal anti-HDEL antibodies were kindly provided by S. Munro (MRC, Cambridge, United Kingdom). Immunocytochemistry was performed essentially as described (Pichler et al., 2000). Briefly, embryos from the SP12::GFP-expressing strain, immobilized on poly-lysine–coated slides, were freeze-cracked and fixed with 3.7% formaldehyde, 75% methanol, 0.5× phosphate-buffered saline for 15 min at –20°C followed by 15 min in absolute methanol. The slides were then rehydrated in PBST. Blocking was performed in PBST containing 5% bovine serum albumin. HDEL antibodies were diluted 1:20 in PBST and incubated with the embryos at 4°C overnight. Secondary CY3-conjugated antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA) were diluted 1:300 and added for 1 h at RT. This treatment did not reduce the fluorescence of SP12::GFP substantially.
RNAi
PCR amplified fragments of the C. elegans genomic DNA corresponding to exons of the genes of interest were used to produce double-stranded RNA for microinjection into the gonad or intestine as described by Fire et al. (1998). Typically 18–25 L4 and young hermaphrodites of the SP12::GFP strain were injected with the appropriate dsRNA and analyzed 18–24 h postinjection. Usually, the concentration of injected dsRNA was 1 μg/μl. For the genes where RNAi resulted in sterility, we reduced the dsRNA concentration 3–8-fold. The brood size of these worms was still very low, and the embryonic lethality approached 100%.
The following C. elegans putative orthologues of the mammalian and yeast genes were subjected to RNAi cdk-1; Cdc48/p97: C06A1.1 (78%), C41C4.8 (79%), and K04G2.3 (34%) (identity to human orthologues is indicated); p47: Y94H6A.9; BiP/Kar2: hsp-3; hsp-4; hsp-1; Jem1: dnj-10; rab-1; NSF/Sec18: nsf-1; Sar1: inx-9 (ZK792.3); sec-23; arf-1; nmy-2. RNAi against α-tubulin (tba-2) was performed using the feeding method described by Kamath and Ahringer (2003).
Multiphoton and Confocal Microscopy
Confocal time-lapse image series were acquired with a Leica TCS SP2 confocal laser scanning microscope system (Deerfield, IL). The laser intensity, scanning speed and time intervals were adjusted empirically not to affect the wild-type embryo viability for at least 1 h of imaging time. Embryos from the dissected worms were mounted on agarose pads in egg salts buffer. For the in utero imaging the worms were immobilized with 12 mM levamizole. Images were processed with either Leica LCS software or Adobe Photoshop 6.0 (San Jose, CA). Live oocytes and embryos were imaged using a multiphoton excitation-based optical workstation (Wokosin et al., 2003) that included a Nikon Eclipse (Nikon, Melville, NY) inverted microscope, a 100× oil immersion lens (1.3 NA), and a Spectra Physics (Spectra Physics, Mountain View, CA) titanium sapphire laser set at a wavelength of 900 nm. The scanning and image collection were controlled via the Bio-Rad Lasersharp software (Bio-Rad, Hercules, CA). Imaging intervals ranged from 2.22 to 8 s, depending on image size and scan rate. Embryos and worms were mounted as described for confocal imaging.
For the quantification of the ER asymmetry during the first zygotic division, we took confocal images of the embryos during pronuclear rotation and centration. ER patches ≥1.5 μm in diameter in direct vicinity of the cortex were counted.
For the 3D reconstruction from confocal images, Z-stacks of confocal images were analyzed for two typical stages: interphase and the onset of mitosis. Any continuous ER structures, along with the nuclear envelope were manually traced. Each plane was assigned a distinct color. The outlines were then superimposed according to their Z-stack order. The 3D reconstruction is an angle view rotated 45° along the X axis. The spacing between Z-stacks in this reconstruction was set in scale with the actual spacing of confocal images.
Preparation of C. elegans Embryos for EM and Image Analysis
Whole young adult worms containing embryos were prepared by high-pressure freezing followed by freeze substitution as described by McDonald (1999). The substitution was carried out in two stages: 1) incubating in the primary medium for 68 h at –90°C and then warming to –60°C, and 2) rinsing and replacing with the secondary medium for an additional 26 h as the samples were slowly warmed to 0°C. The primary medium was 1% glutaraldehyde dissolved in 98% acetone/2% water. The secondary medium was 2% osmium tetroxide dissolved in 98% acetone/2% water (Walther and Ziegler, 2002). Serial longitudinal sections 65 nm thick were collected and stained with saturated aqueous uranyl acetate followed by 0.4% lead citrate. Imaging was performed on a Phillips CM 120 (FEI, Hillsboro, OR) at 80 kV.
Digital images captured on Soft Imaging Systems (Lakewood, CO) CCD camera and software, were automatically stitched together using the “Multiple Image Alignment” tool. Images were imported to Photoshop (Adobe Systems) and RER was highlighted using the brush tool on a second image layer. Sequential layers with different colored highlights were then pasted to a new image and aligned as closely as possible to each other using the x,y translation and rotate tools.
Drug Treatment
Embryos expressing the SP12::GFP fusion protein were treated with pharmacological compounds that interfere with cytoskeletal dynamics using laser ablation basically as previously described (Skop et al., 2001; Wokosin et al., 2003). Briefly, the embryos were sensitized in 1 mg/ml Trypan Blue in egg salts for ∼30 s on a coverslip. The Trypan Blue solution was removed and replaced with egg salts containing the drug of interest. The coverslip was inverted over a circle of Vaseline on a microscope slide, creating a chamber in which the solution on the coverslip could hang. Short bursts of 450-nm nanosecond pulses from a nitrogen-pumped dye laser—part of the optical workstation— were aimed at the viteline membrane causing small perforations to be made through the membrane and eggshell, allowing ingression of the drug. Because the ablation laser was an integral part of the optical workstation, images of the embryo could be collected both before and immediately after the ablation. As a control, dimethyl sulfoxide (DMSO) was used at a 1:100 dilution; Latrunculin A (Calbiochem, La Jolla, CA) was used at a concentration of 200 μM; nocodazole at a concentration of 25 μg/ml; brefeldin A (BFA; Molecular Probes Eugene, OR) at a concentration of 150 μg/ml. To increase exposure to the BFA, very early embryos (before eggshell formation) were incubated in 15 μg/ml BFA in blastomere culture medium (Shelton and Bowerman, 1996) and imaged in hanging drops of this solution.
RESULTS
Generation of an ER Marker for the Early C. elegans Embryo
To study the dynamics of the ER in the early embryo, we used the C. elegans homologue of the signal peptidase SP12 (C34B2.10) and appended it with GFP and generated transgenic lines of worms expressing this construct. SP12 has already been used successfully as a marker for the ER in the body wall muscles and neurons of C. elegans adults (Rolls et al., 2002). Positive lines were selected and one of these lines was tested for the correct ER localization of SP12::GFP by coimmunofluorescence with an antibody recognizing the ER retention signal (–HDEL; Figure 1). As expected, the markers colocalized in early C. elegans embryos. In addition, the expression of the transgene did not appear to interfere with the development or the growth rate of the worms (unpublished data). Therefore, SP12::GFP is a suitable marker for studying ER dynamics in the early C. elegans embryo.
Figure 1.
SP12::GFP localizes to the ER in the early embryo. Immunofluorescence with antibodies against the ER-retention signal HDEL was performed on embryos expressing SP12::GFP.
The ER in the C. elegans Embryo Organizes into Reticular Structures during Prometaphase
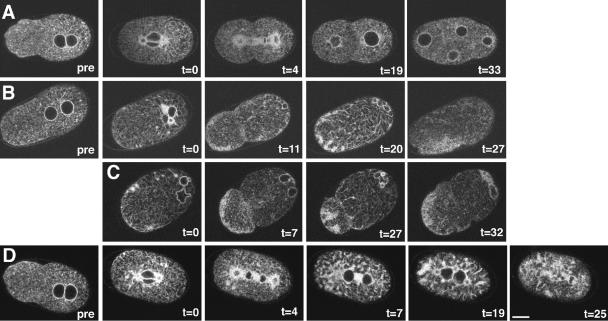
We used time-lapse multiphoton and confocal microscopy to study the ER dynamics in the early embryo and found that the ER undergoes dramatic cyclical changes in organization (Figures 2 and 3, Supplementary Movie 1). During pronuclear migration, the ER was in a dispersed state with small bright foci (Figure 2; t = –3.9, t = –1.2). ER around the centrosomes became ring-like as the two pronuclei moved toward the center of the embryo and the ER started to develop a more macroscopic structure (Figure 2; t = 0.3) as the pronuclei rotated and changed its appearance dramatically as the spindle began to form (Figure 2; t = 2.3). ER reticulations extended out toward and also lined the cortex. During this “highly organized” state dense regions of accumulated ER were also observed. These dense regions were often found directly adjacent to the ER outlining the spindle (Figure 2; t = 3.2). As mitosis progressed, the ER outlining the spindle took on an hourglass shape, with the waist forming at the spindle midzone region (Figure 2; t = 4.3) and the ER reappeared around the reforming anaphase nuclei (Figure 2; t = 4.3). The “reticulation” remained until late anaphase, when the ER rapidly dispersed again at the onset of cleavage (Figure 2; t = 4.5, t = 5.2; and Figure 3). As cytokinesis progressed, a swath of ER was observed at the developing midzone, as if the ER that outlined the spindle moved in to fill the space left by the migrating chromosomes (Figure 2; t = 5.2). Coincident with the transition to the dispersed state, the ER around the centrosomes also became more diffuse. The swath of ER in the midzone narrowed as the furrow ingresses but a faint trail of this swath of ER remained between the scission site and the nuclei (Figure 2; t = 8). This cyclical change in ER organization continued through subsequent cell cycles (Figure 2; t = 15.2 through t = 29.8; and Figure 3), such that the dispersed state coincided with interphase and the reticulate phase coincided with mitosis (Figure 3).
Figure 2.
The ER changes it shape during the cell cycle. A multiphoton time-lapse series from SP12::GFP expressing embryo is shown. The completion of pronuclear centration has been designated t = 0. During pronuclear migration and centration (t = –3.9 and t = –1.2, respectively) the ER was dispersed throughout the cytoplasm and was contiguous with the nuclear envelope. Although the centrosomes were not present in these two focal planes, the ER did surround the centrosomes at theses stages, as it did in later stages. As the spindle begins to form the ER surrounded the centrosomes and the spindle (t = 3.2). Although the ER was absent from the chromosomes during metaphase, it returned to the DNA during anaphase (t = 4.5). At this stage, the ER became dispersed again as the cleavage furrow initiated and a band of ER was found between the reforming nuclei (brackets, t = 5.2, t = 8). This band disappeared after cleavage furrow completion (t = 12.7). The cycle of dispersed and strongly reticulate ER was repeated in subsequent cell divisions (t = 15.2 through t = 29.8). Bar, 10 μm.
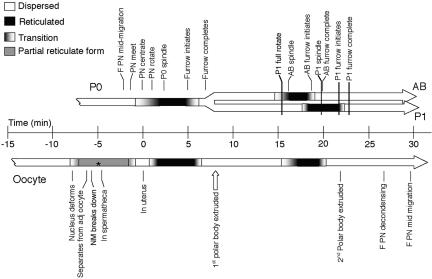
Figure 3.
Time line of changes in ER configuration in relation to developmental events. The top bar of the graph shows the timing of the changes of ER between the dispersed (white) and reticulate (black) states in the early embryo. The transitions (gradient) indicate the period during which the first sign of change in configuration is seen until the entire cell exhibits that state. This shows that the reticulate form is present coincident with the mitotic spindle (correlation coefficients are P0 = 0.41; AB = 0.93; P1 = 0.83; over all 3 cycle = 0.99), whereas the disperse form coincides with the ingression of the cytokinetic furrow (correlation coefficients are P0 = 0.72; AB = 0.99; P1 = 0.99; over all 3 cycles = 0.99) and is maintained throughout interphase. The data for each time point were tabulated from a minimum of 6 to a maximum of 9 embryos. The bottom bar shows these changes in ER organization as they occur in the oocyte, in utero, as it is moves through the spermatheca into the uterus. During the partial reticulate state (gray) the ER takes on a more string-like appearance, particularly near the cortex and nucleus. Asterisk indicates approximate expected time of fertilization based on published descriptions (Kemphues and Strome, 1997). The data for each time point were compiled from a minimum of 3 to a maximum of 9 oocytes. F, female; PN, pronucleus. The time of the 1st polar body extrusion (open arrow) was estimated from a separate set of five embryos expressing a histone::GFP construct in the SP12::GFP background.
The ER was also distributed asymmetrically (Figure 4, WT). Conspicuous cortical ER patches were observed predominantly in the anterior part of the embryo during mitosis. This region corresponds to the region where PAR-3 and associated proteins segregate as the embryo becomes polarized during pronuclear migration (Kemphues and Strome, 1997). This anterior accumulation resulted in a higher ER concentration in the anterior AB daughter cell compared with P1 and was dependent on the nonmuscle myosin NMY-2, which acts upstream of the PAR machinery (Guo and Kemphues, 1996). Knockdown of NMY-2 by RNAi resulted into evenly distributed ER throughout the cell, which was defective in cytokinesis (Figure 4). Cortical patches were rarely present and distributed randomly. Taken together, upon knockdown of NMY-2 the ER was equally distributed both in P0 and later in the subsequent multinucleated cell. A similar symmetric ER distribution was observed after knockdown of PAR-3 and PAR-6 in the early embryo. By contrast, RNAi of PAR-2, which restricts PAR-3 to the anterior of the zygote, did not influence the asymmetric ER distribution. However, cytoplasmic streaming still occurs in par-2 mutants with a similar speed to wild-type embryos but for about one-fifth the duration (Cheeks et al., 2004). Our results indicate that asymmetric ER distribution is disturbed if polarity is not setup correctly.
Figure 4.
The ER is concentrated at the anterior part of the embryo. (A) Confocal time-lapse series of early embryos are depicted. Upon strong reticulation during pronuclear rotation in the first division cycle, cortical ER patches were visible that were concentrated at the anterior. Subsequently the AB cell received more ER than P1. The asymmetric distribution of the ER is dependent on polarization of the zygote. dsRNA was injected into the gonad of SP12::GFP worms. Embryos were analyzed 12–18 h postinjection by confocal microscopy. Knockdown of NMY-2, PAR-3, and PAR-6 by RNAi led to symmetric distribution of the ER, whereas in the case of PAR-2, although the asymmetry of the zygote was lost, the ER remained asymmetrically distributed as in wild type. (B) Quantification of the position of cortical ER patches in fertilized oocytes at pronuclear rotation in RNAi knockdown experiments. For the quantification of the ER asymmetry during the first zygotic division, we took confocal images of the embryos during pronuclear rotation and centration. ER patches ≥1.5 μm in diameter in direct vicinity of the cortex were counted. In the wild type (n = 6) and PAR-2 (n = 6) knockdown most of the cortical patches were found in the anterior portion, whereas in PAR-3 (n = 4) and PAR-6 (n = 3) knockdown experiments, cortical patches were distributed throughout the embryos. The p-value was for all quantifications < 0.001.
Taken together, the ER is highly dynamic in early C. elegans embryos and was present mostly in a dispersed state and in organized, reticulate structures. We found that cycles of reticulation and assembly were repeated in all subsequent divisions observed (Figure 2); reticulate structures always appeared just before nuclear envelope breakdown. Reticulate structures were visible until late anaphase, when they dispersed within seconds at the onset of cleavages.
The ER Forms Sheets at the Onset of Mitosis in the Early Embryo
To characterize the reticulate state further, we analyzed stacks of successive focal planes of early embryonic cells in interphase and at onset of mitosis (Figure 5). The ER structures were highly mobile and dynamic. Therefore, we chose a region of interest around the nuclei in interphase cells and in cells just before nuclear breakdown, when the reticulation began. The nucleus served in all these cases as intrinsic control for a continuous ER structure. We used four-cell-stage embryos, because the ER sheets were less mobile than in two-cell embryos—probably due to space constraints— and it allowed us to analyze dispersed and reticulated ER in the same embryo. In interphase cells, the perinuclear ER could be traced throughout all sections (Figure 5A, 3.74 μm), which is expected because it surrounds the nucleus. Yet, the ER structures in the cytoplasm appeared less continuous. The picture changed dramatically, when we analyzed cells at the onset of mitosis (Figure 5B). The ER appeared more coherent, and this coherence was maintained for most of the ER structures through the stack of 3.74 μm. This result indicates that ER can form sheets that are at least 3.74 μm in diameter.
Figure 5.
The ER forms large sheet structures at onset of mitosis. 3D reconstruction of Z-stacks of confocal images of a four-cell embryo, with one cell in interphase and another one at onset of mitosis. The Z-stacks were collected around the nucleus in order to have an internal control for large ER continuums. The pictures on the right side represent one focal plane of the aligned group. The distance between two focal planes was 0.34 μm, and a stack comprised 12 images.
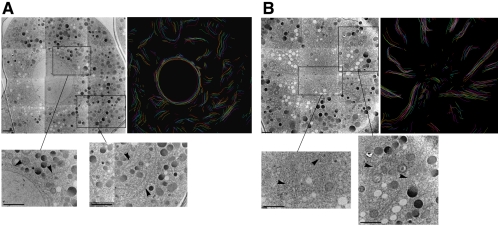
But how do these changes observed at the light microscopic level translate to the ultrastructural level? To address this question, we performed an analysis of serial sections taken from early embryonic cells either in interphase or in mitosis (Figure 6). As expected for dispersed ER, short ER tubules/cisternae of varying length and orientation were detected in serial sections from interphase cells. The unambiguous identification of the curvilinear ER membranes was possible, because the ER was covered with ribosomes (Figure 6A). The ER was still lined with ribosomes in cells in mitosis, yet the ER appeared to be more contiguous, consistent with the sheet-like appearance detected at the light microscopic level. The peripheral ER became longer, more linear, and more radially oriented than that observed in interphase. In the more central region, a higher density of short ER fragments was seen. These fragments may represent the cross-sectional view of a convoluted organization of the ER in this region. We did not detect stacked ER cisternae (karmellae, Snapp et al., 2003) in the embryos, indicating that the ER membranes might have to fuse to form the sheet structures we observed during mitosis.
Figure 6.
Ultrastructure of ER. These electron micrographs show the ultrastructural organization of the ER in the early embryo in either interphase (A) or prometaphase (B). For each set of images, the first image is a composite showing a section through the embryo, the second image is a tracing of multiple sequential sections, with each color representing a single section (see Materials and Methods for additional details), and each inset shows greater detail of the ER structure in either the central region (left inset) or in the more peripheral cytoplasm (right inset). The inset may be either from the same section as the shown composite or from another section in the same series. Arrowheads point to representative ER structures. Tracing in A consists of nine sections, and the tracing in B consists of 14 sections. Scale bar, 2 μm.
The ER Changes Its Appearance in a Cell cycle–dependent Manner
The data in Figures 2 and 3 suggest that the transition of the ER from dispersed to reticulated, or interconnected sheets, occurred in a tightly regulated manner and was reiterated through subsequent cell divisions. To determine the earliest time point at which such a transition could occur we examined ER dynamics in the oocyte. Before fertilization, the ER was dispersed, but underwent dramatic changes as the oocyte moved through the spermatheca into the uterus (Figure 7, Figure 3, Supplementary Movie 2). In the mature oocyte, the maternal nucleus arrests in the prophase of meiosis I (Kemphues and Strome, 1997). At this stage, the oocyte is cylindrical with a large nucleus and dispersed ER (Figure 7A; t = –16.4). As the DNA started to condense, but although the nuclear membrane was still intact, some accumulation of ER strands at the cortex was observed. As the oocyte moved away from its neighboring oocyte, distinct ER strings were observed at the cortex and near the collapsing nucleus (Figure 7A; t =–7.0). By the time the oocyte entered the spermatheca (Figure 7A; t = –6.5; t = –3.2), the ER became dispersed and remained dispersed as the oocyte exited the spermatheca (Figure 7A; t = 1.4). The oocyte is fertilized upon entry into the spermatheca and then undergoes two meiotic divisions (Kemphues and Strome, 1997). After exiting the spermatheca, the ER underwent a cycle of reticulation (Figure 7A; t = 1.4) and dispersion (Figure 7A; t = 5.0), associated with the first of these meiotic divisions. This first reticulation-dispersion cycle is not as pronounced as the subsequent cycles. After the dispersed state, a cloud of ER around the meiotic chromosomes became more focused at the spindle poles (Figure 7A; t = 18.2). The ER returned to the dispersed state as the second polar body extruded (Figure 7A; t = 21.9) and remained in the dispersed state through pronuclear migration. In addition to the sheet structures, meiotic ER contains numerous large foci at the junctions of the sheets (Figure 7A; t = 18.2). These foci grew and then rapidly disappeared before the appearance of the pronuclei. This sequence of events is repeated as the next oocyte prepares to move into the spermatheca (Figure 7A; t = 28.5 through t = 52.7). The striking observation, that the ER cycles in the absence of the formation of daughter cells, suggests that the dynamic changes of the ER might be unrelated to the inheritance of ER to the two daughter cells.
Figure 7.
The cycling of the ER is linked to the cell-cycle. (A) Time-lapse multiphoton of oocyte/embryo development in utero in a worm expressing SP12::GFP. The ER changes its appearance in a cyclical manner as it moves through the reproductive tract. nu indicates the nucleus of the oocyte that will next be moving into the spermatheca. Brackets designate the region occupied by the spermatheca, and the gray arrow shows the direction the oocyte will be moving. White arrows in t = 18.2 show the accumulation of ER at the acentrosomal spindle poles before completion of the second meiotic division. Times are in minutes, with t = 0 set when the fertilized oocyte completely exits the spermatheca into the uterus. Bar, 10 μm. (B) Time-lapse confocal microscopy series of an SP12::GFP-expressing worm in which CDK-1 levels were reduced by RNAi. The fertilized oocytes arrest with dispersed ER at a time where sheets should be present. Bar, 10 μm.
The sheet structures appeared concomitantly with pronuclear centration and rotation and disappeared in late anaphase in the first division cycle, suggesting these structural transitions are linked to the cell cycle. We investigated this possibility with an RNAi experiment where we knocked down a key cell cycle regulator, cell cycle–dependent kinase 1 (Cdk1/Cdc2: NCC-1/CDK-1 in C. elegans, Table 1). After this treatment, the fertilized eggs arrest upon pronuclear meeting with a dispersed ER (Figure 7B). The maternal nucleus did not proceed through meiosis I and II and did not extrude any polar bodies. The nuclei failed to rotate and centrate upon CDK-1 RNAi treatment. The ER did not reach the transition point to organize sheet structures, indicating that the ER cannot cycle in the absence of cell cycle cues. This suggests that the transition between dispersed and strongly reticulate ER is linked to the cell cycle.
Table 1.
Genes targeted by RNAi
| Gene targeted | Phenotype | ER cycling defect | No. of embryos observed |
|---|---|---|---|
| cdk-1 | Ste | Yes | 6 |
| Cdc48/p97 (C06A1.1) | Emb (when together with C41C4.8 (RNAi)) | No | 26 |
| Cdc48/p97 (C41C4.8) | Emb (when together with C06A1.1 (RNAi)) | No | 26 |
| Cdc48/p97 (K04G2.3) | Emb | Yes | 16 |
| p47 (Y94H6A.9) | WT | No | 10 |
| BiP/Kar2 (hsp-3) | WT | No | 9 |
| BiP/Kar2 (hsp-4) | Emb | Yes | 17 |
| BiP/Kar2 (hsp-1) | Ste | NA | NA |
| Jem1 (dnj-10) | Emb, Lva | No | 7 |
| rab-1 | Ste, Emb | No | 8 |
| NSF/Sec18 (nsf-1) | Ste, Emb | No | 12 |
| Sar1 (inx-9) | Ste, Emb | No | 5 |
| sec-23 | Ste | NA | NA |
| arf-1 | Ste, Emb | Yes | 9 |
| ran-1 | Emb | No | 5 |
| ufd-1 | Emb, Lva | No | 11 |
NA, not applicable.
The Changes in ER Structure Are Independent of Microtubules But Not of Microfilaments
Numerous studies implicate the cytoskeleton in ER dynamics in a variety of systems (Voeltz et al., 2002), yet the cytoskeleton does not seem necessary for the maintenance of the ER network. We observed ER surrounding the centrosomes and radiating out toward the cortex, probably situated adjacent to astral microtubules (Figures 1 and 2). Therefore, we tested whether the transitions from dispersed to sheet structures of the ER were dependent on microtubules. We treated embryos with nocodazole, a microtubule-depolymerizing drug. Although the one-cell embryo had major problems with cell division and cytokinesis, it was clear that the ER still cycled (Figure 8, row B). Similar results were obtained when α-tubulin was knocked down by RNAi (Figure 8, row C). These results indicate that the transitions in ER structure are independent of microtubule dynamics and that the formation of reticular structures is independent of the presence of microtubules. We asked next whether microfilaments were required for the cyclical changes in the morphology of the ER. In one-cell embryos treated with latrunculin A, a drug that disrupts microfilaments, the ER did not disperse properly after mitosis (Figure 8, row D). This result indicates that the actin cytoskeleton might have an important role in the transition from the sheet to the dispersed state.
Figure 8.
The actin cytoskeleton is required for ER transitions. Time-lapse multiphoton microscopy was performed on embryos expressing SP12::GFP. (A) ER in a control embryo that was ablated in the presence of DMSO, the carrier for the cytoskeletal drugs. (B) Microtubules were disrupted with nocodazole. Nocodazole did not interfere with the cycling of the ER. (C) Using RNAi to knockdown α-tubulin resulted in a similar phenotype than treatment of the embryo with nocodazole. (D) Disrupting the microfilaments by latrunculin A prevents ER from cycling in a coordinated manner. The ER forms some tubular structures but is unable to disperse completely. In addition the accumulation of ER at the anterior is lost. Image labeled “pre” shows the embryo before ablation. Times are in minutes, with t = 0 set at the stage of the first reticulation (completion of pronuclear rotation). Bar, 10 μm.
The Small GTPase ARF-1 Is Required for the Transition from Dispersed to Sheet-structured ER
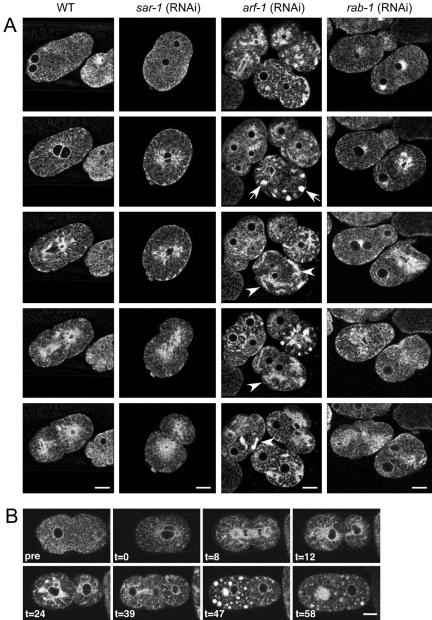
To gain further insight into the dynamic changes in the structure of the ER, we examined components of the protein and membrane transport machinery by reducing the activity of key regulators in the ER-Golgi traffic interface using RNAi. First, we focused on the small GTPases, ARF-1 and SAR-1, which are essential for the formation of COPI (from the Golgi to the ER) and COPII (from the ER to the Golgi) vesicles, respectively. A number of components of the ER-Golgi shuttle led to sterility when knocked down by RNAi (Skop et al., 2004). To circumvent this problem, we adjusted the strength of the RNAi phenotype by lowering the concentration of injected dsRNA by a factor of 5. This led to a few fertilized eggs per injected worm. When we injected dsRNA homologous to sar-1 mRNA, we observed an embryonic lethal phenotype, as previously reported, however the ER behaved like wild-type during the first cell divisions (compare Figure 9A WT and sar-1; RNAi). When we repeated the experiment with Arf-1, which is responsible for COPI vesicle generation at the Golgi apparatus, the transition from dispersed to sheets still occurred but seemed incomplete (Figure 9A, arf-1; RNAi). Large amounts of ER accumulated in bulky foci in the cell that only partially dispersed after anaphase. At stages when we would normally observe sheet structures in wild-type embryos, large aggregates of ER were observed throughout the cell with an absence of sheets. Thus, ARF-1 but not SAR-1 activity seems to be required for the transitions of the ER. To further examine this intriguing possibility, we treated wild-type early embryo with the fungal metabolite BFA. BFA is an inhibitor of the guanine nucleotide exchange factor for ARF-1 and leads to the absorption of at least the cis-Golgi elements into the ER (Lippincott-Schwartz et al., 1989; Peyroche et al., 1999). Treatment of eggs with BFA resulted in a phenotype similar to that observed with RNAi against ARF-1 (Figure 9B), supporting the involvement of this small GTPase in cyclical changes of the ER morphology observed in these embryos. Yet, interfering with ARF-1 function by RNAi or BFA did not alter the asymmetric distribution of the ER or the polarity in the early embryo (Figure 9, Skop et al., 2001).
Figure 9.
RNAi of ARF-1 and BFA interfere with ER cycling. (A) Knockdown by RNAi was performed on SP12::GFP-expressing worms. The embryos were observed by time-lapse confocal microscopy. Although cycling is normal in embryos treated with SAR-1 and RAB-1 dsRNA, knockdown of ARF-1 results in ER clumps and proper sheet formation is inhibited. The arrowheads point to clumped and collapsed ER. (B) Brefeldin A inhibits proper ER cycling. Multiphoton series of an embryo treated with BFA. BFA leads to a compromised dispersal of ER and patches of undispersed ER. The 2–4 cell division initiates but fails (t = 47), as has been previously reported (Skop and White, 2001) and results in large aggregates of ER. The 1–2 cell division is normal even though the distribution of the ER is becoming abnormal at this point, suggesting that the BFA initially disrupts the ER before its effect on cytokinesis. Time is in minutes, with t = 0 ∼1–2 min after drug application, whereas “pre” shows embryos before drug treatment. Bars, 10 μm.
To investigate the role of membrane traffic in ER changes further, we injected worms with dsRNA against RAB-1. RAB-1 is a member of the family of Rab-GTPases, which are required for fusion of transport vesicles with the target membrane. It is assumed that RAB-1 plays this role in the fusion of retrograde COPI vesicles from the Golgi to the ER, although this has not yet been formally shown. Full-strength RNAi led to defects in oogenesis and resulted in sterility, which is a common phenotype when transport components are impaired (unpublished data). Lowering the dsRNA dose resulted in a few fertilized eggs with an ER that appeared wild-type and that cycled in a cell cycle–dependent manner (Figure 9, rab-1; RNAi). These results indicate that the effect of ARF-1 depletion on ER dynamics may not be due to a common vesicle traffic defect in the ER-Golgi shuttle per se, but rather connected to other functions of ARF-1.
In untreated embryos, as mitosis began the ER accumulated at the centrosomes, along the mitotic spindle and was observed around the chromosomes in anaphase (Figure 2). Because RAN-1 is involved in the reformation of the nuclear envelope after mitosis (Askjaer et al., 2002), we wondered whether nuclear envelope formation and dispersal of the ER sheets are connected. Although, as reported before, RNAi of RAN-1 resulted in a defect in nuclear envelope assembly, the transitions between sheet structured and dispersed ER still occurred (unpublished data). Thus, the dynamic of these two different events, nuclear envelope assembly and general ER cycling, appear to be independent from each other.
Homotypic Fusion Events Are Essential for Transitions in the ER Morphology
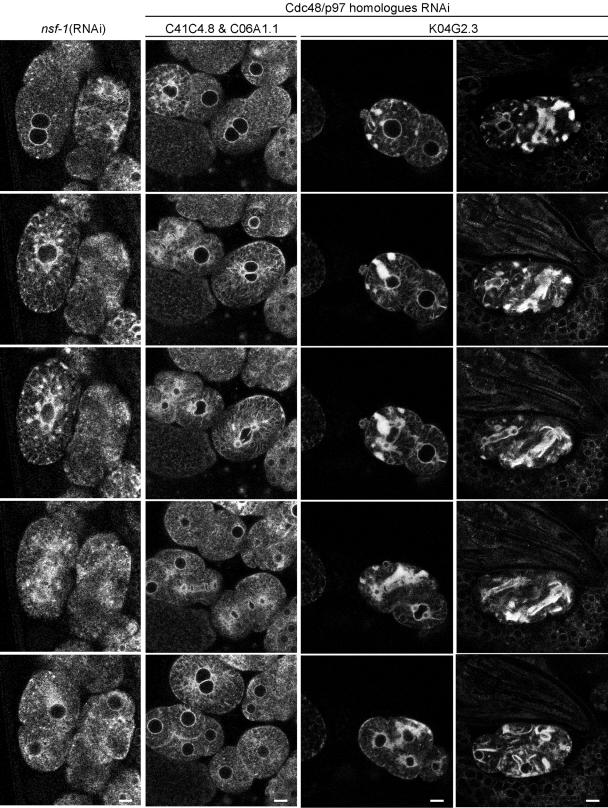
The dynamic changes we observed in the organization of the ER might, at least in part, be due to homotypic ER fusion. The ER sheets could be a way to equipartition ER onto the two daughter cells, without loosing the integrity of the ER, which would occur if the Golgi apparatus was absorbed into the ER during division. Alternatively, the sheet structure might be important for certain stages of the cell cycle because it would allow contents of the ER lumen to readily diffuse. Therefore, back fusion of Golgi elements induced by inhibiting ARF-1 function could lead to a defect in forming sheet structure, such as we observed. If this assumption was correct, one would expect that the function of proteins involved in homotypic membrane fusion should be required for the transition of dispersed to sheet-structured ER. Therefore, we examined the role of AAA-ATPases in ER dynamics. Cdc48/p97 and NSF are AAA-ATPases that play a role in homotypic ER fusion and transport vesicle fusion with the ER, respectively (Latterich et al., 1995; Spang and Schekman, 1998). Cdc48/p97 is involved in ubiquitin-mediated degradation at the ER and in regulation of Golgi morphology (Rabouille et al., 1995; Kondo et al., 1997; Meyer et al., 2000). We tested some of the closest homologues of Cdc48/p97 in C. elegans for their influence on ER morphology by RNAi (Frohlich, 2001). Reduction of the two most highly conserved Cdc48/p97 homologues, C06A1.1 (78% identity to human p97) and C41C4.8 (79% identity), resulted in 30–50% embryonic lethality. When both proteins were knocked down simultaneously, the phenotype was more pronounced and resulted in 100% dead embryos (Figure 10). The third candidate K04G2.3 (34% identity) showed in our hands a 100% embryonic lethal phenotype after RNAi treatment. When we assayed the ER dynamics in embryos that were treated with dsRNA, only RNAi of K04G2.3 affected cycling of the ER (Figure 10). The phenotype was similar to that observed when ARF-1 function was compromised, but was more pronounced. Large ER aggregates formed at times when the sheet structures would normally be present in wild-type embryos. These aggregates persisted in part during the dispersed state of the ER. Thus, homotypic ER membrane fusion might be a prerequisite for the morphological transitions of the ER during the cell cycle. In accordance with these results, RNAi of C. elegans homologues of p47 and UFD-1, cofactors of Cdc48/p97 required for reformation of the Golgi apparatus after mitosis (Rabouille et al., 1995) and ubiquitin-mediated degradation (Meyer et al., 2000), respectively, resulted in no alteration of ER dynamics, although at least in the case of ufd-1 (RNAi) ∼50% of the progeny did not survive (Table 1 and unpublished data).
Figure 10.
Cdc48 is required for ER transitions. Time-lapse confocal microscopy of RNAi treated SP12::expressing embryos. The knockdown of NSF-1 and 2 Cdc48 homologues does not interfere with ER cycling. RNAi of NSF-1 does lead to a more granular ER over time. Knockdown by RNAi of Cdc48 (K04G2.3) causes the ER to collapse. The first row depicts a time-lapse 24 h and the second row 48 h postinjection. Bars, 10 μm.
Our results described above indicated that vesicular ER-Golgi transport might not be involved in the observed morphological changes of the ER. To provide further evidence, we knocked down the N-ethylmaleimide–sensitive factor (NSF/Sec18, NSF-1 in C. elegans). Here again, we had to use a reduced dose of dsRNA in order to get some fertilized eggs. Yet, the ER cycled normally in the embryos treated with low doses of dsRNA (Figure 10). This result supports the previous observation that vesicle transport is not required to permit the transition between the dispersed and sheet-like ER.
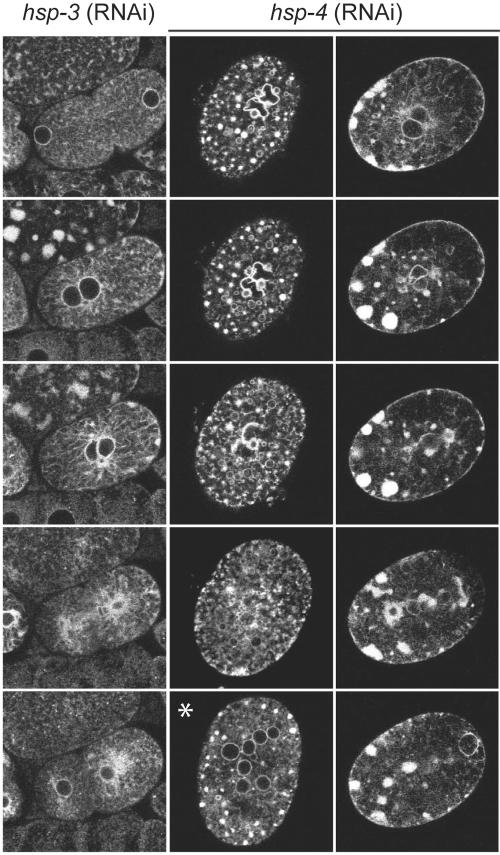
BiP Is Essential to Form Sheet-like ER Structures
The previous results suggested that changes in ER morphology might be dependent on homotypic membrane fusion. Therefore, we wanted to test other factors known to be involved in homotypic membrane fusion. The ER-resident heat-shock protein BiP was identified in the yeast Saccharomyces cerevisiae in a screen for factors involved in karyogamy (Rose et al., 1989). Latterich and Schekman (1994) went on to show that BiP (Kar2p in yeast) is essential for homotypic ER membrane fusion. There are two strong homologues of BiP in C. elegans: HSP-3 and HSP-4 with 66 and 65% identity, respectively. Both seemed to be expressed in early embryos (Hill et al., 2000). hsp-3 (RNAi) did not affect ER dynamics in the early embryos (Figure 11). In contrast, when HSP-4 function was reduced by RNAi, the ER accumulated in multiple foci when it normally would have formed the sheet structure (such as during nuclear rotation, Figure 11, Supplementary Movie 5). The foci dispersed somewhat over time, but clearly the ER morphology was distinctly disrupted. A second example is shown in Figure 11, there the ER is located mostly in clumps at the cortex, and a few tubular structures were observed. When the cell attempted to divide, a portion of the ER dispersed at the end of mitosis. However, most of the ER remained in a less clearly organized state. To exclude the possibility that the phenotype observed with HSP-4 was due to accumulation of unfolded proteins, we used RNAi to reduce the levels of another protein involved in proper folding in the ER, DNJ-10 (a DNAj-like protein). We chose DNJ-10 because the yeast homologue Kar8p is involved in karyogamy. It has been reported, that Kar8p is required for the fusion of the nuclear membrane, but not the ER membrane, the step, which requires BiP/Kar2p action (Brizzio et al., 1999). Although the reduction of DNJ-10 resulted in embryonic lethality, no defects in cycling of the ER were observed in these embryos (unpublished data).
Figure 11.
RNAi of BiP leads to clumping of the ER. Time-lapse confocal microscopy of RNAi treated SP12::expressing embryos. Knockdown of HSP-3 by RNAi did not cause any defects in ER dynamics. In contrast, RNAi of HSP-4 destroys the ER morphology completely. Two examples are depicted by subsequent frames of a time-lapse analysis. The asterisk indicates the third example, a multinucleated single cell. Most of the embryos arrested with this phenotype.
Taken together our results suggest that at least the transition between the dispersed to sheet structured ER requires homotypic membrane fusion.
DISCUSSION
We studied the dynamics of the ER in the early C. elegans embryo using a fusion of the signal peptidase SP12 to GFP. The fusion protein did not interfere with the development and the growth of the worms and the marker protein localized properly to the ER as shown by colocalization with proteins that carry the ER retention signal HDEL. The ER is a highly dynamic endomembrane structure and studies from other organisms indicate that these dynamics are dependent on the cytoskeleton and membrane fusion. In C. elegans embryos, the ER cycles between two major states: the dispersed state and the sheet state. The transition between the diffuse and sheet stage of the ER seemed to be regulated by the cell cycle and occurred not only during the first mitosis but also in subsequent divisions, at least as long as the cells were big enough to allow us to follow the different transitions. Fixation of the embryos, as shown in Figure 1, did not preserve the sheet structure very well. Thus, the different stages of the ER in the early embryo might have been underestimated in previous studies using immunohistochemistry. At the onset of centration and pronuclear rotation in the first division cycle, the reticulation of the ER became more pronounced and sheets of ER were formed, which persisted until anaphase, when the ER dispersed again. These changes in appearance were not dependent on microtubule dynamics, because depolymerization of microtubules did not result in a block of ER transitions. In contrast, the actin cytoskeleton seems to play an important role, because the ER did not cycle properly in the presence of the drug latrunculin A; in particular, the ER did not disperse completely. Actin-based motors might be required to disperse the ER throughout the cell. Inheritance of cortical ER and transport of ER subcompartments are dependent on actin and microtubules in different organisms (Prinz et al., 2000; Wedlich-Soldner et al., 2002; Bannai et al., 2004). In early sea urchin embryos, the ER does not cycle between sheets and dispersed structures; however, it does accumulate at the spindle poles during mitosis. This accumulation is not dependent on the presence of microtubules (Terasaki, 2000). However, the ER dynamics described in other systems are not comparable to the massive rearrangement we observe in C. elegans embryos. More related ER morphologies are observed during syncytial mitosis in the Drosophila embryo (Bobinnec et al., 2003), where the ER forms sheets during mitosis and is dispersed in telophase. The authors speculated that the ER network could participate in separating the sister nuclei in the absence of conventional cytokinesis because the clustering of ER material is reminiscent of the formation of a phragmoplast in plant cytokinesis. Interestingly, although the compact accumulation at the scission site observed in Drosophila is not evident in C. elegans embryos, we did notice accumulations of ER membranes along the ingressing cleavage furrow (Figure 2) and ER was also present in the spindle midzone region. The accumulation of the ER around the spindle in the Drosophila and C. elegans embryos could serve a conserved mechanism: in both systems the nuclear envelope has to reform quickly to prepare for the next mitosis. The ER around the spindle could help to sequester components of nuclear envelope there and facilitate the reformation of the nuclear envelope after anaphase.
Not only did the ER cycle between a sheet and dispersed state, it was also distributed asymmetrically such that the anterior part of the embryo retained a higher concentration of ER. However, knockdown of PAR-2, which is localized to the posterior part of the zygote did not affect the ER concentration although the embryo divided symmetrically. Conversely, RNAi treatment for PAR-3 and PAR-6, anterior polarity markers, lead to symmetrically distribution of the ER. How can we reconcile these results? One explanation would be that the initial establishment of the anterior domain is independent of PAR-2, because PAR-6::GFP still collapses to the anterior pole in the absence of PAR-2 (Cuenca et al., 2003). Knockdown of the nonmuscle myosin NMY-2 by RNAi might destroy/prevent asymmetric ER distribution, because the cortical localization of the PARs is lost (Munro et al., 2004). However, microtubule dynamics do not interfere with the establishment of polarity and therefore the ER is still asymmetrically distributed in tubulin knockdown embryos. Another, and perhaps more likely, scenario could be that cytoplasmic streaming is required to segregate the ER asymmetrically. In par-3 and par-6 mutants, cytoplasmic flow is abolished (Cheeks et al., 2004) and hence the ER is not enriched at the anterior pole. In contrast, the initial cytoplasmic movement in par-2 mutants is indistinguishable from that in wild-type, although the duration is much shorter (Cheeks et al., 2004). In a PAR-2 knockdown the residual cytoplasmic streaming might be sufficient to partition the ER asymmetrically.
To study the influence of components involved in membrane dynamics and secretion, we used a mixed approach of drugs and RNAi. We are aware of the limitation of RNAi; the failure to detect a phenotype might coincide with the failure to knockdown the target RNA sufficiently. However, most of the targeted proteins in this study did exhibit a phenotype (e.g., embryonic lethality), even in the absence of an ER cycling defect. Moreover, the reduced dose of RNAi was often necessary in order to collect at least some fertilized oocytes. Furthermore, we did confirm the RNAi results by drug treatment, whenever possible. Perhaps most importantly, our major conclusions, that the actin cytoskeleton and homotypic membrane fusion are necessary for ER dynamics in C. elegans, are based on positive, and not on negative, RNAi results and/or pharmacological treatment. With these caveats, we are confident that this approach is valid for gaining insight into the regulation of ER dynamics.
There is still considerable debate on how the Golgi is inherited during mitosis. One possibility is that the Golgi is absorbed into the ER and reemerges after mitosis (Zaal et al., 1999). Another possibility is that Golgi inheritance is independent of the ER (Jokitalo et al., 2001; Pecot and Malhotra, 2004). Our results argue for an independent inheritance mechanism, at least in the C. elegans embryo. Interfering with ARF-1 function by either RNAi or BFA treatment, should lead to absorption of Golgi to the ER. If this were the natural sequence of events one would not expect that upon this treatment the cycling of the ER would be disrupted as, indeed, it is. Interestingly, the cycling between the different ER morphologies did not require the vesicle transport machinery but still involved ARF-1 function. However, at least in yeast it has been shown that Arf1p is required for proper actin assembly (Bonangelino et al., 2002). Therefore, the effects observed with RNAi against ARF-1 and BFA could be attributed to the actin phenotype. Alternatively, and perhaps more likely, the defects in ER cycling in the absence of active ARF-1 can be attributed to the fusion of the Golgi with the ER. This interpretation is supported by the our findings that orthologues of proteins involved in homotypic ER fusion in yeast are essential for the transition of ER membranes from dispersed state to sheets in C. elegans embryos. On mixing of ER and Golgi membranes, homotypic ER fusion is no longer possible and thus the formation of ER sheets is blocked. We assessed three homologues of Cdc48/p97 for their effects on ER dynamics. Only one of them blocked ER sheet formation. Therefore, it is not surprising that the C. elegans putative orthologues of the Cdc48/p97 cofactors p47 and Ufd1p/Npl4p had no effect on ER distribution, especially because these cofactors have not been implicated in homotypic ER fusion in yeast. Ufd1p/Npl4p are required for proteasome-mediated ER-associated degradation and p47 is required for reformation of the Golgi apparatus after mitosis (Rabouille et al., 1995; Kondo et al., 1997; Meyer et al., 2000). Although the result on the knockdown of p47 has to be taken with caution, because neither we nor others were able to detect a phenotype after dsRNA treatment using different methods and target sequences (Rual et al., 2004). Finally, the C. elegans orthologue of the yeast BiP, Kar2p, which is required for homotypic ER, but not nuclear envelope fusion (Brizzio et al., 1999) was essential for maintenance of ER integrity and cycling. In contrast, knockdown of the orthologue of yeast Jem1p/Kar8p, which is essential for nuclear envelope, but not ER, fusion, displayed no ER cycling defect. Even though the dsRNA treatment resulted into embryonic and larval lethality. Thus, the shape changes of the ER membranes are likely to require homotypic ER fusion. This mechanism might provide a way to rapidly build the sheet structures. This might be particularly important in biological systems, such as the Drosophila or C. elegans embryo, which have exceedingly short cell cycle times, on the order of a few minutes. In conclusion, homotypic ER fusion could bring about sheet formation, while the actin cytoskeleton would be involved in dispersing the ER at exit of mitosis. This cycling seems to be independent of vesicular transport, because knockdowns by RNAi of COPII components, which form vesicles at the ER, or NSF-1, which is essential for vesicle fusion with the acceptor membrane, have no influence on these particular ER dynamics.
What could be the role of the cycling of the ER between these two states? We do not know the answer yet. One could be tempted to speculate about a role in signaling, to allow rapid protein diffusion between the cortex and the cell center or to adjust the morphology to the secretory capacity. Future experiments will be required to elucidate the function of the different ER morphologies.
Acknowledgments
We acknowledge G. Seydoux, M. Rolls, and S. Munro for the bombardment vector, the SP12 plasmid, and the anti-HDEL antibodies. This work was supported by the Max Planck Society (A.S.), National Institutes of Health (NIH) grant F32 GM063349 (J.M.S) and NIH grant GM57583 (J.G.W.). A.S. is an EMBO Young Investigator.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–08–0726) on February 16, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Allan, V. J., and Vale, R. D. (1991). Cell cycle control of microtubule-based membrane transport and tubule formation in vitro. J. Cell Biol. 113, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askjaer, P., Galy, V., Hannak, E., and Mattaj, I. W. (2002). Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell 13, 4355–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai, H., Inoue, T., Nakayama, T., Hattori, M., and Mikoshiba, K. (2004). Kinesin dependent, rapid, bi-directional transport of ER sub-compartment in dendrites of hippocampal neurons. J. Cell Sci. 117, 163–175. [DOI] [PubMed] [Google Scholar]
- Bobinnec, Y., Marcaillou, C., Morin, X., and Debec, A. (2003). Dynamics of the endoplasmic reticulum during early development of Drosophila melanogaster. Cell Motil. Cytoskeleton 54, 217–225. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C. J., Chavez, E. M., and Bonifacino, J. S. (2002). Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio, V., Khalfan, W., Huddler, D., Beh, C. T., Andersen, S. S., Latterich, M., and Rose, M. D. (1999). Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 609–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeks, R. J., Canman, J. C., Gabriel, W. N., Meyer, N., Strome, S., and Goldstein, B. (2004). C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14, 851–862. [DOI] [PubMed] [Google Scholar]
- Cuenca, A. A., Schetter, A., Aceto, D., Kemphues, K., and Seydoux, G. (2003). Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development 130, 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg, J., Siggia, E. D., Moreira, J. E., Smith, C. L., Presley, J. F., Worman, H. J., and Lippincott-Schwartz, J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138, 1193–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Frohlich, K. U. (2001). An AAA family tree. J. Cell Sci. 114, 1601–1602. [DOI] [PubMed] [Google Scholar]
- Guo, S., and Kemphues, K. J. (1995). par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611–620. [DOI] [PubMed] [Google Scholar]
- Guo, S., and Kemphues, K. J. (1996). A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature 382, 455–458. [DOI] [PubMed] [Google Scholar]
- Henson, J. H., Beaulieu, S. M., Kaminer, B., and Begg, D. A. (1990). Differentiation of a calsequestrin-containing endoplasmic reticulum during sea urchin oogenesis. Dev. Biol. 142, 255–269. [DOI] [PubMed] [Google Scholar]
- Hill, A. A., Hunter, C. P., Tsung, B. T., Tucker-Kellogg, G., and Brown, E. L. (2000). Genomic analysis of gene expression in C. elegans. Science 290, 809–812. [DOI] [PubMed] [Google Scholar]
- Jokitalo, E., Cabrera-Poch, N., Warren, G., and Shima, D. T. (2001). Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R. S., and Ahringer, J. (2003). Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321. [DOI] [PubMed] [Google Scholar]
- Kemphues, K. J., Priess, J. R., Morton, D. G., and Cheng, N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311–320. [DOI] [PubMed] [Google Scholar]
- Kemphues, K. J., and Strome, S. (1997). Fertilization and establishment of polarity in the embryo. In: The Nematode C. elegans, II, ed. T.B.D. Riddle, B. Meyer, and J. Priess, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 335–359. [PubMed]
- Kiebler, M. A., and DesGroseillers, L. (2000). Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron 25, 19–28. [DOI] [PubMed] [Google Scholar]
- Kondo, H., Rabouille, C., Newman, R., Levine, T. P., Pappin, D., Freemont, P., and Warren, G. (1997). p47 is a cofactor for p97-mediated membrane fusion. Nature 388, 75–78. [DOI] [PubMed] [Google Scholar]
- Latterich, M., Frohlich, K. U., and Schekman, R. (1995). Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82, 885–893. [DOI] [PubMed] [Google Scholar]
- Latterich, M., and Schekman, R. (1994). The karyogamy gene KAR2 and novel proteins are required for ER-membrane fusion. Cell 78, 87–98. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S., and Klausner, R. D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, K. (1999). High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 117, 77–97. [DOI] [PubMed] [Google Scholar]
- Meyer, H. H., Shorter, J. G., Seemann, J., Pappin, D., and Warren, G. (2000). A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19, 2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, E., Nance, J., and Priess, J. R. (2004). Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev. Cell 7, 413–424. [DOI] [PubMed] [Google Scholar]
- Pecot, M. Y., and Malhotra, V. (2004). Golgi membranes remain segregated from the endoplasmic reticulum during mitosis in mammalian cells. Cell 116, 99–107. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J., and Jackson, C. L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275–285. [DOI] [PubMed] [Google Scholar]
- Pichler, S., Gonczy, P., Schnabel, H., Pozniakowski, A., Ashford, A., Schnabel, R., and Hyman, A. A. (2000). OOC-3, a novel putative transmembrane protein required for establishment of cortical domains and spindle orientation in the P(1) blastomere of C. elegans embryos. Development 127, 2063–2073. [DOI] [PubMed] [Google Scholar]
- Praitis, V., Casey, E., Collar, D., and Austin, J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz, W. A., Grzyb, L., Veenhuis, M., Kahana, J. A., Silver, P. A., and Rapoport, T. A. (2000). Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Biol. 150, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raul, J. F., Ceron, J., Koreth, J., Hao, I., Nicot, A. S., Hirozane-Kishikawa, T., Vandenhaute, J., Orkin, S. H., Hill, D. E., van den Heuvel, S., and Vidal, M. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille, C., Levine, T. P., Peters, J. M., and Warren, G. (1995). An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell 82, 905–914. [DOI] [PubMed] [Google Scholar]
- Rolls, M. M., Hall, D. H., Victor, M., Stelzer, E. H., and Rapoport, T. A. (2002). Targeting of rough endoplasmic reticulum membrane proteins and ribosomes in invertebrate neurons. Mol. Biol. Cell 13, 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., Misra, L. M., and Vogel, J. P. (1989). KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell 57, 1211–1221. [DOI] [PubMed] [Google Scholar]
- Shelton, C. A., and Bowerman, B. (1996). Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development 122, 2043–2050. [DOI] [PubMed] [Google Scholar]
- Shima, D. T., Cabrera-Poch, N., Pepperkok, R., and Warren, G. (1998). An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J. Cell Biol. 141, 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter, J., and Warren, G. (2002). Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 18, 379–420. Epub 2002 Apr 2002. [DOI] [PubMed] [Google Scholar]
- Skop, A. R., Bergmann, D., Mohler, W. A., and White, J. G. (2001). Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr. Biol. 11, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A. R., Liu, H., Yates, J., 3rd, Meyer, B. J., and Heald, R. (2004). Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305, 61–66. Epub 2004 May 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp, E. L., Hegde, R. S., Francolini, M., Lombardo, F., Colombo, S., Pedrazzini, E., Borgese, N., and Lippincott-Schwartz, J. (2003). Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 163, 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., and Schekman, R. (1998). Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol. 143, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki, M. (1994). Redistribution of cytoplasmic components during germinal vesicle breakdown in starfish oocytes. J. Cell Sci. 107, 1797–1805. [DOI] [PubMed] [Google Scholar]
- Terasaki, M. (2000). Dynamics of the endoplasmic reticulum and golgi apparatus during early sea urchin development. Mol. Biol. Cell 11, 897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki, M., Chen, L. B., and Fujiwara, K. (1986). Microtubules and the endoplasmic reticulum are highly interdependent structures. J. Cell Biol. 103, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki, M., and Jaffe, L. A. (1991). Organization of the sea urchin egg endoplasmic reticulum and its reorganization at fertilization. J. Cell Biol. 114, 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki, M., Runft, L. L., and Hand, A. R. (2001). Changes in organization of the endoplasmic reticulum during Xenopus oocyte maturation and activation. Mol. Biol. Cell 12, 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz, G. K., Rolls, M. M., and Rapoport, T. A. (2002). Structural organization of the endoplasmic reticulum. EMBO Rep. 3, 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, P., and Ziegler, A. (2002). Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 208, 3–10. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer, C. M., and Salmon, E. D. (1998). Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr. Biol. 8, 798–806. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner, R., Schulz, I., Straube, A., and Steinberg, G. (2002). Dynein supports motility of endoplasmic reticulum in the fungus Ustilago maydis. Mol. Biol. Cell 13, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wokosin, D., Squirrell, J., Eliceiri, K., and White, J. (2003). Optical workstation with concurrent, independent multiphoton imaging and experimental laser microbeam capabilities. Rev. Sci. Inst. 74, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaal, K. J. et al. (1999). Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell 99, 589–601. [DOI] [PubMed] [Google Scholar]