Abstract
Originally detected in fixed cells, DNA replication foci (RFi) were later visualized in living cells by using green fluorescent protein (GFP)-tagged proliferating cell nuclear antigen (PCNA) and DNA ligase I. It was shown using fluorescence redistribution after photobleaching (FRAP) assay that focal GFP-PCNA slowly exchanged, suggesting the existence of a stable replication holocomplex. Here, we used the FRAP assay to study the dynamics of the GFP-tagged PCNA-binding proteins: Flap endonuclease 1 (Fen1) and DNA polymerase η (Polη). We also used the GFP-Cockayne syndrome group A (CSA) protein, which does associate with transcription foci after DNA damage. In normal cells, GFP-Polη and GFP-Fen1 are mobile with residence times at RFi (tm) ∼2 and ∼0.8 s, respectively. GFP-CSA is also mobile but does not concentrate at discrete foci. After methyl methanesulfonate (MMS) damage, the mobile fraction of focal GFP-Fen1 decreased and tm increased, but it then recovered. The mobilities of focal GFP-Polη and GFP-PCNA did not change after MMS. The mobility of GFP-CSA did not change after UV-irradiation. These data indicate that the normal replication complex contains at least two mobile subunits. The decrease of the mobile fraction of focal GFP-Fen1 after DNA damage suggests that Fen1 exchange depends on the rate of movement of replication forks.
INTRODUCTION
In mammalian cells, DNA replication during S phase is located at distinct nuclear sites. Replication foci (RFi) each contain on average 10 spatially clustered active replication forks that can be visualized using antibodies against incorporated halogenated deoxyuridines (Nakamura et al., 1986; Nakayasu and Berezney, 1989; van Dierendonck et al., 1989; O'Keefe et al., 1992; Tomilin et al., 1995; Jackson and Pombo, 1998). In fixed cells, the RFi colocalize with the Tritoninsoluble form of the replication protein proliferating cell nuclear antigen (PCNA) (Bravo and McDonald-Bravo, 1987), the p70 subunit of replication protein A (RPA70), protein kinase Cdk2 and cyclin A (Cardoso et al., 1993), DNA polymerase α (Hozak et al., 1993), DNA ligase I (Cardoso et al., 1997), and DNA polymerase η (Polη) (Kannouche et al., 2001). Distinct nuclear sites accumulate green fluorescent protein (GFP)-tagged DNA ligase I (Cardoso et al., 1997), GFP-PCNA (Leonhardt et al., 2000; Somanathan et al., 2001). The foci containing GFP-RPA34 (Sporbert et al., 2002) or GFP-Polη (Kannouche et al., 2001) also can be observed in normally proliferating living S-phase cells, indicating that the RFi seen in fixed cells do not arise during the fixation procedure but reflect real clustering of replication forks and associated molecular machines in the nucleus.
It was initially suggested that the RFi represent specialized subnuclear organelles or nucleoskeleton-attached replication factories in which preassembled holocomplexes (replisomes) duplicate incoming DNA (Hozak et al., 1993). In agreement with this model, recent studies of living cells expressing GFP-tagged PCNA by using the fluorescence redistribution after photobleaching (FRAP) assay (Axelrod et al., 1976; Lippincott-Schwartz et al., 2001) showed that focal GFP-PCNA exchanged very slowly (Sporbert et al., 2002). During S phase, immobile GFP-PCNA foci can assemble and disassemble in the nucleus with lifetimes ranging from 30 min to 3 h (Leonhardt et al., 2000), consistent with significant heterogeneity of replication units and the existence of very large replicons (Liapunova, 1994). However, another replication protein, GFP-RPA34, was found to be mobile at RFi and after bleaching completely redistributed within 2 min (Sporbert et al., 2002). This indicates that despite their existence as morphologically distinct nuclear bodies, replication factories may contain mobile components.
Transcription factories (Jackson et al., 1993) and nucleotide excision repair foci (Jackson et al., 1994; Svetlova et al., 2002) have been visualized in mammalian cells, and recent studies indicate that they are stochastically assembled de novo in each round of transcription or repair from freely diffusible components (Houtsmuller et al., 1999; Dundr et al., 2002; Kimura et al., 2002). Cockayne syndrome group A (CSA) protein is required for the excision of DNA lesions at stalled transcription sites, also called transcription-coupled repair (TCR) (Hanawalt, 2002). After UV-irradiation, a fraction of this protein becomes insoluble in a buffer containing Triton X-100, and this insoluble CSA shows colocalization with active (phosphorylated) RNA polymerase II foci (Kamiuchi et al., 2002). However, possible changes of mobility of CSA protein after DNA damage have not been studied so far.
It is known that strand elongation during DNA replication can be inhibited by DNA lesions induced by UV-irradiation and some chemical carcinogens, e.g., methyl methanesulfonate (MMS) (Merrick et al., 2004), and this leads to the accumulation of S-phase cells with stalled replication forks (Kannouche et al., 2001, 2004). In these cells, clusters of blocked replication forks colocalize with stably bound PCNA and Polη, which is required for translesion DNA synthesis across damaged sites (Masutani et al., 1999). PCNA and Polη associated with normal or stalled replication foci are insoluble in methanol or in a buffer containing Triton X-100 (Celis and Madsen, 1986; Bravo and McDonald-Bravo, 1987; Toschi and Bravo, 1988; Kannouche et al., 2001).
In yeast, Polη (Rad30p)-dependent translesion synthesis requires the monoubiquitination of PCNA performed by the Rad6p/Rad18p complex (Stelter and Ulrich, 2003) and the Rad6 epistasis group (controlling lesion bypass), which in addition to the Rad30 and Rad18 genes also contains the Rad27 gene encoding the structure-specific Flap nuclease Fen1 (Reagan et al., 1995). The yeast Fen1 protein, which is involved in the maturation of Okazaki fragments, suppresses the accumulation of single-stranded DNA (ssDNA) and double-strand breaks (DSBs) at replication forks (Debrauwere et al., 2001). Mammalian Rad18 protein and Polη are required for lesion bypass (Cordeiro-Stone et al., 1997; Masutani et al., 1999; Tateishi et al., 2000; Limoli et al., 2002; Tateishi et al., 2003); mammalian Rad18 also monoubiquitinates Polη at stalled replication forks (Kannouche et al., 2004; Watanabe et al., 2004). How DNA damage induces activation of the Rad18 protein is unknown, but some data suggest that the ATR-Chk1 checkpoint pathway may be involved (Nikiforov et al., 2004). The Fen1 (-/-) mouse is not viable (Kucherlapati et al., 2002), and expression of nuclease-defective Fen1 inhibits S-phase progression after MMS treatment (Shibata and Nakamura, 2002), suggesting that Fen1 may be required for efficient lesion bypass.
Here, we used the FRAP assay to study the mobilities of GFP-tagged human proteins Fen1, Polη, PCNA, and CSA in transfected Chinese hamster cells before and after the induction of DNA damage by MMS or UV. We found that in normally proliferating cells focal GFP-Fen1 and GFP-Polη are very mobile, supporting the concept of self-organization of cellular architecture (Misteli, 2001).
MATERIALS AND METHODS
Cells, Transfections, and Treatments
Chinese hamster V79-4 immortal lung fibroblasts and human A539 cells were obtained from American Type Culture Collection (Manassas, VA). Cells were cultivated in RPMI 1640 medium supplemented with 10% of fetal calf serum (FCS). Transient transfections with the indicated plasmids involved the TRANS-FAST reagent (Promega, Madison, WI), and cells were analyzed 24–48 h after transfection. Stable expressing clones were isolated after transient transfection by selection in growth medium containing 0.6–1 mg/ml G418 (Geneticin). For isolation of a stable clone expressing GFP-Fen1, the corresponding plasmid was introduced into V79-4 cells. A plasmid encoding GFP-CSA protein was introduced into CS3BESV cells (SV40-transformed cells from a patient with Cockayne syndrome group A, cultivated in DMEM with 10% FCS), and UV-sensitivity of the obtained clone (OS-7) was compared with the UV-sensitivity of the normal fibroblasts line WI38VA13 as described previously (Kamiuchi et al., 2002). Expressing clones were identified by GFP fluorescence and then expanded. MMS (Aldrich Chemical, Milwaukee, WI) was added to cells in growth medium to the final concentration of 0.01 or 0.03% for 1 h. Then, cells were washed with phosphate-buffered saline (PBS) and incubated in growth medium for the time intervals indicated.
Sources of Plasmids Encoding GFP-tagged Proteins
Plasmids encoding GFP-Fen1 and GFP-PCNA were constructed for this study as described below. DNA fragments containing the cDNAs of human Fen1 and PCNA were amplified from human polyA-containing RNA by using the single-tube Titan reverse transcription-PCR kit (Roche Diagnostics, Indianapolis, IN). The PolyA-containing RNAs were isolated from cultivated human A539 cells by using the mRNA isolation kit (Roche Diagnostics). The sequences of primers used to amplify human Fen1 cDNA were Fen-C1 CTGTGTTGCCATGGGAATTC and Fen-C2 TTCCCCTTTTAAACTTCCCTGC. The sequences of primers to amplify human PCNA cDNA were PCNA-C1 CTAGCTAGACTTTCCTCCTTCC and PCNA-C2 CTTCTTCATCCTCGATCTTGGG. For the construction of expression plasmids GFP-Fen1 and GFP-PCNA we used C-terminal fusion GFP-TOPO vectors from Invitrogen (Carlsbad, CA) and chemically competent TOP10 cells. Individual clones were screened by PCR, and the presence of the expected inserts was confirmed by sequencing. Plasmid DNAs were purified using a QIAGEN kit (QIAGEN, Valencia, CA). Analysis of Chinese hamster cells transfected with the GFP-Fen1 plasmid involved Western blotting with anti-GFP antibodies (Zymed Laboratories, South San Francisco, CA) and showed presence of the fusion protein of expected size (∼70 kDa). The plasmid encoding GFP-tagged DNA polymerase η (eGFP-Polη) was obtained from A. R. Lehmann (University of Sussex, Brighton, United Kingdom). The plasmid encoding GFP-CSA was constructed by subcloning of the PCR-amplified CSA coding sequence (Kamiuchi et al., 2002) into the vector pEGFP-N3 (BD Biosciences Clontech, Palo Alto, CA).
Immunofluorescence Detection of PCNA and RNA Polymerase II in Fixed Cells
GFP-Fen1–expressing cells grown on glass slides were washed with PBS, extracted overnight at –20°C in 100% methanol, and then fixed in 4% formaldehyde. PCNA was visualized using mouse monoclonal antibodies PC-10 (1:100, 30 min; LabVision/NeoMarkers, Fremont, CA) and anti-mouse IgG coupled to Alexa Fluor 568 (1:400, 30 min; Molecular Probes, Eugene, OR). For the detection of PCNA and RNA polymerase II, GFP-Polη and GFP-CSA–expressing cells were washed in PBS and then extracted (Kamiuchi et al., 2002) for 20 min at room temperature in cytoskeleton (CSK) buffer containing 10 mM Pipes, pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM dithiothreitol (DTT), mM EGTA, and 0.5% Triton X-100 and then washed in PBS and fixed in 4% formaldehyde. PCNA was then visualized in GFP-Polη–expressing cells by using mouse monoclonal antibodies PC-10 (1:100, 30 min; Santa Cruz Biotechnology, Santa Cruz, CA), biotinylated sheep anti-mouse IgG (1:100, 30 min; Sigma-Aldrich, St. Louis, MO), and avidin-Texas Red (1:200, 30 min; Vector Laboratories, Burlingame, CA). RNA polymerase II in GFP-CSA–expressing cells was detected using mouse monoclonal antibodies against phosphorylated RNAP II (clone H5) obtained from BAbCO (Richmond, CA) (1:200, 60 min) followed by the secondary anti-mouse antibodies coupled to Alexa Fluor 568.
Immunoprecipitation and Immunoblotting
For coimmunoprecipitation (CIP), cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (1 ml/100-mm plate) to which (after clearing lysates by centrifugation) 5 μl of undiluted antibodies was added. After incubation for 1 h on ice, immune complexes were isolated using protein A coupled to Sepharose beads (100 μl of 10% suspension per 1 ml of initial lysate), which before SDS electrophoresis, were washed five times with RIPA buffer. Then, pellets were suspended in 2× Laemmli buffer (2% SDS, 10% glycerol, 100 mM DTT, 60 mM Tris-HCl, pH 6.8, and 0.001% bromphenol blue) and heated at 85°C for 10 min. RIPA buffer contained 50 mM Tris-HCl buffer, pH 8, 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium dodecylsulfate, and 0.5% sodium deoxycholate. For CIP, we used mouse monoclonal antibodies (clone 14A1) against human recombinant Fen1 (catalog no. MS-1752; NeoMarkers), mouse monoclonal antibodies (clone JL-8) against GFP (catalog no. 8371-2; BD Biosciences Clontech), and custom-made rabbit polyclonal antibodies against the human Fen1-derived peptide N′-CLTFGS(PO4)PVLMRHLTA-C′ (Genemed Synthesis, South San Francisco, CA). As negative CIP controls, affinity-purified polyclonal antibodies against CSB or CSA proteins (sc-10459 and sc-10997, resp.; Santa Cruz Biotechnology) or preimmune rabbit serum (Genemed Synthesis) was used. On immunoblots, coimmunoprecipitated PCNA was detected using mouse monoclonal antibodies PC-10 (catalog no. MS-106-P1; NeoMarkers). After separation of proteins in denaturing SDS polyacrylamide gels, they were transferred to a membrane and then processed as described previously (Siino et al., 2002; Nazarov et al., 2003). For chemiluminescence detection of Fen1 and GFP-tagged protein, we used the same primary monoclonal mouse monoclonal antibodies described above.
FRAP Assay
The FRAP assay (Axelrod et al., 1976; Lippincott-Schwartz et al., 2001) involved the photobleaching of a small area in the nucleus at maximal argon laser power followed by serial scans at a lower magnification and laser power. All FRAP assays were carried out on cells in four-well Lab-Tek (Naperville, IL) coverglass chambers. Original FRAP curves of relative fluorescence (RF) intensity were calculated as RF(t) = It/Ipre, where Ipre is the prebleach value for each measurement, and It is the fluorescence at the time point t. After correction for the nonspecific fluorescence loss during the image acquisition, the data obtained under identical conditions for at least 10 cells for each variant were averaged, and the SE was calculated for each time point. To determine the representative residence time (tm) of a protein, the relative fluorescence was calculated and further normalized from the original averaged FRAP curves as RFrt (t) = (It–I0)/(Ipost–I0), where I0 is the relative fluorescence intensity immediately after bleaching, and Ipost is the maximal relative fluorescence intensity observed after recovery (taken as 1). Average residence time (tm) for a given protein was then estimated as the time required for recovery of 63.2% of the final fluorescence (Lukas et al., 2004). To determine mobile fractions the relative fluorescence was calculated and further normalized from the original averaged FRAP curves as RFmf (t) = (It–I0)/(Ipre–I0), where Ipre is the relative fluorescence before bleaching (for normalized sets taken as 1), and I0 is the fluorescence intensity just after bleaching. The mobile fraction is then determined as maximal value of RFmf after recovery. In a Zeiss LSM 510, the first scan at low laser intensity after bleaching takes at least 100 ms, but it is tentatively taken throughout this article that the fluorescence intensity at the end of the first scan at low laser intensity is zero point, because the actual zero point fluorescence immediately after bleaching is impossible to measure.
Microscopy
Images of live and fixed cells and FRAP assay were obtained with a Zeiss confocal laser scanning system LSM-510 equipped with a Plan-NEOFLUAR 63/1.3 objective, helium-neon laser of 543 nm wavelength, and/or argon laser (15 mW) of 458/488-nm wavelengths, or a MRC-1024 fluorescence microscopy system (Bio-Rad).
RESULTS
Expression of GFP-tagged Replication/Repair Proteins in Mammalian Cells
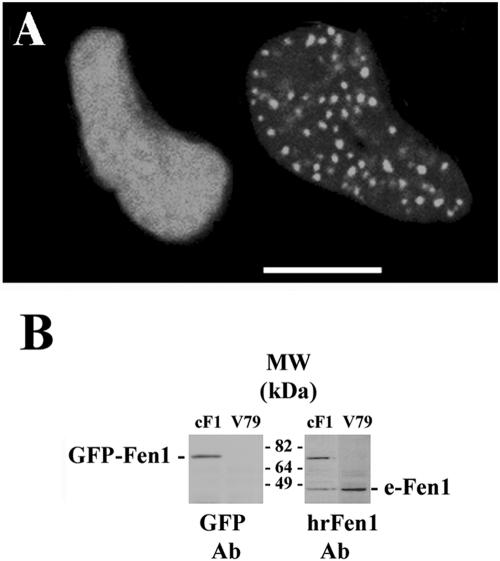
In Chinese hamster cells transiently transfected with the fusion plasmid encoding Fen1-C-GFP, two distinct distributions of the expressed GFP in nuclei were found: one showing only diffuse nuclear fluorescence and the other showing both diffuse and focal GFP nuclear signal (Figure 1A). Similar patterns of GFP labeling were shown in cells from a stable GFP-Fen1–expressing clone which was selected in the growth medium containing G418. Analysis of these cells by Western blotting by using antibodies against GFP showed the presence of a single band of protein of the size expected for the fusion protein (Figure 1B, left) with sensitivity to MMS very similar to that of the parental V79 cells (∼15% survival after 0.01% MMS), indicating that the GFP-tagged Fen1 does not interfere with the repair functions of endogenous Fen1. In stably transformed cells the fusion GFP-Fen1 protein is expressed at about the same level as the endogenous Fen1 (Figure 1B, right blot).
Figure 1.
Expression of GFP-tagged human Fen1 in transfected Chinese hamster V79 cells. (A) Two patterns of GFP-Fen1 distribution in the nuclei of transiently transfected live cells: diffuse (left) and focal (right). Bar, 10 μm. (B) Western blots of total protein from stably transfected V79 cells probed with antibodies against human recombinant Fen1 (right) or GFP (left). Endogenous Fen1 protein is marked as e-Fen1.
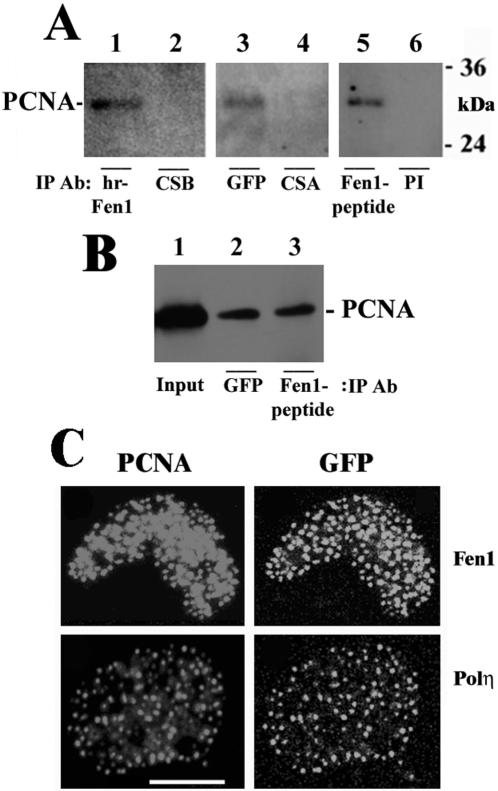
To show that the GFP-Fen1 fusion protein stably expressed in Chinese hamster cells retains its function, we used CIP assay to determine whether it interacts with PCNA. For CIP, we used mouse monoclonal antibodies against human recombinant Fen1 (Figure 2A, lane 2), mouse monoclonal antibodies against GFP (Figure 2A, lane 3), and rabbit polyclonal antibodies against Fen1 peptide (Figure 2A, lane 5). As controls, affinity-purified polyclonal antibodies against CSB or CSA proteins (lanes 2 and 4, respectively) or preimmune rabbit serum (lane 6) were used. It is seen from Figure 2A that PCNA can be coprecipitated from lysates of GFP-Fen1–expressing cells with antibodies against Fen1 (Figure 2A, lanes 2 and 5) as well as with antibodies against GFP (lane 3) but not with antibodies against CSB protein (lane 2), CSA protein (lane 4) or with preimmune rabbit serum (lane 6). PCNA also has been shown to immunoprecipitate by anti-Fen1 and anti-GFP antibodies with equal efficiencies (Figure 2B). These results confirm that Fen1 protein fused at its C terminus to GFP interacts with PCNA.
Figure 2.
Evidence of interaction of GFP-Fen1 protein with endogenous PCNA in Chinese hamster cells. (A) Immunoprecipitation of endogenous PCNA from cell lysates of V79 cells stably expressing GFP-Fen1 with antibodies against human recombinant Fen1 (hr-Fen1, lane 1), GFP (lane 3), or Fen1 peptide (lane 5). As controls, affinity-purified antibodies against CSB protein (lane 2), CSA protein (lane 4) or preimmune serum (lane 6) were used. (B) Comparative PCNA immunoprecipitation with antibodies against GFP (lane 2) and Fen1 peptide (lane 3). Lane 1 shows input. All blots were probed with antibodies against PCNA. (C) Colocalization of endogenous PCNA foci and GFP in V79 cells transiently transfected with GFP-Fen1 plasmid and preextracted with CSK buffer containing 0.5% Triton X-100. Bar, 10 μm.
To confirm that GFP-Fen1 concentrates at replication foci, we examined its colocalization with PCNA foci after preextraction of cells with 100% methanol, which removes soluble PCNA, leaving only molecules involved in DNA replication (Madsen and Celis, 1985), or with CSK buffer containing 0.5% Triton X-100 (Kamiuchi et al., 2002). We found that the focal GFP-Fen1 in transiently transfected cells preextracted with CSK buffer colocalized with PCNA (Figure 2B and Supplementary Figure 1A). Similar colocalization of the focal GFP signal and PCNA was observed in preextracted V79 cells transiently transfected with the plasmid expressing GFP-tagged Polη (Figure 2C and Supplementary Figure 1B), confirming previous observations (Kannouche et al., 2001). The plasmid expressing GFP-Polη used in our study has been shown to be able to complement a repair defect in XP variant cells (Kannouche et al., 2001) and therefore encodes a fully functional fusion protein. Polη, which has a conserved domain for PCNA binding, is known to interact with PCNA in undamaged cells (Haracska et al., 2001). We also confirmed colocalization of GFP-Fen1 and PCNA in S-phase cells of a stable clone preextracted with methanol (Supplementary Figure 1C). Because PCNA is well known to be the main factor in recruiting Fen1 to replication forks (Li et al., 1995; Jonsson et al., 1998; Gomes and Burgers, 2000; Tom et al., 2000), our results indicate that the fusion of the C terminus of Fen1 with the N terminus of GFP obtained in this study does not effect the Fen1 nuclear localization signal (Qiu et al., 2001) nor its binding to PCNA during replication.
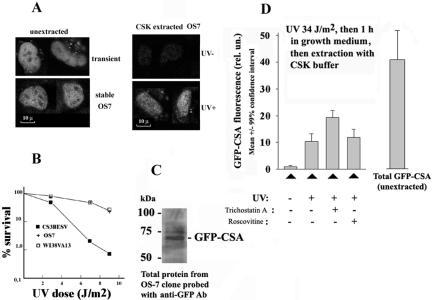
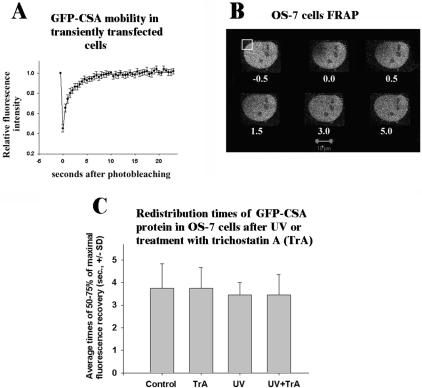
To study the function of the GFP-CSA protein, its plasmid was transfected into human UV-sensitive cells deficient in endogenous CSA protein, resulting in a stable expression clone OS-7 (Figure 3A, left two images) that was found to be UV-resistant (Figure 3B). The expressed fusion protein of expected size was detected in this clone by anti-GFP antibodies (Figure 3C). The CSA protein does not interact with PCNA, and GFP-CSA can be almost completely extracted from unirradiated OS-7 cells with a cytoskeleton buffer containing 0.5% Triton X-100 (Figure 3A, right top image). However, GFP-CSA becomes resistant to extraction with the indicated buffer after UV-irradiation (Figure 3A, right bottom image), confirming previous observations (Kamiuchi et al., 2002). In many preextracted UV-irradiated cells, Triton-insoluble GFP-CSA partially colocalizes with active RNA polymerase II (Supplementary Figure 2). Interestingly, the treatment of cells with an inhibitor of histone deacetylases trichostatin A (but not with an inhibitor of protein kinases roscovitine) leads to a significantly increased amount of Triton-insoluble GFP-CSA in UV-irradiated cells (Figure 3D). This drug does not induce a significant increase of overall expression of GFP-CSA in OS7 cells (our unpublished data), suggesting that the UV-induced insolubilization of CSA protein (Kamiuchi et al., 2002) may involve interactions of this protein with chromatin containing hyperacetylated histones.
Figure 3.
Expression of GFP-CSA protein in human cells (CS3BESV) with mutation of endogenous CSA protein. (A) Images of cells expressing GFP-CSA protein. Left two images show transiently (top) or stably (bottom) transfected living cells, and right two images show unirradiated (top) or UV-irradiated (bottom) stably transfected cells preextracted with CSK buffer containing 0.5% Triton X-100 (Kamiuchi et al., 2002). Bar (A), 10 μm. UV dose was 34 J/m2 followed by 1-h incubation of cells in growth medium. (B) Survival of stably transfected GFP-CSA expressing cells (clone OS-7) after UV, parental cells (CS3BESV), and normal human fibroblasts (line WI38VA13). (C) Immunoblot of proteins from OS-7 cells probed with antibodies against GFP. (D) UV-induced insolubilization of GFP-CSA protein in OS-7 cells and its stimulation by pretreatment with trichostatin A (300 nM, 24 h). Roscovitine (20 μM) was added for 6 h before UV-irradiation. Mean GFP-CSA fluorescence intensity >50 of nuclei of OS-7 cells was measured under identical conditions (amplification gain and magnification) for all variants.
Our GFP-PCNA construct in which GFP is fused to the PCNA C terminus is also likely to be functional because it associates with replication foci and becomes diffusionally immobile (see below), confirming previous observations (Sporbert et al., 2002). It also may be noted that in another study, the C-fusion of PCNA to GFP showed the expected behavior at replication foci (Somanathan et al., 2001), suggesting that GFP linked to C-end of PCNA does not interfere with replication.
Focal GFP-Fen1 and GFP-Polη Are Mobile in Normally Proliferating Living Cells
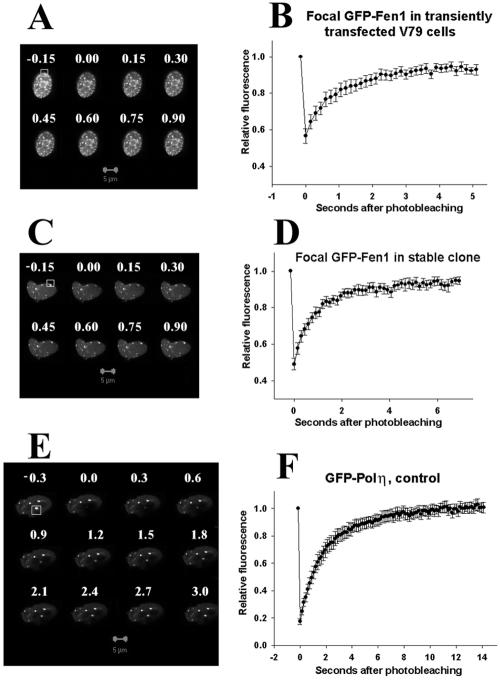
The FRAP analyses of the mobilities of GFP-Fen1 and GFP-Polη are shown in Figure 4. It is seen that fluorescence of the focal GFP-Fen1 in transiently (Figure 4, A and B) or stably expressing (Figure 4, C and D) undamaged Chinese hamster cells is recovered after photobleaching after a few seconds. A similar rapid recovery in transiently transfected V79 cells was found for focal GFP-Polη (Figure 4, E and F), indicating that both these PCNA-interacting proteins dynamically interact with the replication machinery in S-phase cells and are in continuous exchange with the pool of nucleoplasmic protein during DNA synthesis. Using the FRAP assay, we also analyzed the mobilities of diffuse nuclear GFP-Fen1, GFP-Polη, and diffuse and focal GFP-PCNA and calculated the average redistribution times of these proteins (Table 1) and their mobile fractions (Table 2) as described in Materials and Methods. It is seen from Table 1 that in a stably expressing clone, diffuse GFP-Fen1 is redistributed in 0.38 s, whereas focal GFP-Fen1 is redistributed in 0.78 s, thereby explaining the observation of distinct GFP-Fen1 foci in living cells. A similar increase in the redistribution times of the focal component (2.05 s) compared with the diffuse component (1.01 s) is observed in cells transiently transfected with GFP-Polη (Table 1). The mobile fractions of focal GFP-Fen1 and GFP-Polη in undamaged cells are found to be 0.89 and 1.03, respectively (Table 2, first row), indicating that major fractions of these proteins are mobile in the nucleus. The redistribution times of the focal components are equivalent to their representative residence times in foci (Lukas et al., 2004), so GFP-Fen1 associates with replication foci for ∼1 s and GFP-Polη for ∼2 s. Diffuse GFP-PCNA also is mobile and showed a redistribution time of 0.66 s, but the focal GFP-PCNA is very slowly exchanged (Table 1), confirming the observations of other authors (Sporbert et al., 2002).
Figure 4.
FRAP analyses of mobility of GFP-Fen1 in transiently transfected (A and B) or stably expressing cells (C and D) and GFP-Polη in transiently transfected cells (E and F). Bar (A, C, and E), 5 μm; numbers show seconds after photobleaching. In B, D, and F, vertical bars represent SEs of average values from 10 to 30 cells for each variant. Bleached segments are boxed.
Table 1.
Redistribution times of GFP-Fen1, GFP-Polη, GFP-PCNA, and GFP-CSA
| tm (s)
|
||||
|---|---|---|---|---|
| Variant | Diffuse component, normal cells | Focal component, normal cells | Focal component, 2 h after 0.03% MMS | Focal component, 24 h after 0.03% MMS |
| GFP-Fen1 (stable) | 0.38 | 0.78 | 2.03 | 0.79 |
| GFP-Polη (transient) | 1.01 | 2.05 | 1.94 | 2.29 |
| GFP-PCNA (transient) | 0.66 | >10 min | >10 min | >10 min |
| GFP-CSA (stable) | 3.88 | NA | NA | NA |
Normalization of the averaged original recovery curves has been done using equation RF™ (t) = (It – I0)/(Ipost – I0). Value of tm (representative residence time for focal component) was calculated by approximation from the normalized curves as time required for 63.2% recovery of the fluorescence intensity after complete recovery as described previously (Lukas et al., 2003). For mathematical reasons, the SD of tm is always as large as the tm itself (Lukas et al., 2004) and is not shown here.
Table 2.
Mobile fractions of focal GFP-Fen1 and GFP-Polη before and after treatment of cells with MMS
| MF at different times after MMS treatment (h)a
|
||||||
|---|---|---|---|---|---|---|
| Protein | 0 | 0.5 | 2.5 | 5.5 | 8 | 24 |
| GFP-Fen1 (0.01% MMS) | 0.89 ± 0.02 | 1.00 ± 0.02 | 0.78 ± 0.06 | 0.86 ± 0.01 | 0.90 ± 0.02 | 0.97 ± 0.03 |
| GFP-Fen1 (0.03% MMS) | 0.89 ± 0.02 | 0.89 ± 0.04 | 0.40 ± 0.04 | 0.42 ± 0.04 | 0.89 ± 0.04 | 0.94 ± 0.04 |
| GFP-Polη (0.03% MMS) | 1.03 ± 0.04 | 0.82 ± 0.03 | 0.92 ± 0.03 | 0.88 ± 0.04 | 0.93 ± 0.02 | 0.95 ± 0.03 |
MF is calculated as maximal value of the function RFmf = (It–I0)/(Ipre–I0), where Ipre is the initial fluorescence before photobleaching (taken as 1). First row (0 time) shows values obtained for the control cells not treated with MMS
The GFP-CSA protein, which does not interact with PCNA, showed only a diffuse component in the nucleus (Figure 3). The FRAP analysis of its mobility in transiently or stably expressing cells (Figure 5, A and B) indicates that this protein also is mobile, with a redistribution time in stably expressing OS-7 cells of ∼4 s (Table 1). This tm value is ∼10 times higher than that for diffuse GFP-Fen1, although these proteins have approximately the same size (70 kDa), which suggests that GFP-CSA can transiently interact with an immobile nuclear structure, e.g., with chromatin. However, we have found that the redistribution time of GFP-CSA is not significantly changed after treatment of cells with an inhibitor of histone deacetylases, trichostatin A, or UV (Figure 5C). It may be noted that an absence of increased residence time of GFP-CSA after UV-irradiation suggests that a major fraction of this protein is not immobilized at TCR foci, but immobilization of a small fraction of GFP-CSA cannot be excluded.
Figure 5.
FRAP assay of mobility of GFP-CSA in transiently transfected V79 (A) or stably expressing OS-7 cells (B and C). Vertical bars in A show SE; numbers in B show seconds after photobleaching. Bar (B), 10 μm; bleached segment is boxed. Redistribution times in C were calculated from the original FRAP curves as mean times for recovery of 50–75% of the maximal fluorescence recovery ± SD. Trichostatin A (TrA; 300 nM) was added for 24 h, UV-dose was 34 J/m2 followed by 1-h incubation in growth medium.
The function of the mobile Polη associated with normal replication foci is unknown, but it may be involved in the bypass of spontaneous single-strand DNA lesions (Avkin and Livneh, 2002; Kusumoto et al., 2002), which are generated at about ∼1000 per cell per hour (Vilenchik and Knudson, 2003).
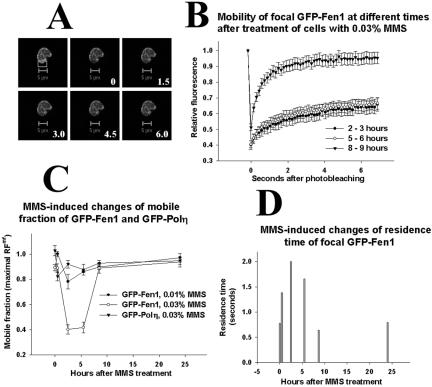
Transient Decrease of Mobility of GFP-Fen1 after Treatment of Cells with Methyl Methanesulfonate
Some DNA lesions induced by MMS in mammalian cells cannot be copied by replicative DNA polymerases, leading to the stalling of replication forks at DNA lesions (Merrick et al., 2004) and to an accumulation of S-phase cells with Polη foci (Kannouche et al., 2001). The treatment of cells with 0.02 and 0.03% MMS (but not with 0.01% MMS) decreases the rate of movement of replication forks (Merrick et al., 2004), which has been shown by the replicative labeling of DNA fibers (Tomilin et al., 1993; Jackson and Pombo, 1998). Using FRAP assay, we examined whether the mobility of focal GFP-Fen1 is effected by MMS treatment. Figure 6, A and B, shows results obtained with stably expressing cells, and it is seen that there is a change in mobility of focal GFP-Fen1 when cells are analyzed 2–3 and 5–6 h after treatment with 0.03% MMS, but normal mobility is observed 8–9 h after DNA damage. Because analysis of each cell takes a few minutes and many cells should be analyzed, we averaged FRAP curves obtained within the indicated time intervals. The mobile fraction of focal GFP-Fen1 is strongly decreased (Figure 6C), and its residence time at replication foci is increased (Figure 6D) at 2–6 h after 0.03% MMS, but when analyzed 8–9 h or at 24 h focal GFP-Fen1 showed normal mobility (Figure 6, C and D). A lower concentration of MMS (0.01%) also induced a slight decrease of the mobile fraction of focal GFP-Fen1 at 2–3 h after treatment (Figure 6C), but it did not effect its residence time. A strong decrease of the mobile fraction of focal GFP-Fen1 at 2–3 h after treatment with 0.03% MMS also was observed with transiently transfected V79 cells (Supplementary Figure 3).
Figure 6.
MMS-induced changes of mobility of GFP-Fen1 in stably expressing V79 cells. In A, bleached segment is boxed; bar is 5 μm, and numbers show seconds after bleaching. Vertical bars in B show SE of average values for 10–30 cells, results obtained at 2–3, 5–6, and 8–9 h after MMS treatment are combined. (C) Mobile fractions of GFP-Fen1 (circles) or GFP-Polη (triangles) at different times after MMS calculated as described in Materials and Methods. (D) Changes of residence time of GFP-Fen1 at replication foci after MMS, calculated as described in Materials and Methods.
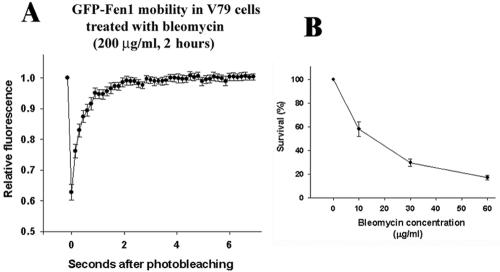
The observed changes of mobility of focal GFP-Fen1 after MMS correlate with the ability of this agent to slowdown the movement of replication forks (Merrick et al., 2004) consistent with the view that they are induced by the stalling of replication forks. However, like ionizing radiation (Painter and Young, 1980; Larner et al., 1999), MMS also suppresses the initiation of new replicons (Merrick et al., 2004). Here, by using FRAP assay, we studied the mobility of focal GFP-Fen1 in transiently transfected Chinese hamster cells after their treatment with radiomimetic bleomycin which suppresses DNA synthesis only through the inhibition of replicon initiation (Noda, 1988; Liapunova et al., 1989). We did not find any changes compared with the control in the recovery of GFP-Fen1 fluorescence after treatment of cells with 200 μg/ml bleomycin (Figure 7A). Bleomycin activity was confirmed by the analysis of γ-H2AX foci (Supplementary Figure 4) and of cell survival (Figure 7B). This result is consistent with the view that the decrease of the mobile fraction of GFP-Fen1 after MMS is caused by an inhibition of movement of replication forks and not by an inhibition of replicon initiation.
Figure 7.
Mobility of GFP-Fen1 after treatment of transiently transfected V79 cells with bleomycin (A) and survival of V79 cells after bleomycin (B). Treatment with bleomycin in B was for 2 h. Vertical bars in A show SE from 10 cells; vertical bars in B show SD from three experiments.
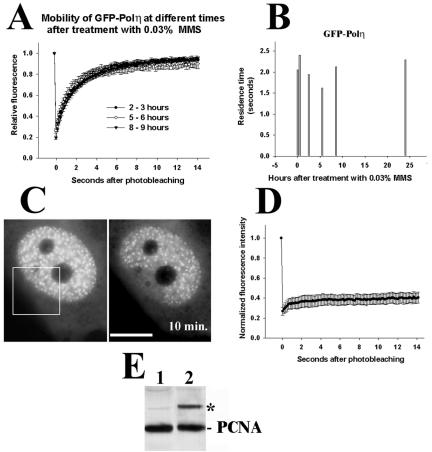
The mobile fraction of focal GFP-Polη is not significantly changed after treatment with 0.03% MMS at 2–3, 5–6, and 8–9 h (Figure 8A). The residence time of GFP-Polη is slightly decreased at 5–6 h after 0.03% MMS but is not changed at 2–3 h (Figure 8B), when a strong decrease of the mobile fraction and an increase of resident time of GFP-Fen1 were observed (Figure 6). The low mobility of GFP-PCNA is not changed after treatment with 0.03% MMS (Figure 8, C and D), although we observed significant monoubiquitination of endogenous PCNA in V79 cells 5 h after lower doses (0.01%) of MMS (Figure 8E, lane 2). It seems, therefore, that GFP-Polη remains very mobile during extensive lesion bypass after MMS damage, which can be functionally important for the efficient polymerase switch at stalled replication forks.
Figure 8.
Mobility of GFP-Polη at different times after MMS treatment (A and B), mobility of GFP-PCNA 5 h after MMS (C and D), and induction of PCNA monoubiquitination by MMS (E). In A–D, treatment was for 1 h with 0.03% MMS, and in E 0.01% MMS was added for 1 h and after washing cells were incubated in growth medium for 5 h. Bar (C), 10 μm. Vertical bars in A show SE calculated for 10–30 cells. Residence times in B were calculated as described in Materials and Methods. Western blot in E was probed with mouse monoclonal antibodies against PCNA (clone PC-10); PCNA means endogenous PCNA.
DISCUSSION
Within the nucleus, DNA synthesis is initiated sequentially at spatially distinct RFi, in which clusters of replication forks (Jackson and Pombo, 1998) accumulate the specific proteins required for DNA synthesis. The accumulation of GFP-tagged variants of these proteins at RFi are seen in living cells, and analyses of their mobilities by using the FRAP assay showed that at least one of them, PCNA, is slowly exchanged (Sporbert et al., 2002). We also found that the focal GFP-PCNA has a low mobility in normally proliferating cells, but two PCNA-binding proteins associated with the RFi in normally proliferating cells, GFP-Fen1 and GFP-Polη, are very mobile with residence times at replication foci ∼0.8 and 2 s, respectively. The redistribution time of focal GFP-PCNA is much longer, and it has been shown that some of the recovery of fluorescence observed in 45 min after bleaching occurs not exactly at the bleached sites but at closely adjacent sites (Sporbert et al., 2002). This may be caused by a slow exchange of PCNA in the moving replication complex with free PCNA molecules or by de novo assembly of PCNA clamps within a single large focus (Sporbert et al., 2002). The lifetime of GFP-PCNA foci in mid-late S-phase cells can reach 3 h (Leonhardt et al., 2000), comparable with the lifetime of mammalian replicons in mid-late S phase, which is ∼3.5 h (Liapunova, 1994).
Together, our results support a dynamic model of the normal replication complex in which relatively long-living and stable DNA-bound PCNA-based holocomplexes bind mobile subunits with low residence time comparable with the time of synthesis of one Okazaki fragment. Our results also directly support the concept of self-organization in cellular architecture (Misteli, 2001). It will be of interest to study the mobilities of other replication proteins, but the existence of at least three mobile components in the replication machinery (RPA34, Fen1, and Polη) indicate that replication complexes are not very different from excision repair, splicing, recombination, and transcription complexes, which are usually assembled from mobile proteins (Houtsmuller et al., 1999; McNally et al., 2000; Phair and Misteli, 2000; Dundr et al., 2002; Essers et al., 2002). High mobility is important for functional plasticity, because it allows different proteins to interact with one and the same target and produce different complexes with common subunits. The very low exchange of PCNA in replication foci may seem to be exceptional, but it should be noted that the half recovery time of transcription-engaged mammalian RNA polymerase II (Pol II) in the FRAP assay in normal living cells is ∼20 min (Kimura et al., 2002). Nonengaged free GFP-Pol II is half-recovered in <0.25 min (Kimura et al., 2002), which is comparable with the redistribution time of the GFP-CSA in undamaged, UV-irradiated or trichostatin A-treated OS-7 cells (3–4 s). This indicates that the majority of GFP-CSA protein is not stably associated with the transcription-engaged Pol II during normal or stalled transcription but may be present in the complex formed by nonengaged Pol II. However, the association with Pol II transcription sites of a minor fraction of CSA protein after UV irradiation (Kamiuchi et al., 2002; Supplementary Figure 2) cannot be excluded using the FRAP assay.
Using the FRAP assay, we also have found that introduction of DNA damage by treatment of cells with MMS leads to a strong but transient decrease of the mobile fraction of GFP-Fen1. The expression of nuclease-defective Fen-1 in mammalian cells causes a prolonged delay of S-phase progression after MMS treatment (Shibata and Nakamura, 2002), indicating involvement of this nuclease in the processing of stalled replication forks. Because Fen1 is known to be recruited to replication through interactions of its conserved C-terminal domain with the interdomain connector loop (IDCL) or C-terminal domain of PCNA (Li et al., 1995; Jonsson et al., 1998; Gomes and Burgers, 2000; Tom et al., 2000), it is possible that DNA damage-induced immobilization of Fen1 reflects modulation of its interactions with PCNA. It is unlikely that Fen1 immobilization is a consequence of the binding to the IDCL of PCNA of the cell cycle inhibitor p21/Cip1 (Zhang et al., 1998), because p21 inhibits Fen1 binding to PCNA (Jonsson et al., 1998). It is also unlikely that MMS-induced immobilization of Fen1 is caused by its phosphorylation at Ser-187 by Cdk1/cyclin A and Cdk2/cyclin A complexes, which also inhibits its binding to PCNA (Henneke et al., 2003). However, we cannot exclude that the Fen1 immobilization is a consequence of DNA damage-induced hyperacetylation of Fen1 protein (Hasan et al., 2001), or of Rad18-dependent monoubiquitination of PCNA (Hoege et al., 2002; Kannouche et al., 2004; Watanabe et al., 2004).
Stalling of DNA polymerase δ at DNA lesions may change contacts of this polymerase with the IDCL of PCNA (Jonsson et al., 1998; Zhang et al., 1998; Ducoux et al., 2001; Lu et al., 2002). Polη also interacts with PCNA through the conserved domain at the Polη C terminus (Haracska et al., 2001; Kannouche et al., 2001), which is very similar to that of Fen1, but the Polη mobile fraction and residence time are not significantly changed after an MMS dose, which induces strong immobilization of GFP-Fen1. This indicates that it is not modulation of Fen1 binding to PCNA but some other factors that can effect Fen1 mobility after DNA damage. If Fen1 detachment from the PCNA clamp during normal lagging strand synthesis is possible only after cutting of the single-stranded tail displaced by Polδ/PCNA complex from the previous Okazaki fragment (Waga and Stillman, 1998; Maga et al., 2001), Fen1 can stay associated with PCNA upon blocking of Polδ/PCNA at the lesion in the template strand and restore its mobility only after elimination of the block. In this scenario, stalling of replication forks at lesions is a direct cause of decreased Fen1 mobile fraction.
MMS-induced changes of GFP-Fen1 mobility found in this study also may be interpreted as an indication of the involvement of this protein in some aspects of lesion bypass. It is known that in yeast the Rad27 gene, which belongs to the Rad6 epistasis group (Reagan et al., 1995), suppresses the accumulation of ssDNA and DSBs at replication forks (Debrauwere et al., 2001). In mammalian cells, nuclease activity of Fen1 promotes S-phase progression after MMS damage (Shibata and Nakamura, 2002), and this activity of Fen1 at replication foci in MMS-treated cells may be directly required for efficient processing of single-strand gaps after DNA polymerase switch and translesion synthesis. This processing may take more time than the elimination of 5′ flaps during normal replication explaining the transient increase of the residence time of mobile fraction of focal GFP-Fen1 in MMS-treated cells (Figure 6D). An increased residence time of Fen1 at replication foci already detectable 30 min after MMS treatment (Figure 6D) may represent the dynamic signal for activation of DNA damage checkpoints required for accumulation of mammalian Rad18 at stalled replication foci (Nikiforov et al., 2004) and Rad18-dependent monoubiquitination of PCNA (Kannouche et al., 2004; Watanabe et al., 2004). Further studies are clearly required to establish actual functional significance of dynamic changes of Fen1 at stalled replication forks.
Acknowledgments
We are grateful to P. C. Hanawalt for support in performing some experiments at the Confocal microscopy facility of the Stanford University, A. R. Lehmann for plasmid encoding GFP-Polη and for helpful discussions; and V. Tomilin for help in the treatment of FRAP curves. This research was supported by the Office of Science (Biological and Environmental Research); U.S. Department of Energy grant DE-FG03-01ER63070; the Russian Fund for Basic Research grants 02-04-49145 and 04-04-49292; the Russian Federal State Contract 10002-251/P-10/143-173/010403-049; and the Russian Academy of Sciences program MCB.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-12-1066) on March 9, 2005.
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
References
- Avkin, S., and Livneh, Z. (2002). Efficiency, specificity and DNA polymerase-dependence of translesion replication across the oxidative DNA lesion 8-oxoguanine in human cells. Mutat. Res. 510, 81-90. [DOI] [PubMed] [Google Scholar]
- Axelrod, D., Koppel, D. E., Schlessinger, J., Elson, E., and Webb, W. W. (1976). Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16, 1055-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo, R., and MacDonald-Bravo, H. (1987). Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J. Cell Biol. 105, 1549-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso, M. C., Leonhardt, H., and Nadal-Ginard, B. (1993). Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 74, 979-992. [DOI] [PubMed] [Google Scholar]
- Cardoso, M. C., Joseph, C., Rahn, H. P., Reusch, R., Nadal-Ginard, B., and Leonhardt, H. (1997). Mapping and use of a sequence that targets DNA ligase I to sites of DNA replication in vivo. J. Cell Biol. 139, 579-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis, J. E., and Madsen, P. (1986). Increased nuclear cyclin/PCNA antigen staining of non S-phase transformed human amnion cells engaged in nucleotide excision DNA repair. FEBS Lett. 209, 277-283. [DOI] [PubMed] [Google Scholar]
- Cordeiro-Stone, M., Zaritskaya, L. S., Price, L. K., and Kaufmann, W. K. (1997). Replication fork bypass of a pyrimidine dimer blocking leading strand DNA synthesis. J. Biol. Chem. 272, 13945-13954. [DOI] [PubMed] [Google Scholar]
- Debrauwere, H., Loeillet, S., Lin, W., Lopes, J., and Nicolas, A. (2001). Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. USA 98, 8263-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dierendonck, J. H., Keyzer, R., van de Velde, C. J., and Cornelisse, C. J. (1989). Subdivision of S-phase by analysis of nuclear 5-bromodeoxyuridine staining patterns. Cytometry 10, 143-150. [DOI] [PubMed] [Google Scholar]
- Ducoux, M., Urbach, S., Baldacci, G., Hubscher, U., Koundrioukoff, S., Christensen, J., and Hughes, P. (2001). Mediation of proliferating cell nuclear antigen (PCNA)-dependent DNA replication through a conserved p21(Cip1)-like PCNA-binding motif present in the third subunit of human DNA polymerase delta. J. Biol. Chem. 276, 49258-49266. [DOI] [PubMed] [Google Scholar]
- Dundr, M., Hoffmann-Rohrer, U., Hu, Q., Grummt, I., Rothblum, L. I., Phair, R. D., and Misteli, T. (2002). A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623-1626. [DOI] [PubMed] [Google Scholar]
- Essers, J., Houtsmuller, A. B., van Veelen, L., Paulusma, C., Nigg, A. L., Pastink, A., Vermeulen, W., Hoeijmakers, J. H., and Kanaar, R. (2002). Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21, 2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, X. V., and Burgers, P. M. (2000). Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 19, 3811-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt, P. C. (2002). Subpathways of nucleotide excision repair and their regulation. Oncogene 21, 8949-8956. [DOI] [PubMed] [Google Scholar]
- Haracska, L., Johnson, R. E., Unk, I., Phillips, B., Hurwitz, J., Prakash, L., and Prakash, S. (2001). Physical and functional interactions of human DNA polymerase η with PCNA. Mol. Cell. Biol. 21, 7199-7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, S., Stucki, M., Hassa, P. O., Imhof, R., Gehrig, P., Hunziker, P., Hubscher, U., and Hottiger, M. O. (2001). Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Mol. Cell 7, 1221-1231. [DOI] [PubMed] [Google Scholar]
- Henneke, G., Koundrioukoff, S., and Hubscher, U. (2003). Phosphorylation of human Fen1 by cyclin-dependent kinase modulates its role in replication fork regulation. Oncogene 22, 4301-4313. [DOI] [PubMed] [Google Scholar]
- Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G., and Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135-141. [DOI] [PubMed] [Google Scholar]
- Houtsmuller, A. B., Rademakers, S., Nigg, A. L., Hoogstraten, D., Hoeijmakers, J. H., and Vermeulen, W. (1999). Action of DNA repair endonuclease ERCC1/XPF in living cells. Science 284, 958-961. [DOI] [PubMed] [Google Scholar]
- Hozak, P., Hassan, A. B., Jackson, D. A., and Cook, P. R. (1993). Visualization of replication factories attached to nucleoskeleton. Cell 73, 361-373. [DOI] [PubMed] [Google Scholar]
- Jackson, D. A., Balajee, A. S., Mullenders, L., and Cook, P. R. (1994). Sites in human nuclei where DNA damaged by ultraviolet light is repaired: visualization and localization relative to the nucleoskeleton. J. Cell Sci. 107, 1745-1752. [DOI] [PubMed] [Google Scholar]
- Jackson, D. A., Hassan, A. B., Errington, R. J., and Cook, P. R. (1993). Visualization of focal sites of transcription within human nuclei. EMBO J. 12, 1059-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D. A., and Pombo, A. (1998). Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140, 1285-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, Z. O., Hindges, R., and Hubscher, U. (1998). Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 17, 2412-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiuchi, S., Saijo, M., Citterio, E., de Jager, M., Hoeijmakers, J. H., and Tanaka, K. (2002). Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc. Natl. Acad. Sci. USA 99, 201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche, P., Broughton, B. C., Volker, M., Hanaoka, F., Mullenders, L. H., and Lehmann, A. R. (2001). Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 15, 158-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche, P. L., Wing, J., and Lehmann, A. R. (2004). Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491-500. [DOI] [PubMed] [Google Scholar]
- Kimura, H., Sugaya, K., and Cook, P. R. (2002). The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159, 777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherlapati, M., et al. (2002). Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proc. Natl. Acad. Sci. USA 99, 9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumoto, R., Masutani, C., Iwai, S., and Hanaoka, F. (2002). Translesion synthesis by human DNA polymerase η across thymine glycol lesions. Biochemistry 41, 6090-6099. [DOI] [PubMed] [Google Scholar]
- Larner, J. M., Lee, H., Little, R. D., Dijkwel, P. A., Schildkraut, C. L., and Hamlin, J. L. (1999). Radiation down-regulates replication origin activity throughout the S phase in mammalian cells. Nucleic Acids Res. 27, 803-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt, H., Rahn, H. P., Weinzierl, P., Sporbert, A., Cremer, T., Zink, D., and Cardoso, M. C. (2000). Dynamics of DNA replication factories in living cells. J. Cell Biol. 149, 271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Li, J., Harrington, J., Lieber, M. R., and Burgers, P. M. (1995). Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 270, 22109-22112. [DOI] [PubMed] [Google Scholar]
- Liapunova, N. A. (1994). Organization of replication units and DNA replication in mammalian cells as studied by DNA fiber radioautography. Int. Rev. Cytol. 154, 261-308. [DOI] [PubMed] [Google Scholar]
- Liapunova, N. A., Lavrushina, O. M., and Terekhov, S. M. (1989). The effect of bleomycin on DNA synthesis in human cells: evidence for grouping of initiation of replicons in the S-period. Mol. Gen. Mikrobiol. Virusol. 1, 34-39. [PubMed] [Google Scholar]
- Limoli, C. L., Giedzinski, E., Bonner, W. M., and Cleaver, J. E. (2002). UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, γ-H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. USA 99, 233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Snapp, E., and Kenworthy, A. (2001). Studying protein dynamics in living cells. Nat. Rev. Mol. Cell. Biol. 2, 444-456. [DOI] [PubMed] [Google Scholar]
- Lu, X., Tan, C. K., Zhou, J. Q., You, M., Carastro, L. M., Downey, K. M., and So, A. G. (2002). Direct interaction of proliferating cell nuclear antigen with the small subunit of DNA polymerase delta. J. Biol. Chem. 277, 24340-24345. [DOI] [PubMed] [Google Scholar]
- Lukas, C., Melander, F., Stucki, M., Falck, J., Bekker-Jensen, S., Goldberg, M., Lerenthal, Y., Jackson, S. P., Bartek, J., and Lukas, J. (2004). Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 23, 2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen, P., and Celis, J. E. (1985). S-phase patterns of cyclin (PCNA) antigen staining resemble topographical patterns of DNA synthesis. A role for cyclin in DNA replication? FEBS Lett. 193, 5-11. [DOI] [PubMed] [Google Scholar]
- Maga, G., Villani, G., Tillement, V., Stucki, M., Locatelli, G. A., Frouin, I., Spadari, S., and Hubscher, U. (2001). Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl. Acad. Sci. USA 98, 14298-14303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani, C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, M., Iwai, S., Takio, K., and Hanaoka, F. (1999). The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700-704. [DOI] [PubMed] [Google Scholar]
- McNally, J. G., Muller, W. G., Walker, D., Wolford, R., and Hager, G. L. (2000). The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262-1265. [DOI] [PubMed] [Google Scholar]
- Merrick, C. J., Jackson, D. A., and Diffley, J. (2004). Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 279, 20067-20075. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2001). The concept of self-organization in cellular architecture. J. Cell Biol. 155, 181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, H., Morita, T., and Sato, C. (1986). Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res. 165, 291-297. [DOI] [PubMed] [Google Scholar]
- Nakayasu, H., and Berezney, R. (1989). Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 108, 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov, I. B., et al. (2003). Dephosphorylation of histone gamma-H2AX during repair of DNA double-strand breaks in mammalian cells and its inhibition by calyculin A. Radiat. Res. 160, 309-317. [DOI] [PubMed] [Google Scholar]
- Nikiforov, A., Svetlova, M., Solovjeva, M., Sasina, L., Siino, J., Nazarov, I., Bradbury, M., and Tomilin, N. (2004) DNA damage-induced accumulation of Rad18 protein at stalled replication forks in mammalian cells involves upstream protein phosphorylation. Biochem. Biophys. Res. Commun. 323, 831-837. [DOI] [PubMed] [Google Scholar]
- Noda, A. (1988). Replicon initiation in normal human cells and in ataxia telangiectasia cells: its differential inhibition by cycloheximide and bleomycin. Cell Biol. Int. Rep. 12, 943-950. [DOI] [PubMed] [Google Scholar]
- O'Keefe, R. T., Henderson, S. C., and Spector, D. L. (1992). Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 116, 1095-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter, R. B., and Young, B. R. (1980). Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc. Natl. Acad. Sci. USA 77, 7315-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair, R. D., and Misteli, T. (2000). High mobility of proteins in the mammalian cell nucleus. Nature 404, 604-609. [DOI] [PubMed] [Google Scholar]
- Reagan, M. S., Pittenger, C., Siede, W., and Friedberg, E. C. (1995). Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 177, 364-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, J., Li, X., Frank, G., and Shen, B. (2001). Cell cycle-dependent and DNA damage-inducible nuclear localization of FEN-1 nuclease is consistent with its dual functions in DNA replication and repair. J. Biol. Chem. 276, 4901-4908. [DOI] [PubMed] [Google Scholar]
- Shibata, Y., and Nakamura, T. (2002). Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J. Biol. Chem. 277, 746-754. [DOI] [PubMed] [Google Scholar]
- Siino, J. S., Nazarov, I. B., Svetlova, M. P., Solovjeva, L. V., Adamson, R. H., Zalenskaya, I. A., Yau, P. M., Bradbury, E. M., and Tomilin, N. V. (2002). Photobleaching of GFP-labeled H2AX in chromatin: H2AX has low diffusional mobility in the nucleus. Biochem. Biophys. Res. Commun. 297, 1318-1323. [DOI] [PubMed] [Google Scholar]
- Somanathan, S., Suchyna, T. M., Siegel, A. J., and Berezney, R. (2001). Targeting of PCNA to sites of DNA replication in the mammalian cell nucleus. J. Cell. Biochem. 81, 56-67. [DOI] [PubMed] [Google Scholar]
- Sporbert, A., Gahl, A., Ankerhold, R., Leonhardt, H., and Cardoso, M. C. (2002). DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell 10, 1355-1365. [DOI] [PubMed] [Google Scholar]
- Stelter, P., and Ulrich, H. D. (2003). Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188-191. [DOI] [PubMed] [Google Scholar]
- Svetlova, M., Solovjeva, L., Pleskach, N., Yartseva, N, Yakovleva, T., Tomilin, N., and Hanawalt, P. (2002). Clustered sites of DNA repair synthesis during early nucleotide excision repair in ultraviolet light-irradiated quiescent human fibroblasts. Exp. Cell Res. 276, 284-295. [DOI] [PubMed] [Google Scholar]
- Tateishi, S., Niwa, H., Miyazaki, J., Fujimoto, S., Inoue, H., and Yamaizumi, M. (2003). Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 23, 474-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi, S., Sakuraba, Y., Masuyama, S., Inoue, H., and Yamaizumi, M. (2000). Dysfunction of human Rad18 results in defective postreplication repair and hypersensitivity to multiple mutagens. Proc. Natl. Acad. Sci. USA 97, 7927-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom, S., Henricksen, L. A., and Bambara, R. A. (2000). Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem. 275, 10498-10505. [DOI] [PubMed] [Google Scholar]
- Tomilin, N., Rosanov, Y., Zenin, V., Bozhkov, V., and Vig, B. (1993). A new and rapid method for visualising DNA replication in spread DNA by immunofluorescence detection of incorporated 5-iododeoxyuridine. Biochem. Biophys. Res. Commun. 190, 257-262. [DOI] [PubMed] [Google Scholar]
- Tomilin, N., Solovjeva, L., Krutilina, R., Chamberland, C., Hancock, R., and Vig, B. (1995). Visualization of elementary DNA replication units in human nuclei corresponding in size to DNA loop domains. Chromosome Res. 3, 32-40. [DOI] [PubMed] [Google Scholar]
- Tomilin, N. V., Solovjeva, L. V., Svetlova, M. P., Pleskach, N. M., Zalenskaya, I. A., Yau, P. M., and Bradbury, E. M. (2001). Visualization of focal nuclear sites of DNA repair synthesis induced by bleomycin in human cells. Radiat. Res. 156, 347-354. [DOI] [PubMed] [Google Scholar]
- Toschi, L., and Bravo, R. (1988). Changes in cyclin/proliferating cell nuclear antigen distribution during DNA repair synthesis. J. Cell Biol. 107, 1623-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik, M. M., and Knudson, A. G. (2003). Endogenous DNA doublestrand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. USA 100, 12871-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga, S., and Stillman, B. (1998). The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721-751. [DOI] [PubMed] [Google Scholar]
- Watanabe, K., Tateishi, S., Kawasuji, M., Tsurimoto, T., Inoue, H., and Yamaizumi, M. (2004). Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23, 3886-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Sun, Y., Hsu, H., Zhang, L., Zhang, Y., and Lee, M. Y. (1998). The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase delta. J. Biol. Chem. 273, 713-719. [DOI] [PubMed] [Google Scholar]