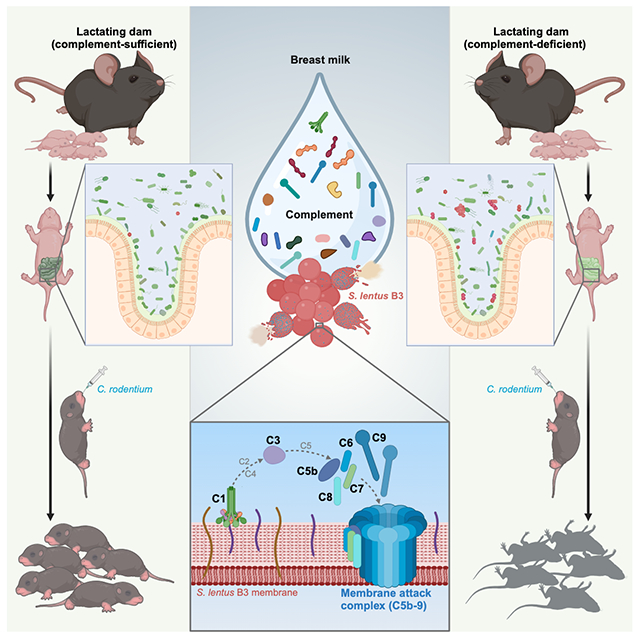
Summary
Breastfeeding offers demonstrable benefits to newborns and infants by providing nourishment, immune protection, and shaping the gut commensal microbiota. Although it has been appreciated for decades that breast milk contains complement components, the physiological relevance of complement in breast milk remains undefined. Here we demonstrate that weanling mice fostered by complement-deficient dams rapidly succumb when exposed to murine pathogen Citrobacter rodentium (CR), whereas pups fostered on complement-containing milk from wild-type dams can tolerate CR challenge. The complement components in breast milk were shown to directly lyse specific members of gram-positive gut commensal microbiota via a C1-dependent, antibody-independent mechanism resulting in the deposition of the membrane attack complex and subsequent bacterial lysis. By selectively eliminating members of the commensal gut community, complement components from breast milk shape neonate and infant gut microbial composition to be protective against environmental pathogens like CR.
Keywords: Staphylococcus lentus, Citrobacter rodentium, MAC deposition, antibody-independent, infant health
Graphical Abstract
In Brief
Breastfeeding offers evident benefits to infant health. This study finds that complement components in breast milk shape the offspring’s evolving gut commensal microbiota, conferring protection against enteric infection.
Introduction
Despite of the remarkable progress made in improving child health in recent decades, the reduction of neonatal, infant, and child mortality remains a priority concern.1 At birth, newborns are antigenically naive and thus lack effective adaptive immunity to most pathogenic challenges.2 During the first weeks of life, neonates rely on innate immune mechanisms, the antibodies that transferred from the mother during gestation, and the molecules and cells in breast milk to provide protection against microbial challenge and to set up a productive dialog with the infant’s evolving microbiota.3–5 Clinical and experimental data strongly indicates that breastfeeding is effective in protecting newborns as milk not only provides high-quality nourishment but also confers a certain level of passive immunity by the transfer of immune cells and protective molecules that include antibodies, cytokines, antimicrobial peptides, and lactoferrin.6,7 While bioactive components in breast milk can directly confer protection against selective pathogenic microbes,8–12 it is becoming increasingly clear that breast milk also has direct and indirect effects on infant health by exerting an influence on the compositional dynamics of the infant’s rapidly evolving gut microbiota.13–15 Although the impact of breastfeeding on the development of a balanced gut microbiota has been established,5,7 the relative contributions of the various immune cells and molecules remain to be defined.
The complement system is composed of more than 30 proteins that are found in the blood and interstitial fluids that, when activated, carries out a complex array of effector and regulatory functions,16–20 the best studied of which focus on host defense against microbial pathogens.21 While initially described from the serum where it makes up 10-15% of the globulin fraction,22 the presence of complement components in breast milk has been noted from multiple mammalian species including humans.23–26 Although a limited number of in vitro studies have examined whether the complement contained in breast milk had the capacity to mediate killing of pathogenic bacteria,27–29 the pathophysiological relevance of complement in breast milk are yet to be fully defined. One of the issues that have not been addressed is the possible contribution of complement components in breast milk in shaping the composition of the infant’s gut microbiota and how this might influence susceptibility of newborns and infants to pathogenic microbes.
Here we show that the mouse pups fostered by dams deficient in complement components in breast milk harbor an altered microbiota that results in a failure to thrive and death upon challenge with the natural murine gut pathogen Citrobacter rodentium (CR). Notably, the complement in breast milk has the activity of directly lysing specific gram-positive commensal bacterial species via a C1-initiated, antibody-independent, and membrane attack complex (MAC)-dependent fashion. The selective killing of certain gram-positive species results in a gut microenvironment that protects the offspring from disease and death when challenged with CR. Our findings provide insights into how the complement contained in breast milk contributes to the establishment of a ‘protective’ gut microbiota during the early stages of development and adds to the list of the protective mechanisms of breast milk that promotes infant health and defense against environmental pathogens.
Results
Complement-deficient weanling mice are susceptible to enteric bacterial infection
The three pathways of complement activation, i.e., the classical pathway, the lectin pathway, and the alternative pathway converge at the central step where the C3 convertase activity is generated.16–18,30 To assess the impact of complement system on host immune responses in infants against bacterial pathogens, weanling (21-day-old) wild-type (WT) C57Bl/6J and complement component C3 knockout (C3−/−) animals were challenged orally with CR at a dose that results in only minor reactions in WT animals.31–33 While WT pups were only minimally affected in their growth (Figure 1A), expression of clinical signs (Figure 1B), and survival (Figure 1C) through 21 days post-inoculation (dpi), CR infection of C3−/− pups caused significant decrements in growth and severe diarrhea by 7 dpi (Figures 1A and 1B), associated with >90% mortality (Figure 1C). The results suggested a key role for C3, and by extension the complement system, in the protective response.
Figure 1. Complement-deficient pups are susceptible to Citrobacter rodentium infection.

(A-C) Body weight changes (A), clinical scores (B), and survival (C) of 21-day-old wild-type (WT), C1qc−/− and C3−/− mice at indicated days post inoculation (dpi) with 2 × 109 CFU of C. rodentium (CR) or PBS vehicle control.
(D) Live CR recovered from the fecal samples of WT, C1qc−/−, and C3−/− pups at 7 dpi.
(E-F) Representative macrographs (E) and lengths (F) of the colon derived from CR-infected pups at 9 dpi. Scale bars, 1 cm.
(G-H) Hematoxylin and eosin staining (G) and histopathology scores (H) of colon sections derived from CR-challenged pups at 9 dpi. Scale bars, 200 μm.
(I) CR burdens in the liver (left) and the spleen (right) derived from CR-challenged pups at 9 dpi.
(J) FITC-dextran concentrations in the sera of CR-infected pups (9 dpi) at 4 h post oral administration of FITC-dextran.
(K) Representative immunofluorescence micrographs of CR in the colon derived from CR-infected pups at 9 dpi, with nuclei counterstained by DAPI. L, colonic lumen. Scale bars, 100 μm.
Data are mean ± s.e.m., with specific n numbers indicated. Data in A-C are combined results from at least three independent experiments; in D-K are representative results of at least two independent experiments. ns, not significant, ** p < 0.01, *** p < 0.001, and **** p < 0.0001, for C1qc−/− versus WT in blue and C3−/− versus WT in red, respectively.
See also Figure S1.
Of the three canonical pathways of C3 activation,34,35 the classical pathway of complement activation, which is initiated via the C1 complex,36–39 was studied first. The C1 complex is composed of 6 C1q, 2 C1r, and 2 C1s; each C1q molecule is composed of 6 heterotrimers made from C1qa, C1qb, and C1qc. The classical pathway is initiated by the C1 complex binding to antigen-bound immunoglobulin molecules or by C1 binding directly to the surface of microbes. A deletion of the gene encoding the C1qc chain (C1qc−/−) blocks production of the heterotrimer and results in a functional C1 deficiency. At weaning and prior to CR challenge, WT, C1qc−/−, and C3−/− animals had nearly identical body weights (Figure S1A). Despite almost identical CR colonization (Figure 1D), the C1qc−/− weanlings displayed the levels of attenuated growth, severe diarrhea, and lethality (Figures 1A–1C), observed in the C3−/− animals. The results suggested that C1-initiated activation was important for the protective effect of complement and, importantly, indicated that the lectin and alternative pathways (the other two mechanistic routes of C3 activation) played no or a minor role in the response. Therefore, our results supported the hypothesis that C3 activation via the C1-dependent cascade plays a critical role in infant defense against CR-induced lethality in weanling mice.
Morphological analysis revealed that the colons derived from CR-challenged C1qc−/− and C3−/− pups were markedly swollen and shortened compared to those from WT controls (Figures 1E and 1F). Histological analysis further illustrated the severe colitis characterized by crypt elongation, goblet cell depletion, and immune cell infiltration in the colons from CR-challenged C1qc−/− and C3−/− animals (Figures 1G and 1H). In contrast, the colons from CR-challenged WT animals showed minimal histological evidence of pathology (Figures 1G and 1H). Additionally, live CR was detected in the liver and the spleen of CR-infected C1qc−/− and C3−/− animals, whereas these organs remained free of live bacteria in the WT controls (Figure 1I). Systemic dissemination of CR in the infected C1qc−/− and C3−/− groups indicated damage to the integrity of the colonic epithelial barrier, a notion that was supported by the substantially increased gut permeability in the C1qc−/− and C3−/− groups as measured by leakage of gut-derived FITC-dextran to the serum (Figure 1J). Moreover, immunofluorescence staining demonstrated that CR predominantly attached to the luminal surface of colonic epithelium in WT pups, whereas the CR in C1qc−/− and C3−/− animals penetrated the epithelial layer, even reaching the colonic crypts (Figure 1K). These results underscored the severe epithelial tissue damage that resulted from CR challenge in the absence of complement components C1 and C3.
Of note, the morphology, length, and histology of the colons derived from unchallenged WT, C1qc−/−, and C3−/− weanling mice were comparable (Figures S1B–S1D). The intestinal epithelial permeability occurred at the low levels typically associated with intact barrier function and the livers and spleens were bacteria-free (Figures S1E–S1G). Moreover, CR challenge of WT, C1qc−/−, and C3−/− adult (6-8-week-old) mice resulted in comparable increases in CR numbers in the gut, only transient body weight loss, and no mortality (Figures S1H–S1J). These mild symptoms observed in mature animals in response to CR challenge, as typically reported for WT adult mice during CR infection,31–33 suggested that the consequences of C1 or C3 deficiency is most impactful during the early stages of development when breastfeeding provides critical passive protection against environmental exposures.
Complement in maternal milk is critical for infant susceptibility to CR
While it has been appreciated for over five decades that complement components are present in colostrum and breast milk,24–26,40 the physiological relevance of this complement remains to be determined. Complement components were readily identified in the whey (the liquid after milk solids are removed) from WT C57Bl/6J mice by mass spectrometry and functional gene enrichment analysis illustrated that the ‘complement and coagulation cascades’ was the top enriched pathway (Figure S2). To assess the potential relevance of complement in breast milk, we utilized a cross-fostering strategy to distinguish the relative contributions to the CR-induced phenotype made by breast milk and by the genetics of C1qc−/− and C3−/− weanlings. Both cross-fostered groups of C1qc−/− pups, which had comparable body weights at 21 days (Figures 2A and 2B), were separated from the dams and challenged with CR. Mirroring the findings in C1qc−/− weanling mice (Figures 1A–1C), C1qc−/− pups fostered by C1qc−/− dams succumbed to CR infection and exhibited decrements in growth and severe diarrhea (Figures 2C–2E). In striking contrast, all the C1qc−/− pups cross-fostered by WT dams survived the CR challenge with no detectable decrease in growth and minimal diarrhea (Figures 2C–2E). In addition, half of WT newborns were cross-fostered by C1qc−/− dams, while the rest lactated by the original WT dams, prior to oral challenge with CR (Figure 2F). While both groups of WT pups had comparable rates of growth through day 21 (Figure 2G), after CR challenge, WT dam-fostered WT pups continued to thrive whereas cross-fostering by C1qc−/− dams rendered WT pups highly susceptible with growth retardation, severe diarrhea, and distinctly elevated mortality (Figures 2H–2J). Similarly, WT and C3−/− newborns were cross-fostered by C3−/− and WT dams, respectively, with outcomes similar as those outlined above, i.e., fostering on WT dams was protective and fostering on C3-deficient dams made both WT and C3−/− pups vulnerable to CR-induced disease (Figures 2K–2T). These results demonstrate that complement-sufficient breast milk protects suckling mice from a CR-triggered lethal outcome.
Figure 2. Complement in maternal milk protects weanling mice from CR infection-caused growth faltering and lethality.

(A, F) Experimental scheme of cross-fostering strategies. Wild-type (WT) C57Bl/6J (black, filled) and C1qc−/− (blue, open) breeding pairs were synchronized to generate pups born on the same day. C1qc−/− (A) and WT (F) pups were divided into two groups at the day of birth and fostered by indicated dams. At postnatal 21 days (P21), the cross-fostered pups were weaned and orally inoculated with 2 × 109 CFU of C. rodentium (CR).
(B, G) Body weight of the cross-fostered C1qc−/− (B) and WT (G) pups at P21 prior to CR infection.
(C-E) Body weight changes (C), clinical scores (D), and survival (E) of the cross-fostered C1qc−/− pups at indicated days post CR inoculation (dpi).
(H-J) Body weight changes (H), clinical scores (I), and survival (J) of the cross-fostered WT pups at indicated dpi.
(K, P) Experimental scheme of cross-fostering strategies. WT (black, filled) and C3−/− (red, open) breeding pairs were synchronized to generate pups born on the same day. C3−/− (K) and WT (P) pups were divided into two groups at the day of birth and fostered by indicated dams. At P21, the cross-fostered pups were weaned and orally inoculated with 2 × 109 CFU of CR.
(L, Q) Body weight of the cross-fostered C3−/− (L) and WT (Q) pups at P21 prior to CR infection.
(M-O) Body weight changes (M), clinical scores (N), and survival (O) of the cross-fostered C3−/− pups at indicated dpi.
(R-T) Body weight changes (R), clinical scores (S), and survival (T) of the cross-fostered WT pups at indicated dpi.
Data are mean ± s.e.m., with specific n numbers indicated. Data in B-E, G-J, L-O, and Q-T are combined results from at least three independent experiments. ns, not significant; * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
See also Figures S2 and S3.
To minimize possible additional microbiota-associated environmental variables beyond breast milk that influence these outcomes, we employed littermates and dam-cohousing strategies. WT and C1qc−/− female mice fostered by the same dams were cohoused since birth until delivering their pups; during lactation, WT and C1qc−/− dams were cohoused to maximize the environmental similarity between WT and C1qc−/− dams (Figure S3A). Consistently, fostering on WT dams was protective while fostering on C1qc−/− dams resulted in pups, regardless of genotype, vulnerable to CR-induced growth retardation and lethality (Figures S3B and S3C). Moreover, we obtained comparable results when WT and C1qc−/− littermate dams were used in the same damcohousing strategy (Figures S3D–S3F), indicating that the susceptibility of pups to CR challenge is largely or wholly determined by the presence of an intact complement system in breast milk.
Complement in breast milk alters microbiota composition that contributes to CR-induced lethality
Breast milk plays an important role in establishing a balanced gut microbiota that indirectly protects against colonization by pathogens.7,8 To assess the impact of gut microbiota on the vulnerability of weanling animals to CR-induced disease, we rederived germ free (GF) WT and C1qc−/− mice for oral CR challenge. As expected, CR rapidly reached and sustained peak loads in weanling (21-day-old) GF WT and C1qc−/− animals (Figure 3A). GF WT weanling mice were not vulnerable to CR challenge (Figures 3B, 3C and S4A–S4D). Intriguingly, CR challenge failed to cause growth retardation, severe symptoms, or mortality in GF C1qc−/− weanling mice (Figures 3B and 3C), mirroring the evidence that the colonic morphology, lengths, and epithelial integrity derived from CR-challenged GF C1qc−/− and WT weanling mice were all comparable (Figures S4A–S4D). Of note, transferring the cecal and colonic microbiota from 21-day-old specific pathogen free (SPF) WT mice to GF C1qc−/− pups exhibited minimal impacts on the growth, diarrheal symptoms, and survival, after CR challenge (Figures 3D–3G). In contrast, the transfer of colonic commensals from SPF C1qc−/− rendered GF C1qc−/− pups highly susceptible to CR challenge with attendant decrements in growth, severe diarrhea, and distinctly elevated mortality (Figures 3D–3G). Shorten, swollen, and inflamed colons with substantially increased gut permeability were consistently observed in GF C1qc−/− pups reconstituted with SPF C1qc−/− gut commensals, but not with SPF WT gut commensals (Figures 3H, 3I, S4E, and S4F). Moreover, GF WT pups reconstituted with SPF C1qc−/− commensal became sensitive to CR challenge, while those received SPF WT commensal remained resistance to CR infection (Figures S4G–S4K), mirroring their GF C1qc−/− equivalents (Figures 3D–3H). Together these results implicate a crucial role for the composition of infant’s gut microbiota that are shaped by the dam’s breast milk in regulating susceptibility to CR-mediated severe disease in weanling animals.
Figure 3. Gut microbiota in suckling mice plays a critical role in CR infection-caused lethality.

(A) Live C. rodentium (CR) recovered from the fecal samples of 21-day-old germ free (GF) wild-type (WT) and C1qc−/− mice at 7 and 14 days post inoculation (dpi) with 2 × 109 CFU of CR.
(B-C) Body weight changes (B) and survival (C) of 21-day-old GF WT and C1qc−/− pups at indicated dpi with 2 × 109 CFU of CR.
(D) Live CR recovered from the fecal samples of 21-day-old GF C1qc−/− pups, reconstituted at postnatal 17 days (P17) with the cecal and colonic microbiota derived from P21 specific pathogen free (SPF) WT or C1qc−/− pups, at 7 dpi with 2 × 109 CFU of CR.
(E-G) Body weight changes (E), survival (F), and clinical scores (G) of 21-day-old GF C1qc−/− pups, reconstituted and infected as in (D), at indicated dpi.
(H) Lengths of the colon derived from GF C1qc−/− pups, reconstituted and infected as in (D), at 12 dpi.
(I) GF C1qc−/− pups were reconstituted and infected as in (D). FITC-dextran concentrations in the sera of GF C1qc−/− pups (12 dpi), at 4 hours after oral administration of FITC-dextran.
(J) Species abundance heatmap of the dominant 35 genera detected using 16S rRNA gene-based high-throughput sequencing among the cecal and colonic contents derived from 21-day-old WT (n=5), C1qc−/− (n=4), and C3−/− (n=5) pups.
Data are mean ± s.e.m., with specific n numbers indicated. Data in A-C are combined results from two independent experiments; in D-G are combined results from three independent experiments; in H and I are representative results of three independent experiments. ns, not significant; * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
See also Figures S4 and S5.
The putative role of gut commensal microbiota in susceptibility of sucking mice to CR-induced disease led us to profile the cecal and colonic microbiota in susceptible and refractory animals. Despite comparable numbers of observed species and similar levels of community richness and diversity (Figures S5A–S5D), 16S ribosomal RNA (rRNA) sequencing revealed that WT, C1qc−/−, and C3−/− weanling mice harbored gut microbiota with distinct overall compositions (Figures 3J, S5E, and S5F). Hence, it is possible that the dramatic difference observed in microbiota composition of WT, C1qc−/−, and C3−/− weanling mice was a key factor in determining the susceptibility to CR infection.
Complement in breast milk eliminates gram-positive Staphylococcus lentus B3 in gut microbiota
The comparable body weights of WT, C1qc−/−, and C3−/− weanling mice, regardless of the complement status of the fostering dams (Figures 2B, 2G, 2L, 2Q and S1A), indicates that there were no substantial differences in the nutritional value between breast milk from WT, C1qc−/−, and C3−/− dams. Indeed, macronutrient content analyses revealed almost identical percentages of crude protein, crude fat, total sugar, water content, and calculated gross energy in breast milk collected from WT, C1qc−/−, and C3−/− dams (Figures S6A–S6F). Moreover, the protein concentrations in the whey from WT, C1qc−/−, and C3−/− dams were comparable (Figures 4A, S6G and S6H). Additionally, the levels of secretory antibodies, especially secretory Immunoglobin A (sIgA) that is reported to regulate the gut microbiota of infants,41 in the breast milk from WT, C1qc−/−, and C3−/− dams were all comparable (Figures S6I–S6N). This indicates that complement components, at least C1 and C3, do not impact the production of secretory antibodies in mouse breast milk. Hence, it seems unlikely that the levels of nutrients or immunoglobins provided by the lactating dams explain the striking differences in gut microbiota observed in WT, C1qc−/−, and C3−/− weanlings (Figure 3J).
Figure 4. Breast milk complement kills gram-positive commensal bacterium Staphylococcus lentus B3.

(A) Representative macrographs of the whole milk and whey (following PBS dilution and centrifugation) derived from wild-type (WT), C1qc−/− and C3−/− dams. li, lipid; W, whey; C, casein.
(B) Experimental scheme of mouse whey bactericidal assays using cultivable commensal bacteria (CB) derived from C1qc−/− pups at postnatal 21 days (P21).
(C) Representative macrographs of bactericidal assays on LB agar plates using WT and C1qc−/− mouse whey to kill CB derived from C1qc−/− pups, with PBS and heat inactivated (HI) whey as negative controls. Indicated is one species, Staphylococcus lentus B3 strain, isolated from the cultivable CB of C1qc−/− pups.
(D) A circular comparative genomic map of S. lentus B3 with other representative Staphylococcus genomes. From outer to inner, circles 1 to 6 show genomic comparisons in nucleotide level with S. lentus strains H29 and NCTC12102, S. aureus MW2, S. epidermids RP62A, S. haemolyticus JCSC1435, and S. saprophyticus ATCC15305; circles 7 and 8 show G+C content and GC skew (G-C/G+C) of S. lentus B3 genome, respectively. The scale is given on the innermost circle.
(E) Representative macrographs of bactericidal assays on LB agar plates using the indicated mouse whey to kill S. lentus B3, with PBS and whey (HI) as negative controls. Left, experimental scheme.
(F-H) Whey bactericidal assays using S. lentus B3 cultured in LB medium, supplemented with whey derived from WT, C1qc−/− and C3−/− dams and HI controls (F) or in the presence of CD59 (G) or Vitronectin (VTN) (H). After 16-hour culture with shaking, the indicated S. lentus B3 cultures were serially diluted and spotted on LB agar plates to determine CFUs. Shown are the concentrations of live S. lentus B3 in indicated LB media.
(I-K) Human whey bactericidal assays using S. lentus B3 cultured in LB medium, supplemented with regular or heat inactivated (HI) human whey (I), or in the presence of CD59 (J) or VTN (K). After 16-hour culture with shaking, the indicated S. lentus B3 cultures were serially diluted and spotted on LB agar plates to determine CFUs. Shown are the concentrations of live S. lentus B3 in indicated LB media.
Data are mean ± s.e.m. and representative of three independent experiments. ns, not significant; * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
The abundance of complement components in mouse whey (Figure S2) led us to examine whether complement directly or indirectly impacted the gut microbiota composition. The cecal and colonic bacteria from weanling C1qc−/− pups were cultivated aerobically on LB agar plates; whey from WT or C1qc−/− dams was spotted on top of the bacterial cultures (Figure 4B). Strikingly, the cultivable commensal bacteria were suppressed by the content of the whey from WT dams, but this inhibitory effect was substantially attenuated when the whey from C1qc−/− dams was used (Figure 4C), suggesting that complement in mouse whey is involved in preventing the growth of certain commensal bacterial species. Indeed, we characterized a cultivable isolate that was resistant to whey from C1qc−/− dams and identified it as Staphylococcus lentus (Figure S7A). We named this gram-positive bacterium S. lentus B3 (Figure 4C), the identification of which was further supported by the comparative genome analyses (Figures 4D and S7B). Likewise, similar findings were obtained when cecal and colonic bacteria from C3−/− weanlings were overlaid with whey from WT or C3−/− dams (Figures S7C and S7D). Of note, the abundance of Staphylococcus was markedly elevated in the gut microbiota of C1qc−/− and C3−/− pups compared to WT controls (Figure 3J), supporting the potential impact of complement in breast milk on selective bacteria (e.g., S. lentus B3) in the offspring’s gut microbiota.
To assess the impact of S. lentus B3 on infant health under steady and infectious conditions, we performed S. lentus B3 monocolonization and simultaneous colonization of S. lentus B3 and CR in GF C1qc−/− pups. Similar to CR (Figures 3B, 3C, S8A, and S8B), S. lentus B3 inoculation alone did not affect the growth and survival of C1qc−/− pups (Figures S8A and S8B). In comparison to monocolonization, simultaneous colonization of S. lentus B3 and CR faintly, but significantly, attenuated the growth of GF C1qc−/− pups (Figure S8A), indicative of synergic S. lentus B3-CR interaction. Although not sufficient to cause mortality in GF C1qc−/− pups, the synergic interaction between S. lentus B3 and CR could substantially worsen the symptoms, which hints that S. lentus B3-CR interaction, in conjunction with other microbes in gut microbiota, contributes to the observed mortality in the CR-challenged SPF C1qc−/− pups (Figure 1). We then assessed whether reshaping the gut microbiota, especially eradicating Staphylococcus abundance in the pups receiving complement-deficient breast milk could affect their susceptibility to CR infection. C1qc−/− pups, fostered by the dams receiving maternal treatments with antibiotics Vancomycin, Neomycin, or Cefoxitin in drinking water, were still vulnerable to CR infection (Figures S8C and S8D). Interestingly, C1qc−/− dams administrated with Fenbendazole diet, a broad-spectrum benzimidazole commonly used in laboratory animals, completely rescued CR infection caused growth faltering and mortality in C1qc−/− pups, associated with dramatically reshaped gut microbiota, particularly reduced Staphylococcus levels, in C1qc−/− pups (Figures S8E–S8G). Likewise, C3−/− pups, fostered by C3−/− dams receiving Fenbendazole treatment, fully reversed the vulnerability to CR infection and displayed substantially altered gut microbiota and attenuated Staphylococcus abundance (Figures S8H–S8J). Of note, eliminating Staphylococcus in the gut microbiota of C1qc−/− and C3−/− pups could avert their vulnerability to CR infection. Additionally, we analyzed the gut microbiota compositions of WT, C1qc−/−, and C3−/− weaning pups (21-day-old) and adult mice (8-week-old). As expected, the numbers of observed species and community richness of bacterial families in the adult gut microbiota are remarkably higher than those in weaning pups (Figures S9A and S9B). Strikingly, in comparison to the high Staphylococcus abundance in C1qc−/− and C3−/− pups, Staphylococcus levels were comparably lower in WT, C1qc−/−, and C3−/− adult mice (Figure S9C), which appears coincidently correlated to their resistance to CR infection (Figure S2). Together, these results hint that Staphylococcus abundance in the mouse gut microbiota appear indicative of the susceptibility to CR infection.
To further characterize the effect of breast milk complement on S. lentus B3 abundance in vivo, we analyzed the microbiota compositions in the breast milk derived from nursing dams and in the stomach and small intestine of suckling pups, when the pups were at postnatal 14 days (P14). Interestingly, Staphylococcus appeared even higher in breast milk derived from WT dams compared to the C1qc−/− and C3−/− equivalents (Figure S10A), whereas Staphylococcus levels in the stomach of 14-day-old WT pups became slightly lower than those in C1qc−/− pups and profoundly lower than C3−/− pups (Figure S10B). Moreover, Staphylococcus abundance in the small intestine of C1qc−/− and C3−/− pups at P14 was also substantially elevated compared to WT pups (Figure S10C). These results are in line with our finding in the cecal and colon microbiota composition in P21 pups where Staphylococcus levels were markedly elevated in C1qc−/− and C3−/− pups compared to WT controls (Figure 3J). Hence, complement in breast milk likely restrains Staphylococcus abundance prior to entering the small intestine, thus impacting the entire gut microbiota in pups.
Complement in breast milk directly kills S. lentus B3 via C1-initiated complement activation
Whey from WT dams readily hindered the growth of S. lentus B3 on LB agar plates or LB medium culture in vitro, whereas the bacterial growth was not inhibited by whey from C1qc−/− or C3−/− dams (Figures 4E and 4F). Moreover, heat-inactivated whey from WT, C1qc−/−, or C3−/− dams failed to inhibit growth of S. lentus B3 (Figures 4E and 4F). The possible role of the complement membrane attack complex (MAC) was explored using CD59, a glycoprotein that blocks the polymerization and thus the pore-forming ability of C942,43 or vitronectin (VTN), a soluble factor that negatively regulates the formation of the C5b67 complex required for MAC formation.44,45 The addition of CD59 or VTN to the in vitro assays dampened the bacterial suppressing capacity of whey from WT dams but had no impact on whey from C1qc−/− or C3−/− dams (Figures 4G and 4H).
The complement in human breast milk also displayed the capability to selectively suppress certain bacteria. Human whey derived from healthy donors hindered the growth of S. lentus B3 through a heat-labile mechanism (Figures 4I and S11A). The bacterial suppressing capacity of human whey was substantially attenuated by CD59 (Figures 4J and S11B) and VTN (Figures 4K and S11C), supporting the involvement of MAC formation. Of note, time kill-kinetics assays further demonstrated bactericidal, rather than bacteriostatic, activities of human whey that were sensitive to CD59 or VTN (Figures S11D to S11F). These results point to a crucial role for an intact complement pathway in mouse and human breast milk for killing S. lentus B3.
The C1 complex is composed of a large subunit (C1q), which acts as a recognition protein or pathogen sensor, and two copies each of the inactive serine proteases C1r and C1s. When C1q binds directly to a pathogen/microbe surface or indirectly to antibodies bound to a pathogen, C1r is auto-activated and cleaves C1s to an active protease, which in turn cleaves the downstream substrates C4 and C2. The proteolytic products of C4 and C2 combine to make heterodimer with enzymatic activity that cleaves C3 (C3 convertase), a crucial step in the enzymatic cascade that leads to the eventual formation of the C5b-9 (MAC).46,47 Incubation of human whey and S. lentus B3 triggered the cleavage of C1s (Figure 5A). In line with C1s activation, the terminal C5b-9 (MAC) was readily detected on the cell surface/membrane of S. lentus B3, when incubated with untreated, but not heat-inactivated, human whey (Figures 5B and 5C). Moreover, as measured by DiBAC4(3), a negatively-charged fluorescent indicator widely used evaluating bacterial membrane potential,48 incubation with human whey disrupted the membrane potential of S. lentus B3 over time (Figure 5D). Of note, both CD59 and VTN substantially attenuated human whey-mediated membrane potential loss in S. lentus B3 (Figures S12A and S12B). These results indicate that the C5b-9 (MAC) generated from the complement components in human whey can directly disrupt the membrane leading to membrane depolarization in S. lentus B3. Indeed, transmission electron microscopy (TEM) revealed numerous MAC pores formed on the cell membrane of S. lentus B3 associated with damaged cell morphology, when incubated with human whey (Figures 5E and 5F). Moreover, MAC formation and S. lentus B3 cell membrane damage were significantly diminished by heat-inactivation of the whey or with the addition of CD59 or VTN (Figures 5E and 5F). Given C1s activation and bactericidal activity of human whey (Figures 4I–4K and S11), these results suggest that human whey-exposed S. lentus B3 was most likely killed via a C1-initiated pathway of complement activation resulting in terminal C5b-9 (MAC) deposition, similar to the killing documented for serum-derived complement for gram-negative Escherichia coli, Salmonella minnesota, and Neisseria gonorrhoeae.49–52 In contrast to S. lentus B3, incubation of human whey with gram-positive S. aureus SH1000 or gram-negative E. coli Nissle 1917 (E. coli N1917) failed to activate C1s or kill these bacteria (Figures S12C–S12F). Furthermore, as illustrated by TEM, S. lentus B3 incubated with WT mouse whey exhibited MAC pores in the bacterial cell membrane and lytic bacterial morphology; in contrast, no pore formation and altered cellular morphology were observed in the presence of C1qc−/− or C3−/− whey (Figures 5G and 5H). Hence, these results provide strong evidence that breast milk complement is activated in C1-dependent cascade and leads to C5b-9 (MAC) deposition into the cell membrane of S. lentus B3, resulting in bacterial killing.
Figure 5. Complement in breast milk lyses Staphylococcus lentus B3 via C1 activation and MAC deposition.

(A) Human whey and Staphylococcus lentus B3 were incubated for indicated time periods, followed by SDS-PAGE separation. The membrane was subjected to Ponceau S staining or immunoblot (IB) for full-length (FL) and cleaved C1s proteins.
(B) C5b-9 levels on S. lentus B3, following incubation with PBS, regular or heat inactivated (HI) human whey, analyzed by flow cytometry.
(C) Representative immunofluorescence micrographs of C5b-9 on S. lentus B3 following incubation with indicated human whey, with DNA and membrane counterstained by Hoechst and FM5-95, respectively. Scale bars, 2 μm.
(D) Representative histograms of DiBAC4(3) fluorescence on S. lentus B3 incubated with PBS or human whey for indicated time periods, analyzed by flow cytometry, with the protonophore CCCP as a positive control.
(E-F) Representative cell membrane (E) and micrographs of bacterial morphology (F) of S. lentus B3 following incubation with indicated human whey in the presence and absence of CD59 or Vitronectin (VTN). White arrows indicate the assembled ring-structured membrane attack complexes (MAC). Scale bars, 100 nm (E) and 500 nm (F).
(G-H) Representative cell membrane (G) and micrographs of bacterial morphology (H) of S. lentus B3 following incubation with whey derived from wild-type (WT), C1qc−/−, and C3−/− dams. White arrows indicate the assembled ring-structured MAC pores. Scale bars, 100 nm (G) and 500 nm (H).
Data in A-G are representative of three independent experiments.
See also Figure S12.
Activation of complement in the breast milk is independent of immunoglobulins
Of note, the cascade of complement activation can be initiated when the C1 complex either recognizes a microbial surface directly or binds to antibodies already bound to a pathogen.53 To examine the role of antibodies in the bactericidal activity of complement in breast milk, we depleted IgG and IgM from human whey (Figure 6A). Strikingly, human whey depleted of IgG and IgM triggered C1s activation (Figure 6B), suggesting that antibodies are not required for the S. lentus B3-elicited complement activation. Incubation with the IgG- and IgM-depleted whey resulted in C5b-9 (MAC) deposition and cell wall disruption in S. lentus B3 (Figures 6C–6E). To further validate that whey-derived S. lentus B3-elicited complement activation is antibody-independent, we employed whey from μMT−/− dams carrying homozygous deletion of immunoglobulin heavy chain of the class μ, with global B cell deficiency54 and absence of immunoglobulins of all isotypes (Figure 6F). Whey from μMT−/− and WT dams resulted in comparable C5b-9 (MAC) pores in the cell membrane and killing of S. lentus B3 (Figures 6G and 6H). Significantly, whey from μMT−/−C1qc−/− dams failed to cause C5b-9 (MAC) formation and cell damage in S. lentus B3 (Figures 6I and 6J). Taken together, these findings suggest an antibody-independent, C1-dependent pathway of complement activation leading to C5b-9 (MAC) deposition and bactericidal activity in human and mouse breast milk.
Figure 6. Complement in breast milk kills Staphylococcus lentus B3 antibody-independently.

(A) Immunoglobulin G (IgG) and IgM depleted (Ig-depleted) and mock-treated human whey samples were SDS-PAGE separated, followed by Ponceau S staining or immunoblot (IB) for IgM and IgG.
(B) Mock-treated and Ig-depleted human whey, incubated with Staphylococcus lentus B3 for indicated time periods, were SDS-PAGE separated, followed by Ponceau S staining or IB for activated C1s proteins.
(C) C5b-9 levels on S. lentus B3, following incubation with PBS or indicated human whey, analyzed by flow cytometry.
(D-E) Representative cell membrane (D) and micrographs of bacterial morphology (E) of S. lentus B3 following incubation with indicated human whey. White arrows indicate the assembled ring-structured membrane attack complexes (MAC) pores. Scale bars, 100 nm (D) and 500 nm (E).
(F) Whey derived from wild-type (WT) C57Bl/6J and μMT−/− dams was SDS-PAGE separated, followed by Ponceau S staining or IB for IgM and IgG.
(G-J) Representative cell membrane (G, I) and micrographs of bacterial morphology (H, J) of S. lentus B3 following incubation with whey derived from indicated dams. White arrows indicate the assembled ring-structured MAC pores. Scale bars, 100 nm (G, I) and 500 nm (H, J).
Data in A-J are representative of three independent experiments.
Discussion
There is an increasing appreciation that the early-life progression in diversity and composition of the gut microbiota is a central contributing factor to human health and that imbalances in the gut microbial composition have short-term and far-reaching impacts on development and disease.55–57 However, the list of factors that contribute to the dynamics of microbiota development in newborns is still evolving. Breastfeeding, besides its well-documented role in providing high-quality nourishment for development and a source of key bacterial species important in establishing the early-life gut microbiota,8,58 contains important factors that directly and indirectly shape the infant’s commensal microbiota.5,7,23 In particular, the multifaceted roles of antibodies in breast milk in conferring effective protection to offspring have been extensively investigated.59 While the presence of complement components in breast milk has been noted for decades, their physiological relevance has not been defined. Previous in vitro studies proposed that complement in breast milk has a bactericidal/bacteriostatic function against selective pathogens E. coli and Helicobacter pylori.28,29 In this study, we demonstrate that complement in mouse breast milk substantially modulates the early gut commensal community structure in offspring by selectively killing certain commensal bacteria, most notably the gram-positive bacteria Staphylococcus. Of note, Staphylococcus is among the first gut colonizers in human newborns during the first week after birth and exhibits declining population sizes after some weeks. The reduction of Staphylococcus levels has been noted to coincide with the cessation of breast-feeding and consumption of solid food in humans.60,61 Consistently, Staphylococcus in mouse gut microbiota evidently shifts from higher abundance in infanthood to declining levels in adulthood, which likely attributes to dietary intake transition from breast milk to formulated chow diet. It appears that during early life, Staphylococcus abundance in gut microbiota of weanling animals is correlated with the susceptibility to bacterial infection and eliminating Staphylococcus averts their vulnerability to CR infection. These results highlight that complement components in breast milk could elegantly control Staphylococcus levels in the offspring’s gut microbiota, which is crucial for newborns and infant health. Moreover, complement-mediated bactericidal activity targeting the isolated Staphylococcus lentus B3 strain is conserved between mouse and human breast milk. Overall, our findings suggest that, beyond controlling pathogens, complement in breast milk possess an evolutionally-conserved capacity to eliminate selective commensal microbes, thus functioning in shaping the gut microbiota during the early stages of development.
We show that complement components C1q and C3 are pivotal for the bactericidal activity of breast milk complement to S. lentus B3. In particular, the C5b-9 (MAC), which is the terminal effector complex of complement activation, is assembled onto the cell membrane of S. lentus B3 and leads to the loss of membrane potential, cell damage, and lysis. It was previously reported that when some gram-positive bacteria exposed to human serum, C5b-9 (MAC) deposition can be observed but does not lead to bacterial killing.62 Distinct to serum complement, when gram-positive S. lentus B3 is exposed to breast milk, the readily-detected C5b-9 (MAC) deposition does result in S. lentus B3 cell lysis thus harboring bactericidal activity. The cascade of complement activation is canonically initiated by an antibody-dependent mechanism that involves C1qr2s2 complex binding to the anti-bacterial IgM or IgG bound to antigenic determinants found on the surface of the microbe.63 Also, the initiation of complement activation can be achieved when C1q directly recognizes a microbial surface, supported by previous evidence from in vitro studies that, in the absence of antibody, serum-derived C1q can bind directly to the surface of a few gram-negative bacteria64–68 or purified surface components of gram-positive bacteria.69,70 In this study, antibody-deficient whey does not impact the C1s activation in response to S. lentus B3, formation of C5b-9 (MAC) pores on S. lentus B3, and bacterial killing. These results strongly support that in breast milk, the cascade of complement activation is initiated antibody-independently to kill certain gram-positive microbes in the infant’s gut microbiota; such selective microbial killing protects the gut from colonization later in life by pathogenic microbes such as Citrobacter rodentium. Together, our findings demonstrate that complement in breast milk plays a critical role in the development and establishment of a health-promoting early-life gut microbiota and adds to the mechanisms by which breastfeeding confers protection and promotes infant health.
Limitations of the Study
Complement in breast milk selectively eliminates certain commensal bacteria thus modifying the gut microbiota compositions in weanling mice. While the role of S. lentus B3 is illustrated here, it is likely that other commensal microbes also contribute to promoting infant health. Further investigations are needed to identify additional microbes similarly eliminated via the antibody-independent action of breast milk complement, which would facilitate studies to define the molecular basis for antibody-independent C1q-microbe interactions that lead to complement activation. Complement in breast milk kills gram-positive S. lentus B3 directly via C5b-9 (MAC) pore formation, whereas the C5b-9 (MAC) deposition during serum or recombinant complement activation does not kill gram-positive bacteria. It warrants further investigation whether additional bioactive component(s) in breast milk may participate in the complement-mediated lysis of gram-positive bacteria. Littermates and dam-cohousing strategies eliminate most possible environmental variables in the experiments performed here using complement global knockout animals. It would be of interest to develop tissue-specific complement conditional knockout mice for studies on breast milk complement biology.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Fengyi Wan (fwan1@jhu.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availability
16S rRNA gene profiling data and S. lentus B3 whole genome sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) database and are publicly available as of the date of publication. This paper also analyzes sequencing data of bacteria S. lentus (H29 strain), S. lentus (NCTC12102 strain), S. aureus (MW2 strain), S. epidermids (RP62A strain), S. haemolyticus (JCSC1435 strain), and S. saprophyticus (ATCC15305 strain), as well as Mus musculus database, all of which are existing and publicly available. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human C1s | Complement Technology | Cat# A204 |
| Rabbit Anti-Human C5b-9 | Bioss Antibodies | Cat# bs-2673R |
| HRP-conjugated Goat Anti-Mouse IgG | SouthernBiotech | Cat# 1030-05 |
| HRP-conjugated Goat Anti-Mouse IgM | SouthernBiotech | Cat# 1021-05 |
| HRP-conjugated Goat Anti-Human IgG | SouthernBiotech | Cat# 2040-05 |
| HRP-conjugated Goat Anti-Human IgM | SouthernBiotech | Cat# 2020-05 |
| Alexa Fluor 488 conjugated Goat Anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-11034 |
| Biotin anti-human IgG | BioLegend | Cat# 410718 |
| Biotin anti-human IgM | BioLegend | Cat# 314504 |
| Bacterial and Virus Strains | ||
| Citrobacter rodentium (DBS100 strain) | ATCC | ATCC# 51459 |
| Escherichia coli (Nissle 1917 strain) | Danino et al.71 | N/A |
| Staphylococcus aureus (SH1000 strain) | Archer et al.72 | N/A |
| Staphylococcus lentus (B3 strain) | This manuscript | N/A |
| Biological Samples | ||
| Human breast milk | Mother’s Milk Bank | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hoechst 33342 | Thermo Fisher Scientific | Cat# H1399 |
| FM5-95 | Thermo Fisher Scientific | Cat# T23360 |
| DAPI (4′,6-Diamidino-2-phenylindole dihydrochloride) | Sigma-Aldrich | Cat# D8417 |
| DTT (Dithiothreitol) | Sigma-Aldrich | Cat# D9779 |
| FITC-dextran | Sigma-Aldrich | Cat# FD4 |
| Oxytocin | Sigma-Aldrich | Cat# O4375 |
| Ponceau S solution | Sigma-Aldrich | Cat# P7170 |
| NuPAGE LDS Sample Buffer | Thermo Fisher Scientific | Cat# NP0007 |
| DiBAC4(3) [Bis-(1,3-Dibutylbarbituric Acid) Trimethine Oxonol] | Cayman Chemical | Cat# 33924 |
| CCCP (Carbonyl cyanide m-chlorophenyl hydrazone) | Cayman Chemical | Cat# 25458 |
| Recombinant Human CD59 Protein | Sino Biological | Cat# 12474-H08H |
| Uranyl Acetate | Electron Microscopy Sciences | Cat# 22400 |
| 2.5% Glutaraldehyde in 0.1M Sodium Cacodylate Buffer | Electron Microscopy Sciences | Cat# 16537-15 |
| Neomycin sulfate | Santa Cruz Biotechnology | Cat# sc-3573 |
| Vancomycin Hydrochloride | Santa Cruz Biotechnology | Cat# sc-204938A |
| Cefoxitin | Sagent Pharmaceuticals | NDC 25021-109-10 |
| Sterilizable Fenbendazole Diet | Envigo | Cat# TD.01432 |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1) | Sigma-Aldrich | Cat# P2069 |
| Tryptic Soy Broth | Millipore | Cat# 22092 |
| MacConkey agar | Criterion | Cat# C6131 |
| Lennox L Broth | Research Products International | Cat# L24066 |
| Lennox L Agar | Research Products International | Cat# L24030 |
| Bovine Serum Albumin | Research Products International | Cat# A30075 |
| Trypsin Protease | Thermo Fisher Scientific | Cat# 90057 |
| Critical Commercial Assays | ||
| NucleoSpin Tissue Kit | Macherey-Nagel | Cat# 740952.50 |
| Mouse Immunoglobulin Isotyping ELISA kit | Thermo Fisher Scientific | Cat# 88-50630-88 |
| BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23225 |
| TruSeq Nano DNA Library Prep Kit | Illumina | Cat# 20015965 |
| GeneJET Gel Extraction Kit | Thermo Fisher Scientific | Cat# K0691 |
| Ion Plus Fragment Library Kit | Thermo Fisher Scientific | Cat# 4471252 |
| Streptavidin Magnetic Beads | Thermo Fisher Scientific | Cat# 88816 |
| BeadBug prefilled tubes with 0.1mm Silica glass beads | Sigma-Aldrich | Cat# Z763721 |
| Milk Bacterial DNA isolation Kit | Norgen Bioteck | Cat# 21550 |
| PureLink Microbiome DNA Purification Kit | Thermo Fisher Scientific | Cat# A29789 |
| Phusion High-Fidelity PCR Master Mix | New England Biolabs | Cat# M0531S |
| Deposited Data | ||
| 16S rRNA gene profiling data | NCBI | Accession: BioProject PRJNA796457 |
| S. lentus B3 whole genome sequencing data | NCBI | Accession: BioProject PRJNA796456 |
| S. lentus H29 | GenBank | GenBank accessions: CP059679 |
| S. lentus NCTC12102 | GenBank | GenBank accessions: UHDR01000002 |
| S. aureus MW2 | NCBI | NCBI Reference Sequence: NC_003923 |
| S. epidermids RP62A | NCBI | NCBI Reference Sequence: NC_002976 |
| S. haemolyticus JCSC1435 | NCBI | NCBI Reference Sequence: NC_007168 |
| S. saprophyticus ATCC15305 | NCBI | NCBI Reference Sequence: NC_007350 |
| SwissProt 2020 Mus musculus database | UniProtKB | https://www.uniprot.org |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57Bl/6J mice | Jackson Laboratory | Strain# 000664 |
| Mouse: C1qc−/−: C57BL/6NJ-C1qcem1(IMPC)J/J | Jackson Laboratory | Strain# 029409 |
| Mouse: C3−/−: B6.129S4-C3tm1Crr/J | Jackson Laboratory | Strain# 029661 |
| Mouse: μMT−/−: B6.129S2-Ighmtm1Cgn/J | Jackson Laboratory | Strain# 002288 |
| Software and Algorithms | ||
| GraphPad Prism 9.4.0 | GraphPad | https://www.graphpad.com/ |
| FlowJo 10.9.0 | BD | https://www.flowjo.com/solutions/flowjo |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| Proteome Discoverer v2.4 | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html |
| Mascot v.2.6.2 | Matrix Science | https://www.matrixscience.com/mascot_support_v2_6.html |
| Scaffold 4 | Proteome Software | https://www.proteomesoftware.com |
| ShinyGO v0.741 | South Dakota State University | http://bioinformatics.sdstate.edu/go/ |
| FV31S-SW_V2.1 | Olympus | https://www.olympus-lifescience.com/en/downloads/detail-iframe/?0[downloads][id]=847252002 |
| Fiji | Image J | https://imagej.net/software/fiji/downloads |
| Trimmomatic software V0.32 | Bolger et al.73 | http://www.usadellab.org/cms/index.php?page=trimmomatic |
| QIIME2 | QIIME 2 development team74 | https://library.qiime2.org/about/ |
Experimental model and study participant details
Animals
Wild-type C57Bl/6J mice (stock no. 000664) and C1qc−/− mice (stock no. 029409) were purchased from The Jackson Laboratory (Bar Harbor, ME). C3−/− and μMT−/− mice were kindly shared by S. Lajoie and M. Mugnier (Johns Hopkins University), respectively. Wild-type C57Bl/6J and C1qc−/− mice (8-20 weeks old) were crossed to generate C1qc−/− pups. C1qc−/− and μMT−/− mice (8-20 weeks old) were bred to generate F1 C1q+/−μMT+/− progeny; and F1 C1q+/−μMT+/− mice (8-20 weeks old) were used to generate F2 C1q−/−μMT−/− progeny. All mice were fed sterilized food and water ad libitum and maintained in a specific pathogen free (SPF) mouse facility. C57Bl/6J and C1qc−/− mice were rederived as germ free (GF) using a standard protocol and maintained in the GF mouse facilities at Johns Hopkins University in flexible-film isolators. Sterility was verified at regular intervals using aerobic cultures, anaerobic cultures, and PCR. Mouse genotyping was conducted per the Jackson Laboratory genotyping protocol for the corresponding stock numbers. Except that breast milk samples were collected from female mice during lactation periods, male and female mice (age ranging from weanling [21-day-old] to adulthood [6-8-week-old] as indicated in Method details) were used for all experiments. Littermates of the same sex were randomly assigned to different experimental groups. All animal experiments were performed according to protocols approved by the Johns Hopkins University’s Animal Care and Use Committee and in direct accordance with the NIH guidelines for housing and care of laboratory animals.
Microbe strains
C. rodentium (CR, DBS100 strain, ATCC No. 51459) was purchased from the American Type Culture Collection (ATCC, Manassas, VA). Escherichia coli (Nissle 1917 strain) was kindly shared by T. Danino (Columbia University).71 Staphylococcus lentus (B3 strain) was isolated in this study. All of them were grown from single colonies on Luria–Bertani (LB) plates in LB broth at 37°C overnight with shaking. S. aureus (SH1000 strain) was kindly shared by N. Archer (Johns Hopkins University)72 and cultured in Tryptic Soy Broth (TSB) with chloramphenicol.
Human breast milk samples
The randomly selected and de-identified human breast milk samples from healthy donors were kindly provided by Mother’s Milk Bank in San Jose (www.mothersmilk.org). The donors are healthy lactating women from variant racial background and approved using a specialized clinical review typically including an oral or written interview, screening for general health and medication use, and serological testing.
Method details
Cross-fostering experiments
The breeding pairs (one male and one female) of wild-type C57Bl/6J, C1qc−/−, and C3−/− were synchronized to generate pups born on the same day for subsequent cross-fostering with the dams switched at the day of birth. After 21 days of cross-fostering, pups were separated and used for CR infection experiments.
Reconstitution of GF mice with SPF commensal microbiota
Wild-type and C1qc−/− donor mice (21 days old) maintained in SPF mouse facility were sacrificed in a BACTRON anaerobic chamber (Sheldon, Cornelius, OR). The cecal and colonic contents were collected and diluted into 5 mL sterile PBS, and the suspension was passed through 40 μm nylon filter to remove particulate matter. Fresh slurries were orally gavaged (200 μL per mouse) into GF C1qc−/− recipients (17 days old). Four days later, the transferred GF C1qc−/− mice were challenged with an oral infection with CR or vehicle control.
Bacterial infection in mice
CR infection in mice was conducted as described previously.75 In brief, pups (males and females), separated from their dams on 21 days postnatal (P21), were inoculated by oral gavage with 200 μL of PBS containing 2 × 109 colony-forming units (CFU) of CR or PBS vehicle control. Adult mice (6-8 weeks old) were infected as described above; the body weight, clinical scores, and survival of infected mice were monitored daily. For fecal CR burden analysis, stool was collected from live animals at various time periods post-inoculation. The stool and tissue were homogenized and diluted in sterile PBS at 10 mL per gram of stool or tissue, plated on MacConkey agar plates, and CFUs were enumerated at the following day.
Histology and immunofluorescence
Histology and immunofluorescence staining of colon tissue sections were performed as previously described.76 In brief, after euthanizing mice, the colon was removed under aseptic conditions, washed once with ice-cold PBS, fixed in 10% buffered formalin for 24 hours, and processed for paraffin embedding. Sections (5 μm) were cut and processed for Hematoxylin and Eosin (H&E) staining. Histopathology scores were determined in a blinded fashion using the previously described criteria.75
FITC-dextran assays
FITC-dextran assays for intestinal permeability were performed as previously described.75 In brief, mice were administrated with 150 μL of 80 mg/mL FITC-dextran (4,000 Da, FD4, Sigma) in PBS by oral gavage. After 4 hours, mice were anaesthetized, and blood was collected by cardiac puncture. After centrifugation at 1,000 × g at 4°C for 15 minutes and the levels of FITC-dextran in the plasma was measured using a BioTek Synergy HT microplate reader (BioTek, Winooski, VT) at excitation 485 nm and emission 528 nm.
Flow cytometry and confocal microscopy
S. lentus (B3 strain), incubated for 2 hours with indicated human whey, were washed twice with PBS-1% BSA and incubated with 2 μg/mL rabbit anti-C5b-9 (bs-2673R, Bioss) in 100 μL PBS-1% BSA at 4°C. After 1 hour incubation, bacteria was washed twice with PBS-1% BSA, followed by incubation with 2 μg/mL Alexa Fluor488 conjugated goat anti-rabbit IgG in 100 μL PBS-1%BSA (Thermo Fisher Scientific) for 45 minutes at 4°C. For flow cytometry, bacteria were washed and resuspended with PBS, then analyzed on Cytek NL-3000 flow cytometer (Cytek Biosciences, Fremont, CA). Data were analyzed using the FlowJo software (version 10, BD Life Sciences). For confocal microscopy, S. lentus (B3 strain) was incubated with indicated human whey and stained with anti-C5b-9 and Alexa Fluor 488 conjugated antibodies as described above. Following counterstaining of bacterial DNA and membrane with Hoechst 33342 (1 μg/mL) and FM5-95 (5 μg/mL), respectively, Bacterial samples were mounted with coverslip, sealed, and imaged immediately with Olympus FV3000RS confocal microscope equipped with UPLSAPO 100X SI OIL objective (Olympus, Tokyo, Japan).
Microbiota composition
Total genomic DNA from the cecal and colonic contents, collected from indicated pups (21 days old), was extracted as previously described.77,78 Briefly, cecal and colonic contents were transferred to BeadBug™ prefilled microtubes (2 mL) with 500 μL of 0.1 mm acid washed Silica glass beads (Z763721, Sigma-Aldrich), suspended with 500 μL of extraction buffer (200 mM Tris-HCl, pH 8.0, 200 mM NaCl, 20 mM EDTA), 210 μL of 20% SDS, 500 μL of phenol:chloroform:isoamyl alcohol solution (25:24:1, v/v, Sigma-Aldrich). Samples were disrupted on a VWR Bead Mill Homogenizer (VWR) at maximum speed for 10 minutes to ensure uniform and efficient cell lysis, followed by DNA extraction with phenol:chloroform:isoamyl alcohol solution and DNA precipitation with isopropanol (Sigma-Aldrich), respectively. DNA concentration and purity were monitored on 1% agarose gels, followed by normalization to the same concentration (1 ng/μL). The V4 region of 16S rRNA gene were amplified using Phusion® High-Fidelity PCR Master Mix (New England Biolabs), with the published primer pair of 515F (5’-GTGCCAGCMGCCGCGGTAA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’)79 with the barcode. PCR products were separated by agarose gel electrophoresis for detection, mixed in equidensity ratios, and purified with GeneJETTM Gel Extraction Kit (Thermo Fisher Scientific). Sequencing libraries were generated using Ion Plus Fragment Library Kit (Thermo Fisher Scientific) following manufacturer’s recommendations. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Fisher Scientific), followed by sequencing on an Ion S5™ XL platform and 400-bp single end (SE) reads were generated at Novogene (Sacramento, CA). A mean sequence depth of 160,414 tags per sample was obtained. SE reads were assigned to samples based on their unique barcodes and quality trimmed using the Trimmomatic software V0.32.73 Microbiome bioinformatics were performed with the QIIME 2 2023.5.74 Raw sequence data were demultiplexed and quality filtered using the q2-demux plugin followed by denoising with Deblur80 (via q2-deblur). All amplicon sequence variants (ASVs) were aligned with MAFFT81 (via q2-alignment) and used to construct a phylogeny with FastTree282 (via q2-phylogeny). Taxonomy was assigned to ASVs using the q2-feature-classifier83 classify-sklearn naive Bayes taxonomy classifier against the Greengenes 13_8 99% operational taxonomic units (OTUs) reference sequences.84 Alpha diversity metrics (observed features, Chao1 index, Shannon index, and Simpson index) and beta diversity metrics (weighted UniFrac85 and unweighted UniFrac86) were estimated using q2-diversity after samples were rarefied. All these indices were visualized by R scripts V4.3.0 and GraphPad Prism V7.04 (GraphPad Software, Boston, MA). To further evaluate the differences in species complexity of samples from each group (beta diversity), the non-metric multidimensional scaling (NMDS) clustering was conducted by QIIME2 software74 with weighted UniFrac distance. A permutational multivariate anova (PERMANOVA) test87 was performed to determine the statistical significance for each beta group of NMDS ordination. Significance testing of differences in relative abundance at different taxonomic levels was performed using ANCOM.88
Collection of breast milk whey
Manually express milk from the teat of mice were collected as described previously.89 Briefly, the dam was separated from the litter (5-15 days) for approximately 2 hours prior to the collection of milk. The dam was injected intraperitoneally with 2 IU/kg of oxytocin. After 5-10 minutes to allow the oxytocin to stimulate milk production, dams were anesthetized, and the mammary tissue was gently massaged until a visible bead of milk formed at the base of the teat. The collected mouse breast milk samples and human breast milk samples (obtained from Mother’s Milk Bank) were centrifuged 16,000 × g at 4°C for 15 minutes, and the whey phase was collected and stored at −80°C. For complement inactivation, mouse and human whey samples were heated in a water-bath at 56°C for 30 minutes. Mouse whey protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Dry matter, water content, crude fat, total sugar, crude protein, and gross energy in mouse milk were analyzed as previously described.90
Mass spectrometry and protein functional annotation
Mass spectrometry was conducted by the Mass Spectrometry and Proteomics Core Facility at The Johns Hopkins University School of Medicine. In brief, mouse whey proteins were digested with trypsin (Pierce, Dallas, TX) at 1:50 enzyme to protein ratio overnight at 37°C. Tryptic peptides were analyzed by reverse-phase chromatography tandem mass spectrometry on an EasyLC1100 UPLC interfaced with a Orbitrap-Fusion Lumos mass spectrometer (Thermo Fisher Scientific). Fragmentation spectra were processed by Proteome Discoverer v2.4 (PD2.4, Thermo Fisher Scientific) and searched with Mascot v.2.6.2 (Matrix Science, London, UK) against the SwissProt 2020 Mus musculus database. Peptide identifications from the Mascot searches were processed and imported into Scaffold (Proteome Software Inc.), validated by Protein Prophet to filter at a 95% confidence on peptides and proteins. The list of whey proteins was subjected to ontology enrichment analysis using the ShinyGO (v0.741) tool,91 with the species Mouse and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways selected.
Immunoblot
Mouse whey proteins were mixed with NuPAGE LDS Sample Buffer (Invitrogen, Waltham, MA) and heated at 95°C for 10 minutes, followed by SDS-PAGE under reduced and denaturing conditions. The separated proteins were electro-transferred onto nitrocellulose membranes (Santa Cruz Biotechnology, Dallas, TX) and stained with Ponceau S solution to visualize the transferred material. The membrane was de-stained and immunoblotted with specific antibodies and the Super Signaling system (Thermo Scientific) was used to detect antibody binding according to manufacturer’s instructions. The immunoblots were imaged using a FluorChem E System (Protein Simple, Santa Clara, CA), as previously described.92
Breast milk bactericidal assays
Bactericidal activity of mouse milk on commensal bacteria (CB) was determined on LB agar plates. Briefly, cecal and colonic contents isolated from P21 pups were resuspended in sterile PBS and 20 μL of the suspension was spotted on LB agar plates, and cultured at 25°C. After 30 minutes, 5 μL of mouse whey or PBS vehicle control was layered on the top of the bacteria. The LB agar plates were incubated at 37°C for 16 hours. The milk antimicrobial activity was detected by measuring the colony growth in the whey-covered area. Bactericidal activities of human and mouse milk on the isolated S. lentus B3 were examined on LB agar plates and LB medium culture. For LB agar plate assays, 20 μL of 1 × 105 CFU/ml S. lentus B3 solution, diluted from an overnight culture with sterile LB medium, was spotted on LB agar and cultured at room temperature. After 30 minutes, 5 μL of whey or PBS vehicle control was added to the center of bacterial culture, and the LB agar plates were incubated and imaged as described above. For LB medium culture assays, 1 × 104 CFU/ml S. lentus B3, S. aureus SH1000, or E. coli N1917 in LB medium, supplemented with 2.5% (v/v) mouse whey, 40% (v/v) human whey, or PBS vehicle control was cultured at 37°C with shaking at 150 rpm. After 16 hours, bacterial concentrations were determined by serial dilutions and plating on LB agar plates. CFUs were enumerated after overnight culture and normalized to the culture volume. For time kill-kinetics assays in LB medium, 1 × 104 CFU/mL S. lentus B3, was incubated with 40% (v/v) human whey or PBS vehicle control at 37°C with shaking at 150 rpm. At indicated time points, 100 μL of bacterial culture was plated on LB agar dishes and cultured at 37°C overnight, followed by plate imaging.
Whole genome sequencing, genome assembly, and comparative analysis
Genomic DNA was extracted from overnight culture of S. lentus B3 using NucleoSpin Tissue Kit (Macherey-Nagel, Bethlehem, PA) following manufacturer’s instructions. DNA concentration and purity were monitored on 1% agarose gels, followed by normalization to the same concentration (1 ng/μL). Sequencing libraries were generated using TruSeq Nano DNA Library Prep Kit (Illumina Inc., San Diego, CA) following manufacturer’s recommendations. The library quality was assessed on an Agilent TapeStation 4200, followed by sequencing on a NovaSeq6000 S4 platform and 150-bp paired end (PE) reads were generated at Psomagen (Rockville, MD). The quality control of raw sequencing data, 30,126,990 total reads and 4.549 Gb, was conducted using the Trimmomatic v0.36 with settings to trim low-quality bases (Q<15) from both ends of each read. Valid reads were then fed to the de novo genome assembler SPAdes v3.9.0 to reconstruct the draft genome of S. lentus B3 with default parameters.93 The complete genomes of the S. lentus strains H29 and NCTC12102 (GenBank accessions: CP059679 and UHDR01000002) were used to assemble and order contigs to build the S. lentus B3 genome. The GenomeComp v1.294 was used for linear comparison of the aforementioned three S. lentus genomes, whereas the Proksee95 was employed for circular genomic comparison between S. lentus B3 and other representative Staphylococcus genomes, including S. lentus (H29 and NCTC12102), S. aureus MW2 (NC_003923), S. epidermids RP62A (NC_002976), S. haemolyticus JCSC1435 (NC_007168) and S. saprophyticus ATCC15305 (NC_007350).
Enzyme-linked immunosorbent assays
Immunoglobulins in the milk from the dams were measured using a Mouse Immunoglobulin Isotyping ELISA kit (Thermo Fisher Scientific) following manufacturer’s instructions. Results were read at a BioTek Synergy HT microplate reader (BioTek) at optical density 450 nm.
Maternal antibiotics treatment
For maternal antibiotics treatment, dams were fed sterile drinking water containing Neomycin (1 mg/mL), Vancomycin (500 μg/mL), or Cefoxitin (500 μg/mL), starting from day 7 after giving birth. For Fenbendazole treatment, dams were administrated with Sterilizable Fenbendazole Diet (150 ppm, Envigo, Indianapolis, IN) during lactation period. All antibiotic administration was halted on day 19 after birth. Following removal of maternal antibiotics treatment for 48 hours, pups were inoculated with CR by oral gavage.
Membrane depolarization
Fluorescent probe Bis-(1,3-Dibutylbarbituric Acid) Trimethine Oxonol [DiBAC4(3), Cayman Chemical, Ann Arbor, MI] was used to assess membrane depolarization, as previously described.48 In brief, S. lentus B3 was incubated with whey or PBS control, in the presence or absence of CD59 (200 μg/mL) or Vitronectin (100 μg/mL), at 37 °C with 150 rpm shaking. At indicated time periods, bacteria were washed with PBS and stained with 500 μL of 1 μg/mL DiBAC4(3). The protonophore carbonyl cyanide m-chlorophenyl hydrazine (CCCP, Cayman Chemical) was included in bacteria suspensions at a final concentration of 10 μM for 30 minutes for a positive control. Bacterial fluorescence was measured on Cytek NL-3000 flow cytometer (Cytek Biosciences), and data were analyzed using the FlowJo software (BD Life Sciences).
Immunoglobin depletion in human milk whey
Human whey samples were mock treated or incubated with biotin-conjugated antibodies specific for human IgG and IgM (BioLegend) together with Streptavidin magnetic beads (Thermo Scientific), followed by magnetic column separation. The complete IgG/IgM depletion in human milk whey samples was validated by SDS-PAGE separation and immunoblot, prior to the usage in the indicated assays.
Transmission electron microscopy
S. lentus B3 derived from a log-phase culture was incubated with mouse or human breast-milk-derived whey from the indicated dams for 2 hours, followed by fixation with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer for 30 minutes. The cells were washed, suspended with DPBS, and subjected to negative staining, as previously described.96 Briefly, 10 μL of cell suspension was placed onto copper EM grids and stained with 1% uranyl acetate for 1 minute. Following three washes with distilled water, the grids were dried on filter paper for at least 30 minutes and mounted on a Hitachi H7600 Transmission Electron Microscope (Chiyoda, Japan). Specimens were illuminated with a focused beam of electrons in a vacuum chamber at 80 KV electron energy.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.0.1 (GraphPad Software). Standard errors of means (s.e.m.) were plotted in graphs. Statistical significance was determined by Student’s t-test, one-way ANOVA with Bonferroni post-hoc test, or log-rank (Mantel-Cox) test (survival curves). Significant differences were considered: ns, non-significant difference; * at p < 0.05; ** at p < 0.01; *** at p < 0.001; and **** at p < 0001.
Supplementary Material
Highlights.
Weanling mice with complement-deficient milk are susceptible to enteric infection
Complement in milk selectively culls certain gram-positive microbes in infant gut
Breast milk complement is activated by a C1-dependent, antibody-independent pathway
Early-life gut microbiota regulates neonate susceptibility to enteric infection
Acknowledgements
We appreciate S. Lajoie and M. Mugnier for sharing mouse strains; N. Archer and T. Danino for sharing bacterial strains; M. Power for measuring milk macronutrients; H. Ding, R. Cole, and B. Smith for facilitating the studies using germ free animals, mass spectrometry, and electron microscopy, respectively; C. Sears for sharing reagents; and Mother’s Milk Bank for providing human milk samples. This work was supported in part by grants from National Institutes of Health (GM111682, AI137719, CA244350 to F.W.); Department of Defense (W81XWH-19-1-0479 to F.W.); American Association of Immunologists (Career in Immunology Fellowship to X.D.); and American Heart Association (19PRE34380234 to Y.L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- 1.Perin J, Mulick A, Yeung D, Villavicencio F, Lopez G, Strong KL, Prieto-Merino D, Cousens S, Black RE, and Liu L (2022). Global, regional, and national causes of under-5 mortality in 2000-19: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet Child Adolesc Health 6, 106–115. 10.1016/S2352-4642(21)00311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basha S, Surendran N, and Pichichero M (2014). Immune responses in neonates. Expert Rev Clin Immunol 10, 1171–1184. 10.1586/1744666X.2014.942288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camacho-Morales A, Caba M, Garcia-Juarez M, Caba-Flores MD, Viveros-Contreras R, and Martinez-Valenzuela C (2021). Breastfeeding Contributes to Physiological Immune Programming in the Newborn. Front Pediatr 9, 744104. 10.3389/fped.2021.744104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurley WL, and Theil PK (2011). Perspectives on immunoglobulins in colostrum and milk. Nutrients 3, 442–474. 10.3390/nu3040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fehr K, Moossavi S, Sbihi H, Boutin RCT, Bode L, Robertson B, Yonemitsu C, Field CJ, Becker AB, Mandhane PJ, et al. (2020). Breastmilk Feeding Practices Are Associated with the Co-Occurrence of Bacteria in Mothers’ Milk and the Infant Gut: the CHILD Cohort Study. Cell Host Microbe 28, 285–297 e284. 10.1016/j.chom.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Hanson LA, and Korotkova M (2002). The role of breastfeeding in prevention of neonatal infection. Semin Neonatol 7, 275–281. 10.1016/s1084-2756(02)90124-7. [DOI] [PubMed] [Google Scholar]
- 7.Le Doare K, Holder B, Bassett A, and Pannaraj PS (2018). Mother’s Milk: A Purposeful Contribution to the Development of the Infant Microbiota and Immunity. Front Immunol 9, 361. 10.3389/fimmu.2018.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyons KE, Ryan CA, Dempsey EM, Ross RP, and Stanton C (2020). Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 12. 10.3390/nu12041039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris NL, Spoerri I, Schopfer JF, Nembrini C, Merky P, Massacand J, Urban JF Jr., Lamarre A, Burki K, Odermatt B, et al. (2006). Mechanisms of neonatal mucosal antibody protection. J Immunol 177, 6256–6262. 10.4049/jimmunol.177.9.6256. [DOI] [PubMed] [Google Scholar]
- 10.Zheng W, Zhao W, Wu M, Song X, Caro F, Sun X, Gazzaniga F, Stefanetti G, Oh S, Mekalanos JJ, and Kasper DL (2020). Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 577, 543–548. 10.1038/s41586-019-1898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gopalakrishna KP, Macadangdang BR, Rogers MB, Tometich JT, Firek BA, Baker R, Ji J, Burr AHP, Ma C, Good M, et al. (2019). Maternal IgA protects against the development of necrotizing enterocolitis in preterm infants. Nat Med 25, 1110–1115. 10.1038/s41591-019-0480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson JJ, Archer-Hartmann S, Yarawsky AE, Miller JLC, Seveau S, Shao TY, Severance AL, Miller-Handley H, Wu Y, Pham G, et al. (2022). Pregnancy enables antibody protection against intracellular infection. Nature 606, 769–775. 10.1038/s41586-022-04816-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumler AJ, and Sperandio V (2016). Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535, 85–93. 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahrstrom CT, Pariente N, and Weiss U (2016). Intestinal microbiota in health and disease. Nature 535, 47. 10.1038/535047a. [DOI] [PubMed] [Google Scholar]
- 15.Kim YG, Sakamoto K, Seo SU, Pickard JM, Gillilland MG 3rd, Pudlo NA, Hoostal M, Li X, Wang TD, Feehley T, et al. (2017). Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319. 10.1126/science.aag2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricklin D, Reis ES, and Lambris JD (2016). Complement in disease: a defence system turning offensive. Nat Rev Nephrol 12, 383–401. 10.1038/nrneph.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricklin D, Hajishengallis G, Yang K, and Lambris JD (2010). Complement: a key system for immune surveillance and homeostasis. Nat Immunol 11, 785–797. 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesargikar PN, Spiller B, and Chavez R (2012). The complement system: history, pathways, cascade and inhibitors. Eur J Microbiol Immunol (Bp) 2, 103–111. 10.1556/EuJMI.2.2012.2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liszewski MK, Java A, Schramm EC, and Atkinson JP (2017). Complement Dysregulation and Disease: Insights from Contemporary Genetics. Annu Rev Pathol 12, 25–52. 10.1146/annurev-pathol-012615-044145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolev M, Le Friec G, and Kemper C (2014). Complement--tapping into new sites and effector systems. Nat Rev Immunol 14, 811–820. 10.1038/nri3761. [DOI] [PubMed] [Google Scholar]
- 21.Heesterbeek DAC, Angelier ML, Harrison RA, and Rooijakkers SHM (2018). Complement and Bacterial Infections: From Molecular Mechanisms to Therapeutic Applications. J Innate Immun 10, 455–464. 10.1159/000491439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavaillon JM, Sansonetti P, and Goldman M (2019). 100th Anniversary of Jules Bordet’s Nobel Prize: Tribute to a Founding Father of Immunology. Front Immunol 10, 2114. 10.3389/fimmu.2019.02114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudry G, Charton E, Le Huerou-Luron I, Ferret-Bernard S, Le Gall S, Even S, and Blat S (2021). The Relationship Between Breast Milk Components and the Infant Gut Microbiota. Front Nutr 8, 629740. 10.3389/fnut.2021.629740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korhonen H, Marnila P, and Gill HS (2000). Milk immunoglobulins and complement factors. Br J Nutr 84 Suppl 1, S75–80. 10.1017/s0007114500002282. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Boeren S, Hageman JA, van Hooijdonk T, Vervoort J, and Hettinga K (2015). Bovine milk proteome in the first 9 days: protein interactions in maturation of the immune and digestive system of the newborn. PLoS One 10, e0116710. 10.1371/journal.pone.0116710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakajima S, Baba AS, and Tamura N (1977). Complement system in human colostrum: presence of nine complement components and factors of alternative pathway in human colostrum. Int Arch Allergy Appl Immunol 54, 428–433. [PubMed] [Google Scholar]
- 27.Rainard P. (2003). The complement in milk and defense of the bovine mammary gland against infections. Vet Res 34, 647–670. 10.1051/vetres:2003025. [DOI] [PubMed] [Google Scholar]
- 28.Ogundele MO (1999). Complement-mediated bactericidal activity of human milk to a serum-susceptible strain of E. coli 0111. J Appl Microbiol 87, 689–696. 10.1046/j.1365-2672.1999.00911.x. [DOI] [PubMed] [Google Scholar]
- 29.Korhonen H, Syvaoja EL, Ahola-Luttila H, Sivela S, Kopola S, Husu J, and Kosunen TU (1995). Bactericidal effect of bovine normal and immune serum, colostrum and milk against Helicobacter pylori. J Appl Bacteriol 78, 655–662. 10.1111/j.1365-2672.1995.tb03112.x. [DOI] [PubMed] [Google Scholar]
- 30.Dunkelberger JR, and Song WC (2010). Complement and its role in innate and adaptive immune responses. Cell Res 20, 34–50. 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 31.Mullineaux-Sanders C, Sanchez-Garrido J, Hopkins EGD, Shenoy AR, Barry R, and Frankel G (2019). Citrobacter rodentium-host-microbiota interactions: immunity, bioenergetics and metabolism. Nat Rev Microbiol 17, 701–715. 10.1038/s41579-019-0252-z. [DOI] [PubMed] [Google Scholar]
- 32.Collins JW, Keeney KM, Crepin VF, Rathinam VA, Fitzgerald KA, Finlay BB, and Frankel G (2014). Citrobacter rodentium: infection, inflammation and the microbiota. Nat Rev Microbiol 12, 612–623. 10.1038/nrmicro3315. [DOI] [PubMed] [Google Scholar]
- 33.Caballero-Flores G, Pickard JM, and Nunez G (2021). Regulation of Citrobacter rodentium colonization: virulence, immune response and microbiota interactions. Curr Opin Microbiol 63, 142–149. 10.1016/j.mib.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gros P, Milder FJ, and Janssen BJ (2008). Complement driven by conformational changes. Nat Rev Immunol 8, 48–58. 10.1038/nri2231. [DOI] [PubMed] [Google Scholar]
- 35.Ricklin D, Reis ES, Mastellos DC, Gros P, and Lambris JD (2016). Complement component C3 - The “Swiss Army Knife” of innate immunity and host defense. Immunol Rev 274, 33–58. 10.1111/imr.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thielens NM, Tedesco F, Bohlson SS, Gaboriaud C, and Tenner AJ (2017). C1q: A fresh look upon an old molecule. Mol Immunol 89, 73–83. 10.1016/j.molimm.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid KBM (2018). Complement Component C1q: Historical Perspective of a Functionally Versatile, and Structurally Unusual, Serum Protein. Front Immunol 9, 764. 10.3389/fimmu.2018.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kishore U, Thielens NM, and Gaboriaud C (2016). Editorial: State-of-the-Art Research on C1q and the Classical Complement Pathway. Front Immunol 7, 398. 10.3389/fimmu.2016.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bajic G, Degn SE, Thiel S, and Andersen GR (2015). Complement activation, regulation, and molecular basis for complement-related diseases. EMBO J 34, 2735–2757. 10.15252/embj.201591881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogundele M. (2001). Role and significance of the complement system in mucosal immunity: particular reference to the human breast milk complement. Immunol Cell Biol 79, 1–10. 10.1046/j.1440-1711.2001.00976.x. [DOI] [PubMed] [Google Scholar]
- 41.Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ, and Kaetzel CS (2014). Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A 111, 3074–3079. 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rollins SA, Zhao J, Ninomiya H, and Sims PJ (1991). Inhibition of homologous complement by CD59 is mediated by a species-selective recognition conferred through binding to C8 within C5b-8 or C9 within C5b-9. J Immunol 146, 2345–2351. [PubMed] [Google Scholar]
- 43.Davies A, Simmons DL, Hale G, Harrison RA, Tighe H, Lachmann PJ, and Waldmann H (1989). CD59, an LY-6-like protein expressed in human lymphoid cells, regulates the action of the complement membrane attack complex on homologous cells. J Exp Med 170, 637–654. 10.1084/jem.170.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh B, Su YC, and Riesbeck K (2010). Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol Microbiol 78, 545–560. 10.1111/j.1365-2958.2010.07373.x. [DOI] [PubMed] [Google Scholar]
- 45.Milis L, Morris CA, Sheehan MC, Charlesworth JA, and Pussell BA (1993). Vitronectin-mediated inhibition of complement: evidence for different binding sites for C5b-7 and C9. Clin Exp Immunol 92, 114–119. 10.1111/j.1365-2249.1993.tb05956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaboriaud C, Ling WL, Thielens NM, Bally I, and Rossi V (2014). Deciphering the fine details of c1 assembly and activation mechanisms: “mission impossible”? Front Immunol 5, 565. 10.3389/fimmu.2014.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallis R, Mitchell DA, Schmid R, Schwaeble WJ, and Keeble AH (2010). Paths reunited: Initiation of the classical and lectin pathways of complement activation. Immunobiology 215, 1–11. 10.1016/j.imbio.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kashyap DR, Wang M, Liu LH, Boons GJ, Gupta D, and Dziarski R (2011). Peptidoglycan recognition proteins kill bacteria by activating protein-sensing two-component systems. Nat Med 17, 676–683. 10.1038/nm.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harriman GR, Podack ER, Braude AI, Corbeil LC, Esser AF, and Curd JG (1982). Activation of complement by serum-resistant Neisseria gonorrhoeae. Assembly of the membrane attack complex without subsequent cell death. J Exp Med 156, 1235–1249. 10.1084/jem.156.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joiner KA, Brown EJ, and Frank MM (1984). Complement and bacteria: chemistry and biology in host defense. Annu Rev Immunol 2, 461–491. 10.1146/annurev.iy.02.040184.002333. [DOI] [PubMed] [Google Scholar]
- 51.Tomlinson S, Taylor PW, Morgan BP, and Luzio JP (1989). Killing of gram-negative bacteria by complement. Fractionation of cell membranes after complement C5b-9 deposition on to the surface of Salmonella minnesota Re595. Biochem J 263, 505–511. 10.1042/bj2630505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bloch EF, Knight EM, Carmon T, McDonald-Pinkett S, Carter J, Boomer A, Ogunfusika M, Petersen M, Famakin B, Aniagolu J, et al. (1997). C5b-7 and C5b-8 precursors of the membrane attack complex (C5b-9) are effective killers of E. coli J5 during serum incubation. Immunol Invest 26, 409–419. 10.3109/08820139709022698. [DOI] [PubMed] [Google Scholar]
- 53.Murphy K, Weaver C, and Janeway C (2017). Janeway’s immunobiology, Ninth edition. Edition (W.W. Norton & Company; ). [Google Scholar]
- 54.Kitamura D, Roes J, Kuhn R, and Rajewsky K (1991). A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350, 423–426. 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 55.Sarkar A, Yoo JY, Valeria Ozorio Dutra S, Morgan KH, and Groer M (2021). The Association between Early-Life Gut Microbiota and Long-Term Health and Diseases. J Clin Med 10. 10.3390/jcm10030459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gaufin T, Tobin NH, and Aldrovandi GM (2018). The importance of the microbiome in pediatrics and pediatric infectious diseases. Curr Opin Pediatr 30, 117–124. 10.1097/MOP.0000000000000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Derrien M, Alvarez AS, and de Vos WM (2019). The Gut Microbiota in the First Decade of Life. Trends Microbiol 27, 997–1010. 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, Adisetiyo H, Zabih S, Lincez PJ, Bittinger K, et al. (2017). Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr 171, 647–654. 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atyeo C, and Alter G (2021). The multifaceted roles of breast milk antibodies. Cell 184, 1486–1499. 10.1016/j.cell.2021.02.031. [DOI] [PubMed] [Google Scholar]
- 60.Adlerberth I, Lindberg E, Aberg N, Hesselmar B, Saalman R, Strannegard IL, and Wold AE (2006). Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res 59, 96–101. 10.1203/01.pdr.0000191137.12774.b2. [DOI] [PubMed] [Google Scholar]
- 61.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, et al. (2015). Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 17, 690–703. 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 62.Berends ET, Dekkers JF, Nijland R, Kuipers A, Soppe JA, van Strijp JA, and Rooijakkers SH (2013). Distinct localization of the complement C5b-9 complex on Gram-positive bacteria. Cell Microbiol 15, 1955–1968. 10.1111/cmi.12170. [DOI] [PubMed] [Google Scholar]
- 63.Diebolder CA, Beurskens FJ, de Jong RN, Koning RI, Strumane K, Lindorfer MA, Voorhorst M, Ugurlar D, Rosati S, Heck AJ, et al. (2014). Complement is activated by IgG hexamers assembled at the cell surface. Science 343, 1260–1263. 10.1126/science.1248943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Betz SJ, and Isliker H (1981). Antibody-independent interactions between Escherichia coli J5 and human complement components. J Immunol 127, 1748–1754. [PubMed] [Google Scholar]
- 65.Clas F, and Loos M (1981). Antibody-independent binding of the first component of complement (C1) and its subcomponent C1q to the S and R forms of Salmonella minnesota. Infect Immun 31, 1138–1144. 10.1128/iai.31.3.1138-1144.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tenner AJ, Ziccardi RJ, and Cooper NR (1984). Antibody-independent C1 activation by E. coli. J Immunol 133, 886–891. [PubMed] [Google Scholar]
- 67.Alberti S, Marques G, Camprubi S, Merino S, Tomas JM, Vivanco F, and Benedi VJ (1993). C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect Immun 61, 852–860. 10.1128/iai.61.3.852-860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mintz CS, Arnold PI, Johnson W, and Schultz DR (1995). Antibody-independent binding of complement component C1q by Legionella pneumophila. Infect Immun 63, 4939–4943. 10.1128/iai.63.12.4939-4943.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Loos M, Clas F, and Fischer W (1986). Interaction of purified lipoteichoic acid with the classical complement pathway. Infect Immun 53, 595–599. 10.1128/iai.53.3.595-599.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilkinson BJ, Kim Y, and Peterson PK (1981). Factors affecting complement activation by Staphylococcus aureus cell walls, their components, and mutants altered in teichoic acid. Infect Immun 32, 216–224. 10.1128/iai.32.1.216-224.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, Hasty J, and Bhatia SN (2015). Programmable probiotics for detection of cancer in urine. Sci Transl Med 7, 289ra284. 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Archer NK, Wang Y, Ortines RV, Liu H, Nolan SJ, Liu Q, Alphonse MP, Dikeman DA, Mazhar M, Miller RJ, et al. (2020). Preclinical Models and Methodologies for Monitoring Staphylococcus aureus Infections Using Noninvasive Optical Imaging. Methods Mol Biol 2069, 197–228. 10.1007/978-1-4939-9849-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37, 852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xia X, Liu Y, Hodgson A, Xu D, Guo W, Yu H, She W, Zhou C, Lan L, Fu K, et al. (2019). EspF is crucial for Citrobacter rodentium-induced tight junction disruption and lethality in immunocompromised animals. PLoS Pathog 15, e1007898. 10.1371/journal.ppat.1007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Y, Fu K, Wier EM, Lei Y, Hodgson A, Xu D, Xia X, Zheng D, Ding H, Sears CL, et al. (2022). Bacterial Genotoxin Accelerates Transient Infection-Driven Murine Colon Tumorigenesis. Cancer Discov 12, 236–249. 10.1158/2159-8290.CD-21-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, et al. (2014). Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510, 417–421. 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, et al. (2016). Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351. 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624. 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, and Knight R (2017). Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2. 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katoh K, Misawa K, Kuma K, and Miyata T (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066. 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Price MN, Dehal PS, and Arkin AP (2010). FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, and Gregory Caporaso J (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90. 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, and Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6, 610–618. 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lozupone CA, Hamady M, Kelley ST, and Knight R (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73, 1576–1585. 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lozupone C, and Knight R (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–8235. 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anderson MJ Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online, pp. 1–15. 10.1002/9781118445112.stat07841. [DOI] [Google Scholar]
- 88.Mandal S, Van Treuren W, White RA, Eggesbo M, Knight R, and Peddada SD (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26, 27663. 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Willingham K, McNulty E, Anderson K, Hayes-Klug J, Nalls A, and Mathiason C (2014). Milk collection methods for mice and Reeves’ muntjac deer. J Vis Exp. 10.3791/51007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Power ML, Schulkin J, Drought H, Milligan LA, Murtough KL, and Bernstein RM (2017). Patterns of milk macronutrients and bioactive molecules across lactation in a western lowland gorilla (Gorilla gorilla) and a Sumatran orangutan (Pongo abelii). Am J Primatol 79, 1–11. 10.1002/ajp.22609. [DOI] [PubMed] [Google Scholar]
- 91.Ge SX, Jung D, and Yao R (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fu K, Sun X, Wier EM, Hodgson A, Liu Y, Sears CL, and Wan F (2016). Sam68/KHDRBS1 is critical for colon tumorigenesis by regulating genotoxic stress-induced NF-kappaB activation. Elife 5. 10.7554/eLife.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang J, Wang J, Yao ZJ, Jin Q, Shen Y, and Chen R (2003). GenomeComp: a visualization tool for microbial genome comparison. J Microbiol Methods 54, 423–426. 10.1016/s0167-7012(03)00094-0. [DOI] [PubMed] [Google Scholar]
- 95.Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen CY, Graham M, Van Domselaar G, and Stothard P (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res 51, W484–W492. 10.1093/nar/gkad326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Scarff CA, Fuller MJG, Thompson RF, and Iadanza MG (2018). Variations on Negative Stain Electron Microscopy Methods: Tools for Tackling Challenging Systems. J Vis Exp. 10.3791/57199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rRNA gene profiling data and S. lentus B3 whole genome sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) database and are publicly available as of the date of publication. This paper also analyzes sequencing data of bacteria S. lentus (H29 strain), S. lentus (NCTC12102 strain), S. aureus (MW2 strain), S. epidermids (RP62A strain), S. haemolyticus (JCSC1435 strain), and S. saprophyticus (ATCC15305 strain), as well as Mus musculus database, all of which are existing and publicly available. These accession numbers for the datasets are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat anti-human C1s | Complement Technology | Cat# A204 |
| Rabbit Anti-Human C5b-9 | Bioss Antibodies | Cat# bs-2673R |
| HRP-conjugated Goat Anti-Mouse IgG | SouthernBiotech | Cat# 1030-05 |
| HRP-conjugated Goat Anti-Mouse IgM | SouthernBiotech | Cat# 1021-05 |
| HRP-conjugated Goat Anti-Human IgG | SouthernBiotech | Cat# 2040-05 |
| HRP-conjugated Goat Anti-Human IgM | SouthernBiotech | Cat# 2020-05 |
| Alexa Fluor 488 conjugated Goat Anti-Rabbit IgG (H+L) | Thermo Fisher Scientific | Cat# A-11034 |
| Biotin anti-human IgG | BioLegend | Cat# 410718 |
| Biotin anti-human IgM | BioLegend | Cat# 314504 |
| Bacterial and Virus Strains | ||
| Citrobacter rodentium (DBS100 strain) | ATCC | ATCC# 51459 |
| Escherichia coli (Nissle 1917 strain) | Danino et al.71 | N/A |
| Staphylococcus aureus (SH1000 strain) | Archer et al.72 | N/A |
| Staphylococcus lentus (B3 strain) | This manuscript | N/A |
| Biological Samples | ||
| Human breast milk | Mother’s Milk Bank | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Hoechst 33342 | Thermo Fisher Scientific | Cat# H1399 |
| FM5-95 | Thermo Fisher Scientific | Cat# T23360 |
| DAPI (4′,6-Diamidino-2-phenylindole dihydrochloride) | Sigma-Aldrich | Cat# D8417 |
| DTT (Dithiothreitol) | Sigma-Aldrich | Cat# D9779 |
| FITC-dextran | Sigma-Aldrich | Cat# FD4 |
| Oxytocin | Sigma-Aldrich | Cat# O4375 |
| Ponceau S solution | Sigma-Aldrich | Cat# P7170 |
| NuPAGE LDS Sample Buffer | Thermo Fisher Scientific | Cat# NP0007 |
| DiBAC4(3) [Bis-(1,3-Dibutylbarbituric Acid) Trimethine Oxonol] | Cayman Chemical | Cat# 33924 |
| CCCP (Carbonyl cyanide m-chlorophenyl hydrazone) | Cayman Chemical | Cat# 25458 |
| Recombinant Human CD59 Protein | Sino Biological | Cat# 12474-H08H |
| Uranyl Acetate | Electron Microscopy Sciences | Cat# 22400 |
| 2.5% Glutaraldehyde in 0.1M Sodium Cacodylate Buffer | Electron Microscopy Sciences | Cat# 16537-15 |
| Neomycin sulfate | Santa Cruz Biotechnology | Cat# sc-3573 |
| Vancomycin Hydrochloride | Santa Cruz Biotechnology | Cat# sc-204938A |
| Cefoxitin | Sagent Pharmaceuticals | NDC 25021-109-10 |
| Sterilizable Fenbendazole Diet | Envigo | Cat# TD.01432 |
| Phenol:Chloroform:Isoamyl Alcohol (25:24:1) | Sigma-Aldrich | Cat# P2069 |
| Tryptic Soy Broth | Millipore | Cat# 22092 |
| MacConkey agar | Criterion | Cat# C6131 |
| Lennox L Broth | Research Products International | Cat# L24066 |
| Lennox L Agar | Research Products International | Cat# L24030 |
| Bovine Serum Albumin | Research Products International | Cat# A30075 |
| Trypsin Protease | Thermo Fisher Scientific | Cat# 90057 |
| Critical Commercial Assays | ||
| NucleoSpin Tissue Kit | Macherey-Nagel | Cat# 740952.50 |
| Mouse Immunoglobulin Isotyping ELISA kit | Thermo Fisher Scientific | Cat# 88-50630-88 |
| BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23225 |
| TruSeq Nano DNA Library Prep Kit | Illumina | Cat# 20015965 |
| GeneJET Gel Extraction Kit | Thermo Fisher Scientific | Cat# K0691 |
| Ion Plus Fragment Library Kit | Thermo Fisher Scientific | Cat# 4471252 |
| Streptavidin Magnetic Beads | Thermo Fisher Scientific | Cat# 88816 |
| BeadBug prefilled tubes with 0.1mm Silica glass beads | Sigma-Aldrich | Cat# Z763721 |
| Milk Bacterial DNA isolation Kit | Norgen Bioteck | Cat# 21550 |
| PureLink Microbiome DNA Purification Kit | Thermo Fisher Scientific | Cat# A29789 |
| Phusion High-Fidelity PCR Master Mix | New England Biolabs | Cat# M0531S |
| Deposited Data | ||
| 16S rRNA gene profiling data | NCBI | Accession: BioProject PRJNA796457 |
| S. lentus B3 whole genome sequencing data | NCBI | Accession: BioProject PRJNA796456 |
| S. lentus H29 | GenBank | GenBank accessions: CP059679 |
| S. lentus NCTC12102 | GenBank | GenBank accessions: UHDR01000002 |
| S. aureus MW2 | NCBI | NCBI Reference Sequence: NC_003923 |
| S. epidermids RP62A | NCBI | NCBI Reference Sequence: NC_002976 |
| S. haemolyticus JCSC1435 | NCBI | NCBI Reference Sequence: NC_007168 |
| S. saprophyticus ATCC15305 | NCBI | NCBI Reference Sequence: NC_007350 |
| SwissProt 2020 Mus musculus database | UniProtKB | https://www.uniprot.org |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57Bl/6J mice | Jackson Laboratory | Strain# 000664 |
| Mouse: C1qc−/−: C57BL/6NJ-C1qcem1(IMPC)J/J | Jackson Laboratory | Strain# 029409 |
| Mouse: C3−/−: B6.129S4-C3tm1Crr/J | Jackson Laboratory | Strain# 029661 |
| Mouse: μMT−/−: B6.129S2-Ighmtm1Cgn/J | Jackson Laboratory | Strain# 002288 |
| Software and Algorithms | ||
| GraphPad Prism 9.4.0 | GraphPad | https://www.graphpad.com/ |
| FlowJo 10.9.0 | BD | https://www.flowjo.com/solutions/flowjo |
| Adobe Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| Proteome Discoverer v2.4 | Thermo Fisher Scientific | https://www.thermofisher.com/us/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html |
| Mascot v.2.6.2 | Matrix Science | https://www.matrixscience.com/mascot_support_v2_6.html |
| Scaffold 4 | Proteome Software | https://www.proteomesoftware.com |
| ShinyGO v0.741 | South Dakota State University | http://bioinformatics.sdstate.edu/go/ |
| FV31S-SW_V2.1 | Olympus | https://www.olympus-lifescience.com/en/downloads/detail-iframe/?0[downloads][id]=847252002 |
| Fiji | Image J | https://imagej.net/software/fiji/downloads |
| Trimmomatic software V0.32 | Bolger et al.73 | http://www.usadellab.org/cms/index.php?page=trimmomatic |
| QIIME2 | QIIME 2 development team74 | https://library.qiime2.org/about/ |


