Abstract
E2F transcription factors play an essential role in cell proliferation and apoptosis and their activity is frequently deregulated in human cancers. In a yeast two-hybrid screen we identified a novel E2F-binding protein. Due to its strong phosphorylation we named it EAPP (e2F-associated phosphoprotein). EAPP is localized in the nucleus and interacts with E2F-1, E2F-2, and E2F-3, but not with E2F-4. Examination of a number of human cell lines revealed that EAPP levels are elevated in most transformed cells. Moreover, EAPP mRNA was detected in all investigated human tissues in varying amounts. EAPP is present throughout the cell cycle but disappears during mitosis. In transfection assays with reporters controlled by either an artificial E2F-dependent promoter or the murine thymidine kinase promoter, EAPP increased the activation caused by E2F-1 but not by E2F-4. Surprisingly, the promoter of the p14ARF gene, which was also activated by E2F-1, became repressed by EAPP. Overexpression of EAPP in U2OS cells resulted in a significant increase of cells in S-phase, whereas RNAi-mediated knock down of EAPP reduced the fraction of cells in S-phase. Taken together, these data suggest that EAPP modulates E2F-regulated transcription, stimulates proliferation, and may be involved in the malignant transformation of cells.
INTRODUCTION
Studies of transcriptional regulatory mechanisms have demonstrated the critical role that promoter-binding proteins have on the expression of a given gene. Many of these transcription factors are composed of independent domains, which define DNA-binding specificity, activate transcription or mediate interaction with other proteins. The E2F transcription factor family is believed to integrate cell cycle progression with transcription through its cyclical interactions with important cell cycle regulators such as the retinoblastoma-tumor suppressor gene product (pRB), cyclins, and cyclin-dependent kinases (Lam and La Thangue, 1994; Slansky and Farnham, 1996). In mammalian cells, seven E2F (E2F-1 to E2F-7) and two DP proteins have been identified. With the exception of E2F-7, E2F activity arises from heterodimeric transcription factors, where each heterodimer consists of one member of the E2F branch bound to a member of the DP branch of the family, potentially allowing the formation of a series of E2F complexes within the cell (De Bruin et al., 2003; Stevens and La Thangue, 2003).
Although DP proteins stabilize DNA binding of E2F and influence the entry of E2F-4 and E2F-5 into the nucleus, the regulatory functions seem to be carried out mainly by the E2F part of the heterodimer. E2F proteins can be broadly divided into three classes: activators (E2F-1, -2, and -3), pocket protein-dependent repressors (E2F-4 and -5) and E2F-6 and -7, which, contrary to other E2F family proteins, lack a transactivation domain as well as a binding site for pocket proteins and which act exclusively as repressors of E2F-dependent transcription (Trimarchi and Lees, 2002; De Bruin et al., 2003; Di Stefano et al., 2003).
E2F regulates genes, the products of which are essential for progression through the mammalian cell cycle (Dyson, 1998). E2F-1, -2, and –3 can act as oncogene products (Xu et al., 1995), and E2F-1 is also noted for its role as a tumor suppressor (Field et al., 1996; Yamasaki et al., 1996). This can be explained by the ability of E2F-1 to induce apoptosis (for a review see Bell and Ryan, 2004), e.g., by activating the expression of the tumor suppressor protein p14ARF (Bates et al., 1998). Transcriptional activation by adenovirus E1A or by other viral proteins results, at least in part, from the release of E2F from complexes with pRB and the pRB-related proteins p107 and p130 (pocket proteins). The binding of cyclin A/cdk2 to the N-terminal domains of E2F-1, -2, and -3 and the subsequent phosphorylation of the associated DP protein (Krek et al., 1994; Xu et al., 1994) seems to cause a loss of DNA binding ability and thus transcriptional activity. Transcription factors of the Sp family have been found to bind E2F-1, -2, and -3 adjacent to the cyclin A/cdk2 complex and this interaction seems to be necessary for the activity of certain promoters (Karlseder et al., 1996; Rotheneder et al., 1999).
Chromatin immunoprecipitation (ChIP) experiments, microarray data, and Northern blot analysis have demonstrated that a large number of genes appear to be E2F regulated (Ma et al., 2002; Wells et al., 2002). E2F transcription factors may therefore play a pivotal role in the transcriptional regulation of several cellular processes far beyond the originally described cell cycle and proliferation.
The pocket protein family members pRb, p107, and p130 act as the main regulators of E2F activity. However, other protein-protein interactions have been described for E2Fs in recent years. Such interactions, sometimes resulting in posttranslational modifications, may have significant implications in the stability, half-life, and functional activity of E2Fs.
We have identified a novel E2F-binding protein through a yeast two-hybrid interaction screen using the N-terminal domain of E2F-1 as the bait. This protein is highly phosphorylated and we have therefore called it EAPP (e2F-associated phosphoprotein). In this study we examine the expression and localization of EAPP and its role as a putative cofactor of E2F. Using techniques like immunofluorescence, coimmunoprecipitation, and GST-pulldown assays, flow cytometry, and transient transfections and reporter assays with a variety of E2F-dependent promoters, we provide evidence that EAPP can modulate E2F-dependent transcription and influence cell proliferation.
MATERIALS AND METHODS
Plasmids
pACT, pAS2, the pACT cDNA library plasmids (Harper et al., 1993), pGAL-Luc (Pestell et al., 1996), p3xE2F-Luc, p3xE2F-mut-Luc (Krek et al., 1994), pTK-Luc, pCIneo-HA (Doetzlhofer et al., 1999), E1β-Luc (Bates et al., 1998), and pSUPER (Brummelkamp et al., 2002) have been described. pCIneoHA-E2F-1, -2, -3 (aa 1–396) and -4 were created by recloning the cDNAs from the respective pGex-vectors (Karlseder et al., 1996) into pCIneoHA (Doetzlhofer et al., 1999). pGex-4T1-EAPP, pCIneoHA-EAPP, and pCIneo9xMyc-EAPP were constructed by ligating the EAPP cDNA obtained by PCR with the oligonucleotides: 5′AAAAACTCGAGCCCGGGAATTCCATATGAACCGGCTTCCGGATGAC 3′ and 5′ TTACCCGGGGCGGCCGCTACAGACAATTCAGGAAAGAGTTTC 3′ from a human EST clone [IMAGE p998O239962Q2], into the EcoRI/NotI cut vectors. pAS2-E2F1 (1–125) was generated by cloning the 5′ part of the E2F-1 cDNA obtained by PCR (oligonucleotides: 5′ AAAAACCATGGCCTTGGCCGGGGC 3′ and 5′ AAAGGATCCAGCTGTTCTCCCCCGGGGA 3′) and digested with NcoI/BamHI into pAS2. pM-E2F1 (1–157) was generated by cloning the 5′ BamHI/SalI fragment of the E2F-1 cDNA into pM (Clontech, Palo Alto, CA). pVP16-EAPP (aa 1–266) by cloning the EAPP cDNA into the EcoRI/SalI cut pVP16 (Clontech). pEGFP-C1-EAPP was made by ligating the EAPP cDNA into the BglII/SalI cut pEGFP-C1 vector. pCIneoHA-mEAPP was created by cloning a PCR fragment derived from EST-clone [IMAGE p998C034718Q2]; with the oligonucleotides: 5′ TATAGAATTCATGAACCGACTCCAGGATG 3′and 5′ GCTCTAGATTATTTATTTAGCGGCCGCTGTTTTAATAACAACATC 3′, via EcoRI/NotI into pCIneoHA. pSUPER-EAPP was generated by cloning a double-stranded oligonucleotide containing the sequence ATAGTGATGCTGTCTTGAA of the EAPP cDNA as an inverted repeat into pSUPER.
Yeast Two-Hybrid Screen
A yeast two-hybrid system including a B-cell cDNA library cloned into the XhoI site of pACT was made available to us and was used as described (Staudinger et al., 1993). The 5′ part of the E2F-1 cDNA was cloned into the pAS2 yeast two-hybrid vector and transformed into the yeast strain Y190.
Cell Culture, Transfection, and Luciferase Assays
MRC-5, SAOS-2, MCF7, T98G, HELA, U2OS, and 293 cells were cultured in DMEM supplemented with 10% fetal calf serum and antibiotics. T98G cells were growth arrested by incubation in culture medium with low serum (0.2%) for 96 h and restimulated by addition of fresh medium containing 20% serum. Transient transfections were performed as described (Ogris et al., 1993) using the calcium phosphate method. About 3 × 106 cells per 100-mm dish were seeded the day before transfection. Calcium phosphate coprecipitates contained equal amounts of vector DNA added up to a total of 20 μg DNA with sheared salmon sperm DNA. After about 16 h the medium was replaced and after additional 32 h the cells were harvested. Growth arrest and stimulation were controlled by fluorescence-activated cell sorter analysis (FACS) with a Partec PAS-II sorter. Elutriation of cells was carried out as described (Mikulits et al., 1997). For luciferase reporter assays, cells were cultivated and transfected in six-well plates, washed twice with phosphate-buffered saline (PBS), pelleted, and resuspended in 300 μl assay buffer (basic buffer: 25 mM Tricine, 0.5 mM EDTA, 0.54 mM Na-tripolyphosphate, 16.3 mM MgSO4*7H2O, 0.1% Triton X-100, adjusted to pH 7.8; immediately before usage as assay buffer 0.56 vol % of 1 M dithiothreitol [DTT], 1.2 vol % of 0.1 M ATP at pH 7.8 and 4.6 vol % of 1 mM D-luciferin at pH 7.5 were added to the basic buffer) and after 3–5 min at RT the suspension was centrifuged. Two hundred microliters of the supernatant were used for measurement of luciferase activity in a Berthold Autolumat LB 953. Five microliters of the supernatant were used for protein concentration measurement, and thirty microliters of the supernatant were used for β-galactosidase assays.
GST-pulldown Assays and In Vitro;translation
Recombinant proteins were expressed in Escherichia coli and purified as described (Karlseder et al., 1996). In vitro translation was performed with the Promega TnT system (Madison, WI) as described by the manufacturer. Beads coated with GST fusion proteins (1 μg) were incubated in lysis buffer (20 mM Tris-HCl pH 8; 100 mM NaCl; 1 mM EDTA; 0.5% NP-40; 1 mM phenylmethylsulfonyl fluoride; 2 mM DTT; Boehringer's Complete Protease Inhibitor Cocktail) with 300 μg whole cell extract from transfected 293 cells for 4 h. In case of in vitro–translated 35S-methionine–labeled samples we used 4 μl of Promega TNT Reticulocyte lysate for incubation with GST fusion proteins in a total volume of 60 μl lysis buffer. Samples were washed three times with lysis buffer, and bound proteins were eluted by boiling in SDS-PAGE loading buffer, resolved by electrophoresis, and visualized by Western blotting and immunostaining or by autoradiography.
Coimmunoprecipitations
Whole-cell extracts of transfected 293 cells were prepared as described (Pagano et al., 1992) and equal amounts (1000 μg) were incubated in 200 μl lysis buffer with 10 μl antibody-agarose conjugate (murine polyclonal anti-EAPP antiserum, murine monoclonal anti-HA-tag 12CA5 antibody, and murine preimmune serum) for 4 h at 4°C. After three washes with lysis buffer the bound proteins were resolved by electrophoresis and visualized by Western blotting and immunostaining (anti-Myc antibody [9E10]).
Dephosphorylation and Phosphorylation Experiments
Twenty-microgram extracts from HA-EAPP–expressing cells were incubated with 20U CIP (Boehringer Ingelheim, Ingelheim, Germany) in a volume of 20 μl for 30 min at 30°C. After separation by SDS-PAGE, proteins were analyzed by immunostaining with an anti-HA antibody (16B12). For in vitro phosphorylation 0.5 μg of bacterially expressed GST-fusion proteins bound to glutathione agarose beads were incubated with 100 μg extract from T98G cells and 5 μCi γ-32P[ATP] for 30 min at 30°C. After three washes with lysis-buffer the samples were boiled in loading buffer, separated by SDS-PAGE, and visualized by autoradiography.
Northern Analysis
EAPP and β-actin cDNAs were labeled with 32P by random priming and used as probes for hybridization of a commercial human tissue blot (Clontech 7760-1) as described (Mudrak et al., 1994).
Immunofluorescence
Cells were grown on coverslips and washed with PBS before use. The cells were fixed with 2% formaldehyde in PBS at room temperature (RT) for 5 min, washed twice with PBS, and permeabilized with 0.1% Triton-X 100 in PBS for 10 min. After two additional washing steps with PBS the coverslips were covered with primary antibodies diluted in PBS (1:2000 anti-HA 16B12 or 1:2000 anti-EAPP serum) and sealed in a humid chamber over night at 4°C. The coverslips were then washed three times with 0.1% Tween-20 in PBS (PBS-T) for 10 min at RT and afterward incubated with secondary antibody solution (1:2000 anti-mouse Alexa-488 in PBS) for 1 h at RT. Finally the cells were washed once with PBS-T for 10 min at RT, once with 0.05 μg/ml 4′, 6′-diamidino-2-phenylindole (DAPI) in PBS-T 10 min at RT and once with PBS-T. Mounted coverslips were examined with a Zeiss Axiovert 135 TV fluorescence microscope (Oberkochen, Germany) and grayscale images were captures with a Philips CCD camera (Mahwah, NJ).
Antibodies
Anti-HA (BAbCO, HA.11, Richmond, CA), anti-Myc (9E10), anti-β-actin (Sigma, Diesenhofen, Germany; AC-74), anti-Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA, PEP2), anti-E2F-1 (Santa Cruz, KH95) anti-mouse Alexa-488 (Molecular Probes, Eugene, OR) are commercially available. Anti-EAPP polyclonal serum was produced by immunizing female BALB/c mice with purified GST-EAPP (aa 1–266).
RESULTS
Identification and Cloning of a Novel E2F-binding Protein
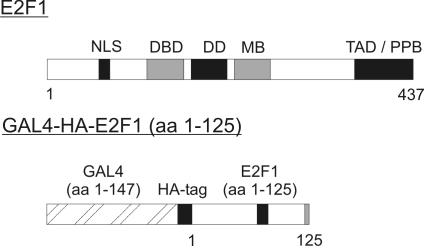
To identify new E2-binding proteins, we undertook a search for cellular proteins that interact with the amino terminal domain of E2F-1 using yeast two-hybrid interaction cloning. Cells expressing the correct fusion protein consisting of the GAL4 DNA-binding domain, an HA-tag, and the N-terminal 125 amino acids of E2F-1 (Figure 1) were transformed with a human B-cell library, cloned into the pACT plasmid. We selected the candidate interacting colonies on the basis of their ability to grow in appropriate selection medium and to turn on the LacZ reporter gene (unpublished data). Sequencing of one of the selected clones revealed a cDNA highly homologous to the cDNA of an uncharacterized human protein in the database (Accession no. BC001245). We initially obtained a cDNA encoding the N-terminal 266 amino acids and later on the full-length cDNA encoding all 285 amino acids of the human protein by PCR using an EST-clone as a template. We also cloned the murine cDNA that encodes 281 amino acids and which is highly homologous to the human protein with 86% identity in the amino acid sequence (Figure 2). Both cDNAs were checked by sequencing and cloned into expression vectors. The sequences were submitted to GenBank with the accession nos. AY869694 for the human cDNA and AY882557 for the murine cDNA.
Figure 1.
Schematic drawing of E2F1 and the GAL4-HA-E2F1 (aa 1–125) fusion-protein used for the two-hybrid assay. (NLS, nuclear localization signal; DBD, DNA-binding domain; DD, dimerization domain; MB, marked box; TAD, transactivation domain; PPB, pocket protein-binding domain).
Figure 2.
Amino acid sequence and alignment of human EAPP (aa 1–285) and murine EAPP (aa 1–281).
EAPP Is a Nuclear Phosphoprotein
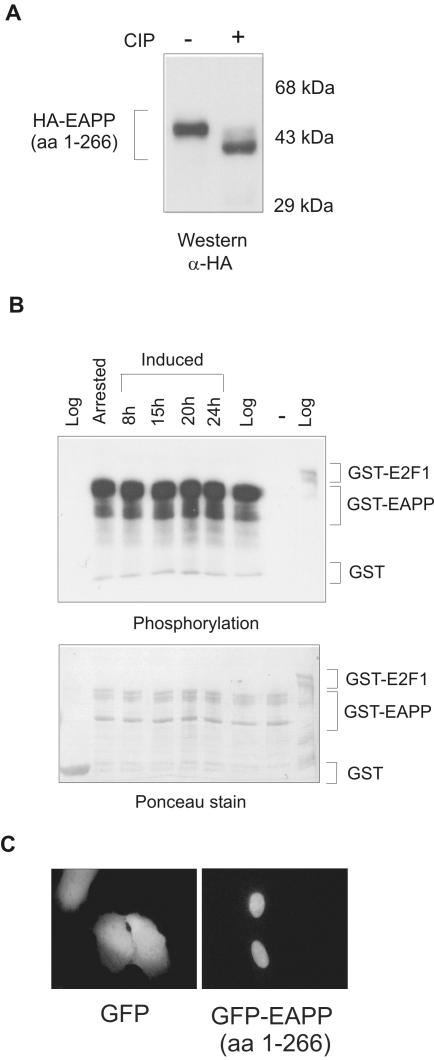
Western analyses revealed that the HA-tagged human protein migrates at ∼45 kDa in SDS PAGE (Figure 3A). This is unusual for a protein with a calculated molecular weight of ∼36 kDa and suggested that it might carry posttranslational modifications. Computer analysis of the amino acid sequence predicted several phosphorylation sites, primarily on serine, but also on threonine and tyrosine residues (unpublished data). Hence, to determine if this protein is truly phosphorylated, we performed phosphorylation and dephosphorylation experiments with EAPP (1–266). Addition of calf intestine alkaline phosphatase (ALP; CIP) brought about a significant shift in the migration of the HA-tagged protein upon SDS-PAGE, indicating phosphorylation of several residues of the protein (Figure 3A). Therefore we named this protein EAPP. Moreover, bacterially produced GST-EAPP incubated with extracts from serum-starved, reinduced, or growing T98G cells and [γ-32P]ATP became much stronger phosphorylated than GST-E2F-1 that served as a positive control. GST, which was used as a negative control, was not phosphorylated. The degree of phosphorylation of GST-EAPP did not vary significantly between extracts from growing and resting cells. As GST-EAPP incubated with [γ-32P]ATP but without cellular extract did not exhibit any phosphorylation, autophosphorylation of the protein seemed unlikely (Figure 3B). Thus, these results demonstrate that EAPP can be phosphorylated and suggest that phosphorylation contributes to the unusual migration in SDS-PAGE.
Figure 3.
Phosphorylation and intracellular localization of EAPP. (A) HA-tagged EAPP was transiently expressed in U2OS cells. Extracts from these cells were treated with CIP (calf intestine ALP) and analyzed by SDS-PAGE, Western blotting, and immunostaining with an anti-HA antibody. (B) Glutathione-agarose bound GST, GST-EAPP, and GST-E2F-1 fusion proteins were incubated with extracts from logarithmically growing, serum-starved, and reinduced T98G cells and [γ-32P]ATP. GST served as a negative and GST-E2F-1 as a positive control for phosphorylation. Proteins were incubated for 30 min at 30°C, washed three times with GST-lysis buffer, boiled with protein loading buffer, and separated by SDS-PAGE. The gel was blotted to a nitrocellulose membrane, stained with Ponceau S, and exposed to an x-ray film. The top panel is the autoradiography and shows the phosphorylation. The bottom panel is the Ponceau stain that shows the amounts of fusion proteins. (C) GFP and GFP-EAPP (1–266) were transiently expressed in U2OS cells. Forty-eight hours after transfection, images were taken. GFP is distributed throughout the cell, whereas GFP-EAPP appears predominantly in the nucleus.
To examine the intracellular localization of EAPP, we expressed a GFP-EAPP fusion protein in U2OS cells and compared its localization to that of GFP by direct fluorescence microscopy. Contrary to GFP, which was evenly distributed within the cell, GFP-EAPP could be found almost exclusively in the nucleus (Figure 3C). The repetition of the above-described experiments with EAPP (1–285) had the same outcome (unpublished data).
Interaction of E2F and EAPP
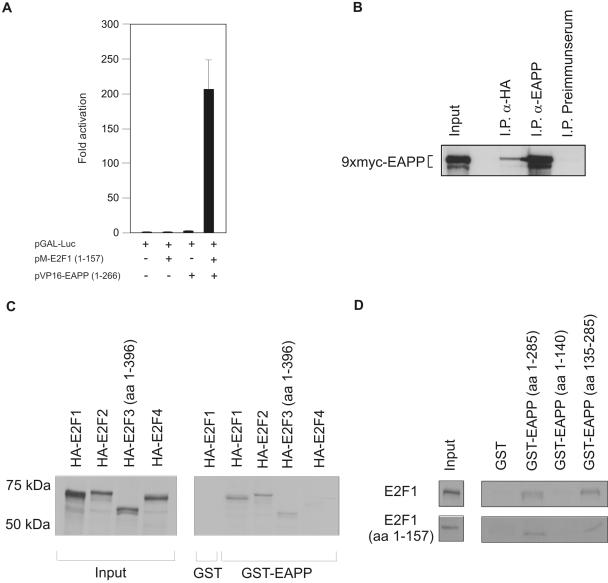
To confirm the interaction of E2F-1 and EAPP observed in the yeast two-hybrid screen, we performed mammalian two-hybrid assays. The N-terminal part of E2F-1 (amino acids 1–157) lacking the transactivation domain was fused to the DNA-binding domain of GAL4, whereas EAPP was fused to the VP16 activation domain. pGAL-Luc, a plasmid carrying the luciferase gene controlled by a GAL4-dependent promoter, served as a reporter for protein-protein interaction. Transfection of the reporter or the reporter together with either the GAL-E2F-1 (pM-E2F-1;1–157) or the VP16-EAPP (pVP16-EAPP) plasmid into U2OS cells did not result in significant activation of the GAL-dependent promoter. Cotransfection of all three vectors, however, gave rise to strong luciferase activity, confirming the interaction of EAPP and E2F-1 (Figure 4A).
Figure 4.
Interaction of E2F proteins with EAPP. (A) Mammalian two-hybrid assay in U2OS cells. A Gal4-dependent reporter vector (pGAL-Luc) was cotransfected with an expression vector for a GAL-E2F-1 fusion-protein (pM-E2F1; 1–157), a VP16-EAPP fusion-protein (pVP16-EAPP; 1–266), or both. Each bar represents six independent samples adjusted for β-Gal activity. SD is indicated. (B) Coimmunoprecipitation of EAPP and E2F-1. 9xMyc-EAPP and HA-E2F-1 were coexpressed in U2OS cells. Immunoprecipitation (IP) experiments were carried out with the anti-EAPP antibody (positive control), an anti-HA antibody (12CA5), and a murine preimmune serum (negative control). Bound proteins were separated by SDS-PAGE and coprecipitated Myc-EAPP was visualized by immunostaining with an anti-Myc antibody (9E10). Input means that 1/50 of the amount of cell extract used for the IPs was loaded directly onto the gel. (C) Pulldown assays with GST or GST-EAPP and in vitro–translated 35S-labeled HA-tagged E2F proteins. GST incubated with HA-E2F-1 was used as a negative control. Input means that 1/4 of the amount of in vitro–translated proteins used for the pulldown assays was loaded directly onto the gel. Input- and GST-EAPP bound proteins were visualized by autoradiography. (D) Pulldown assays with GST, GST-EAPP, GST-EAPP(1–140), and GST-EAPP(135–285), and in vitro–translated 35S-labeled E2F-1 (wt or aa 1–157). Input means that one fourth of the amount of in vitro–translated proteins used for the pulldown assays was loaded directly onto the gel. Input- and GST-EAPP bound proteins were visualized by autoradiography.
As another approach, Myc-tagged EAPP and HA-tagged E2F-1 were coexpressed in U2OS cells. Extracts from these cells were used for immunoprecipitation experiments with an anti-HA antibody to precipitate HA-E2F-1, with an anti-EAPP antibody (as a positive control) or preimmune serum (used as a negative control). The precipitated proteins were separated by SDS-PAGE followed by Western blotting. Analyses with an anti-Myc tag antibody revealed the presence of Myc-EAPP in the anti-EAPP and the anti-HA precipitates, but not in the precipitate of the preimmune serum, again confirming the interaction (Figure 4B).
As a third approach pulldown assays were performed with in vitro–translated and 35S-labeled E2F-1, -2, -3, and -4 and GST-EAPP. The results obtained showed that EAPP binds to E2F-1, -2 and -3, but not to E2F-4 (Figure 4C). This is not surprising, because E2F-4 does not contain the N-terminal domain that is present in E2F-1, -2, and -3 and that was used for the two-hybrid screen. In addition, pulldown assays with either the full-length EAPP or the N-terminal (aa 1–140) or C-terminal (aa 135–285) half fused to GST and either full-length or C-terminally truncated E2F-1 (aa 1–157) revealed that the interaction is mediated by the C-terminal part of EAPP (Figure 4D). All GST pulldown assays were performed in the presence of RNase A and ethidium bromide to avoid possible bridging effects of nucleic acids.
Expression of EAPP
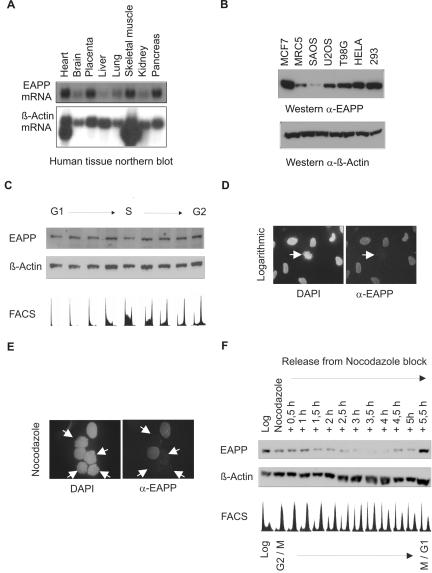
Probing of a human mRNA tissue blot with 32P-labeled EAPP cDNA revealed that EAPP mRNA is present in all examined tissues albeit at different levels. Heart, placenta, skeletal muscle, and pancreas harbored high levels of EAPP mRNA, whereas brain, lung, and kidney had intermediate levels, and liver had rather low levels (Figure 5A). To investigate protein levels of EAPP we raised murine polyclonal antibodies. First we checked EAPP levels in a variety of human cell lines by Western blotting and immunostaining with the anti-EAPP antibody. In MRC-5, human diploid fibroblasts, the amount of EAPP is low; all other checked cells are transformed and exhibit, with one exception (SAOS-2), much more EAPP (Figure 5B). To look for possible cell cycle–dependent fluctuations of EAPP levels, we elutriated U2OS cells and examined the EAPP protein levels. The amount of EAPP remained rather constant throughout the cell cycle (Figure 5C). Surprisingly, immunofluorescence experiments suggested that EAPP disappears in mitotic cells (Figure 5D). This cannot be the result of a posttranslational modification of EAPP, which might mask the antibody recognition site, as ectopically expressed HA-EAPP detected with an anti-HA antibody also disappeared during mitosis (unpublished data). After treatment with nocodazole, EAPP could not be detected in cells with already condensed chromosomes, whereas others still showed the presence of this protein (Figure 5E). To examine the disappearance of EAPP in mitotic cells in more detail, we arrested U2OS cells with nocodazole for 18 h and released them from the G2/M block by washing away the drug. Extracts were prepared from arrested and released cells at intervals of 30 min after the removal of nocodazole and subjected to Western analysis using the anti-EAPP antibody. This demonstrated the disappearance and reappearance of EAPP after the release (Figure 5F).
Figure 5.
Expression of EAPP. (A) Northern analysis of human tissues. A human mRNA tissue blot was probed first with radiolabeled human EAPP cDNA (top panel) and then reprobed with β-Actin cDNA (bottom panel). EAPP mRNA appears as a single band of ∼1.4 kB. (B) Western analyses of human cell lines. The top panel shows the amount of EAPP, and the bottom panel of β-actin. (C) Growing U2OS cells were elutriated and the fractions were examined by FACS, followed by SDS-PAGE, Western blotting, and immunostaining with the anti-EAPP antibody. The membrane was reprobed with anti-β-actin antibody to show equal loading. (D) Immunofluorescence of endogenous EAPP in growing cells. In the left picture interphase cells surround a mitotic cell with condensed DAPI-stained chromosomes. EAPP is not detectable in this cell (right picture). (E) Immunofluorescence of endogenous EAPP in cells arrested for 18 h with nocodazole. The arrows indicate cells with already condensed chromosomes. (F) U2OS cells were arrested with nocodazole for 18 h and then released for 5.5 hours. Every 30 min extracts from released cells were prepared and examined by SDS-PAGE, Western blotting, and immunostaining with the anti-EAPP antibody. The membrane was reprobed with anti-β-actin antibody to show equal loading. FACS analysis of each fraction was carried out in parallel (bottom panel).
EAPP Modulates E2F-dependent Transcription
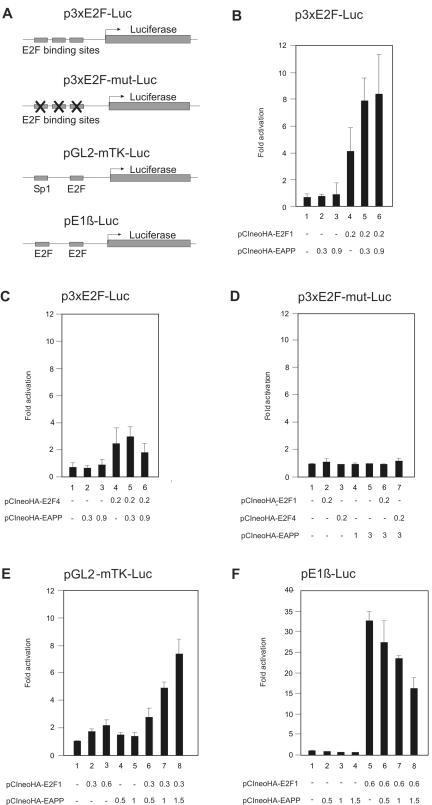
Reporter gene assays are a sensitive method to detect changes in the activity of transcription factors. Most natural promoters are controlled by the combined activity of more than one kind of transcription factors, which are themselves influenced by a variety of signals. Hence, to study the role of EAPP in E2F-dependent transcription, we used an artificial promoter-reporter construct comprising just three E2F binding sites. Any effect of EAPP on transcription should be mediated by E2F factors in this system. Expression of EAPP slightly elevated promoter activity, in a dose-dependent manner, above the level caused by endogenous proteins. As expected, E2F-1 expression resulted in strong activation, again depending on the dosage. Promoter activity could be further increased by expression of both, E2F-1 and EAPP (Figure 6A). Expression of E2F-4 also caused activation of this promoter, but, contrary to E2F-1–mediated activation, it could not be further enhanced by EAPP (Figure 6B). This is in accordance with the interaction data showing that E2F-4 does not bind to EAPP (Figure 4C). A promoter with all three E2F-binding sites mutated was responsive neither to E2F-1, E2F-4, or EAPP, nor to combinations of these proteins (Figure 6D). A similar experiment was done with the murine thymidine kinase promoter controlling the reporter gene expression. This promoter has just one E2F-binding site but an Sp1-binding site in addition (Ogris et al., 1993). Here the activation by E2F-1 alone was not as pronounced but the synergistic effect of E2F-1 and EAPP was much stronger than with the artificial E2F promoter (Figure 6E). To investigate the promoter of a gene involved in E2F-1–induced apoptosis, we carried out reporter assays with the p14ARF promoter. Overexpression of E2F-1 resulted in strong activation as described (Bates et al., 1998). However, contrary to the other examined E2F-controlled promoters, coexpression of EAPP caused repression of p14ARF promoter activity (Figure 6F). These results suggest that EAPP can act not only as an activator but also as a repressor of transcription.
Figure 6.
Reporter-assays with E2F controlled promoters in U2OS cells. The numbers refer to the amount of expression vectors in μg used for each experiment. The data are means and SDs of at least three independent experiments adjusted for β-Gal activity. (A) Schematic drawings of the used promoter-reporter constructs. (B) p3xE2F-Luc, carrying the luciferase reporter controlled by an artificial E2F-dependent promoter, was cotransfected with expression vectors for HA-E2F-1, HA-EAPP, or both. (C) p3xE2F-Luc was cotransfected with expression vectors for HA-E2F-4, HA-EAPP, or both. (D) p3 × E2F-mut-Luc, a reporter vector with all three E2F-binding sites mutated, was cotransfected with expression vectors for HA-E2F-1, HA-E2F-4, HA-EAPP, or combinations of them. (E) pTK-Luc, carrying the murine thymidine kinase promoter, was cotransfected with expression vectors for HA-E2F-1, HA-EAPP, or both. (F) pE1β-Luc, carrying the p14ARF promoter, was cotransfected with expression vectors for HA-E2F-1, HA-EAPP, or both.
EAPP Levels Influence S-phase Entry
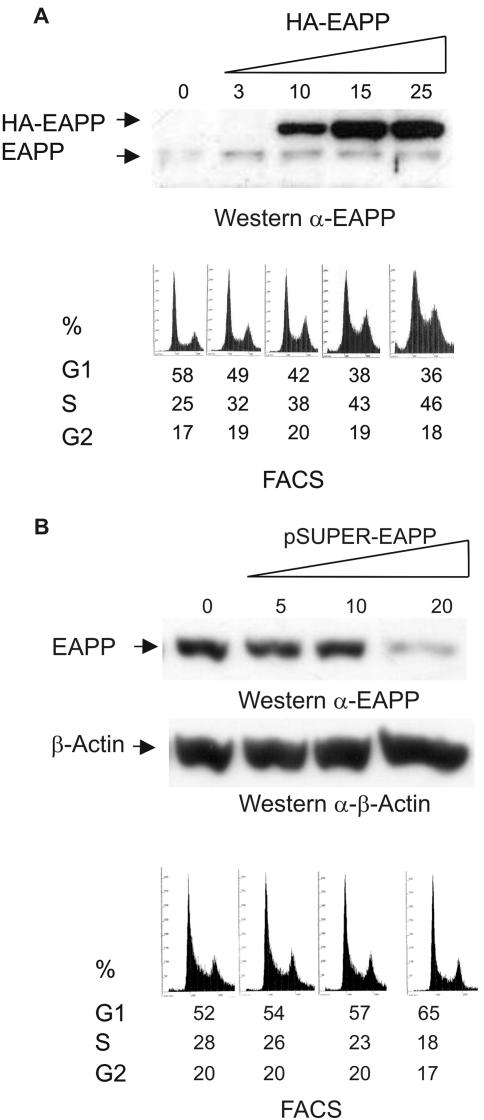
The reporter assays described above suggested an increase of E2F activity and subsequently of the expression of E2F controlled genes as a result of higher EAPP levels. To examine the possible consequences for the affected cells, we transiently increased or lowered EAPP levels in U2OS cells, checked the levels by Western blotting and immunostaining with the anti-EAPP antibody, and determined the fractions of cells in the respective phases of the cell cycle by FACS analyses. Overexpression of EAPP resulted in a higher fraction of cells in S-phase (Figure 7A), whereas RNAi-induced knockdown of EAPP caused a reduction of S-phase cells (Figure 7B). Only 20–30% of the cells are actually transfected; this suggests that the effect on the individual cell is even more severe and might even result in G1 arrest in the case of EAPP knockdown.
Figure 7.
EAPP overexpression and knock down in U2OS cells. Cells were transfected with either the empty vector or increasing amounts of an expression vector for either HA-EAPP (A) or EAPP siRNA (B). Forty-eight hours after transfection the cells were harvested and analyzed. The top panels are immunoblots showing endogenous and ectopic amounts of EAPP and β-Actin. The numbers refer to the amount of HA-EAPP or EAPP siRNA expression vector in μg used for the respective transfection. The bottom panels are FACS analysis showing the fractions of cells in G1-, S-, and G2-phase of the cell cycle.
DISCUSSION
Transcription factors of the E2F family play an important role in the control of cell cycle and proliferation in many different species, including mammals, flies, nematodes, amphibians, and plants. Their activity is regulated by a variety of mechanisms, frequently mediated by proteins binding to individual members or a subgroup of the family.
Herein we describe the identification, cloning, and characterization of a novel protein that interacts with a subset of E2F factors and influences E2F-dependent promoter activity. We isolated this protein as an E2F-interacting factor in a yeast two-hybrid screen using the amino-terminal domain of E2F-1 as the bait. It is strongly phosphorylated and consequently we named it EAPP. The overall phosphorylation of EAPP does not seem to change significantly throughout the cell cycle as judged by in vitro phosphorylation experiments and migration in SDS-PAGE. This does not rule out that the phosphorylation status of individual sites changes in certain stages of the cell thereby influencing the activity of the protein.
The interaction with E2F-1 was confirmed in vivo and in vitro by mammalian two-hybrid and coimmunoprecipitation assays. GST-pulldown assays with in vitro–translated E2Fs showed that EAPP also interacts with E2F-2 and -3, which contain a similar amino-terminal domain, but not with E2F-4. The N-terminal domain of E2F-1, -2, and -3 not only contains the nuclear localization signal but also binding sites for cyclin A (Krek et al., 1994), transcription factors of the Sp1 family (Karlseder et al., 1996), p45skp2 (Marti et al., 1999), p53 (Hsieh et al., 2002), and EBP1 and EBP2 (Jordan et al., 1996). The interaction with EAPP might interfere with the binding of one or more of these proteins, thereby influencing E2F activity.
Regulated degradation of proteins is essential for the progression of the cell cycle. Without Cdk1 inactivation by cyclin B destruction chromosomes do not decondense and cells do not divide (for reviews see Peters, 1998, 2002). The disappearance of EAPP during mitosis indicates that it might interfere with the completion of the cell cycle and therefore has to be removed in this phase. That it takes 3 h after the release from nocodazole until EAPP completely disappears could be explained by the observation that not all cells seem to arrest exactly at the same stage after nocodazole addition (Figure 5E). A significant fraction of cells might lag behind after the release from nocodazole. Whether EAPP becomes destroyed by ubiquitin-dependent proteolysis like cyclin B remains to be investigated. The quick reappearance suggests that EAPP is needed in G1, presumably, but perhaps not exclusively, to enhance S-phase stimulating E2F activity.
Interestingly, EAPP enhanced the E2F dependent activity of growth stimulated promoters like the thymidine kinase promoter, but inhibited the promoter of the p14ARF gene. p14ARF can act as a mediator of E2F-induced apoptosis (Bates et al., 1998). The observed down-regulation of the p14ARF promoter by EAPP does not have to be mediated by E2F. This promoter is also activated by transcription factors of the Sp1 family (Parisi et al., 2002; Berkovich et al., 2003) and repressed by T-box factors (Lingbeek et al., 2002) and p53 (Robertson and Jones, 1998; Stott et al., 1998). Any of these factors could mediate the repressing effect of EAPP. The inactivation of p53 seems to increase p14ARF expression (Robertson and Jones, 1998; Stott et al., 1998), which can result in p53-independent apoptosis (Hemmati et al., 2002; Eymin et al., 2003). If the repression of the p14ARF promoter by EAPP is p53 independent, overexpression of EAPP might offer a cell with inactivated p53 an escape from p14ARF-mediated apoptosis. E2F-1 induced expression of p14ARF also results in binding of this protein to E2F-1 (Eymin et al., 2001; Mason et al., 2002), thereby promoting the binding of p45skp2 (Marti et al., 1999) and subsequently degradation of E2F-1 via proteasome pathways (Martelli et al., 2001). Overexpression of EAPP could interfere with this negative feedback control of E2F-1 activity. Concordant with this model we found a slight increase of E2F-1 levels in cells transiently overexpressing EAPP (unpublished data). Transcription of the E2F-1 gene is regulated by E2F-binding sites (Neuman et al., 1994). The elevated E2F-1 level could therefore be the result of EAPP-enhanced E2F activity, or of both, increased E2F-1 promoter activity, and reduced p14ARF-mediated E2F-1 degradation. Thus, EAPP on the one hand seems to enhance transcription of growth-correlated, E2F-controlled genes like thymidine kinase, resulting in the observed S-phase induction, and on the other hand it inhibits the expression of the tumor suppressor p14ARF. This implies that EAPP could play a role in malignant transformation. In line with this speculation, compared with diploid fibroblasts, EAPP levels were elevated in almost all investigated transformed human cell lines.
How could EAPP stimulate E2F activity? One possibility would be an increase of the DNA-binding ability of the activating E2Fs. We examined this in electrophoretic mobility shift assays (EMSA) and found neither an increase in DNA binding, nor EAPP as a component of the E2F complexes (unpublished data). This does not rule out that EAPP acts in this way, because weakly interacting proteins are often not detectable in EMSAs and binding of proteins to the naked DNA of an oligonucleotide might differ from binding to DNA organized as nucleosomes in the context of a promoter. Alternative mechanisms would be increased transactivation activity caused by EAPP binding or by EAPP-mediated posttranslational modification of E2F proteins or chromatin rearrangement caused by EAPP bound proteins. E2F-1 activity can be activated or repressed by modifications (for a review see Mundle and Saberwal, 2003). Although there are no indications that EAPP itself is an enzyme, it might act as a bridging factor for modifying factors. We have found kinase activity in immunoprecipitations of EAPP capable of phosphorylating added GST-E2F-1 (unpublished data). The increase of the S-phase fraction in EAPP-overexpressing cells and the inhibition (or slow down) of S-phase entry in EAPP knockdown cells indicate that EAPP is required for cell cycle progression. Whether the observed S-phase–enhancing activity of EAPP is mediated only by E2F or if it also involves other factors remains to be determined.
EAPP is conserved not only among mammals. Open reading frames corresponding to the EAPP gene can be found in the genomes of many species. RNA interference experiments in Caenorhabditis elegans suggest that inhibiting the expression of the corresponding gene is embryonic lethal (Kamath et al., 2003).
Taken together, EAPP might play an important role in the fine-tuning of both major E2F-1 activities, the regulation of the cell cycle and the induction of apoptosis. By stimulating S-phase entry and at the same time inhibiting p14ARF expression, overexpression of EAPP could contribute to the malignant transformation of a cell.
Acknowledgments
We are grateful to Stephen J. Elledge for the yeast two hybrid system, Wilhelm Krek for the p3xE2F-Luc and p3xE2F-mut-Luc plasmids, Karen H. Vousden for the E1β-Luc vector, and Richard G. Pestell for the pGAL-Luc vector. We thank Marie-Joëlle Miron, Erhard Wintersberger, Egon Ogris, and Christian Seiser for critical review of the manuscript; Ingrid Mudrak for help with the generation of antibodies; and Thomas Sauer for assistance with the FACS experiments. This work was supported by the Austrian FWF (grants P15490 and P15632) and the Herzfelder Stiftung.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–11–0975) on February 16, 2005.
Abbreviations used: EAPP, E2F-associated phosphoprotein; GST, glutathione-S-transferase; GFP, green fluorescent protein; CIP, calf intestine ALP.
References
- Bates, S., Phillips, A. C., Clark, P. A., Stott, F., Peters, G., Ludwig, R. L., and Vousden, K. H. (1998). p14ARF links the tumour suppressors RB and p53. Nature 395, 124–125. [DOI] [PubMed] [Google Scholar]
- Bell, L. A., and Ryan, K. M. (2004). Life and death decisions by E2F-1. Cell Death Differ. 11, 137–142. [DOI] [PubMed] [Google Scholar]
- Berkovich, E., Lamed, Y., and Ginsberg, D. (2003). E2F and Ras synergize in transcriptionally activating p14ARF expression. Cell Cycle 2, 127–133. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- de Bruin, A., Maiti, B., Jakoi, L., Timmers, C., Buerki, R., and Leone, G. (2003). Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 278, 42041–42049. [DOI] [PubMed] [Google Scholar]
- Di Stefano, L., Jensen, M. R., and Helin, K. (2003). E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22, 6289–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetzlhofer, A., Rotheneder, H., Lagger, G., Koranda, M., Kurtev, V., Brosch, G., Wintersberger, E., and Seiser, C. (1999). Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19, 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson, N. (1998). The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- Eymin, B., Karayan, L., Seite, P., Brambilla, C., Brambilla, E., Larsen, C. J., and Gazzeri, S. (2001). Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 20, 1033–1041. [DOI] [PubMed] [Google Scholar]
- Eymin, B., Leduc, C., Coll, J. L., Brambilla, E., and Gazzeri, S. (2003). p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene 22, 1822–1835. [DOI] [PubMed] [Google Scholar]
- Field, S. J., Tsai, F. Y., Kuo, F., Zubiaga, A. M., Kaelin, W. G., Jr., Livingston, D. M., Orkin, S. H., and Greenberg, M. E. (1996). E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85, 549–561. [DOI] [PubMed] [Google Scholar]
- Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K., and Elledge, S. J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hemmati, P. G., Gillissen, B., von Haefen, C., Wendt, J., Starck, L., Guner, D., Dorken, B., and Daniel, P. T. (2002). Adenovirus-mediated overexpression of p14(ARF) induces p53 and Bax-independent apoptosis. Oncogene 21, 3149–3161. [DOI] [PubMed] [Google Scholar]
- Hsieh, J. K., Yap, D., O`Connor, D. J., Fogal, V., Fallis, L., Chan, F., Zhong, S., and Lu, X. (2002). Novel function of the cyclin A binding site of E2F in regulating p53-induced apoptosis in response to DNA damage. Mol. Cell. Biol. 22, 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, K. L., Evans, D. L., Steelman, S., and Hall, D. J. (1996). Isolation of two novel cDNAs whose products associate with the amino terminus of the E2F1 transcription factor. Biochemistry 35, 12320–12328. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S. et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Karlseder, J., Rotheneder, H., and Wintersberger, E. (1996). Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 16, 1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek, W., Ewen, M. E., Shirodkar, S., Arany, Z., Kaelin, W. G., Jr., and Livingston, D. M. (1994). Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 78, 161–172. [DOI] [PubMed] [Google Scholar]
- Lam, E. W., and La Thangue, N. B. (1994). DP and E2F proteins: coordinating transcription with cell cycle progression. Curr. Opin. Cell Biol. 6, 859–866. [DOI] [PubMed] [Google Scholar]
- Lingbeek, M. E., Jacobs, J. J., and van Lohuizen, M. (2002). The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J. Biol. Chem. 277, 26120–26127. [DOI] [PubMed] [Google Scholar]
- Ma, Y., Croxton, R., Moorer, R. L., Jr., and Cress, W. D. (2002). Identification of novel E2F1-regulated genes by microarray. Arch. Biochem. Biophys. 399, 212–224. [DOI] [PubMed] [Google Scholar]
- Martelli, F., Hamilton, T., Silver, D. P., Sharpless, N. E., Bardeesy, N., Rokas, M., DePinho, R. A., Livingston, D. M., and Grossman, S. R. (2001). p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. USA 98, 4455–4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti, A., Wirbelauer, C., Scheffner, M., and Krek, W. (1999). Interaction between ubiquitin-protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1(1), 14–19. [DOI] [PubMed] [Google Scholar]
- Mason, S. L., Loughran, O., and La Thangue, N. B. (2002). p14(ARF) regulates E2F activity. Oncogene 21, 4220–4230. [DOI] [PubMed] [Google Scholar]
- Mikulits, W., Dolznig, H., Edelmann, H., Sauer, T., Deiner, E. M., Ballou, L., Beug, H., and Mullner, E. W. (1997). Dynamics of cell cycle regulators: artifact-free analysis by recultivation of cells synchronized by centrifugal elutriation. DNA Cell Biol. 16, 849–859. [DOI] [PubMed] [Google Scholar]
- Mudrak, I., Ogris, E., Rotheneder, H., and Wintersberger, E. (1994). Coordinated trans activation of DNA synthesis- and precursor-producing enzymes by polyomavirus large T antigen through interaction with the retinoblastoma protein. Mol. Cell. Biol. 14, 1886–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundle, S. D., and Saberwal, G. (2003). Evolving intricacies and implications of E2F1 regulation. FASEB J. 17, 569–574. [DOI] [PubMed] [Google Scholar]
- Neuman, E., Flemington, E. K., Sellers, W. R., and Kaelin, W. G., Jr. (1994). Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter [published erratum appears in Mol. Cell. Biol. 1995 Aug; 15(8):4660]. Mol. Cell. Biol. 14, 6607–6615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogris, E., Rotheneder, H., Mudrak, I., Pichler, A., and Wintersberger, E. (1993). A binding site for transcription factor E2F is a target for trans activation of murine thymidine kinase by polyomavirus large T antigen and plays an important role in growth regulation of the gene. J. Virol. 67, 1765–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano, M., Draetta, G., and Jansen Durr, P. (1992). Association of cdk2 kinase with the transcription factor E2F during S phase. Science 255, 1144–1147. [DOI] [PubMed] [Google Scholar]
- Parisi, T., Pollice, A., Di Cristofano, A., Calabro, V., and La Mantia, G. (2002). Transcriptional regulation of the human tumor suppressor p14(ARF) by E2F1, E2F2, E2F3, and Sp1-like factors. Biochem. Biophys. Res. Commun. 291, 1138–1145. [DOI] [PubMed] [Google Scholar]
- Pestell, R. G., Albanese, C., Watanabe, G., Lee, R. J., Lastowiecki, P., Zon, L., Ostrowski, M., and Jameson, J. L. (1996). Stimulation of the P-450 side chain cleavage enzyme (CYP11A1) promoter through ras- and Ets-2-signaling pathways. Mol. Endocrinol. 10, 1084–1094. [DOI] [PubMed] [Google Scholar]
- Peters, J. M. (1998). SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr. Opin. Cell Biol. 10, 759–768. [DOI] [PubMed] [Google Scholar]
- Peters, J. M. (2002). The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9, 931–943. [DOI] [PubMed] [Google Scholar]
- Robertson, K. D., and Jones, P. A. (1998). The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol. Cell. Biol. 18, 6457–6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotheneder, H., Geymayer, S., and Haidweger, E. (1999). Transcription factors of the Sp1 family: interaction with E2F and regulation of the murine thymidine kinase promoter. J. Mol. Biol. 293, 1005–1015. [DOI] [PubMed] [Google Scholar]
- Slansky, J. E., and Farnham, P. J. (1996). Introduction to the E2F family: protein structure and gene regulation. Curr. Top. Microbiol. Immunol. 208, 1–30. [DOI] [PubMed] [Google Scholar]
- Staudinger, J., Perry, M., Elledge, S. J., and Olson, E. N. (1993). Interactions among vertebrate helix-loop-helix proteins in yeast using the two-hybrid system. J. Biol. Chem. 268, 4608–4611. [PubMed] [Google Scholar]
- Stevens, C., and La Thangue, N. B. (2003). E2F and cell cycle control: a double-edged sword. Arch. Biochem. Biophys. 412, 157–169. [DOI] [PubMed] [Google Scholar]
- Stott, F. J. et al. (1998). The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17, 5001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi, J. M., and Lees, J. A. (2002). Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Wells, J., Graveel, C. R., Bartley, S. M., Madore, S. J., and Farnham, P. J. (2002). The identification of E2F1-specific target genes. Proc. Natl. Acad. Sci. USA 99, 3890–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G., Livingston, D. M., and Krek, W. (1995). Multiple members of the E2F transcription factor family are the products of oncogenes. Proc. Natl. Acad. Sci. USA 92, 1357–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M., Sheppard, K. A., Peng, C. Y., Yee, A. S., and Piwnica Worms, H. (1994). Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol. Cell. Biol. 14, 8420–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, L., Jacks, T., Bronson, R., Goillot, E., Harlow, E., and Dyson, N. J. (1996). Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85, 537–548. [DOI] [PubMed] [Google Scholar]