Summary
In Arctic regions, glaciers are major sources of iron to rivers and streams; however, estuaries are considered iron sinks due to the coagulation and flocculation processes that occur at higher salinities. It is unknown how iron dynamics in a glacial influenced river and estuary environment affect microbial mechanisms for iron acquisition. Microbial taxonomic and functional sequencing was performed on samples taken throughout the year from the Kenai River and the estuary, Alaska. Despite distinct iron, sodium, and other nutrient concentrations, the river and estuary did not have statistically different microbial communities nor was time of sampling significant. However, ferrous iron transport (Feo) system genes were more abundant in river environments, while siderophore genes were more abundant and diverse in estuary environments. Siderophore transport and iron storage genes were found in all samples, but gene abundance and distribution were potentially influenced by physical drivers such as discharge rates and nutrient distributions. Differences in iron metabolism between river and estuary ecosystems indicate environmental conditions drive microbial mechanisms to sequester iron. This could have implications for iron transport as the Arctic continues to warm.
Keywords: aquatic microbiology, metagenomics/community genomics, metals
Graphical Abstract
Microbial taxonomic and functional sequencing was performed on samples taken throughout the year from a glacial influenced river and the estuary in Alaska. Ferrous iron transport (Feo) system genes were more abundant in river environments, while siderophore genes were more abundant and diverse in estuary environments. Differences in iron metabolism between river and estuary ecosystems indicate environmental conditions drives microbial mechanisms to sequester iron.
Introduction
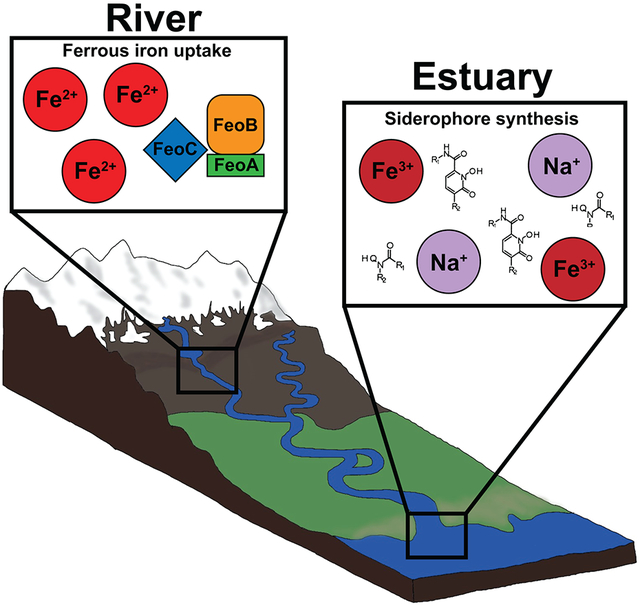
In high-latitude regions, glaciers and ice sheets are major sources of nutrients to oceans, rivers, and other bodies of water (Markussen et al., 2016; Laufer-Meiser et al., 2021). Deglaciation can alter downstream bodies of water changing biogeochemical, physical, and biological properties (van den Broeke et al., 2016). Alaskan water systems, such as the Gulf of Alaska, are especially susceptible to deglaciation with 47% of freshwater discharge coming from glaciers and icefields (Neal et al., 2010). Potential disturbances on aquatic biogeochemical cycling in Alaska could have catastrophic effects to aquatic food chains. Iron is one such element that can be significantly affected by deglaciation since glacial runoff is a main source of iron to downstream waters (Bhatia et al., 2013; Dierssen et al., 2002; Reisdorph and Mathis, 2015).
All living organisms require iron for various metabolic processes such as amino acid synthesis, nucleic acid synthesis, the citric acid cycle, and many others (Messenger and Barclay, 1983; Sandy and Butler, 2009). Most rivers, both glaciated and non-glaciated, have been found to have high concentrations of dissolved iron (ferrous and ferric) and are major sources of iron to oceans (Pokrovsky and Schott, 2002; Klunder et al., 2012; Li et al., 2019; Tashiro et al., 2020). Previous studies determined that dissolved iron in rivers is likely mixed iron oxide-organic matter colloids that are stabilized by dissolved organic matter, which prevents iron colloids from aggregating and coagulating (Pokrovsky and Schott, 2002). However, once rivers mix with saltwater environments, the negative charged iron colloids interact with seawater cations and form insoluble precipitates (Eckert and Sholkovitz, 1976; Boyle et al., 1977). Flocculation in estuaries then causes the iron to sink into the sediment and become less bioavailable for organisms (Markussen et al., 2016). Typically, less than 15% of the total iron within estuaries is in the bioavailable dissolved ferrous form (Schroth et al., 2014).
Microorganisms have several mechanisms to obtain iron. For example, some microbes sequester iron with a ferrous iron transport (Feo) system that allows soluble ferrous iron to freely diffuse through the cell membrane (Lau et al., 2016; Sestok et al., 2018). While some use intracellular proteins called ferritins to oxidized excessive ferrous iron (that can be potentially toxic) thus allowing it to be removed or stored as bioavailable ferric iron (Smith, 2004). However, when iron is in an insoluble form, microorganisms can obtain iron through organic ligands such as siderophores (Hunter and Boyd, 2007; Gledhill and Buck, 2012). Siderophores are low molecular weight chelators and could play a role in increasing iron solubility in aquatic ecosystems (Boyd et al., 2010; Gledhill and Buck, 2012; Yarimizu et al., 2014). For example, most iron (III) is bound to organic ligands in the ocean (Liu and Millero, 2002) and siderophores were found to play a role in supplying bioavailable iron in a creek that interacted with anoxic groundwater (Duckworth et al., 2009). Siderophores must enter cells through outer membrane receptors specific to the siderophore (Andrews et al., 2003; Wandersman and Delepelaire, 2004). The Exb gene family is a group of three outer membrane proteins, TonB, ExbB, and ExbD that transport siderophores into the periplasmic space (Andrews et al., 2003; Faraldo-Gómez and Sansom, 2003; Wandersman and Delepelaire, 2004). The Exb gene family is commonly referred to as the TonB complex and is the only known system for transporting siderophores in Gram-negative bacteria, and therefore is an indicator for siderophore uptake (Garber et al., 2020; Faraldo-Gómex and Sansom, 2003; Caza and Kronstad, 2013).
Siderophore production has been shown to be influenced by environmental conditions (Gärdes et al., 2013; Saha et al., 2016). One study showed vibrioferrin gene (pvsB) decreased with depth in the ocean while aerobactin (iucC) and petrobactin (asbE) increased (Gärdes et al., 2013). The results suggested sunlight might influence siderophore gene abundance. Studies have identified bacteria that produce photoactive siderophores such as vibrioferrin make up 1-9% of bacterial populations in the surface ocean (Saha et al., 2016). These vibrioferrin producing organisms also likely form mutualistic relationships with phytoplankton where bacteria provide photochemically reduced iron in exchange for algae metabolites (Saha et al., 2016). Environmental factors such as sunlight can influence microbial siderophore production; however, it is unknown which of these factors connect to potential shifts in iron acquisition between fresh and saltwater ecosystems.
The knowledge of what environmental factors influence iron-related gene distributions can greatly aid in understanding estuary and aquatic processes that ultimately deliver bioavailable iron to the ocean. This study seeks to determine the differences in iron related genes and associated taxonomy between fresh and saltwater environments, and to identify spatial, temporal, and physico-chemical influences on microbial siderophore gene abundance and distribution within river and estuary environments.
Experimental Procedures
Kenai River site description and sampling
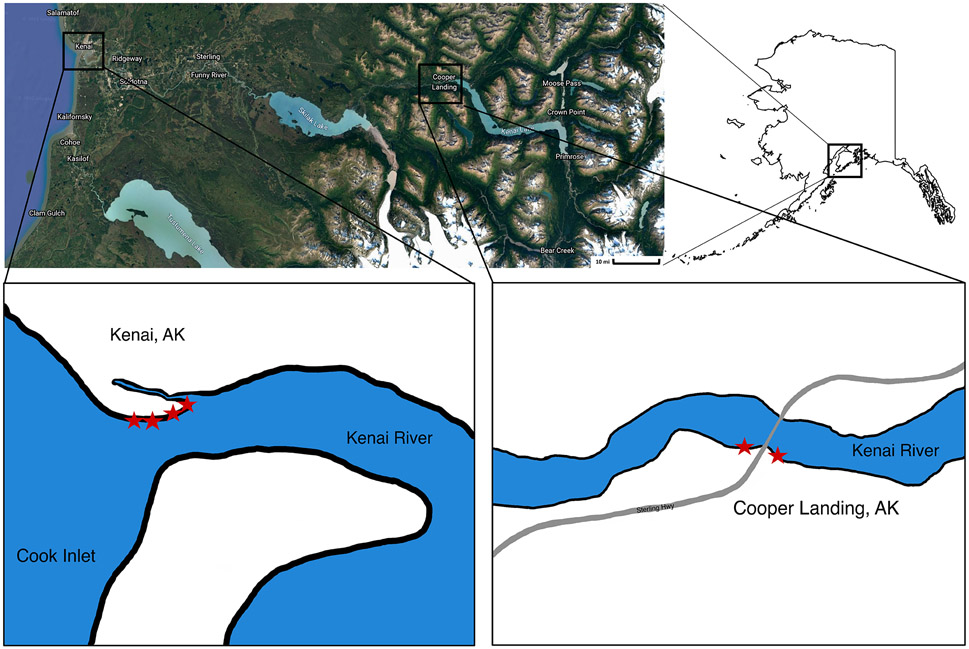
The Kenai River, the longest river on the Kenai Peninsula, is primarily glacial fed and empties into the Cook Inlet of the Gulf of Alaska. Sampling was conducted in October 2019 and in 2020 when the estuary was frozen (February), when the ice broke up (May), and in summer (August). At each time point, four samples were collected along the Kenai River Estuary (KE) at Kenai Beach, Alaska (60.550185, −151.262079) during high tide (Fig. 1) and two samples were collected in Cooper Landing (CL), Alaska (60.491982, −149.810386). The CL samples are upstream of KE (Fig. 1). At each site about 1L of water was filtered through a 0.22 μm pore-sized filter. A total of four filters were collected at each site and 50 mL of surface sediment. All collected samples were stored on dry ice during the return to the laboratory. A total of 24 water samples and 18 sediment samples were collected as sediment was not collected in October 2019. Water filters and sediment were stored at −80°C until DNA extraction.
Figure 1.
Map of sample locations (indicated by stars) on the Kenai Peninsula in southcentral Alaska. Lower left box is Kenai Estuary (KE) at Kenai Beach, Alaska (60.550185, - 151.262079) and lower right box is Cooper Landing (CL), Alaska (60.491982, - 149.810386).
Physico-chemical analysis
Temperature and pH were measured in-situ with a YSI meter. Water was collected at each site and filtered through 0.22 μm pore-sized filters and stored at 4°C. Ion chromatography (IC) was used to identify major ions at the University of Idaho Analytical Sciences for 2019 samples and University of Georgia Center for Applied Isotope Studies laboratories for 2020 samples. Another set of filtered water samples were stored with 1% nitric acid then analyzed at the above laboratories for trace metals with inductively coupled plasma mass spectrometry (ICP-MS).
16S rRNA gene sequencing and bioinformatic analysis
DNA was extracted from water filters and sediment using DNeasy PowerWater and PowerLyzer Soil kits from Qiagen following the manufacturer’s protocol (Qiagen, Valencia, CA, USA). Microbial community composition was analyzed by amplifying the 16S rRNA gene sequences in triplicate from extracted DNA of sediment and water samples. Primers 515F and 806R with Golay barcodes were used to target the V4 conserved region (Caporaso et al., 2011). Triplicate PCR products were verified using gel electrophoresis, combined, and purified using AMPure XP beads. Samples were multiplexed and sequenced using Illumina MiSeq Platform using the 300 cycle MiSeq Reagent kit V2 at the Advanced Instrumentation for Microbiome Studies (AIMS) core facility. Raw sequences from Illumina were processed using QIIME2 (Bolyen et al., 2019). Sequences were demultiplexed, quality filtered (Phred score of 20), and denoised using DADA2 (Callahan et al., 2016). Amplicon sequence variants (ASVs) were assigned taxonomy with the feature-classifier (Bokulich, 2018) using the Silva 138 reference database (Quast et al., 2013).
Further processing to ensure high-quality sequence data was performed in R (Version 4.0.4) (Team, 2016). Samples were rarified to 9,500 total reads and samples with less were removed from further analyses. ASVs that had less than five reads were also removed. An outlier analysis was also performed using the interquartile range method in the outliers package in R and any sample that was more than 2 standard deviations from the mean was removed (Komsta, 2022).
Metagenomic gene sequencing and bioinformatic analysis
Metagenomic sequencing was performed on water samples collected throughout 2020. The October 2019 samples did not yield enough DNA for metagenomic analysis. Extracted DNA from the filtered water was fragmented by the Covaris ultra-sonicator, shearing to 500 bp and size checked with an Agilent TapeStation. DNA was then ligated and multiplexed with the Ovation Ultralow Library System V2 (Tecan, CA). Final products were quantified using Qubit and Agilent TapeStation. Using Illumina MiSeq Platform at AIMS, samples were sequenced using the 600 cycle MiSeq Reagent kit V3. Multiplexing of four to six samples at a time were run to produce about 2 GB of data per sample.
Illumina raw reads were checked with FastQC (Andrews, 2010) and quality controlled using Trimmomatic (Bolger et al., 2014). Sequenced data were merged and assembled into longer contigs using MetaSPAdes (Nurk et al., 2017). Prodigal (Hyatt et al., 2010) was used to detect genes and transcribe them to proteins after assembly. The proteins were annotated with DIAMOND (Buchfink et al., 2015). MEGAN was used to visualize both taxonomic and functional results (Huson et al., 2007). Annotations were mapped to the SEED (Overbeek et al., 2014) database for functional annotation in MEGAN (Huson et al., 2007).
Metagenomic assemblies were also processed using FeGenie (Garber et al., 2020) to identify iron related genes. FeGenie uses Hidden Markov Models (HMM) to find iron acquisition, iron gene regulation, iron oxidation and reduction, iron storage, and magnetosome-related gene categories (Garber et al., 2020). The percentage of each iron gene abundance was calculated by the number of genes detected divided by total reads for each sample.
Metagenome Assembled Genomes
Assembled metagenomic contigs were binned using MetaBAT2 (Kang et al., 2019) and quality filtered to 75% completeness and 10% contamination thresholds using CheckM (Parks et al., 2015). MAGs were classified with Genome Taxonomy Database (GTDB-Tk v2) (Chaumeil et al., 2022) and annotated with Rapid Annotation using Subsystem Technology (RASTtk) (Aziz et al., 2008; Brettin et al., 2015).
Vibrioferrin (pvsB) gene quantification
To determine if photoreactive siderophores such as Vibrioferrin were quantifiable and potentially missed with the sequencing depth, the vibrioferrin biosynthesis gene (pvsB) was targeted. Extracted DNA from water and sediment samples were amplified in triplicate for the pvsB siderophore gene using real-time qPCR. Primers previously developed to target potential bacterial-algal mutualism in Marinobacter species (marpvsB) and all other non-Marinobacter species (vibXpvsB) were used as previously described (Gärdes et al., 2013). Marinobacter hydrocarbonoclasticus ATCC 49840 (Gauthier et al., 1992) and Vibrio parahaemolyticus ATCC 17802D-5 (Sakazaki et al., 1963; Fujino et al., 1974) were used as standards for the corresponding vibrioferrin primer to calculate corresponding copy numbers. Samples out of range from the standards were removed from analysis.
Statistical Analysis
Principal coordinate analysis (PCoA) with a Bray-Curtis dissimilarity matrix was performed with the vegan-package and ggplot2 (Dixon, 2003; Wickham, H., 2016; Andersen et al., 2018). Multiple response permutation procedure (MRPP) was conducted to test significant differences between groups of samples using Vegan (version 2.4-2) (Dixon, 2003). Indicator taxa analysis based on location of sample (KE or CL) or groups defined in the PCoA plot was performed with the vegan package. Taxa was considered a significant indicator if the p-value was less than 0.05 and indicator value was above 0.9. Iron related genes, physico-chemical data, and vibrioferrin gene copy number were analyzed using an ANOVA test and checked with Tukey multiple pairwise-comparisons with the ggpubr package (Kassambara, A, 2020). A p-value less than 0.05 was considered significant. All figures were created using R (Version 4.0.4) (Team, 2016).
Results
Physico-chemical properties
Significant differences in physico-chemical properties between KE and CL were found. KE samples had significantly more total iron and sodium than CL samples (Table 1). Anions (chloride, nitrate, and sulfate) were significantly higher in KE samples. While other anions bromide, fluoride, and phosphate showed no significant difference between locations. Other nutrients including calcium, potassium, magnesium, molybdenum, and vanadium were also elevated in KE samples. Temperature and pH showed no significant difference between KE and CL.
Table 1:
Physico-chemical data for each sample collected in Kenai Estuary (KE) and Cooper Landing (CL). All data are average ± SE.
| February KE (N=4) | February CL (N=2) | May KE (N=4) | May CL (N=2) | August KE (N=4) | August CL (N=2) | October KE (N=4) | October CL (N=2) | |
|---|---|---|---|---|---|---|---|---|
| Iron (ppm) | 0.17 ± 0.07 | 0.10 ± 0.00 | 1.15 ± 0.12 | 0.28 ± 0.00 | 0.74 ± 0.18 | 0.26 ± 0.01 | 0.70 ± 0.27 | 0.10 ± 0.01 |
| Sodium (ppm) | 6615.00 ± 410.94 | 9.80 ± 0.00 | 1968.09 ± 52.24 | 2.63 ± 0.65 | 1488.87 ± 212.74 | 2.21 ± 0.17 | 5708.50 ± 1130.46 | 9.80 ± 0.00 |
| pH | 7.31 ± 0.18 | 7.31 ± 0.11 | 6.91 ± 0.02 | 7.2 ± 0.14 | 7.49 ± 0.23 | 7.21 ± 0.16 | 7.8 ± 0.25 | 7.6 ± 0.02 |
| Temperature (°C) | (−)1.35 ± 0.05 | 0.95 ± 0.15 | 8.48 ± 0.68 | 6.35 ± 0.25 | 13.73 ± 0.03 | 13.5 ± 0.2 | 6.8 ± 0.30 | 6.35 ± 0.15 |
| Barium (ppm) | 0.02 ± 0.0 | 0.02 ± 0.0 | 0.01 ± 0.0 | 0.01 ± 0.0 | 0.01 ± 0.0 | 0.01 ± 0.0 | 0.02 ± 0.0 | 0.02 ± 0.0 |
| Calcium (ppm) | 257.25 ± 17.61 | 12.74 ± 0.0 | 150.36 ± 21.67 | 8.12 ± 0.42 | 80.41 ± 27.47 | 9.61 ± 2.06 | 192.57 ± 36.17 | 12.25 ± 0.49 |
| Cadmium (ppm) | 0.02 ± 0.0 | 0.02 ± 0.0 | 0.00 ± 0.0 | 0.0001 ± 0.0 | 0.00009 ± 0.0 | 0.0001 ± 0.0 | 0.02 ± 0.0 | 0.02 ± 0.0 |
| Cobalt (ppm) | 0.01 ± 0.0 | 0.01 ± 0.0 | 0.0005 ± 0.0 | 0.0004 ± 0.0 | 0.0004 ± 0.0 | 0.0004 ± 0.0 | 0.01 ± 0.0 | 0.01 ± 0.0 |
| Chromium (ppm) | 0.06 ± 0.0 | 0.05 ± 0.0 | 0.0046 ± 0.0 | 0.0002 ± 0.0 | 0.004 ± 0.0 | 0.0005 ± 0.0 | 0.05 ± 0.0 | 0.05 ± 0.0 |
| Copper (ppm) | 0.02 ± 0.0 | 0.02 ± 0.0 | 0.02 ± 0.01 | 0.03 ± 0.03 | 0.03 ± 0.01 | 0.001 ± 0.00 | 0.02 ± 0.0 | 0.02 ± 0.0 |
| Potassium (ppm) | 247.45 ± 17.61 | 1.18 ± 0.2 | 198.77 ± 32.62 | 0.92 ± 0.07 | 102.97 ± 39.04 | 0.86 ± 0.15 | 213.40 ± 41.60 | 1.42 ± 0.25 |
| Magnesium (ppm) | 766.85 ± 54.47 | 1.13 ± 0.05 | 1003.96 ± 162.46 | 1.22 ± 0.07 | 531.72 ± 200.08 | 1.06 ± 0.01 | 539.00 ± 98.25 | 0.96 ± 0.03 |
| Manganese (ppm) | 0.036 ± 0.03 | 0.005 ± 0.0 | 0.022 ± 0.02 | 0.001 ± 0.0 | 0.003 ± 0.0 | 0.003 ± 0.0 | 0.03 ± 0.01 | 0.005 ± 0.0 |
| Molybdenum (ppm) | 0.25 ± 0.0 | 0.25 ± 0.0 | 74.90 ± 10.09 | 3.47 ± 0.10 | 38.60 ± 13.57 | 3.92 ± 0.58 | 0.25 ± 0.0 | 0.25 ± 0.0 |
| Nickel (ppm) | 0.05 ± 0.0 | 0.05 ± 0.0 | 0.006 ± 0.0 | 0.002 ± 0.0 | 0.004 ± 0.0 | 0.002 ± 0.0 | 0.05 ± 0.0 | 0.05 ± 0.0 |
| Vanadium (ppm) | 0.02 ± 0.0 | 0.02 ± 0.0 | 0.13 ± 0.01 | 0.01 ± 0.00 | 0.07 ± 0.02 | 0.005 ± 0.0 | 0.02 ± 0.0 | 0.02 ± 0.0 |
| Zinc (ppm) | 0.04 ± 0.00 | 0.02 ± 0.00 | 0.005 ± 0.00 | 0.009 ± 0.01 | 0.01 ± 0.00 | 0.006 ± 0.00 | 0.045 ± 0.01 | 0.02 ± 0.00 |
| Fluoride (ppm) | 4.07 ± 2.20 | 0.43 ± 0.04 | 4.15 ± 1.93 | 0.24 ± 0.12 | 61.59 ± 12.90 | 0.69 ± 0.07 | 0.29 ± 0.21 | 0.0 ± 0.00 |
| Chloride (ppm) | 16901.91 ± 7003.54 | 1.4 ± 0.4 | 18592.22 ± 3356.14 | 2.06 ± 0.07 | 9918.38 ± 3412.75 | 1.54 ± 0.17 | 8450 ± 1862.57 | 2.20 ± 1.41 |
| Nitrite-n (ppm) | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Bromide (ppm) | 4.06 ± 1.55 | 1.77 ± 1.02 | 4.08 ± 1.27 | 2.36 ± 2.11 | 1.25 ± 0.39 | 0.10 ± 0.01 | 37.3 ± 10.22 | 0.00 ± 0.00 |
| Nitrate-n (ppm) | 25.00 ± 8.61 | 1.38 ± 0.73 | 28.02 ± 4.60 | 3.81 ± 2.59 | 27.67 ± 6.42 | 1.11 ± 0.01 | 0.23 ± 0.08 | 0.15 ± 0.15 |
| o-phosphate-p (ppm) | 17.6 ± 2.10 | 0.21 ± 0.02 | 22.66 ± 13.62 | 0.29 ± 0.29 | 5.37 ± 4.25 | 0.13 ± 0.08 | 0.08 ± 0.08 | 0.0 ± 0.0 |
| Sulfate (ppm) | 2200.21 ± 833.50 | 15.22 ± 6.81 | 2139.60 ± 369.78 | 20.33 ± 3.39 | 1202.60 ± 402.01 | 18.96 ± 0.12 | 1340 ± 299.89 | 13.5 ± 0.50 |
Samples collected in February at KE contained the lowest iron concentration (0.17 mg/L) with the highest sodium concentration (6615 mg/L) (Table 1). Iron concentrations increased in May (1.15 mg/L) and then decreased throughout summer in August (0.74 mg/L). CL samples had the lowest average iron and sodium concentrations compared to samples collected in KE, however similar temporal shifts were observed in CL samples. February CL samples contained the lowest iron concentrations (0.10 mg/L) that increased in May (0.28 mg/L) and decreased slightly but not significantly throughout summer in August (0.26 mg/L).
Microbial community structure
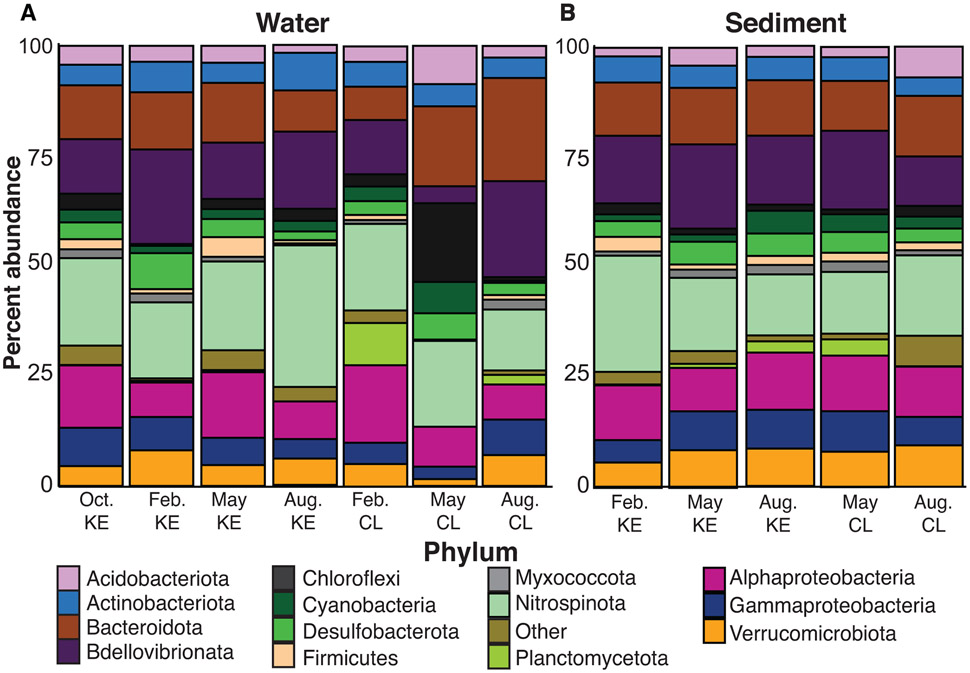
Gene sequencing of the 16S rRNA gene produced 7,152,113 sequences from 42 samples. After rarefying, 26 samples including water and sediment remained with 7,131,303 sequences. There was a total of 25,533 ASVs that were assigned to a total of 74 phyla which included over 10 candidate phyla. An average of 2,858 ASVs were detected at each site. The bacterial phylum Proteobacteria (41.9%) including Alpha- (19.9%), Gamma- (79.9%), and Zeta- (<1%) were the largest percentage of bacterial sequences for all sites (Fig. 2). The next most abundant phyla were Bacteroidota (16.6%), Actinobacteriota (7.4%), and Verrucomicrobiota (5.8%). KE samples contained higher relative abundance of Proteobacteria (33.7%), Bacteridota (18.2%), Actinobacteriota (7.3%), and Verrucomicrobiota (6.5%) than CL samples. This differed from CL samples which had higher relative abundance of Firmicutes (10.3%), Planctomycetota (6.5%), and Acidobacteriota (3.5%) (Fig. 2).
Figure 2.
Percent abundance of phyla detected from 16S rRNA gene sequencing. Samples shown are location of sampling; Cooper Landing (CL) and Kenai Estuary (KE) and month of sampling; October, February, May, and August. (A) Water samples (B) Sediment samples.
October samples had slightly higher relative abundance of Actinobacteriota (7.8%) and Verrucomicrobiota (7.4%) than other months. Both phyla decreased in February, resulting in February having the lowest Verrucomicrobiota abundance (4.2%). February samples contained the highest relative abundance of Proteobacteria (37.6%) then decreased to a low of 26.4% in May. February also contained the overall lowest amount of Cyanobacteria (1.8%). May samples showed an increase in Firmicutes (7.8%), almost three times more compared to other sampling months. The samples taken in August had higher Proteobacteria (30.7%), Bacteroidota (18.5%), and Actinobacteriota (7.6%) compared to May. August samples also contained the overall highest abundance of Cyanobacteria (7.8%).
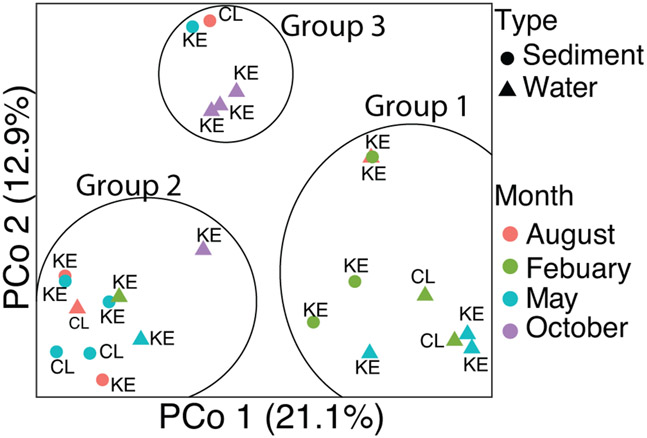
The PCoA plot had three distinct groups but there was no statistically significant correlation with physico-chemical measurements. In addition, both time and location were not significantly different based on MRPP results (Fig. 3). However, most of February samples were clustered in Group 1, most of August samples were in Group 2, and most of October samples were in Group 3. May samples were spread across all groups. Indicator taxa analysis identified taxa that were predominantly found within each group. Indicator genera for Group 1 were Woeseia and Nitrosopumilus. Three other unclassified genera within the Kangiellaceae and Rhodobacteraceae families and an unclassified genus within the Gammaproteobacteria class were also predominantly only found in Group 1. Group 2 contained the genera Planktophila and Bradymonadales as well as unclassified genera in the families Sporichtyaceae, Comamonadaceae, and an unclassified Actinobacteria. Group 3 contained the genera Geobacter, Terrimonas, Desulfoporsinus, and an unclassified genus in the Microbacteriaceae family. No significant indicator taxa was detected when comparing KE to CL.
Figure 3.
Principal coordinate analysis (PCoA) plot of the surface water and sediment microbial community structure of the Kenai River. Colors indicate month of sampling collection. Shapes indicate the type of sample; sediment or water.
FeGenie detected gene families
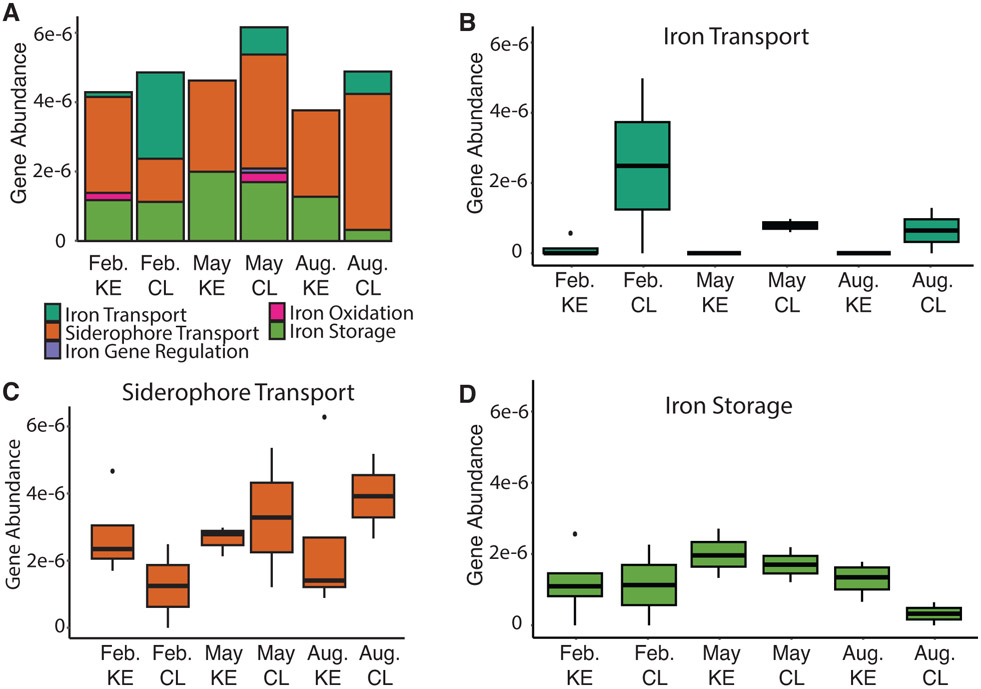
Over 38 million metagenomic reads were obtained after trimming and quality filtering. After assembly, the 18 water samples from 2020 had an average N50 score of 413 with a range from 403 to 535 and a total of 1,057,665 sequences. Prodigal detected a total of 2,716,040 proteins. Of those proteins, FeGenie detected a total of 190 iron-related genes from iron acquisition (67.4%), iron storage (30.5%), iron oxidation (1.6%), and iron gene regulation (0.5%) categories (Fig. 4A). One sample (collected from KE in May) did not have any iron genes detected and was removed from further analysis. FeGenie detected genes fluctuated based on month and location of sample. CL samples contained the highest abundance of total iron related genes with the most detected in May (Fig. 4A).
Figure 4.
The abundance of FeGenie detected iron genes from metagenomic water samples collected during 2020. The percentage of each sample was out of the total sequences after trimming and quality filtering. Samples shown are location of sampling; Cooper Landing (CL) and Kenai Estuary (KE) and month of sampling; February, May, and August. (ns = not significant) (A) Iron gene category percent abundance for each sample. (B) Iron acquisition: Iron transport gene abundance. (C) Iron acquisition: Siderophore transport gene abundance. (D) Iron storage gene abundance.
Within the iron acquisition category, siderophore transport (60%) and iron (II)/(III) transport (7.4%) were the only two detected. The most abundant protein families detected within siderophore transport were ExbB (40.5%) and ExbD (42.2%). These proteins play a role in the TonB complex and transport siderophores across gram negative bacteria membranes. There was no significant difference between CL and KE siderophore transport gene abundance (Fig. 4C). However, siderophore transport gene abundance increased throughout the year in CL samples, while KE samples slightly decreased throughout the year (Fig. 4C). The decrease resulted in the lowest abundance of siderophore transport genes of the KE samples in August which also contained the lowest abundance of total iron genes (Fig. 4A,C).
Within iron transport, 71.4% of genes belonged to the ferrous iron transport (Feo) system. Iron (II)/(III) transport genes were highest in February in CL and decreased to the lowest abundance in August (Fig. 4B). Iron transport genes in KE was only detected in February. Although slight but not significant differences between samples were observed.
The second most abundant iron related genes detected were iron storage and were found in all samples. KE samples contained a higher abundance of iron storage genes compared to CL samples (Fig. 4A). Samples taken in May contained the highest abundance of iron storage genes for both KE and CL. The lowest abundance of iron storage genes for KE and CL were February and August, respectively (Fig. 4D).
Siderophore synthesis genes
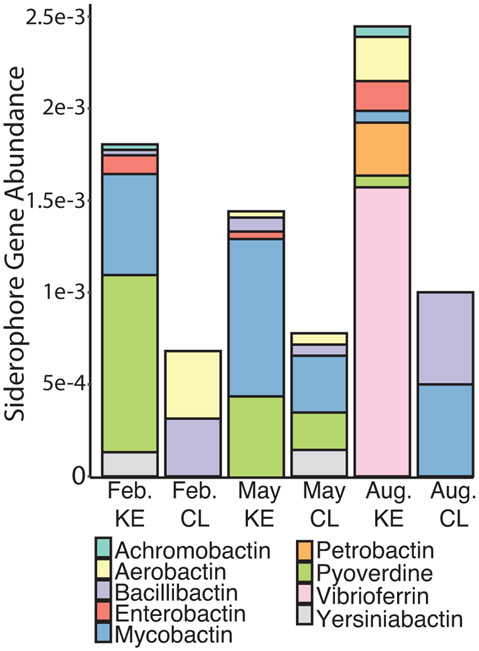
FeGenie contains limited HMM for siderophore synthesis therefore SEED was used to further annotate. Annotations based on SEED database had 1105 iron acquisition and metabolism genes from all water samples. Of the total genes detected 257 belonged to the siderophore category making up 23.3% of detected genes. A total of nine siderophore gene types were detected, with the most abundant being mycobactin (27.9%), pyoverdine (20.5%), and vibrioferrin (19.3%) (Fig. 5).
Figure 5.
Siderophore gene abundance for each sample annotated with the SEED database. Samples shown are location of sampling; Cooper Landing (CL) and Kenai Estuary (KE) and month of sampling; February, May, and August.
Siderophore synthesis genes were detected in both CL and KE samples. CL samples had the lowest abundance and diversity of siderophore synthesis genes. Of the nine siderophore synthesis genes, only five were detected in CL samples (Fig. 5). In KE samples, siderophore synthesis gene abundance and diversity increased and contained all nine detected siderophores.
Siderophore synthesis gene abundance and diversity also differed by month of sampling. February CL contained the lowest abundance of siderophore synthesis genes from all samples. May KE samples contained the lowest abundance and diversity of siderophore synthesis genes and the highest total iron concentration of the KE samples (Fig. 5). August KE contained the highest abundance and diversity of all samples, containing seven of the nine siderophores (Fig. 5). August CL had the highest abundance of siderophore genes while May CL had the highest diversity of the CL samples.
Vibrioferrin gene detection and quantification
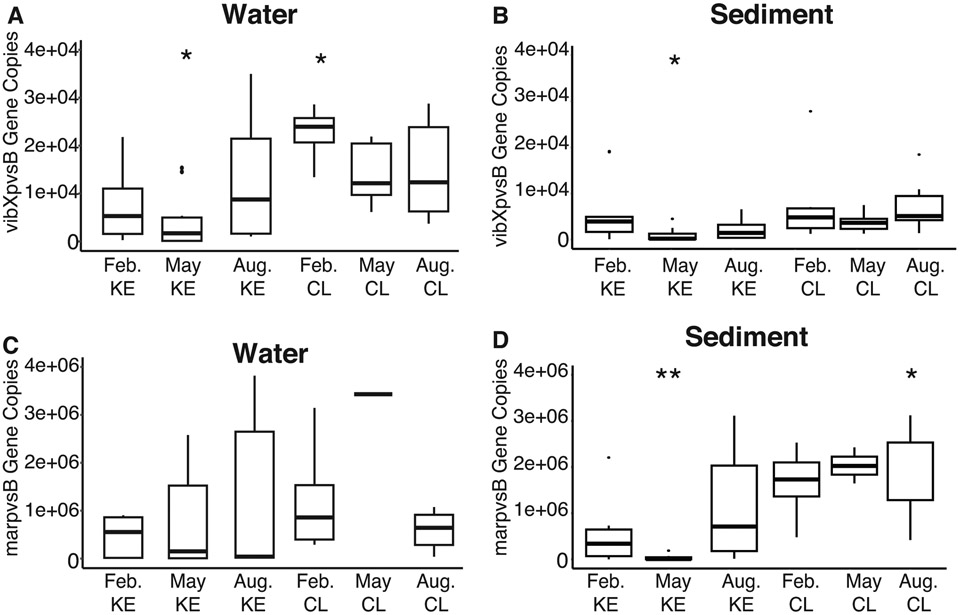
Vibrioferrin gene copies were found in all sediment and water samples. Marinobacter spp. and Vibrio spp. specific primers detected vibXpvsB and marpvsB gene copy numbers in both surface water and sediment DNA samples. February CL water samples had significantly more vibXpvsB gene copy numbers compared to the other water samples (Fig. 6A). May KE had significantly lower vibXpvsB gene copy numbers in both water and sediment samples (Fig. 6 A,B). No significant differences between water samples were found between marpvsB gene copies (Fig. 6C). However, sediment samples did contain differences between samples. August CL samples contained significantly higher marpvsB gene copies while May KE contained significantly lower compared to sediment samples (Fig. 6D).
Figure 6.
Vibrioferrin (pvsB) gene copy number detected in each sample. Asterisk (*) indicates significant p-value. The number of asterisks represent smaller p-values (*0.05, **0.01). (A) vibXpvsB gene detected in water samples. (B) vibXpvsB gene copy number detected in sediment samples. (C) marpvsB gene copy number detected in water samples. (D) marpvsB gene copy number detected in sediment samples.
Metagenome Assembled Genomes (MAGs)
KE water surface samples had 16,222 input contigs with 12,856 being binned. There were 31 bins detected with six remaining after filtering (Table 2). All the MAGs except one were in the bacteria domain while the other was an archaea in the Thermoproteota phyla. The bacterial MAGs all belonged to the Proteobacteria phyla including Gamma- (80%) and Alpha- (20%) aligning with other taxonomic results (Fig. 2). Ferrous iron transporters (Feo) and ferric siderophore transport system genes were found in half of the KE bins. A Gammaproteobacteria Rheinheimer sp. (KE Bin028) contained ferric siderophore transport system genes and no Feo genes. While an Alphaproteobacteria Brevundiimonas sp. (KE Bin016) contained Feo genes and no siderophore transport genes.
Table 2:
Metagenome assembled genomes (MAGs) for Kenai Estuary (KE) and Copper Landing (CL) samples.
| Genome | Completion (%) |
Contamination (%) |
Contigs | Length (Mb) |
GC (%) | Taxonomy | FEO | FSTS |
|---|---|---|---|---|---|---|---|---|
| KE Bin011 | 81.4 | 0.97 | 142 | 1.07 | 32.1 | Archaea; Thermoproteota; Nitrosopumilus | 0 | 0 |
| KE Bin012 | 94.2 | 2.22 | 308 | 3.13 | 37.4 | Bacteria; Gammaproteobacteria; Colwellia | 2 | 1 |
| KE Bin016 | 87.0 | 1.96 | 427 | 2.33 | 67.1 | Bacteria; Alphaproteobacteria; Brevundimonas | 1 | 0 |
| KE Bin025 | 95.9 | 1.38 | 200 | 4.15 | 56.4 | Bacteria; Gammaproteobacteria; SG8-40 | 2 | 1 |
| KE Bin028 | 85.5 | 2.42 | 505 | 2.60 | 48.2 | Bacteria; Gammaproteobacteria; Rheinheimera | 0 | 2 |
| KE Bin031 | 96.3 | 6.83 | 317 | 3.82 | 48.8 | Bacteria; Gammaproteobacteria; GCA-001735895 | 0 | 0 |
| CL Bin001 | 92.7 | 1.36 | 177 | 2.73 | 61.5 | Bacteria; Chloroflexota; BD1 | 3 | 0 |
| CL Bin002 | 87.5 | 6.62 | 422 | 2.65 | 50.5 | Bacteria; Gammaproteobacteria; Limnohabitans | 2 | 1 |
| CL Bin013 | 82.7 | 0.1 | 287 | 2.16 | 53.6 | Bacteria; Planctomycetota; SYAC01 | 1 | 0 |
| CL Bin014 | 87.8 | 1.28 | 205 | 1.65 | 47.8 | Bacteria; Actinobacteriota; BACL27 | 1 | 0 |
| CL Bin016 | 78.5 | 2.16 | 434 | 2.22 | 45.1 | Bacteria; Gammaproteobacteria; Polynucleobacter | 0 | 1 |
| CL Bin017 | 80.6 | 1.14 | 492 | 2.22 | 51.7 | Bacteria; Planctomycetota; Phycisphaerales | 1 | 1 |
| CL Bin023 | 96.6 | 1.21 | 32 | 1.18 | 31.0 | Archaea; Thermoproteota; Nitrosarchaeum | 0 | 0 |
FEO: Ferrous iron transport gene number
FSTS: Ferric siderophore transport gene number
CL water surface samples had 13,145 input contigs with 10,699 being binned. There were 34 bins detected with 7 remaining after filtering (Table 2). One of the seven MAGs was in the archaea domain within the Thermoproteota phyla. The other six MAGs were within the domain bacteria including Chloroflexota, Actinobacteriota, Planctomycetota, and Proteobacteria. Most of the CL bins contained iron metabolism genes for ferrous iron transporters. While less than half of the CL bins had ferric siderophore transport system genes. CL Bin028 (Gammaproteobacteria: Polynucleobacter sp.) was the only CL bin that had ferric siderophore transporters and no ferrous iron transporters.
Discussion
Iron is a critical cofactor for many metabolic processes and in Arctic regions iron availability can be dependent on glacial processes. In particular, river systems influenced by glacial retreat can have significantly altered iron transport dynamics. Modeled and long-term monitoring of sediment transport from retreating glaciers shows an increase of sediment discharge as the glaciers retreat due to more exposed weathered rock (Delaney et al., 2020; Koppes et al., 2015). This sediment can have high concentrations of iron (Hopwood et al., 2014; Li et al., 2019). In freshwater ecosystems, organisms typically use bioavailable ferrous iron to meet metabolic needs. However, most iron is lost when rivers connect with saltwater in many estuaries (Schroth et al., 2014; Zhang et al., 2015). For example, despite increased sediment discharge in a Greenlandic fjord, iron export did not increase suggesting that in-fjord processes were still effective at removing iron (Hopwood 2016). Decreased iron solubility caused by estuary mixing processes may cause microorganisms to produce siderophores, a possible evolutionary advantage in environments where associated cations facilitate iron loss through aggregation and flocculation (Vraspir and Butler, 2009; Hider and Kong, 2010; Saha et al., 2016). Most siderophores have been identified across the world’s ocean, including the North Pacific however, little is known about siderophores in Alaskan tributaries such as river and estuary ecosystems (Gärdes et al., 2013; Chen et al., 2019; Bundy et al., 2018). The connection between iron transport and microbial produced siderophores is not constrained in high-latitude regions that are experiencing rapid glacial retreat. This project identified iron related genes and associated taxonomy in river and estuary ecosystems. Results from this study identified key spatial, temporal, and environmental parameters that could influence siderophore gene abundance, distribution, and diversity. This information is key in understanding global warming effects on iron biogeochemistry.
Microbial taxonomic diversity
Incremental changes between microbial communities could contribute to differences in nutrients and iron related genes. Proteobacteria including Pseudomonas and Vibrio, Cyanobium within Cyanobacteria, Micrococcus within Actinobacteria, and Bacteroidota were more abundant in KE samples (Fig. 2). These microbes have previously been identified as siderophore producing microorganisms (Debeljak et al., 2019; Hopkinson and Morel 2009). The increase in siderophore synthesis gene abundance and diversity in KE samples could be associated with the increase in known siderophore producing microbes. Previous studies have also found Pseudomonas, Vibrio, and Micrococcus were slightly halophilic having salt tolerance of 2 to 5% (Surendran et al., 1983). All these organisms were found more abundant in KE samples than CL samples. CL samples contained a higher abundance of Bacillus within the Firmicutes phylum (Fig. 2). In addition to increases in Firmicutes, CL samples contained the highest abundance of the siderophore bacillibactin genes (Fig. 5). This catecholic siderophore has been identified as a component to iron acquisition in Bacillus subtilis and Bacillus cereus (May et al., 2001; Segond et al., 2014; Mengjie Zhou et al., 2018). Acknowledging that the short V3 region of the 16S rRNA gene did not allow for taxonomic assignment to the species level and there are species level differences, indicator taxa identified in Group 1 and Group 3 have close relatives that are known to produce siderophores. For example, in Group 1 there are microbes within the Rhodobacteraceae that are known to produce siderophores that may encourage epiphytic lifestyles with macroalgae (Dogs et al., 2017). Also, in Group 1 the genera Nitrosopumilus has species such as maritimus that can’t make siderophores but can uptake the siderophore desferrioxamine (Shafiee et al., 2019). In Group 3 species within the genera Geobacter, Terrimonas and the family microbacteriaceae all have been shown to produce siderophores (Kumari et al, 2020; Corretto et al., 2020; Holmén et al, 1999). Indicator taxa within Group 2 is unknown if they produce or transport siderophores; however, one of them, Bradymonadales, is a bacterial predator and likely doesn’t produce siderophores (Mu et al., 2020). Microbial community composition seems to contribute to iron related genes, physio-chemical properties, and siderophore gene abundance and diversity in both fresh and saltwater.
The most abundant phyla detected within the microbial communities and MAGs, was Proteobacteria with Gammaproteobacteria being the most abundant class. Gammaproteobacteria have shown dominance of TonB ferric siderophore uptake systems for catecholate, hydroxamate, and citrate siderophores compared to Flavobacteria, Alphaproteobacteria, and Cyanobacteria (Fig. 2, Table 2) (Hopkinson and Barbeau 2012; Desai et al., 2012). One detected Gammaproteobacteria, Colwellia, has previously been found highly abundant in environments with iron limiting conditions (Mason et al., 2014). Colwellia species lack genes for siderophore synthesis but do contain siderophore transport mechanisms giving them the ability to uptake siderophores produced by other microbes (Mason et al., 2014). Colwellia was identified in KE samples (KE Bin012) and had an indicator value of 0.7 (p <0.05) for Group 1. This MAG contained ferric siderophore transport system genes (Table 2). Other Proteobacteria such as Escherichia coli, possess transport systems for hydroxamate exogenous siderophores, not typically produced by this bacterium (Hider and Kong, 2010). Another phylum, Cyanobacteria, were found in all samples with August having the highest abundance (Fig. 2). Some Cyanobacteria can produce hydroxamate siderophores (Jiang et al., 2015; Řezanka et al., 2018; Årstøl and Hohmann-Marriott, 2019). Synechocystis sp. PCC 6803 is a cyanobacterium that cannot produce siderophores, however, was found to have three ExbB-ExbD clusters indicating a potential evolutionary advantage to maintaining siderophore transport systems (Jiang et al., 2015). Gram-negative bacteria seem to maintain siderophore transport systems for iron acquisition. The presence of siderophore transport systems with the absence of siderophore production genes in these MAGs implies potential microbial relationships in obtaining iron. A few microorganisms could have the ability to produce siderophores that others nearby can uptake with siderophore transport genes. This would also be energy efficient for surrounding microorganisms to not waste materials and energy to produce siderophores.
Iron acquisition genes
Data showed potential shifts in iron acquisition genes between environments with low and high sodium. Iron acquisition in these two aquatic environments relies on two different strategies: iron transport using the Feo system and siderophores depending on the form of iron (Vraspir and Butler, 2009; Lau et al., 2016; Sestok et al., 2018; Chen et al., 2019). The Feo system is thought to be conserved in many species and is dedicated to ferrous iron transport (Lau et al., 2016; Sestok et al., 2018). Samples with low sodium concentrations taken in freshwater environments (CL) all contained iron transport genes with the most abundant being the Feo system (Fig. 4B). The CL samples containing Feo system genes contained low siderophore genes compared to KE samples (Fig. 5). Additionally, more of the CL MAGs contained ferrous iron transport genes compared to ferric siderophore transport genes (Table 2). It is possible ferrous iron is more available in CL freshwater environments which could contribute to the higher abundance of iron transport genes. While the reverse was shown in KE samples where more siderophore genes were present in samples lacking iron transport genes. Studies have found the Feo uptake system is a distinct pathway from ferric iron uptake and is important for uptake when ferrous iron is bioavailable (Andrews et al., 2003; Cartron et al., 2006). The abundance of Feo genes in freshwater samples and absence in saltwater could represent potential metabolic switches. As sodium increases in KE samples, iron becomes less soluble, forcing microorganisms to rely on siderophores. Differences in iron acquisition genes between river and estuary systems could imply sodium concentrations and associated ions influence how microbes acquire iron in aquatic environments.
Siderophore diversity and distribution
Although iron transport genes were higher in environments with lower sodium, siderophore and transport genes were found in all samples regardless of sodium concentrations (Fig. 4). Siderophores cannot diffuse through cell porins thus need to enter through outer membrane receptors (Andrews et al., 2003; Wandersman and Delepelaire, 2004). The Exb gene family (commonly referred to as the TonB complex) is composed of three proteins TonB, ExbB, and ExbD found in the cytoplasmic membrane (Wandersman and Delepelaire, 2004). TonB uses a proton-motive force to activate surface receptor proteins on the outer membrane (Faraldo-Gómez and Sansom, 2003). Siderophores bound to iron (III) are moved across the inner membrane into the cytoplasm with an ABC transporter (Noinaj et al., 2010). The ferric iron is then reduced to ferrous iron for use by the cell (Noinaj et al., 2010). Outer membrane transporters can vary based on the microorganism and the siderophore being received. Microorganisms living in aquatic environments likely have evolved multiple siderophore iron acquisition gene clusters and gram-negative bacteria maintained TonB complexes in response to iron needs (Noinaj et al., 2010; Thode et al., 2018).
Iron concentrations and siderophore genes were lowest in February while iron transport genes were the highest. When the estuary was frozen Kenai River experienced a low discharge rate (~711 ft3/s) and lower temperature (U.S. Geological Survey, 2020). The low discharge is most likely contributing to the low iron concentration since glacial runoff is often a significant source of iron to waters (Bhatia et al., 2013). February was the only time that iron transport genes were detected in KE samples (Fig. 4B). In addition to increases in iron transport genes, Zetaproteobacteria were identified in February and May samples which also contained iron oxidation genes (Fig. 4A). Zetaproteobacteria are marine iron oxidizing bacterium that play a role in iron cycling (Makita et al., 2017; McAllister et al., 2019). Iron concentrations in estuaries have previously been influenced by salinity, temperature, pH, and many other physico-chemical properties but can also be influenced by biotic interactions such as iron oxidizing bacteria (Daneshvar, 2015). It is also possible that under these low discharge and temperature conditions iron could be in a soluble ferrous form (Tashiro et al., 2020; Morita et al., 2017).
Seasonal impact on iron storage genes
Iron storage genes relating to ferritin like domains were found in all samples and highest in May and KE samples (Fig. 4D). May had the highest iron concentrations and Kenai River experienced an increase in discharge rates from February (~711 ft3/s) to May (~3,450 ft3/s) (U.S. Geological Survey, 2020). The increased temperature in May caused ice break up, increased flow, and presumably transport of more nutrients to the estuary (Prowse, 1993; Prowse and Beltaos, 2002). This also explains May samples spread across all three groups in the PCoA plot showing the microbial community is similar to all other months of sampling (Fig. 3). Excessive amounts of iron can be toxic to bacterial cells. Ferritins or ferritin-like compounds are often used to remove excess ferrous ions from the cytoplasm (Smith, 2004). Excessive ferrous iron is oxidized by ferritins (intracellular proteins) and stored away from cellular processes as non-toxic, bioavailable ferric iron (Smith, 2004). The increase in iron concentration in May likely caused an increase in microorganisms that have more ferritin genes to combat excessive iron influxes caused by increases in discharge rates. Ferritins also function to store the iron to be used at a later time when iron availability becomes limited (Briat, 1992; Smith, 2004). The function of ferritins could also explain why iron storage genes were higher in estuary (KE) environments compared to freshwater (CL). Estuary environments experience rapid flocculation of iron requiring microbes to store iron when it is available.
Seasonal influence on siderophores
The increase in discharge rate in May could also explain the changes in vibrioferrin gene copies. May KE samples containing the highest total iron concentration had significantly lower vibrioferrin gene copies in sediment and water samples (Fig. 6 A,B,D). The increase in iron concentrations may indicate microorganisms did not need siderophores such as vibrioferrin to sequester iron. Seasonal discharge rate fluctuations and temperatures can alter nutrient availability causing microorganisms to need multiple iron related genes.
Siderophore gene abundance and diversity is likely influenced by changes in sunlight in estuary environments. Light is often a driving factor in aquatic nutrient cycles (Lueder et al. 2020). Alaska experiences increases in daylight during summer months and is presumably contributing to the increase in siderophores. Siderophore gene abundance and diversity was highest in August, during summer when Alaska experiences more light, compared to February and May (Fig. 5). Photolysis, the breakdown of molecules by light, has been shown to affect siderophore iron complex groups specifically containing α- or β- hydroxy carboxylate functional groups (Barbeau et al., 2002; Amin et al., 2009). Vibrioferrin and petrobactin, both containing α-hydroxy carboxylic acid groups were only detected in August in the metagenomes. However, qPCR, which is more sensitive and provides more accurate quantifications, detected vibrioferrin in all samples. These siderophores have shown light sensitivity when complexed with Fe (III) (Barbeau et al., 2002; Amin et al., 2009). When exposed to sunlight the siderophore-iron complex undergoes decarboxylation, reduction of Fe(III), and release of Fe(II) (Amin et al., 2009; Chen et al., 2019). The new photolytic Fe(II) can be oxidized to Fe(III), biologically taken up, or oxidized by another ligand (Barbeau et al., 2001). Light and siderophores both play a role in the marine iron cycle providing soluble iron to many organisms.
Biotic relationships between bacteria and phytoplankton are potentially contributing to siderophore production and influencing iron cycling in estuaries. Most phytoplankton do not produce siderophores but require high amounts of iron (Hopkinson and Morel, 2009; Hider and Kong, 2010). The low solubility of iron in estuaries could limit the amount of iron phytoplankton can sequester and since phytoplankton are typically primary producers in aquatic food chains, limited iron could pose a potential problem. Phytoplankton productivity and ocean biomass have previously been controlled by iron limitation in summer (Martin et al., 1994; Behrenfeld, 1999; Boyd et al., 2000). As shown photochemical reactivity of iron-siderophore complexes contributes Fe(II) to marine ecosystems. Bacterial photoreactive siderophores such as vibrioferrin can provide iron to phytoplankton in exchange for organic molecules used for growth (Amin et al., 2009). Vibrioferrin gene (marpvsB) copies relating to Marinobacter spp. that are often associated in-algal relationships were found in all water and sediment samples (Fig. 6 C,D). August KE water samples contained the highest of these vibroferrin genes further suggesting biotic relationships between microorganisms are essential for iron acquisition.
Vibrioferrin was one of the most abundant siderophore genes detected. Although vibrioferrin was highly abundant, it is considered one of the weakest iron chelators of known marine siderophores since it lacks the six donor groups required for the preferred octahedral coordination with Fe(III) (Amin et al., 2009; Hider and Kong, 2010; Lueder et al., 2020). However, since vibrioferrin contains two α-hydroxy acid groups, it is exceptionally susceptible to photolysis. Studies have found vibrioferrin has a higher rate of photolysis compared to other photoreactive siderophores such as petrobactin (Amin et al., 2009). This photosensitive siderophore could be favorable in high-light environments since it is readily reduced by light producing bioavailable iron (Amin et al., 2009). For example, in exchange for photoreduced iron, diatoms have been shown to provide Marinobacter with organic carbon (Amin et al., 2009; Amin et al., 2012). Siderophore production also seems to be influenced by biotic interactions that benefit microbial communities. Environmental conditions such as sunlight provide an adaptive advantage to iron sequestration through microbial siderophores.
Siderophore genes were most abundant and diverse in estuary environments with higher sodium concentrations compared to the lower sodium river environments. All KE samples containing higher sodium concentrations contained siderophore genes emphasizing the potential need to increase iron solubility as sodium concentrations increase (Fig. 5). The estuary (KE) environment containing higher sodium concentrations also had significantly higher iron concentrations (p<0.05). Identifying siderophore gene expression levels in future work will help clarify if higher siderophore genes found in estuary environments could lead to higher iron concentrations. More organisms producing siderophores could result in less iron flocculation and higher concentrations since the strongest ligand complexes have been found to be resistant to flocculation (Bundy et al., 2015). Bioavailable iron rather than total iron is likely driving this process, however data regarding bioavailable iron is lacking and needs to be investigated.
Iron related genes specifically siderophore genes can have significant implications for Alaska food chains. Photoreactive siderophores produced by bacteria such as vibrioferrin likely provide soluble Fe(II) to primary producers in exchange for metabolites. Environmental conditions seem to be influencing siderophore genes and play a role in aquatic iron cycling. Since >99% of dissolved iron in oceans is bound to organic ligands, results suggest that siderophores in an estuary may facility iron transport to the oceans (Gledhill and Buck 2012). This project has provided a better understanding of iron dynamics in an Alaskan river and estuary. Future projects should investigate bioavailable iron and active gene expression of siderophores in estuaries.
Conclusions
Microbial community composition and associated iron related genes within a glacial influenced estuary and river are critical missing pieces in iron biogeochemistry. This study determined that microbial iron acquisition gene abundance, distribution, and diversity were distinct between freshwater river and estuary environments. Microbes in freshwater systems mainly use Feo system genes. However, microbes in estuary systems rely on siderophores to obtain needed iron. This is likely due to iron lost because of flocculation that occurs in the presence of sodium cations in estuary systems. Siderophore transport genes were abundant in all samples, indicating these genes might be conserved and used in biotic interactions regardless if the microbe inhabits estuary or river environments. Temporal and environmental influences including sunlight, discharge rate, and temperature were determined to correlate to iron cycling genes in Alaskan estuaries and river systems. Understanding iron dynamics in estuaries offers insight into key water ecosystems, organismal adaptation to nutrient limitation, and changes in biogeochemical cycling influenced by glacial systems.
Acknowledgments
Funding was provided by the Alaska Space Grant Program and the North Pacific Research Board Graduate Student Research Award to MB. Sequencing was provided by the Advanced Instrumentation for Microbiome Sciences (AIMS) core facility, which is supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM130443. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
Data Availability
All sequence data are available in NCBI under BioProject PRJNA984928 (metagenomes) and PRJNA984929 (amplicon).
References
- Amin SA, Green DH, Hart MC, Kupper FC, Sunda WG, and Carrano CJ (2009) Photolysis of iron-siderophore chelates promotes bacterial-algal mutualism. Proceedings of the National Academy of Sciences 106: 17071–17076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin SA, Parker MS, and Armbrust EV (2012) Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76: 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KS, Kirkegaard RH, Karst SM, and Albertsen M (2018) ampvis2: an R package to analyze and visualize 16S rRNA amplicon data, Bioinformatics. [Google Scholar]
- Andrews S. (2010) FastQC: a quality control tool for high throughput sequence data, Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- Andrews SC, Robinson AK, and Rodríguez-Quiñones F (2003) Bacterial iron homeostasis. FEMS Microbiol Rev 27: 215–237. [DOI] [PubMed] [Google Scholar]
- Årstøl E and Hohmann-Marriott MF (2019) Cyanobacterial Siderophores—Physiology, Structure, Biosynthesis, and Applications. Marine Drugs 17: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartel AA, DeJongh M, Disz T, Edwards RA, et al. (2008) The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeau K, Rue EL, Bruland KW, and Butler A (2001) Photochemical cycling of iron in the surface ocean mediated by microbial iron(iii)-binding ligands. Nature 413: 409–413. [DOI] [PubMed] [Google Scholar]
- Barbeau K, Zhang G, Live DH, and Butler A (2002) Petrobactin, a Photoreactive Siderophore Produced by the Oil-Degrading Marine Bacterium Marinobacter hydrocarbonoclasticus. J Am Chem Soc 124: 378–379. [DOI] [PubMed] [Google Scholar]
- Behrenfeld MJ (1999) Widespread Iron Limitation of Phytoplankton in the South Pacific Ocean. Science 283: 840–843. [DOI] [PubMed] [Google Scholar]
- Bhatia MP, Kujawinski EB, Das SB, Breier CF, Henderson PB, and Charette MA (2013) Greenland meltwater as a significant and potentially bioavailable source of iron to the ocean. Nature Geosci 6: 274–278. [Google Scholar]
- Bokulich NA (2018) Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd PW, Ibisanmi E, Sander SG, Hunter KA, and Jackson GA (2010) Remineralization of upper ocean particles: Implications for iron biogeochemistry. Limnol Oceanogr 55: 1271–1288. [Google Scholar]
- Boyd PW, Watson AJ, Law CS, Abraham ER, Trull T, Murdoch R, et al. (2000) A mesoscale phytoplankton bloom in the polar Southern Ocean stimulated by iron fertilization. Nature 407: 695–702. [DOI] [PubMed] [Google Scholar]
- Boyle EA, Edmond JM, and Sholkovitz ER (1977) The mechanism of iron removal in estuaries. Geochimica et Cosmochimica Acta 41: 1313–1324. [Google Scholar]
- Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, et al. (2015) RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5: 8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF (1992) Iron assimilation storage in prokaryotes. Microbiology, 138: 2475–2483. [DOI] [PubMed] [Google Scholar]
- van den Broeke MR, Enderlin EM, Howat IM, Kuipers Munneke P, Noël BPY, van de Berg WJ, et al. (2016) On the recent contribution of the Greenland ice sheet to sea level change. The Cryosphere 10: 1933–1946. [Google Scholar]
- Buchfink B, Xie C, and Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nature Methods 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Bundy RM, Abdulla HAN, Hatcher PG, Biller DV, Buck KN, and Barbeau KA (2015) Iron-binding ligands and humic substances in the San Francisco Bay estuary and estuarine-influenced shelf regions of coastal California. Marine Chemistry 173: 183–194. [Google Scholar]
- Bundy RM, Boiteau RM, McLean C, Turk-Kubo KA, McIlvin MR, Saito MA, et al. (2018) Distinct Siderophores Contribute to Iron Cycling in the Mesopelagic at Station ALOHA. Front Mar Sci 5:61. [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, and Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Noah Fierer N, and Knight R (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108: 4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron ML, Maddocks S, Gillingham P, Craven CJ, and Andrews SC (2006) Feo – Transport of Ferrous Iron into Bacteria. Biometals 19: 143–157. [DOI] [PubMed] [Google Scholar]
- Caza M, and Kronstad JW (2013). Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol. 3:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P-A, Mussig AJ, Hugenholtz P, and Parks DH (2022) GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38: 5315–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Guo Y, Lu Y, Wang B, Sun J, Zhang H, and Wang H (2019) Chemistry and Biology of Siderophores from Marine Microbes. Marine Drugs 17: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corretto E, Antonielli L, Sessitsch A, Höfer C, Puschenreiter M, Widhalm S, Swarnalakshmi K, Brader G (2020) Comparative genomics of Microbacterium species to reveal diversity, potential for secondary metabolites and heavy metal resistance. Front. Microbiol 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar E. (2015) Dissolved Iron Behavior in the Ravenglass Estuary Waters, An Implication on the Early Diagenesis. ujg 3: 1–12. [Google Scholar]
- Debeljak P, Toulza E, Beier S, Blain S, and Obernosterer I (2019) Microbial iron metabolism as revealed by gene expression profiles in contrasted Southern Ocean regimes. Environ Microbiol 21: 2360–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney I, and Adhikari S (2020). Increased subglacial sediment discharge in a warming climate: Consideration of ice dynamics, glacial erosion, and fluvial sediment transport. Geophys Res Lett 47: e2019GL085672 [Google Scholar]
- Desai DK, Desai FD, and LaRoche J (2012). Factors influencing the diversity of iron uptake systems in aquatic microorganisms. Front Microbiol 3: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierssen HM, Smith RC, and Vernet M (2002) Glacial meltwater dynamics in coastal waters west of the Antarctic peninsula. Proceedings of the National Academy of Sciences 99: 1790–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. (2003) VEGAN, a package of R functions for community ecology. Journal of Vegetation Science 14: 927–930. [Google Scholar]
- Dogs M, Wemheuer B, Wolter L, Bergen N, Daniel R, Simon M, Brinkhoff T, (2017) Rhodobacteraceae on the marine brown alga Fucus spiralis are abundance and show physiological adaptation to an epiphytic lifestyle. Syst. Appl. Microbiol 40:370–382 [DOI] [PubMed] [Google Scholar]
- Duckworth OW, Holmström SJM, Peña J, Sposito G (2009). Biogeochemistry of iron oxidation in a circumneutral freshwater habitat. Chemical Geology 260:149–158 [Google Scholar]
- Eckert JM and Sholkovitz ER (1976) The flocculation of iron, aluminium and humates from river water by electrolytes. Geochimica et Cosmochimica Acta 40: 847–848. [Google Scholar]
- Faraldo-Gómez JD and Sansom MSP (2003) Acquisition of siderophores in Gram-negative bacteria. Nat Rev Mol Cell Biol 4: 105–116. [DOI] [PubMed] [Google Scholar]
- Fujino T, Sakazaki R, and Tamura K (1974) Designation of the type strain of Vibrio parahaemolyticus and description of 200 strains of the species. International Journal of Systematic and Evolutionary Microbiology, 24: 447–449. [Google Scholar]
- Garber AI, Nealson KH, Okamoto A, McAllister SM, Chan CS, Barco RA, and Merino N (2020) FeGenie: A Comprehensive Tool for the Identification of Iron Genes and Iron Gene Neighborhoods in Genome and Metagenome Assemblies. Front Microbiol 11: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärdes A, Triana C, Amin SA, Green DH, Romano A, Trimble L, and Carrano CJ (2013) Detection of photoactive siderophore biosynthetic genes in the marine environment. Biometals 26: 507–516. [DOI] [PubMed] [Google Scholar]
- Gauthier MJ, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, and Bertrand JC (1992) Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol 42: 568–576. [DOI] [PubMed] [Google Scholar]
- Gledhill M and Buck KN (2012) The organic complexation of iron in the marine environment: a review. Front Microbiol 3: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider RC and Kong X (2010) Chemistry and biology of siderophores. Nat Prod Rep 27: 637. [DOI] [PubMed] [Google Scholar]
- Holmén B, Sison J, Nelson D, Casey W, (1999) Hydroxamate siderophores, cell growth and Fe(III) cycling in two anaerobic iron oxide media containing Geobacter metallireducens. Geochim. Cosmo. Acta 63:227–239 [Google Scholar]
- Hopkinson BM and Barbeau KA (2012) Iron transporters in marine prokaryotic genomes and metagenomes. Environ Microbial 14: 114–128. [DOI] [PubMed] [Google Scholar]
- Hopkinson BM and Morel FMM (2009) The role of siderophores in iron acquisition by photosynthetic marine microorganisms. Biometals 22: 659–669. [DOI] [PubMed] [Google Scholar]
- Hopwood MJ, Connelly DP, Arendt KE, Juul-Pedersen T, Stinchcombe MC, Meire L, Esposito M, Krishna R. (2016) Seasonal changes in Fe along a glaciated Greenlandic fjord. Front Earth Sci 4. [Google Scholar]
- Hopwood MJ, Statham PJ, Tranter M, Wadham JL. (2014) Glacial flours as a potential source of Fe(II) and Fe(III) to polar waters. Biogeochemistry 118: 443–452. [Google Scholar]
- Hunter KA and Boyd PW (2007) Iron-binding ligands and their role in the ocean biogeochemistry of iron. Environ Chem 4: 221. [Google Scholar]
- Huson DH, Auch AF, Qi J, and Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Research 17: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, and Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H-B, Lou W-J, Ke W-T, Song W-Y, Price NM, and Qiu B-S (2015) New insights into iron acquisition by cyanobacteria: an essential role for ExbB-ExbD complex in inorganic iron uptake. ISME J 9: 297–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, and Wang Z (2019) MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7: e7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A (2020) ggplot2 Based Publication Ready Plots. [Google Scholar]
- Klunder MB, Bauch D, Laan P, Baar H.J.W. de, van Heuven S, and Ober S (2012) Dissolved iron in the Arctic shelf seas and surface waters of the central Arctic Ocean: Impact of Arctic River water and ice-melt. Journal of Geophysical Research: Oceans 117. [Google Scholar]
- Komsta L. (2022). Outliers: Tests for Outliers. R package version 0.15, https://CRAN.R-project.org/package=outliers. [Google Scholar]
- Koppes M, Hallet B, Rignot E, Mouginot J, Wellner JS, Boldt K. (2015). Observed latitudinal variations in erosion as a function of glacier dynamics. Nature 526:100–103. [DOI] [PubMed] [Google Scholar]
- Kumari A, Patel A, Banjare U, Pandey K, (2020) Growth promoting characteristics of endophytic bacteria isolated from Costus igneus plant. Plant Achieves 20: 2991–3001 [Google Scholar]
- Lau CKY, Krewulak KD, and Vogel HJ (2016) Bacterial ferrous iron transport: the Feo system. FEMS Microbiology Reviews 40: 273–298. [DOI] [PubMed] [Google Scholar]
- Laufer-Meiser K, Michaud AB, Maisch M, Byrne JM, Kappler A, Patterson MO, et al. (2021) Potentially bioavailable iron produced through benthic cycling in glaciated Arctic fjords of Svalbard. Nat Commun 12: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ding Y, Hood E, Raiswell R, Han T, He X, Kang S, Wu Q, Yu Z, Mika S, Liu S, Li Q. (2019) Dissolved iron supply from Asian glaciers: Local controls and a regional perspective. Global Biogeochem Cycles 33: 1223–1237. [Google Scholar]
- Liu X and Millero FJ (2002) The solubility of iron in seawater. Marine Chemistry 77: 43–54. [Google Scholar]
- Lueder U, Jørgensen BB, Kappler A, and Schmidt C (2020) Photochemistry of iron in aquatic environments. Environ Sci: Processes Impacts 22: 12–24. [DOI] [PubMed] [Google Scholar]
- Makita H, Tanaka E, Mitsunobu S, Miyazaki M, Nunoura T, Uematsu K, et al. (2017) Mariprofundus micogutta sp. nov., a novel iron-oxidizing zetaproteobacterium isolated from a deep-sea hydrothermal field at the Bayonnaise knoll of the Izu-Ogasawara arc, and a description of Mariprofundales ord. nov. and Zetaproteobacteria classis nov. Arch Microbiol 199: 335–346. [DOI] [PubMed] [Google Scholar]
- Markussen TN, Elberling B, Winter C, and Andersen TJ (2016) Flocculated meltwater particles control Arctic land-sea fluxes of labile iron. Scientific Reports 6: 24033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JH, Coale KH, Johnson KS, Fitzwater SE, Gordon RM, Tanner SJ, et al. (1994) Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature 371: 123–129. [Google Scholar]
- Mason OU, Han J, Woyke T, Jansson JK (2014) Single-cell genomics reveals features of a Colwellia species that was dominant during the Deepwater Horizon oil spill. Front Microbiol 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JJ, Wendrich TM, and Marahiel MA (2001) The dhb Operon of Bacillus subtilisEncodes the Biosynthetic Template for the Catecholic Siderophore 2,3-Dihydroxybenzoate-Glycine-Threonine Trimeric Ester Bacillibactin. Journal of Biological Chemistry 276: 7209–7217. [DOI] [PubMed] [Google Scholar]
- McAllister SM, Moore RM, Gartman A, Luther GW, Emerson D, and Chan CS (2019) The Fe(II)-oxidizing Zetaproteobacteria : historical, ecological and genomic perspectives. FEMS Microbiology Ecology 95:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Mengjie, Liu Fawang, Yang Xiaoyan, Jin Jing, Dong Xin, Zeng Ke-Wu, et al. (2018) Bacillibactin and Bacillomycin Analogues with Cytotoxicities against Human Cancer Cell Lines from Marine Bacillus sp. PKU-MA00093 and PKU-MA00092. Marine Drugs 16: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messenger AJ and Barclay R (1983) Bacteria, iron and pathogenicity. Biochemical Education 11: 54–63. [Google Scholar]
- Morita Y, Yamagata K, Oota A, Ooki A, Isoda Y, and Kuma K (2017) Subarctic wintertime dissolved iron speciation driven by thermal constraints on Fe(II) oxidation, dissolved organic matter, and stream reach. Geochimica et Cosmochimica Acta 215: 33–50. [Google Scholar]
- Mu D, Wang S Liang Q Du Z Tian R Ouyang Y, Wang X, Zhou A Gong Y Chen C, Van Nostrand J, Yang Y, Zhou J, Du. Z (2020) Bradymonabacteria, a novel bacterial predator group with versatile survival strategies in saline environments. Microbiome 8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal EG, Hood E, and Smikrud K (2010) Contribution of glacier runoff to freshwater discharge into the Gulf of Alaska. Geophysical Research Letters 37:. [Google Scholar]
- Neefs J-M, Van de Peer Y, De Rijk P, Chapelle S, and De Wachter R (1993) Compilation of small ribosomal subunit RNA structures. Nucl Acids Res 21: 3025–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noinaj N, Guillier M, Barnard TJ, and Buchanan SK (2010) TonB-Dependent Transporters: Regulation, Structure, and Function. Annu Rev Microbiol 64: 43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov A, and Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27: 824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucl Acids Res 42: D206–D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, and Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovsky OS and Schott J (2002) Iron colloids/organic matter associated transport of major and trace elements in small boreal rivers and their estuaries (NW Russia). Chemical Geology 190: 141–179. [Google Scholar]
- Prowse TD (1993) Suspended sediment concentration during river ice breakup. Can J Civ Eng 20: 872–875. [Google Scholar]
- Prowse TD and Beltaos S (2002) Climatic control of river-ice hydrology: a review. Hydrol Process 16: 805–822. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisdorph SC and Mathis JT (2015) Assessing net community production in a glaciated Alaskan fjord. Biogeosciences 12: 5185–5198. [Google Scholar]
- Řezanka T, Palyzová A, and Sigler K (2018) Isolation and identification of siderophores produced by cyanobacteria. Folia Microbiologica 63: 569–579. [DOI] [PubMed] [Google Scholar]
- Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, and Tribedi P (2016) Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res 23: 3984–3999. [DOI] [PubMed] [Google Scholar]
- Sakazaki R, Iwanami S, and Fukumi H (1963) Studies on the enteropathogenic, facultatively halophilic bacteria Vibrio Parahaemollyticus. I. Morphological, cultural, and biochemical properties and its taxonomical position. Jpn J Med Sci Biol 16: 161–188. [DOI] [PubMed] [Google Scholar]
- Sandy M and Butler A (2009) Microbial Iron Acquisition: Marine and Terrestrial Siderophores. Chem Rev 109: 4580–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth AW, Crusius J, Hoyer I, and Campbell R (2014) Estuarine removal of glacial iron and implications for iron fluxes to the ocean. Geophys Res Lett 41: 3951–3958. [Google Scholar]
- Segond D, Abi Khalil E, Buisson C, Daou N, Kallassy M, Lereclus D, et al. (2014) Iron Acquisition in Bacillus cereus: The Roles of IlsA and Bacillibactin in Exogenous Ferritin Iron Mobilization. PLoS Pathog 10: e1003935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sestok AE, Linkous RO, and Smith AT (2018) Toward a mechanistic understanding of Feo-mediated ferrous iron uptake. Metallomics 10: 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiee R, Snow J, Zhang Q, (2019) Iron requirements and uptake strategies of the globally abundant marine ammonia-oxidixing archaeon, Nitrosopumilus maritimus SCM1. ISME J. 13:2295–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL (2004) The Physiological Role of Ferritin-Like Compounds in Bacteria. Critical Reviews in Microbiology 30: 173–185. [DOI] [PubMed] [Google Scholar]
- Surendran PK, Mahadeva Iyer K, and Gopakumar K (1983) Salt tolerance of bacteria isolated from tropical marine fish and prawn. Fishery Technology 20: 105–110. [Google Scholar]
- Tashiro Y, Yoh. M, Shiraiwa T, Onishi T, Shesterkin V, and Kim V (2020) Seasonal variations of dissolved iron concentrations in active layer and rivers in permafrost areas, Russian Far East. Water 12: 2579. [Google Scholar]
- Team, R.C. (2016) Vienna: R Foundation for Statistical Computing, 2016.
- Thode SK, Rojek E, Kozlowski M, Ahmad R, and Haugen P (2018) Distribution of siderophore gene systems on a Vibrionaceae phylogeny: Database searches, phylogenetic analyses and evolutionary perspectives. PLoS ONE 13: e0191860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Geological Survey (2020) USGS Water Data for the Nation. National Water Information System data available on the World Wide Web. [Google Scholar]
- Vraspir JM and Butler A (2009) Chemistry of Marine Ligands and Siderophores. Annu Rev Mar Sci 1: 43–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman C and Delepelaire P (2004) Bacterial Iron Sources: From Siderophores to Hemophores. Annu Rev Microbiol 58: 611–647. [DOI] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis., Springer-Verlag; New York. [Google Scholar]
- Yarimizu K, Polido G, Gardes A, Carter ML, Hilbern M, and Carrano CJ (2014) Evaluation of photo-reactive siderophore producing bacteria before, during and after a bloom of the dinoflagellate Lingulodinium polyedrum. Metallomics 6: 1156–63. [DOI] [PubMed] [Google Scholar]
- Zhang R, John SG, Zhang J, Ren J, Wu Y, Zhu Z, et al. (2015) Transport and reaction of iron and iron stable isotopes in glacial meltwaters on Svalbard near Kongsfjorden: From rivers to estuary to ocean. Earth and Planetary Science Letters 424: 201–211. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence data are available in NCBI under BioProject PRJNA984928 (metagenomes) and PRJNA984929 (amplicon).