Abstract
Recent cohort studies suggested that SARS-CoV-2 infection is associated with changes in brain structure. However, the potential causal relationship remains unclear. We performed a two-sample Mendelian randomization analysis to determine whether genetic susceptibility of COVID-19 is causally associated with changes in cortical and subcortical areas of the brain. This 2-sample MR (Mendelian Randomization) study is an instrumental variable analysis of data from the COVID-19 Host Genetics Initiative (HGI) meta-analyses round 5 excluding UK Biobank participants (COVID-19 infection, N=1,348,701; COVID-19 severity, N=1,557,411), the Enhancing NeuroImaging Genetics through Meta Analysis (ENIGMA) Global and regional cortical measures, N=33,709; combined hemispheric subcortical volumes, N=38,851), and UK Biobank (left/right subcortical volumes, N=19,629). A replication analysis was performed on summary statistics from different COVID-19 GWAS study (COVID-19 infection, N=80,932; COVID-19 severity, N=72,733). We found that the genetic susceptibility of COVID-19 was not significantly associated with changes in brain structures, including cortical and subcortical brain structure. Similar results were observed for different (1) MR estimates, (2) COVID-19 GWAS summary statistics, and (3) definitions of COVID-19 infection and severity. This study suggests that the genetic susceptibility of COVID-19 is not causally associated with changes in cortical and subcortical brain structure.
Keywords: COVID-19, Mendelian randomization, brain structure, cortical area, subcortical area
Introduction
COVID-19 has long-lasting adverse effects on multiple organ systems, including brain [1]–[3]. A recent cohort study of MRI (magnetic resonance imaging) suggests that SARS-CoV-2 infection was associated with changes in brain structure [3]. However, this study has limitations inherent in observational epidemiological studies including potential reverse causation and confounding bias [4]. Tian et al. [5] reported that the cortical thickness was recovered in COVID-19 cases after a 10-month recovery period, suggesting that the associations between COVID-19 and brain structures are time-dependent. In this study, we employed 2-sample MR (Mendelian randomization) studies to investigate whether genetic susceptibility of COVID-19 is causally associated with changes in cortical and subcortical areas of the brain, including global and 34 regional cortical thickness and surface area as well as 7 combined hemispheric and 17 left/right subcortical volume.
Methods
This non-overlapping 2-sample MR study was based on GWAS (genome-wide association study) summary statistics of COVID-19 infection and severity (COVID-19 infection, N=1,348,701; COVID-19 severity, N=1,557,411, available from https://www.covid19hg.org/results/r5/ ) and GWAS summary statistics of brain structure (Global and regional cortical measures [6], N=33,709, available from http://enigma.ini.usc.edu/research/download-enigma-gwas-results; combined hemispheric subcortical volumes [7], N=38,851, available from http://enigma.ini.usc.edu/research/download-enigma-gwas-results; left/right subcortical volumes [8], N=19,629, available from https://github.com/BIG-S2/GWAS ). The GWAS summary-level data for COVID-19 were obtained from COVID-19 Host Genetics Initiative meta-analyses round 5 (release date: January 18, 2021) [9], which excludes participants from UK Biobank and ensures no overlapping samples between COVID-19 and brain structure GWAS studies. Moreover, the release date (January 18, 2021) of the round 5 dataset aligns with the time frame of Douaud’s study [3] (before August 19, 2021). Therefore, utilizing the round 5 dataset, rather than the latest round dataset, could minimize the influence of strain variations, potentially resulting in different results. COVID-19 infection was based on testing positive for COVID-19 versus population controls and COVID-19 severity was based on hospitalization of patients with COVID-19 versus population controls [10], [11]. The original GWAS investigations of brain structures were conducted with participants in Enhancing NeuroImaging Genetics through Meta Analysis (ENIGMA) and UK Biobank. The outcome measures included global and 34 regional cortical measures (thickness and surface area) [6] as well as 7 combined hemispheric [7] and 17 left/right subcortical volumes [8] (amygdala, hippocampus, accumbens, putamen, pallidum, thalamus, insula, caudate, and brain stem (combined volumes)). Included GWAS studies have obtained ethical approval from the corresponding ethics review boards (Supplementary Table 1).
Instrumental SNPs (single nucleotide polymorphisms) were extracted from GWAS summary statistics of COVID-19 at P<5*10−6, ensuring the enough SNPs for estimates. All SNPs were clumped at a window size of 10,000kb and linkage disequilibrium (LD) r2=0.001. We searched appropriate proxies for those SNPs that are not available in the brain structure (r2≥0.8). We harmonized COVID-19 and brain structure data and removed palindromic SNPs with minor allele frequency above 0.42 (Supplementary Tables 2–6). The Steiger filtering method [12] was implemented to discard SNPs exhibiting reverse causation. The MR-PRESSO method [13] was performed to remove outlier SNPs that may cause horizontal pleiotropy. We conducted inverse-variance weighted (IVW) [14] for the MR main estimate, and MR-Egger [15] and weighted median [16] are used as complementary methods to assess the causality for sensitivity analyses. The beta coefficient and 95% confidence intervals were used to describe the strength of the effect of COVID-19 susceptibility to the changes in brain structures. We performed extensive sensitivity analyses. First, we employed Cochran’s Q statistic and MR-Egger intercept to evaluate heterogeneity among SNPs and pleiotropy, respectively. Second, we conducted the leave-one-out analysis to evaluate single SNP effects.
We validated the findings using summary statistics from AncestryDNA (available from https://rgc-covid19.regeneron.com ) [17] with different definitions of COVID infection and severity [10], [11], which included COVID-19 infection, including 1) Covid vs. population; 2) Covid vs. negative; 3) Covid not hospitalized vs. controls, and COVID-19 severity, including 1) hospitalized Covid vs. controls; 2) severity covid vs. controls; 3) hospitalized covid vs. not hospitalized covid.
Results
We found no significant causal effects of COVID-19 on global cortical thickness (COVID-19 infection: beta 0.003, 95% CI −0.006 to 0.012; COVID-19 severity: beta 0.001, 95% CI −0.001 to 0.004) or on surface area (COVID-19 infection: beta 628.9, 95% CI −731.9 to 1989.7; COVID-19 severity: beta −71.8 95% CI −442.4 to 298.9) (Table 1). Consistently, our study couldn’t find causal associations of COVID-19 infection or severity with changes in 34 regional cortical thickness and surface area (Figure 1). The findings remained null on the combined hemispheric subcortical volumes with genetic susceptibility of COVID-19 infection and severity. Similar null results were obtained for left/right subcortical volumes. Sensitivity analyses suggested that these null results were not due to horizontal pleiotropy (Egger intercept P value > .05) and heterogeneity (Cochran Q P > .05). The null finding was not sensitive to specific SNP selections (leave-one-out analysis, data not shown). These findings were validated in independent GWAS studies of COVID-19 that used different definitions for COVID-19 infection and severity (Figure 2).
Table.
Results of MR analyses of COVID-19 on global cortical and combined hemispheric subcortical structures
| Outcome | MR method | COVID-19 infection (#SNPs: 16) | COVID-19 severity (#SNPs:30) | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| MR P | Beta (95% CI) | Q P | Egger P | MR P | Beta (95% CI) | Q P | Egger P | ||
|
| |||||||||
| Global cortical brain structure | |||||||||
|
| |||||||||
| IVW | 0.46 | 0.003(−0.006,0.012) | 0.97 | 0.26 | 0.001(−0.001,0.004) | 0.99 | |||
| thickness | Weighted median | 0.19 | 0.008(−0.004,0.019) | 0.68 | 0.001(−0.003,0.004) | ||||
| MR Egger | 0.7 | −0.009(−0.052,0.034) | 0.96 | 0.59 | 0.43 | 0.002(−0.003,0.008) | 0.99 | 0.73 | |
|
|
|||||||||
| IVW | 0.37 | 628.9(−731.9,1989.7) | 1 | 0.7 | −71.8(−442.4,298.9) | 0.86 | |||
| Surface | Weighted median | 0.42 | 672.5(−974.2,2319.3) | 0.86 | −47.8(−582.3,486.8) | ||||
| area | MR Egger | 0.75 | −994.0(−6878.8,4890.7) | 1 | 0.6 | 0.21 | −555.1(−1392.0,281.9) | 0.9 | 0.22 |
|
| |||||||||
| Combined hemispheric subcortical structures | |||||||||
|
| |||||||||
| IVW | 0.91 | 0.005(−0.077,0.086) | 0.99 | 0.48 | 0.01(−0.018,0.039) | 1 | |||
| accumbens | Weighted median | 0.93 | 0.005(−0.098,0.107) | 0.61 | 0.01(−0.028,0.048) | ||||
| MR Egger | 0.8 | −0.027(−0.227,0.172) | 0.99 | 0.74 | 0.95 | 0.002(−0.059,0.063) | 0.99 | 0.77 | |
|
|
|||||||||
| IVW | 0.61 | 0.021(−0.061,0.103) | 0.96 | 0.58 | 0.007(−0.018,0.033) | 0.95 | |||
| amygdala | Weighted median | 0.62 | 0.027(−0.079,0.132) | 0.17 | 0.024(−0.011,0.06) | ||||
| MR Egger | 0.56 | 0.067(−0.148,0.282) | 0.94 | 0.67 | 0.82 | 0.007(−0.051,0.065) | 0.93 | 0.99 | |
|
|
|||||||||
| IVW | 0.87 | −0.007(−0.094,0.08) | 0.98 | 0.74 | 0.006(−0.027,0.038) | 0.97 | |||
| brainstem | Weighted median | 0.51 | −0.038(−0.15,0.075) | 0.74 | −0.007(−0.05,0.035) | ||||
| MR Egger | 0.79 | −0.033(−0.265,0.198) | 0.97 | 0.82 | 0.48 | −0.026(−0.094,0.043) | 0.98 | 0.33 | |
|
|
|||||||||
| IVW | 0.73 | 0.015(−0.07,0.1) | 0.99 | 0.86 | −0.002(−0.027,0.023) | 0.9 | |||
| caudate | Weighted median | 0.74 | 0.018(−0.086,0.121) | 0.9 | 0.002(−0.033,0.038) | ||||
| MR Esser | 0.93 | −0.01(−0.225,0.205) | 0.97 | 0.81 | 0.92 | 0.003(−0.056,0.062) | 0.86 | 0.85 | |
|
|
|||||||||
| IVW | 0.27 | −0.045(−0.125,0.035) | 0.98 | 0.64 | −0.006(−0.032,0.02) | 1 | |||
| pallidum | Weighted median | 0.18 | −0.069(−0.169,0.031) | 0.33 | −0.017(−0.053,0.018) | ||||
| MR Egger | 0.23 | −0.138(−0.346,0.07) | 1 | 0.37 | 0.33 | −0.029(−0.086,0.028) | 1 | 0.39 | |
|
|
|||||||||
| IVW | 0.49 | 0.027(−0.051,0.105) | 0.98 | 0.9 | 0.002(−0.025,0.029) | 0.97 | |||
| putamen | Weighted median | 0.62 | 0.024(−0.073,0.122) | 0.68 | 0.008(−0.029,0.044) | ||||
| MR Egger | 0.66 | 0.052(−0.165,0.268) | 0.96 | 0.82 | 0.5 | 0.022(−0.039,0.083) | 0.96 | 0.48 | |
|
|
|||||||||
| IVW | 0.76 | 0.011(−0.062,0.085) | 0.99 | 0.71 | 0.005(−0.021,0.031) | 1 | |||
| thalamus | Weighted median | 0.51 | 0.031(−0.061,0.123) | 0.57 | 0.01(−0.024,0.043) | ||||
| MR Egger | 0.72 | 0.037(−0.16,0.235) | 0.98 | 0.79 | 0.79 | 0.008(−0.052,0.068) | 0.99 | 0.9 | |
Figure 1.
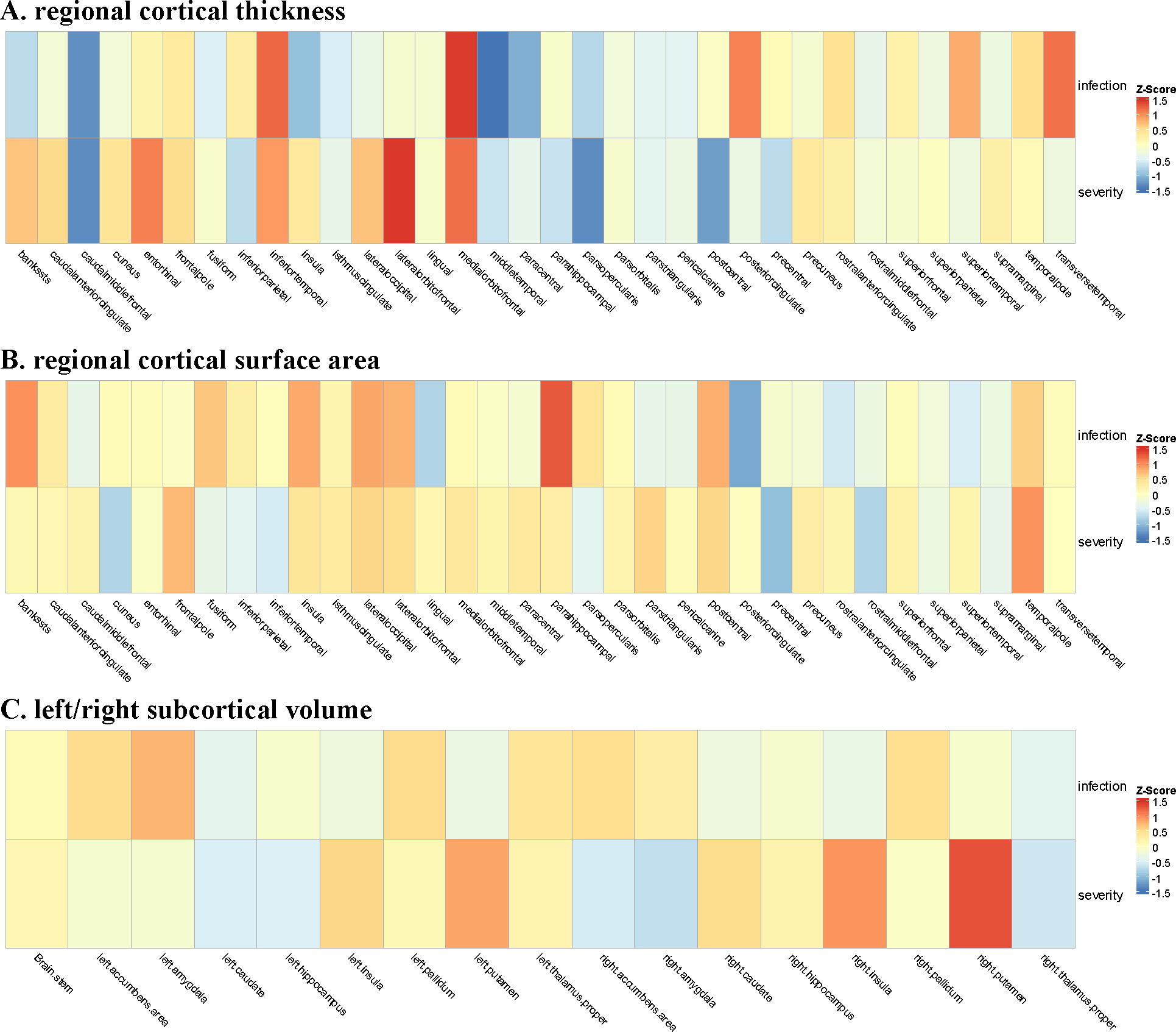
Inverse variance weighted effect estimates (Z Score) for the association between COVID-19 and regional cortical thickness and surface area as well as left/right subcortical volume
Figure 2.
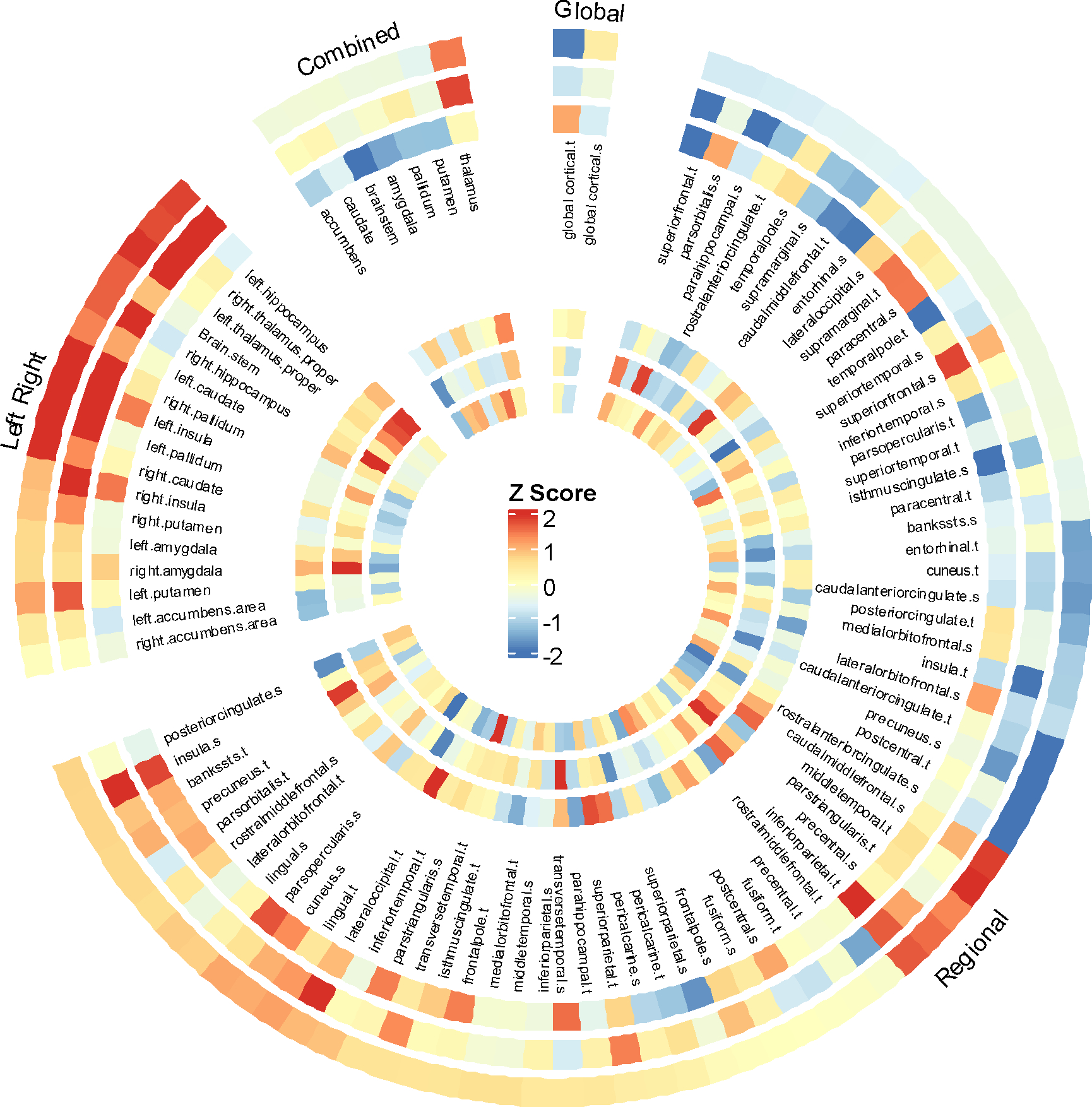
IVW effect estimates (Z Score) using different GWAS summary statistics (COVID-19 GWAS study of AncestryDNA) based on different definitions for COVID-19 infection and severity. The outer ring to the inner ring: 1) Covid vs. population; 2) Covid not hospitalized vs. controls; 3) Covid vs. negative, 4) hospitalized covid vs. not hospitalized covid, 5) hospitalized Covid vs. controls, 6) severity covid vs. controls.
Discussion
Our non-overlapping 2-sample MR analysis suggests that genetic susceptibility to COVID-19 including both infection and severity, is not causally associated with brain structure changes. Several previous MR studies investigated potential causal associations between COVID-19 and neuropsychiatric phenotypes, [18]–[21]. Recently, a 2-sample MR study [22] suggested that there are nominally significant associations between the genetically predicted COVID-19 phenotype and changes in cortical structures based on COVID-19 HGI GWAS round 7 (release date April 8, 2022) [10]. Our study was based on COVID-19 HGI GWAS round 5 (release date January 18, 2021), when the COVID-19 infection was mainly caused by the original viral variant of SARS-CoV2 and there were no wide use of COVID-19 vaccines (first approval in December 2020) and antivirals (first approval December 2021), therefore our findings were less confounded by viral strains and preventive and therapeutic factors. Since both COVID-19 GWAS study (including release round 7) and the cortex GWAS study included the UK Biobank cohort, there were significant overlaps in exposure and outcome samples, which may lead to overfitting [19]. In our study, we conducted a non-overlapping 2-sample MR analysis in order to mitigate overfitting. Recent observational cohort studies have shown an association between COVID-19 and subsequent brain structure changes [3], [5]. These observed associations are dependent on time elapsed from initial COVID-19 infection and on the SARS-CoV-2 virus strains [4]. On the other hand, our study showed no causal relationships between genetic susceptibility to COVID-19 infection and its severity and genetic susceptibility of brain structure changes. Taken together, our MR study and existing cohort studies collectively indicate that the observed brain structures changes following COVID-19 infections might have been mediated by modifiable factors other than genetic susceptibility, which are potentially reversible by targeting those modifiable mediators. Long COVID including neuropsychiatric sequelae of COVID-19 is prevalent and of a global concern [23], future works are necessary to identify and target modifiable mediators of COVID-19 infections and brain structure changes.
Our study has limitations. First, we did not examine the causal effects of genetic susceptibility to COVID-19 on brain structure alterations in specific populations stratified by age, gender, race/ethnicity, virus variants, or comorbidities, due to the lack of such information in the summary statistics data. Second, the GWAS summary statistics from COVID-19 HGI and AncestryDNA were derived from individuals of European ancestry. Further research is warranted to examine the causal associations of COVID-19 with brain structure changes in ethnic and racial minorities in the US who were disproportionately affected by the pandemic [24]. Third, potential selection bias may introduce spurious effect sizes between COVID-19 and brain structure alteration. Finally, considering olfaction and cognitive disorders have improved over time and the cortical thickness was recovered after a 10-month recovery period among COVID-19 patients [5], it is needed to assess the effect of the genetically predicted COVID-19 on brain structure changes at different time points. However, our study did not directly investigate whether the alteration of brain structure in patients with COVID-19 depends on the time period, as we don’t have summary statistics of brain structure at a single time point from COVID-19 infection. Finally, our study was based the COVID-19 Host Genetics Initiative meta-analyses round 5 (release date: January 18, 2021). While this dataset provided a unique opportunity to assess the causal associations of COIVD-19 infection and brain structures with limited confounding effects introduced by a wide variety of SARS-CoV-2 variants, COVID-19 vaccines, and anti-viral treatments, future works are needed to assess the causal associations of genetic susceptibility to COVID-19 infections and severity caused by different viral strains with brain structure changes while controlling for covid-19 vaccines and anti-viral usage.
Supplementary Material
Highlights.
A non-overlapping 2-sample Mendelian randomization study was conducted to investigate relationships between COVID-19 and changes in brain structures.
The genetically predicted COVID-19 is not associated with changes in cortical thickness and surface area.
The genetically predicted COVID-19 is not associated with changes in subcortical volume.
Future studies will need to investigate various COVID-19 variants to determine if any risks are present.
Acknowledgment
The authors thank the participants from all cohorts who contributed to this study. This work has been supported by the NIH National Institute of Aging (grants nos. AG057557, AG061388, AG062272, AG076649).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
Ethics approval statement
This study relied on multiple publicly available GWAS summary statistics on COVID-19 and brain structure. Ethical approval was obtained in all original studies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability of statement
The summary statistics in this study can be obtained from the original genome-wide association studies. Any other data generated during the analysis can be requested from the authors.
Reference
- [1].Helms J et al. , “Neurologic features in severe SARS-CoV-2 infection,” New England Journal of Medicine, vol. 382, no. 23, pp. 2268–2270, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wang L, Davis PB, Volkow ND, Berger NA, Kaelber DC, and Xu R, “Association of COVID-19 with new-onset Alzheimer’s disease,” Journal of Alzheimer’s Disease, no. Preprint, pp. 1–4, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Douaud G et al. , “SARS-CoV-2 is associated with changes in brain structure in UK Biobank,” Nature, vol. 604, no. 7907, pp. 697–707, Apr. 2022, doi: 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kremer S and Jäger HR, “Brain changes after COVID-19—how concerned should we be?,” Nat Rev Neurol, vol. 18, no. 6, pp. 321–322, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tian T et al. , “Long-term follow-up of dynamic brain changes in patients recovered from COVID-19 without neurological manifestations,” JCI Insight, vol. 7, no. 4, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Grasby KL et al. , “The genetic architecture of the human cerebral cortex,” Science (1979), vol. 367, no. 6484, p. eaay6690, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Satizabal CL et al. , “Genetic architecture of subcortical brain structures in 38,851 individuals,” Nat Genet, vol. 51, no. 11, pp. 1624–1636, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao B et al. , “Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits,” Nat Genet, vol. 51, no. 11, pp. 1637–1644, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kousathanas A et al. , “Whole-genome sequencing reveals host factors underlying critical COVID-19,” Nature, vol. 607, no. 7917, pp. 97–103, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].C.−19 H. G. I. aganna@ broadinstitute. org, “The COVID-19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic,” European Journal of Human Genetics, vol. 28, no. 6, pp. 715–718, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Leong A, Cole JB, Brenner LN, Meigs JB, Florez JC, and Mercader JM, “Cardiometabolic risk factors for COVID-19 susceptibility and severity: a Mendelian randomization analysis,” PLoS Med, vol. 18, no. 3, p. e1003553, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hemani G, Tilling K, and Davey Smith G, “Orienting the causal relationship between imprecisely measured traits using GWAS summary data,” PLoS Genet, vol. 13, no. 11, p. e1007081, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Verbanck M, Chen C-Y, Neale B, and Do R, “Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases,” Nat Genet, vol. 50, no. 5, pp. 693–698, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bowden J, Del Greco M F, Minelli C, Davey Smith G, Sheehan N, and Thompson J, “A framework for the investigation of pleiotropy in two‐sample summary data Mendelian randomization,” Stat Med, vol. 36, no. 11, pp. 1783–1802, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bowden J, Davey Smith G, and Burgess S, “Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression,” Int J Epidemiol, vol. 44, no. 2, pp. 512–525, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bowden J, Davey Smith G, Haycock PC, and Burgess S, “Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator,” Genet Epidemiol, vol. 40, no. 4, pp. 304–314, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Roberts GHL et al. , “Expanded COVID-19 phenotype definitions reveal distinct patterns of genetic association and protective effects,” Nat Genet, vol. 54, no. 4, pp. 374–381, 2022. [DOI] [PubMed] [Google Scholar]
- [18].Cao H, Baranova A, Song Y, Chen J-H, and Zhang F, “Causal associations and genetic overlap between COVID-19 and intelligence,” QJM: An International Journal of Medicine, p. hcad122, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Baranova A, Cao H, and Zhang F, “Causal effect of COVID‐19 on Alzheimer’s disease: a Mendelian randomization study,” J Med Virol, vol. 95, no. 1, p. e28107, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Baranova A, Cao H, Teng S, Su K, and Zhang F, “Shared genetics and causal associations between COVID‐19 and multiple sclerosis,” J Med Virol, vol. 95, no. 1, p. e28431, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ding P, Gurney M, Perry G, and Rong Xu, “Association of COVID-19 with risk and progression of Alzheimer’s disease: non-overlapping two-sample Mendelian randomization analysis of 2.6 million subjects,” Journal of Alzheimer’s Disease, vol. in press, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhou S et al. , “Causal effects of COVID-19 on structural changes in specific brain regions: a Mendelian randomization study,” BMC Med, vol. 21, no. 1, p. 261, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davis HE, McCorkell L, Vogel JM, and Topol EJ, “Long COVID: major findings, mechanisms and recommendations,” Nat Rev Microbiol, vol. 21, no. 3, pp. 133–146, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Aburto JM, Tilstra AM, Floridi G, and Dowd JB, “Significant impacts of the COVID-19 pandemic on race/ethnic differences in US mortality,” Proceedings of the National Academy of Sciences, vol. 119, no. 35, p. e2205813119, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The summary statistics in this study can be obtained from the original genome-wide association studies. Any other data generated during the analysis can be requested from the authors.


