Malaria has seen a worrying resurgence across tropical regions, with cases and mortality rising after a 15-year decline earlier this century. The major impact falls on sub-Saharan Africa, with an estimated 593 000 deaths in 2021.1 Treatment of blood stage infection caused by Plasmodium falciparum depends on rapid clinical efficacy of artemisinin-based combination therapies. Artemisinin partial resistance (driven by Kelch13 mutations) has recently emerged in eastern Africa, threatening future treatment efficacy.2 In southeast Asia, the earlier emergence of artemisinin resistance was soon followed by resistance to piperaquine, with regional treatment failure rates averaging 50%.3 Piperaquine resistance has not previously been documented elsewhere.
In this issue of The Lancet Infectious Diseases, Celia Florimond and colleagues4 report that P falciparum piperaquine resistance has emerged in French Guiana and neighbouring countries in the Guiana Shield.
This drug, partnered with dihydroartemisinin, is sporadically prescribed but is used as self-medication by gold miners in remote forested regions. Piperaquine resistance was observed in 40 (47%) of 86 in vitro-cultured parasites, and three (50%) of six patients experienced piperaquine–dihydroartemisinin treatment failure despite having adequate piperaquine intake. Based on the evidence provided, we agree with the authors’ recommendation to discontinue piperaquine use in the region.
Previous studies by this group had noted a novel mutation, C350R (ie, Cys350Arg), in the P falciparum chloroquine resistance transporter (pfCRT), which emerged on the background of the South American 7G8 variant.5 Here, the authors show that by 2008 this mutation was present in two-thirds of the clinical isolates sampled from French Guiana, Suriname, and Guyana. These C350R isolates showed low genetic relatedness, suggesting multiple independent origins. In a piperaquine survival assay, 40 (71%) of 56 isolates with pfCRTC350R showed parasite survival rates of more than 10%, a standardised threshold of in-vitro resistance. None of the 30 pfCRTC350 isolates were resistant. Genome-wide association studies pinpointed C350R as the sole point mutation associating with elevated survival. This report also focused on plasmepsin 2 and plasmepsin 3 (pfpm2 and pfpm3) gene amplification, which in southeast Asia is a robust molecular marker of piperaquine resistance.6,7 Unlike in Asia, multi copy pfpm2 and pfpm3 did not emerge before the spread of mutant piperaquine-resistant pfCRT in the Guiana Shield. In French Guiana, multi copy pfpm2 and pfpm3 isolates were not resistant in the absence of C350R, whereas one third of the piperaquine-resistant mutant pfCRT isolates harboured a single copy of pfpm2 and pfpm3. The highest levels of resistance were observed with parasites harbouring C350R and multi copy pfpm2 and pfpm3. In this therapeutic efficacy study, the three individuals who experienced piperaquine-dihydroartemisinin treatment failure all harboured parasites with C350R, of which two also had multi copy pfpm2 and pfpm3. All patient isolates harboured wild-type Kelch13. This report provides worrying evidence that piperaquine resistance alone can suffice to cause artemisinin-based combination therapy treatment failure, even without artemisinin partial resistance. This is a major difference from southeast Asia, where piperaquine resistance (via other mutations in pfCRT as well as multi copy pfpm2 and pfpm3) emerged in artemisinin-resistant parasites harbouring mutant Kelch13.3
Complementary to this field-based analysis by Florimond and colleagues,4 we examined the contribution of pfpm2 and pfpm3 copy number on a pfCRTC350R background, using the S170 isolate from French Guiana. We observed spontaneous deamplification of pfpm2 and pfpm3 in this isolate (appendix p 4), as earlier seen with piperaquine-resistant Cambodian parasites and reflecting genomic instability of this locus.8,9 Phenotypic profiling of isogenic S170 clones with one or two tandem copies of pfpm2 and pfpm3 revealed significantly higher survival in multi copy clones following a 48 h or 72 h exposure to a range of piperaquine concentrations (figure; appendix p 5). Based on recent data from southeast Asia,11 we suspect that pfpm2 and pfpm3 would revert to a single copy in isolates from the Guiana Shield if piperaquine were discontinued. Of note, pfpm2 and pfpm3 copy status in our isogenic S170 clones only affected piperaquine and no other first-line antimalarial drugs tested (appendix pp 6–7).
Figure: Piperaquine resistance levels of isogenic French Guiana S170 clones expressing one versus two copies of pfpm2 and pfpm3 on the background of pfCRTC350R.
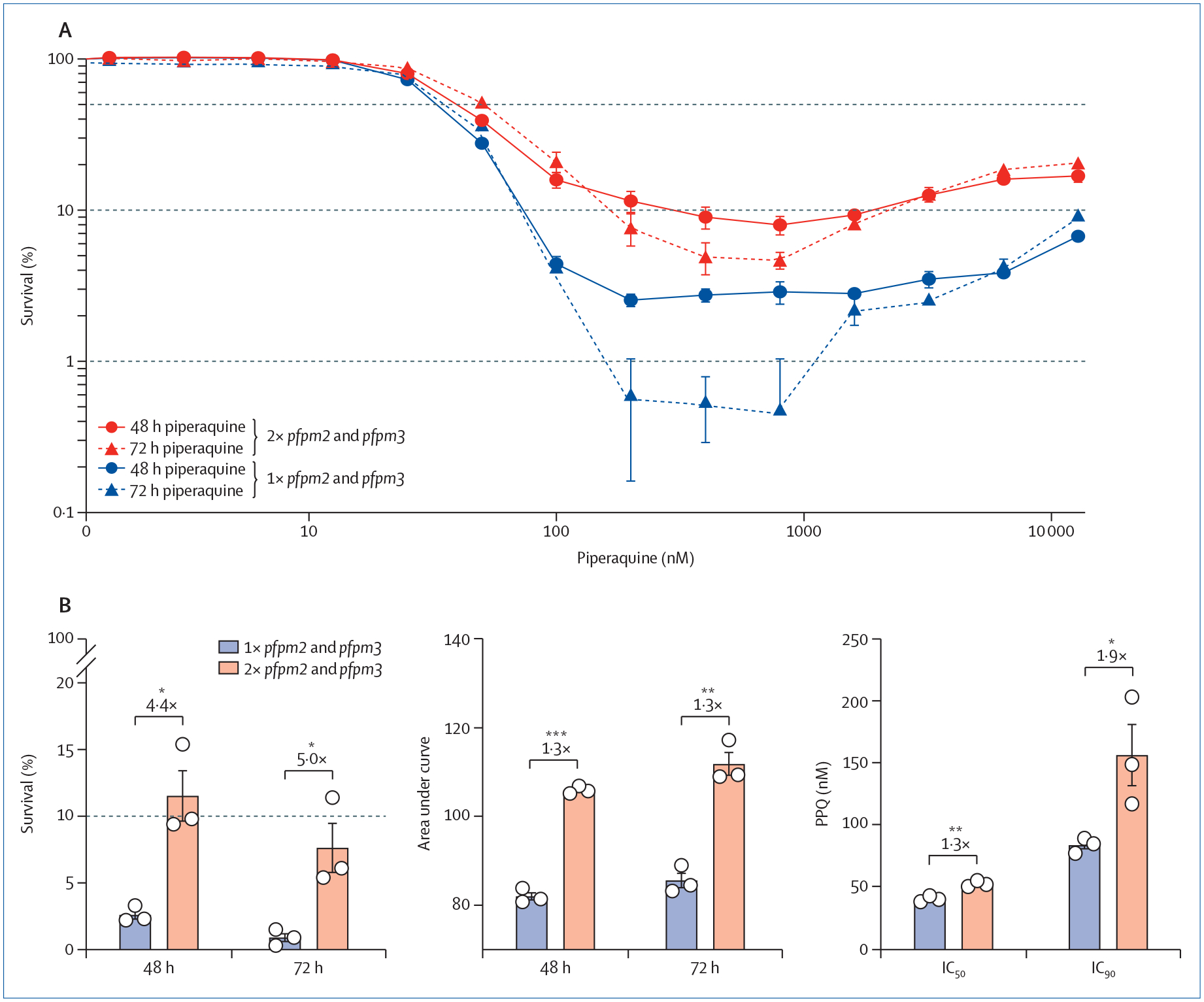
(A) Survival of parasite clones exposed to a range of piperaquine concentrations for 48 h or 72 h. The 10% cutoff represents the resistance threshold at 200 nM piperaquine. Control piperaquine-sensitive Dd2 or 3D7 parasites show <0·4% survival at 200 nM piperaquine.8,11 (B) Resistance indices following 200 nM piperaquine exposure for 48 h or 72 h, calculated as percent survival, area under the curve, and 72 h IC50 and IC90 values. Significance was tested using two-tailed unpaired Student’s t-test. *p<0·05, **p<0·01, and ***p<0·001. Fold-shifts are as indicated. Mean with SEM data for isogenic clones with one vs two copies of pfpm2 and pfpm3 (N=3 per group) were obtained in two to four independent experiments with technical replicates. 1× pfpm2 and pfpm3=one copy of pfpm2 and pfpm3. 2× pfpm2 and pfpm3=two copies of pfpm2 and pfpm3.
The prospects for the pfCRTC350R mutation being lost over time following removal of piperaquine are less clear. The impact of this mutation on asexual blood stage parasite fitness has not been determined, unlike piperaquine-resistant southeast Asian pfCRT variants (such as Phe145Ile) that show significant growth defects.10 Notably, earlier gene-editing studies showed that C350R sensitises parasites to amodiaquine, chloroquine, and lumefantrine,5 suggesting therapeutic strategies to eliminate piperaquine-resistant parasites in the Guiana Shield. One option could be to implement first-line combination therapies that exert opposing selective pressures on phenotypically diverse parasite populations. To inform these strategies, we propose that future studies should quantify the impact of mutant pfCRT and its interplay with multi copy pfpm2 and pfpm3 on asexual blood stage growth in piperaquine-resistant South American parasites. These data can help predict whether these resistance-conferring mutations persist upon the removal of piperaquine pressure.
Previous studies from this group identified a low prevalence of mutant K13 Cys580Tyr that mediated artemisinin partial resistance in isolates from Guyana, as confirmed using gene editing.12 This mutation was not observed by Florimond and colleagues4 among more than 350 samples. Nonetheless, several isolates, despite being K13 wild-type, showed survival levels of greater than 2% in the ring-stage survival assay (>1% is a standardised threshold for resistance), suggesting a possible non-K13 mediated reduction in artemisinin susceptibility. These authors also found a wide variation in piperaquine response in pfCRTC350R parasites with multi copy pfpm2 and pfpm3, suggesting that additional factors can modulate piperaquine resistance. Molecular surveillance of this region including targeted gene sequencing, genetic population structure analyses and genome-wide association studies, combined with therapeutic efficacy studies, are vital to detecting the emergence and spread of drug-resistant malaria. With the recent increase in malaria cases worldwide in some South American countries including Venezuela, and artemisinin partial resistance now threatening Africa, efforts must be increased to curb the impact of malaria on critically under-resourced nations and communities.
Supplementary Material
Acknowledgments
SM acknowledges financial support from the Human Frontier Science Program (HFSP) Long term fellowship (LT000976/2016-L). DAF acknowledges financial support from the National Institutes of Health (R01 AI109023, R37 AI050234, and R01 AI124678) and the Bill & Melinda Gates Foundation (INV-006599).
Footnotes
See Online/Articles https://doi.org/10.1016/S1473-3099(23)00502-9
See Online for appendix
References
- 1.WHO. WHO world malaria report 2022. Dec 8, 2022. https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022 (accessed Aug 25, 2023).
- 2.Conrad MD, Asua V, Garg S, et al. Evolution of partial resistance to artemisinins in malaria parasites in Uganda. N Engl J Med 2023; 389: 722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Pluijm RW, Imwong M, Chau NH, et al. Determinants of dihydroartemisinin–piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect Dis 2019; 19: 952–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Florimond C, de Laval F, Early AM, et al. Impact of piperaquine resistance in Plasmodium falciparum on malaria treatment effectiveness in French Guiana: a descriptive epidemiological study. Lancet Infect Dis; published online Oct 16. 10.1016/S1473-3099(23)00502-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelleau S, Moss EL, Dhingra SK, et al. Adaptive evolution of malaria parasites in French Guiana: reversal of chloroquine resistance by acquisition of a mutation in PfCRT. Proc Natl Acad Sci USA 2015; 112: 11672–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witkowski B, Duru V, Khim N, et al. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect Dis 2017; 17: 174–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amato R, Lim P, Miotto O, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect Dis 2017; 17: 164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross LS, Dhingra SK, Mok S, et al. Emerging southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat Commun 2018; 9: 3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mok S, Yeo T, Hong D, et al. Mapping the genomic landscape of multidrug resistance in Plasmodium falciparum and its impact on parasite fitness. Sci Adv; in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhingra SK, Small-Saunders JL, Ménard D, Fidock DA. Plasmodium falciparum resistance to piperaquine driven by PfCRT. Lancet Infect Dis 2019; 19: 1168–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imwong M, Suwannasin K, Srisutham S, et al. Evolution of multidrug resistance in Plasmodium falciparum: a longitudinal study of genetic resistance markers in the Greater Mekong subregion. Antimicrob Agents Chemother 2021; 65: e0112121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathieu LC, Cox H, Early AM, et al. Local emergence in Amazonia of Plasmodium falciparum k13 C580Y mutants associated with in vitro artemisinin resistance. eLife 2020; 9: e51015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


