Abstract
The majority of excitatory synapses in the mammalian brain form on filopodia and spines, actin-rich membrane protrusions present on neuronal dendrites. The biochemical events that induce filopodia and remodel these structures into dendritic spines remain poorly understood. Here, we show that the neuronal actin- and protein phosphatase-1–binding protein, neurabin-I, promotes filopodia in neurons and nonneuronal cells. Neurabin-I actin–binding domain bundled F-actin, promoted filopodia, and delayed the maturation of dendritic spines in cultured hippocampal neurons. In contrast, dimerization of neurabin-I via C-terminal coiled-coil domains and association of protein phosphatase-1 (PP1) with neurabin-I through a canonical KIXF motif inhibited filopodia. Furthermore, the expression of a neurabin-I polypeptide unable to bind PP1 delayed the maturation of neuronal filopodia into spines, reduced the synaptic targeting of AMPA-type glutamate (GluR1) receptors, and decreased AMPA receptor-mediated synaptic transmission. Reduction of endogenous neurabin levels by interference RNA (RNAi)-mediated knockdown also inhibited the surface expression of GluR1 receptors. Together, our studies suggested that disrupting the functions of a cytoskeletal neurabin/PP1 complex enhanced filopodia and impaired surface GluR1 expression in hippocampal neurons, thereby hindering the morphological and functional maturation of dendritic spines.
INTRODUCTION
The majority of excitatory synapses in the mammalian brain exist on specialized membrane protrusions known as dendritic spines (Harris, 1999). Spines house the machinery for neurotransmitter signaling and compartmentalize the biochemical and cell biological events that elicit the synaptic modifications associated with learning and memory (Hering and Sheng, 2001). During brain development, dendrites extend filopodia that contact complementary presynaptic sites and differentiate into mature spines capable of synaptic transmission (Ziv and Smith, 1996; Fiala et al., 1998; Ziv and Garner, 2001; Yuste and Bonhoeffer, 2004). Although central to the development of synapses and neural circuits, the molecular mechanisms that regulate the extension and remodeling of dendritic filopodia into functional postsynaptic spines remain poorly understood.
In the adult brain, filopodia and spines continue to undergo morphological alterations that have been linked with learning and memory (Yuste and Bonhoeffer, 2001; Lamprecht and LeDoux, 2004). For example, long-term potentiation (LTP) elicits dramatic changes in spine shape and number (Lang et al., 2004; Matsuzaki et al., 2004), and, during LTP, filopodia are frequently seen as precursors of nascent spines (Maletic-Savatic et al., 1999). Several human mental retardation syndromes have been linked with altered morphology and number of dendritic spines (Kaufmann and Moser, 2000; Irwin et al., 2001; Fiala et al., 2002). These are compelling reasons for investigating the molecular basis of filopodia and spine formation and the events that generate mature functional synapses. It is widely anticipated that such studies will provide critical insights into the events that dictate brain development, learning and memory, and neurological disease.
Filopodia and spines are rich in F-actin and remodeling of the actin cytoskeleton controls the formation and motility of filopodia as well as the maturation of dendritic spines (Fischer et al., 1998; Matus, 2000; Colicos et al., 2001; Star et al., 2002). Actin reorganization in dendritic spines is highly dynamic and responsive to synaptic signals (Halpain et al., 1998; Matus, 2000; Colicos et al., 2001; Star et al., 2002). Indeed, actin remodeling in dendritic spines is essential for the maintenance of LTP (Kim and Lisman, 1999; Krucker et al., 2000; Fukazawa et al., 2003) and several actin-associated proteins have been shown to alter spine structure (Hering and Sheng, 2001; Penzes et al., 2001; Ehlers, 2002). Our studies focused on the structurally related actin-binding proteins, neurabin-I and spinophilin/neurabin-II, that are highly concentrated in dendritic spines and specifically in the actin-rich postsynaptic density (Allen et al., 1997; Terry-Lorenzo et al., 2000). Recent studies have implicated both neurabin isoforms in the regulation of neuronal morphology (Feng et al., 2000; Oliver et al., 2002; Zito et al., 2004).
Neurabin-I and spinophilin share multiple protein interaction domains (Nakanishi et al., 1997; Satoh et al., 1998), including an N-terminal actin-binding domain, a PDZ (PSD-95, Dlg-large, and ZO-1 homology) domain, a protein phosphatase-1 (PP1)-binding motif (Allen et al., 1997; McAvoy et al., 1999), and C-terminal coiled-coil domains. Our previous studies suggested that multiple domains in neurabin-I (NrbI), in particular the actin-binding domain, are required to elicit morphological changes in cultured cells (Oliver et al., 2002). The N-terminal actin binding domain, NrbI(1–287), also stimulated spine actin dynamics, altered the motility and morphology of dendritic spines, and increased synapse formation in cultured rat hippocampal slices (Zito et al., 2004). By comparison, no changes in cell morphology were reported when similar N-terminal fragments of spinophilin/Neurabin-II (Spino) were expressed in either cultured cells or neurons (Grossman et al., 2002; Barnes et al., 2004). The study by Zito et al. (2004) focused solely on spine alteration mediated by the neurabin-I actin–binding domain. However, beyond their ability to bind actin, neurabin-I and spinophilin form multimers via their C-terminal domains and act as a scaffold for the prominent signaling enzyme PP1 (Allen et al., 1997; McAvoy et al., 1999; Oliver et al., 2002). Yet, little knowledge has existed regarding the role of PP1 binding or dimerization of neurabin I in postsynaptic assembly or function.
The present studies compared the ability of NrbI and Spino to induce filopodia in cultured cells and established the functional and structural similarities in the two neurabin isoforms. Subsequent experiments focused on the role of NrbI and its ability to target PP1 to the actin cytoskeleton in controlling the formation of neuronal filopodia and dendritic spines and the maturation of functional synapses. We expressed wild-type and mutant neurabins and utilized shRNA to knockdown endogenous NrbI to demonstrate that NrbI promotes spine formation, increases the incorporation of AMPA-type glutamate receptors at the postsynaptic membrane, and enhances excitatory synaptic transmission. Our studies, for the first time, highlight a role of the neuronal NrbI/PP1 complex in development of dendritic spines and the formation of glutamatergic synapses in mammalian brain.
MATERIALS AND METHODS
Plasmids
Plasmids encoding the neurabin-I (NrbI) polypeptides, GFP-NrbI-FL, GFP-NrbI(1–552), GFP-NrbI-(286–1095), and GFP-NrbI-FL(F460A) were previously described (Oliver et al., 2002; Terry-Lorenzo et al., 2002b). GFP-α-actinin was a kind gift from Carol Otey (Edlund et al., 2001). The spinophilin (Spino) polypeptides, GFP-Spino-FL and GFP-Spino(178–817), were gifts from Sharon Milgram (University of North Carolina). The plasmid encoding GFP-NrbI(1–287) was provided by Karen Zito, Cold Spring Harbor Laboratories (Zito et al., 2004). QuickChange mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce stop codons in GFP-NrbI-FL and GFP-Spino-FL to generate C-terminal truncations. Site-directed mutagenesis introduced the F460A substitution in GFP-NrbI-FL and F451A in GFP-Spino-FL. GFP-Spino(151–289) was created by digesting the GFP-Spino(1–289) expression plasmid with XmnI and ScaI and religating the blunt ends.
To create gyraseB fusions of spinophilin, a BsiWI site was introduced in GFP-Spino-FL cDNA after codon 586 using Quick Change mutagenesis to create Bsi-Spino. The gyraseB sequence was amplified by PCR using GGTGGCGTACGATCTAGAAGCAATTCT and GAGGGTCGTACGTTATGTGTACATGTCGACTTTGGATTCGG with pKS-ATG-GyrB (Farrar et al., 2000) as template. Amplified cDNA was digested with BsiWI and inserted into BsiSpino to produce GFP-Spino-GyrB.
To create His-tagged fusions, GFP-NrbI-FL was digested with BglII and KpnI to excise NrbI amino acids 1–287, and this fragment was ligated intoBglII/KpnI-digested pRSET-B (Invitrogen, Carlsbad, CA) to yield His-tagged NrbI(1–287). GFP-NrbI-FL was digested with BglII and PvuII to excise amino acids 1–146, and this fragment was ligated into BglII/PvuII-digested pRSET-B to yield His-NrbI(1–146). To make GFP- and His-tagged NrbI(146–287), a BglII site was introduced into GFP- and His-NrbI(1–287) at the position encoding amino acid 147 by PCR with CACAGAGACTCGAAAGATCTTTGAGAGAAGTGGG and its inverse complement. The cDNA encoding NrbI amino acids 1–146 was then excised using a BglII/KpnI digest. The resulting fragment was ligated into BglII/KpnI-digested pEGFP-C2 (Clontech, Palo Alto, CA) to express GFP-NrbI(1–146) and pRSET-A (Invitrogen) to generate His-NrbI(1–146).
The cDNA encoding mRFP in pRSET-B (Campbell et al., 2002) was digested with BamHI and HindIII and insert into pCMV4. The resulting plasmid was digested with BstYI and XbaI and inserted into pFlag-CMV-2 (Eastman Kodak, Rochester, NY) digested with BglII and XbaI to generate a plasmid encoding FLAG-mRFP.
pZOFF-EGFP was engineered to coexpress shRNAs along with EGFP in order to identify transfected cells and to analyze the cellular effects of reducing endogenous protein levels by RNA interference (see Figure 6A). pZOFF-EGFP was constructed by inserting the H1 expression cassette from pSUPER (Brummelkamp et al., 2002) into pEGFP-C1 (Clontech). pEGFP-C1 was prepared by digestion with BglII and BamHI to remove part of the multiple cloning site region; subsequently, he H1 cassette was ligated between the MluI and DraIII sites of pEGFP-C1. Oligonucleotides for generation of shRNAs were cloned into the BglII-HindIII sites in the H1 expression cassette.
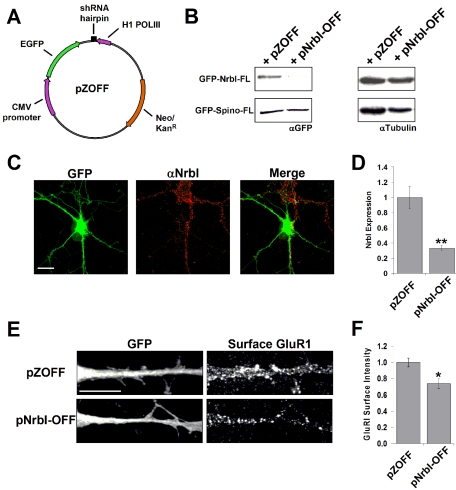
Figure 6.
Endogenous neurabin-I regulates AMPAR surface expression. (A) pNrbI-OFF was constructed by insertion of the NrbI shRNA hairpin loop in the pZOFF vector. In this vector shRNA expression is driven by an H1 PolIII promoter and GFP is separately expressed under the control of a CMV promoter. (B) Left panels, GFP-NrbI-FL or GFP-Spino-FL was expressed into COS7 cells, which were transfected with pZOFF vector alone or pNrbI-OFF, and protein levels were analyzed using anti-GFP Western blots. Right panels, equal protein loading was verified by immunoblotting with an anti-tubulin antibody. (C) On DIV15, hippocampal neurons were transfected with pNrbI-OFF, and on DIV26, cells were immunostained with anti-NrbI antibody and imaged for both GFP and NrbI staining. Scale bar, 10 μm. (D) Quantification of NrbI expression in cultured hippocampal neurons transfected on DIV14 and immunostained on DIV19. Data are presented as a ratio of the anti-NrbI fluorescence intensity in GFP-positive cells relative to pZOFF containing control cells ± SEM (n >10 for each condition; **p < 0.01 using the Student's t test). (E) Hippocampal neurons were transfected on DIV14 with pZOFF and pNrbI-OFF. On DIV19, surface GluR1 AMPA receptor (AMPAR) subunits were labeled on live cells as described in Materials and Methods. Cells were imaged for GFP and GluR1 staining. (F) Quantification of surface GluR1 levels is shown as a fraction of the surface GluR1 fluorescence intensity in GFP-positive cells relative to pZOFF containing control cells ± SEM (n >30 for each condition; *p < 0.05 using the Student's t test). Scale bar, 10 μm.
pNrbI-OFF was created by ligating the annealed oligos, GATCCCCGATGTCTCTCGAGAAGGCTTTCAAGAGAAGCCTTCTCGAGAGACATCTTTTTGGAAA and AGCTTTTCCAAAAAGATGTCTCTCGAGAAGGCTTCTCTTGAAAGCCTTCTCGAGAGACATCGGG into HindIII- and BglII-digested pZOFF-EGFP. The resulting shRNA targeted rat NrbI base pairs 1291–1309 (amino acids 430–437). Efficacy and selectivity of pNrbI-OFF was analyzed by cotransfecting either GFP-NrbI-FL or GFP-Spino-FL with pZOFF or pNrbI-OFF, and monitoring protein expression by western analysis with mouse anti-GFP antibody (Chemicon, Temecula, CA). Other mRNA sequences, such as those encoding amino acids 65–72, 708–715, and 1049–1056, were also evaluated but none proved to be more efficacious.
Culture, Transfection, and Imaging of Cells
COS7 cells were maintained in DMEM plus 10% (vol/vol) fetal bovine serum. To express GFP-NrbI polypeptides, cells at 50% confluency were transfected with DNA (1.5 μg) and Lipofectamine (6 μL, Invitrogen). For forced dimerization experiments, cells were treated with 700 nM coumermycin (Sigma, St. Louis, MO) or novobiocin (Sigma) immediately after transfection. After overnight incubation, all cells were fixed with 4% (vol/vol) paraformaldehyde. Some cells were permeabilized with 0.1% (vol/vol) Triton X-100, blocked with 2% bovine serum albumin for 1 h, and incubated with 1:200 dilution of rhodamine-phalloidin (Molecular Probes, Eugene, OR) for 5 min. Coverslips were mounted in phosphate-buffered saline (PBS)/glycerol (1:1) with 25 mg/ml DABCO, sealed, and visualized with 40× and 60× objectives on an Olympus spinning disk inverted confocal microscope (Lake Success, NY) using Ultra View acquisition software (PerkinElmer LAS, Boston, MA).
Filopodia Counting in COS7 Cells
We utilized a double-blind approach to quantify filopodia, with 0–5 filopodia per cell scoring 0, 5–20 filopodia scoring 1, and greater than 20 filopodia scoring 2. At least eight independent counts of 50 cells each on minimum of 4 slides were undertaken to calculate the average score, which was normalized to the control GFP-expressing cells. Counts were performed using both mRFP (fill) and GFP fluorescence and yielded identical results.
Culture, Transfection, and Imaging of Hippocampal Neurons
Dissociated hippocampal cultures (Scott et al., 2001) were transfected using 1.5 μg of DNA and 1 μl of Lipofectamine 2000 (Invitrogen) in 50 μl Opti-MEM. Cells were imaged at 40, 60, or 100× magnifications using a Nikon spinning disk confocal microscope with a cooled CCD camera (Princeton Instruments, Princeton, NJ).
Immunostaining of Hippocampal Neurons
Filamentous F-Actin was stained with Alexa568-phalloidin (Molecular Probes) in cells fixed with 4% (vol/vol) paraformaldehyde containing 4% (wt/vol) sucrose as recommended in the manufacturer's protocol. A rabbit anti-GluR1 antibody (Mammen et al., 1997) was incubated with a 1:250 dilution of Alexa568-anti-rabbit secondary antibody (Molecular Probes) in neurobasal medium (Life Technologies, Rockville, MD) for 30 min, and the antibody complex was applied to live cultured hippocampal neurons for 20 min at 4°C. Cells were rinsed in PBS containing 2 mM CaCl2, fixed, and mounted for imaging (Ehlers, 2000; Scott et al., 2001). For NrbI immunostaining, 1:100 dilution of mouse anti-NrbI antibody (Transduction Laboratories, Lexington, KY) and 1:200 dilution of Alexa647-anti-mouse secondary antibody (Molecular Probes) were used. The anti-NrbI antibody recognized NrbI(1–146) and NrbI(1–107) in Western immunoblots but did not cross-react with Spino(1–152) (unpublished data).
Quantification was undertaken by tracing the GFP-containing cell using Metamorph software (Universal Imaging, West Chester, PA), transferring the trace to the parallel immuno-stained cell and determining average staining intensity within the trace after subtracting the background. All pooled images were analyzed using identical acquisition parameters and were normalized to the average value obtained from control cells from sister cultures.
Morphometric Analyses of Hippocampal Neurons
For these studies, mRFP was used as an unbiased cell-fill. Because protrusions often crossed several z planes, we took 0.5–1-μm z series stacks from the bottom to the top of all dendrites and used the Metamorph function to generate image projections for quantitative analyses. Images were not further processed and were of high quality similar to that of the original single planes. The number of planes, typically 8–12, was chosen to cover the entire dendrite from top to bottom. In each experiment, appropriate controls were used to ensure there was no bleed-through between different wavelength channels. All morphological experiments were repeated at least three times with an n of 7–12 for individual experiments.
DIV7 neurons (7 days in vitro; 30,000/well) were transfected and after 24 h, fixed in 4% (vol/vol) PFA containing 4% (wt/vol) sucrose. Images at 40× magnification were generated on a spinning disk confocal microscope. Protrusions 0.7–10 μm in length were counted in a double-blind manner and expressed per unit length of dendrite. Protrusions with a bulbous head wider than the base were scored as spines. All other protrusions were scored as nonspines. These nonspine protrusions were typically longer than mushroom spines and possessed narrow or tapered heads, much like filopodia. Between 150–300 protrusions were scored for every neuron. Measurements of length and width of the protrusions were performed as described previously (Sala et al., 2001).
Purification of Recombinant Neurabin-I Proteins
His-NrbI(1–287), His-NrbI(1–146), and His-NrbI(146–287) were expressed in Escherichia coli BL21 DE3 pLys+ in LB-ampicillin (500 ml) grown to OD600 =0.6 and induced with 1 mM IPTG for 3 h at 37°C. All recombinant proteins were purified on TALON metal affinity resin (BD Biosciences, San Diego, CA).
Actin Binding and Bundling Assays
Actin-binding and bundling were assayed using the BK001 actin-binding kit (Cytoskeleton, Denver, CO) following the manufacturer's instructions. Briefly, purified His-NrbI (10 μL) was mixed with polymerized F-actin (40 μL) and incubated at 24°C for 30 min before centrifugation at 150,000 × g (to detect F-actin-binding) or 10,000 × g (to detect F-actin bundling; Wang et al., 2001). Presence of the test protein in the supernatants and pellets was analyzed on 12% (wt/vol) SDS-PAGE followed by either Western immunoblotting or protein staining with Coomassie blue.
F-actin mixed with equimolar amounts of purified His-NrbI proteins in F-actin polymerizing buffer (Cytoskeleton) was incubated for 30 min to 1 h, and aliquots of these mixtures were negatively stained with 1% uranyl acetate, placed on copper grids, and imaged by transmission electron microscopy (Moody et al., 1985) by the Duke Comprehensive Cancer Center EM facility.
Electrophysiology of Cultured Neurons
Whole-cell voltage clamp recordings were performed on hippocampal neurons (DIV 16) grown at high density (50,000 cells/well). Neurons were held at −60 mV using a MultiClamp 700A amplifier (Axon Instruments, Foster City, CA) controlled with a Pentium PC running MultiClamp Commander and pClamp (Axon Instruments). Extracellular solution contained (in mM): 150 NaCl, 5 KCl, 10 HEPES, 1 MgCl2, 30 d-glucose, 2 CaCl2, 0.001 TTX, 0.03 bicuculline (330 mOsm/l, ph 7.4). Electrodes were pulled from glass capillary tubes (Narishige, Tokyo, Japan). Recording pipettes were filled with a solution containing (in mM): 30 CsSO4, 70 K2SO4, 25 HEPES, 25 N-methyl-d-glucamine, 0.1 CaCl2, 1 EGTA, 2 Na2ATP, 0.1 leupeptin (300 mOsm/l, ph 7.2) with resistance ranging from 3 to 5 MΩ. Recordings with series resistances greater than 10 MΩ were discarded. (Data were analyzed using MiniAnalysis software (Synaptosoft, Decatur, GA). More than 300 mEPSC events with amplitudes greater than 5 pA and rise times <5 ms were collected for each neuron.
RESULTS
Multiple Neurabin Domains Regulate Cell Morphology
To date, there has been only limited structural and functional analysis of neurabin-I (NrbI; Oliver et al., 2002) or spinophilin/neurabin-II (Spino; Barnes et al., 2004) and its role in regulating cell morphology. Thus, we analyzed the ability of NrbI and Spino polypeptides to alter the morphology of cultured cells, focusing on the structural homology between the two neuronal actin-binding proteins (Figure 1A). We examined COS7 cells expressing green fluorescent protein (GFP) fusions of NrbI and Spino polypeptides with selective deletions of known protein-interaction domains (Figure 1B). As previously noted, the full-length (FL) proteins, GFP-NrbI-FL (Oliver et al., 2002) and GFP-Spino-FL (Barnes et al., 2004), localized to the cell periphery, consistent with their association with the cortical actin cytoskeleton. Cells expressing these polypeptides showed no notable change in morphology. However, expression of shorter polypeptides containing the N-terminal actin-binding domains, specifically GFP-NrbI (1–287) and GFP-Spino (1–289), elicited numerous filopodia in COS7 cells (Figure 1B). By comparison, GFP-NrbI(286–1095), which lacks actin-binding (Nakanishi et al., 1997), failed to localize to the cell periphery and had no effect on COS7 morphology (Figure 1B). Similar results were obtained in A549, NIH3T3, HEK 293, and HeLa cells (unpublished data), indicating that the functional effect of the Nrb1 and spinophilin actin-binding domains were not cell-specific.
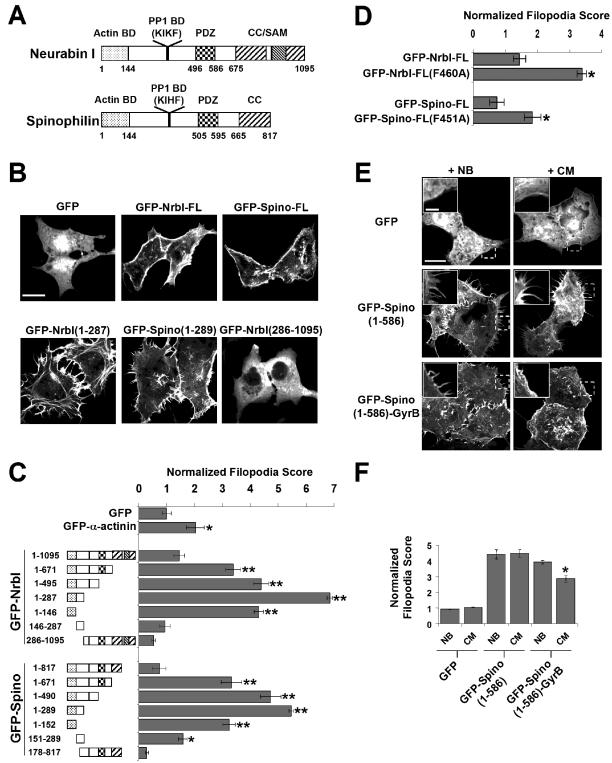
Figure 1.
Multiple protein-interaction domains in neurabin-I and spinophilin regulate the morphology of COS7 cells. (A) The schematic representation of neurabin-I (NrbI) and spinophilin/neurabin-II (Spino) highlights the actin-binding domain (Actin BD), PP1-binding motifs (PP1 BD; KIKF in NrbI and KIHF in Spino), PDZ domain, and the coiled-coil (CC) and sterile alpha motif (SAM) domains, which mediate oligomerization. The numbers indicate amino acids that flank these protein interaction domains. (B) COS7 cells were transfected with GFP-NrbI and GFP-Spino plasmids, fixed after 16 h, and imaged. Scale bar, 20 μm. (C) Filopodia induced by GFP-neurabins were quantified and normalized to control GFP-expressing cells. Results are shown ± SEM; *p < 0.05 and **p < 0.005 using Student's t test. (D) Filopodia induced by NrbI and Spino proteins unable to bind PP1 because of the F-to-A substitutions within the PP1-binding motif were compared with control GFP-expressing cells as described above. *p < 0.05 using Student's t test. (E) COS7 cells expressing GFP-Spino(1–586) or GFP-Spino(1–586)-GyrB were incubated with either novobiocin (NB) or coumermycin (CM) for 16 h before imaging. Scale bar, 20 μm. Inset represent higher magnification of regions of these cells marked by a dashed box. Scale bar, 5 μm. (F) Data from the experiment shown in E were quantified as described for C. *p < 0.05 using Student's t test.
Quantitative analysis of filopodia induction showed that, compared with α-actinin, a known actin-binding protein, GFP-NrbI and GFP-Spino polypeptides containing the N-terminal actin-binding domain induced extensive filopodia (Figure 1C). NrbI and Spino polypeptides lacking the actinbinding domain, specifically GFP-NrbI(286–1095) and GFP-Spino(178–817), failed to elicit filopodia. (Figure 1C). Furthermore, GFP-neurabins lacking C-terminal sequences were more effective than GFP-NrbI-FL and GFP-Spino-FL in inducing filopodia. These data suggest that association of cellular proteins with one or more C-terminal protein-interaction domains negatively regulates the ability of the actinbinding domain to elicit filopodia. Interestingly, GFP-NrbI(1–146) and GFP-Spino(1–152) were less effective than GFP-NrbI(1–287) and GFP-Spino(1–289) in filopodia induction, suggesting that as yet unidentified proteins proposed to bind NrbI(196–210) (Zito et al., 2004) and Spino(151–284) (Barnes et al., 2004), may stimulate the activity of the minimal actin-binding domain (Nakanishi et al., 1997; Satoh et al., 1998) in inducing filopodia.
We next examined the role of neurabin-bound PP1 by substituting a single amino acid, phenylalanine (F), with alanine (A) in the conserved KIXF PP1-binding motif in both Spino-FL and NrbI-FL. The F-to-A substitution abolishes PP1 binding to both Spino and NrbI (Hsieh-Wilson et al., 1999; Terry-Lorenzo et al., 2002b). Compared with GFP-NrbI-FL or GFP-Spino-FL, GFP-NrbI-FL-F460A, and GFP-Spino-FL-F451A elicited increased filopodia(Figure 1D). This demonstrated that the loss of PP1-binding enhanced the ability of neurabins to increase filopodia.
Because the C-terminal coiled-coil domain not only mediates NrbI and Spino dimerization (Oliver et al., 2002), but also recruits cellular proteins such as TGN38 (Stephens and Banting, 1999) and Lfc (Ryan et al., 2003), the mechanism by which this domain regulates cell morphology was unclear. To address the role of dimerization, we generated GFP-Spino(1–586)-GyrB, which contains the antibiotic-interaction domain of the bacterial enzyme gyrase B (GyrB) fused to the C-terminus of GFP-Spino(1–586), a truncation that lacks a coiled-coil domain. This fusion protein displays drug-induced dimerization in the presence of the bivalent antibiotic, coumermycin (CM), which binds two GyrB domains simultaneously (Farrar et al., 2000). Novobiocin (NB), a monovalent analogue of CM that binds a single GyrB domain, fails to induce dimerization and serves as a control. As expected, treatment of cells expressing GFP-Spino(1–586) with NB or CM had no effect on its ability to induce filopodia in COS7 cells (Figure 1, E and F). Moreover, cells expressing GFP-Spino(1–586)-GyrB displayed similar numbers of filopodia, indicating that the C-terminal fusion of GyrB had little effect on the ability of GFP-Spino(1–586) to induce filopodia. Treatment of GFP-Spino(1–586)-GyrB–expressing cells with CM, however, significantly inhibited filopodia in COS7 cells (Figure 1E). Quantitative analyses showed that CM-treated cells expressing GFP-Spino(1–586)-GyrB possessed 20–25% fewer filopodia than same cells treated with NB (Figure 1F). These studies support the conclusion that it is not the binding of other cellular proteins, but rather the dimerization of GFP-Spino(1–586)-GyrB, and by inference, of full-length spinophilin, that inhibits filopodial outgrowth. Taken together, these data show that the actin-binding domains of NrbI and spinophilin promote filopodia, whereas dimerization via C-terminal coiled-coil domains and association of PP1 inhibit filopodial outgrowth.
Neurabin-I Induces Filopodia on Hippocampal Neurons
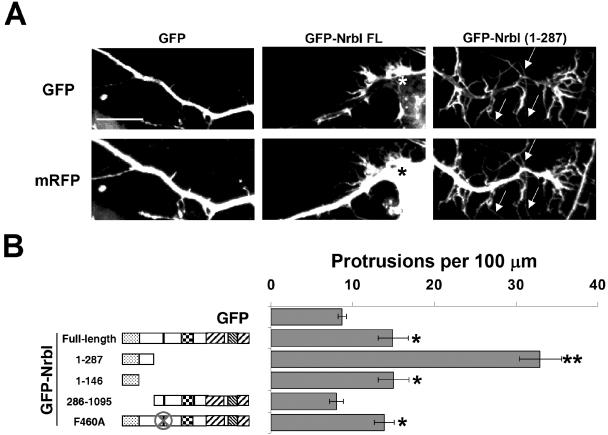
Unlike spinophilin, which is widely expressed in mammalian tissues (Allen et al., 1997; Satoh et al., 1998), NrbI is found exclusively in neurons (Nakanishi et al., 1997). To define the structural requirements in NrbI that modulate neuronal morphology, we expressed GFP-NrbI polypeptides in primary cultures of rat hippocampal neurons and examined neuronal morphology. In these experiments, we cotransfected neurons with monomeric red fluorescent protein (mRFP) to provide an unbiased cell fill. Neurons transfected with GFP on day 7 in vitro (DIV7) and imaged on DIV8 (Figure 2A) showed few dendritic protrusions and no obvious spines, as is typical for cultured neurons of this age (Takahashi et al., 2003). In contrast to COS7 cells, which were largely unaffected by the expression of full-length neurabin-I, GFP-NrbI-FL promoted a significant increase (∼50%) in filopodia in hippocampal neurons (Figure 2, A and B). These filopodia frequently appeared as “bursts” with five or more projections emanating from a single site on the dendrite (asterisk in Figure 2A) that consistently displayed high concentrations of GFP-NrbI-FL. By comparison, neurons expressing GFP-NrbI(1–287) exhibited prolific outgrowth of highly elongated filopodia along the cell body (unpublished data) and dendrites (Figure 2, A and B). As seen in COS7 cells, the N-terminal actin-binding domain, GFP-NrbI(1–146), was necessary and sufficient for NrbI-induced filopodia formation in neurons (Figure 2B). However, unlike in COS7 cells, the disruption of PP1-binding to GFP-NrbI-FL(F460A) resulted in no further enhancement of GFP-NrbI-FL's ability to promote neuronal filopodia (Figure 2B).
Figure 2.
Neurabins induce dendritic filopodia in hippocampal neurons. (A) Cultured hippocampal neurons (DIV7) were cotransfected with plasmids encoding mRFP along with GFP, GFP-NrbI-FL, or GFP-NrbI(1–287). Representative dendrites of DIV8 neurons cotransfected with the GFP fusion protein (top panels) and mRFP (bottom panels) are shown. Arrows indicate filopodia. Asterisk (*) indicates a filopodial “burst.” Scale bar, 10 μm. (B) Quantification of dendritic protrusions expressed per length of dendrite is shown for experiments conducted as described in A. The data for the number of protrusions per 100 μm of dendrite are shown ± SEM; *p < 0.05, **p < 0.005 compared with GFP controls, Student's t test.
The Actin-binding Domain of Neurabin-I Reorganizes the Neuronal Actin Cytoskeleton
Recent studies showed that GFP-NrbI(1–287) modulates neuronal actin dynamics in hippocampal slices (Zito et al., 2004). However, the mechanism underlying actin rearrangement by GFP-NrbI(1–287) remains unresolved. To address this question, we expressed GFP-NrbI in cultured hippocampal neurons and imaged the actin cytoskeleton stained with Alexa568-phalloidin. Consistent with prior studies (Zhang and Benson, 2001), F-actin in the dendrites of control DIV8 neurons, expressing GFP alone, was either diffuse or appeared as patches and puncta (arrowheads in Figure 3A). Expression of GFP-NrbI(1–287) resulted in marked rearrangement of the cytoskeleton in dendrites with the appearance of linear bundles of F-actin and the increased concentration of F-actin at the dendritic plasma membrane (arrows in Figure 3A). Interestingly, expression of GFP-NrbI-FL elicited an intermediate pattern with the presence of both linear bundles and F-actin puncta (Figure 3A). Unlike the much smaller GFP-NrbI (1–287), which distributed throughout the entire length of filopodia, GFP-NrbI-FL was largely concentrated at the base of these structures, suggesting that the subcellular localization of NrbI(1–287) and NrbI-FL might account for their differing effects on neuronal morphology.
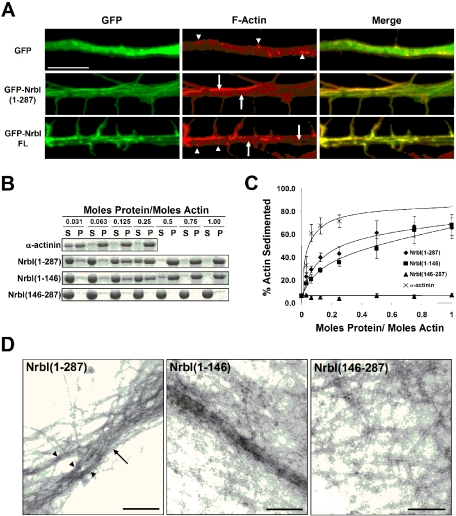
Figure 3.
The N-terminal domain of neurabin-I reorganizes the neuronal actin cytoskeleton in vivo and bundles actin in vitro. (A) Cultured hippocampal neurons (DIV8) expressing GFP-neurabins were fixed and F-actin was stained with Alexa-568-phalloidin. Representative GFP fluorescence, phalloidin staining, and merged images are shown. Arrowheads highlight F-actin puncta and arrows indicate linear bundles of actin filaments. Scale bar, 5 μm. (B) Purified F-actin was mixed with α-actinin or His-NrbI proteins at the increasing molar ratios and low-speed centrifugation separated F-actin filaments in the supernatant (S) from bundled F-actin in the pellet (P). Supernatants and pellets were subjected to SDS-PAGE, and the gels were stained with Coomassie blue. Coomassie-stained actin is shown. (C) Quantification of results in B is shown as the percent of total F-actin sedimented at various His-NrbI/actin molar ratios (±SEM; n = 4). (D) The bundles of F-actin filaments induced by His-NrbI proteins were examined by electron microscopy. Arrow points to a “loose” F-actin bundle consisting of several smaller bundles (arrowheads). Scale bars, 200 nm.
Initiation of filopodia is characterized by the enhanced bundling of filamentous actin in the growing protrusion (Svitkina et al., 2003). Previously, Nakanishi et al. (1997) demonstrated that NrbI binds the sides of actin filaments and generates F-actin bundles. They hypothesized that NrbI requires both F-actin binding by the N-terminal domain and dimerization via the C-terminal coiled-coil domains to bundle actin filaments. In contrast, our results show that GFP-NrbI(1–287), which lacks the oligomerization domains, is the most effective NrbI polypeptide in rearranging neuronal actin cytoskeleton and inducing filopodia. Thus, we evaluated the ability of NrbI(1–287) to bundle F-actin in vitro. Using low-speed centrifugation to sediment bundled F-actin, we established that α-actinin, a known actin-bundling protein, bundled F-actin in a dose-dependent manner (Figure 3, B and C; EC50 = 0.034 ± 0.010 mol of α-actinin per mole actin). Both His-NrbI(1–287) and His-NrbI(1–146) also bundled F-actin, albeit less efficiently than α-actinin (Figure 3, B and C; EC50 for NrbI(1–287) was 0.10 ± 0.020 mol NrbI per mole actin, and for NrbI(1–146) was 0.26 ± 0.078). Of note, NrbI(1–287), which was more potent than NrbI(1–146) in generating filopodia (Figure 1) also was more potent in bundling actin (Figure 3C). In contrast, NrbI(146–286), which does not induce filopodia (Figure 1), failed to bind or bundle F-actin (Figure 3, B and C). Electron microscopy showed that NrbI(1–287) and NrbI(1–146) promoted loosely packed 50- to 100-nm F-actin bundles (Figure 3D). By comparison, actin filaments were widely dispersed in the presence of NrbI(146–286). These findings demonstrate that the N-terminal domain of neurabin is sufficient to bundle actin, suggesting that the induction of dendritic filopodia may be due to the intrinsic actin-bundling activity of NrbI(1–287).
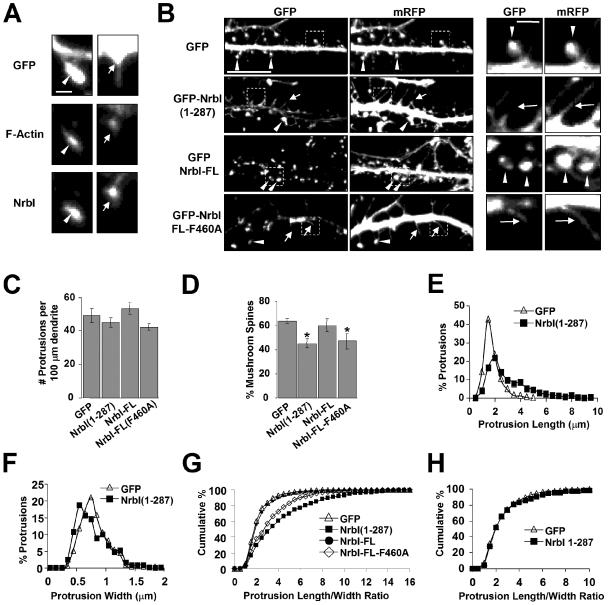
Neurabin-I Regulates Dendritic Spine Morphology
Prior studies suggested that mature mushroom spines arise after the stabilization of highly motile filopodia-like precursors (Ziv and Smith, 1996; Matus, 2000; Yuste and Bonhoeffer, 2004). Immunocytochemistry of control GFP-expressing neurons with an anti-NrbI antibody showed that endogenous NrbI was concentrated in the actin-rich heads of spines and at the base of dendritic filopodia (Figure 4A), as was also noted using immunoelectron microscopy in hippocampal slices (Muly et al., 2004; Zito et al., 2004). Because NrbI localizes to both filopodia and spines and is capable of remodeling the actin cytoskeleton to induce filopodia, we hypothesized that NrbI would play a role in the maturation of dendritic filopodia into spines. To test this, we transfected GFP-NrbI constructs in hippocampal neurons at DIV12, when no spines are present. After growing these cultures to DIV26, control GFP expressing and untransfected neurons possessed numerous mushroom-shaped dendritic spines (Figure 4B). Neurons expressing GFP-NrbI-FL also possessed numerous spines (Figure 4B). By comparison, DIV26 neurons expressing NrbI(1–287) showed few spines and instead possessed numerous protrusions characterized by longer necks and smaller heads, termed “long-necked spines” or filopodia (Figure 4B). Interestingly, the total number of protrusions was unchanged by GFP-NrbI(1–287) expression (Figure 4C). NrbI-FL-F460A, which lacked PP1 binding, also resulted in a significant loss of spines and a concomitant increase in thin, elongated filopodia (Figure 4B). Quantitative analyses showed that expression of either NrbI(1–287) or NrbI-FL-F460A resulted in a decrease in the number of mature mushroom spines (Figure 4D) accompanied by an increase in the number of filopodia. Morphometric analyses showed that NrbI(1–287) increased the average length of dendritic protrusions (Figure 4E) and decreased the width of protrusions (Figure 4F). Because the total number of protrusions was unchanged (Figure 4C), these data suggest that NrbI(1–287) and NrbI-FL-F460A expression prevents or delays the filopodia-to-spine conversion, as reflected by the increase in length-to-width ratio of individual dendritic protrusions in neurons expressing the mutant neurabins (Figure 4G).
Figure 4.
Neurabin-I regulates spine development in hippocampal neurons. (A) Hippocampal neurons (DIV35) expressing GFP were stained with Alexa568-phalloidin and immunostained for endogenous NrbI using an anti-NrbI antibody. An actin-rich NrbI-containing spine (left) and a dendritic filopodia with NrbI localized at the base (right) are shown. Scale bar, 1 μm. (B) Hippocampal neurons (DIV12) were cotransfected with plasmids that expressed mRFP and GFP-NrbI fusion proteins, fixed at DIV26, and imaged. Arrowheads indicate spines and arrows point to filopodia. Scale bar, 10 μm. Right panels show the enlarged images of the areas indicated by the dotted squares. Scale bar, 1 μm. (C) Using the mRFP channel, the number of protrusions per unit length of dendrite was measured following the expression of the indicated GFP-NrbI proteins. (D) Dendritic protrusions of neurons transfected with indicated NrbI proteins were defined as either mushroom-shaped spines or nonspines/filopodia (see Materials and Methods for details) and expressed as the percentage of protrusions that are spines. *p < 0.05, Student's t test. (E) Length of individual protrusions was measured and the results were presented as histograms. (F) Histogram of protrusion width. (G) The length/width ratios of dendritic protrusions expressing the indicated GFP-fusion proteins were calculated, and results are presented as cumulative frequency distributions. (H) Hippocampal neurons were cotransfected with mRFP and GFP-NrbI(1–287) at DIV28 and fixed at DIV30. Results from pooled experiments were quantified, and length/width ratios are expressed as cumulative frequency distributions.
Two mechanisms could account for the delayed conversion of filopodia to spines in DIV26 neurons by GFP-NrbI(1–287). Either NrbI(1–287) slows or stops the conversion process, or it reverts previously mature spines into filopodia. To distinguish between these two events, we transfected GFP-NrbI(1–287) into older DIV28 neurons that already possessed numerous mushroom-shaped spines and few filopodia and imaged the cells 2 d later. Although GFP-NrbI(1–287) effectively concentrated in the actin-rich heads of DIV30 spines (unpublished data), the length-to-width ratio of dendritic protrusions in DIV30 neurons expressing GFP-NrbI(1–287) was indistinguishable from neurons expressing GFP alone (Figure 4H). These findings strongly suggest that NrbI(1–287), which induces filopodia in younger neurons, cannot revert mature spines into filopodia, but instead inhibits the transition of filopodia into spines. The results also hint at a defined window of time (between DIV 12 and DIV 26) in neuronal development during which NrbI regulates spine morphology.
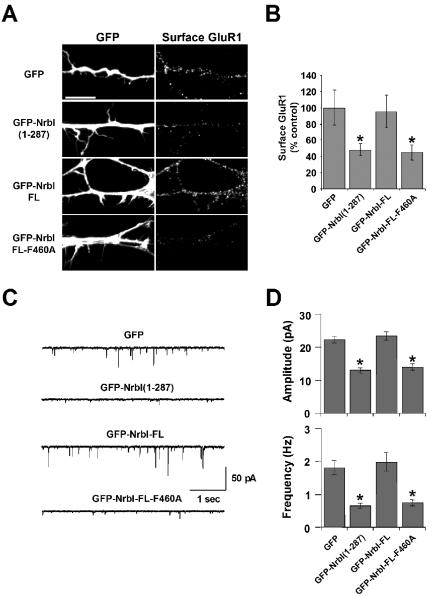
Neurabin-I Increases the Synaptic Accumulation of AMPA Receptors and Potentiates Excitatory Synaptic Transmission
AMPA-type glutamate receptors (AMPARs) are the principal neurotransmitter receptors mediating excitatory transmission and recruitment of AMPARs to postsynaptic membrane is a hallmark of spine development (Ziv and Garner, 2001; Bredt and Nicoll, 2003). Although GFP-NrbI(1–287) preferentially localizes to the postsynaptic density in hippocampal slices and GFP-NrbI(1–287)-expressing spines make synapses, electrophysiological studies showed that these synapses are functionally down-regulated (Zito et al., 2004). These observations suggest that the increased number of long-necked spines induced by NrbI(1–287) may be correlated with a reduced postsynaptic targeting of AMPARs. To test this possibility, we utilized live-cell antibody labeling to analyze the surface expression of GluR1 AMPARs in cultured hippocampal neurons expressing GFP-neurabins. In neurons expressing GFP alone, dendrites were decorated with numerous surface GluR1 puncta corresponding to excitatory synapses (Figure 5A). Expression of GFP-NrbI-FL, which had no effect on spine morphology, also had no measurable effect on GluR1 surface expression. By comparison, GFP-NrbI(1–287), which retarded spine maturation in the dissociated cultures, resulted in a marked reduction in GluR1 surface expression (Figure 5A). GFP-NrbI-FL-F460A also resulted in significant reduction in surface GluR1 staining. Quantitative analyses showed that both GFP-NrbI(1–287) and GFP-NrbI-FL-F460A resulted in a 50–60% reduction in the surface expression of GluR1 (Figure 5B).
Figure 5.
Neurabin-I regulates surface AMPA Receptor expression and excitatory synaptic transmission. (A) Hippocampal neurons were transfected on DIV14 with plasmids encoding the indicated GFP-NrbI proteins. On DIV17, surface GluR1 AMPA receptor (AMPAR) subunits were labeled on live cells by incubating with anti-GluR1 antibody directed against an extracellular epitope before fixation and immunostaining with fluorophore-conjugated secondary antibody. Cells were then imaged for GFP and surface GluR1 staining. Scale bar, 10 μm. (B) Quantification of surface GluR1 levels of pooled experiments shown in A with ± SEM (n = 12–15 for each transfection; *p < 0.05, Student's t test). (C) Representative traces of mEPSCs recorded from DIV16 hippocampal neurons expressing GFP (top) or the indicated GFP-NrbI proteins are shown. (D) mEPSC amplitudes (top) and frequencies (bottom) are shown with SEM determined using Student's t test (n = 8–11 for each construct; *p < 0.05).
To test the functional effect of reduced GluR1 at the plasma membrane, we monitored AMPAR-mediated synaptic transmission by measuring miniature excitatory postsynaptic currents (mEPSC) in neurons expressing GFP-NrbI proteins (Figure 5C). Both mEPSC amplitude and frequency were decreased in neurons expressing GFP-NrbI(1–287) and GFP-Nrb-FL-F460A relative to cells that expressed either GFP or GFP-NrbI-FL (Figure 5D; GFP, 22.3 ± 1.0 pA, 1.8 ± 0.2 Hz; GFP-Nrb-FL, 23.5 ± 1.3 pA, 2.0 ± 0.3 Hz; GFP-Nrb-FL-F460A, 14.1 ± 1.0 pA, 0.8 ± 0.1 Hz; and GFP-NrbI(1–287), 13.1 ± 0.8 pA, 0.7 ± 0.1 Hz). These functional deficits closely paralleled the reductions in surface GluR1 expression (Figure 5A). Together, these data indicate that the delayed morphological maturation of dendritic spines was accompanied by a reduction in excitatory synaptic transmission.
Neurabin-I Facilitates Synaptic Targeting of AMPARs
The above studies utilized overexpression of wild-type and mutant NrbIs in cultured neurons, and thus the results could be ascribed to either the intrinsic activity of mutant NrbI proteins or disruption of endogenous NrbI-mediated events. To establish the requirement for endogenous neurabin in spine maturation, we used RNA interference with short hairpin RNAs (shRNA; Paddison and Hannon, 2002) to “knockdown” the expression of endogenous NrbI in cultured neurons. The vector, pZOFF, was designed to express shRNAs of interest driven by H1 RNA polymerase-III, whereas GFP expression by a separate cassette served as a marker of successful transfection (Figure 6A). To test shRNA efficacy, pZOFF and pNrbI-OFF (containing shRNA directed against NrbI amino acids 430–437) were transfected in COS7 cells expressing GFP-NrbI-FL or GFP-Spino-FL. Immunoblotting with anti-GFP antibody showed that pNrbI-OFF but not pZOFF significantly reduced GFP-NrbI-FL levels (Figure 6B, left panel). Neither plasmid had an effect on the expression of GFP-Spino-FL (Figure 6B, left panel). Equal protein loading was established by immunoblotting with an antitubulin antibody (Figure 6B, right panel). Immunocytochemical analysis of transfected neurons showed that pNrbI-OFF, but not pZOFF, dramatically reduced endogenous NrbI levels relative to nearby untransfected neurons (Figure 6C and unpublished data [pZOFF]). Quantification of several independent experiments showed ∼70% reduction in endogenous NrbI levels after 5 d of NrbI shRNA expression (Figure 6D). Live-cell staining for surface GluR1 in the same age cultures showed that neurons expressing pNrbI-OFF displayed significantly reduced surface GluR1 (Figure 6E). Quantification of multiple independent experiments showed a 30% reduction in surface GluR1 after 5 d of NrbI shRNA expression (Figure 6F). These studies demonstrate that endogenous NrbI regulates the synaptic targeting of AMPARs.
DISCUSSION
Induction and remodeling of dendritic spines play key roles both in the development of the mammalian brain and in the subsequent activity-dependent changes of adult neuronal circuits. Recent studies suggested that the number and shape of dendritic spines are regulated by a number of proteins (Hering and Sheng, 2001; Ehlers, 2002) that either directly or indirectly modify the actin cytoskeleton (Nakayama et al., 2000; Tashiro et al., 2000; Penzes et al., 2001; Meng et al., 2002; Hering and Sheng, 2003; Ishikawa et al., 2003; Takahashi et al., 2003). The present work focused on neurabin-I (NrbI), a neuronal actinand PP1-binding protein that was recently implicated in regulating spine number and morphology (Oliver et al., 2002; Zito et al., 2004). Spinophilin, also known as neurabin-II, is a structural homologue of neurabin-I that is highly expressed in neurons and may also regulate spine development, as hippocampal neurons from mice lacking spinophilin displayed an increased number of spines both in vivo and in vitro (Feng et al., 2000).
Multiple Neurabin Domains Regulate Cell Morphology
We first analyzed the morphological changes induced by neurabin-I and spinophilin in cultured COS7 cells. One highlight of the data are the unique ability of the isolated N-terminal actin-binding domains, NrbI(1–287) and Spino(1–289), to induce extended filopodia in these cells. Interestingly, NrbI(1–287) was a more potent inducer of filopodia than Spino(1–289), consistent with its higher affinity for the actin cytoskeleton. Spinophilin is uniquely phosphorylated at several serines within the actin-binding domain by both c-AMP–dependent protein kinase (PKA; Hsieh-Wilson et al., 2003) and Ca2+-calmodulin–dependent protein kinase type II (CaMKII) (Grossman et al., 2004). These covalent modifications reduce spinophilin's affinity for F-actin and, as actin binding is essential for modifying cell morphology, one possibility is that ongoing phosphorylation of spinophilin limits its ability to reorganize the actin cytoskeleton to induce filopodia. More generally, these data and findings presented here suggest quantitatively distinct roles for spinophilin and neurabin-I in regulating the actin cytoskeleton.
As deletion of C-terminal sequences enhanced the ability of both neurabin-I and spinophlin to modify cell morphology, we also analyzed other protein-interaction domains. To assess the role of dimerization, we substituted C-terminal coiled-coil domains with the bacterial enzyme gyrase B. Forced dimerization of GFP-Spino(1–586)-GyrB by coumermycin inhibited filopodia providing direct experimental evidence that dimerization attenuates the ability of the actinbinding domain to modify the actin cytoskeleton and induce filopodia. However, we cannot exclude the possibility that in the context of WT NrbI, other proteins known to bind the C-terminal coiled-coiled domain (Stephens and Banting, 1999; Ryan et al., 2003) may also regulate actin cytoskeleton.
A point mutation that abolished PP1-binding also enhanced filopodia induction by full-length neurabin-I and spinophlin, suggesting a role for neurabin-bound PP1 in regulating the actin cytoskeleton. Finally, the PDZ domain of neurabin-I and spinophlin is known to bind the RacGEF kalirin-7 (Penzes et al., 2001) and p70 S6 Kinase (Burnett et al., 1998), two proteins implicated in regulating neuronal morphology. We found that deletion of the PDZ domain (compare GFP-NrbI(1–671) with GFP-NrbI (1–495) or GFP-Spino(1–671) with GFP-Spino(1–490) in Figure 1C) enhanced the ability of both neurabins to induce filopodia in COS7 cells. Although much more research is required to define the relative roles of the various domains within neurabin-I and spinophilin in controlling actin reorganization, our studies strongly suggest that multiple domains in the two neurabin isoforms regulate cell morphology.
Actin Bundling by Neurabin-I Modifies Cell Morphology
Recent studies in cultured cells established that filopodia arise as a result of localized F-actin bundling in a lamellipodial actin network and are initiated by actin-bundling proteins concentrated at specific points on the cell surface (Svitkina et al., 2003). Although wild-type NrbI-FL was previously reported to bundle actin (Nakanishi et al., 1997), our studies show that the isolated N-terminal actin-binding domain, NrbI(1–287), also bundles F-actin filaments in vitro. However, the 50–100-nm actin bundles elicited by NrbI N-terminal fragments in our experiments are much smaller and “looser” than the 200-nm bundles previously noted to be induced by NrbI-FL (Nakanishi et al., 1997). This suggests that the C-terminal region of NrbI is capable of modulating NrbI's actin-bundling capability. Furthermore, our examination of actin organization in intact cells indicated that both NrbI-FL and NrbI(1–287) promotes elongated actin fibers consisted with bundled F-actin in the dendrites of cultured hippocampal neurons. In contrast to GFP-NrbI-FL, which was highly concentrated at the base of filopodia, NrbI(1–287) was distributed throughout the length of filopodia. This suggests that both differing subcellular localization and differential bundling activity may account for NrbI(1–287)'s enhanced ability to induce filopodia in neurons and COS7 cells compared with wild-type NrbI-FL.
Neurabin-PP1 Complex Regulates Maturation of Filopodia to Spines
A growing body of evidence suggests that dendritic filopodia are morphological precursors of spines (Ziv and Smith, 1996; Matus, 2000; Bonhoeffer and Yuste, 2002; Yuste and Bonhoeffer, 2004). Although we found that Nrb(1–287) promoted dendritic filopodia formation, long-term expression of Nrb(1–287) in hippocampal neurons delayed the onset of dendritic spine formation. This contrasts with the normal development of spines containing wild-type NrbI-FL. Because our studies showed that expression of the non-PP1 binding mutant NrbI-FL-F460A (Hsieh-Wilson et al., 1999; Terry-Lorenzo et al., 2002b), like NrbI(1–287), delays the onset of dendritic spines in cultured hippocampal neurons, biochemical events catalyzed by NrbI-bound PP1 may control the ability of NrbI-FL to facilitate spine formation. For instance, PP1 may be capable of regulating the transition from linear bundles of F-actin in filopodia to a latticed actin cytoskeleton such as that found in the mushroom-shaped heads of spines (Rao and Craig, 2000). Indeed, PP1 is already known to regulate the actin cytoskeleton in cultured cells (Fernandez et al., 1990).
In vitro studies demonstrated that spinophilin and neurabin-I are potent regulators of PP1 activity (Allen et al., 1997; McAvoy et al., 1999; Terry-Lorenzo et al., 2002a), and deficits in PP1 signaling were noted in hippocampal neurons from the spinophilin null mice (Feng et al., 2000). The ability of NrbI-FL-F460A to delay spine formation is consistent with the function of neurabin-I and spinophilin as PP1 regulatory subunits (Cohen, 2002) that selectively target the PP1γ1 isoform to the actin-rich postsynaptic density in dendritic spines (MacMillan et al., 1999; McAvoy et al., 1999; Cohen, 2002; Terry-Lorenzo et al., 2002a). The importance of PP1 targeting was further evidenced by the fact that NrbI-FL-F460A displayed an enhanced ability to promote filopodia relative to wild-type NrbI-FL in COS7 cells. Because NrbI-FL-F460A likely interferes with the proper targeting of PP1 to substrates within filopodia and spines, the simplest interpretation of these results is that PP1 bound to neurabin negatively regulates filopodia formation induced by the actin-bundling domain. Such an antagonistic relationship between the effects of actin bundling and PP1 binding are further supported by our observation of reciprocal effects of these neurabin domains on synaptic recruitment of AMPARs and excitatory synaptic transmission. The targeting of PP1 does not always dampen NrbI's effects on cell morphology, however, as NrbI-FL stimulated numerous filopodia in young (DIV7) neurons, but mutation of the PP1-binding site had no further effect. Together, these studies suggest that the actin-bound neurabin/PP1 complex acts at defined stages during neuronal development to control spine morphogenesis.
To regulate the spine actin cytoskeleton, the NrbI/PP1 complex may dephosphorylate multiple postsynaptic substrates, such actin-depolymerizing factor (ADF)/cofilin (Meberg et al., 1998), or CaMKII (Lisman and Zhabotinsky, 2001). Interestingly, increased phosphorylation of ADF/cofilin is required for actin recruitment to spines during LTP (Fukazawa et al., 2003) and net ADF/cofilin dephosphorylation in mice lacking LIMK-1, an ADF/cofilin kinase, altered spine morphology (Meng et al., 2002). CaMKIIβ also regulates filopodial growth, motility, and spine formation (Fink et al., 2003; Jourdain et al., 2003). In hippocampal slices, pharmacological inhibition of PP1 activity caused a rapid increase in dendritic spines and filopodia (Jourdain et al., 2003), suggesting that the ongoing synaptic PP1 activity associated with the neurabin/PP1 complex maintains normal spine number and morphology. One interesting possibility is that NrbI acts in conjunction with other PP1 complexes, such as the postsynaptic Phactr-1 complex, which contains both PP1 and G-actin (Allen et al., 2004).
The Neurabin/PP1 Complex Regulates Synaptic Localization of GluR1 Subunits
As filopodia are transformed into spines, postsynaptic protein complexes are assembled and neurotransmitter receptors are recruited to postsynaptic sites (Ziv and Garner, 2001; Bredt and Nicoll, 2003). In this regard, it is interesting to note that PP1 association with unspecified postsynaptic targeting subunits is required for hippocampal long-term depression (Morishita et al., 2001). This synaptic depression is associated with PP1-catalyzed dephosphorylation of GluR1 subunits (at Ser845), which triggers AMPAR internalization (Beattie et al., 2000; Ehlers, 2000). Although previous electrophysiological studies suggested that expression of NrbI(1–287) in hippocampal slices down-regulated AMPAR and NMDAR currents (Zito et al., 2004), our data showed that delayed morphological maturation of dendritic spines by both NrbI(1–287) and NrbI-FL-F460A resulted in parallel decreases in surface expression of GluR1 subunits and diminished glutamatergic transmission. The shRNA-mediated knockdown of endogenous NrbI levels also decreased surface GluR1 subunits, providing independent corroboration for a role for NrbI in facilitating the synaptic recruitment of AMPARs. It is unlikely, however, that the NrbI/PP1 complex directly dephosphorylates GluR1, as defective PP1-mediated down-regulation of AMPAR and LTD in hippocampal slices from spinophilin null mice (Feng et al., 2000) suggests that the Spino/PP1 complex is the likely GluR1 (Ser845) phosphatase. Thus, we speculate that the PSD-associated NrbI/PP1 complex acts to dephosphorylate other substrates that regulate AMPAR recruitment to synapses. Interestingly, as noted in NrbI(1–287)-expressing hippocampal slices (Zito et al., 2004) and spinophilin null mice (Feng et al., 2000), NMDAR currents may also be affected by altered PP1 targeting. This may suggest a coordinate regulation of the NrbI/PP1 complex and NMDAR-associated yotiao/PP1 complex (Westphal et al., 1999) to regulate excitatory neurotransmission. Finally, in contrast to the spinophilin/PP1 complex, which down-regulates AMPAR (Feng et al., 2000), our studies suggest the NrbI-bound PP1 facilitates increased synaptic targeting of GluR1 and AMPAR-mediated synaptic transmission. Because NrbI and spinophilin heterodimerize in neurons (MacMillan et al., 1999), this hints at a complex interplay between spinophilin/PP1 and NrbI/PP1 to regulate protein composition and function of excitatory synapses.
In summary, results presented here demonstrate an important role for the neurabin-I/PP1 complex in the formation of filopodia in young neurons and the transformation of neuronal filopodia into dendritic spines. The ability of neurabin-I to promote synaptic targeting of GluR1 subunits and enhance AMPAR currents in cultured neurons implies that the neurabin-I/PP1 complex also plays a key role in the functional maturation of excitatory synapses. It will be important for future studies to identify the synaptic targets of the neurabin-I/PP1 complex and elucidate the signaling mechanisms that coordinate the action of this actin-associated phosphatase complex with that of other postsynaptic PP1 complexes. More broadly, by revealing molecular events integrating actin remodeling and PP1 activity at the postsynaptic membrane, our results provide new insights into the spatial and temporal events required for spine development and excitatory synaptic transmission in the mammalian brain.
Acknowledgments
We thank Chi Zhang and Haiwei Zhang for help with hippocampal cultures and Peter Reinhart for electrophysiology equipment. The authors thank Matthew Brush, Guoping Feng, Dani Gitler, Juliet Hernandez, April Horton, Sangmi Lim, Wade Morishita, Rob Wechsler-Reya, Suzanne Sikes, Fan Wang, and Xiang Yu for critical comments. We thank Karen Zito and Karel Svoboda for valuable discussions. This work was supported by National Institutes of Health (NIH) grant NS41063 (S.S.), NIH grant NS39471 (C.C.G.), and NIH grants NS39402, MH64748; the Christopher Reeve Paralysis Foundation; and Alzheimer's Association (M.D.E.). M.D.E. is also a Ruth K. Broad Scholar in Neuroscience. T.A.B. is supported by the National Alliance for Research on Schizophrenia and Depression.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1054) on March 2, 2005.
Abbreviations used: AMPAR, AMPA-type glutamate receptor; DIV, day in vitro; LTD, long-term depression; LTP, long-term potentiation; mEPSC, miniature excitatory postsynaptic current; NrbI, Neurabin-I; PP1, protein phosphatase 1; Spino, Spinophilin/NeurabinII; shRNA, short hairpin RNA.
References
- Allen, P. B., Greenfield, A. T., Svenningsson, P., Haspeslagh, D. C., and Greengard, P. (2004). Phactrs 1–4, a family of protein phosphatase 1 and actin regulatory proteins. Proc. Natl. Acad. Sci. USA 101, 7187–7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, P. B., Ouimet, C. C., and Greengard, P. (1997). Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc. Natl. Acad. Sci. USA 94, 9956–9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, A. P., Smith, F. D., 3rd, VanDongen, H. M., VanDongen, A. M., and Milgram, S. L. (2004). The identification of a second actin-binding region in spinophilin/neurabin II. Brain Res. Mol. Brain Res. 124, 105–113. [DOI] [PubMed] [Google Scholar]
- Beattie, E. C., Carroll, R. C., Yu, X., Morishita, W., Yasuda, H., von Zastrow, M., and Malenka, R. C. (2000). Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat. Neurosci. 3, 1291–1300. [DOI] [PubMed] [Google Scholar]
- Bonhoeffer, T., and Yuste, R. (2002). Spine motility. Phenomenology, mechanisms, and function. Neuron 35, 1019–1027. [DOI] [PubMed] [Google Scholar]
- Bredt, D. S., and Nicoll, R. A. (2003). AMPA receptor trafficking at excitatory synapses. Neuron 40, 361–379. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553. [DOI] [PubMed] [Google Scholar]
- Burnett, P.E., Blackshaw, S., Lai, M. M., Qureshi, I. A., Burnett, A. F., Sabatini, D. M., and Snyder, S. H. (1998). Neurabin is a synaptic protein linking p70 S6 kinase and the neuronal cytoskeleton. Proc. Natl. Acad. Sci. USA 95, 8351–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. E., Tour, O., Palmer, A. E., Steinbach, P. A., Baird, G. S., Zacharias, D. A., and Tsien, R. Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, P. T. (2002). Protein phosphatase 1—targeted in many directions. J. Cell Sci. 115, 241–256. [DOI] [PubMed] [Google Scholar]
- Colicos, M. A., Collins, B. E., Sailor, M. J., and Goda, Y. (2001). Remodeling of synaptic actin induced by photoconductive stimulation. Cell 107, 605–616. [DOI] [PubMed] [Google Scholar]
- Edlund, M., Lotano, M. A., and Otey, C. A. (2001). Dynamics of alpha-actinin in focal adhesions and stress fibers visualized with alpha-actinin-green fluorescent protein. Cell Motil. Cytoskeleton 48, 190–200. [DOI] [PubMed] [Google Scholar]
- Ehlers, M. D. (2000). Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron 28, 511–525. [DOI] [PubMed] [Google Scholar]
- Ehlers, M. D. (2002). Molecular morphogens for dendritic spines. Trends Neurosci. 25, 64–67. [DOI] [PubMed] [Google Scholar]
- Farrar, M. A., Olson, S. H., and Perlmutter, R. M. (2000). Coumermycininduced dimerization of GyrB-containing fusion proteins. Methods Enzymol. 327, 421–429. [DOI] [PubMed] [Google Scholar]
- Feng, J., Yan, Z., Ferreira, A., Tomizawa, K., Liauw, J.A., Zhuo, M., Allen, P. B., Ouimet, C. C., and Greengard, P. (2000). Spinophilin regulates the formation and function of dendritic spines. Proc. Natl. Acad. Sci. USA 97, 9287–9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, A., Brautigan, D. L., Mumby, M., and Lamb, N. J. (1990). Protein phosphatase type-1, not type-2A, modulates actin microfilament integrity and myosin light chain phosphorylation in living nonmuscle cells. J. Cell Biol. 111, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala, J. C., Feinberg, M., Popov, V., and Harris, K. M. (1998). Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J. Neurosci. 18, 8900–8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala, J. C., Spacek, J., and Harris, K. M. (2002). Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res. Brain Res. Rev. 39, 29–54. [DOI] [PubMed] [Google Scholar]
- Fink, C. C., Bayer, K. U., Myers, J. W., Ferrell, J. E., Jr., Schulman, H., and Meyer, T. (2003). Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron 39, 283–297. [DOI] [PubMed] [Google Scholar]
- Fischer, M., Kaech, S., Knutti, D., and Matus, A. (1998). Rapid actin-based plasticity in dendritic spines. Neuron 20, 847–854. [DOI] [PubMed] [Google Scholar]
- Fukazawa, Y., Saitoh, Y., Ozawa, F., Ohta, Y., Mizuno, K., and Inokuchi, K. (2003). Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron 38, 447–460. [DOI] [PubMed] [Google Scholar]
- Grossman, S. D., Futter, M., Snyder, G. L., Allen, P. B., Nairn, A. C., Greengard, P., and Hsieh-Wilson, L. C. (2004). Spinophilin is phosphorylated by Ca2+/calmodulin-dependent protein kinase II resulting in regulation of its binding to F-actin. J. Neurochem. 90, 317–324. [DOI] [PubMed] [Google Scholar]
- Grossman, S. D., Hsieh-Wilson, L. C., Allen, P.B., Nairn, A. C., and Greengard, P. (2002). The actin-binding domain of spinophilin is necessary and sufficient for targeting to dendritic spines. Neuromolecular Med. 2, 61–69. [DOI] [PubMed] [Google Scholar]
- Halpain, S., Hipolito, A., and Saffer, L. (1998). Regulation of F-actin stability in dendritic spines by glutamate receptors and calcineurin. J. Neurosci. 18, 9835–9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, K. M. (1999). Structure, development, and plasticity of dendritic spines. Curr. Opin. Neurobiol. 9, 343–348. [DOI] [PubMed] [Google Scholar]
- Hering, H., and Sheng, M. (2001). Dendritic spines: structure, dynamics and regulation. Nat. Rev. Neurosci. 2, 880–888. [DOI] [PubMed] [Google Scholar]
- Hering, H., and Sheng, M. (2003). Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J. Neurosci. 23, 11759–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Wilson, L. C., Allen, P. B., Watanabe, T., Nairn, A. C., and Greengard, P. (1999). Characterization of the neuronal targeting protein spinophilin and its interactions with protein phosphatase-1. Biochemistry 38, 4365–4373. [DOI] [PubMed] [Google Scholar]
- Hsieh-Wilson, L. C., Benfenati, F., Snyder, G. L., Allen, P. B., Nairn, A. C., and Greengard, P. (2003). Phosphorylation of spinophilin modulates its interaction with actin filaments. J. Biol. Chem. 278, 1186–1194. [DOI] [PubMed] [Google Scholar]
- Irwin, S. A. et al. (2001). Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet. 98, 161–167. [DOI] [PubMed] [Google Scholar]
- Ishikawa, Y., Katoh, H., and Negishi, M. (2003). A role of Rnd1 GTPase in dendritic spine formation in hippocampal neurons. J. Neurosci. 23, 11065–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain, P., Fukunaga, K., and Muller, D. (2003). Calcium/calmodulin-dependent protein kinase II contributes to activity-dependent filopodia growth and spine formation. J. Neurosci. 23, 10645–10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, W. E., and Moser, H. W. (2000). Dendritic anomalies in disorders associated with mental retardation. Cereb. Cortex 10, 981–991. [DOI] [PubMed] [Google Scholar]
- Kim, C. H., and Lisman, J. E. (1999). A role of actin filament in synaptic transmission and long-term potentiation. J. Neurosci. 19, 4314–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krucker, T., Siggins, G. R., and Halpain, S. (2000). Dynamic actin filaments are required for stable long-term potentiation (LTP) in area CA1 of the hippocampus. Proc. Natl. Acad. Sci. USA 97, 6856–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamprecht, R., and LeDoux, J. (2004). Structural plasticity and memory. Nat. Rev. Neurosci. 5, 45–54. [DOI] [PubMed] [Google Scholar]
- Lang, C., Barco, A., Zablow, L., Kandel, E. R., Siegelbaum, S. A., and Zakharenko, S. S. (2004). Transient expansion of synaptically connected dendritic spines upon induction of hippocampal long-term potentiation. Proc. Natl. Acad. Sci. USA 101, 16665–16670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman, J. E., and Zhabotinsky, A. M. (2001). A model of synaptic memory: a CaMKII/PP1 switch that potentiates transmission by organizing an AMPA receptor anchoring assembly. Neuron 31, 191–201. [DOI] [PubMed] [Google Scholar]
- MacMillan, L. B., Bass, M. A., Cheng, N., Howard, E. F., Tamura, M., Strack, S., Wadzinski, B. E., and Colbran, R. J. (1999). Brain actin-associated protein phosphatase 1 holoenzymes containing spinophilin, neurabin, and selected catalytic subunit isoforms. J. Biol. Chem. 274, 35845–35854. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic, M., Malinow, R., and Svoboda, K. (1999). Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science 283, 1923–1927. [DOI] [PubMed] [Google Scholar]
- Mammen, A. L., Huganir, R. L., and O'Brien, R. J. (1997). Redistribution and stabilization of cell surface glutamate receptors during synapse formation. J. Neurosci. 17, 7351–7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki, M., Honkura, N., Ellis-Davies, G. C., and Kasai, H. (2004). Structural basis of long-term potentiation in single dendritic spines. Nature 429, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus, A. (2000). Actin-based plasticity in dendritic spines. Science 290, 754–758. [DOI] [PubMed] [Google Scholar]
- McAvoy, T., Allen, P. B., Obaishi, H., Nakanishi, H., Takai, Y., Greengard, P., Nairn, A. C., and Hemmings, H. C., Jr. (1999). Regulation of neurabin I interaction with protein phosphatase 1 by phosphorylation. Biochemistry 38, 12943–12949. [DOI] [PubMed] [Google Scholar]
- Meberg, P. J., Ono, S., Minamide, L. S., Takahashi, M., and Bamburg, J. R. (1998). Actin depolymerizing factor and cofilin phosphorylation dynamics: response to signals that regulate neurite extension. Cell Motil. Cytoskeleton 39, 172–190. [DOI] [PubMed] [Google Scholar]
- Meng, Y. et al. (2002). Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 35, 121–133. [DOI] [PubMed] [Google Scholar]
- Moody, C. J., Marston, S. B., and Smith, C. W. (1985). Bundling of actin filaments by aorta caldesmon is not related to its regulatory function. FEBS Lett. 191, 107–112. [DOI] [PubMed] [Google Scholar]
- Morishita, W., Connor, J. H., Xia, H., Quinlan, E. M., Shenolikar, S., and Malenka, R. C. (2001). Regulation of synaptic strength by protein phosphatase 1. Neuron 32, 1133–1148. [DOI] [PubMed] [Google Scholar]
- Muly, E. C., Allen, P., Mazloom, M., Aranbayeva, Z., Greenfield, A. T., and Greengard, P. (2004). Subcellular distribution of neurabin immunolabeling in primate prefrontal cortex: comparison with spinophilin. Cereb. Cortex 14, 1398–1407. [DOI] [PubMed] [Google Scholar]
- Nakanishi, H., Obaishi, H., Satoh, A., Wada, M., Mandai, K., Satoh, K., Nishioka, H., Matsuura, Y., Mizoguchi, A., and Takai, Y. (1997). Neurabin: a novel neural tissue-specific actin filament-binding protein involved in neurite formation. J. Cell Biol. 139, 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, A. Y., Harms, M. B., and Luo, L. (2000). Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. 20, 5329–5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, C. J., Terry-Lorenzo, R. T., Elliott, E., Bloomer, W. A., Li, S., Brautigan, D. L., Colbran, R. J., and Shenolikar, S. (2002). Targeting protein phosphatase 1 (PP1) to the actin cytoskeleton: the neurabin I/PP1 complex regulates cell morphology. Mol. Cell. Biol. 22, 4690–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddison, P. J., and Hannon, G. J. (2002). RNA interference: the new somatic cell genetics? Cancer Cell 2, 17–23. [DOI] [PubMed] [Google Scholar]
- Penzes, P., Johnson, R. C., Sattler, R., Zhang, X., Huganir, R. L., Kambampati, V., Mains, R. E., and Eipper, B. A. (2001). The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229–242. [DOI] [PubMed] [Google Scholar]
- Rao, A., and Craig, A. M. (2000). Signaling between the actin cytoskeleton and the postsynaptic density of dendritic spines. Hippocampus 10, 527–541. [DOI] [PubMed] [Google Scholar]
- Ryan, X. P., Alldritt, J., Allen, P., Nairn, A. C., Wu, G., and Greengard, P. (2003). Interaction of the Rho-GEF LFC with neurabin and spinophilin: a linkage between microtubules and actin. Society for Neuroscience 2003 Abstract Viewer/Itinerary Planner, Washington, DC, 870.7.
- Sala, C., Piech, V., Wilson, N. R., Passafaro, M., Liu, G., and Sheng, M. (2001). Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron 31, 115–130. [DOI] [PubMed] [Google Scholar]
- Satoh, A. et al. (1998). Neurabin-II/spinophilin. An actin filament-binding protein with one pdz domain localized at cadherin-based cell-cell adhesion sites. J. Biol. Chem. 273, 3470–3475. [DOI] [PubMed] [Google Scholar]
- Scott, D. B., Blanpied, T. A., Swanson, G. T., Zhang, C., and Ehlers, M. D. (2001). An NMDA receptor ER retention signal regulated by phosphorylation and alternative splicing. J. Neurosci. 21, 3063–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star, E. N., Kwiatkowski, D. J., and Murthy, V. N. (2002). Rapid turnover of actin in dendritic spines and its regulation by activity. Nat. Neurosci. 5, 239–246. [DOI] [PubMed] [Google Scholar]
- Stephens, D. J., and Banting, G. (1999). Direct interaction of the trans-Golgi network membrane protein, TGN38, with the F-actin binding protein, neurabin. J. Biol. Chem. 274, 30080–30086. [DOI] [PubMed] [Google Scholar]
- Svitkina, T. M., Bulanova, E. A., Chaga, O. Y., Vignjevic, D. M., Kojima, S., Vasiliev, J. M., and Borisy, G. G. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Sekino, Y., Tanaka, S., Mizui, T., Kishi, S., and Shirao, T. (2003). Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J. Neurosci. 23, 6586–6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro, A., Minden, A., and Yuste, R. (2000). Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb. Cortex 10, 927–938. [DOI] [PubMed] [Google Scholar]
- Terry-Lorenzo, R. T., Carmody, L. C., Voltz, J. W., Connor, J. H., Li, S., Smith, F. D., Milgram, S. L., Colbran, R. J., and Shenolikar, S. (2002a). The neuronal actin-binding proteins, neurabin I and neurabin II, recruit specific isoforms of protein phosphatase-1 catalytic subunits. J. Biol. Chem. 277, 27716–27724. [DOI] [PubMed] [Google Scholar]
- Terry-Lorenzo, R. T., Elliot, E., Weiser, D. C., Prickett, T. D., Brautigan, D. L., and Shenolikar, S. (2002b). Neurabins recruit protein phosphatase-1 and inhibitor-2 to the actin cytoskeleton. J. Biol. Chem. 277, 46535–46543. [DOI] [PubMed] [Google Scholar]
- Terry-Lorenzo, R. T., Inoue, M., Connor, J. H., Haystead, T. A., Armbruster, B. N., Gupta, R. P., Oliver, C. J., and Shenolikar, S. (2000). Neurofilament-L is a protein phosphatase-1-binding protein associated with neuronal plasma membrane and post-synaptic density. J. Biol. Chem. 275, 2439–2446. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Miller, A. L., Mooseker, M. S., and Koleske, A. J. (2001). The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc. Natl. Acad. Sci. USA 98, 14865–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal, R. S., Tavalin, S. J., Lin, J. W., Alto, N. M., Fraser, I. D., Langeberg, L. K., Sheng, M., and Scott, J. D. (1999). Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science 285, 93–96. [DOI] [PubMed] [Google Scholar]
- Yuste, R., and Bonhoeffer, T. (2001). Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu. Rev. Neurosci. 24, 1071–1089. [DOI] [PubMed] [Google Scholar]
- Yuste, R., and Bonhoeffer, T. (2004). Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat. Rev. Neurosci. 5, 24–34. [DOI] [PubMed] [Google Scholar]
- Zhang, W., and Benson, D. L. (2001). Stages of synapse development defined by dependence on F-actin. J. Neurosci. 21, 5169–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito, K., Knott, G., Shepherd, G. M., Shenolikar, S., and Svoboda, K. (2004). Induction of spine growth and synapse formation by regulation of the spine actin cytoskeleton. Neuron 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Ziv, N. E., and Garner, C. C. (2001). Principles of glutamatergic synapse formation: seeing the forest for the trees. Curr. Opin. Neurobiol. 11, 536–543. [DOI] [PubMed] [Google Scholar]
- Ziv, N. E., and Smith, S. J. (1996). Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17, 91–102. [DOI] [PubMed] [Google Scholar]