Abstract
Burkitt lymphoma (BL) is an aggressive B-cell lymphoma that significantly contributes to childhood cancer burden in sub-Saharan Africa. Plasmodium (P.) falciparum, which causes malaria, is geographically associated with BL, but the evidence remains insufficient for causal inference. Inference could be strengthened by demonstrating that mendelian genes known to protect against malaria —such as the sickle cell trait variant, HBB-rs334(T)—also protect against BL. We investigated this hypothesis among 800 BL cases and 3,845 controls in four East African countries using genome-scan data to detect polymorphisms in 22 genes known to affect malaria risk. We fit generalized linear mixed models to estimate odds ratios (OR) and 95% confidence intervals (95% CI), controlling for age, sex, country, and ancestry. The ORs of the loci with BL and P. falciparum infection among controls were correlated (Spearman’s ρ = 0.37, p = 0.039). HBB-rs334(T) was associated with lower P. falciparum infection risk among controls (OR = 0.752, 95% CI 0.628–0.9; p = 0.00189) and BL risk (OR=0.687, 95% CI 0.533–0.885; p=0.0037). ABO-rs8176703(T) was associated with decreased risk of BL (OR=0.591, 95% CI 0.379–0.992; p=0.00271), but not of P. falciparum infection. Our results increase support for the etiological correlation between P. falciparum and BL risk.
Keywords: Plasmodium falciparum, Burkitt lymphoma, non-Hodgkin lymphoma, malaria resistance loci, East Africa
Graphical Abstract

The association between Burkitt lymphoma (BL) with malaria resistance was analyzed in a case-control study conducted in children from four countries in Africa. Results indicate that the odds of BL were lower in children with genetic resistance due to the sickle cell trait or a variant linked to blood group O and apparent immunity indicated by loa grade asymptomatic infection.
INTRODUCTION
Burkitt lymphoma (BL) is a B-cell lymphoma that occurs worldwide, but with a marked excess in sub-Saharan African (SSA) children in whom it is one of the leading cancers [1]. This geographic pattern, which mirrors that of Plasmodium (P.) falciparum infection[2], which causes the deadliest malaria, suggested a possible causal role of malaria in BL. This hypothesis is supported by case-control associations of BL with serological [3, 4] [5, 6] and molecular markers of P. falciparum infection [7, 8]. It is also supported by immunological data indicating that P. falciparum infection impairs control of Epstein-Barr virus (EBV) [9], a known oncogenic factor for BL [10, 11]. EBV and P. falciparum parasites induce activation induced cytosine deaminase (AID), an enzyme that is involved in somatic hypermutation and IG::MYC translocation, genomic instability considered an early or primary abnormality in BL [11, 12]. However, this evidence remains insufficient for causal inference [13, 14].
A pathogenic variant in the β-hemoglobin gene, HBB-rs334 (Glu6Val), which causes the sickle cell disease in homozygotes, is associated with a 90% decrease in risk for severe malaria in heterozygotes (carriers), i.e., those with sickle cell trait (SCT) [15]. Demonstration that SCT—or others malaria protective loci— also protect against BL could strengthen inferences about the causal role of malaria in BL. SCT was associated with a reduced risk of BL in three studies [16–18] but not in three others [19–21]. All of these studies were small, lacked data to control for genetic ancestry [22], and did not evaluate effects of other loci known to affect malaria risk [23].
In 2017, we tested the SCT-BL hypothesis in the Epidemiology of Burkitt Lymphoma in East-African Children and Minors (EMBLEM) study in Uganda [24] using a larger sample (200 BL cases and 800 controls) and controlling for ancestry using genetic data. We found significant protection against BL among those with SCT, as well as suggestive protective associations with IL10-rs1800896(C), IL1A-rs2856838(A), and SEMA3C-rs4461841(C) [24]. Here, we expanded that analysis to 800 BL cases and 3,845 BL-free controls in Uganda, Tanzania, Kenya, and Malawi to evaluate associations of malaria loci with BL. In addition to SCT, we evaluated 31 other loci in 22 malaria-related genes. We included α3.7-thalassemia because co-inheritance of α3.7-thalassemia with SCT has been previously found to affect SCT phenotype, including malaria resistance [25], osmolality, and hemoglobin S (HbS) percentage [26]. We also controlled for contemporaneous asymptomatic P. falciparum detection, which we previously found to have an inverse association with BL, independent from prior history of and treatment for malaria [27, 28]. Asymptomatic P. falciparum detection indicates that naturally acquired immunity (NAI) is sufficient to control malaria symptoms but not to prevent infection [29, 30]. Thus, asymptomatic P. falciparum is an indicator of NAI. We tested whether contemporaneous asymptomatic P. falciparum infection (or NAI) was protective against BL when controlling for the identified genetic associations. We hypothesized that in children with basic NAI (i.e., sufficient to prevent symptomatic malaria [29, 30]), there exists another threshold above which NAI reduces P. falciparum infection to levels below the threshold sufficient to increase the risk of BL.
Methods
Study population
Our study was conducted using samples from the EMBLEM study conducted in Uganda, Tanzania, and Kenya [27], and from the Childhood and Infections Cancer study conducted in Malawi [4]. Briefly, the EMBLEM study was conducted in six geographical areas (Figure 1a). The average child in these areas’ experiences 100–400 P. falciparum infections per year and a 39% risk of BL per 100 P. falciparum infections versus those with 50 lifetime infections [31]. For both BL cases and controls, eligibility was limited to usual residents (≥4 months prior to enrollment) of a village in the defined study areas who were aged 0–15 years. Eligible BL cases were enrolled at six participating hospitals with capacity to diagnose and treat BL. BL diagnosis was confirmed by local histology/cytology, whenever possible, or by clinical diagnosis, with supporting imaging and laboratory results for those lacking histology or cytology. The controls were a random sample of apparently healthy community children residing in 295 villages that were randomly selected from a list of all villages in the study areas constructed from national census files [27]. The primary controls were selected to match the age and sex frequency of BL cases diagnosed in their area over the past 10 years before the current study [27]. Other community controls were enrolled using household survey methods without any frequency-matching in 12 villages in Uganda (i.e., all children in the selected households who were eligible were enrolled) [32]. A small set of health-facility controls were enrolled from the same 12 villages in Uganda. These controls were selected to match the age and sex frequency of BL cases diagnosed in their area over the past 10 years before the current study.
Figure 1. a) Map of the study sites: a locator map for Africa and a zoom out of EMBLEM study sites in Uganda, Kenya, and Tanzania and the Infections and Childhood Cancer Study site in Malawi.

Red crosses mark location of hospitals where cases were enrolled, and a green shade marks the 6 regions where cases and controls were enrolled in EMBLEM. Stars mark location of country capitals to show the relative distance between the participating hospitals versus enrollment hospitals. b) Flow chart showing participants enrolled and reasons for exclusion from the final set included in case-control analysis. c) Flow chart showing the process followed to identify polymorphisms associated with protection from or risk of severe malaria in epidemiological studies. Significantly associated polymorphisms were considered “malaria index” and the identified genes labeled “malaria index genes”. The genotypes are analyzed are based on genotype results from the Omni 5M array or from imputed results. Non-index polymorphisms were extracted and analyzed for hypothesis generation. Genotypes for HBA1 and HBA2 were determined using droplet digital PCR were included in the analysis. The 3.7 kb deletion (-α3.7/), which has been reported to interact with the sickle cell trait, is the most prevalent in our population (details in Supplementary data).
In Malawi, participants were enrolled at Queen Elizabeth hospital, a tertiary cancer care facility in Blantyre [4]. Children with BL were designated as cases, while children with non-BL cancers were used as controls. No matching on age or sex was performed as was done in EMBLEM. In both studies (EMBLEM and Malawi), pre-treatment venous blood was collected in EDTA tubes at enrollment. In EMBLEM, the samples were processed into plasma and buffy coats, then stored at −80°C until shipment to the National Cancer Institute (NCI). In Malawi, samples were stored as whole blood at −20°C until shipment to NCI. In EMBLEM, P. falciparum was detected using microscopy of Giemsa stained thick-film blood smears and antigen capture rapid diagnostic tests (RDTs) [27]. In Malawi, P. falciparum was detected using P. falciparum-specific polymerase chain reaction (PCR) [8].
Ethical Approvals
Ethical approvals for the EMBLEM study were granted by the Uganda Virus Research Institute (GC/127), Uganda National Council for Science and Technology (H816), Tanzania National Institute for Medical Research (NIMR/HQ/R.8c/Vol. IX/1023), Moi University/Moi Teaching and Referral Hospital (000536), and National Cancer Institute (10-C-N133). Ethical approvals for the Childhood Infections and Cancer study were granted by Malawi College of Medicine (P.03/04/277R), Oxford University, and National Health Sciences Research Committee (2405). Guardians of participants provided written informed consent. Children aged ≥7 years old gave written assent. Ethical approval for genetic research was granted primarily to establish whether BL is a phenotype associated with malaria, in order to strengthen evidence efforts to control BL by targeting malaria before agnostically searching for other genetic factors associated with BL.
Selection and genotyping of malaria loci
DNA extracted from buffy coat aliquots using the Qiagen QIAsymphony automated instrument were genotyped at approximately 4.6 million single nucleotide positions (SNPs) using the Infinium Omni5Exome-4 v1.3 BeadChip (Illumina, San Diego, CA, USA) at the NCI Cancer Genomics Research (CGR) Laboratory (Figure S1). We used standard genome-wide association study quality control pipelines, including sample completion call rate >0.95; contamination rate <0.1; exclusion of unexpected, duplicated samples; and SNPs with less than a 95% completion rate. Phased genome scan data were imputed using the African Genome Resources panel with 6 230 African genomes on the Sanger imputation server (https://imputation.sanger.ac.uk/). We retained SNPs imputed with high confidence (INFO ≥ 0.9) and with a minor allele frequency (MAF) ≥ 0.01, and those passing the Hardy-Weinberg equilibrium tests in the controls p<10−6. We performed technical validation of the Illumina chip results for thirteen SNPs, including the classical HBB-rs334 for SCT, in 900 samples with 100% concordance [24]. We genotyped copy numbers of HBA 3.7 kb deletions using droplet digital polymerase chain reaction (ddPCR) (Bio-Rad QX200 ddPCR system) (Figure S2).
Statistical analysis
After QC filtering, we had an analytic set with 800 BL cases and 3,845 controls (Figure 1b). We analyzed associations between BL with 32 loci in 22 genes known to affect malaria risk (Figure 1c). We excluded seven loci that did not pass the quality control metrics mentioned above (Table S1). The analyzed loci included the classical SCT allele (HBB-rs334(T), which is known to reduce malaria risk [33] and previously shown to reduce BL risk in four [16–18, 24] of seven studies [16–21, 24]. We reported HBB-rs334 results according to the minus strand (i.e., sickle mutation is A to T) following the practice in sickle cell disease scientific literature.
We calculated principal components (PC) using 787,731 genotyped uncorrelated (r2 < 0.3) SNPs outside the HLA region to assess ancestry patterns. The PCs showed extensive population substructure among participants across the four countries (Figure S3), with minimal relatedness in controls from Kenya and Tanzania but a high degree of relatedness in the Uganda controls (Figure S4 and Figure S5). Only two related BL cases, previously reported in Uganda [34], were identified. We used country-specific PCs and a genetic relationship matrix (GRM), based on the probability that two individuals i and j share 0, 1, or 2 alleles identical by descent (IBD) [35], to adjust for ancestry. We fit a generalized linear mixed model, controlling for sex, age, P. falciparum detection, country, and ancestry as fixed effects, and GRM as a random variable. P. falciparum was included in the models as an indicator for current P. falciparum exposure with NAI (detection of infection). Statistical significance for the loci was based on two-sided Wald tests with a p-value < 0.05 considered significant to detect concordant effects of the malaria loci against BL. We performed sensitivity analyses to assess robustness of effects. In Sensitivity 1, we adjusted for loci significantly associated with BL; In sensitivity 2, we excluded 24 participants with sickle cell disease HBB-rs334(TT). Additionally, we ran analyses when dropping country, ancestry, and P. falciparum detection (Sensitivity 3–5) from the primary model. We examined the independent contribution of P. falciparum detection, which is an indicator of NAI, by comparing unrestricted models (with P. falciparum) to restricted models (without P. falciparum) using likelihood ratio tests. Because co-inheritance of α3.7 thalassemia with HBB-rs334(T) modifies the phenotype of SCT [25], we analyzed the associations between BL and α3.7 thalassemia genotypes stratified by HBB-rs334 genotypes (AA versus T/A).
We leveraged available genome scan data to explore associations between BL and 2,735 loci in the 22 genes with linked malaria (Figure 1c; Table S1). We applied a modified Bonferroni correction based on the effective degrees of freedom as previously described to assess statistical significance [36] (see Supplementary methods and Table S2). Analyses were performed using computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
RESULTS
Study participants’ characteristics are shown in Table 1. The median age was 7 years for both BL cases and controls, with similar distributions (Figure S6); 64% of the participants were 6 years or older. The BL cases in the analytic set included 592 (74%) who were confirmed by local histology or cytology and 208 (26%) based on clinical diagnosis, usually with a consistent opinion from a fine needle aspirate read by local clinicians. Compared to the histologically diagnosed cases, those clinically diagnosed were more likely to be younger (6.5 years versus 7.4 years, p=0.0009), more likely to report a history of fever other than malaria in the past 6 months (attributed to BL symptoms: 63% versus 42%, p<0.001), and to report a history of treatment for outpatient malaria in the past 12 months (54% versus 40%, p=0.001), attributed to less well developed immunity to malaria due to their younger age (Table S3). However, no differences were observed by sex, presence of current fever, or history of fever due to malaria in the past 6 months or 12 months, and history of admission for severe malaria. There were more males among BL cases than among the controls (63.2% versus 52.1%, p<0.0001). HIV status was known for 770 (96.2%) of 800 BL cases and unknown for 30 (3.8%). The BL cases with HIV resembled those without regarding sex, mean age, having current fever or histories of fever in the past 12 months, fever due to malaria or unelated to malaria in the past 6 months, and history of outpatient treatment for malaria (Table S4). However, compared to those who were HIV-negative, HIV positive BL cases had a lower prevalence of asymptomatic P. falciparum infection (12% versus 37%, p=0.013) and were more likely to report an admission in the past 12 months when compared to HIV-negative BL cases (26% versus 9%, p=0.029). As previously reported [27], HIV infection was associated with being a BL case (OR~4), but the number of HIV positive participants was small [24 BL cases, 15 controls (Table S5)] and their impact on the overall findings was considered negligible. BL cases tended to live in areas likely to have heavier P. falciparum exposure, such as rural villages or villages close to surface water (i.e., <500m) (Table S1). Despite this heightened exposure, and consistent with our earlier findings [27], BL cases were less likely to have asymptomatic P. falciparum detection than controls (282 [32.5%] versus 1 857 [48.3%], p<0.0001) (Table 1). When P. falciparum was detected among EMBLEM participants (but not in Malawi, which used PCR rather than blood microscopy/RDT), BL cases displayed lower geometric parasite density compared to their control counterparts (Table S5). Similarly, EMBLEM BL cases were less likely than controls to report a fever that was diagnosed as or treated as malaria in the past 6 months before enrollment (Table S5). Also of relevance and arguing against poor recall of malaria-related fevers, EMBLEM BL cases were more likely than controls to report fever that was diagnosed as due to other causes other than malaria (Table S5); these fevers were interpreted as indicating presence of B-symptoms, which are defined as fever of unknown origin in patients with neoplasia [37].
Table 1.
Participant demographic and clinical characteristics at enrollment
| Characteristic | BL Cases, n (%) | Controls, n (%) | p value |
|---|---|---|---|
|
| |||
| All subjects | 800 (100) | 3 845 (100) | |
| Median age, (IQR) * | 7·00 (4–10) | 7.00 (5–10) | 0·07 |
| Age group ±, years | 0·12 | ||
| 0–2 | 67 (8·4) | 342 (8·9) | |
| 3–5 | 219 (27·4) | 914 (23·8) | |
| 6–8 | 239 (29·9) | 1,130 (29·4) | |
| 9–11 | 165 (20·6) | 820 (21·3) | |
| 12–15 | 110 (13·7) | 639 (16·6) | |
| Sex | <0·0001 | ||
| Males | 503 (62.9) | 2,005 (52·1) | |
| Females | 297 (37·1) | 1,840 (47·8) | |
| Country | <0·0001 | ||
| Uganda | 249 (31·1) | 2 004 (52·1) | |
| Tanzania | 97 (12·1) | 753 (19·6) | |
| Kenya | 229 (28·6) | 878 (22·8) | |
| Malawi | 225 (28·1) | 210 (5·5) | |
| P. falciparum infection ¶ | <0·0001 | ||
| Negative | 499 (63·3) | 1 943 (50·5) | |
| Positive | 282 (35·2) | 1 857 (48·3) | |
| Unknown (missing) | 19 (2·4) | 45 (1·2) | |
Abbreviation: BL Burkitt lymphoma; SD standard deviation
The age distribution of cases and controls was not normally distributed, so median and inter-quartile ranges are used to describe central tendency (See supplementary Figures S3)
The age groups were selected to reflect the age distribution of Burkitt lymphoma in the study area..
Asymptomatic P. falciparum infection was determined using thick film microscopy or rapid diagnostic test results as in the EMBLEM study (Uganda, Tanzania, and Kenya) and using quantitative polymerase chain reaction tests in Malawi. As such, while parasitemia positivity is combined, the TFM/RDT results are at a lower sensitivity, while the PCR results are more sensitive (see methods and supplementary methods for more detailed discussion).
Differences for continuous variables were assessed by unpaired t-test. Differences in categorical variables were assessed by Chi-squared test.
Effects of loci against P. falciparum infection and Burkitt lymphoma were correlated
The effects of the 32 loci against BL and against P. falciparum detection in the controls were found to be correlated (ρ = 0.37, p = 0.039; Figure 2). Two loci showed statistically significant protection against BL in models adjusting for age, sex, country, P. falciparum detection, ancestry, and genetic relatedness—namely, HBB-rs334(T) with an OR=0.687 (95% CI 0.533–0.885; p=0.0037) and ABO-rs8176703(T) with an OR=0.591 (95% CI 0.379–0.992; p=0.00271; Figure 2 and Table S6, Table S7). The associations of BL with HBB-rs334 and with ABO-rs8176703 persisted in models with both loci mutually adjusted for each other (Table S7), and in sensitivity analyses excluding children with sickle cell disease, country, population-specific PCs, or P. falciparum detection (Table S7). The association of the sickle cell HBB-rs334(A/T) with BL was similar in individuals without HIV infection (OR = 0.486) and those with HIV infection (OR=0.590) compare with the reference group (HBB-rs334(A/A), but the results were only significant in those without HIV, which had a large sample size. The protective effects of HBB-rs334(T) against severe malaria have been known to vary in different countries [38] and, following a similar pattern, the ORs for BL with HBB-rs334(T) in the four countries ranged from 0.21 to 0.94 (Figure S7), but with overlapping CIs. This pattern was similar for ABO-rs8176703(T) (Figure S7), with overlapping CIs.
Figure 2. Forest plots showing associations between 32 malaria-risk index SNPs and endemic Burkitt lymphoma (left) or P. falciparum infection (right).
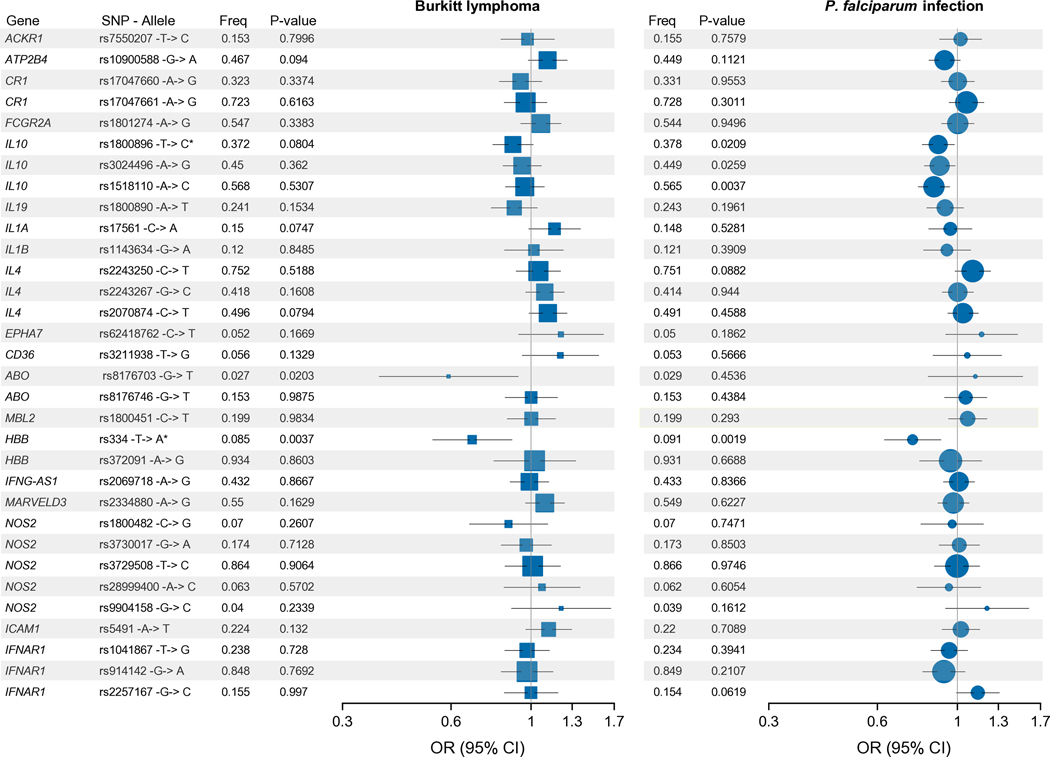
Forest plots show OR values (95% confidence intervals) and p-values from the mixed-effects logistic regression model analyses for association with BL among 4 581 BL cases and controls using model: BL case status~ SNP_ij+factor(P. falciparum)+ age+sex+pop-specific PC1+ pop-specific PC2+ pop-specific PC3+ factor(country)+[genetic relation matrix, random effect], and for association with P. falciparum infection among 3800 controls using model P. falciparum ~ SNP_ij+ age+sex+pop-specific PC1+ pop-specific PC2+ pop-specific PC3+ factor(country)+[genetic relation matrix, random effect]. p < 0.05 was considered statistically significant, * effect of SNP on BL concordant with effects against malaria in the literature and infection among the controls (see methods). The area of the squares/circles is proportional to the frequency (freq) of the variant in the population. The minor alleles are reported based on the sequence on the plus strand as reported in dbGAP, however results for HBB-rs334 are reported in the test based on the minus strand to be consistent with conventional usage across scientific literature on sickle cell disease.
In addition to its protective effect against BL, HBB-rs334 was also protective against P. falciparum detection among controls [OR= 0.752 (95% CI 0.628–0.9; p = 0.00189); Figure 2 and Table S8]. The OR for P. falciparum detection with ABO-rs8176703(T) was 1.122 (95% CI 0.83 – 1.517, p=0.543). The effects of three IL10 loci against BL and against P. falciparum detection among controls were concordant, but statistically significant only for P. falciparum infection: IL10-rs1800896(C) (OR for infection = 0.886, 95% CI 0.799 – 0.982, p= 0.0208), IL10-rs3024496(G) (OR=0.894, 95% CI 0.810 – 0.987, p=0.0259), and IL10-rs1518110(C) (OR= 0.862, 95% CI 0.779 – 0.953, p= 0.0037; Figure 2 and Table S8).
Association of Burkitt lymphoma with α3.7-thalassemia
Among BL-free children, 47% had normal alpha genotypes (αα/αα), 41% were heterozygous for the α3.7 thalassemia deletion (-α3.7/αα), 10% were homozygous for the deletion (-α3.7/-α3.7), and approximately 1% had five copies of the normal gene (ααα/αα). Compared to homozygous normal participants, the ORs for BL ranged from 0.778 in those with two copies of the thalassemia deletion gene to 1.655 in those with five copies of the normal gene, although none of these results reached statistical significance (all p>0.05, as shown in Table S9).
Association of asymptomatic P. falciparum infection with Burkitt lymphoma
In agreement with previously published results [27, 28], the OR for BL was 0.526 (95% CI 0.438–0.633, p=9.64×10−12; Table 2) among those with P. falciparum detection. This decreased OR for BL persisted (OR=0.524, 95% CI 0.435–0.630, 7.29 ×10−12) in models further adjusting for the HBB-rs334(T) and ABO-rs8176703(T) alleles. Using the LRT, we confirmed that unrestricted models (i.e., with P. falciparum detection) were statistically better than restricted models (i.e., without the P. falciparum detection) for estimating associations with BL (Table 2). However, as noted above, the association of P. falciparum detection with BL was inverse in EMBLEM countries (Uganda, Tanzania, and Kenya) and positive in Malawi. This was most likely a reflection of different detection methods — PCR versus blood microscopy/RDT —with different analytic sensitivities (10 parasites/ul versus 100–200 parasites/ul) [39]. Because parasites can remain detectable at low levels for up 365 days [40, 41], detection only by PCR likely reflects both heavier exposure and chronic low-grade infection as risk factors for BL.
Table 2.
Associations between childhood Burkitt lymphoma status and Plasmodium falciparum infection at enrollment
| Characteristic | OR (95% CI) * | p-value | OR (95% CI) ± | p-value |
|---|---|---|---|---|
| All Countries combined | 0·526 (0·438–0·633) | 9.64E-12 | 0·524 (0·435–0·630) | 7·29E-12 |
| Likelihood ratio test χ2 (p-value) ¶ | 6·11 (0·0134) | 6·16 (0·0131) | ||
| Uganda§ | 0·410 (0·299–0·562) | 3·09E-08 | 0·400 (0·291–0·549) | 1·493E-08 |
| Likelihood ratio test χ2 (p-value) ¶ | 3·12 (0·0774) | 2·96 (0·0853) | ||
| Kenya§ | 0·302 (0·203–0·449) | 3·53E-09 | 0·302 (0·203–0·450) | 3·81E-09 |
| Likelihood ratio test χ2 (p-value) ¶ | 4·55 (0·0329) | 4·73 (0·0296) | ||
| Tanzania§ | 0·451 (0·289–0·726) | 0·0010 | 0·452 (0·281–0·727) | 0·0011 |
| Likelihood ratio test χ2 (p-value) ¶ | 1·45 (0·2282) | 1·.43 (0·2315) | ||
| Malawi§§ | 1·793 (1·203–2·673) | 0·0041 | 1·829 (1·221–2·739) | 0·0034 |
| Likelihood ratio test χ2 (p-value) ¶ | 2·61 (0·1061) | 2·57 (0·1090) |
Model: BL case status~factor(P. falciparum)+ age+sex+pop-specific PC1+ pop-specific PC2+ pop-specific PC3+ factor(country)+[genetic relation matrix, random effect]. Country was not included in country-specific models.
Model: BL case status ~ factor(P. falciparum)+age+sex+pop-specific PC1+ pop-specific PC2+ pop-specific PC3+ factor(country)+[genetic relation matrix, random effect] + HBB-rs334(T)+ABO-rs8176703 (T). Country was not included in country-specific analyses.
χ2 goodness-of-fit (p-value) testing differences in nested models with P. falciparum (unrestricted model) versus without P. falciparum (restricted model).
P. falciparum infection was determined by thick film microscopy or rapid diagnostic tests. Analytic sensitivity of giemsa-stained blood films and rapid diagnostic test is 100–200 parasites/μl and may detect 55% of the infectious reservoir population (See methods)
P. falciparum infection was determined by polymerase chain reaction (PCR) in Malawi. Analytic sensitivity of PCR is 10 parasites per ul of blood and may detect 83% of the infectious reservoir population, including submicroscopic infections (See methods).
Exploratory analysis of associations of loci in malaria genes with Burkitt lymphoma
In exploratory analyses, using a modified Bonferroni procedure to correct for multiple comparisons after accounting for correlation between loci in the same gene (Table S2) [36], we observed suggestive associations for BL with 10 loci in two genes (ATP2B4 and PECAM1) (Table S10). Nine of the loci, all with elevated ORs for BL, were in ATP2B4 in a haplotype spanning a ~12 kb region, including ATP2B4-rs11808688(G) (OR=1.276 (95% CI 1.106–1.472; p = 0.0008), which has been associated with significantly decreased ATP2B4 gene expression in whole blood (z-score = −11.84, p = 2.40×10−32) [42]. The tenth locus associated with BL was PECAM1-rs45495798(A) with an OR for BL=1.366 (95% CI 1.160– 1.656; p= 0.0015, Table S10).
DISCUSSION
This study was conducted to investigate whether carriage of one copy of the abnormal HBB gene (the cause of SCT), which has been shown to be protective against BL in Uganda [24], remained protective in a large study with children from multiple countries. This association has been reported before [16–18], but not confirmed three other studies [19–21]. Our study controlled for multiple confounders, including P. falciparum, other malaria loci that affect malaria risk, and ancestry, which was not possible before. SCT is a reliable marker of resistance against P. falciparum burden [43] that has ben useful in identifying malaria phenotypes whose risk decreased in those with SCT-mediated genetic resistance against malaria [44]. For example inverse associations of SCT with invasive bacteremia [45], iron-deficiency anemia [46], and hypertension in African individuals [47] provided strong inferential evidence that malaria is linked to those phenotypes. Our findings here (i.e., that SCT reduces the risk for both BL and detection of P. falciparum) thereby demonstrate that BL is a phenotype of P. falciparum infection. Demonstration of this correlation has important translational value because it could justify closer coordination between malaria and pediatric cancer programs to increase awareness about the link between BL and malaria particularly at primary care centers where most patients are seen first [48].
Our observation that carriers of ABO-rs8176703(T) were at a lower risk of BL is intriguing because this variant has shown protection against severe malaria in Ghana [49]. However, in contrast to the SCT allele, this variant did not concordantly protect against P. falciparum detection among controls. Thus, it is not clear whether the effect of this ABO locus on BL reflects a protective effect on P. falciparum-susceptibility in those with blood group O erythrocytes [15, 50]. Such a mechanism would not be supported by the low linkage disequilibrium (r2=0.051) between ABO-rs8176703(T) and rs8176719(delG), the allele most correlated with blood group O status in the LWK population in the 1000 Genomes reference panel. However, such a mechanism is supported by the strong normalized disequilibrium coefficient (D′ =1.00) between them, which implies that both alleles residing on the same haplotype.
Our results for α-thalassemia with BL were not significant; however, this may not be surprising because although α-thalassemia genotypes are selected by malaria [51], the protective effect of α-thalassemia on malaria is less than that of SCT on malaria. Therefore, additional analysis of this relationship may require larger sample sizes to be observed against BL.
Our findings suggest that the effects of the 32 loci against BL and P. falciparum detection among controls are concordant but not equal. The inequality of effects fits the classical malaria model for BL [12], which implies a complex interplay of multiple factors, including EBV [2]. It also fits with different mechanisms of genetic control of malaria. For example, the effects of loci in IL10 against BL and P. falciparum infection among controls were concordant in direction but significant only for P. falciparum infection. This unequal pattern of effects suggests that some loci may have stronger effects on aspects of malaria pathophysiology (e.g., such as fever and cytokine storm), which was the basis of their selection, but weaker effects on direct P. falciparum infection control, which is the relevant biological risk factor for BL. It is possible that some host genes, such as IL10 loci, are selected because they reduce severe malaria symptoms due to fever and inflammation without directly reducing parasitemia. If so, such loci may protect individuals from severe malaria death without an equal impact on recurrent asymptomatic P. falciparum infections, which persist as multiclonal infections from super- or supra-infection [52, 53].
Our finding of strong inverse association between BL and P. falciparum detection agrees with our previous results [27, 28] and those reported by others [54, 55]. We note that this seeming paradox points to the importance of host immune responses against P. falciparum‒i.e., natural acquired immunity to host survival by mediating clearance of a parasite clones [56] before developing BL and to a possible mechanism of BL pathogenesis. Specifically, children who develop BL, typically at approximately age 7 as in the current study, survive malaria in no small part thanks to NAI, i.e., an immune response that senses, reacts to, and controls parasitemia [56, 57]. NAI initially develops quickly against severe malaria, after the first one or two infections, [58, 59] then increases progressively to suppress symptoms (i.e., anti-disease immunity), and then P. falciparum infection (i.e., anti-parasite immunity) [60, 61]. NAI is mediated by circulating memory B cells, which represent a T-helper 2 (Th2) response characterized by up-regulation of antibody production to fight extracellular organisms [62], including P. falciparum. NAI against P. falciparum transmission is durable [63] and the half-life of the produced antibodies can range between several years to life-long persistence [64, 65]. It is possible that children prone to BL develop strong Th-2 type anti-inflammatory response, mediated by cytokines such as interleukin 10 [66], both controls parasitemia to limit severe disease while increasing tolerance of low levels of parasitemia without triggering severe symptoms [62].
We note that NAI could also be modulated by co-infection with EBV, which encodes BCRF-1, a viral interleukin-10-homolog [67] with capacity to down-regulate Th1-cytokines while upregulating Th-2 cytokines [68]. Because EBV infection occurs by 6 months in ~35% of children in areas where BL is endemic [69, 70], it has been suggested that EBV may protect against severe malaria via interleukin 10 mimicry [71], while the low levels of P. falciparum parasitemia promote BL by upregulating EBV infection [72]. Interestingly, in a recent study of BL cases reported localized disease with prominent granulomatous reaction and an unexplained high propensity for regression despite receiving none or minimal treatment [73]. These cases, exhibiting tumor cellular features characterized by Th1 lymphocytes and M1 macrophages, which is different from the usual Th2 phenotype described above, highlighted the importance of host immunity in BL pathogenesis. Is it possible that while most BL cases develop under a Th2-type response, a small fraction of BL cases with Th1 type response might be transient or have a good prognosis [73]. Additional research is needed to identify the close interaction between oncogenic factors, host immunity, including NAI that could be permissive for BL development or conversion to Th1-type response, which promotes BL regression or indolence.
The strengths of our study include the largest study conducted to date and the first multicenter study of BL, with confounder data, including P. falciparum detection, prior history of malaria and treatment, and genetically defined ancestry and relatedness. The limitations include incomplete histological verification of BL cases (about one quarter of cases were not confirmed), which could introduce biases. However, our results are within range of current capacity in poor resource settings where histological services are not always readily available. We also note that our analysis did not include malaria loci on the X chromosome (e.g., G6PD and CD40LG) as well as those that could not be imputed from GWAS data (e.g., EPB41, GYPB, and FREM3/GYPE). Our results for SNPs in the 22 malaria genes are exploratory but provide a useful baseline. Our inferences about NAI are not based on immune markers, but identify NAI as an important concept to be explored in BL. Finally, parasite evolution may be a factor. Recent discoveries indicate that certain parasite strains, such as those with P. falciparum sickle-cell associated alleles (Pfsa), have evolved to infect and cause severe malaria in individuals with SCT [74]. These discoveries suggest that parasite factors should be considered in future studies of BL.
In conclusion, we report that SCT is protective both against BL and against P. falciparum detection in controls when controlling for confounders. In addition, we observed protective associations between ABO-rs8176703(T) loci and BL, but not against infection. Our results increase support for the hypothesis that BL is a phenotype of P. falciparum. This inference opens a window to encourage closer coordination between malaria and pediatric oncology programs to improve the control of BL in malaria-endemic areas in Africa.
Supplementary Material
Acknowledgement
We thank the study population and communities for their participation. We thank Ms. Janet Lawler-Heavener at Westat Inc, (Rockville, MD, USA) and Mr. Erisa Sunday at the African Field Epidemiology Network (Kampala, Uganda) for managing the study. We are grateful to Daniel Shriner at NHGRI (Bethesda, Maryland) for serving as Mateus H. Gouveia’s mentor and for assistance with statistical genetics and computational analyses. We are grateful to the leadership of the collaborating countries and institutions for hosting local field offices and laboratories and supporting the fieldwork. We thank Ms. Laurie Buck, Dr. Carol Giffen, Mr. Greg Rydzak and Mr. Jeremy Lyman at Information Management Services Inc. (Calverton, MD, USA) for coordinating data, and preparing data analysis files, and David Check of the Division of Cancer Epidemiology and Genetics, National Cancer Institute (Bethesda, Maryland) for drawing forest plots.
Funding statement
The study was funded by the Intramural Research Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI) (Contracts HHSN261201100063C and HHSN261201100007I), Cancer Genomics Research Laboratory (75N910D00024), National Institute of Allergy and Infectious Diseases (SJR), the Sickle Cell Branch, National Heart, Lung and Blood Institute; and Center for Research on Genomics and Global Health (Z01HG200362), National Human Genome Research Institute, National Institutes of Health (NIH), Department of Health and Human Services.
Support
The study was funded by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute (NCI) (Contracts HHSN261201100063C and HHSN261201100007I), the Intramural Research Program, National Institute of Allergy and Infectious Diseases (SJR), and the Intramural Research Program, Sickle Cell Branch, National Heart, Lung and Blood Institute; Center for Research on Genomics and Global Health (Z01HG200362), National Human Genome Research Institute; National Institutes of Health (NIH), Department of Health and Human Services. The authors acknowledge the research contributions of the Cancer Genomics Research Laboratory for their expertise, execution, and support of this research in the areas of project planning, wet laboratory processing of specimens, and bioinformatics analysis of generated data. from the NCI, NIH, under NCI Contract No. 75N910D00024. We acknowledge support to FB from the NCI Continuing Umbrella of Research Experiences (NCI-iCURE). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The content of this manuscript is the sole responsibility of the authors.
Footnotes
Ethical approvals and patient consent
Ethical approvals for the EMBLEM study were granted by the Uganda Virus Research Institute (GC/127), Uganda National Council for Science and Technology (H816), Tanzania National Institute for Medical Research (NIMR/HQ/R.8c/Vol. IX/1023), Moi University/Moi Teaching and Referral Hospital (000536), and National Cancer Institute (10-C-N133). Ethical approvals for the Childhood Infections and Cancer study were granted by Malawi College of Medicine (P.03/04/277R), Oxford University, and National Health Sciences Research Committee (2405). Guardians of participants provided written informed consent. Children aged ≥7 years old gave written assent.
Permission to reproduce material from other sources: not applicable.
Clinical trial registration. Not applicable.
DISCLOSURE OF CONFLICT OF INTEREST
None declared
Conflict of interest: The authors declare no competing financial interests.
Data availability
The genetic data will be deposited in dbGaP at: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001705.v1.p1. Access to covariate data for EMBLEM and Malawi studies can be requested directly from the corresponding author. The study forms are available for download from the EMBLEM website at: https://emblem.cancer.gov/resources/index.html.
REFERENCES
- 1.Mbulaiteye SM, Devesa SS. Burkitt Lymphoma Incidence in Five Continents. Hemato 2022;3:434–453. [Google Scholar]
- 2.López C, Burkhardt B, Chan JKC, et al. Burkitt lymphoma. Nat Rev Dis Primers 2022;8:78. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter LM, Newton R, Casabonne D, et al. Antibodies against malaria and Epstein-Barr virus in childhood Burkitt lymphoma: a case-control study in Uganda. Int J Cancer 2008;122:1319–1323. [DOI] [PubMed] [Google Scholar]
- 4.Mutalima N, Molyneux E, Jaffe H, et al. Associations between Burkitt lymphoma among children in Malawi and infection with HIV, EBV and malaria: results from a case-control study. PloS one 2008;3:e2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aka P, Vila MC, Jariwala A, et al. Endemic Burkitt lymphoma is associated with strength and diversity of Plasmodium falciparum malaria stage-specific antigen antibody response. Blood 2013;122:629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derkach A, Otim I, Pfeiffer RM, et al. Associations between IgG reactivity to Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) antigens and Burkitt lymphoma in Ghana and Uganda case-control studies. Ebiomedicine 2019;39:358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston WT, Mutalima N, Sun D, et al. Relationship between Plasmodium falciparum malaria prevalence, genetic diversity and endemic Burkitt lymphoma in Malawi. Scientific reports 2014;4:3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arisue N, Chagaluka G, Palacpac NMQ, et al. Assessment of Mixed Plasmodium falciparum sera5 Infection in Endemic Burkitt Lymphoma: A Case-Control Study in Malawi. Cancers (Basel) 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Proceedings of the IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Epstein-Barr Virus and Kaposi’s Sarcoma Herpesvirus/Human Herpesvirus 8. Lyon, France, 17–24 June 1997. IARC Monogr Eval Carcinog Risks Hum 1997;70:1–492.9705682 [Google Scholar]
- 10.Thomas N, Dreval K, Gerhard DS, et al. Genetic subgroups inform on pathobiology in adult and pediatric Burkitt lymphoma. Blood 2023;141:904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robbiani DF, Deroubaix S, Feldhahn N, et al. Plasmodium Infection Promotes Genomic Instability and AID-Dependent B Cell Lymphoma. Cell 2015;162:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torgbor C, Awuah P, Deitsch K, et al. A multifactorial role for P. falciparum malaria in endemic Burkitt’s lymphoma pathogenesis. PLoS Pathog 2014;10:e1004170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mbulaiteye SM, Talisuna AO, Ogwang MD, et al. African Burkitt’s lymphoma: could collaboration with HIV-1 and malaria programmes reduce the high mortality rate? Lancet 2010;375:1661–1663. [DOI] [PubMed] [Google Scholar]
- 14.Bouvard V, Baan RA, Grosse Y, et al. Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol 2012;13:339–340. [DOI] [PubMed] [Google Scholar]
- 15.Billo MA, Johnson ES, Doumbia SO, et al. Sickle cell trait protects against Plasmodium falciparum infection. Am J Epidemiol 2012;176 Suppl 7:S175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams AO. Haemoglobin genotypes, ABO blood groups, and Burkitt’s tumour. J Med Genet 1966;3:177–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hesseling PB, Jam DT, Palmer DD, et al. Burkitt’s lymphoma patients in Northwest Cameroon have a lower incidence of sickle cell trait (Hb AS) than healthy controls. S Afr Med J 2016;106:10693. [DOI] [PubMed] [Google Scholar]
- 18.Ahamed SG, Ibrahim UA, Kagu MB. Synergistic Protective Effect of Sickle Cell Trait and Blood Group-O on the Risk of Endemic Burkitt’s Lymphoma. Gulf J Oncolog 2018;1:11–16. [PubMed] [Google Scholar]
- 19.Pike MC, Morrow RH, Kisuule A, Mafigiri J. Burkitt’s lymphoma and sickle cell trait. Br J Prev Soc Med 1970;24:39–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nkrumah FK, Perkins IV. Sickle cell trait, hemoglobin C trait, and Burkitt’s lymphoma. Am J Trop Med Hyg 1976;25:633–636. [DOI] [PubMed] [Google Scholar]
- 21.Mulama DH, Bailey JA, Foley J, et al. Sickle cell trait is not associated with endemic Burkitt lymphoma: an ethnicity and malaria endemicity-matched case-control study suggests factors controlling EBV may serve as a predictive biomarker for this pediatric cancer. Int J Cancer 2014;134:645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouveia MH, Bergen AW, Borda V, et al. Genetic signatures of gene flow and malaria-driven natural selection in sub-Saharan populations of the “endemic Burkitt Lymphoma belt”. PLoS genetics 2019;15:e1008027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackinnon MJ, Mwangi TW, Snow RW, et al. Heritability of malaria in Africa. PLoS Med 2005;2:e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Legason ID, Pfeiffer RM, Udquim KI, et al. Evaluating the Causal Link Between Malaria Infection and Endemic Burkitt Lymphoma in Northern Uganda: A Mendelian Randomization Study. EBioMedicine 2017;25:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams TN, Mwangi TW, Wambua S, et al. Negative epistasis between the malaria-protective effects of alpha+-thalassemia and the sickle cell trait. Nature genetics 2005;37:1253–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta AK, Kirchner KA, Nicholson R, et al. Effects of alpha-thalassemia and sickle polymerization tendency on the urine-concentrating defect of individuals with sickle cell trait. J Clin Invest 1991;88:1963–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peprah S, Ogwang MD, Kerchan P, et al. Risk factors for Burkitt lymphoma in East African children and minors: A case-control study in malaria-endemic regions in Uganda, Tanzania and Kenya. Int J Cancer 2020;146:953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peprah S, Ogwang MD, Kerchan P, et al. Inverse association of falciparum positivity with endemic Burkitt lymphoma is robust in analyses adjusting for pre-enrollment malaria in the EMBLEM case-control study. Infect Agent Cancer 2021;16:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith T, Felger I, Tanner M, Beck HP. Premunition in Plasmodium falciparum infection: insights from the epidemiology of multiple infections. Trans R Soc Trop Med Hyg 1999;93 Suppl 1:59–64. [DOI] [PubMed] [Google Scholar]
- 30.Smith T, Beck HP, Kitua A, et al. Age dependence of the multiplicity of Plasmodium falciparum infections and of other malariological indices in an area of high endemicity. Trans R Soc Trop Med Hyg 1999;93 Suppl 1:15–20. [DOI] [PubMed] [Google Scholar]
- 31.Broen K, Dickens J, Trangucci R, et al. Burkitt lymphoma risk shows geographic and temporal associations with Plasmodium falciparum infections in Uganda, Tanzania, and Kenya. Proc Natl Acad Sci U S A 2023;120:e2211055120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maziarz M, Nabalende H, Otim I, et al. A cross-sectional study of asymptomatic Plasmodium falciparum infection burden and risk factors in general population children in 12 villages in northern Uganda. Malaria journal 2018;17:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allison AC. The distribution of the sickle-cell trait in East Africa and elsewhere, and its apparent relationship to the incidence of subtertian malaria. Trans R Soc Trop Med Hyg 1954;48:312–318. [DOI] [PubMed] [Google Scholar]
- 34.Gouveia MH, Otim I, Ogwang MD, et al. Endemic Burkitt Lymphoma in second-degree relatives in Northern Uganda: in-depth genome-wide analysis suggests clues about genetic susceptibility. Leukemia 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leal TP, Furlan VC, Gouveia MH, et al. NAToRA, a relatedness-pruning method to minimize the loss of dataset size in genetic and omics analyses. Comput Struct Biotechnol J 2022;20:1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos E, Chen G, Shriner D, et al. Replication of genome-wide association studies (GWAS) loci for fasting plasma glucose in African-Americans. Diabetologia 2011;54:783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horowitz HW. Fever of unknown origin or fever of too many origins? N Engl J Med 2013;368:197–199. [DOI] [PubMed] [Google Scholar]
- 38.Band G, Le QS, Clarke GM, et al. Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa, Asia and Oceania. Nature Communications 2019;10:5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bousema T, Okell L, Felger I, Drakeley C. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nature Reviews Microbiology 2014;12:833–840. [DOI] [PubMed] [Google Scholar]
- 40.Jeffery GM, Eyles DE. The Duration in the Human Host of Infections with a Panama Strain of Plasmodium Falciparum. The American Journal of Tropical Medicine and Hygiene 1954;3:219–224. [DOI] [PubMed] [Google Scholar]
- 41.Wångdahl A, Bogale RT, Eliasson I, et al. Malaria parasite prevalence in Sub-Saharan African migrants screened in Sweden: a cross-sectional study. Lancet Reg Health Eur 2023;27:100581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westra HJ, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nature genetics 2013;45:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uyoga S, Macharia AW, Ndila CM, et al. The indirect health effects of malaria estimated from health advantages of the sickle cell trait. Nature Communications 2019;10:856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rockett KA, Clarke GM, Fitzpatrick K, et al. Reappraisal of known malaria resistance loci in a large multicenter study. Nature Genetics 2014;46:1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor SM, Parobek CM, Fairhurst RM. Haemoglobinopathies and the clinical epidemiology of malaria: a systematic review and meta-analysis. The Lancet Infectious Diseases 2012;12:457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muriuki JM, Mentzer AJ, Mitchell R, et al. Malaria is a cause of iron deficiency in African children. Nature Medicine 2021;27:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Etyang AO, Smeeth L, Cruickshank JK, Scott JA. The Malaria-High Blood Pressure Hypothesis. Circ Res 2016;119:36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mawalla WF, Morrell L, Chirande LF, et al. Treatment Delays in Children and Young Adults with Lymphoma: report from an East Africa Lymphoma Cohort Study. Blood Adv 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmann C, Thye T, Vens M, et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature 2012;489:443–446. [DOI] [PubMed] [Google Scholar]
- 50.Rowe JA, Handel IG, Thera MA, et al. Blood group O protects against severe Plasmodium falciparum malaria through the mechanism of reduced rosetting. Proc Natl Acad Sci U S A 2007;104:17471–17476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flint J, Hill AV, Bowden DK, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature 1986;321:744–750. [DOI] [PubMed] [Google Scholar]
- 52.Zhong D, Koepfli C, Cui L, Yan G. Molecular approaches to determine the multiplicity of Plasmodium infections. Malaria Journal 2018;17:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wesolowski A, Taylor AR, Chang HH, et al. Mapping malaria by combining parasite genomic and epidemiologic data. BMC Med 2018;16:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de-The G, Geser A, Day NE, et al. Epidemiological evidence for causal relationship between Epstein-Barr virus and Burkitt’s lymphoma from Ugandan prospective study. Nature 1978;274:756–761. [DOI] [PubMed] [Google Scholar]
- 55.Asito AS, Piriou E, Odada PS, et al. Elevated anti-Zta IgG levels and EBV viral load are associated with site of tumor presentation in endemic Burkitt’s lymphoma patients: a case control study. Infect Agent Cancer 2010;5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bull PC, Lowe BS, Kortok M, et al. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat Med 1998;4:358–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Donnell RA, de Koning-Ward TF, Burt RA, et al. Antibodies against merozoite surface protein (MSP)-1(19) are a major component of the invasion-inhibitory response in individuals immune to malaria. J Exp Med 2001;193:1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goncalves BP, Huang CY, Morrison R, et al. Parasite burden and severity of malaria in Tanzanian children. N Engl J Med 2014;370:1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodriguez-Barraquer I, Arinaitwe E, Jagannathan P, et al. Quantifying Heterogeneous Malaria Exposure and Clinical Protection in a Cohort of Ugandan Children. J Infect Dis 2016;214:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner L, Lavstsen T, Mmbando BP, et al. IgG antibodies to endothelial protein C receptor-binding cysteine-rich interdomain region domains of Plasmodium falciparum erythrocyte membrane protein 1 are acquired early in life in individuals exposed to malaria. Infection and immunity 2015;83:3096–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eldh M, Hammar U, Arnot D, et al. Multiplicity of Asymptomatic Plasmodium falciparum Infections and Risk of Clinical Malaria: A Systematic Review and Pooled Analysis of Individual Participant Data. J Infect Dis 2020;221:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kidd P Th1/Th2 balance: the hypothesis, its limitations, and implications for health and disease. Altern Med Rev 2003;8:223–246. [PubMed] [Google Scholar]
- 63.Wipasa J, Suphavilai C, Okell LC, et al. Long-lived antibody and B Cell memory responses to the human malaria parasites, Plasmodium falciparum and Plasmodium vivax. PLoS Pathog 2010;6:e1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fowkes FJ, McGready R, Cross NJ, et al. New insights into acquisition, boosting, and longevity of immunity to malaria in pregnant women. J Infect Dis 2012;206:1612–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Drakeley CJ, Corran PH, Coleman PG, et al. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci U S A 2005;102:5108–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howard M, O’Garra A. Biological properties of interleukin 10. Immunol Today 1992;13:198–200. [DOI] [PubMed] [Google Scholar]
- 67.Burdin N, Peronne C, Banchereau J, Rousset F. Epstein-Barr virus transformation induces B lymphocytes to produce human interleukin 10. J Exp Med 1993;177:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsu DH, de Waal Malefyt R, Fiorentino DF, et al. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science 1990;250:830–832. [DOI] [PubMed] [Google Scholar]
- 69.Reynaldi A, Schlub TE, Chelimo K, et al. Impact of Plasmodium falciparum Coinfection on Longitudinal Epstein-Barr Virus Kinetics in Kenyan Children. J Infect Dis 2016;213:985–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piriou E, Asito AS, Sumba PO, et al. Early age at time of primary Epstein-Barr virus infection results in poorly controlled viral infection in infants from Western Kenya: clues to the etiology of endemic Burkitt lymphoma. J Infect Dis 2012;205:906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watier H, Auriault C, Capron A. Does Epstein-Barr virus infection confer selective advantage to malaria-infected children? Lancet 1993;341:612–613. [DOI] [PubMed] [Google Scholar]
- 72.Lam KM, Syed N, Whittle H, Crawford DH. Circulating Epstein-Barr virus-carrying B cells in acute malaria. Lancet 1991;337:876–878. [DOI] [PubMed] [Google Scholar]
- 73.Granai M, Lazzi S, Mancini V, et al. Burkitt lymphoma with a granulomatous reaction: an M1/Th1-polarised microenvironment is associated with controlled growth and spontaneous regression. Histopathology 2022;80:430–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Band G, Leffler EM, Jallow M, et al. Malaria protection due to sickle haemoglobin depends on parasite genotype. Nature 2022;602:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic data will be deposited in dbGaP at: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001705.v1.p1. Access to covariate data for EMBLEM and Malawi studies can be requested directly from the corresponding author. The study forms are available for download from the EMBLEM website at: https://emblem.cancer.gov/resources/index.html.


