Abstract
Four of the seven members of the FXYD protein family have been identified as specific regulators of Na,K-ATPase. In this study, we show that FXYD3, also known as Mat-8, is able to associate with and to modify the transport properties of Na,K-ATPase. In addition to this shared function, FXYD3 displays some uncommon characteristics. First, in contrast to other FXYD proteins, which were shown to be type I membrane proteins, FXYD3 may have a second transmembrane-like domain because of the presence of a noncleavable signal peptide. Second, FXYD3 can associate with Na,K- as well as H,K-ATPases when expressed in Xenopus oocytes. However, in situ (stomach), FXYD3 is associated only with Na,K-ATPase because its expression is restricted to mucous cells in which H,K-ATPase is absent. Coexpressed in Xenopus oocytes, FXYD3 modulates the glycosylation processing of the β subunit of X,K-ATPase dependent on the presence of the signal peptide. Finally, FXYD3 decreases both the apparent affinity for Na+ and K+ of Na,K-ATPase.
INTRODUCTION
The Na,K-ATPase is an ubiquitous enzyme consisting of an α and a β subunit, which is responsible for the creation and maintenance of the Na+ and K+ gradients across the cell membrane by transporting three Na+ out and two K+ into the cell. This function is crucial for cell survival and body homeostasis because the Na+ gradient is used as an energy source to transport ions or solutes and is at the origin of the vectorial Na+ reabsorption in the kidney and of action potentials in excitable tissues.
Regulation of the activity and expression of Na,K-ATPase is tight and governed by a variety of mechanisms. Shortterm regulation involves protein kinases and results in modulation of the cell surface expression of the Na,K-ATPase, whereas long-term regulation, mediated by mineralocorticoid or thyroid hormone, leads to a change in the total number of Na,K-ATPase units (Therien and Blostein, 2000; for review, see Feraille and Doucet, 2001). Moreover, the existence of multiple α and β isoforms permits the production of isozymes with different transport properties (Crambert et al., 2000). Finally, recent experimental evidence shows that members of the FXYD protein family specifically associate with and modulate the transport properties of Na,K-ATPase (for review, see Crambert and Geering, 2003).
The FXYD family contains seven members that share a common signature sequence encompassing the transmembrane and adjacent regions (Sweadner and Rael, 2000). So far, all characterized FXYD proteins exhibit a similar structure with a single transmembrane domain and a type I orientation that is achieved, in some, but not all cases, by the cleavage of an N-terminal signal peptide. Among the seven members (FXYD1–7), four FXYD proteins have so far been shown to regulate the Na,K-ATPase in a tissue- and isozyme-specific way. All affect the apparent affinity for extracellular K+ of the Na,K-ATPase, which in the case of FXYD7 (the brain-specific FXYD protein) is thought to be physiologically relevant in neuronal excitability (Béguin et al., 2002). In addition, FXYD1 (Crambert et al., 2002), FXYD2 (Arystarkhova et al., 1999; Béguin et al., 2001; Pu et al., 2001), and FXYD4 (Béguin et al., 2001; Garty et al., 2002) affect the apparent affinity for internal Na+ of Na,K-ATPase in a way that is consistent with the physiological demands of the tissues in which they are expressed.
These previous results suggested that at least one common function of all FXYD proteins could be the regulation of Na,K-ATPase transport properties. In this study, we have investigated another member of this family, FXYD3, also known as Mat-8. FXYD3 was previously identified as a phospholemman-like protein inducing a chloride conductance when overexpressed in Xenopus laevis oocytes (Morrison et al., 1995). FXYD3 is expressed at high levels in uterus, stomach, and colon (Morrison et al., 1995). A striking feature of FXYD3 is its overexpression in human breast cancer tumors and in tumors initiated by c-neu or v-Ha-ras oncongenes but not in tumors initiated by the c-myc oncogene (Morrison and Leder, 1994).
In this study, we have investigated the biosynthesis of FXYD3, its association with Na,K- and H,K-ATPases, the modulation of Na,K-ATPase function, and its cellular distribution in the stomach. Our results show that FXYD3 has structural and functional characteristics that are distinct from other FXYD proteins.
MATERIALS AND METHODS
Cloning and Site-directed Mutagenesis
An expressed sequence tag clone corresponding to mouse FXYD3 (IMAGE clone 1051256) was obtained from UK HGMP Resource Centre (Cambridge, United Kingdom) and verified by sequencing. The coding sequence was PCR amplified using a sense primer tailed with an EcoRI site and an antisense primer tailed with a XbaI restriction site. These two sites were used to subclone mouse FXYD3 into a pSD5 vector. cDNAs of rat Na,K-ATPase α1 and β1 subunits (kindly provided by J. Lingrel, University of Cincinnati, Cincinnati, OH), Xenopus FLAG β1 (Béguin et al., 1997), rabbit, gastric H,K-ATPase α and β subunits (kindly provided by G. Sachs, UCLA, Los Angeles, CA), rat, colonic H,K-ATPase α subunit (kindly provided by F. Jaisser, INSERM U478, Paris, France), Bufo bladder β subunit (kindly provided by F. Jaisser), human, sarcoendoplasmic reticulum Ca2+-ATPase 2a (SERCA2a) (kindly provided by D. H. MacLennan, University of Toronto, Toronto, Ontario, Canada), rat glucose transporter-2 (Glut-2) (kindly provided by B. Thorens, University of Lausanne, Lausanne, Switzerland), and human FLAG-Aquaporin-2 (kindly provided by E. Féraille, Fondation pour Recherches Medicales, Geneva, Switzerland) were subcloned into a pSD5 vector. The FXYD3 mutant V5N in which Val5 was replaced by Asn to create an N-glycosylation consensus site was prepared by the PCR-based method described by Nelson and Long (1989). The truncated mutant lacking 22 N-terminal amino acids (Nt-Δ22) was obtained by a single PCR by using a sense primer tailed with an EcoRI site and bearing the start codon ATG at the position of the Asp22 codon and an antisense primer tailed with a NotI restriction site. The PCR product was then digested and ligated into a pSD5 vector. To prepare the chimera containing the N-terminal domain of rat FXYD4 (Met1-Arg38) and the transmembrane and the C-terminal domain of mouse FXYD3 (Val39-Cys88), the sequence coding for the N-terminal domain of rat FXYD4 (kindly provided by H. Garty, Weizmann Institute of Science, Rehovoth, Israel) was PCR amplified by using a sense primer tailed with an EcoRI site and an antisense primer tailed with 15 base pairs coding for the first amino acids of the FXYD3 transmembrane domain. In parallel, the sequence coding for the transmembrane and C-terminal domain of mouse FXYD3 was PCR amplified by using a sense primer tailed with 15 base pairs coding for the last amino acids of the N-terminal part of FXYD4 and with an antisense primer tailed with a NotI restriction site. Both PCR products were mixed and used for a third PCR by using the sense primer tailed with an EcoRI site and the antisense primer tailed with a NotI site. The final product was digested by EcoRI and NotI and introduced into a pSD5 vector. All constructs were sequenced before in vitro transcription (Melton et al., 1984).
Immunodetection of FXYD3
We used two different antibodies to detect FXYD3. First, an antibody (kindly provided by H. Garty) produced against a C-terminal glutathione S-transferase (GST) fusion peptide of mouse FXYD4 (74GKATPLIIPGSANT), which cross-reacted with mouse FXYD3 expressed in Xenopus oocytes, probably due to the high sequence similarity (57%) in this region. Second, an antibody directed against a C-terminal GST fusion peptide of mouse FXYD3 (71HR-PGEGPPLITPGSAHNC). The corresponding cDNA region was cloned in frame into the pGEX-4T1 vector. After purification using glutathione beads, and dialysis against phosphate-buffered saline (PBS), the fusion protein was used for immunization of rabbits (Cocalico Biologicals, Reamstown, PA). This antibody did not cross-react with FXYD4.
Protein Expression in Xenopus Oocytes, Metabolic Labeling, and Immunoprecipitations
Stage V to VI oocytes were prepared from X. laevis as described previously (Geering et al., 1996). To study protein expression and FXYD3 association, cRNA-injected oocytes were incubated in modified Barth's solution (MBS) containing 0.7–1 mCi/ml [35S]methionine for 6 or 24 h and subjected to 24- to 48-h chase periods in MBS containing 10 mM unlabeled methionine. FXYD3 was expressed in excess over coexpressed Na,K-ATPase to ensure stoichiometric association. After the various pulse-chase periods, microsomes or digitonin [0.5% (wt/vol)] extracts were prepared and subjected to nondenaturing immunoprecipitations as described previously (Jaunin et al., 1993). In brief, samples were diluted to a volume of 400 μl with digitonin washing buffer (DWB) (100 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 7.4, and 0.2% digitonin), and then 2 mM (final concentration) phenylmethysulfonyl fluoride and 2% (final concentration) bovine serum albumin (BSA) were added. After overnight incubation at 4°C with antibodies, the immunocomplexes were recovered on protein A or protein G-Sepharose beads and washed six times with DWB and once with DWB without digitonin. Antibodies against the Na,K-ATPase α subunit (Girardet et al., 1981), the gastric H,K-ATPase (Claeys et al., 1994), the colonic H,K-ATPase α subunit (kindly provided by F. Jaisser), the SERCA2a (Affinity Bioreagents, Golden, CO), anti-Glut-2 (kindly provided by B. Thorens), or anti-FLAG (M2) antibodies were used. Immunoprecipitates were resolved either by SDS-PAGE (5–13% polyacrylamide) or on SDS-tricine (15% polyacrylamide) gels (prepared according to the manufacturer's instructions).
Electrophysiological Measurements
Electrophysiological measurements were performed 3 d after oocyte injections with rat α1 and β1 cRNAs alone or together with mouse FXYD3 cRNA by using the two-electrode voltage-clamp technique. Measurements of the apparent external K+ affinity were carried out as described previously (Béguin et al., 2001) in the presence of 1 μM ouabain, which inhibits the endogenous Na,K-pump but not the expressed ouabain-resistant rat Na,K-ATPase. The maximal pump current and the apparent K+ affinity (K½ K+) measured in the presence of external Na+ (100 mM) were obtained by fitting the Hill equation to the data by using a Hill coefficient of 1.6 (Jaisser et al., 1994). Measurements of the apparent Na+ affinity of Na,K-ATPase were performed as described previously (Hasler et al., 1998) by coexpressing rat α1 and rat β1 cRNAs together with rat epithelial Na+ channel α, β, and γ subunit cRNAs in the presence or absence of FXYD3 cRNA. The maximal current (Imax) and theapparent Na+ affinity (K½ Na+)) were fitted by using the Hill equation and using a Hill coefficient of 3.
Protein Expression in Human Embryonic Kidney (HEK) Cells
Mouse FXYD3 was subcloned into the mammalian expression vector pRK5 and transiently transfected into HEK cells by using Effectene technology (QIAGEN, Basel, Switzerland). Two days after transfection, protein extracts were prepared using a lysis buffer containing 1% Triton X-100, 100 mM NaCl, 5 mM EDTA, and 50 mM Tris-HCl, pH 7.4. The protein concentration was determined by the method of Lowry et al. (1951).
Microsome Preparations from Stomach Epithelium
Mouse stomachs were removed, cut longitudinally, and washed five times in PBS. The gastric mucosa was scraped off with a glass coverslip. Gastric mucosa was homogenized on ice by using a Potter glass/Teflon (15 × 5 s at 200 rpm) in a homogenization buffer containing 250 mM sucrose, 5 mM EDTA, and 30 mM histidine, pH 7.4 (1 ml/stomach). Homogenates were centrifuged 15 min at 10,000 × g at 4°C. Supernatants were pooled and centrifuged at 100,000 × g for 1 h at 4°C. Pellets were resuspended in homogenization buffer (50 μl/stomach), and the protein concentration was measured by the method of Lowry et al. (1951). Thirty micrograms of protein was resolved on SDS-tricine gels, transferred to nitrocellulose, and revealed with an anti-FXYD4 antibody.
Immunofluorescence Microscopy
Mouse stomach slices were obtained from The Binding Site (Birmingham, United Kingdom). The slices were first rehydrated in PBS for 5 min and then blocked in PBS containing 5% BSA for 30 min at room temperature. The first antibody was diluted in the same buffer (monoclonal anti-H,K-ATPase 1/300, monoclonal anti-Na,K-ATPase 1/100, polyclonal anti-FXYD3 1/500, and anti-FXYD3 preimmune serum 1/500) and applied for 1 h at room temperature. After three washes in PBS, anti-rabbit fluorescein isothiocyanate (FITC) (SigmaAldrich, St. Louis, MO), anti-mouse FITC (Sigma-Aldrich), or anti-mouse rhodamine (Chemicon International, Temecula, CA) were applied for 1 h at room temperature at a 1/100 dilution. The slices were washed in PBS and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Slices were analyzed by using an Axio-Phot fluorescence microscope.
Similar results were obtained by using monoclonal H,K-ATPase antibodies HK9 (kindly provided by M. Caplan, Yale University School of Medicine, New Haven, CT) or an autoimmune antibody produced in mice after thymectomy (Claeys et al., 1994) by using anti-Na,K-ATPase antibodies 6H (kindly provided by M. Caplan) or 9-A5 (Affinity Bioreagents) and by using the anti-FXYD3 or the anti-FXYD4 antibodies described above.
RESULTS
The Signal Peptide of FXYD3 Is Not Cleaved
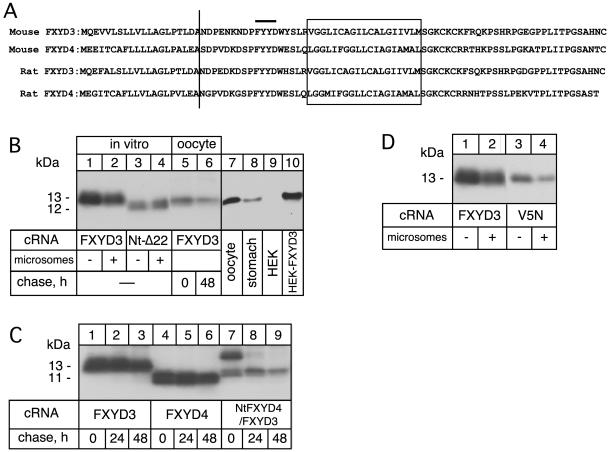
We have previously shown that cleavage of a signal peptide in rat FXYD4, which among the FXYD proteins is most closely related to FXYD3, leads to its type I membrane orientation with the N terminus exposed to the extracytoplasmic side (Béguin et al., 2001). Similar to FXYD4, SignalIP 3.0 program (Bendtsen et al., 2004) predicts a signal peptide cleavage site with high probability between amino acid positions 20 and 21 (LDA-ND) in mouse FXYD3 (Figure 1A). To verify this prediction, we expressed wild-type FXYD3 and an N-terminally truncated mutant (Nt-Δ22) in reticulocyte lysates in the presence or absence of dog pancreatic microsomes. As shown in Figure 1B, both in the absence and presence of microsomes, the wild-type FXYD3 proteins migrated at a higher molecular mass (around 13 kDa) than the Nt-Δ22 mutant (12 kDa, lanes 1–4), indicating lack of signal peptide cleavage in FXYD3. Similar results were obtained after expression of wild-type FXYD3 and the Nt-Δ22 mutant in Xenopus oocytes (our unpublished data). Moreover, FXYD3 expressed in oocytes had the same molecular mass as that expressed in vitro, either after a 24-h pulse or a 48-h chase period (lanes 5 and 6), indicating that the predicted cleavage does not occur in oocytes either. Finally, Western blot analysis revealed that in transfected HEK cells (lane 10) and in situ in the stomach (lane 8), FXYD3 has the same molecular mass as when expressed in oocytes, suggesting a lack of the predicted signal peptide cleavage in all expression systems.
Figure 1.
Biosynthesis of FXYD3. (A) Sequence comparison between mouse and rat FXYD3 and FXYD4. The vertical bar indicates the putative cleavage site of the signal peptide. The FXYD motif is indicated. The transmembrane domain is boxed. (B) Wild-type FXYD3 and the Nt-Δ22 cRNAs were translated in vitro in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of canine pancreatic microsomes and [35S]methionine. Mouse FXYD3 cRNA (2 ng) was injected into Xenopus oocytes and pulse labeled with [35S]methionine for 24 h (lane 5) followed by a 48-h chase period (lane 6). In vitro-translated samples or oocyte microsomes were subjected to SDS-tricine gel electrophoresis and revealed by fluorography. Proteins from microsomes of FXYD3-expressing oocytes (50 μg) (lane 7) or from mouse stomach microsomes (50 μg) (lane 8) or from nontransfected or FXYD3-transfected HEK cells (50 μg) (lanes 9 and 10) were resolved on SDS-tricine gels and subjected to Western blot analysis by using an anti-FXYD4 antibody (1/250). Indicated is the molecular mass of FXYD3 and the Nt-Δ22 mutant. (C) Mouse FXYD3, FXYD4, or FXYD4/FXYD3 cRNAs (2 ng) were injected into Xenopus oocytes and pulse labeled with [35S]methionine for 6 h followed by a 24- and a 48-h chase period. Oocyte microsomes were subjected to SDS-tricine gel electrophoresis and revealed by fluorography. (D) Mouse FXYD3 and V5N mutant cRNAs were translated in vitro in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of canine pancreatic microsomes. In vitro-translated samples were subjected to SDS-tricine gel electrophoresis and revealed by fluorography. One of two similar experiments is shown.
FXYD4, which has a cleavable signal peptide, shows >55% sequence homology with FXYD3 in the N-terminal domain and a nearly 50% overall sequence homology. To investigate which part of the FXYD3 protein is responsible for the lack of signal peptide cleavage, we produced a chimera in which the N-terminal domain of FXYD3 was replaced by that of FXYD4. After expression in oocytes, FXYD4 migrated with a lower molecular mass (11 kDa) compared with FXYD3 after only a 6-h pulse period. Thus, despite possessing a similar number of amino acids as FXYD3, FXYD4 migrates at a lower molecular mass protein, as would be expected from the cleavage of its signal peptide (Figure 1C, lanes 1–6). Interestingly, the FXYD4/FXYD3 chimera exhibited two forms after a 6-h pulse (lane 7), a slow-migrating band corresponding approximately to the uncleaved FXYD3 protein (14 kDa), and a doublet resembling in its molecular mass the cleaved form (∼12 kDa). After 24- and 48-h chase periods, only the doublet remained stably expressed (lanes 8 and 9). The presence of a doublet is characteristic for both FXYD3 and FXYD4 (Béguin et al., 2001) and may be due to an unknown posttranslational modification. The reason for the slight difference in the migration of both the cleaved and uncleaved species of the chimera NtFXYD4/FXYD3 is not known, but it is likely due to a particular conformation of this chimera that leads to aberrant migration. Thus, these results show that in contrast to FXYD3, the FXYD4/FXYD3 chimera is cleaved, but at a slower rate than FXYD4. This indicates that the N-terminal sequence is in part, but not entirely, responsible for the lack of cleavage of a signal peptide in FXYD3.
The lack of cleavage of the signal peptide of FXYD3 raises a question concerning its structure. Indeed, the noncleaved hydrophobic domain may either be embedded in the membrane, forming an extratransmembrane-like domain, or it may be exposed to the endoplasmic reticulum (ER) lumen. To discriminate between these two possibilities, we introduced a putative N-glycosylation site at position 5 by replacing valine 5 with an asparagine residue and thus creating an N-glycosylation consensus site (Figure 1A). If the uncleaved N terminus is exposed to the ER lumen, FXYD3 should become glycosylated at this site (Nilsson and von Heijne, 1993; Hasler et al., 2000). As shown in Figure 1D, this mutant (V5N), when expressed in vitro in the absence or presence of microsomes, had the same molecular mass as the wild-type FXYD3, indicating that the mutation does not lead to a cleavage of the N-terminal signal peptide nor to glycosylation of FXYD3. Similar results were obtained after expression of the V5N mutant in Xenopus oocytes (our unpublished data). This result strongly suggests that the N-terminal domain of the FXYD3 is not exposed to the ER lumen but rather remains partly or entirely embedded in the membrane.
FXYD3 Interacts with Na,K- and H,K-ATPases after Expression in Xenopus Oocytes and Influences Their Glycosylation Processing
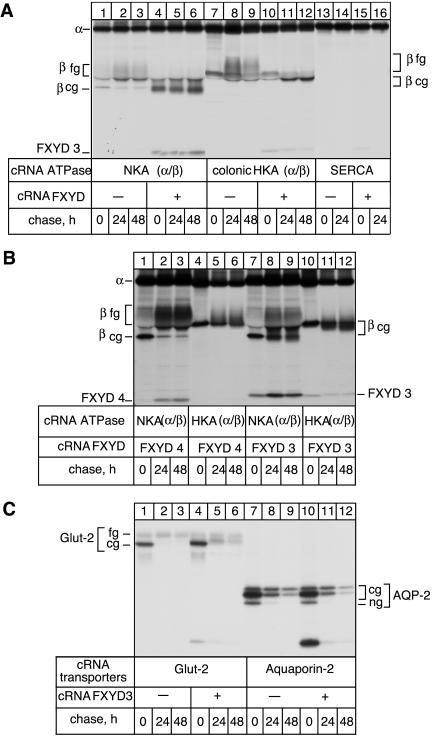
The ability of FXYD3 to interact with Na,K-ATPase or other proteins was first tested in Xenopus oocytes, by coexpressing the Na,K-ATPase (rat α1 + rat β1), the gastric H,K-ATPase (rabbit gαHK + rabbit gβHK), the colonic H,K-ATPase (rat cαHK + Bufo bladder β), the Ca-ATPase (human SERCA2a), the rat Glut-2 transporter, and the human epitope-flagged Aquaporin-2 with or without mouse FXYD3. After metabolic labeling and various chase periods, digitonin extracts of oocytes were prepared and subjected to immunoprecipitation under nondenaturing conditions with antibodies against Na,K-ATPase α subunit, H,K-ATPase α subunit, SERCA or Glut-2, or a FLAG antibody (for the Aquaporin-2), respectively. As shown in Figure 2A, in addition to the β subunit, the antibodies against the α subunit of the Na,K-ATPase (lanes 4–6) and to a lesser extent the colonic H,K-ATPase (lanes 10–12) coimmunoprecipitated FXYD3 both during the pulse and the chase periods. On the other hand, FXYD3 was not coimmunoprecipitated with the Ca-ATPase (lanes 15–16). Thus, in contrast to other FXYD proteins studied so far, like the γ subunit (Béguin et al., 1997), CHIF (Béguin et al., 2001), FXYD7 (Béguin et al., 2002), and phospholemman (Crambert et al., 2002), FXYD3 seems to stably associate not only with the Na,K-ATPase but also with H,K-ATPases. To confirm this result, we expressed in the same experiment FXYD4 or FXYD3 together with Na,K-ATPase or gastric H,K-ATPase and performed nondenaturing immunoprecipitations under identical conditions. As reported previously (Béguin et al., 2001), under the nondenaturing conditions used, FXYD4 was coimmunoprecipitated with Na,K-ATPase antibodies (Figure 2B, lanes 1–3) but not with gastric H,K-ATPase antibodies (lanes 4–6). On the other hand, FXYD3 coimmunoprecipitated with Na,K-ATPase antibodies (lanes 7–9) as well as with gastric H,K-ATPase antibodies (lanes 10–12) during the pulse and the chase period, although to a somewhat lesser extent. Moreover, coimmunoprecipitation of gastric H,K-ATPase could be achieved by using FXYD3 antibodies under nondenaturing conditions (our unpublished data). FXYD3 was also coimmunoprecipitated with Glut-2 (Figure 2B, lanes 4–6) and Aquaporin-2 (lanes 10–12) but mainly during the pulse period and not during the chase periods, indicating a weak and unstable association that may be considered as nonspecific. The reason for the recovery of very high amounts of FXYD3 with Aquaporin-2 during the 6-h pulse period is not known.
Figure 2.
Association of FXYD3 with P-type ATPases and other transporters in Xenopus oocytes, and effect on the glycosylation processing. (A) Oocytes were injected with rat Na,K-ATPase (NKA) cRNA (10 ng of α1 and 1 ng of β1 cRNAs, lanes 1–6) or rat colonic H,K-ATPase (colonic HKA) cRNA (10 ng of α1 and 1 ng of Bufo bladder β cRNAs, lanes 7–12), or human SERCA2a cRNA (5 ng, lanes 13–16) in the absence or presence of 2 ng of FXYD3 cRNA. After cRNA injection, oocytes were labeled with [35S]methionine for 6 h followed by a 24- and 48-h chase period, and digitonin extracts were prepared. Nondenaturing immunoprecipitation with antibodies against the Na,K-ATPase α subunit, the colonic H,K-ATPase, or SERCA2a was performed, and the immunoprecipitates were resolved by SDS-PAGE and revealed by fluorography. (B) Oocytes were injected with rat Na,K-ATPase (NKA) cRNA (10 ng of α1 and 1ngof β1 cRNAs, lanes 1–3 and 7–9) or rabbit, gastric H,K-ATPase cRNA (HKA) (10 ng of α1 and 1 ng of β1 cRNAs, lanes 4–6 and 10–12) in the presence of 2 ng of FXYD4 (lanes 1–6) or FXYD3 (lanes 7–12). Nondenaturing immunoprecipitations were performed with antibodies against the Na,K-ATPase α subunit or the gastric H,K-ATPase. (C) Oocytes were injected with 5 ng of Glu-2 transporter cRNA (lanes 1–6) or 5 ng of FLAG-Aquaporin-2 cRNA (lanes 7–12) in the absence (lanes 1–3 and 7–9) or presence of FXYD3 (lanes 4–6 and 10–12). Digitonin extracts were prepared after the pulse and the chase periods and were immunoprecipitated under nondenaturing conditions with a Glut-2 antibody (lanes 1–6) or a FLAG antibody (lanes 7–12). cg, core glycosylated form; fg, fully glycosylated form. One of 2–20 experiments is shown.
Interestingly, the presence of FXYD3 modified the processing of the β subunits of the β subunit-containing P-type ATPases, which remained in their core-glycosylated forms even after a 48-h chase (Figure 2A, compare lanes 1–3 with lanes 4–6; lanes 7–9 with 10–12). Similarly, the glycosylation processing of Glut-2, which partially remained in its coreglycosylated form during the chase period, was impeded in the presence of FXYD3 (Figure 2B, compare lanes 1–3 with 4–6), despite the lack of association with FXYD3. Alternatively, the presence of FXYD3 may prevent the degradation of the core-glycosylated form of Glut-2. The glycosylation processing of Aquaporin-2, which remained in its core-glycosylated, endoglycosidase-H–sensitive form when expressed in Xenopus oocytes (lanes 7–9), was not affected by the presence of FXYD3 (lanes 10–12).
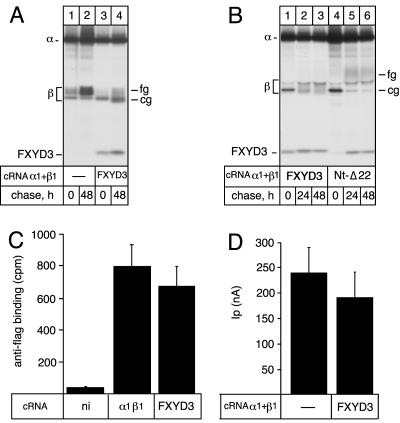
An effect on the glycosylation processing normally reflects ER retention of a protein. We thus investigated whether the presence of FXYD3 impedes the exit of the Na,K-ATPase out of the ER and, in consequence, its cell surface expression. For this purpose, we expressed the rat Na,K-ATPase α1 with an epitope-flagged β1 subunit in the presence or absence of FXYD3. As shown in Figure 3A, the presence of a FLAG epitope in the β subunit did not change the association efficiency of Na,K-ATPase with FXYD3. Moreover, the glycosylation processing of the flagged β subunit was retarded in the presence of FXYD3 similar to that of the wild-type β subunit. On the other hand, both the number of Na,K-ATPase at the cell surface, as assessed by binding of an iodinated FLAG-antibody on intact oocytes (Figure 3C), and the Na,K-pump current amplitude (Figure 3D) were not significantly different in the absence or presence of FXYD3, indicating that the Na,K-ATPase is not retained in the ER but normally expressed and functional at the cell surface 3 d after expression in oocytes.
Figure 3.
Effect of FXYD3 on the glycosylation processing and cell surface expression of the Na,K-ATPase in Xenopus oocytes. (A and B) Rat Na,K-ATPase α1 (10 ng) and Xenopus FLAGβ1 (2 ng) cRNAs (A) or rat α1 and β1 (B) cRNAs were injected into oocytes in the absence or presence of mouse FXYD3 (2 ng) or Nt-Δ22 mutant (4 ng) cRNAs, and pulse labeled for 6 h followed by various chase periods. Microsomes were prepared and subjected to nondenaturating immunoprecipitations by using an antibody against Na,K-ATPase α subunit. Immunoprecipitates were resolved on 5–13% SDS-PAG and revealed by fluorography. (C) Three days after injection as described in A, intact noninjected (ni) or cRNA-injected oocytes were subjected to a radioimmunolabeling assay by using a 125I-labeled anti-FLAG antibody. Shown are means ± SE from seven to nine oocytes. (D) Three days after injection as described in B, the K+-activated Na,K-pump current was measured on intact oocytes by the two-electrode voltage-clamp technique as described in Materials and Methods. Shown are means ± SE of five to six oocytes from two batches of oocytes.
To check whether the specific topology of FXYD3 was involved in its effect on the glycosylation processing of membrane proteins, we compared the glycosylation pattern of the β subunit of Na,K-ATPase in the presence of wild-type FXYD3 or of the Nt-Δ22 mutant. As shown in Figure 3B, the absence of the putative signal peptide did not abrogate the ability of FXYD3 to interact with the Na,K-ATPase but the association was delayed compared with the wild-type FXYD3 (compare lanes 1–3 with 4–6). Nearly no Nt-Δ22 mutant coimmunoprecipitated with Na,K-ATPase after the pulse period but amounts similar to wild-type FXYD3 were coimmunoprecipitated after the chase periods. Moreover, the effect on the glycosylation processing of the β subunit of Na,K-ATPase was abolished in the presence of the Nt-Δ22 mutant, indicating a link between the presence of the signal peptide and the effect of FXYD3 on the protein glycosylation processing.
Location of FXYD3 in the Stomach and Interaction with Na,K-ATPase In Situ
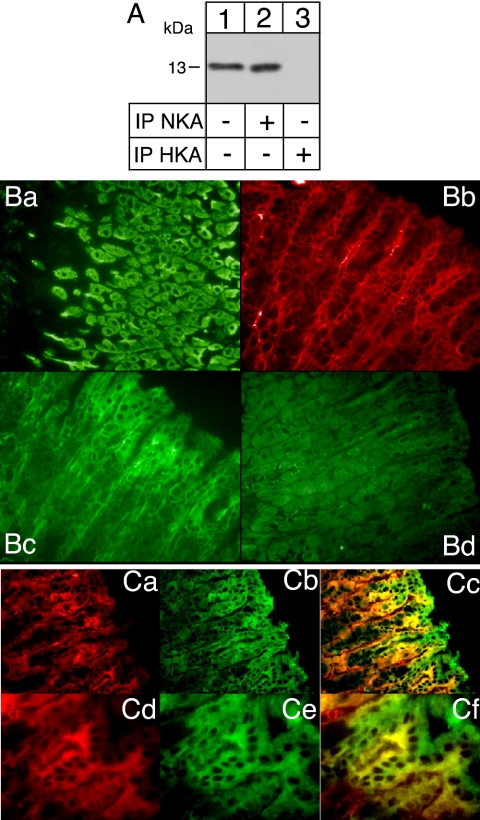
As shown in Figure 2, when expressed in Xenopus oocytes, FXYD3 is able to interact with Na,K-ATPase as well as with gastric and colonic H,K-ATPases. In view of its known tissue localization, we determined which ATPase interacts with FXYD3 in situ. We prepared microsomes from mouse gastric epithelium and performed nondenaturing immunoprecipitations by using an antibody against the Na,K-ATPase α subunit or against the H,K-ATPase α subunit. Western blot analysis of gastric epithelial microsomes revealed a protein with a molecular mass of 13 kDa (Figure 4A, lane 1) similar to that obtained after in vitro or oocyte expression and corresponding to the FXYD3 protein. Nondenaturing immunoprecipitations of the Na,K-ATPase (Figure 4A, lane 2), but not of the H,K-ATPase (Figure 4A, lane 3), revealed a similar protein indicating that in situ, FXYD3 is specifically associated with the Na,K-ATPase. To better understand the reason for this specificity, we investigated the localization of FXYD3 in the gastric gland. This analysis revealed that FXYD3 is mainly expressed in cells forming the upper part of the gland (compare Figure 4Bc and 4Cb to preimmune staining in 4Bd), namely, the mucus cells, and is absent from the H,K-ATPase–expressing cells, namely, the parietal cells (compare H,K-ATPase labeling in Figure 4Ba and FXYD3 labeling in Figure 4Bc). On the other hand, Na,K-ATPase is expressed at the basolateral side of the cells all along the gastric gland (Figure 4Bb, 4Ca, and 4Cd) and shows colocalization with FXYD3 (Figure 4Cc and 4Cf) in the mucus cells. Moreover, FXYD3 is expressed at both sides of the epithelial mucus cells, suggesting that it may have other functions in addition to its role as an Na,K-ATPase–associated protein.
Figure 4.
FXYD3 is associated with Na,K-ATPase in stomach epithelial cell. (A) Aliquots (40 μg of protein) from stomach epithelial microsomes were directly loaded on a tricine-gel (lane 1) or first immunoprecipitated (400 μg of protein) with an antibody against Na,K-ATPase α subunit (lane 2) or against gastric H,K-ATPase α subunit (6H) (lane 3) under nondenaturating conditions. Proteins were transferred onto nitrocellulose, and Western blot analysis was performed using an anti-FXYD4 antibody. (B and C) Mouse stomach slices were used for simple or double immunostaining with an anti FXYD3 antibody (Bc, Cb, and Ce, green) and with either a monoclonal anti-H,K-ATPase antibody (HK9 in Ba) or monoclonal anti-Na,K-ATPase antibodies (6H, in Bb; 9-A5, in Ca and Cd, red). After merging (Cc and Cf), colocalization is indicated in yellow. Magnification, 40× (Ba, Bb, Bd, Ca, Cb, and Cc) and 63× (Cd, Ce, and Cf).
Functional Effect of FXYD3 Association on Na,K-ATPase Transport and Enzymatic Properties
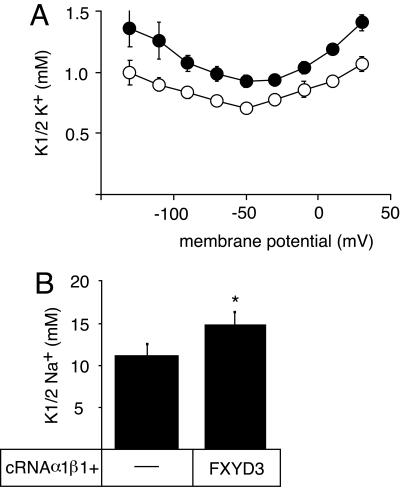
We investigated the effects of FXYD3 on the Na,K-ATPase transport properties by electrophysiological means after coexpression in Xenopus oocytes. As shown in Figure 5A, FXYD3 had a small but significant effect on the apparent affinity for external K+. In the presence of external Na+ (90 mM), the K½ K+ values increased by 15–40%, depending on the membrane potential. Moreover, the apparent affinity for intracellular Na+ was decreased by ∼20% in the presence of FXYD3 (Figure 5B).
Figure 5.
Effects of FXYD3 on the transport and enzymatic properties of Na,K-ATPase. (A) Three days after injection of rat Na,K-ATPase α1 (10 ng) and β1 (1 ng) cRNAs in the absence or presence of FXYD3 (2 ng) cRNA, K+-activation constants (K½ K+) of the Na,K-ATPase were determined in the presence of 90 mM external Na+. Open circles, Na,K-ATPase alone; closed circles, Na,K-ATPase plus FXYD3. Shown are means ± SE of 10 oocytes from two different batches. Phosphate-buffered saline <0.05 at all membrane potentials. (B) Two days after injection of rat α1 (10 ng) and β1 (1 ng) cRNAs in the absence or presence of FXYD3 (2 ng) cRNA, the epithelial Na+ channel subunit cRNAs (α, β, and γ, 0.3 ng each) were injected. After overnight incubation, Na+ activation constants (K½ Na+) were determined at–50 mV. Shown are means ± SE from 11 oocytes from three different batches. *p < 0.01. For electrophysiological measurements, endogenous, oocyte Na,K-ATPase was inhibited by the presence of 1 μM ouabain.
DISCUSSION
Four of seven members of the FXYD protein family have been identified as modulators of the Na,K-ATPase. In this study, we provide evidence that a fifth member of this family, FXYD3 or Mat-8, also interacts with and modulates Na,K-ATPase functions. However, FXYD3 exhibits structural and functional features that differ from the other members of the FXYD protein family characterized so far.
FXYD3, a Double-spanning Membrane Protein?
Analysis of the mouse FXYD3 sequence by the SignalIP 3.0 program (Bendtsen et al., 2004) predicts the presence of an N-terminal signal peptide of 20 amino acids similar to FXYD1 and FXYD4. However, in contrast to FXYD1 (Palmer et al., 1991) and FXYD4 (Béguin et al., 2001), the signal peptide is not cleaved in FXYD3 after in vitro or in vivo expression as suggested by the difference in the gel migration of the wild-type FXYD3 and a truncated mutant lacking 22 N-terminal amino acids. This lack of cleavage is not an artifact of the expression systems used because in cRNA-injected oocytes, in transfected mammalian cells (HEK 293) and in situ (stomach microsomes), a shorter form, which could correspond to a processed product of FXYD3, is never observed. Further evidence for the lack of signal peptide cleavage in FXYD3 is provided by the fact that Morrison et al. (1995), who first cloned and studied FXYD3, could detect the protein expressed in Xenopus oocytes and in breast tumor cells by using an antibody directed against amino acids 2–17, encompassing the putative signal peptide. Interestingly FXYD4, which is most closely related to FXYD3, also displays an unusual cleavage of its signal peptide, because it is only processed in vivo but not in vitro in the presence of microsomes (Béguin et al., 2001).
A typical signal peptide is composed of three regions: a positively charged N-terminal region (n), a central hydrophobic core (h), and a neutral but polar C-terminal domain (c). Whether a signal peptide is cleaved or not mainly depends on the amino acids at position 3 and 1 of the cleavage site (von Heijne, 1984). Although FXYD3 contains small uncharged amino acids at these positions necessary for cleavage, only replacement of the entire N-terminal signal peptide by that of FXYD4 permits cleavage. These data indicate that cleavage may depend on other features. This is supported by the observation by Rehm et al. (2001) who showed that, despite the presence of a classical N-terminal signal peptide in terms of its sequence and cleavage site, type I human cytomegalovirus US11 polypeptide shows delayed cleavage of its signal peptide. In addition to the n-domain of the signal peptide, the delayed cleavage is dependent on the transmembrane domain close to the C terminus, suggesting an interaction of the hydrophobic part of the signal peptide with the transmembrane domain that may impede signal peptide cleavage. Interestingly, the transmembrane domain of FXYD3 is more hydrophobic in the C-terminal part than that of FXYD4. An–IVL-sequence in FXYD3 is replaced by –AMA- in FXYD4, which could favor interaction between the signal peptide and the transmembrane domain, and impede cleavage. On the other hand, inefficient cleavage of HIV-1 gp120 protein was correlated with a prolonged association with the molecular chaperone calnexin that leads to retention of gp120 in the ER (Li et al., 1996).
Concerning FXYD3, the reason for the lack of cleavage is not fully understood, but it seems that the N-terminal domain is involved because its replacement by that of FXYD4 leads to cleavage. The identification of the domain or amino acids involved in this phenomenon needs further investigation.
The lack of cleavage of the signal peptide raises the question about the membrane topology of FXYD3. To determine whether the signal peptide is embedded in the membrane or exposed to the ER lumen, we inserted an N-glycosylation consensus motif in the h-domain of the putative signal peptide Nilsson and von Heijne, 1993; Hasler et al., 2000). The observed lack of glycosylation of FXYD3 indicates that the hydrophobic part is not accessible to oligosaccharyltransferase and may therefore be embedded in the membrane, forming a second transmembrane domain. Alternatively, FXYD3 may be retained in the membrane only by its signal peptide, whereas the putative transmembrane domain would have flipped to the ER lumen leading to a type II orientation. This is unlikely because such a FXYD protein would not preserve the functional interaction with Na,K-ATPase, which requires the transmembrane domain of FXYD proteins (Li et al., 2004). Furthermore, the mutant lacking the signal peptide is still membrane bound.
Interaction of FXYD3 with P-Type ATPases and Effect on the Glycosylation Processing of Plasma Membrane Protein
A common feature of FXYD proteins characterized so far is their specific interaction with Na,K-ATPase (for review, see Crambert and Geering, 2003). FXYD3 shares this property but shows a broader specificity than other FXYD proteins. After expression in Xenopus oocytes, FXYD3 can interact with all X,K-ATPase. On the other hand, FXYD3 does not associate with other P-type ATPases such as SERCA. FXYD3 cRNA is most abundantly expressed in stomach (Morrison et al., 1995), which coexpresses Na,K-ATPase and H,K-ATPase. We show by coimmunoprecipitation experiments that FXYD3 is specifically associated with Na,K-ATPase in this organ. The reason for this specificity in situ is due to the predominant expression of FXYD3 in mucus cells of the gastric gland (devoid of H,K-ATPase) and its lack of expression in parietal cells.
From these observations it may be concluded that the specificity of interaction of FXYD proteins is restricted to X,K-ATPase. Some FXYD proteins, such as FXYD7, are able to discriminate between Na,K-ATPase isozymes (Béguin et al., 2002), and some, such as FXYD3, are able to associate with all known X,K-ATPases, at least in a heterologous expression system. It is possible that this specificity is related to the physiological context in which the FXYD proteins are expressed. Indeed, FXYD7 is present in cells (neurons and glial cells) that contain many different Na,K-ATPase isozymes, necessitating a high specificity of FXYD7 for one isozyme. On the other hand, FXYD3 is expressed in cells containing only one Na,K-ATPase isozyme and no other X,K-ATPases, rendering discrimination between different X,K-ATPases unnecessary.
An interesting property of FXYD3 is its effect on the glycosylation processing of the β subunits of X,K-ATPases. Indeed, all X,K-ATPases expressed in Xenopus oocytes in the presence of FXYD3 contain a core-glycosylated β subunit even after a 48-h chase period. Generally, the presence of core-glycosylated species in glycoproteins indicates that proteins are retained in the ER and cannot pass through the Golgi apparatus. This is not the case for Na,K-ATPase associated with FXYD3, which is expressed at the cell surface. Interestingly, FXYD3 has the same effect on the glycosylation processing of Glut-2, despite the lack of stable association with FXYD3, indicating that association of FXYD3 is not directly linked to the effect on the glycosylation processing. It is possible that the presence of FXYD3 (perhaps at high concentrations) may interfere with the sorting pathway of proteins targeted to the plasma membrane and impede their full glycosylation. Interestingly, the effect of FXYD3 on the glycosylation processing depends on the presence of the uncleaved signal peptide.
Physiological Relevance of the Regulation of Na,K-ATPase by FXYD3
Similar to other FXYD proteins studied, association of FXYD3 with Na,K-ATPase decreases its apparent K+ affinity. This K+ effect is a current feature of FXYD proteins, probably reflecting a common interaction site with Na,K-ATPase that involves transmembrane segment 9 of the Na,K-ATPase α subunit (Li et al., 2004). Although it is most closely related to FXYD4 at the sequence level, the effect of FXYD3 on the apparent Na+ affinity of Na,K-ATPase is opposite to that of FXYD4. Mutational analysis in the transmembrane domain, which has been shown to be involved in both the stable association with and the functional effects of FXYD proteins on Na,K-ATPase (Lindzen et al., 2003; Zouzoulas et al., 2003; Crambert et al., 2004), may help to identify amino acids responsible for the differential functional effect of FXYD3 and FXYD4.
It is difficult to draw conclusions on the physiological relevance of the Na+ and K+ effects of FXYD3 on Na,K-ATPase. FXYD3 is highly up-regulated in specific types of cancer such as breast or prostate tumors. Based on experimental evidence, FYXD3 has been implicated in both antiproliferating (Maxwell et al., 2003) and proliferating processes (Grzmil et al., 2004). Further experiments are needed to show whether regulation of the functional properties of Na,K-ATPase by FXYD3 leading to a modification of the ionic cellular milieu may be important for triggering or impeding cell proliferation.
Finally, because FXYD3 not only colocalizes with Na,K-ATPase in basolateral membranes of mucus cells of the stomach but also seems to be expressed in apical membranes, it cannot be excluded that FXYD3 has other yet unknown functions in addition to being an Na,K-ATPase–modulating protein.
Acknowledgments
We thank Danièle Schaer and Sophie Roy for skillful technical help. This study was supported by grant 31-64793.01 to K. G. from the Swiss National Science Foundation.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–10–0878) on March 2, 2005.
References
- Arystarkhova, E., Wetzel, R. K., Asinovski, N. K., and Sweadner, K. J. (1999). The γ subunit modulates Na+ and K+ affinity of the renal Na,K-ATPase. J. Biol. Chem. 274, 33183–33185. [DOI] [PubMed] [Google Scholar]
- Béguin, P., Crambert, G., Guennoun, S., Garty, H., Horisberger, J.-D., and Geering, K. (2001). CHIF, a member of the FXYD protein family, is a regulator of Na,K-ATPase distinct from the γ-subunit. EMBO J. 20, 3993–4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguin, P., Crambert, G., Monnet-Tschudi, F., Uldry, M., Horisberger, J.-D., Garty, H., and Geering, K. (2002). FXYD7 is a brain-specific regulator of Na,K-ATPase α1-β isozymes. EMBO J. 21, 3264–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguin, P., Wang, X. Y., Firsov, D., Puoti, A., Claeys, D., Horisberger, J. D., and Geering, K. (1997). The γ subunit is a specific component of the Na,K-ATPase and modulates its transport properties. EMBO J. 16, 4250–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J. D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Claeys, D., Karapetian, O., Saraga, E., Schreyer, M., Louis, J., Acha-Orbea, H., Blum, A. L., and Kraehenbuhl, J. P. (1994). Mouse mammary tumor virus superantigens and murine autoimmune gastritis. Gastroenterology 107, 924–933. [DOI] [PubMed] [Google Scholar]
- Crambert, G., Fuzesi, M., Garty, H., Karlish, S., and Geering, K. (2002). Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc. Natl. Acad. Sci. USA 99, 11476–11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crambert, G., and Geering, K. (2003). FXYD proteins: new tissue-specific regulators of the ubiquitous Na,K-ATPase. Sci. STKE 166, RE1. [DOI] [PubMed] [Google Scholar]
- Crambert, G., Hasler, U., Beggah, A. T., Yu, C., Modyanov, N. N., Horisberger, J. D., Lelievre, L., and Geering, K. (2000). Transport and pharmacological properties of nine different human Na, K-ATPase isozymes. J. Biol. Chem. 275, 1976–1986. [DOI] [PubMed] [Google Scholar]
- Crambert, G., Li, C., Swee, L. K., and Geering, K. (2004). FXYD7: mapping of functional sites involved in endoplasmic reticulum export, association with and regulation of Na,K-ATPase. J. Biol. Chem. 279, 30888–30895. [DOI] [PubMed] [Google Scholar]
- Feraille, E., and Doucet, A. (2001). Sodium-potassium-adenosinetriphosphatase-dependent sodium transport in the kidney: hormonal control. Physiol. Rev. 81, 345–418. [DOI] [PubMed] [Google Scholar]
- Garty, H., Lindzen, M., Scanzano, R., Aizman, R., Fuzesi, M., Goldshleger, R., Farman, N., Blostein, R., and Karlish, S.J.D. (2002). A functional interaction between CHIF and Na-K-ATPase: implication for regulation by FXYD proteins. Am. J. Physiol. 283, F607–F615. [DOI] [PubMed] [Google Scholar]
- Geering, K., Beggah, A., Good, P., Girardet, S., Roy, S., Schaer, D., and Jaunin, P. (1996). Oligomerization and maturation of Na,K-ATPase: functional interaction of the cytoplasmic NH2 terminus of the β subunit with the α subunit. J. Cell Biol. 133, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardet, M., Geering, K., Frantes, J. M., Geser, D., Rossier, B. C., Kraehenbuhl, J.-P., and Bron, C. (1981). Immunochemical evidence for a transmembrane orientation of both the (Na+,K+)-ATPase subunits. Biochem. J. 20, 6684–6691. [DOI] [PubMed] [Google Scholar]
- Grzmil, M., Voigt, S., Thelen, P., Hemmerlein, B., Helmke, K., and Burfeind, P. (2004). Up-regulated expression of the MAT-8 gene in prostate cancer and its siRNA-mediated inhibition of expression induces a decrease in proliferation of human prostate carcinoma cells. Int. J. Oncol. 24, 97–105. [PubMed] [Google Scholar]
- Hasler, U., Greasley, P. J., von Heijne, G., and Geering, K. (2000). Determinants of topogenesis and glycosylation of type II membrane proteins. Analysis of Na,K-ATPase β1 and β 3 subunits by glycosylation mapping. J. Biol. Chem. 275, 29011–29022. [DOI] [PubMed] [Google Scholar]
- Hasler, U., Wang, X., Crambert, G., Beguin, P., Jaisser, F., Horisberger, J. D., and Geering, K. (1998). Role of β-subunit domains in the assembly, stable expression, intracellular routing, and functional properties of Na,K-ATPase. J. Biol. Chem. 273, 30826–30835. [DOI] [PubMed] [Google Scholar]
- Jaunin, P., Jaisser, F., Beggah, A. T., Takeyasu, K., Mageat, P., Rossier, B. C., Horisberger, J.-D., and Geering, K. (1993). Role of the transmembrane and extracytoplasmic domain of β subunits in the assembly, intracellular transport, and functional expression of Na,K-pumps. J. Cell Biol. 123, 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaisser, F., Jaunin, P., Geering, K., Rossier, B. C., and Horisberger, J. D. (1994). Modulation of the Na,K-pump function by β subunit isoforms. J. Gen. Physiol. 103, 605–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Grosdidier, A., Crambert, G., Horisberger, J. D., Michielin, O., and Geering, K. (2004). Structural and functional interaction sites between Na,K-ATPase and FXYD proteins. J. Biol. Chem. 279, 38895–38902. [DOI] [PubMed] [Google Scholar]
- Li, Y., Bergeron, J. J., Luo, L., Ou, W. J., Thomas, D. Y., and Kang, C. Y. (1996). Effects of inefficient cleavage of the signal sequence of HIV-1 gp 120 on its association with calnexin, folding, and intracellular transport, Proc. Natl. Acad. Sci. USA 93, 9606–9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindzen, M., Aizman, R., Lifshitz, Y., Lubarski, I., Karlish, S.J.D., and Garty, H. (2003). Structure-function relations of interactions between Na,K-ATPase, the γ subunit, and corticosteroid hormone-induced factor. J. Biol. Chem. 278, 18738–18743. [DOI] [PubMed] [Google Scholar]
- Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) Protein measurements with the Folin phenol reagent. J. Biol. Chem. 193, 265–275. [PubMed] [Google Scholar]
- Maxwell, P. J., Longley, D. B., Latif, T., Boyer, J., Allen, W., Lynch, M., McDermott, U., Harkin, D. P., Allegra, C. J., and Johnston, P. G. (2003). Identification of 5-fluorouracil-inducible target genes using cDNA microarray profiling. Cancer Res. 63, 4602–4606. [PubMed] [Google Scholar]
- Melton, D. A., Krieg, P. A., Rebagliati, M. R., Maniatis, T., Zinn, K., and Green, M. R. (1984). Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12, 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, B. W., and Leder, P. (1994). Neu and ras initiate murine mammary tumors that share genetic markers generally absent in c-myc and int-2initiated tumors. Oncogene 9, 3417–3426. [PubMed] [Google Scholar]
- Morrison, B. W., Moorman, J. R., Kowdley, G. C., Kobayashi, Y. M., Jones, L. R., and Leder, P. (1995). Mat-8, a novel phospholemman-like protein expressed in human breast tumors, induces a chloride conductance in Xenopus oocytes. J. Biol. Chem. 270, 2176–2182. [DOI] [PubMed] [Google Scholar]
- Nelson, R. M., and Long, G. L. (1989). A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal. Biochem. 180, 147–151. [DOI] [PubMed] [Google Scholar]
- Nilsson, I. M., and von Heijne, G. (1993). Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268, 5798–5801. [PubMed] [Google Scholar]
- Palmer, C. J., Scott, B. T., and Jones, L. R. (1991). Purification and complete sequence determination of the major plasma membrane substrate for cAMPdependent protein kinase and protein kinase C in myocardium. J. Biol. Chem. 266, 11126–11130. [PubMed] [Google Scholar]
- Pu, H. X., Cluzeaud, F., Goldshlegger, R., Karlish, S.J.D., Farman, N., and Blostein, R. (2001). Functional role and immunocytochemical localization of the γa and γb forms of the Na,K-ATPase γsubunit. J. Biol. Chem. 276, 20370–20378. [DOI] [PubMed] [Google Scholar]
- Rehm, A., Stern, P., Ploegh, H. L., and Tortorella, D. (2001). Signal peptide cleavage of a type I membrane protein, HCMV US11, is dependent on its membrane anchor. EMBO J. 20, 1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweadner, K. J., and Rael, E. (2000). The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics 68, 41–56. [DOI] [PubMed] [Google Scholar]
- Therien, A. G., and Blostein, R. (2000). Mechanisms of sodium pump regulation. Am. J. Physiol. 279, C541–C566. [DOI] [PubMed] [Google Scholar]
- von Heijne, G. (1984). How signal sequences maintain cleavage specificity. Patterns of amino acids near signal-sequence cleavage sites. J. Mol. Biol. 173, 243–251. [DOI] [PubMed] [Google Scholar]
- Zouzoulas, A., Therien, A. G., Scanzano, R., Deber, C. M., and Blostein, R. (2003). Modulation of Na,K-ATPase by the γ subunit: studies with transfected cells and transmembrane mimetic peptides. J. Biol. Chem. 278, 40437–40441. [DOI] [PubMed] [Google Scholar]