SUMMARY
The evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to challenge the efficacy of vaccination efforts against coronavirus disease 2019 (COVID-19). The Omicron XBB lineage of SARS-CoV-2 has presented dramatic evasion of neutralizing antibodies stimulated by mRNA vaccination and COVID-19 convalescence. XBB.1.16, characterized by two mutations relative to the dominating variant XBB.1.5, i.e., E180V and K478R, has been on the rise globally. In this study, we compare the immune escape of XBB.1.16 with XBB.1.5, alongside ancestral variants D614G, BA.2, and BA.4/5. We demonstrate that XBB.1.16 is strongly immune evasive, with extent comparable to XBB.1.5 in bivalent-vaccinated healthcare worker sera, 3-dose-vaccinated healthcare worker sera, and BA.4/5-wave convalescent sera. Interestingly, the XBB.1.16 spike is less fusogenic than that of XBB.1.5, and this phenotype requires both E180V and K478R mutations to manifest. Overall, our findings emphasize the importance of the continued surveillance of variants and the need for updated mRNA vaccine formulations.
Graphical Abstract
In brief
SARS-CoV-2 continues to exhibit increasing immune escape. Here, Faraone et al. demonstrate that XBB.1.16 escapes antibodies in bivalent-vaccinated, 3-dose-vaccinated, and BA.4/5-wave convalescent sera to a similar extent compared with XBB.1.5. They also show that the XBB.1.16 spike is less fusogenic than XBB.1.5 but exhibits enhanced infectivity.
INTRODUCTION
The continued evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus behind the coronavirus disease 2019 (COVID-19) pandemic, presents an ever-present challenge to the efficacy of vaccination efforts.1-6 The nature of the COVID-19 pandemic changed dramatically with the emergence of the Omicron BA.1 variant due to the marked evolutionary distance between it and previous variants of concern.7 The vast array of mutations accrued in BA.1 led to markedly decreased pathogenicity,8-12 increased transmissibility,9 and dramatic immune escape3-5,13-17 by the virus. Since then, the Omicron lineage of SARS-CoV-2 has continued to evolve, resulting in a series of subvariants that have generated their own smaller waves of infection. Many of these subvariants have been characterized by increasing extents of immune escape.1,18-26 Up until September of 2022, vaccines against SARS-CoV-2 were largely designed against the prototype version of the virus that emerged early in the pandemic, but in response to the increasing extents of immune escape exhibited by the Omicron lineage of SARS-CoV-2, new versions of vaccines have been rolled out, of note the Pfizer and Moderna bivalent vaccines that include the BA.4/5 spike.27 Despite these efforts, the continued evolution of Omicron subvariants could endanger the efficacy of this vaccine formulation.1
Early 2023 was marked by the emergence of a recombinant Omicron subvariant XBB that resulted from the recombination of two BA.2-derived subvariants, BJ.1 and BM.1.1.17,28,.29 In February 2023, an XBB-lineage subvariant named XBB.1.5 emerged and quickly surged across the globe to become the dominant infecting variant.30 The XBB lineage is characterized by dramatic immune escape, demonstrating a strong selective advantage even among vaccinated individuals.20,24,31-34 In March of 2023, a new XBB-lineage subvariant, XBB.1.16, rose quickly in circulation in India, causing widespread breakthrough infection.35,36 It has since spread to several countries and is projected to continue growing in the proportion of infecting variants.35-37 The spike (S) protein of XBB.1.16 has two mutations distinct from XBB.1.5: E180V in the N-terminal region and T478R in the receptor-binding domain (RBD) (Figure 1A). XBB.1.16 has rapidly overtaken XBB.1.5 as the main circulating variant in India,35 making it critical that we further assess this emerging variant. In this study, we investigate the neutralization escape of XBB.1.16 and its defining mutations (XBB.1.5-E180V and XBB.1.5-K478R) from vaccinated sera (monovalent and bivalent), convalescent sera (BA.4/5 wave), and a monoclonal antibody (mAb) S309 in comparison to related and parental variants XBB.1.5, BA.4/5, BA.2, and D614G. We also examine the ability of S to trigger membrane fusion, as well as S protein expression and processing. Our results provide insights into the biology of XBB.1.16 and emphasize the importance of continued surveillance of SARS-CoV-2 variants for updating COVID-19 vaccines.
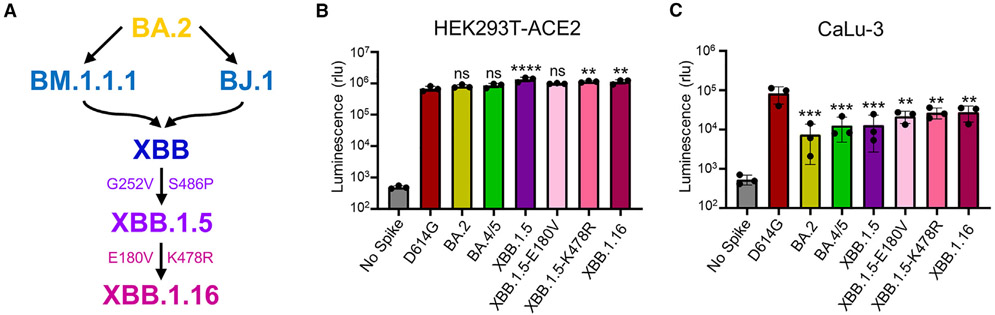
Figure 1. Infectivity of lentiviral pseudotype bearing the S protein of interest in HEK293T-ACE2 and CaLu-3 cells.
(A) Graphic depiction of the evolutionary relationship between XBB.1.16 and earlier lineage variants XBB.1.5, XBB, BM.1.1.1, BJ.1, and BA.2, with defining mutations displayed. Curved arrows connecting BM.1.1.1 and BJ.1 to XBB represent recombination.
(B and C) Infectivity of lentiviral pseudotypes bearing SARS-CoV-2 S protein of interest as determined in (B) HEK293T-ACE2 cells and (C) human lung epithelial cell line CaLu-3.
Bars in (B) and (C) represent means ± standard deviation for 3 replicates represented by individual dots (n = 3). p values are displayed as **p < 0.01, ***p < 0.001, and ****p < 0.0001.
RESULTS
Omicron subvariant XBB.1.16 maintains reduced infectivity in CaLu-3 cells relative to D614G
First, we examined the infectivity of lentiviral pseudotypes bearing each of the variant S proteins of interest in HEK293T cells expressing human ACE2 (HEK293T-ACE2) and human lung epithelial cell line CaLu-3, respectively. In 293T-ACE2 cells, the infectivity of XBB.1.16 remained comparable to XBB.1.5, with a 1.2-fold decrease relative to XBB.1.5 (p > 0.05), but was higher than D614G, with a 2.0-fold increase (p < 0.01) (Figure 1B). The K478R single mutation exhibited a 1.7-fold increase (p < 0.01) relative to D614G, while the E180V single mutation exhibited a 1.4-fold increase (p > 0.05) (Figure 1B), which agreed with results from a recent study.38 In CaLu-3 cells, XBB.1.16 exhibited a 2.1-fold higher infectivity compared with XBB.1.5 (p > 0.05) (Figure 1C). The two single mutations had similar infectivity compared with XBB.1.16, with 1.7- (p > 0.05) and 1.2-fold (p > 0.05) increases, respectively, relative to XBB.1.5 (Figure 1C). Compared with D614G, all Omicron subvariants exhibited significantly reduced infectivity (p < 0.01), with the XBB variants having overall higher titers than BA.2 in both HEK293T-ACE2 and CaLu-3 cells (p > 0.05) (Figures 1B and 1C).
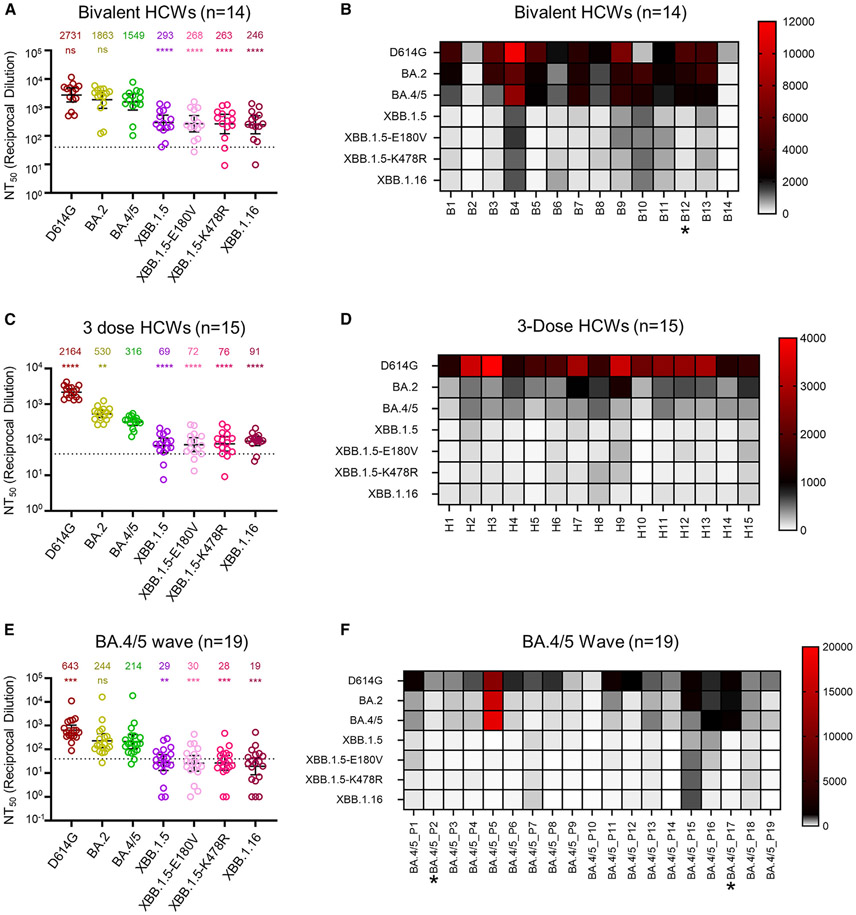
XBB.1.16 demonstrates comparable escape of neutralizing antibodies in bivalent-vaccinated sera compared with XBB.1.5
We next investigated the resistance of XBB.1.16 to neutralizing antibodies induced by mRNA vaccines or SARS-CoV-2 Omicron infection using our previously described pseudotyped lentivirus neutralization assay.39 The first cohort we studied consisted of The Ohio State University Wexner Medical Center healthcare workers (HCWs) that had received a bivalent mRNA booster vaccination containing both the SARS-CoV-2 S proteins of prototype and BA.4/5 (n = 14). XBB.1.16 exhibited a marked reduction in neutralizing antibody titers relative to BA.5, which was the major predominant strain of summer 2022, with a 6.3-fold decrease (p < 0.0001) (Figures 2A and 2B). This escape was similar to what we had observed for XBB.1.5,20 which showed a 5.3-fold drop in neutralizing antibody titer compared with BA.5 (p < 0.0001) (Figures 2A and 2B). Two single mutations, i.e., XBB.1.5-E180V and XBB.1.5-K478R, exhibited similar escape, with neutralizing antibody titers 5.8- (p < 0.0001) and 5.9-fold (p < 0.0001) lower than BA.5, respectively (Figures 2A and 2B). Overall, the escape of neutralizing antibodies in bivalent-vaccinated sera by XBB.1.16 is comparable to XBB.1.5.
Figure 2. Neutralization of XBB.1.16 by bivalent-vaccinated, 3-dose-vaccinated, and BA.4/5 convalescent sera.
Neutralizing antibody titers of three different cohorts of human sera against lentiviral pseudotypes bearing XBB.1.16 or other S proteins of interest were determined.
HCWs receiving 3 doses of monovalent mRNA vaccine plus 1 dose of bivalent mRNA vaccine (n = 14) (A), HCWs receiving 3 doses of monovalent mRNA vaccine (n = 15) (C and D), and first responders and household contacts that tested positive for COVID-19 during the BA.4/5 wave of infection (E and F) are all from Columbus, OH (USA). Bars represent geometric means with 95% confidence intervals. Geometric mean NT50 values are displayed for each subvariant on the top (A, C, and E). Heatmap depictions of neutralizing antibody titers were made for individuals in the (B) bivalent-vaccinated, (D) 3-dose-vaccinated, and (F) BA.4/5-wave-infected cohorts. Statistical significance in (A), (C), and (E) was determined using log10-transformed NT50 values to better approximate normality. Comparisons between multiple groups were made using a one-way ANOVA with Bonferroni post-test, with all compared to BA.4/5 in each panel. Dashed lines represent the assay’s limit of detection, i.e., 40 (the lowest dilution factor of serum samples). Asterisk in (B) indicates the individual infected by SARS-CoV-2 within 6 months before the sera sample collection, and asterisks in (F) indicate the individuals who had received three doses of mRNA vaccine before infection. p values are displayed as “ns” p > 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
XBB.1.16 maintains escape of neutralizing antibodies in 3-dose-vaccinated sera
We then investigated the sensitivity of these XBB.1.16 variants to neutralizing antibodies in a cohort of HCWs that received 3 doses of monovalent mRNA vaccine (n = 15). Again, XBB.1.16 exhibited marked escape, with neutralizing antibody titers 3.5-fold lower than BA.5 (p < 0.0001) (Figures 2C and 2D). This was similar to the escape exhibited by XBB.1.5, with neutralizing antibody titers 4.6-fold lower than BA.5 (p < 0.0001) (Figures 2C and 2D). The single mutations contributed comparably to the escape, with neutralizing antibody titers 4.4- (p < 0.0001) and 4.2-fold (p < 0.0001) lower than BA.5 for XBB.1.5-E180V and XBB.1.5-K478R, respectively (Figures 2C and 2D). Notably, the geometric mean antibody titers of 3-dose vaccinees were 3- to 5-fold lower than those in the bivalent vaccination cohort for Omicron variants (Figures 2A-2D). However, overall XBB.1.16 had comparable escape of neutralizing antibodies in 3-dose-vaccinated sera relative to XBB.1.5.
Marked resistance of Omicron subvariant XBB.1.16 to BA.4/5 convalescent sera
The last cohort we investigated were first responders and their household contacts that tested positive for COVID-19 during the BA.5 wave of infection in Columbus, OH (USA; n = 19). XBB.1.16 exhibited strong resistance, with neutralizing antibody titers 11.3-fold lower than BA.5 (p < 0.001) (Figures 2E and 2F); this escape appeared higher than XBB.1.5, which showed neutralizing antibody titers 7.4-fold lower than BA.5 (p < 0.01) (Figures 2E and 2F). Two single mutations, XBB.1.5-E180V and XBB.1.5-K478R, contributed similarly to this escape, with neutralizing antibody titers 7.1- (p < 0.001) and 7.6-fold lower (p < 0.001), respectively, than BA.5. Overall, the neutralizing antibody titers for the XBB-derived variants largely fell below the limit of detection (NT50 = 40) for the BA.5-wave cohort, in part because of their generally low titers compared with that of 3-dose and bivalent booster vaccinees.
Maintained efficacy of mAb S309 against Omicron subvariant XBB.1.16
Omicron subvariants are known to exhibit marked escape of mAbs.5 We chose to focus on mAb S309, which we have previously shown to still be effective against most of the Omicron variants, including XBB.1.5, except for CH.1.1, CA.3.1, and BA.2.75.2.20 We found that S309 effectively neutralized XBB.1.16, with an inhibitory concentration at 50% (IC50) of 3.84 ± 1.04 μg/mL, which was comparable to the IC50 of XBB.1.5, i.e., 2.91 ± 0.82 μg/mL (Figures 3A and 3B). The single mutations did not exhibit distinct phenotypes, with IC50 values of 3.06 ± 0.72 and 4.49 ± 1.38 μg/mL for XBB.1.5-E180V and XBB.1.5-K478R, respectively (Figures 3A and 3B).
Figure 3. Neutralization of XBB.1.16 by monoclonal antibody S309.
Neutralization of lentiviral pseudotypes by a class III monoclonal antibody S309 was determined.
Plot curves are shown (A), and calculated IC50 values (best fit ±95% confidence interval) are listed (B); dashed line in (A) represents 50% relative infectivity.
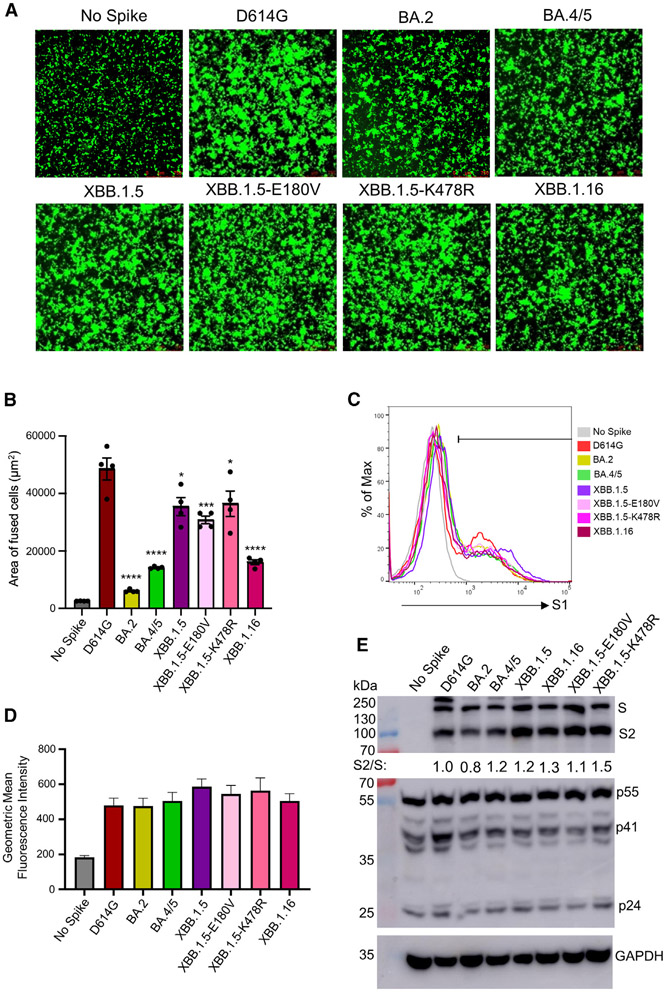
XBB.1.16 S demonstrates dramatically reduced fusogenicity with no change in processing
In addition to neutralization, we also assessed the S protein biology of XBB.1.16. Our experiments included investigation of the S fusogenicity, expression, and processing into S1 and S2. As demonstrated previously,3,13,18-20 all Omicron subvariants exhibited reduced fusion relative to D614G in HEK293T-ACE2 cells (Figures 4A and 4B). Interestingly, fusogenicity of XBB.1.16Swas lower than XBB.1.5 (p < 0.01), but more comparable to BA.4/5, with a 3.0-fold (p < 0.0001) decrease relative to D614G (Figures 4A and 4B). The cell surface expression of XBB.1.16 was comparable to other Omicron subvariants (Figures 4C and 4D), as we have demonstrated previously for XBB.1.5.20 Lastly, we examined the expression and processing of S protein in virus producer cells through immunoblotting. The total level of XBB.1.16 S expression in the transfected cells was comparable to that of XBB.1.5 and other S proteins examined (Figure 4E, top). While the overall processing of XBB.1.16 S was comparable to that of XBB.1.5 (S2/S ratio of 1.3 versus 1.2), XBB.1.5-K478R exhibited a slight increase (ratio of 1.5), and XBB.1.5-E180V had a slight decrease (ratio of 1.1) (Figure 4E, top).
Figure 4. Expression, fusion, and processing of S proteins of XBB.1.16.
(A and B) Fusogenicity of the S proteins. HEK293T-ACE2 cells were transfected with S proteins of interest and GFP and incubated for 24 h. Syncytia formation was imaged using fluorescence microscopy (A), with total areas of fused cells quantified (B). Bars represent means ± standard error, and dots represent two biological replicates with two random areas for each replicate. Significance relative to D614G was determined using a one-way repeated measures ANOVA with Bonferroni’s multiple testing correction (n = 4). “No Spike” refers to the negative control, which was transfected with GFP and empty pcDNA3.1 plasmid. Significance between XBB.1.5 and XBB.1.16 was determined by unpaired two-sided Student’s t tests.
(C and D) The expression level of XBB.1.16 S on the plasma membrane was determined by flow cytometry on HEK293T cells transfected with S and probed with polyclonal anti-S1 antibody. Three technical replicates were performed, and no significant difference was detected between the variants. The geometric mean fluorescence values were averaged and plotted, and an overlaid representative histogram was selected to represent the comparison between each of the variants. Bars represent means ± standard error.
(E) The S processing was determined through the lysis of HEK293T cells producing lentiviral pseudotypes and the probing for the S2 subunit of S specifically through immunoblotting. Relative S processing was determined by quantifying the band intensities using NIH ImageJ and calculating a ratio between S2 and S. Ratios were normalized to D614G and are listed below the anti-S2 blot (D614G = 1.0). The lysate was also probed with anti-p24 (HIV structural proteins, transfection control) and GAPDH (loading control) antibodies. p values are displayed as *p < 0.05, ***p < 0.001, and ****p < 0.0001.
DISCUSSION
Our study highlights the continued need for monitoring and characterizing new variants of SARS-CoV-2. We demonstrated that, similar to XBB.1.5, XBB.1.16 is immune evasive, especially against sera from those who received 3 doses of monovalent mRNA vaccine and in BA.4/5 convalescent individuals (Figure 2). These results are in agreement with another study demonstrating that sera from BA.2 convalescent individuals was unable to effectively neutralize XBB.1.16, though sera from hamsters infected with XBB.1 and XBB.1.5 could do so,38 emphasizing the need for a better understanding of immune imprinting upon mRNA vaccination. Of note, another recent study demonstrated that vaccinating mice with a second Omicron-based booster (BA.5, BQ.1, or XBB) greatly enhanced protection against Omicron variants, while receiving only one Omicron booster after a course of prototype S vaccinations was not as effective.40 In addition, sera from individuals who had repeated breakthrough Omicron infections also more effectively neutralized Omicron variants.40 These studies together suggested that repeated exposure to Omicron variants, either through vaccination or natural infection, may be required to grant improved protection.40,41
While vaccination is a critical aspect of protection against COVID-19, mAb treatments have been shown to be important in treatment of infected individuals, especially in the period of pre-Omicron waves. Omicron subvariants have presented mounting resistance toward the majority of mAb treatments, with most mAbs being rendered completely ineffective over the course of the pandemic.1,5,24,38,42-44 S309 is an antibody characterized by retaining activity against early Omicron subvariants, though we have demonstrated previously that subvariants CH.1.1, CA.3.1, and BA.2.75.2 can escape neutralization.20 Fortunately, we found in this work that XBB.1.16 remained sensitive to S309 neutralization (Figure 3). This was not unexpected, based on the location of the two unique mutations: neither E180V nor K478R was expected to impact interactions with S309’s target region of the RBD.20,45 Continued monitoring and investigation will be required to determine mutations that drive the neutralizing Ab (nAb) escape of Omicron subvariants.
Another important aspect of our study was the investigation of S protein biology, which could provide indicators of changes in viral entry, tropism, and/or viral spread.8-12 We found that the decrease in fusogenicity exhibited by XBB.1.16 S did not appear to be driven by E180V or K478R alone, suggesting a combinatorial effect imparted by the coexistence of these two mutations (Figures 4A and 4B). This change in fusion could be attributed to a possible decrease in affinity of XBB.1.16 S for human ACE2 relative to XBB.1.5.38 However, the modest change in fusogenicity among these XBB subvariants did not appear to correlate with the efficiency of S processing, a situation that is distinguishable from that was shown between Omicron BA.1 and D614G13 (Figure 4). These results suggested that XBB variants, including XBB. 1.5 and XBB.1.16, are still in the lineage of Omicron that likely favors the upper tract for infection.10,12 Further investigations of new Omicron XBB subvariants, including XBB.1.16 studied here, for cellular and tissue tropism will be informative, including in patients infected with SARS-CoV-2.
Overall, our study highlights the importance of the continued surveillance of new SARS-CoV-2 variants and emphasizes the benefits of booster COVID-19 vaccination. While our findings support the use of bivalent booster mRNA vaccine formulation, consideration must be given to include XBB variant S proteins in future vaccine designs, as recently recommended by the US Food and Drug Administration.46 Given the timeline of the pandemic, repeated exposures to the virus have become more likely, which underscores the development of new and more effective COVID-19 vaccines and therapeutics.
Limitations of the study
While authentic SARS-CoV-2 is the ideal model to test new variant biology, we used pseudotyped virus for all experiments because some of the new live viruses are not available. Furthermore, we have previously validated our pseudotyping system in comparison to live SARS-CoV-2 and found that it recapitulated the results accurately.39 Pseudotyped virus is also a very common way to analyze vaccine response and S protein biology in the field. Another limitation is the relatively small size of our cohorts used for the neutralization assays. However, similar studies with comparable sizes of cohorts have proven to be important to characterizing these new variants in a timely manner and informing public health decisions.46 The small size of the cohorts does prevent us from performing robust subgroup analysis, such as if the two individuals in the BA.4/5-convalescent cohort that received mRNA vaccines have a significantly different response than their colleagues. We also wish to acknowledge that sample times after vaccination and infection throughout the cohorts vary.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for reagents and resources can be requested from the lead contact, Dr. Shan-Lu Liu (liu.6244@osu.edu).
Materials availability
Plasmids generated for this study can be made available upon request from the lead contact.
Data and code availability
Accession numbers have not been provided to the lab yet from the NIH SeroNet program because data must first be published before deposition. De-identified information is available upon request made to the corresponding author and will be deposited with the NIH SeroNet Coordinating Center.
NT50 values and de-identified patient information can be shared by the lead contact upon request. Any other additional data can be provided for reanalysis if requested from the lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Vaccinated and patient cohorts
Neutralizing antibody titers in the sera of various individuals were determined for three cohorts. The first cohort included healthcare workers (HCWs) at the OSU Medical Center that had received 3 doses of homologous mRNA vaccine. The total number of HCWs in this cohort was 15, with 3 having received three doses of monovalent Moderna mRNA-1273 vaccine and 12 having received three doses of monovalent Pfizer BioNTech BNT162b2 vaccine. Samples were collected between 14 and 86 days post-third dose administration and collected under the approved IRB protocols 2020H0228, 2020H0527, and 2017H0292. HCWs in this cohort ranged from 26 to 61 years of age (median 33) and included 10 male and 5 female individuals.
The second cohort included HCWs that received a bivalent mRNA vaccine formulation in addition to receiving at least two doses of monovalent vaccine. The total number of HCWs in this cohort was 14, with 12 having received 3 doses of monovalent vaccine (Moderna or Pfizer formulations) and 1 dose of the bivalent vaccine (Pfizer formulation), 1 HCW having received 2 doses of monovalent vaccine (Pfizer formulation) and 1 dose of the bivalent vaccine (Pfizer formulation), and 1 HCW having received 4 doses of monovalent vaccine (Pfizer formulation) and 1 dose of the bivalent vaccine (Pfizer formulation). Samples were collected between 23 and 108 days post-bivalent booster administration. Samples were collected under the approved IRB protocols 2020H0228, 2020H0527, and 2017H0292. HCWs in this cohort ranged from 25 to 48 years of age (median 36) and included 8 male and 6 female individuals.
The third cohort was comprised of first responders and their household contacts that were infected with SARS-CoV-2 during the BA.4/5 wave in Columbus, OH. A positive test was confirmed for each individual via a nasal swab and the virus RNA was isolated and sequenced to determine the infecting variant. The total number of individuals in this cohort was 19, 4 of which were infected with BA.4, 7 infected with BA.5, and 8 individuals whose sequencing results were not conclusive but the timing of infection lines up with when BA.4/5 was dominant in Columbus, OH (late July 2022 through late September 2022). The cohort included 17 unvaccinated individuals and 2 individuals that received 3 homologous doses of monovalent vaccine (Pfizer (n = 1) or Moderna (n = 2)). Samples were collected under approved IRB protocols 2020H0527, 2020H0531, and 2020H0240. Individuals in this cohort ranged from 27 to 58 years of age (median 44) and included 4 male and 14 female individuals (age and gender of one individual unknown).
The full demographic information of cohorts can be found in a previous study.20
Cell lines and maintenance
Cell lines used in this study included human embryonic kidney (HEK) 293T cells (ATCC CRL-11268, RRID: CVCL_1926), HEK293T cells stably expressing ACE2 (BEI NR-52511, RRID: CVCL_A7UK), and human lung adenocarcinoma cell line CaLu-3 (RRID: CVCL_0609). HEK293T cells were maintained in DMEM (Gibco, 11965-092) supplemented with 10% FBS (Sigma, F1051) and 0.5% penicillin-streptomycin (HyClone, SV30010) and CaLu-3 cells were maintained in EMEM (ATCC, 30–2003) supplemented with 10% FBS and 0.5% penicillin-streptomycin. To passage, cells were washed in PBS (Sigma, D5652-10X1L) then incubated in 0.05% trypsin (Corning, 25-052-CI) until complete detachment.
METHOD DETAILS
Plasmids
Spike plasmids were constructed in the backbone of pcDNA3.1 and bear an N-and C-terminal FLAG tags. D614G, BA.2, BA.4/5, and XBB spikes were made through restriction enzyme cloning (BamHI and KpnI) by GenScript Biotech (Piscataway, NJ) and the remaining spikes (XBB.1.16, XBB.1.5, XBB.1.5-E180V, and XBB.1.5-K478R) were generated through PCR-based site-directed mutagenesis. The lentiviral vector used for production of pseudotyped virus was an HIV-1-derived vector pNL4-3 carrying an intronic Gaussia luciferase reporter gene that we have described in a previous study.4
Pseudotyped lentivirus production and infectivity
Pseudotyped virus was produced as previously described.4 In short, HEK293T cells were co-transfected with the pNL4-3-inGLuc vector and spike of interest in a 2:1 ratio using a polyethyleneimine transfection (Transporter 5 Transfection Reagent, Polysciences). Pseudovirus was collected from the media of producer cells 48- and 72- hours post transfection and used to infect HEK293T-ACE2 or CaLu-3 cells. Luminescence derived from secreted luciferase was used as a readout for infectivity and captured by a BioTek Cytation plate reader.
Virus neutralization assay
Neutralization assays using the pseudotyped vectors were performed as described previously.4 In brief, the sera of interest were serially diluted 4-fold with a starting dilution of 1:40 and with one well left as a no sera control (final dilutions 1:40, 1:160, 1:640, 1:2560. 1:10240, and no serum control). Monoclonal antibody S309 (kind gift of Dr. Tongqing Zhou at NIH) was diluted 4-fold from a starting concentration of 12 μg/mL. Pseudoviruses were normalized via dilution in DMEM (to ensure similar infectious viruses), incubated for 1 h, and incubated with target HEK293T-ACE2 cells; luminescence readings were taken 48 and 72 h post infection. Neutralization titers at 50% (NT50) values were determined through least squares fit nonlinear regression using GraphPad Prism 9 (San Diego, CA).
Syncytia formation
Syncytia formation was used to determine the ability of the spikes of interest to mediate viral membrane fusion. As described previously, HEK293T-ACE2 cells were co-transfected with the spike of interest and GFP, incubated for 24 h and imaged using fluorescence microscopy (Leica DMi8 confocal microscope) to observe fusion. Two wells were imaged for each sample with two randomly selected images taken. Software within the Leica X Applications Suite was used to quantify the average area of fused cells by outlining the perimeters of syncytia and calculating the area inside. The “No Spike” control refers to a well that was transfected with GFP and empty pcDNA3.1 plasmid backbone to serve as a negative control.
Spike protein processing
HEK293T cells producing pseudotyped virus were used to determine the extent to which the spikes were being processed into S1/S2 subunits. Lysates were collected at 72 h post-transfection through a RIPA buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, Nonidet P-40, 0.1% SDS) lysis supplemented with protease inhibitor cocktails (Sigma, P8340). The lysis was performed for 40 min and subjected to SDS-PAGE and western blotting. Blots were probed with anti-S2 (Sino Biological, 40590; RRI-D:AB_2857932), anti-p24 (NIH HIV Reagent Program, ARP-1513), and anti-GAPDH (Santa Cruz, Cat# sc-47724, RRID: AB_627678). Secondary antibodies used for blotting included Anti-Rabbit-IgG-HRP (Sigma, A9169; RRID:AB_258434) and Anti-Mouse (Sigma, Cat# A5278, RRID: AB_258232). Blots were performed using Immobilon Crescendo Western HRP substrate (Millipore, WBLUR0500) and exposed on a GE Amersham Imager 600. Quantification of band intensity was performed with NIH ImageJ (Bethesda, MD). ImageJ quantification was used to calculate an S2/S ratio that was normalized to D614G (D614G = 1.0).
Spike protein surface expression
To assess S surface expression at the plasma membrane, HEK293T cells were transfected with 2.5 μg of the S protein of interest. 48 h post-transfection, cells were incubated in PBS + 5mM EDTA for 10 min at 37°C to detach and fixed in 3.7% formaldehyde for 10 min at room temperature. Cells from a single well were split into 3 tubes and stained with 1:200 dilution anti-S1 (Singo Biological, 40591; AB_2893171) and 1:200 dilution anti-Rabbit-IgG-FITC (Sigma, F9887, RRID:AB_259816). Flow cytometry was performed on Life Technologies Attune NxT flow cyometer and data were analyzed using FlowJo v.10.8 (Ashland, OR).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using GraphPad Prism 9 and are described in the figure legends. NT50 values were determined by least-squares fit non-linear regression in GraphPad Prism 9. Error bars in (Figures 1B and 1C) represent means ± standard deviation. Error bars in (Figures 2A, 2C, and 2E) represent geometric means with 95% confidence intervals. Statistical significance for neutralization experiments was determined using log10 transformed NT50 values to better approximate normality (Figures 2A, 2C, and 2E), comparisons between multiple groups were made using a one-way ANOVA with Bonferroni post-test. And the comparison between XBB.1.5 and XBB.1.16 for syncytia formation assay was performed using unpaired two-sided Student’s t tests (Figure 4B). Bars in (Figure 3) represent best fit for IC50 ± 95% confidence interval (n = 1). Bars in (Figures 4B and 4D) represent means ± standard error. Significance in (Figure 4D) was determined using a one-way ANOVA with Bonferroni post-test to compare 3 technical replicate geometric mean fluorescence values.18,47,48
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Bivalent HCW Sera | 20 | N/A |
| 3-dose HCW Sera | 1, 30 | N/A |
| Omicron BA.4/5-wave Infected First Responders and Household Contacts Sera | 1, 30 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Transporter 5 Transfection Reagent | Polysciences | Cat# 26008-5 |
| Dulbecco’s Modified Eagles Medium (DMEM) | Sigma-Aldrich | Cat#: 11965-092 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat#: F1051 |
| 0.05% Trypsin +0.53 mM EDTA | Corning | Cat# 25-052-CI |
| Penicillin-Streptomycin | HyClone | Cat#: SV30010 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| Coelenterazine | GoldBio | Cat#: CZ2.5, CAS: 55779-48-1 |
| Deposited data | ||
| NT50 Values and De-identified patient data | SeroNet Coordinating Center, NCI, NIH | N/A |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat#: CRL-11268, RRID: CVCL_1926 |
| HEK293T-ACE2 | BEI Resources | Cat#: NR-52511, RRID: CVCL_A7UK |
| CaLu-3 | ATCC | RRID: CVCL_0609 |
| Recombinant DNA | ||
| pNL4-3-inGluc | David Derse, NIH47 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_D614G | GenScript Biotech48 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2 | GenScript Biotech30 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5 | GenScript Biotech1 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB.1.5 | GenScript Biotech20 | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB.1.5-E180V | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB.1.5-K478R | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_XBB.1.16 | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism | Version 9.0.0 | GraphPad |
| Other | ||
| Cytation 5 Imaging Reader | BioTek | N/A |
Highlights.
XBB.1.16 exhibits high extents of immune escape comparable to XBB.1.5
XBB.1.16 escapes neutralizing antibodies in bivalent- and 3-dose-vaccinated sera
XBB.1.16 spike is less fusogenic than XBB.1.5 due to the E180V and K478R mutations
Higher infectivity of XBB.1.16 in 293T-ACE2 cells is driven by the K478R mutation
ACKNOWLEDGMENTS
We thank the Clinical Research Center/Center for Clinical Research Management of The Ohio State University Wexner Medical Center and The Ohio State University College of Medicine in Columbus, OH (USA), specifically Francesca Madiai, Dina McGowan, Breona Edwards, Evan Long, and Trina Wemlinger, for logistics, collection, and processing of samples. We thank Tongqing Zhou at NIH for providing the S309 mAb. In addition, we thank Sarah Karow, Madison So, Preston So, Daniela Farkas, and Finny Johns in the clinical trials team of The Ohio State University for sample collection and other supports. We thank Mirela Anghelina, Patrick Stevens, and Soledad Fernandez for their assistance in providing the sample information of the first responders and their household contacts. S.-L.L., D.J., A.R.P., R.J.G., L.J.S., and E.M.O. were supported by the National Cancer Institute of the NIH under award no. U54CA260582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was also supported by a fund provided by an anonymous private donor to OSU. R.J.G. was additionally supported by the Robert J. Anthony Fund for Cardiovascular Research and the JB Cardiovascular Research Fund, and L.J.S. was partially supported by NIH R01 HD095881.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Qu P, Faraone J, Evans JP, Zou X, Zheng Y-M, Carlin C, Bednash JS, Lozanski G, Mallampalli RK, Saif LJ, et al. (2022). Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. N. Engl. J. Med 386, 2526–2528. 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faraone JN, Qu P, Evans JP, Zheng Y-M, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Lozanski G, et al. (2023). Neutralization escape of Omicron XBB, BR.2, and BA.2.3.20 subvariants. Cell Rep. Med 4, 101049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans JP, Zeng C, Qu P, Faraone J, Zheng Y-M, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, et al. (2022). Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe 30, 1093–1102.e3. 10.1016/j.chom.2022.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia H, Zou J, Kurhade C, Cai H, Yang Q, Cutler M, Cooper D, Muik A, Jansen KU, Xie X, et al. (2022). Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe 30, 485–488.e3. 10.1016/j.chom.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, Bolland WH, Porrot F, Staropoli I, Lemoine F, et al. (2022). Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675. 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 6.Davis-Gardner ME, Lai L, Wali B, Samaha H, Solis D, Lee M, Porter-Morrison A, Hentenaar IT, Yamamoto F, Godbole S, et al. (2023). Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA Bivalent Booster. N. Engl. J. Med 388, 183–185. 10.1056/NEJMc2214293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Toole Á, Scher E, Underwood A, Jackson B, Hill V, McCrone JT, Colquhoun R, Ruis C, Abu-Dahab K, Taylor B, et al. (2021). Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 7, veab064. 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuai H, Chan JFW, Hu B, Chai Y, Yuen TTT, Yin F, Huang X, Yoon C, Hu JC, Liu H, et al. (2022). Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature 603, 693–699. 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 9.Yuan S, Ye Z-W, Liang R, Tang K, Zhang AJ, Lu G, Ong CP, Man Poon VK, Chan CC-S, Mok BW-Y, et al. (2022). Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science 377, 428–433. 10.1126/science.abn8939. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, et al. (2022). Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 603, 700–705. 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerber PP, Fatihi S, Rathore S, et al. (2022). Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 603, 706–714. 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hui KPY, Ho JCW, Cheung MC, Ng KC, Ching RHH, Lai KL, Kam TT, Gu H, Sit KY, Hsin MKY, et al. (2022). SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 603, 715–720. 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 13.Zeng C, Evans JP, Qu P, Faraone J, Zheng YM, Carlin C, Bednash JS, Zhou T, Lozanski G, Mallampalli R, et al. (2021). Neutralization and Stability of SARS-CoV-2 Omicron Variant. Preprint at bio-Rxiv2021.12.16.472934. 10.1101/2021.12.16.472934. [DOI] [Google Scholar]
- 14.Wang X, Zhao X, Song J, Wu J, Zhu Y, Li M, Cui Y, Chen Y, Yang L, Liu J, et al. (2022). Homologous or heterologous booster of inactivated vaccine reduces SARS-CoV-2 Omicron variant escape from neutralizing antibodies. Emerg. Microb. Infect 11, 477–481. 10.1080/22221751.2022.2030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt F, Muecksch F, Weisblum Y, Da Silva J, Bednarski E, Cho A, Wang Z, Gaebler C, Caskey M, Nussenzweig MC, et al. (2022). Plasma Neutralization of the SARS-CoV-2 Omicron Variant. N. Engl. J. Med 386, 599–601. 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Then E, Lucas C, Monteiro VS, Miric M, Brache V, Cochon L, Vogels CBF, Malik AA, De la Cruz E, Jorge A, et al. (2022). Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med 28, 481–485. 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobsen H, Strengert M, Maaß H, Ynga Durand, M.A., Katzmarzyk M, Kessel B, Harries M, Rand U, Abassi L, Kim Y, et al. (2022). Diminished neutralization responses towards SARS-CoV-2 Omicron VoC after mRNA or vector-based COVID-19 vaccinations. Sci. Rep 12, 19858. 10.1038/s41598-022-22552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu P, Evans JP, Zheng YM, Carlin C, Saif LJ, Oltz EM, Xu K, Gumina RJ, and Liu SL (2022). Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe 30, 1518–1526.e4. 10.1016/j.chom.2022.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu P, Evans JP, Faraone JN, Zheng YM, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Lozanski G, et al. (2023). Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell Host Microbe 31, 9–17.e3. 10.1016/j.chom.2022.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu P, Faraone JN, Evans JP, Zheng Y-M, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Panchal AR, et al. (2023). Enhanced Evasion of Neutralizing Antibody Response by Omicron XBB. Cell Rep. 42, 112443, CH.1.1 and CA.3.1 Variants. 10.1016/j.celrep.2023.112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Q, Wu L, Xu Z, Wang X, Xie Y, Chai Y, Zheng A, Zhou J, Qiao S, Huang M, et al. (2023). An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB. Cell Rep. Med 4, 100991. 10.1016/j.xcrm.2023.100991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J, Hachmann NP, Collier ARY, Lasrado N, Mazurek CR, Patio RC, Powers O, Surve N, Theiler J, Korber B, and Barouch DH (2023). Substantial Neutralization Escape by SARS-CoV-2 Omicron Variants BQ.1.1 and XBB.1. N. Engl. J. Med 388, 662–664. 10.1056/NEJMc2214314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hachmann NP, Miller J, Collier ARY, Ventura JD, Yu J, Rowe M, Bondzie EA, Powers O, Surve N, Hall K, and Barouch DH (2022). Neutralization Escape by SARS-CoV-2 Omicron Subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med 387, 86–88. 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Iketani S, Li Z, Liu L, Guo Y, Huang Y, Bowen AD, Liu M, Wang M, Yu J, et al. (2023). Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 186, 279–286.e8. 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurhade C, Zou J, Xia H, Liu M, Chang HC, Ren P, Xie X, and Shi PY (2023). Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nat. Med 29, 344–347. 10.1038/s41591-022-02162-x. [DOI] [PubMed] [Google Scholar]
- 26.Zou J, Kurhade C, Patel S, Kitchin N, Tompkins K, Cutler M, Cooper D, Yang Q, Cai H, Muik A, et al. (2023). Neutralization of BA.4-BA.5, BA.4.6, BA.2.75.2, BQ.1.1, and XBB.1 with Bivalent Vaccine. N. Engl. J. Med 388, 854–857. 10.1056/NEJMc2214916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coronavirus (COVID-19) Update (2023). FDA Authorizes Changes to Simplify Use of Bivalent mRNA COVID-19 Vaccines. [Google Scholar]
- 28.Tamura T, Ito J, Uriu K, Zahradnik J, Kida I, Anraku Y, Nasser H, Shofa M, Oda Y, Lytras S, et al. (2023). Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat. Commun 14, 2800. 10.1038/s41467-023-38435-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.TAG-VE statement on Omicron sublineages BQ.1 and XBB. (2022).
- 30.XBB.1 (2023). 5 Updated Risk Assessment, 20 June 2023. [Google Scholar]
- 31.Zhang X, Chen LL, Ip JD, Chan WM, Hung IF, Yuen KY, Li X, and To KK (2022). Omicron sublineage recombinant XBB evades neutralising antibodies in recipients of BNT162b2 or CoronaVac vaccines. Lancet Microbe. 10.1016/s2666-5247(22)00335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue C, Song W, Wang L, Jian F, Chen X, Gao F, Shen Z, Wang Y, Wang X, and Cao Y (2023). Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. Preprint at bioRxiv, 2023.2001.2003.522427. 10.1101/2023.01.03.522427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uraki R, Ito M, Furusawa Y, Yamayoshi S, Iwatsuki-Horimoto K, Adachi E, Saito M, Koga M, Tsutsumi T, Yamamoto S, et al. (2023). Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis 23, 30–32. 10.1016/s1473-3099(22)00816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faraone JN, Qu P, Evans JP, Zheng Y-M, Carlin C, Anghelina M, Stevens P, Fernandez S, Jones D, Lozanski G, et al. (2023). Neutralization Escape of Omicron XBB. BR.2, and BA.2.3.20 Subvariants. Cell Reports Medicine. 10.1016/j.xcrm.2023.101049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karyakarte RP, Das R, Rajmane MV, Dudhate S, Agarasen J, Pillai P, Chandankhede PM, Labhshetwar RS, Gadiyal Y, Kulkarni PP, et al. (2023). Chasing SARS-CoV-2 XBB.1.16 Recombinant Lineage in India and the Clinical Profile of XBB.1.16 Cases in Maharashtra, India. Preprint at medRxiv, 2023.2004.2022.23288965. 10.1101/2023.04.22.23288965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.XBB.1.16 Initial Risk Assessment, 17 April 2023 (2023). [Google Scholar]
- 37.COVID Data Tracker, Variant Proportions, Centers for Disease Control and Prevention. (2022). [Google Scholar]
- 38.Yamasoba D, Uriu K, Plianchaisuk A, Kosugi Y, Pan L, Zahradnik J, Consortium T.G.t.P.J., Ito J, and Sato K (2023). Virological characteristics of the SARS-CoV-2 Omicron XBB.1.16 variant. Preprint at bioRxiv, 2023.2004.2006.535883. 10.1101/2023.04.06.535883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng C, Evans JP, Pearson R, Qu P, Zheng YM, Robinson RT, Hall-Stoodley L, Yount J, Pannu S, Mallampalli RK, et al. (2020). Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight 5, e143213. 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yisimayi A, Song W, Wang J, Jian F, Yu Y, Chen X, Xu Y, Yang S, Niu X, Xiao T, et al. (2023). Repeated Omicron infection alleviates SARS-CoV-2 immune imprinting. Preprint at bioRxiv, 2023.2005.2001.538516. 10.1101/2023.05.01.538516. [DOI] [Google Scholar]
- 41.Wang Q, Guo Y, Tam AR, Valdez R, Gordon A, Liu L, and Ho DD (2023). Deep immunological imprinting due to the ancestral spike in the current bivalent COVID-19 vaccine. Preprint at bioRxiv, 2023.2005.2003. 539268. 10.1101/2023.05.03.539268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Guo Y, Iketani S, Nair MS, Li Z, Mohri H, Wang M, Yu J, Bowen AD, Chang JY, et al. (2022). Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 608, 603–608. 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Y, Jian F, Wang J, Yu Y, Song W, Yisimayi A, Wang J, An R, Chen X, Zhang N, et al. (2023). Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 614, 521–529. 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arora P, Zhang L, Krüger N, Rocha C, Sidarovich A, Schulz S, Kempf A, Graichen L, Moldenhauer AS, Cossmann A, et al. (2022). SARS-CoV-2 Omicron sublineages show comparable cell entry but differential neutralization by therapeutic antibodies. Cell Host Microbe 30, 1103–1111.e6. 10.1016/j.chom.2022.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto D, Park Y-J, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. (2020). Crossneutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 46.Recommendation for the 2023-2024 Formula of COVID-19 Vaccines in the U.S (2023). [Google Scholar]
- 47.Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, and Derse D (2010). Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 6, e1000788. 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng C, Evans JP, Faraone JN, Qu P, Zheng YM, Saif L, Oltz EM, Lozanski G, Gumina RJ, and Liu SL (2021). Neutralization of SARS-CoV-2 Variants of Concern Harboring Q677H. mBio 12, e0251021. 10.1128/mBio.02510-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Accession numbers have not been provided to the lab yet from the NIH SeroNet program because data must first be published before deposition. De-identified information is available upon request made to the corresponding author and will be deposited with the NIH SeroNet Coordinating Center.
NT50 values and de-identified patient information can be shared by the lead contact upon request. Any other additional data can be provided for reanalysis if requested from the lead contact.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work is available from the lead contact upon request.