Abstract
Bacteria contribute to many physiological functions of coral holobionts, including responses to bleaching. The bacterial genus, Endozoicomonas, dominates the microbial flora of many coral species and its abundance appears to be correlated with coral bleaching. However, evidences for decoupling of bleaching and Endozoicomonas abundance changes have also been reported. In 2020, a severe bleaching event was recorded at reefs in Taiwan, providing a unique opportunity to re-examine bleaching-Endozoicomonas association using multiple stony corals in natural environments. In this study, we monitored tissue color and microbiome changes in three coral species (Montipora sp., Porites sp., and Stylophora pistillata) in Kenting National Park, following the bleaching event. All tagged Montipora sp. and Porites sp. recovered from bleaching within 1 year, while high mortality occurred in S. pistillata. Microbiome analysis found no correlation of Endozoicomonas relative abundance and bleaching severity during the sampling period, but found a stronger correlation when the month in which bleaching occurred was excluded. Moreover, Endozoicomonas abundance increased during recovery months in Montipora sp. and Porites sp., whereas in S. pistillata it was nearly depleted. These results suggest that Endozoicomonas abundance may represent a gauge of coral health and reflect recovery of some corals from stress. Interestingly, even though different Endozoicomonas strains predominated in the three corals, these Endozoicomonas strains were also shared among coral taxa. Meanwhile, several Endozoicomonas strains showed secondary emergence during coral recovery, suggesting possible symbiont switching in Endozoicomonas. These findings indicate that it may be possible to introduce Endozoicomonas to non-native coral hosts as a coral probiotic.
Keywords: Endozoicomonas, coral health, symbiont switching, coral microbiome, thermal bleaching, coral recovery
Introduction
Ocean warming due to climate change has raised great concerns about its impact on coral reefs globally. Bleaching refers to disruption of symbiosis between corals and photosynthetic dinoflagellates of the family Symbiodiniaceae, a phenomenon commonly observed in thermally stressed corals [1]. Since its first documentation in the 1980s, coral bleaching has been reported with increasing frequency, with three pan-tropical coral bleaching events in 1998, 2010, and 2016 [2-6]. Regional bleaching episodes have also been recorded from tropical reefs in Australia [7-10] and the Caribbean [11, 12] to subtropical regions such as Okinawa, Japan [13-15], and Taiwan [5, 16-18]. As photosynthates from dinoflagellate symbionts constitute the major carbon source in stony corals [19, 20], severe bleaching can devastate coral physiology. Subsequent coral mortality can also change the structure of coral reef ecosystems, impacting all associated species.
In addition to dinoflagellates, corals are associated with a great diversity of bacteria—collectively termed the coral microbiome. Bacteria are thought to participate in many physiological functions of coral holobionts, including ontogeny [21, 22], metabolism [23, 24], immunity [25], and stress tolerance [23, 26]. In both laboratory and field studies, coral bleaching has been associated with decreases in Gammaproteobacteria [27-29]. Increases in Vibrio bacteria have also been documented in several stony corals during bleaching [1, 27, 30-32]. In fact, early studies demonstrated that Vibrio shiloi and Vibrio coralliilyticus induce bleaching in Oculina patagonia and Pocillopora damicornis, respectively [33, 34]. However, our knowledge of functional associations between the coral microbiome and bleaching, especially for nonpathogenic symbionts, is still limited.
Endozoicomonas (Gammaproteobacteria; Oceanospirillales; and Endozoicomonadaceae) constitutes a dominant bacterial taxon in several corals [35-38]. Based on genomic evidence, recent studies have proposed that Endozoicomonas regulates various biological functions in corals, such as metabolism, signaling, and nutrient cycling, suggesting that these bacteria are potentially beneficial for corals [39, 40]. Using denaturing gradient gel electrophoresis and cloning techniques, Bourne et al. [30] first identified a correlation between Endozoicomonas abundance (identified as Spongiobacter sp. in that study) and zooxanthella density in Acropora millepora. Occurrence of microbiome changes prior to visible signs of bleaching led to the hypothesis that Endozoicomonas is an early indicator of coral health/stress [30]. Thereafter, several studies reported that Endozoicomonas abundance decreases during natural [28, 29, 41] or experimental bleaching in other corals [26, 27, 42]. However, decoupling of coral bleaching and Endozoicomonas abundance changes has also been observed. For instance, Núñez-Pons et al. [38] monitored microbiome changes in three stony corals (Montipora capitata, Porites compressa, and Pocillopora acuta) in Hawaii following a natural bleaching event in 2016 and found no significant correlation between Endozoicomonas relative abundance and bleaching severity in those corals. In laboratory experiments, dynamics of Endozoicomonas abundance in Pocillopora verrucosa and Euphyllia glabrescens were also independent of the bleaching induced by excess dissolved organic carbon (DOC) and dark treatments, respectively [43, 44]. These contradictory findings suggest that the association between bleaching and decreased Endozoicomonas abundance is probably more complicated and may depend on specific combinations of Endozoicomonas bacteria and host corals.
Interestingly, in A. millepora, Bourne et al. [30] also found high microbiome similarity between unbleached and recovered (post-bleaching) corals, suggesting that Endozoicomonas recovery accompanies coral recovery. As Endozoicomonas is rare in seawater [45] and shows strong host-specificity in symbiosis with corals [35], it may be that the taxonomic composition of Endozoicomonas remains relatively stable during bleaching recovery. Supporting this hypothesis, Pootakham et al. [28] showed clear evidence that, in Porites lutea, dominant Endozoicomonas bacteria returned to pre-bleaching densities during coral recovery without changing their relative dominance. However, as similar studies on other corals are still limited, whether the same phenomenon applies to other stony corals remains unknown. In 2020, an extremely warm summer caused intense coral bleaching in coral reefs around Taiwan, with 57%–84% of shallow (3 m depth) corals in Kenting, southern Taiwan, showing partial or complete bleaching [46]. Given that Kenting National Park is a marine protected area, this bleaching event provided an opportunity to study bleaching-Endozoicomonas correlation in multiple coral species with minimal anthropogenic disturbance. By monitoring tagged corals for 1 year following the bleaching event, we examined two hypotheses: (i) Endozoicomonas abundance in corals negatively correlates with bleaching severity; (ii) the taxonomic composition of Endozoicomonas bacteria does not change after natural bleaching.
Materials and methods
Coral and seawater sampling
In 2020 and 2021, we collected corals six times from reefs located in Kenting National Park in southern Taiwan. Sampling covered the bleaching event in August 2020 (herein, the bleaching month) and five times during the following year: September 2020, October 2020, November 2020, April 2021, and August 2021 (recovery months) to examine coral recovery. Fifteen bleached colonies of Montipora sp. (N = 5), Porites sp. (N = 5), and Stylophora pistillata (N = 5) were tagged in August 2020 and were monitored for tissue color changes in subsequent fieldwork. Colonies of the same species were selected at distances >5 m from one another to maximize genetic randomness. Photos of tagged colonies were taken to estimate bleaching extents based on categories defined in Fisch et al. [47]. At each sampling time, three fragments (2–3 cm2) from each tagged, living colony and 1 liter of seawater were collected, comprising a total of 243 samples (237 coral tissues + 6 seawater samples). Immediately after sampling, coral tissues were washed once with 0.22-μm-filtered natural seawater and were preserved in 99% ethanol. Coral tissue and seawater samples were transported at 4°C and were then stored at −20°C until DNA extraction.
Deoxyribonucleic acid extraction and 16S amplicon sequencing
Genomic DNA was extracted from coral tissues and seawater samples (filtered through 0.22-μm membranes) using a modified cetyltrimethylammonium bromide method [48]. To construct 16S amplicon libraries, we first performed polymerase chain reaction (PCR) to amplify the V6-V8 hypervariable region of 16S rRNA gene using the 968F (5'-AACGCGAAGAACCTTAC-3′) and 1391R (5'-ACGGGCGGTGWGTRC-3′) primers with following PCR conditions: initial step at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 52°C for 20 s, and 72°C for 45 s, and a final step at 72°C for 10 min. PCR products were tagged using DNA-tagging PCR, following the protocol in Chen et al. [49]. The 16S amplicon libraries that were constructed were submitted to Yourgene Health Co., Ltd (New Taipei City, Taiwan) for sequencing using a Miseq reagent kit v3 (300-bp paired-end sequencing; 600 cycles) on an Illumina MiSeq system.
Amplicon sequence analysis
Quantitative Insights Into Microbial Ecology 2 (QIIME2) was used to analyze the 16S rRNA amplicon sequences [50]. Briefly, raw reads from Illumina MiSeq sequencing were first reoriented, primers were trimmed, and sequences were demultiplexed using the cutadapt plugin [51]. Demultiplexed reads were then truncated to 235 bp from both ends and were denoised using the DADA2 plugin [52]. To refine the sparseness of the amplicon sequence variant (ASV) abundance table, ASVs acquired from QIIME2 were reclustered to k-mer taxonomic units (KTUs) using the “ktusp” function of the KTU algorithm in the R environment (v4.2.1). This procedure has been proposed to improve biological explanations of microbiome data [53]. Taxonomy of each KTU was assigned based on the SILVA 138 SSU reference database using the “kaxonomy” function (annotation parameter: consensus = 0.5) in the KTU algorithm. KTUs affiliated with chloroplasts or mitochondria were removed, as were those affiliated with unclassified kingdoms or phyla. Libraries with <1000 remaining sequences (seven libraries) were removed from subsequent analyses.
Statistical and biodiversity analyses of coral microbial communities
To examine microbiome structure, we first rarefied coral tissue and seawater libraries to 1000 sequences/library. Coral tissue libraries of the same colony at the same sampling time were then pooled and re-rarefied to 1000 sequences to remove pseudo-replicates, yielding a total of 85 merged libraries for subsequent analyses (30 for Montipora sp., 30 for Porites sp., 19 for S. pistillata, and 6 for seawater). However, as library merging tends to inflate estimates of species richness (Supplementary Table S1), pseudo-replicate removal in alpha diversity was conducted by calculating alpha diversity on a per-sample basis and by averaging over coral colonies for each sampling time. Alpha diversity was estimated with the Chao1 and Shannon indexes. Statistical analyses for group comparisons of alpha diversity were conducted using the Kruskal–Wallis test with Dunn’s post hoc test. Beta diversity of microbial communities was assessed using Bray–Curtis dissimilarity and was visualized with principal coordinate analysis (PCoA). Heterogeneity among communities was analyzed using Analysis Of Similarities (ANOSIM; permutations = 1000) in Mothur [54]. To examine Endozoicomonas abundance changes and composition shifts, low-abundance Endozoicomonas KTUs (<20 sequences across merged libraries) were removed to minimize stochastic errors. This resulted in 19 Endozoicomonas KTUs. Abundance changes of Endozoicomonas during the sampling period were analyzed using the Kruskal–Wallis test and Dunn’s post hoc test for coral species. Correlations between Endozoicomonas relative abundance and bleaching extent for each coral colony was examined using Pearson’s correlation test. A significance level of α = 0.05 was set for all analyses in this study. Given that sample numbers in this study were not high (N = 3 or 5 for each group), no p-value adjustment was conducted for multiple comparisons.
Species identification and phylogenetic analysis of Endozoicomonas k-mer taxonomic units
Species-level taxonomy of 19 non-rare Endozoicomonas KTUs were identified by Blast search against the National Center for Biotechnology Information (NCBI) r/RNA/ITS database. A phylogenetic tree was reconstructed using the Maximum Likelihood method and the Tamura-Nei model based on representative sequences of these Endozoicomonas KTUs with 500 bootstrap replicates in MEGA11 [55]. 16S rRNA genes from nine Endozoicomonas bacteria (trimmed to the V6–V8 region) were included in the phylogenetic analysis as references, with an Aliivibrio fischeri 16S rRNA gene sequence (NR_029255.1) as the outgroup taxon.
Results
Coral physiology
In August 2020, intense coral bleaching was observed at Kenting National Park in southern Taiwan. Bleached colonies of Montipora sp., Porites sp., and S. pistillata were tagged and subsequently monitored for 1 year. Both Montipora sp. and Porites sp. showed signs of color recovery commencing in October 2020, and all tagged Montipora sp. and Porites sp. colonies visually recovered from bleaching in 2021 (mean bleaching extent: 1.65; Fig. 1). In contrast, S. pistillata showed no signs of color recovery following thermal bleaching. Mortality of S. pistillata was detected in November 2020, and all tagged S. pistillata colonies were dead and covered with algae in August 2021 (Fig. 1).
Figure 1.
Morphological changes of corals following the 2020 bleaching event. (A) Representative photos of three coral species during the sampling period. Numbers of collected samples at each sampling time are indicated under photos, with numbers in parentheses indicating numbers of colonies * numbers of fragments. (B) Bleaching extents of three coral species during the sampling period. Data are presented as means ± standard deviations, and numbers of libraries at each sampling time are indicated. Coral species are labeled by genus.
Sequencing overview and bacterial composition
Due to deaths of S. pistillata, only three colonies of S. pistillata were sampled in November 2020 for microbiome analysis and one colony in April 2021, while no S. pistillata colonies were sampled in August 2021. After data processing, seven libraries were removed from subsequent analyses due to low sequencing depth (<1000 bacterial sequences/library). This yielded a total of 2 280 673 bacterial sequences from 236 coral tissue and seawater libraries (1052–40 839 sequences/library), which were clustered into 10 044 KTUs affiliated with 115 bacterial classes. After data rarefication (1000 sequences/library) and pseudo-replicate removal, 5873 KTUs remained across 85 merged libraries. In both seawater and merged coral tissue libraries, Gammaproteobacteria, Alphaproteobacteria, and Cyanobacteriia were the dominant bacterial classes (Fig. 2). The merged dataset is available in Supplementary Table S2.
Figure 2.
Bacterial composition at the class level. Data are presented for each coral colony/seawater at each sampling time, with numbers of collected samples indicated in parentheses. Bacterial classes with <1% relative abundances across all merged libraries are presented as “others”. M: Montipora sp.; P: Porites sp.; S: S. pistillata; SW: seawater.
Alpha diversity
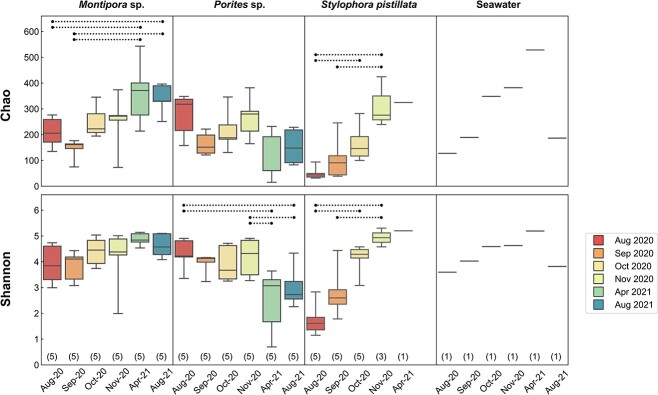
During the sampling period, different patterns of alpha diversity changes were found among sample types (Fig. 3). In Montipora sp., both bacterial species richness (Chao1 index) and evenness (Shannon index) showed weak trends of increase during the sampling period, with significant differences identified in the Chao1 index for samples collected in August and September 2020 compared to those collected in 2021 (Supplementary Table S3). In contrast, Porites sp. showed decreasing trends in Chao1 and Shannon indexes during the sampling period. Significant differences were found in the Shannon index for samples collected in August and November 2020 compared to those in 2021. In S. pistillata, both species richness and evenness showed pronounced increases during the sampling period, which became significant in October 2020, compared to the bleaching month.
Figure 3.
Alpha diversity of bacterial communities in coral tissue and seawater libraries. Chao1 and Shannon indexes were used to estimate alpha diversity. Boxes and whiskers indicate quartiles and full data ranges, respectively. Numbers of libraries at each sampling time are indicated in parentheses. Dashed lines indicate statistically significant differences (Dunn’s post hoc test; p < 0 .05).
Beta diversity
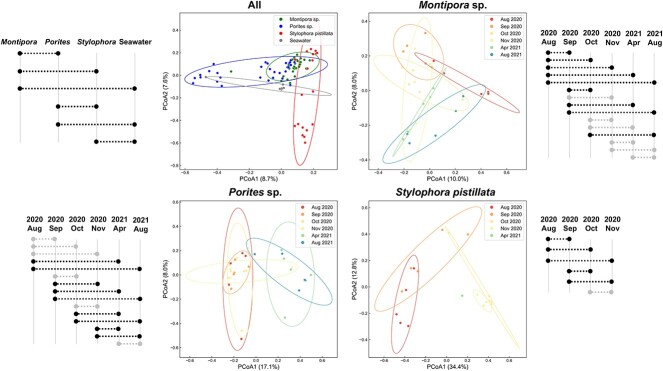
Analysis of beta diversity showed significant differences among coral species and seawater (Fig. 4; ANOSIM; 1000 permutations; p < 0.05; Supplementary Table S4). In all three coral species, significant changes were identified during the study. In Montipora sp. and S. pistillata, multiple comparisons yielded significant differences in combinations both within and across years (for Montipora sp.), whereas in Porites sp., significant differences were found mostly in cross-year comparisons.
Figure 4.
PCoA plots of bacterial composition at the KTU level. Pairwise comparisons are indicated with dashed lines on the side of each PCoA plot with statistically significant differences highlighted (ANOSIM; p < 0 .05).
Endozoicomonas composition
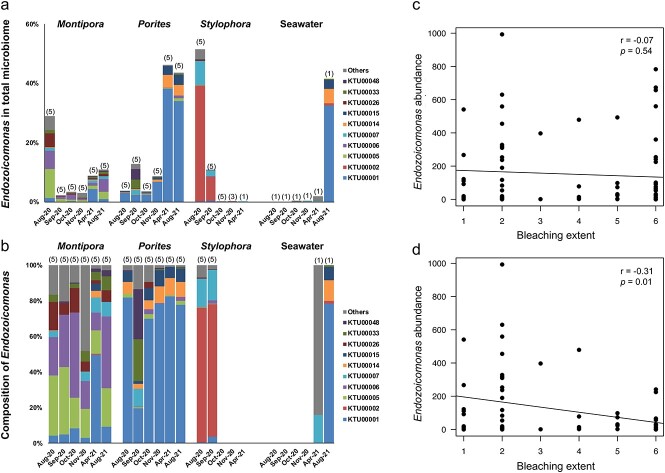
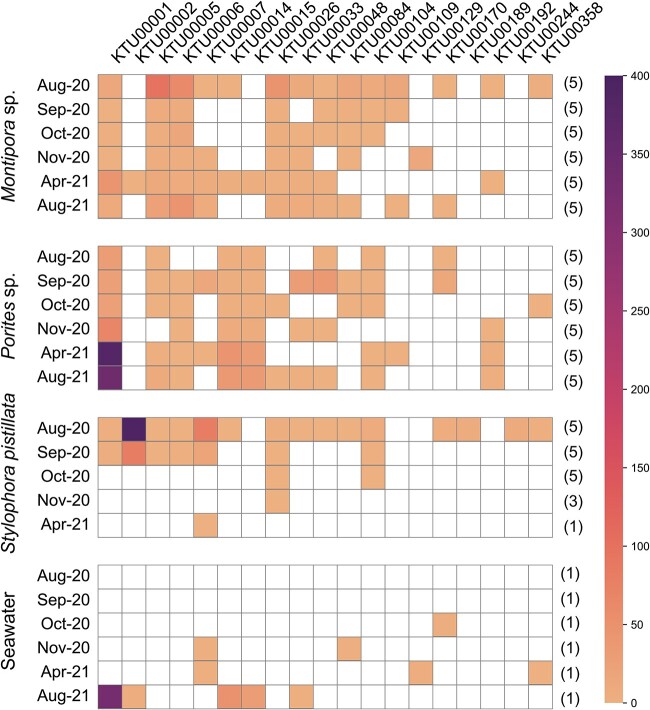
Nineteen KTUs were retained after removal of low-abundance Endozoicomonas KTUs. Total Endozoicomonas abundance varied significantly among sampling times for Porites sp. and S. pistillata (Kruskal–Wallis test; p < 0 .05; Supplementary Table S5). For Montipora sp. and S. pistillata, decreasing Endozoicomonas relative abundance occurred during the bleaching month and the first few recovery months. For Porites sp., the bleaching month showed the lowest Endozoicomonas relative abundance during this study. During recovery months, both Montipora sp. and Porites sp. showed increases in Endozoicomonas relative abundance, whereas Endozoicomonas became almost undetectable in S. pistillata after October 2020. Taxonomically, dominant Endozoicomonas KTUs varied between coral species (Fig. 5). During the sampling period, taxonomic composition of dominant Endozoicomonas remained relatively stable in the three coral species, with structural fluctuations occurring only sporadically (Fig. 5B). When comparing Endozoicomonas relative abundances and bleaching extents in the three coral species, no significant correlation was found during the entire sampling period (Fig. 5C). However, a significant, negative correlation was found when the bleaching month was excluded from the analysis (Fig. 5D). Disregarding abundances, several KTUs were present in corals in recovery months but not in the bleaching month (Montipora sp.: three KTUs; Porites sp.: eight KTUs; S. pistillata: zero KTUs; Fig. 6). A phylogenetic analysis and Blast search against the NCBI database showed that most Endozoicomonas KTUs in our data could not be clearly affiliated with known Endozoicomonas bacteria, with 11 Endozoicomonas KTUs showing <95% sequence identity to their corresponding best match sequences in the NCBI database (Fig. 7).
Figure 5.
Endozoicomonas bacterial composition and correlation with bleaching extent. (A) Percentages of Endozoicomonas bacteria versus total microbiome. (B) Percentages of Endozoicomonas bacteria versus total Endozoicomonas communities; Data are presented as averages for each sampling time, with numbers of libraries indicated in parentheses. KTUs with <20 sequences across merged libraries are not included and KTUs with <1% of the total Endozoicomonas community across merged libraries are presented as “others”. (C) Correlation of Endozoicomonas relative abundance and bleaching extent during the entire sampling period. (D) Correlation of Endozoicomonas relative abundance and bleaching extent with the bleaching month removed. Statistical significance of data correlation was examined using Pearson’s correlation test.
Figure 6.
Heatmap of Endozoicomonas KTUs in coral tissue and seawater libraries. Data are presented as averaged relative abundances of Endozoicomonas KTUs for each sampling time, with numbers of libraries indicated in parentheses. Data of zero abundance are highlighted in white. KTUs with abundances <20 sequences across merged libraries were not included to avoid bias.
Figure 7.
Phylogenetic tree of Endozoicomonas KTUs in this study. Sequences of publicly available Endozoicomonas bacteria were included as references, with A. fischeri (NR_029255.1) serving as an outgroup taxon. Best matches against the NCBI database are provided following KTU numbers with sequence identities in parentheses. Bootstrap values are presented at the node of each branch.KTUs with 100%, >97%, and <95% sequences identities to their best matches in the NCBI database are differently highlighted.
Discussion
Tissue color recovery after bleaching
In this study, we monitored tagged colonies of Montipora sp., Porites sp., and S. pistillata at Kenting National Park in southern Taiwan following an intense bleaching event in 2020. Bleaching is a common coral response to thermal stress, but it is also considered a means to rapidly adapt to changing environments [56]. According to this concept, bleaching averts intracellular accumulation of ROS generated by Symbiodiniaceae at elevated temperatures, allowing a coral to survive thermal stressors [57, 58]. When stresses abate, corals can re-establish the coral–Symbiodiniaceae symbiosis by either repopulating remnant photosynthetic dinoflagellates within their tissues or by capturing planktonic symbionts from ambient seawater [56, 59]. Commensurate with this notion, encrusting Montipora sp. and massive Porites sp. in this study showed recovery of color several months after the bleaching event (Fig. 1). Branched S. pistillata, however, was covered with algae and died the next year. Lower survival rates in branched corals compared to massive or encrusting corals have been reported previously in Mombasa and Okinawa following the extremely warm summer in 1998 El Nino [14, 60]. Given that branched corals have lower metabolic rates than massive corals, Gates and Edmunds [61] suggested that branched corals may have reduced capacity to respond to environmental changes. Loya et al. [14] also hypothesized that lower mass-transfer efficiency associated with branched morphology compared to encrusting and massive morphologies may be responsible for higher post-bleaching mortality in branched corals. Furthermore, lower tissue masses of branched corals may mean that branched corals have smaller energy reservoirs available for post-stress recovery [60]. This morphology-dependent survivorship may explain the variation in recovery among corals in this study. However, given that we did not measure tissue thickness or metabolic rates in our corals and that only three coral species were examined in this study, a definitive conclusion awaits further investigation.
Bacterial community
Consistent with observations in other corals [26, 27, 62], Gammaproteobacteria, Alphaproteobacteria, and Cyanobacteriia dominated all coral samples collected in this study (Fig. 2). However, three coral species showed different microbiome dynamics following the bleaching event. For example, significant microbiome changes in Montipora sp. and S. pistillata were found in most pairwise comparisons throughout the sampling period, whereas microbiome changes in Porites sp. occurred primarily in comparisons across 2020 and 2021, but not within individual years (Fig. 4). Composition of microbiomes and their responses to environmental factors reportedly vary by coral species, sites, stress histories, and even between different compartments of a given coral colony [27, 28, 45, 63-65]. Our findings provide further evidence of the complexity of coral microbiomes. Unfortunately, in this study, microbiomes were not examined prior to the bleaching event. Therefore, whether microbiome changes in our corals, especially Montipora sp. and Porites sp., represent restoration of “pre-bleaching” bacterial communities cannot be definitively concluded. The lack of negative controls in this study and a relatively high number of PCR cycles (30 cycles) may also have resulted in some biases. Although we do not believe that these biases significantly affected the structure of dominant coral microbiomes, this possibility should be considered.
Endozoicomonas abundance changes
Healthy corals host more abundant Endozoicomonas than bleached corals [27, 28, 30, 41, 66, 67]. Consistent with these observations, increases in Endozoicomonas relative abundance paralleled tissue color recovery in our corals, and a significant negative correlation between bleaching extent and Endozoicomonas abundance was found during recovery months (September 2020 to August 2021; Fig. 5D). However, the correlation was insignificant when data from the bleaching month were included (Fig. 5C), suggesting a more complicated association between coral bleaching and Endozoicomonas dynamics. In A. millepora in the Great Barrier Reef, Bourne et al. [30] identified shifts in coral microbiomes prior to visible signs of bleaching, including a decrease in Endozoicomonas. Although the low Endozoicomonas abundance in our Porites sp. in the bleaching month supports this hypothesis, both Montipora sp. and S. pistillata showed the highest Endozoicomonas relative abundances in the bleaching month. These findings suggest that Endozoicomonas decreases can also happen (or at least continue) after a bleaching event, raising the possibility that changes in Endozoicomonas abundance may not be an “early” indicator of coral stress in all coral taxa.
In an early study on Hawaiian corals, it was proposed that Montipora verrucosa possesses a lower lipid metabolic rate compared to Porites compressa [68]. In addition, branched corals have lower metabolic rates than corals of massive or encrusting morphotype [14, 61]. Although in this study we did not analyze lipid reserves, we assumed that Porites sp. in our study experienced earlier starvation due to faster consumption of its lipid reserve after bleaching, whereas Montipora sp. and S. pistillata showed delayed responses due to slower metabolism. According to this concept, Endozoicomonas abundance in a coral, instead of serving as an “early” stress indicator, more likely represents a “gauge” of coral health. This hypothesis helps to explain the correlation between Endozoicomonas deprivation and coral mortality in our S. pistillata and the decoupling of coral bleaching and Endozoicomonas abundance changes observed in other field surveys [28, 38] and laboratory experiments [43, 44]. Nevertheless, given that only three coral species were examined in this study and neither coral microbiomes before the bleaching event nor lipid reserves were available for our corals, further investigation is needed to test the global applicability and robustness of this hypothesis.
Endozoicomonas composition shifts
Despite dynamics of Endozoicomonas abundance, taxonomic composition of dominant Endozoicomonas bacteria was largely stable in our corals throughout the sampling period, suggesting strong selection upon Endozoicomonas communities by coral hosts (Fig. 5B). However, most Endozoicomonas KTUs were common to multiple corals (Fig. 6). Although no negative controls were included in our microbiome sequencing, all our Endozoicomonas KTUs showed best matches to bacteria isolated/identified from seawater or marine invertebrates, indicating their presences in multiple corals were not likely due to contamination (Fig. 7). Furthermore, although most of our Endozoicomonas KTUs are likely novel Endozoicomonas species/strains (Fig. 7), two Endozoicomonas KTUs showed 100% sequencing identity to the E. acroporae strain Acr-14 and E. atrinae strain WP70, previously isolated from an Acropora coral and a comb pen shell, respectively [69, 70]. These findings suggest certain levels of flexibility in symbiosis between Endozoicomonas bacteria and corals and possibly also among dissimilar marine invertebrates. In addition, variations in levels of dominance among Endozoicomonas bacteria were detectable (Figs 5B and 6). For instance, KTU00005 was the dominant Endozoicomonas in Montipora sp. in the bleaching month, but it was surpassed by KTU00006 or KTU00001 during recovery. In Porites sp., the dominant KTU00001 showed a remarkable decrease in September 2020, when KTU00033 and KTU00048 became dominant. Species shuffling in the genus Endozoicomonas has been reported in P. verrucosa and E. glabrescens in response to DOC and dark treatments, respectively [43, 44] and in Acropora muricata during a reciprocal transplant experiment [45]. The present findings provide further evidence for Endozoicomonas shuffling in corals during recovery from a natural bleaching event. As Endozoicomonas bacteria show great genomic diversity [71], species shuffling in this bacterial genus may imply significant functional shifts in coral holobionts.
Interestingly, in our corals, we found a great variation in taxonomic composition of Endozoicomonas beside the dominant taxa (Fig. 6). Symbiont switching is defined as acquisition of novel symbionts from the environment, which was recently identified in algal symbionts of corals following natural disturbances [72, 73]. However, there is still no evidence of switching in coral-associated Endozoicomonas. Given that detection of rare bacteria can be strongly affected by sequencing depth, in our analysis, we filtered out low-abundance KTUs in an attempt to minimize this potential bias. Still, we found that several secondary Endozoicomonas appeared in recovery months in our corals, particularly in Porites sp., suggesting possible species switching in coral-associated Endozoicomonas community. Furthermore, most secondary Endozoicomonas KTUs were absent in seawater samples but were primary to other corals (Fig. 6), implying bacterial swapping among sympatric corals. Coral mucus harbors an abundant bacterial community [25, 74], in which dominance of Endozoicomonas is evident, as in A. muricata and Porites astreoides [27, 75]. Therefore, horizontal Endozoicomonas transfer between corals may be due to mucus secretion. Contamination with coral mucus may also explain the similarity of Endozoicomonas communities between the seawater sample in August 2021 and those in Porites sp. (Fig. 6). However, absence of certain Endozoicomonas bacteria in the bleaching month may also be due to the difficulty in detecting rare bacterial taxa. Accordingly, whether emergence of secondary Endozoicomonas in our corals is truly de novo requires further examination.
In this study, we analyzed bacterial community changes, particularly for Endozoicomonas, in three coral species following a bleaching event in 2020. These results challenge the early hypothesis that decreases of Endozoicomonas are linked to coral bleaching and suggest instead that Endozoicomonas abundance is likely correlated with host health. Furthermore, our findings provide evidence of species shuffling and possible switching within coral-associated Endozoicomonas following a disturbance. Flexibility in coral–Endozoicomonas symbiosis suggests the possibility of inoculating non-native coral hosts with Endozoicomonas. However, different dynamics of the same Endozoicomonas bacterium were observed between coral species, implying that effectiveness of Endozoicomonas as a coral probiotic may depend on coral species. Transient presence of secondary Endozoicomonas bacteria also suggests that repeat inoculation might be necessary for a long-term effect. Together, these findings show the potential of employing Endozoicomonas as a coral probiotic, which warrants further study.
Supplementary Material
Acknowledgements
This work was supported by research funding of Academia Sinica, Taiwan. We thank Dr Steven D. Aird for editing and commenting on the manuscript.
Contributor Information
Po-Shun Chuang, Biodiversity Research Center, Academia Sinica, Taipei 115, Taiwan.
Sheng-Ping Yu, Biodiversity Research Center, Academia Sinica, Taipei 115, Taiwan.
Po-Yu Liu, School of Medicine, College of Medicine, National Sun Yat-sen University, Kaohsiung 804, Taiwan.
Ming-Tsung Hsu, Biodiversity Research Center, Academia Sinica, Taipei 115, Taiwan.
Yu-Jing Chiou, Biodiversity Research Center, Academia Sinica, Taipei 115, Taiwan; Centre for Marine Science and Innovation, School of Biological Earth and Environmental Sciences, University of New South Wales, Sydney 2052, Australia.
Chih-Ying Lu, Biodiversity Research Center, Academia Sinica, Taipei 115, Taiwan; Molecular and Biological Agricultural Sciences Program, Taiwan International Graduate Program, National Chung Hsing University and Academia Sinica, Taipei 115, Taiwan; Graduate Institute of Biotechnology, National Chung Hsing University, Taichung 402, Taiwan.
Sen-Lin Tang, Biodiversity Research Center, Academia Sinica, Taipei 115, Taiwan.
Author contributions
Po-Shun Chuang (Conceptualization, Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing—original draft), Sheng-Ping Yu (Data curation, Formal analysis, Investigation, Software), Po-Yu Liu (Data curation, Formal analysis, Methodology, Software, Validation, Visualization, Writing—original draft), Ming-Tsung Hsu (Investigation), Yu-Jing Chiou (Investigation), Chih-Ying Lu (Investigation), and Sen-Lin Tang (Conceptualization, Funding acquisition, Methodology, Project. administration, Resources, Supervision, Writing—review & editing)
Conflicts of interest
On behalf of all authors, the corresponding author states that there are no conflicts of interest.
Funding
This work was supported by the research funding of Academia Sinica, Taiwan.
Data availability
Demultiplexed MiSeq data generated in this study are available on the NCBI Sequence Read Archive database under BioProject PRJNA1010003.
References
- 1. Rosenberg E, Kushmaro A, Kramarsky-Winter E et al. The role of microorganisms in coral bleaching. ISME J 2009;3:139–46. 10.1038/ismej.2008.104. [DOI] [PubMed] [Google Scholar]
- 2. Hoegh-Guldberg O. Climate change, coral bleaching and the future of the world's coral reefs. Mar Freshw Res 1999;50:839–66. 10.1071/MF99078. [DOI] [Google Scholar]
- 3. Hughes TP, Kerry JT, Álvarez-Noriega M et al. Global warming and recurrent mass bleaching of corals. Nature 2017;543:373–7. 10.1038/nature21707. [DOI] [PubMed] [Google Scholar]
- 4. Oliver JK, Berkelmans R, Eakin CM. Coral bleaching in space and time. In: van Oppen M, Lough, J. (eds) Coral Bleaching: Patterns, Processes, Causes and Consequences, 2018, 27–49. Ecological Studies, vol 233. Springer, Cham. 10.1007/978-3-319-75393-5_3. [DOI] [Google Scholar]
- 5. Ribas-Deulofeu L, Denis V, Château P-A et al. Impacts of heat stress and storm events on the benthic communities of Kenting National Park (Taiwan). PeerJ 2021;9:e11744. 10.7717/peerj.11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wilkinson C. The 1997-1998 mass bleaching event around the world. 1998. In: Wilkinson. C. (ed) Status of Coral Reefs of the World, 1998, 15–38. Australian Institute of Marine Science. Townsville, Australia.
- 7. Berkelmans R, De’ath G, Kininmonth S et al. A comparison of the 1998 and 2002 coral bleaching events on the great barrier reef: spatial correlation, patterns, and predictions. Coral Reefs 2004;23:74–83. 10.1007/s00338-003-0353-y. [DOI] [Google Scholar]
- 8. Hughes T, Kerry J, Simpson T. Large-Scale Bleaching of Corals on the Great Barrier Reef. Ecology 2018;99(2):501. 10.1002/ecy.2092. [DOI] [PubMed] [Google Scholar]
- 9. Moore JA, Bellchambers LM, Depczynski MR et al. Unprecedented mass bleaching and loss of coral across 12 of latitude in Western Australia in 2010–11. PLoS One 2012;7:e51807. 10.1371/journal.pone.0051807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pratchett MS, Heron SF, Mellin C et al. Recurrent Mass-Bleaching and the Potential for Ecosystem Collapse on Australia’s Great Barrier Reef. In: Canadell JG, Jackson RB. (eds) Ecosystem Collapse and Climate Change, 2021, 265–89. Ecological Studies, vol 241. Springer, Cham. 10.1007/978-3-030-71330-0_10. [DOI] [Google Scholar]
- 11. Alemu IJB, Clement Y. Mass coral bleaching in 2010 in the southern Caribbean. PLoS One 2014;9:e83829. 10.1371/journal.pone.0083829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eakin CM, Morgan JA, Heron SF et al. Caribbean corals in crisis: record thermal stress, bleaching, and mortality in 2005. PLoS One 2010;5:e13969. 10.1371/journal.pone.0013969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kayanne H, Suzuki R, Liu G. Bleaching in the Ryukyu Islands in 2016 and associated degree heating week threshold. Galaxea J Coral Reef Stud 2017;19:17–8. 10.3755/galaxea.19.1_17. [DOI] [Google Scholar]
- 14. Loya Y, Sakai K, Yamazato K et al. Coral bleaching: the winners and the losers. Ecol Lett 2001;4:122–31. 10.1046/j.1461-0248.2001.00203.x. [DOI] [Google Scholar]
- 15. Yamazato K. Coral bleaching in Okinawa, 1980 vs 1998. J Jpn Coral Reef Soc 1999;1999:83–7. 10.3755/jcrs.1999.83. [DOI] [Google Scholar]
- 16. Chen CA, Dai C-F. Local phase shift from Acropora-dominant to Condylactis-dominant community in the Tiao-Shi reef, Kenting National Park, southern Taiwan. Coral Reefs 2004;23:508–8. 10.1007/s00338-004-0423-9. [DOI] [Google Scholar]
- 17. Hung T-C, Huang C-C, Shao K-T. Ecological survey of coastal i water adjacent to nuclear power I plants in Taiwan. Chem Ecol 1998;15:129–42. 10.1080/02757549808037625. [DOI] [Google Scholar]
- 18. Keshavmurthy S, Chen T-R, Liu P-J et al. Learning from the past is not enough to survive present and future bleaching threshold temperatures. Sci Total Environ 2022;852:158379. 10.1016/j.scitotenv.2022.158379. [DOI] [PubMed] [Google Scholar]
- 19. Muscatine L, R. McCloskey L, E. Marian R. Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration 1. Limnol Oceanogr 1981;26:601–11. 10.4319/lo.1981.26.4.0601. [DOI] [Google Scholar]
- 20. Palardy JE, Rodrigues LJ, Grottoli AG. The importance of zooplankton to the daily metabolic carbon requirements of healthy and bleached corals at two depths. J Exp Mar Biol Ecol 2008;367:180–8. 10.1016/j.jembe.2008.09.015. [DOI] [Google Scholar]
- 21. Bernasconi R, Stat M, Koenders A et al. Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front Microbiol 2019;10:1529. 10.3389/fmicb.2019.01529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Oppen MJ, Blackall LL. Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol 2019;17:557–67. 10.1038/s41579-019-0223-4. [DOI] [PubMed] [Google Scholar]
- 23. Santoro EP, Borges RM, Espinoza JL et al. Coral microbiome manipulation elicits metabolic and genetic restructuring to mitigate heat stress and evade mortality. Sci Adv 2021;7:eabg3088. 10.3389/fevo.2023.1079271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thurber RV, Willner-Hall D, Rodriguez-Mueller B et al. Metagenomic analysis of stressed coral holobionts. Environ Microbiol 2009;11:2148–63. 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 25. Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 2006;322:1–14. 10.3354/meps322001. [DOI] [Google Scholar]
- 26. Pootakham W, Mhuantong W, Yoocha T et al. Heat-induced shift in coral microbiome reveals several members of the Rhodobacteraceae family as indicator species for thermal stress in Porites lutea. MicrobiologyOpen 2019;8:e935. 10.1002/mbo3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee ST, Davy SK, Tang S-L et al. Successive shifts in the microbial community of the surface mucus layer and tissues of the coral Acropora muricata under thermal stress. FEMS Microbiol Ecol 2015;91:fiv142. 10.1093/femsec/fiv142. [DOI] [PubMed] [Google Scholar]
- 28. Pootakham W, Mhuantong W, Putchim L et al. Dynamics of coral-associated microbiomes during a thermal bleaching event. MicrobiologyOpen 2018;7:e00604. 10.1002/mbo3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zou Y, Chen Y, Wang L et al. Differential responses of bacterial communities in coral tissue and mucus to bleaching. Coral Reefs 2022;41:951–60. 10.1007/s00338-022-02261-8. [DOI] [Google Scholar]
- 30. Bourne D, Iida Y, Uthicke S et al. Changes in coral-associated microbial communities during a bleaching event. ISME J 2008;2:350–63. 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- 31. Morrow K, Muller E, Lesser M. How does the coral microbiome cause, respond to, or modulate the bleaching process? In: van Oppen M, Lough J. (eds) Coral Bleaching: Patterns, Processes, Causes and Consequences, 2018, 153–88. Ecological Studies, vol 233. Springer, Cham. 10.1007/978-3-319-75393-5_7. [DOI] [Google Scholar]
- 32. Zhou J, Lin ZJ, Cai ZH et al. Opportunistic bacteria use quorum sensing to disturb coral symbiotic communities and mediate the occurrence of coral bleaching. Environ Microbiol 2020;22:1944–62. 10.1111/1462-2920.15009. [DOI] [PubMed] [Google Scholar]
- 33. Ben-Haim Y, Rosenberg E. A novel vibrio sp. pathogen of the coral Pocillopora damicornis. Mar Biol 2002;141:47–55. 10.1007/s00227-002-0797-6. [DOI] [Google Scholar]
- 34. Kushmaro A, Rosenberg E, Fine M et al. Bleaching of the coral Oculina patagonica by vibrio AK-1. Mar Ecol Prog Ser 1997;147:159–65. 10.3354/meps147159. [DOI] [Google Scholar]
- 35. Bayer T, Neave MJ, Alsheikh-Hussain A et al. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol 2013b;79:4759–62. 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cai L, Tian R-M, Zhou G et al. Exploring coral microbiome assemblages in the South China Sea. Sci Rep 2018;8:2428. 10.1038/s41598-018-20515-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neave MJ, Rachmawati R, Xun L et al. Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J 2017;11:186–200. 10.1038/ismej.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Núñez-Pons L, Cunning R, Nelson CE et al. Hawaiian coral holobionts reveal algal and prokaryotic host specificity, intraspecific variability in bleaching resistance, and common interspecific microbial consortia modulating thermal stress responses. Sci Total Environ 2023;889:164040. 10.1016/j.scitotenv.2023.164040. [DOI] [PubMed] [Google Scholar]
- 39. Ding J-Y, Shiu J-H, Chen W-M et al. Genomic insight into the host–endosymbiont relationship of Endozoicomonas montiporae CL-33T with its coral host. Front Microbiol 2016;7:251. 10.3389/fmicb.2016.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pogoreutz C, Oakley CA, Rädecker N et al. Coral holobiont cues prime Endozoicomonas for a symbiotic lifestyle. ISME J 2022;16:1883–95. 10.1038/s41396-022-01226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun F, Yang H, Shi Q et al. Changes in coral bacterial communities during a natural bleaching event linked to El Niño in the South China Sea. Reg Stud Mar Sci 2022;53:102383. 10.1016/j.rsma.2022.102383. [DOI] [Google Scholar]
- 42. Tout J, Siboni N, Messer LF et al. Increased seawater temperature increases the abundance and alters the structure of natural vibrio populations associated with the coral Pocillopora damicornis. Front Microbiol 2015;6:432. 10.3389/fmicb.2015.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pogoreutz C, Rädecker N, Cárdenas A et al. Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol 2018;8:2240–52. 10.1002/ece3.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shiu J-H, Yu S-P, Fong C-L et al. Shifting in the dominant bacterial group endozoicomonas is independent of the dissociation with coral symbiont algae. Front Microbiol 2020;11:1791. 10.3389/fmicb.2020.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tandon K, Chiou Y-J, Yu S-P et al. Microbiome restructuring: dominant coral bacterium Endozoicomonas species respond differentially to environmental changes. Msystems 2022;7:e00359–22. 10.1128/msystems.00359-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye Z-M, Mayfield AB, Fan T-Y. Variable responses to a marine heat wave in five fringing reefs of southern Taiwan. Appl Sci 2023;13:5554. 10.3390/app13095554. [DOI] [Google Scholar]
- 47. Fisch J, Drury C, Towle EK et al. Physiological and reproductive repercussions of consecutive summer bleaching events of the threatened Caribbean coral Orbicella faveolata. Coral Reefs 2019;38:863–76. 10.1007/s00338-019-01817-5. [DOI] [Google Scholar]
- 48. Wilson K. Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol 2001;56:2.4.1–5. 10.1002/0471142727.mb0204s56. [DOI] [PubMed] [Google Scholar]
- 49. Chen C-P, Tseng C-H, Chen CA et al. The dynamics of microbial partnerships in the coral Isopora palifera. ISME J 2011;5:728–40. 10.1038/ismej.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bolyen E, Rideout JR, Dillon MR et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852–7. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–2. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 52. Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu PY, Yang SH, Yang SY. KTU: K-mer taxonomic units improve the biological relevance of amplicon sequence variant microbiota data. Methods Ecol Evol 2022;13:560–8. 10.1111/2041-210X.13758. [DOI] [Google Scholar]
- 54. Kozich JJ, Westcott SL, Baxter NT et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79:5112–20. 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38:3022–7. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kinzie RA III, Takayama M, Santos SR et al. The adaptive bleaching hypothesis: experimental tests of critical assumptions. Biol Bull 2001;200:51–8. 10.2307/1543084. [DOI] [PubMed] [Google Scholar]
- 57. Downs C, Fauth JE, Halas JC et al. Oxidative stress and seasonal coral bleaching. Free Radic Biol Med 2002;33:533–43. 10.1016/S0891-5849(02)00907-3. [DOI] [PubMed] [Google Scholar]
- 58. Lesser MP. Coral bleaching: causes and mechanisms. In: Dubinsky Z, Stambler N. (eds) Coral Reefs: An Ecosystem in Transition, 2011, 405–19. Springer, Dordrecht. 10.1007/978-94-007-0114-4_23. [DOI]
- 59. Nakamura T, Yamasaki H, Van Woesik R. Water flow facilitates recovery from bleaching in the coral Stylophora pistillata. Mar Ecol Prog Ser 2003;256:287–91. 10.3354/meps256287. [DOI] [Google Scholar]
- 60. McClanahan T. The relationship between bleaching and mortality of common corals. Mar Biol 2004;144:1239–45. 10.1007/s00227-003-1271-9. [DOI] [Google Scholar]
- 61. Gates RD, Edmunds PJ. The physiological mechanisms of acclimatization in tropical reef corals. Am Zool 1999;39:30–43. 10.1093/icb/39.1.30. [DOI] [Google Scholar]
- 62. Bayer T, Arif C, Ferrier-Pagès C et al. Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar Ecol Prog Ser 2013a;479:75–84. 10.3354/meps10197. [DOI] [Google Scholar]
- 63. Gardner SG, Camp EF, Smith DJ et al. Coral microbiome diversity reflects mass coral bleaching susceptibility during the 2016 El Niño heat wave. Ecol Evol 2019;9:938–56. 10.1002/ece3.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hernandez-Agreda A, Leggat W, Bongaerts P et al. Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. MBio 2018;9:e00812–8. 10.1128/mbio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ziegler M, Seneca FO, Yum LK et al. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 2017;8:14213. 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meyer JL, Paul VJ, Teplitski M. Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS One 2014;9:e100316. 10.1371/journal.pone.0100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. O’Brien PA, Smith HA, Fallon S et al. Elevated CO2 has little influence on the bacterial communities associated with the pH-tolerant coral, massive Porites spp. Front Microbiol 2018;9:2621. 10.3389/fmicb.2018.02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Grottoli A, Rodrigues L, Juarez C. Lipids and stable carbon isotopes in two species of Hawaiian corals, Porites compressa and Montipora verrucosa, following a bleaching event. Mar Biol 2004;145:621–31. 10.1007/s00227-004-1337-3. [DOI] [Google Scholar]
- 69. Hyun D-W, Shin N-R, Kim M-S et al. Endozoicomonas atrinae sp. nov., isolated from the intestine of a comb pen shell Atrina pectinata. Int J Syst Evol Microbiol 2014;64:2312–8. 10.1099/ijs.0.060780-0. [DOI] [PubMed] [Google Scholar]
- 70. Sheu S-Y, Lin K-R, Hsu M-y et al. Endozoicomonas acroporae sp. nov., isolated from Acropora coral. Int J Syst Evol Microbiol 2017;67:3791–7. 10.1099/ijsem.0.002194. [DOI] [PubMed] [Google Scholar]
- 71. Tandon K, Lu C-Y, Chiang P-W et al. Comparative genomics: dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP). ISME J 2020;14:1290–303. 10.1038/s41396-020-0610-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Boulotte NM, Dalton SJ, Carroll AG et al. Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J 2016;10:2693–701. 10.1038/ismej.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Quigley KM, Willis BL, Kenkel CD. Transgenerational inheritance of shuffled symbiont communities in the coral Montipora digitata. Sci Rep 2019;9:13328. 10.1038/s41598-019-50045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Garren M, Azam F. New method for counting bacteria associated with coral mucus. Appl Environ Microbiol 2010;76:6128–33. 10.1128/AEM.01100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Apprill A, Weber LG, Santoro AE. Distinguishing between microbial habitats unravels ecological complexity in coral microbiomes. MSystems 2016;1:e00143–16. 10.1128/mSystems.00143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Demultiplexed MiSeq data generated in this study are available on the NCBI Sequence Read Archive database under BioProject PRJNA1010003.