Abstract
Soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) complexes form bundles of four parallel α-helices. The central `0' layer of interacting amino acid side chains is highly conserved and contains one arginine and three glutamines, leading to the classification of SNAREs into R, Qa, Qb, and Qc-SNAREs. Replacing one of the glutamines with arginine in the yeast exocytotic SNARE complex is either lethal or causes a conditional growth defect that is compensated by replacing the R-SNARE arginine with glutamine. Using the yeast SNARE complex mediating traffic from the endoplasmic reticulum to the Golgi apparatus, we now show that functionally interacting SNAREs can be mapped by systematically exchanging glutamines and arginines in the `0' layer. The Q→ R replacement in the Qb-SNARE Bos1p has the strongest effect and can be alleviated by an Q→ R replacement in the R-SNARE Sec22p. Four Q residues in the central layer caused growth defects above 30°C that were rescued by Q→ R substitutions in the Qa and Qc SNAREs Sed5p and Bet1p, respectively. The sec22(Q)/sed5(R) mutant is temperature sensitive and is rescued by a compensating R→ Q replacement in the R-SNARE Ykt6p. This rescue is attributed to the involvement of Sed5p and Ykt6p in a different SNARE complex that functions in intra-Golgi trafficking.
INTRODUCTION
Intracellular membrane traffic requires the ordered formation of vesicles, transport of these vesicles to the appropriate target compartment, and finally attachment and the fusion of the vesicles with the target membranes. Attachment and fusion are mediated by supramolecular assemblies of conserved proteins. These proteins are not only responsible for executing the docking and fusion reaction but also for ensuring that only appropriate membranes fuse with each other. Many components of intracellular fusion machines have been identified in recent years, but we are only beginning to understand how they cooperate to organize fusion in a highly specific and controlled manner (Rizo and Südhof, 1998; Jahn et al., 2003; Bonifacino and Glick, 2004).
The soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) proteins are among the best characterized components of intracellular fusion machines. They are represented by a superfamily of small membrane proteins that is characterized by a conserved stretch of 60–70 amino acids termed SNARE motif. Each SNARE protein contains at least one SNARE motif that is usually located adjacent to the C-terminal transmembrane domain. SNARE motifs spontaneously assemble into stable complexes. They form coiled-coil–like bundles made of four α-helices in which each helix is represented by a different SNARE motif. The crystal structures of two SNARE complexes containing only distantly related SNARE motifs revealed an extraordinary degree of structural conservation (Sutton et al., 1998; Antonin et al., 2002). The core of the helix bundles consist of 16 layers of interacting amino acid side chains that are arranged perpendicular to the axis of the helix bundle. These amino acids are highly conserved. Most layers contain hydrophobic amino acids with the notable exception of a highly polar layer in the middle of the bundle (`0' layer). This unique layer consists of three glutamines and one arginine connected by highly polar bonds. Sequence comparisons revealed that all known SNARE motifs can be classified into four subfamilies, referred to as Qa-, Qb-, Qc-, and R-SNAREs, that define their position in the SNARE complex (Fasshauer et al., 1998; Bock and Scheller, 1999; Jahn et al., 2003; Uemura et al., 2004). Accordingly, it seems that all SNARE complexes have a QaQbQcR-composition.
Detailed studies on purified SNAREs, in solution or after reconstitution into artificial vesicles, have led to the development of a well documented model that explains how SNAREs may mediate the fusion of bilayers (Fasshauer, 2003; Jahn et al., 2003). According to this view, the parallel alignment of SNARE domains anchored in different membranes results in a tight connection between these membranes that overcomes membrane repulsion and initiates fusion. Assembly would be initiated at the N-terminal ends of the SNARE motifs and progresses toward the C-terminal membrane anchors, thus pulling the membranes closely together. Indeed, many randomly selected mutants with strong phenotypes carry mutations in the “layer” residues, supporting the view that correct packing is critical for function (Banfield et al., 1995; Lewis et al., 1997; Sacher et al., 1997; Stone et al., 1997; Littleton et al., 1998; Saifee et al., 1998; Dilcher et al., 2001). During fusion, the assembled SNAREs change from a strained “trans-” to a relaxed “cis-” configuration. For the SNAREs to be used for another round of fusion, the SNARE complex is disassembled by the chaperone-ATPase N-ethylmaleimide–sensitive factor (NSF) together with cofactors (Malhotra et al., 1988; Mayer et al., 1996). According to this scenario, it is essential that at least one SNARE on each membrane carries a transmembrane anchor to transmit the energy released during complex assembly to the lipid bilayers (Hanson et al., 1997; Grote et al., 2000; McNew et al., 2000; Rohde et al., 2003).
Although most investigators agree that SNAREs play an important role during bilayer fusion, it is controversial to which extent they contribute to the specificity of intracellular fusion reactions. Apparently, each fusion step in the secretory pathway requires a unique set of SNAREs, but it is unclear whether this specificity is encoded in the SNAREs themselves or whether it is brought about by upstream regulatory factors. During assembly of SNARE complexes in vitro, SNAREs belonging to the same subfamily can substitute rather broadly for each other, resulting in non-“cognate” complexes (Fasshauer et al., 1998; Yang et al., 1999). In contrast, using fusion of liposomes reconstituted with recombinant yeast SNAREs, it was suggested that only those SNAREs are capable of catalyzing fusion that are also interacting with each other in vivo (Parlati et al., 2000). However, in these experiments the “QabcR-rule” for functional SNARE complexes was violated, and it remains to be clarified whether SNAREs capable of forming complexes in solution are also capable of catalyzing liposome fusion.
On the other hand it is becoming apparent that in vivo individual SNAREs participate in several fusion steps, each involving different SNARE partners. For example, in yeast the SNARE complex involved in transport from the endoplasmic reticulum to the cis-Golgi is thought to consist of Sec22p (R-SNARE), Sed5p (Qa-SNARE), Bos1p (Qb), and Bet1p (Qc) (Søgaard et al., 1994; Sacher et al., 1997; Cao and Barlowe, 2000; Parlati et al., 2000). In mammalian cells, the homologous proteins include Sec22b (R), syntaxin 5 (Qa), membrin (Qb), and Bet1 (Qc) (Xu et al., 2000). In both yeast and mammals Sed5p/syntaxin 5 is also involved in trafficking steps within the Golgi apparatus where it is assumed to interact with a homologous set of SNAREs, including Ykt6p/Ykt6 (R, a SNARE that lacks a transmembrane domain but contains a lipid anchor), Gos1p/GS28 (Qb), and Sft1p/GS15 (Qc) (Parlati et al., 2002; Xu et al., 2002).
Syntaxin 5 and its yeast homolog Sed5p are not the only SNARE proteins involved in more than one trafficking step. The R-SNARE Sec22p participates both in anterograde and retrograde traffic between the Golgi and the endoplasmic reticulum (Lewis et al., 1997). In yeast, its SNARE partners in the retrograde step have been tentatively identified as Ufe1p (Qa), Sec20p (Qb), and Use1p (Qc) (Lewis et al., 1997; Burri et al., 2003; Dilcher et al., 2003), with a homologous complex also being present in mammals. Other examples of such “multifunctional” SNAREs in yeast include Ykt6p (McNew et al., 1997; Dilcher et al., 2001; Kweon et al., 2003), Vti1p (Fischer von Mollard and Stevens, 1999), and Snc1/2p (Gurunathan et al., 2000). Conversely, it is becoming apparent that certain SNAREs can functionally substitute for each other such as Ykt6p for Sec22p (in the step endoplasmic reticulum [ER]-to-Golgi; Liu and Barlowe, 2002) and for Nyv1p (Kweon et al., 2003), albeit with less efficiency.
The fact that individual SNAREs are involved in multiple trafficking steps makes it difficult to unambiguously assign sets of SNAREs to each intracellular trafficking step. In yeast, mutations rendering SNAREs nonfunctional may result in complex phenotypes, a problem particularly prevalent in the early steps of the secretory pathway. As discussed above, many of these mutations affect “layer” amino acids, resulting in impaired packing of all SNARE complexes in which this particular SNARE is participating. Interestingly, it has been shown that such phenotypes can be rescued if mutations are introduced into one of the partner SNAREs, which restore correct packing of the affected layer. In the yeast exocytotic SNARE complex, substitution of the `0' layer glutamine with an arginine in the Qa-position (Sso2p) leads to a strong growth and secretion defect that is associated with an accumulation of transport vesicles in the cytoplasm (Katz and Brennwald, 2000; Ossig et al., 2000). This pheno-type can be rescued by replacing the adjacent arginine of Snc2p with a glutamine.
Here, we have investigated in more detail whether such rotational substitutions can be used to map functionally interacting SNAREs. We have focused on the SNAREs involved in anterograde traffic from the endoplasmic reticulum to the Golgi because two of the SNAREs (Sed5p and Sec22p) are assumed to operate in multiple trafficking steps. Yeast is an ideal system to address these questions because shuffling of residues between the subunits of the SNARE complexes is easy to perform and in vivo effects can be examined conveniently. Our findings suggest that pheno-types caused by the presence of two arginines in the `0' layer can be rescued by compensating glutamine substitutions, but they also show that the severity of the phenotype strongly depends on the relative positions of the two arginines. Furthermore, we found that Gln-to-Arg substitution in Sed5p resulted in defects of both ER-to-Golgi and intra-Golgi trafficking steps that were selectively rescued by sec22(Q) and ykt6(Q), respectively.
MATERIALS AND METHODS
Yeast Strains, Nucleic Acid, and Genetic Techniques
Molecular cloning techniques were performed as described by (Sambrook et al., 1989). Propagation of plasmids was carried out in Escherichia coli strains DH5α or XL1Blue by using standard media. Enzymes used for manipulation of DNA were supplied from New England Biolabs (Beverly, MA).
The plasmids used in this study are listed in Table 1 and were constructed as follows. `0' layer mutations were generated by QuikChange (Stratagene, La Jolla, CA) site-directed mutagenesis PCR by using primers coding for the desired amino acid exchange and additional silent mutations to create a new restriction site for control. All point mutations were confirmed by sequencing. bet1S86R was constructed with the oligonucleotides “for-bet1R GAT GAG ATC CGC GGC CGC AAT CAA ACT” and “rev-bet1R AGT TTG ATT GCG GCC GCG GAT CTC ATC” to generate the arginine mutation in position 86 in addition to silent mutations to introduce a SacII site. Analogously, the bet1S86Q mutation, which was made using the oligonucleotides for-bet1Q GAT GAG ATC CGC GGC CAG AAT CAA ACT and rev-bet1Q AGT TTG ATT CTG GCC GCG GAT CTC ATC, also contains an additional SacII site due to silent mutations. PCR reactions were carried out using pSPT18-SLY12 as DNA-template, and the resulting amplification products were subcloned into pRS316 (CEN6-URA3, (Sikorski and Hieter, 1989) by using HindIII-SacI sites (pCG27 and pCG39). In parallel, the 1.8-kb BET1 fragment (enclosing 440 bp upstream of ATG and 790 bp downstream of STOP) was subcloned into pRS316 via the same restriction sites (pCG36). A XbaI restriction site was added by PCR at the 3′ end of the 1.8-kb bet1S86R fragment, and the insert was cloned into pUA4 via XbaI-HindIII (pCG64). Bet1S86Q was subcloned into pCG60 and into pCG165 by using the XhoI-KpnI restriction sites (pCG170 and pCG172). The sed5Q283R construct was generated from the DNA-template pUA43 by using the oligonucleotide pair for-sed5R CCA TGG TGC AGG AAC GGG GCG AAG and rev-sed5R CTT CGC CCC GTT CCT GCA CCA TGG, which introduces silent base substitutions to generate a new BsgI site in addition to the mutated codon at position 283, and subcloned into pRS316 (pCG32) and pRS317 (pCG163) via SalI. In parallel, the 3-kb insert of pUA43 was moved to pRS316 (pCG34) by using the same restriction site as before. Plasmid pCG62 was obtained by cloning the insert of pCG32 (SalI) into the XhoI site of pUA4. For the bos1Q186R mutation pUA37 served as template and we used the primers for-bos1R CGA AAA TAT TGT GGA ACG AAA CAA AAT TTT and rev-bos1R AAA ATT TTG TTT CGT TCC ACA ATA TTT TCG, introducing a SspI restriction site. The bos1 arginine mutant was subcloned into pRS315 and pRS316 via PvuII (pCG69; pCG161). Ufe1Q289R, containing a new BglII site (via silent mutations), was obtained from template pTN75 with the oligonucleotides for-ufe1R CAT TTA ACG GTG AGA TCT CAA AAT ATA AA and rev-ufe1R TTT ATA TTT TGA GAT CTC ACC GTT AAA TG (pCG5). Ykt6R165Q from pBK87 was moved to pRS316 by using the BamHI-EcoRI restriction sites (pCG60). YKT6 was subcloned from pUA65 into pRS316 using the XhoI-BamHI restriction sites (pCG165).
Table 1.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pUA4 | sec22-R157Q in pRS313 (CEN6-HIS3) | H. D. Schmitt |
| pUA8 | SEC22 in pRS313 (CEN6-HIS3) | H. D. Schmitt |
| pSPT18-SLY12 | 1.8-kb BET1 HindIII/NcoI in pSPT18 | Dascher et al. (1991) |
| pCG36 | 1.8-kb BET1 Hind III/SacI in pRS316 (CEN6-URA3) | This study |
| pCG27 | 1.8-kb containing bet1-S86R HindIII/SacI in pRS316 (CEN6-URA3) | This study |
| pCG39 | 1.8-kb containing bet1-S86Q in pRS316 (CEN6-URA3) | This study |
| pCG64 | 1.8- + 1.5-kb bet1-S86R XbaI/SacI, sec22-R157Q in pRS313 (CEN6-HIS3) | This study |
| pUA43 | SED5 in pRS325 (2 μ-LEU2) | H. D. Schmitt |
| pCG34 | 3-kb SED5 in pRS316 (CEN6-URA3) | This study |
| pCG32 | 3-kb sed5-Q283R in pRS316 (CEN6-URA3) | This study |
| pCG51 | 3-kb sed5-Q283R in pRS315 (CEN6-LEU2) | This study |
| pCG163 | 3-kb sed5-Q283R in pRS317 (CEN6-LYS2) | This study |
| pCG62 | 3- + 1.5-kb sed5-Q283R SalI in XhoI, sec22-R157Q in pRS313 (CEN6-HIS3) | This study |
| pUA37 | BOS1 in pRS315 (CEN6-LEU2) | Andag et al. (2001) |
| pCG69 | 1.8-kb bos1-Q186R in pRS315 (CEN6-LEU2) | This study |
| pCG161 | 1.8-kb bos1-Q186R in pRS316 (CEN6-URA3) | This study |
| pTN75 | UFE1 in pRS316 (CEN6-URA3) | H. D. Schmitt |
| pCG5 | 1.8-kb ufe1-Q289R in pRS316 (CEN6-URA3) | This study |
| pUA65 | YKT6 in pRS315 (CEN6-LEU2) | H. D. Schmitt |
| pCG165 | YKT6 BamHI/XhoI in pRS316 (CEN6-URA3) | This study |
| pBK87 | 1.1-kb ykt6-R165Q in pRS315 (CEN6-LEU2) | Dilcher et al. (2001) |
| pCG60 | ykt6-R165Q BamHI/EcoRI in pRS316 (CEN6-URA3) | This study |
| pCG170 | 1.8-kb bet1-S86Q XhoI/KpnI in pCG60 (CEN6-URA3) | This study |
| pCG172 | 1.8-kb bet1-S86Q XhoI/KpnI in pCG165 (CEN6-URA3) | This study |
The Saccharomyces cerevisiae strains used in this study are listed in Table 2. Standard techniques were used for sporulation and tetrad analysis of yeast strains (Sherman et al., 1981). Transformation of plasmids into yeast was carried out using chemically competent yeast cells (Ito et al., 1983). For transformation of yeast cells with linear DNA segments, a lithium acetate method was used (Gietz et al., 1992). Yeast strains were grown in rich medium (YEPD) or in standard synthetic minimal medium (SD) or synthetic complete medium (SC). Strain YCG15 [sed5(R)] was generated by transformation of the diploid strain Y21581 with plasmid pCG32-sed5(R) (URA3) and subsequent tetrad dissection. Strain YCG74 was obtained from strain YCG15 [sed5(R)], which was transformed with pCG51-sed5(R) (LEU2), and the transformants were selected for loss of pCG32-sed5(R) (URA3) on 5-fluoroorotic acid-containing medium. Deletion of the SEC22 open reading frame was carried out by using the LEU2 cassette for gene replacement. The PCR amplification of the LEU2 from pRS315 with the primers CTG ACA GTG ACA CCC CGT TAC ACA CTC ACA ATT AAG TAG GGG GCG CCT GAT TCA AGA AAT ATC and CAA ATT GAT CGG GAT TTG TGA TGT GGG ATG ATG GGG TGA CGT GGG AAA TGG TTC AAG AAG GTA yielded a 2-kb product with flanking sequences homologous to 41 bp upstream of the ATG and 42 bp downstream of the stop codon in SEC22, which was transformed into the heterozygous diploid deletion strains (Y21396 and Y21581) and selected for the insertion by growth on appropriate selective medium. In a second transformation, plasmid-encoded wild-type copies of BET1 and SED5 (pCG36 or pCG34) were introduced into the diploid strains. After sporulation and tetrad dissection, spores with the desired genotype were selected (confirmed by PCR; strains YCG7 and YCG2). The `0' layer swaps were obtained by transformation of pCG64-bet1(R)/sec22(Q) or pCG62-sed5(R)/sec22(Q) into the sec22Δ/bet1Δ and sec22Δ/sed5Δ strains (YCG7 and YCG2), respectively, and subsequent exchange of the wild-type plasmids for the plasmids carrying the mutations by using 5-fluoroorotic acid, thus leading to strains YCG8 [bet1(R)/sec22(Q)] and YCG3 [sed5(R)/sec22(Q)]. Strain YCG9 was obtained by transformation of pCG69-bos1(R) and pUA4-sec22(Q) into the diploid strain Y22689 and tetrad dissection. The bos1Δ/sec22Δ strain YCG38 was generated by crossing the strains YCG9 and Y15177. After tetrad dissection the genotype was confirmed by PCR. Strain YCG84 (sed5Δ/bet1Δ/sec22Δ) was obtained by crossing strains YCG2 and YCG8 followed by dissection of tetrads and confirmation of the genotype by PCR. Strain YCG103 [bet1(R)/sed5(R)] was generated from strain YCG84, carrying pCG34-SED5 (URA3), which was transformed with plasmid pCG163-sed5(R) (LYS2), and appropriate transformants were subsequently shifted to 5-fluoroorotic acid to select for cells lacking the wild-type version of SED5.
Table 2.
Yeast strains used in this study
| Strain | Abbreviation | Genotype | Source |
|---|---|---|---|
| BY4742 | Wild type | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf |
| Y15177 | sec22Δ | MATα his3Δ1 leu2Δ0 ura3Δ0 sec22::KanMX4 | Euroscarf |
| Y21396 | BY4743 Mata/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 | Euroscarf | |
| ura3Δ0/ura3Δ0 BET1/BET1::KanMX4 | |||
| YCG7 | MATa his3Δ1 leu2Δ0 ura3Δ0 | This study | |
| bet1::KanMX4 sec22::LEU2 +pCG36-BET1 | |||
| YCG8 | sec22(Q)/ | MATa his3Δ1 leu2Δ0 ura3Δ0 | This study |
| bet1(R) | bet1::KanMX4 sec22::LEU2 +pCG64-bet1S86R-sec22R157Q | ||
| Y21581 | BY4743 Mata/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/LYS2 MET15/met15Δ0 | Euroscarf | |
| ura3Δ0/ura3Δ0 SED5/SED5::KanMX4 | |||
| YCG15 | sed5(R) | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | This study |
| sed5::KanMX4 +pCG32-sed5Q283R | |||
| YCG74 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | This study | |
| sed5::KanMX4 +pCG51-sed5Q283R | |||
| YCG2 | MATα his3Δ1 leu2Δ0 ura3Δ0 | This study | |
| sed5::KanMX4 sec22::LEU2 +pCG34-SED5 | |||
| YCG3 | sec22(Q)/ | MATα his3Δ1 leu2Δ0 ura3Δ0 | This study |
| sed5(R) | sed5::KanMX4 sec22::LEU2 +pCG62-sed5Q283R-sec22R157Q | ||
| Y22689 | BY4743 Mata/α, his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2D0/LYS2 MET15/met15Δ0 | Euroscarf | |
| ura3Δ0/ura3Δ0 BOS1/bos1::KanMX4 | |||
| YCG9 | MATa met15Δ0 ura3Δ0 bos1::KanMX4 | This study | |
| +pCG8-bos1Q186R +pUA4-sec22R157Q | |||
| YCG38 | sec22(Q)/ | MATα his3Δ1 ura3Δ0 leu2Δ0 bos1::KanMX4 sec22::KanMX4 | This study |
| bos1(R) | +pCG8-bos1Q186R +pUA4-sec22R157Q | ||
| YCG84 | MATα lys2Δ0 sed5::KanMX4 sec22::LEU2 bet1::KanMX4 +pCG64-bet1S86R- | This study | |
| sec22R157Q +pCG34-SED5 | |||
| YCG103 | bet1(R)/ | MATα lys2Δ0 sed5::KanMX4 sec22::LEU2 bet1::KanMX4 | This study |
| sed5(R) | +pCG64-bet1S86R-sec22R157Q +pCG32-sed5Q283R |
Growth Assays
Overnight yeast cultures grown to the logarithmic phase at 25°C in selective media were harvested and diluted into fresh selective medium to an OD600 of 0.1. For the growth curves, the cells were incubated at 25 or 37°C in a water bath under constant shaking at 150 rpm. The first samples were taken 90 min after dilution. Further samples were taken every 60 min, and OD600 was measured. Each growth point represents the average of three samples. For drop tests, the initial cell suspension was diluted 16-fold. Ten microliters of each dilution was spotted onto YEPD plates, and the plates were incubated at the indicated temperatures.
Trafficking of Carboxypeptidase Y, Protein Labeling, and Immunoprecipitation
An 2.5 OD600 of yeast cells grown overnight to the logarithmic phase on synthetic medium (25°C, 150 rpm) was harvested, resuspended in fresh SD medium, and incubated for 15 min at the desired temperature (normally 37°C). The samples were pulse labeled with 0.1 mCi of 35S-Translabel mix (MP Biomedicals, Irvine, CA) at the respective temperature for 5 min, followed by addition of 30 μl of 10× chase solution [100 mM methionine, 10 mM cysteine, 2% glucose (wt/vol), and 4% yeast extract (wt/vol)]. Samples were taken either immediately after addition of the chase solution or after an additional incubation time of 30 min. Carboxypeptidase Y (CPY) was immunoprecipitated using polyclonal antibodies. The labeled proteins were separated by SDS-PAGE, and the gel was incubated with Amplify (Amersham Biosciences, Piscataway, NJ) for 30 min. Fluorography on a Kodak BIO MAX film was performed at –80°C for several days.
Yeast Cell Morphology
Overnight yeast cultures grown to the logarithmic phase at 25°C in selective media were harvested, diluted into prewarmed fresh YEPD to an OD600 of ∼0.3, and grown for two further generations at 25°C under constant shaking at 150 rpm to an OD600 = 0.8–1.2, before shifting the cells to the restrictive temperature of 37°C for 1 h (strain YCG15; 30°C). Ten OD600 of cells were harvested, washed three times with 100 mM cacodylate, pH 6.8, and fixed in 100 mM cacodylate containing 2% glutaraldehyde. After washing twice with 50 mM potassium phosphate buffer, pH 7.5, the cell walls were digested at 30°C for 40 min with 125 μg of zymolyase 100 T (Seikagaku America, Rockville, MD) in phosphate buffer. The samples were washed twice with cacodylate buffer and further fixed with 2% glutaraldehyde at 4°C overnight. After resuspension in cacodylate buffer containing 2% OsO4, the samples were incubated at 0°C for 60 min, followed by a 2 h incubation in uranyl acetate. Small agarose blocks were prepared, dehydrated consecutively by increasing ethanol concentrations and embedded in Agar 100. Ultrathin sections were prepared and stained with 1% aqueous uranyl acetate and Reynold's lead citrate.
RESULTS
Search for sec22(Q) Compensating Mutants
Before investigating the effects of R→ Q substitutions in the ER-to-Golgi SNARE complex, we checked whether glutamine in the `0' layer position of Bet1p is functional. Unlike mammalian Bet1, yeast Bet1 protein (like other fungal homologues) contains a serine instead of a glutamine at the `0' layer position (Dascher et al., 1991; Gupta and Brent Heath, 2002). Cells expressing Bet1p(Q) as the only copy showed normal growth behavior (our unpublished data).
We then investigated the effect of the `0' layer R→ Q mutation in Sec22p [referred to as Sec22p(Q)]. The resulting mutant strain [expressing sec22(Q) in a genomic sec22Δ background] showed a temperature-sensitive growth phenotype, i.e., growth was strongly reduced at 37°C (Figure 1), but it was normal at 25°C (our unpublished data). These data show that replacement of the only arginine with glutamine in the ER-to-Golgi SNARE complex is less well tolerated than in the exocytotic complex. Here, no growth phenotype was observed when the corresponding R→ Q substitution was introduced into Snc2p (Katz and Brennwald, 2000; Ossig et al., 2000). The phenotype of sec22(Q) resembles that of a previously characterized sec22 `0' layer mutant, sec22-3, in which the arginine residue at position 157 is replaced by a glycine [sec22(G)] (Novick et al., 1980; Sacher et al., 1997).
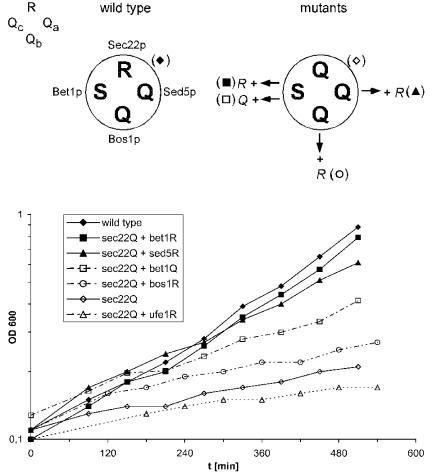
Figure 1.
Growth phenotypes of `0' layer Q→ R mutants analyzed for their ability to suppress the growth defect of the sec22(Q) mutation. The schematic drawings above the panel illustrate the arrangement of the amino acid side chains involved in the `0' layer of the ER-to-Golgi SNARE complex from wild-type and mutant yeast used in the growth assays. The ER-to-Golgi SNARE complex is composed of four proteins: R-SNARE Sec22p and the three Q-SNAREs Sed5p, a member of the syntaxin protein family, at position-Qa, Bos1p at Qb, and Bet1p at Qc. Note that, in contrast to mammalian Bet1 protein, yeast Bet1p carries a serine in the central layer. Bottom, growth curves of yeast cells (sec22Δ) expressing sec22(Q) as the sole version of SEC22 as well as the `0' layer mutants that were analyzed for their rescuing activity. The `0' layer mutants of SED5, BET1, BOS1, and UFE1 were expressed from plasmids in the presence of the normal copies on the chromosomes. Both bet1(R) and sed5(R) suppressed the growth defect caused by the sec22(Q) mutation to almost wild-type level, although the effect of the latter mutation rescued to a slightly lower extent. Yeast strain Y15177 (sec22::KanMX4) was transformed with the following sets of plasmids: pUA8-SEC22 and pRS316 (empty vector; filled diamonds), pUA4-sec22Q and pRS316 (open diamonds), pUA4 and pCG32-sed5R (filled triangles), pUA4 and pCG69-bos1R (open circles), pUA4 and pCG27-bet1R (filled squares), and pUA4 and pCG39-bet1Q (open squares). For control, strain Y15177 expressing pUA4-sec22Q was transformed with pCG5-ufe1R, coding for a `0' layer mutation in the syntaxin involved in retrograde transport to the ER (open triangles). Growth at 37°C was monitored by measuring the optical density at 600 nm. Selective minimal medium was used to prevent the loss of plasmids.
The temperature-sensitive phenotype of sec22(Q) mutants allowed us to test whether complementary Q→ R exchanges in the SNAREs interacting with Sec22p in ER-to-Golgi traffic suppress the effect of the sec22(Q) mutation. Corresponding mutations were introduced into the genes encoding for Bet1p, Bos1p, and Sed5p. All mutants were expressed from centromeric low copy number vectors under the control of their endogenous promoters in sec22(Q) cells still carrying chromosomal BET1, BOS1, and SED5 copies. Whereas bos1(R) did not improve growth of sec22(Q) cells at 37°C (Figure 1; ○), both bet1(R) and sed5(R) were able to suppress the temperature sensitivity (Figure 1, ▪, ▴). Because it was shown previously that the phenotype of sec22-3 is suppressed by overexpression of Bet1p (Newman et al., 1990), we expressed wild-type versions of Bet1p and Sed5p from the same vectors used for the mutants. However, no rescue was observed, with only a slight improvement observed for BET1, indicating that the rescue is not due to a mere increase of the protein levels. Thus, complementary mutations in two of the SNAREs implicated in ER-to-Golgi traffic in yeast (Lian et al., 1994; Søgaard et al., 1994; Cao and Barlowe, 2000; Parlati et al., 2002; Liu et al., 2004) compensate for the defect caused by the R→ Q substitution in the `0' layer of Sec22p.
As discussed in Introduction, Sec22p also is involved in traffic from the Golgi to the ER, where it is supposed to interact with the SNAREs Ufe1p (Qa), Sec20p (Qb), and Use1p (Qc) (Lewis et al., 1997; Burri et al., 2003; Dilcher et al., 2003). We therefore tested whether a Q→ R exchange in Ufe1p [ufe1(R)] influenced the growth behavior of the sec22(Q) strain. However, no rescue was observed (Figure 1). This suggests that the growth defect of sec22 `0' layer mutation is primarily due to a defect in forward transport.
Effects of the Presence of Two Arginines at Varying Positions in the `0' Layer of the ER-to-Golgi SNARE Complex
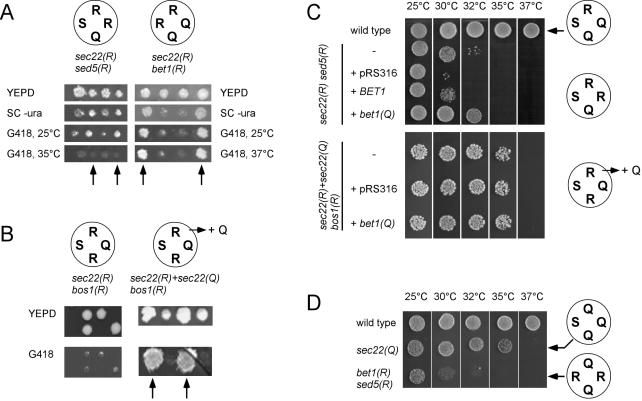
In the next series of experiments, we asked whether the presence of two arginines in the `0' layer of the ER-to-Golgi SNARE complex affects cell viability, and if so, whether the relative position of the two arginines toward each other influences the phenotype. Because BET1, BOS1, and SED5 all are essential genes, we chose the following strategy. Q→ R mutants were introduced into diploid strains heterozygous for the corresponding gene deletion and expressing wild-type Sec22p. Tetrad dissection was used to obtain cells expressing the mutant genes in a background in which the corresponding wild-type genes are deleted. Figure 2A shows the growth phenotypes of spores from representative tetrads. To identify the cells carrying the chromosomal deletions, cells were grown in the presence of G418 (arrows).
Figure 2.
Phenotypes of yeast cells containing ER-to-Golgi SNARE complexes with 2R:2Q stoichiometry depend on the positions of the arginines. (A) Cells expressing ER-to-Golgi SNARE mutations that introduce a second arginine into the SNARE complex either at position Qa or Qc are viable. Four viable spores were obtained from a heterozygous SED5 or BET1 knockout strain (strains Y21581 and Y21396), which harbors a plasmid-encoded sed5(R) (pCG32) or bet1(R) (pCG27) mutation. Sed5(R) or bet1(R) mutant cells (arrows) were resistant to Geneticin (G418+) as the KanMX cassette was used to replace the SED5 gene. They were Ura+ because they express the sed5(R) or bet1(R) mutation on plasmids carrying the URA3 marker. Replica plating also showed that the sed5(R) cells are temperature sensitive at 35°C. Unlike the sed5(R) mutation, however, the bet1(R) mutants showed no growth defect. (B) The bos1(R) mutant as the only source of BOS1 is not viable. Left, two of the tetrads obtained from a heterozygous BOS1 knockout strain (Y22689) expressing bos1(R) (pCG69). All viable spores are Geneticin sensitive, indicating that they carry the wild-type BOS1 gene. Subsequent analysis of the viable spores indicated that the bos1(R) plasmid was not lost during sporulation. However, cotransformation of bos1(R) and sec22(Q) into Y22689 yielded tetrads with four viable spores. Two spores carried the marker used for the deletion of BOS1 (right, arrows). Analysis of their auxotrophic markers demonstrated that all Geneticin-resistant spores contained both plasmids, indicating that the bos1(R) mutants require sec22(Q) for viability. (C) Introducing an additional copy of bet1(Q) raises the symmetry in the `0' layer, but it has only mild effects on the growth phenotype. Cells were grown on YEPD for 60 h (top) or 65 h (bottom). In a 2R:2Q ratio where the second arginine is contributed by sed5(R) (strain YCG74) additional expression of bet1(Q) resulted in moderate improvement of growth at 32°C. When bet1(Q) was introduced into strain YCG9 [sec22(R) + sec22(Q) bos1(R)], where the second arginine is shifted to the Qb-position, no improvement of growth was observable (bottom). (D) Two arginines at positions Qa and Qc are tolerated. Yeast cells (strain YCG103), expressing bet1(R), sec22(Q), and sed5(R) were viable but sensitive to higher temperature (37°C). The resulting ER-to-Golgi SNARE complex contained two opposing arginines at positions-Qa and -Qc. Strains were spotted on YEPD plates and grown for 48 h.
Cells expressing Sed5p with the Q→ R mutation exhibited a strong phenotype. Although tetrads with four viable spores were obtained from heterozygous knockout strains expressing sed5(R) (Figure 2 A, left), it was apparent that these cells have serious growth defects. They grow poorly on YEPD medium (our unpublished data; see below) and were not viable at 37°C. This phenotype is reminiscent of that of the strain carrying the corresponding `0' layer configuration in the exocytotic SNARE complex (Ossig et al., 2000). In contrast, no growth defects were observable for cells expressing Bet1p with the Q→ R mutation [bet1(R); Figure 2A, right]. This differs from the phenotype obtained for the corresponding mutation in the exocytotic SNARE complex, which was shown to be temperature sensitive (Katz and Brennwald, 2000).
For Bos1p, no bos1Δ cells could be obtained carrying the bos1 Q→ R mutation (Figure 2B, left). Again, this observation parallels the results obtained previously with the exocytotic SNARE complex (Katz and Brennwald, 2000). However, these results do not exclude that the Bos1p(R) variant is completely nonfunctional, e.g., due to misfolding or degradation, a view that gains support by the failure of Bos1p(R) to rescue the Sec22p(Q) phenotype (Figure 1). We therefore tested whether viable spores are obtained by introducing the Sec22p(Q) variant into the Bos1p(R)-expressing diploid strain, thus restoring a 3Q:1R configuration in the `0' layer. Tetrad analysis revealed that viable bos1(R) cells were only obtained when the plasmid encoding Sec22p(Q) was present in addition to the chromosomally encoded Sec22p(R) (Figure 2B, right).
As discussed above, wild-type Bet1p contains a serine instead of a glutamine in the `0' layer position, raising the possibility that the stability of the complex is reduced, particularly when a second arginine is introduced. Therefore, we tested whether additional expression of bet1(Q) improved viability of the strains containing two arginines in the `0' layer. However, with exception of a slight improvement of the Sed5p(R)/Sec22p(R)-expressing strain, no differences were observed (Figure 2C).
The failure of obtaining viable spores for the Sec22p(R)/Bos1p(R) combination could indicate that two arginine residues occupying opposite positions in the `0' layer are not tolerated. To address this question, we constructed cells expressing Sec22p(Q) as well as Sed5p(R) and Bet1p(R) as the only versions of these proteins. In contrast to the results obtained with the Bos1p(R) mutant, these cells carrying the arginines at positions Qa and Qc were viable, although temperature sensitive (Figure 2D).
Analysis of Cells with the `0' Layer Arginine Moved from Sec22p to Bet1p or Bos1p
The experiments presented in Figure 1 show that Q-SNAREs carrying a Q→ R substitution are able to suppress the growth defect of cells expressing Sec22p(Q) when expressed in addition to the chromosomal wild-type copies of these genes. However, in these experiments it is possible that the presence of the corresponding wild-type copies contributes to the rescue. Therefore, we examined the effects of these mutual amino acid exchanges on a genomic deletion background of the corresponding genes.
We created cells expressing only the mutated versions of BET1 or BOS1 in a sec22(Q) background. These cells carried deletions of SEC22 (sec22::LEU2 or sec22::KanMX4) and either BET1 (bet1::KanMX4) or BOS1 (bos1::KanMX4) at their chromosomal sites as well as centromeric plasmids containing sec22(Q) and either bet1(R) or bos1(R).
Cells with the arginine moved from the R-SNARE to the putative Qc-position [sec22(Q)/bet1(R) expressing cells] grow well at 37°C (Figure 3A). To analyze ER-to-Golgi transport in these cells, we performed pulse-chase experiments with [35S]methionine to monitor maturation of CPY. CPY first occurs as a p1 precursor in the ER; is then modified to a larger form, p2, in the Golgi; and finally is transported to vacuoles where it is processed by proteolysis to its mature form, m (Stevens et al., 1982). In wild-type cells, without chase only the ER form is observed, whereas after 30 min the protein is completely converted into the mature form (Figure 3B, left lanes). In sec22(Q) cells, CPY remains in the p1 ER form after 30 min of chase as expected for a defect in ER-to-Golgi transport (Figure 3B, middle lanes). If these cells express Bet1p(R) instead of the wild-type protein, processing of CPY was as in wild-type cells, indicating that transport of CPY to the vacuole is normal.
Figure 3.
Exchanging the arginine between Sec22p and Bet1p [sec22(Q)/bet1(R)] does not affect growth or ER-to-Golgi transport. (A) Cells expressing wild-type BET1 and sec22(Q) as the only source of SEC22 (4Q ratio, strain Y15177 +pUA4-sec22R157Q) show reduced growth at 35°C and failed to grow at 37°C when incubated for 48 h (see Figure 1). Growth was normal in cells where the 3Q:1R ratio was restored by introducing an arginine residue at the `0' layer position of Bet1p (second row, strain YCG8). (B) Processing of CPY, analyzed by SDS-PAGE and autofluorography. CPY was immunoprecipitated from cell extracts after a 15-min preincubation and pulse-chase labeling at 37°C (see Materials and Methods). In sec22(Q)/bet1(R) mutant cells, like in wild-type cells, all of the labeled CPY was transported to the vacuole. After a chase period of 30 min, only the mature form of CPY (mCPY) was detected, whereas the ER form (p1CPY) accumulated in sec22(Q) mutant cells.
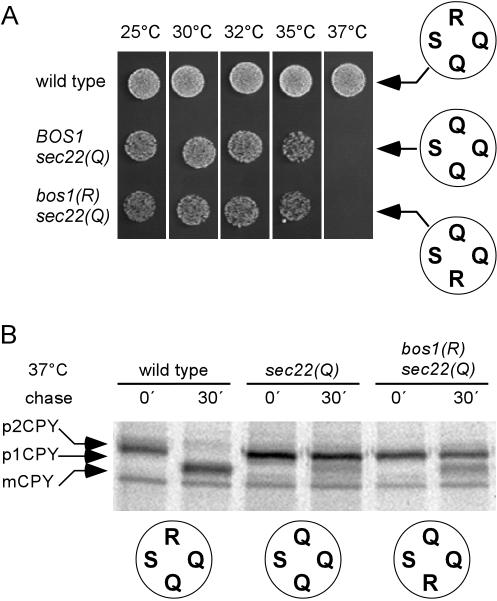
When the arginine was moved to the Qb-position, a different phenotype was observed. These cells showed no growth at 37°C and grew poorly at lower temperatures. In fact, growth behavior was even worse than that of a strain expressing bos1(Q) (Figure 4A). Pulse-chase experiments revealed that CPY processing was partially inhibited after 30 min with only a fraction being converted into the mature form (Figure 4B). In contrary to our original expectations, these data suggest that the three glutamine positions in the original complex are structurally not equivalent. This is consistent with the observation that overexpression of Bos1p(R) is not able to rescue the Sec22p(Q) phenotype (Figure 1).
Figure 4.
Exchanging the arginine between Sec22p and Bos1p [sec22(Q)/bos1(R)] renders the mutant cells temperature sensitive and causes a block in anterograde traffic from ER to Golgi. (A) Cells with the ratio in the central layer of the ER-to-Golgi SNARE complex restored to 3Q:1R, but with the arginine moved to Qb, are viable but show a stronger growth defect than cells with a 4Q ratio. When incubated for 48 h on YEPD plates sec22(Q)/bos1(R) mutants (YCG38) failed to grow at 37°C and grow poorly even at medium or low temperatures. (B) Analysis of CPY processing in sec22(Q)/bos1(R) mutants. The cells were pulse labeled at 37°C (see legend to Figure 3). In these mutant cells, a block in an early step in transport of CPY was observed, resembling the transport defect seen in the sec22(Q) mutant. After a 30-min chase most of the labeled CPY accumulated in the ER form (p1CPY), and only a minor fraction was detected in the vacuolar mature form (mCPY).
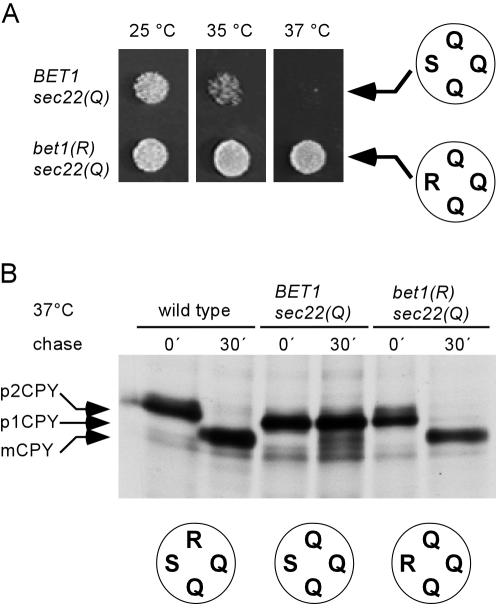
Analysis of Cells with the `0' Layer Arginine Moved from Sec22p to Sed5p
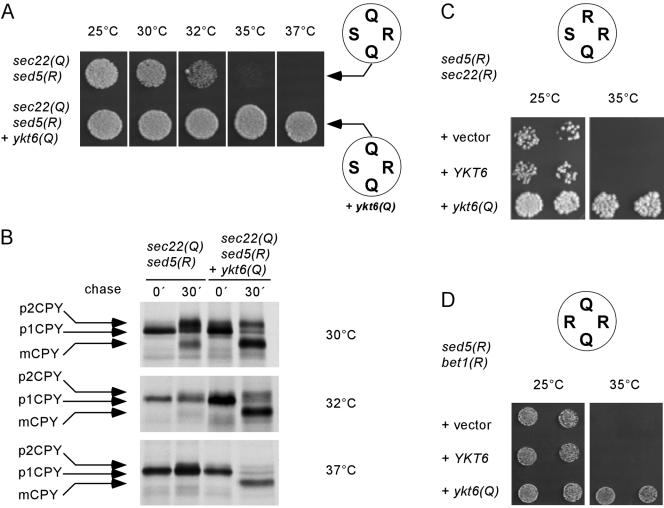
Finally, we investigated the growth behavior of cells in which the 3Q:1R ratio was restored but the arginine was moved to the Qa-position [sed5(R)]. Again, no rescue to wild-type growth levels was observed. The cells were temperature sensitive, exhibiting a growth defect at 35°C (Figure 5A), in contrast to normal growth observed when wild-type Sed5p is still present. As outlined in Introduction, Sed5p seems to be involved in a second trafficking step within the Golgi apparatus where it is supposed to function in a SNARE complex with different SNAREs, including the R-SNARE Ykt6p (McNew et al., 1997; Dilcher et al., 2001). Thus, it is conceivable that the growth defect can be attributed to a nonfunctional intra-Golgi SNARE complex, whereas ERto-Golgi transport is functioning normally. To test for this possibility, we performed two types of experiments. First, we asked whether coexpression of an ykt6(Q) mutant allele results in restoration of a wild-type phenotype. As shown in Figure 5A, these cells grew normally at 37°C. Furthermore, we tested for processing of CPY at different temperatures. The predominant form of CPY accumulating in the Sec22p(Q)/Sed5p(R) cells at the semipermissive temperature of 30°C is the Golgi p2 form, whereas processing is completely blocked at higher temperatures. Additional expression of Ykt6p(Q) resulted in enhanced CPY processing at 30°C, with complete conversion to the mature form being observed at 37°C (Figure 5B).
Figure 5.
Effects of arginine at the Qa-position (Sed5p). (A) Cells expressing sec22(Q)/sed5(R) as the sole versions of SEC22 and SED5 (strain YCG3) failed to grow at 35°C and showed reduced growth after 48 h at 32°C. This growth defect was suppressed by introducing a plasmid-encoded version of the intra-Golgi R-SNARE Ykt6p. (B) Swapping the arginine to the Qa-position causes defects in CPY trafficking to the vacuole. CPY processing was analyzed after growing the strains at the indicated temperatures (see Materials and Methods). In sec22(Q)/sed5(R) mutant cells grown at 30°C most of the CPY accumulated in its Golgi form (p2CPY), indicating that the intra-Golgi transport is affected. At elevated temperatures CPY accumulated exclusively in the ER form. The expression of ykt6(Q) in addition to wild-type YKT6, an intra-Golgi R-SNARE, restored transport of CPY to the vacuole as indicated by the appearance of the mature form (mCPY). (C and D) ykt6(Q) partially rescues the temperature-sensitive phenotype of the 2R:2Q mutants sec22(R)/sed5(R) (YCG15) and bet1(R)/sed5(R) (YCG103). In C, cells were grown on YEPD for 4 d and in D, for 48 h. To rule out gene dosage effects, we analyzed transformants carrying either the empty vector or vectors with the wild-type YKT6 gene or the ykt6(Q) mutant allele as insert.
Second, we examined whether coexpression of Ykt6p(Q) leads to growth improvement in strains containing two arginines in the `0' layer of the ER-to-Golgi SNARE complex, with one of the arginines being contributed by Sed5p(R). Two strains characterized by a 2R:2Q ratio in the `0' layer with Sed5p(R) as an artificial R-SNARE and either Sec22p(R) or Bet1p(R) as second R-SNARE were analyzed. In both cases, Ykt6p(Q) would be expected to rescue the intra-Golgi trafficking defect while having no effect on the ER-to-Golgi trafficking defect. As shown in Figure 5, C and D, the growth was improved by introducing a plasmid-encoded ykt6(Q) allele. Coexpression of Bet1p(Q) did not change the pheno-types (Supplemental Figure 1).
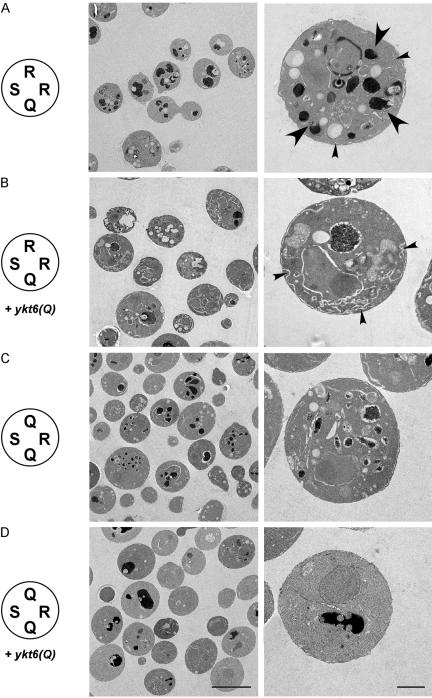
To confirm that Sed5p is indeed involved in two separate trafficking steps, interacting with the R-SNAREs Sec22p and Ykt6p, respectively, we performed electron microscopy on the cells expressing Sed5p(R) as the only version of Sed5p (Figure 6). Cells were shifted to restrictive temperatures before harvesting and fixation to uncover morphological changes associated with the transport defects. Sed5p(R)-expressing cells show a strong accumulation of intracellular membranes in the presence of wild-type Sec22p(R), with numerous vacuolar structures being visible (large arrows). Furthermore, elongated cisternae were observed close to the cell periphery bearing similarities to typical ER membranes (small arrows). In sharp contrast, no aberrant vacuolar morphology was observed when Ykt6p(Q) was expressed in these cells, suggesting that the trafficking defect leading to the accumulation of these structures (presumably intra-Golgi) was rescued. Instead, massive accumulation of flat cisternae and small vesicles was observed, indicating an accumulation of ER-derived membranes (Figure 6B). Conversely, accumulation of vacuolar structures but no obvious cisternal changes were observed in the presence of Sec22p(Q), again in agreement with the notion that in these cells the defect is associated with an intra-Golgi and not with an ER-to-Golgi trafficking step. Finally, expression of Ykt6p(Q) in this strain did not lead to accumulation of intracellular membranes, with the cell morphology being similar to that of wild-type cells (Figure 6D).
Figure 6.
Morphology of mutant yeast cells expressing sed5(R) as the only version of SED5 as well as compensating mutations in the R-SNAREs SEC22 or YKT6, analyzed by electron microscopy. (A) sed5(R) as sole copy of SED5 (strain YCG15) caused accumulation of large amounts of intracellular membranes (small arrowheads) and fragmentation of vacuoles (large arrowheads). (B) Coexpression of plasmid-encoded ykt6(Q) in the sed5(R) mutant resulted in healthier cells that showed accumulation of membrane cisternae (small arrowheads) but displayed normal vacuole morphology. (C) sec22(Q)/sed5(R) double mutants (strain YCG3) displayed accumulation of fragmented vacuolar structures but lacked anomalous membrane cisternae. (D) sec22(Q)/sed5(R) mutant cells expressing ykt6(Q) in addition to the chromosomal YKT6 gene showed normal (wild-type) morphology.
DISCUSSION
In this study, we have explored how substitutions in the central `0' layer affect the function of the SNARE complex involved in anterograde traffic from the ER to the Golgi apparatus. By systematically exchanging glutamines for arginines and vice versa in all four SNAREs, we found that this technique can be used to map functionally interacting SNARE complexes and to document cooperation of individual SNAREs with different SNARE partners in distinct trafficking steps. However, our study also reveals that despite the rotational symmetry of the `0' layer, the positions of the four side chains are not equivalent, resulting in phenotypes that are dependent on the position of the exchanges, both in mutants carrying two arginines, as well as in mutants in which the arginine has been moved to another position. A summary of the results is shown in Table 3.
Table 3.
Summary of phenotypes caused by `0' layer mutations tested in this study
| ER—Golgi SNAREs
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Sec22 | Sed5 | Bos1 | Bet1 | Other SNAREs | Growth | Transport of CPY (37°C) | Morphology (electron microscopy) | |
| R | Q | Q | S | wt | Vacuolar form | Wild type (reference) | ||
| Q | Q | Q | S | Ts- | ER—cis-Golgi form | — | ||
| Q | Q+R | Q | S | wt | Vacuolar form | — | ||
| Q | Q | Q+R | S | Ts- | ER—cis-Golgi form | — | ||
| Q | Q | Q | S+R | wt | Vacuolar form | — | ||
| Q | Q | Q | S+Q | Ts- | ER—cis-Golgi form | — | ||
| 2R:2Q | ||||||||
| R | R | Q | S | Ts-* | — | Accumulation of ER-like membranes; fragmented vacuoles | ||
| R | R | Q | S | + ykt6(Q) | Ts- | — | Accumulation of ER-like membranes | |
| R | R | Q | S+Q | Ts-* | — | — | ||
| R | R | Q | S+Q | + ykt6(Q) | Ts- | — | — | |
| R | Q | R | S | lethal | — | — | ||
| R+Q | Q | R | S | Ts- | — | — | ||
| R+Q | Q | R | S+Q | Ts- | — | — | ||
| R | Q | Q | R | wt | — | — | ||
| Q | R | Q | R | Ts-* | — | — | ||
| Q | R | Q | R | + ykt6(Q) | wt | — | — | |
| Swaps | ||||||||
| Q | R | Q | S | Ts-* | ER and Golgi form | Fragmented vacuoles | ||
| Q | R | Q | S | + ykt6(Q) | wt | Vacuolar form | Resembling wild type | |
| Q | Q | R | S | Ts- | ER and vacuolar form | — | ||
| Q | Q | Q | R | wt | Vacuolar form | — | ||
Ts-, restrictive temperature 37°C; Ts-*, restrictive temperature 33°C; wt, wild-type growth.
SNARE complexes are beautiful examples of four-helix coiled-coils in which binding between helices is primarily mediated by 16 layers of four interacting side chains each. Mutations in various layers were identified in mutant screens, or they were introduced by site-directed mutagenesis. In many cases, these layer mutations result in an impairment of the respective fusion step (Banfield et al., 1995; Lewis et al., 1997; Sacher et al., 1997; Stone et al., 1997; Littleton et al., 1998; Saifee et al., 1998; Dilcher et al., 2001). Several layers are highly asymmetric, i.e., large side chains interact with small side chains. However, due to its unusual charged/hydrophilic interactions that are surrounded by tight hydrophobic packing, the `0' layer stands out and thus has attracted considerable interest. Although some `0' layer substitutions have only mild phenotypes, most are rather severe, which may explain the high degree of evolutionary conservation.
Q→ R-substitutions have hitherto been carried out in three different yeast Qa-SNAREs, each resulting in defects of the corresponding fusion steps. In Sso2p, a Qa-SNARE functioning in exocytosis, a Q→ R exchange resulted in a severe and partially temperature-sensitive growth defect that was fully rescued by replacing the R-SNARE Snc2p with the corresponding R→ Q variant (Katz and Brennwald, 2000; Ossig et al., 2000). These experiments were carried out in strains containing deletions of the SNC1 and SSO1 genes, avoiding functional redundancy by these isoforms. Similarly, a Q→ R exchange in the Qa-SNARE Vam3p functioning in yeast vacuole fusion resulted in a strong impairment, but not a complete block, of vacuole fusion as measured by a cell-free assay (Wang et al., 2001). In our experiments, the strong growth defect obtained by the Q→ R substitution in the Qa-SNARE Sed5p also was rescued by corresponding R→ Q exchanges. However, for a full rescue the exchanges needed to be carried out in two different R-SNAREs, which is due to the involvement of Sed5p in two separate trafficking steps requiring different SNARE partners (see below). Together, these data are consistent with each other, allowing the conclusions 1) that Q→ R-exchanges in Qa-SNAREs generally lead to strong inhibition of the respective fusion step, and 2) that this defect can be fully rescued by performing an R→ Q substitution in the corresponding R-SNARE. Thus, the R- and Qa-positions seem to be structurally and functionally equivalent.
In contrast, it is evident that the Qb-position is not equivalent to the Qa- or the R-position. No viable cells were obtained when Bos1p was replaced with Bos1p(R), and only partial rescue was observed when the R→ Q variant of Sec22p was expressed in these cells. This parallels findings with the exocytotic SNARE complex made by (Katz and Brennwald, 2000), although here a full rescue was observed upon expression of the R→ Q variant of Snc1p.
Q→ R-replacements in the Qc-position again yield milder phenotypes. In our hands, the strain expressing the S→ R version of Bet1p displayed no obvious phenotype, leading to the surprising result that two arginines in this configuration are better tolerated than the 4“Q” (or 3Q:1S) configuration. One might argue that the unusual serine in this position (which is smaller than glutamine) may result in weaker interactions, thus making substitutions in this position better tolerable. Although the presence of Bet1p(Q) led to slight improvements under some conditions, no major impact was seen, supporting the view that the Qc-position may accommodate more structural diversity than the other positions. Q→ R substitutions in the Qc-position of Sec9p (Katz and Brennwald, 2000) and in the Qc-SNARE Vti1p (Dilcher et al., 2001) resulted in growth defects both of which were rescued by the expression of the R→ Q variants of the R-SNAREs Snc1p and Ykt6p, respectively (in each case against the background of the corresponding wild-type proteins).
In contrast to the `0' layer substitutions of the Q-SNAREs, a variety of phenotypes is observed when R-SNAREs are replaced by corresponding R→ Q substitution mutants, resulting in a 4“Q” (or 3Q:1S) configuration in the `0' layer of the respective SNARE complexes. Whereas in the exocytotic complex defects were only uncovered in combination with other layer mutations (Ossig et al., 2000), the phenotype is much more severe in the ER-to-Golgi complex (this study). An even more severe effect was observed by swapping R for Q in Ykt6p where no viable cells were obtained (Dilcher et al., 2001). It should be noted, however, that in contrast to YKT6 (an essential gene), deletions of both SNC1/2 or SEC22 result in viable, although growth-impaired, cells, which at least for SEC22 has recently been attributed to another R-SNARE (Ykt6p) taking over. In our hands, however, ykt6(Q) is not capable of substituting for sec22(Q) in compensating for the sed5(R) defect in the ER-to-Golgi trafficking step (Figure 6; our unpublished observations). Furthermore, both Sec22p and Ykt6p operate in more than one trafficking step, complicating the interpretation of phenotypes caused by such single substitutions.
Two of the SNAREs of the ER-to-Golgi complex are thought to operate in additional trafficking steps. As outlined in Introduction, Sec22p is thought to function as R-SNARE in retrograde traffic, whereas Sed5p also functions in an intra-Golgi trafficking step (probably retrograde vesicle traffic) (Tsui et al., 2001; Parlati et al., 2002). A Q→ R exchange in Sed5p affected both trafficking steps, which are rescued separately by the Q-variants of Sec22p (ER-to-Golgi) and Ykt6p (intra-Golgi), as revealed by our electron microscopic analysis of the respective strains. Thus Q→ R and R→ Q substitutions of Qa- and R-SNAREs, respectively, are useful tools for mapping functionally interacting SNAREs and for dissecting the trafficking steps involved.
It remains to be established, however, why the pheno-types of 2Q:2R complexes are strongly dependent on the position of the arginines. The transport defects may be caused by a lowered stability of the complex, by a retarded zippering kinetics, by interference with the disassembly reaction, or by a combination of these effects. In chromaffin cells where phases of exocytosis can be resolved with high kinetic resolution, substitution of glutamine in soluble N-ethylmaleimide–sensitive factor attachment protein (SNAP)-25 resulted in a delayed refilling of releasable vesicle pools with no apparent impact on the kinetics of release-ready vesicles (Wei et al., 2000; Graham et al., 2001). Furthermore, certain `0' layer substitutions prevent NSF-driven disassembly in vitro (Scales et al., 2001), and the cochaperone α-SNAP has recently been shown to bind in the region of the `0' layer (Marz et al., 2003). Thus, it is possible that layer mutations affect steps leading up to fusion, whereas the fusion step itself remains unaffected.
In summary, our data provide direct evidence that Sec22p, Bet1p, Bos1p, and Sed5p form a functional SNARE complex in vivo that requires correct packing of the `0' layer. Furthermore, they provide genetic evidence for a functional interaction between Sed5p and Ykt6p in a different SNARE complex.
Acknowledgments
We are indebted to Gabi Fischer von Mollard and Dieter Gallwitz for plasmids and antibodies and Hannegret Frahm for excellent technical assistance. This work was supported by the Gottfried Wilhelm Leibnitz Program of the Deutsche Forschungsgemeinschaft (to R. J.).
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–09–0830) on February 23, 2005.
Abbreviations used: CPY, carboxypeptidase Y; ER, endoplasmic reticulum; SNARE, soluble N-ethylmaleimide–sensitive factor attachment protein receptor.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Antonin, W., Fasshauer, D., Becker, S., Jahn, R., and Schneider, T. R. (2002). Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol. 9, 107–111. [DOI] [PubMed] [Google Scholar]
- Banfield, D. K., Lewis, M. J., and Pelham, H.R.B. (1995). A SNARE-like protein required for traffic through the Golgi-complex. Nature 375, 806–809. [DOI] [PubMed] [Google Scholar]
- Bock, J. B., and Scheller, R. H. (1999). SNARE proteins mediate lipid bilayer fusion. Proc. Natl. Acad. Sci. USA 96, 12227–12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J. S., and Glick, B. S. (2004). The mechanisms of vesicle budding and fusion. Cell 116, 153–166. [DOI] [PubMed] [Google Scholar]
- Burri, L., Varlamov, O., Doege, C. A., Hofmann, K., Beilharz, T., Rothman, J. E., Söllner, T. H., and Lithgow, T. (2003). A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 100, 9873–9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., and Barlowe, C. (2000). Asymmetric requirements for a Rab GTPase and SNARE proteins in fusion of COPII vesicles with Acceptor membranes. J. Cell Biol. 149, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher, C., Ossig, R., Gallwitz, D., and Schmitt, H. D. (1991). Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the ras-superfamily. Mol. Cell. Biol. 11, 872–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher, M., Köhler, B., and Fischer von Mollard, G. (2001). Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J. Biol. Chem. 276, 34537–34544. [DOI] [PubMed] [Google Scholar]
- Dilcher, M., Veith, B., Chidambaram, S., Hartmann, E., Schmitt, H. D., and Fischer von Mollard, G. (2003). Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 22, 3664–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer, D. (2003). Structural insights into the SNARE mechanism. Biochim. Biophys. Acta 1641, 87–97. [DOI] [PubMed] [Google Scholar]
- Fasshauer, D., Sutton, R. B., Brünger, A. T., and Jahn, R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard, G., and Stevens, T. H. (1999). The Saccharomyces cerevisiae v-SNARE Vti1p is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell 10, 1719–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St Jean, A., Woods, R. A., and Schiestl, R. H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, M. E., Washbourne, P., Wilson, M. C., and Burgoyne, R. D. (2001). SNAP-25 with mutations in the zero layer supports normal membrane fusion kinetics. J. Cell Sci. 114, 4397–4405. [DOI] [PubMed] [Google Scholar]
- Grote, E., Baba, M., Ohsumi, Y., and Novick, P. J. (2000). Geranylgeranylated snares are dominant inhibitors of membrane fusion. J. Cell Biol. 151, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, G. D., and Heath, I. B. (2002). Predicting the distribution, conservation, and functions of SNAREs and related proteins in fungi. Fungal Genet. Biol. 36, 1–21. [DOI] [PubMed] [Google Scholar]
- Gurunathan, S., Chapman-Shimshoni, D., Trajkovic, S., and Gerst, J. E. (2000). Yeast exocytic v-SNAREs confer endocytosis. Mol. Biol. Cell 11, 3629–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, P. I., Roth, R., Morisaki, H., Jahn, R., and Heuser, J. E. (1997). Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90, 523–535. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn, R., Lang, T., and Südhof, T. C. (2003). Membrane fusion. Cell 112, 519–533. [DOI] [PubMed] [Google Scholar]
- Katz, L., and Brennwald, P. (2000). Testing the 3Q:1R `rule': mutational analysis of the ionic `zero' layer in the yeast exocytic snare complex reveals no requirement for arginine. Mol. Biol. Cell 11, 3849–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon, Y., Rothe, A., Conibear, E., and Stevens, T. H. (2003). Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell 14, 1868–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, M. J., Rayner, J. C., and Pelham, H.R.B. (1997). A novel SNARE complex implicated in vesicle fusion with the endoplasmic-reticulum. EMBO J. 16, 3017–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, J. P., Stone, S., Jiang, Y., Lyons, P., and Ferro-Novick, S. (1994). Ypt1p implicated in v-SNARE activation. Nature 372, 698–701. [DOI] [PubMed] [Google Scholar]
- Littleton, J. T., Chapman, E. R., Kreber, R., Garment, M. B., Carlson, S. D., and Ganetzky, B. (1998). Temperature-sensitive paralytic mutations demonstrate that synaptic exocytosis requires SNARE complex assembly and disassembly. Neuron 21, 401–413. [DOI] [PubMed] [Google Scholar]
- Liu, Y., and Barlowe, C. (2002). Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell 13, 3314–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. T., Flanagan, J. J., and Barlowe, C. (2004). Sec22p export from the endoplasmic reticulum is independent of SNARE pairing. J. Biol. Chem. 279, 27225–27232. [DOI] [PubMed] [Google Scholar]
- Malhotra, V., Orci, L., Glick, B. S., Block, M. R., and Rothman, J. E. (1988). Role of an N-ethylmaleimide sensitive transport component in promoting fusion of transport vesicles with cisternae of the Golgi stack. Cell 54, 221–227. [DOI] [PubMed] [Google Scholar]
- Marz, K. E., Lauer, J. M., and Hanson, P. I. (2003). Defining the SNARE complex binding surface of α-SNAP-Implications for SNARE complex disassembly. J. Biol. Chem. 278, 27000–27008. [DOI] [PubMed] [Google Scholar]
- Mayer, A., Wickner, W., and Haas, A. (1996). Sec18p (NSF)-driven release of sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell 85, 83–94. [DOI] [PubMed] [Google Scholar]
- McNew, J. A., Søgaard, M., Lampen, N. M., Machida, S., Ye, R. R., Lacomis, L., Tempst, P., Rothman, J. E., and Söllner, T. H. (1997). Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J. Biol. Chem. 272, 17776–17783. [DOI] [PubMed] [Google Scholar]
- McNew, J. A., Weber, T., Parlati, F., Johnston, R. J., Melia, T. J., Söllner, T. H., and Rothman, J. E. (2000). Close is not enough: snare-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J. Cell Biol. 150, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, A. P., Shim, J., and Ferro-Novick, S. (1990). BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol. Cell. Biol. 10, 3405–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, P., Field, C., and Schekman, R. (1980). Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21, 205–215. [DOI] [PubMed] [Google Scholar]
- Ossig, R., Schmitt, H. D., de Groot, B., Riedel, D., Keränen, S., Ronne, H., Grubmüller, H., and Jahn, R. (2000). Exocytosis requires asymmetry in the central layer of the SNARE complex. EMBO J. 19, 6000–6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati, F., McNew, J. A., Fukuda, R., Miller, R., Söllner, T. H., and Rothman, J. E. (2000). Topological restriction of SNARE-dependent membrane fusion. Nature 407, 194–198. [DOI] [PubMed] [Google Scholar]
- Parlati, F., Varlamov, O., Paz, K., McNew, J. A., Hurtado, D., Söllner, T. H., and Rothman, J. E. (2002). Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc. Natl. Acad. Sci. USA 99, 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo, J., and Südhof, T. C. (1998). Mechanics of membrane-fusion. Nat. Struct. Biol. 5, 839–842. [DOI] [PubMed] [Google Scholar]
- Rohde, J., Dietrich, L., Langosch, D., and Ungermann, C. (2003). The transmembrane domain of Vam3 affects the composition of cis- and trans-SNARE complexes to promote homotypic vacuole fusion. J. Biol. Chem. 278, 1656–1662. [DOI] [PubMed] [Google Scholar]
- Sacher, M., Stone, S., and Ferro-Novick, S. (1997). The synaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J. Biol. Chem. 272, 17134–17138. [DOI] [PubMed] [Google Scholar]
- Saifee, O., Wei, L. P., and Nonet, M. L. (1998). The Caenorhabditis elegans unc-64 locus encodes a syntaxin that interacts genetically with synaptobrevin. Mol. Biol. Cell 9, 1235–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Scales, S. J., Yoo, B. Y., and Scheller, R. H. (2001). The ionic layer is required for efficient dissociation of the SNARE complex by α-SNAP and NSF. Proc. Natl. Acad. Sci. USA 98, 14262–14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., Fink, G. R., and Hicks, J. B. (1981). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Sikorski, R. S., and Hieter, P. (1989). A system of yeast shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard, M., Tani, K., Ye, R. R., Geromanos, S., Tempst, P., Kirchhausen, T., Rothman, J. E., and Söllner, T. (1994). A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell 78, 937–948. [DOI] [PubMed] [Google Scholar]
- Stevens, T., Esmon, B., and Schekman, R. (1982). Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell 30, 439–448. [DOI] [PubMed] [Google Scholar]
- Stone, S., Sacher, M., Mao, Y., Carr, C., Lyons, P., Quinn, A. M., and Ferro-Novick, S. (1997). Bet1p activates the v-SNARE Bos1p. Mol. Biol. Cell 8, 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, R. B., Fasshauer, D., Jahn, R., and Brunger, A. T. (1998). Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Tsui, M. M., Tai, W. C., and Banfield, D. K. (2001). Selective formation of sed5p-containing snare complexes is mediated by combinatorial binding interactions. Mol. Biol. Cell 12, 521–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., Ueda, T., Ohniwa, R. L., Nakano, A., Takeyasu, K., and Sato, M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49–65. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Dulubova, I., Rizo, J., and Südhof, T. C. (2001). Functional analysis of conserved structural elements in yeast syntaxin Vam3p. J. Biol. Chem. 276, 28598–28605. [DOI] [PubMed] [Google Scholar]
- Wei, S., Xu, T., Ashery, U., Kollewe, A., Matti, U., Antonin, W., Rettig, J., and Neher, E. (2000). Exocytotic mechanism studied by truncated and zero layer mutants of the C-terminus of SNAP-25. EMBO J. 19, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, D., Joglekar, A. P., Williams, A. L., and Hay, J. C. (2000). Subunit structure of a mammalian ER/Golgi SNARE complex. J. Biol. Chem. 275, 39631–39639. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Martin, S., James, D. E., and Hong, W. J. (2002). GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Mol. Biol. Cell 13, 3493–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B., Gonzalez, L., Prekeris, R., Steegmaier, M., Advani, R. J., and Scheller, R. H. (1999). SNARE interactions are not selective. Implications for membrane fusion specificity. J. Biol. Chem. 274, 5649–5653. [DOI] [PubMed] [Google Scholar]