Summary
The regulation of polymorphonuclear leukocyte (PMN) function by mechanical forces encountered during their migration across restrictive endothelial cell junctions is not well understood. Using genetic, imaging, microfluidic and in vivo approaches, we demonstrated that the mechanosensor Piezo1 in PMN plasmalemma induced spike like Ca2+ signals during trans-endothelial migration. Mechanosensing increased the bactericidal function of PMN entering tissue. Mice genetically deleted of Piezo1 in PMN were defective in clearing bacteria and the lungs were predisposed to severe infection. Adoptive transfer of Piezo1-activated PMN into lungs of Pseudomonas aeruginosa infected mice, or exposing PMN to defined mechanical forces in microfluidic systems, improved bacterial clearance phenotype of PMN. Piezo1 transduced the mechanical signals activated during transmigration to upregulate NADPH oxidase 4, crucial for the increased PMN bactericidal activity. Thus, Piezo1 mechanosensing of increased PMN tension while traversing the narrow endothelial adherens junctions is a central mechanism activating the host-defense function of transmigrating PMN.
Graphical Abstract
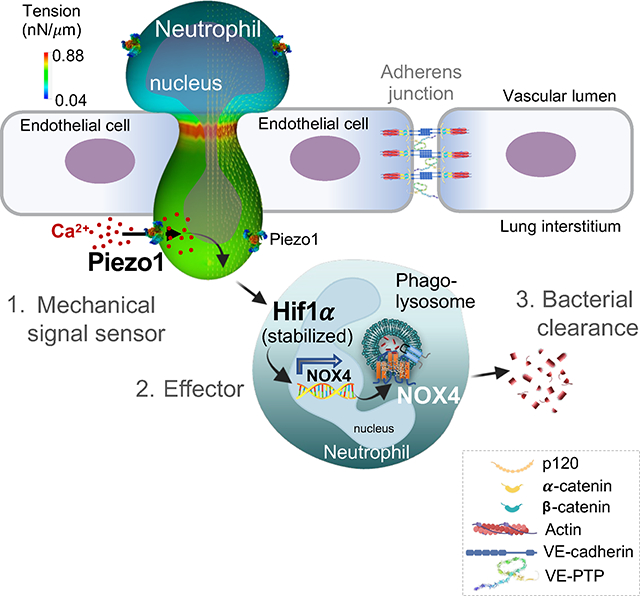
In brief
PMN migration through restrictive endothelial junction generates mechanical tension on the PMN plasma membrane, but its impact on PMN function or immune response is unknown. Mukhopadhyay et. al., showed that Piezo1 mediated mechanosensing during transmigration, enhanced bacterial killing function of PMN in tissue via NOX4 and prevented inflammatory tissue injury.
Introduction
Polymorphonuclear leukocytes (PMN) generate reactive oxygen species (ROS) and serve as the first line of defense against pathogens such as bacteria and fungi 1,2. PMN are particularly important for the lung’s defense function in response to air borne irritants and pathogens 3–5. Circulating PMN receiving chemoattractant signals from infected tissue are mobilized in the lung interstitium and airspace upon transmigrating through the restrictive endothelial adherens junctions (AJs) and alveolar epithelial barrier 6,7. Due to the large PMN diameter ranging from 8.7 to13 μm in humans 8–11 relative to the narrow slit-like “pores” of AJs (mean diameter of 4nm in most continuous endothelial barriers) 7,12, the PMN plasma membrane breaching the AJs experiences unusually high tension and stretching during the transmigration event 13. We surmised that physical constraints at the junctions may influence PMN transmigration and functional properties such as their fitness for host-defense and bacterial killing 14.
The trans-endothelial junctional route is responsible for approximately 90% of PMN migration across continuous endothelial barriers 15,16. An essential host-defense function of PMN is to activate the phagocytic machinery and kill bacterial pathogens through proteases and ROS production 17. Phagocytosis is a multifaceted process consisting of pathogen recognition, engulfment, and elimination 18,19. Bacterial killing is driven largely by nicotinamide adenine dinucleotide phosphate (NADPH) oxidases 20 and maturation of phagosomes into phagolysosomes, the organelles with oxidative and lytic properties 18,19. PMN express several NADPH oxidase isoforms NOX1, NOX2 and NOX4 using NADPH to generate ROS 20. The function of redox signaling in the mechanism of PMN innate immune function via NOX2 was described in patients with chronic granulomatous disease 21,22,23 who are highly susceptible to bacterial and fungal infections 24,25,26. While the canonical mechanism of Nox2 activation in PMN bactericidal function is understood 27, less is known about other NOX isoforms in the PMN anti-bactericidal function.
Here we used multiple approaches to investigate the role of mechanical signals generated during PMN migration across the restrictive endothelial junction barrier in activating the PMN host-defense function. We demonstrated that mechanical forces acting on PMN sensed by Piezo1 28,29 during transmigration induced the expression of Nox4, a constitutively active NADPH oxidase producing hydrogen peroxide 30,31,32. In contrast Nox2 was not altered. These findings show the immune regulatory role of restrictive endothelial junctions in promoting PMN bactericidal function through activation of Piezo1-Nox4 signaling pathway in the PMN entering tissue. Thus, mechanical activation of Piezo1 expressed in PMN during their passage through the restrictive space may be a useful therapeutic strategy for treating inflammatory diseases such as acute respiratory distress syndrome (ARDS) through amplifying the PMN bactericidal function.
Results
Adherens junctions (AJs) enhance bactericidal function of transmigrating PMN
To determine whether transmigration of PMN via junctions of the endothelial cell monolayer lining blood vessels 33,34,35 is responsible for a PMN phenotype shifts, we distinguished between transmigrated and non-transmigrated PMN in mice using the approach in 36 (Figure 1a). We used genetically targeted CatchupIVM-red mice co-expressing Cre recombinase along with the fluorescent protein td-Tomato under the control of PMN-specific locus Ly6G 37 (Suppl. Figure 1a). Following insufflation of lipopolysaccharide (LPS) in mice to induce PMN transmigration from lung microvessels into extravascular space, we observed that tdTomato+ PMN accumulated in both vessels and lung tissue (Figure 1a–c; Suppl. Figure 1b, c). The vascular resident non-transmigrating PMN were intravitally stained with α-Ly6G antibody conjugated to Brilliant-violent (BV421) fluorophore injected i.v. 2 min prior to euthanasia (Figure 1b, Suppl Figure 1c). Using fluorescence-activated cell sorting (FACS), we demonstrated that both peripheral blood and lung intravascular PMN isolated from control mice were positive for td-Tomato stained with α-Ly6G-BV421 antibody (Suppl. Figure 1c–e). Exposure of mice to LPS, however, increased accumulation of td-Tomato+ PMN as evident by the cells that did not stain with α-Ly6G-BV421 antibody (Figure 1c, Suppl. Figure 1c). The latter staining enabled the distinction between the transmigrated PMN population and the PMN remaining in microvessels (Suppl. Figure 1c). Analysis of PMN transmigration in mice exposed to LPS showed that recruitment of PMN into lung tissue occurred as early as 15 min post-LPS (the earliest time it could be assessed) and the number of PMN in lung tissue continued to increase over the next 60 min (Figure 1c; Suppl. Figure 1c).
Figure 1. PMN transmigration in endothelial AJs promotes PMN bactericidal activity.
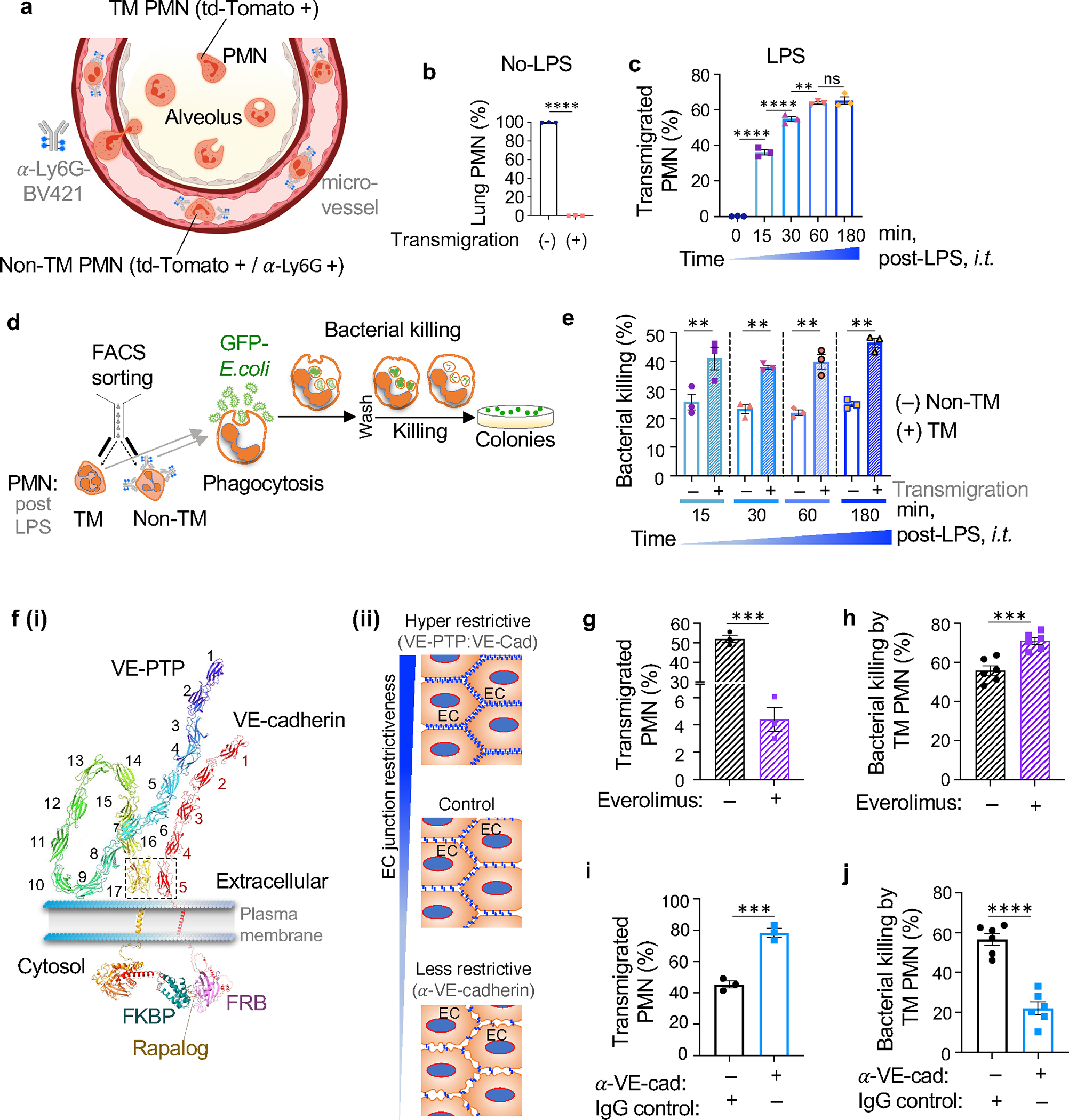
a. Schematic of the method used to separate transmigrated and non-transmigrated PMN across lung micro-vessels. All PMN from Ly6GCre td-Tomato +/− mice expressed td-Tomato as shown in Suppl Fig 1a. Intravascular PMN were additionally labelled by α-Ly6G-BV421 antibody injected i.v. 2m prior to euthanasia. The number of extravasated td-Tomato PMN relative to intravascular PMN stained with α-Ly6G-BV421 was calculated to quantify trans-vascular PMN migration. b-c. Percentage transmigrated (TM) and non-transmigrated (non-TM) PMN assessed by FACS analysis in naïve mice and mice exposed to 5mg/ml insufflated (i.t.) LPS for different times (as shown in Suppl. Figure 1c). d. Workflow to assess bactericidal activity of transmigrated vs. non-transmigrated PMN. Flow sorted transmigrated and non-transmigrated lung PMN were incubated with GFP-E.coli for 1.5h determine bacteria phagocytosis and number of live bacteria was determined at 3.5h after bacteria washout. e. Bactericidal activity represented as % of killed GFP-E.coli normalized to phagocytosed GFP-E.coli (Suppl Figure 1f) of transmigrated vs. non-transmigrated lung PMN obtained at different times post-LPS challenge as in c. At each time point, transmigrated PMN induced 2-fold increase in bacterial killing. Data are obtained from 3 mice per group from 3 independent experiments. f(i-ii). (i) Model demonstrating the method of increasing the restrictiveness of endothelial AJs through expression of engineered VE-cadherin-FK506 and VE-PTP-FRB fusion proteins in mice as in 38; this method stabilized VE-cadherin-VE-PTP interaction, and increased junction barrier restrictiveness on treating with the drug everolimus. (ii) Schema of the approaches used to either increase or decrease restrictiveness of AJs. g-h. Percent of transmigrated PMN into lungs (g) and bactericidal activity of transmigrated PMN (h) in transgenic mice treated with everolimus (“+”) or vehicle control (“−“). Bactericidal activity of PMN transmigrating through restrictive AJs of mice treated with everolimus was greater than control. i-j. Percent of transmigrated PMN into lungs (i) and bactericidal activity of transmigrated lung PMN (j) of C57B6 mice treated with isotype-matched antibody as well as functional blocking α-VE-cadherin antibody used to disassemble AJs. PMN bactericidal activity was reduced in lungs after α-VE-cadherin antibody-induced opening of AJs. Data are obtained from 3–6 mice per group from 3 independent experiments. Additional information is provided as Suppl. Figure 1.
We next studied E. coli clearance via phagosomes in transmigrated PMN (Figure 1d). Assessment of FACS-sorted lung extravasated PMN showed that they possessed twice the GFP-E. coli clearance capacity as compared to non-transmigrated PMN, and unlike non-transmigrated PMN, the response persisted up to 180 min post LPS (Figure 1e). In contrast, we observed no difference in phagocytosis between transmigrated and non-transmigrated PMN at any time post-LPS (Suppl. Figure 1f). The increased clearance of GFP-E.coli seen in Figure 1e was unlikely due to inactivation of Ly6G used to detect phagocytosis since exposure of PMN to α-Ly6G antibody for periods up to 15 min had no effect on anti-microbial activity of PMN (Suppl. Figure 1g). These results together show that transmigration of PMN through the restrictive endothelial AJs augmented the bactericidal function of the PMN.
To address genetically the role of restrictiveness of AJs in activating PMN bactericidal function, we enhanced the AJ barrier using a transgenic mouse model 38 expressing an engineered construct containing both VE-cadherin-FK506 and VE-PTP-FRB fusion proteins under the control of the endogenous VE-cadherin promoter (Figure f (i)). Endothelial junctions became far more restrictive after treatment of mice with the rapamycin analog everolimus that irreversibly stabilized the interaction between VE-cadherin-FK506 and the interacting VE-cadherin associated protein tyrosine phosphatase VE-PTP-FRB 38,39. This mechanism of increasing endothelial barrier restrictiveness is based on the described interaction between extracellular [fibronectin type III (FNIII)] domain 17 of VE-PTP and domain 5 (EC-V) of VE-cadherin 40. Using this model to study the role of increased endothelial barrier restrictiveness on transmigrating PMN [Figure 1f(ii)], we observed markedly reduced number of transmigrating PMN (Figure 1g) whereas bactericidal function of PMN exiting the barrier was enhanced (Figure 1h). This finding is consistent with the crucial role of endothelial junctional barrier in promoting PMN bactericidal function. Conversely, the opening of AJs using anti-VE-cadherin blocking antibody (clone 11D4.1) to make the barrier more permissive 41 increased the number transmigrating PMN (Figure 1i) while reducing their bactericidal activity (Figure 1j). The number of phagocytosed bacteria (Suppl. Figure 1f) was not different between the experimental groups (Suppl. Figure 1h–i) in contrast to their bacterial clearance mechanism (Figure 1h, j). In control experiments, we showed that neither treating PMN with high concentrations of everolimus nor adding supernatant of endothelial monolayers incubated with α-VE-cadherin blocking antibody independently altered the antimicrobial function of PMN (Suppl. Figure 1j–k). Together these experiments showed that the restrictive nature of the endothelial junctions is a key determinant of augmented bactericidal function of transmigrated PMN.
PMN migration through restricted microfluidic pore system promotes bactericidal function
We next used a microfluidic approach to test whether PMN transmigration via known pore diameters could induce changes in PMN phenotype similar to transmigration in AJs above. PMN were perfused in this system through either 5μm or 200μm diameter pores (Figure 2a). PMN passing through 5μm pores showed enhanced bacterial killing as compared to PMN migrating through the 200μm pores (Figure 2b) whereas no difference was seen in bacterial phagocytosis in either pore dimensions following PMN transmigration (Suppl. Figure 2a). We next tested whether the chemoattractant N-formyl-methionyl-leucyl-phenylalanine (fMLP)-induced transmigration of PMN through the Transwell membrane containing pores of different diameters also influenced the antimicrobial activity of PMN. Results showed that PMN migration across pores in this system was a direct function of the pore diameter (Figure 2d). PMN transmigrating through smaller diameter pores effectively killed E. coli as compared to transmigration through larger pores (Figure 2d,e). In control experiments (Suppl. Figure 2b), PMN exposed to 1μM fMLP in the absence of passage through pores showed no such difference in killing of E. coli, highlighting the importance of restrictive pores in mediating the bactericidal killing function of PMN.
Figure 2. PMN transmigration via restrictive pores in a microfluidic system enhances bactericidal activity of PMN.
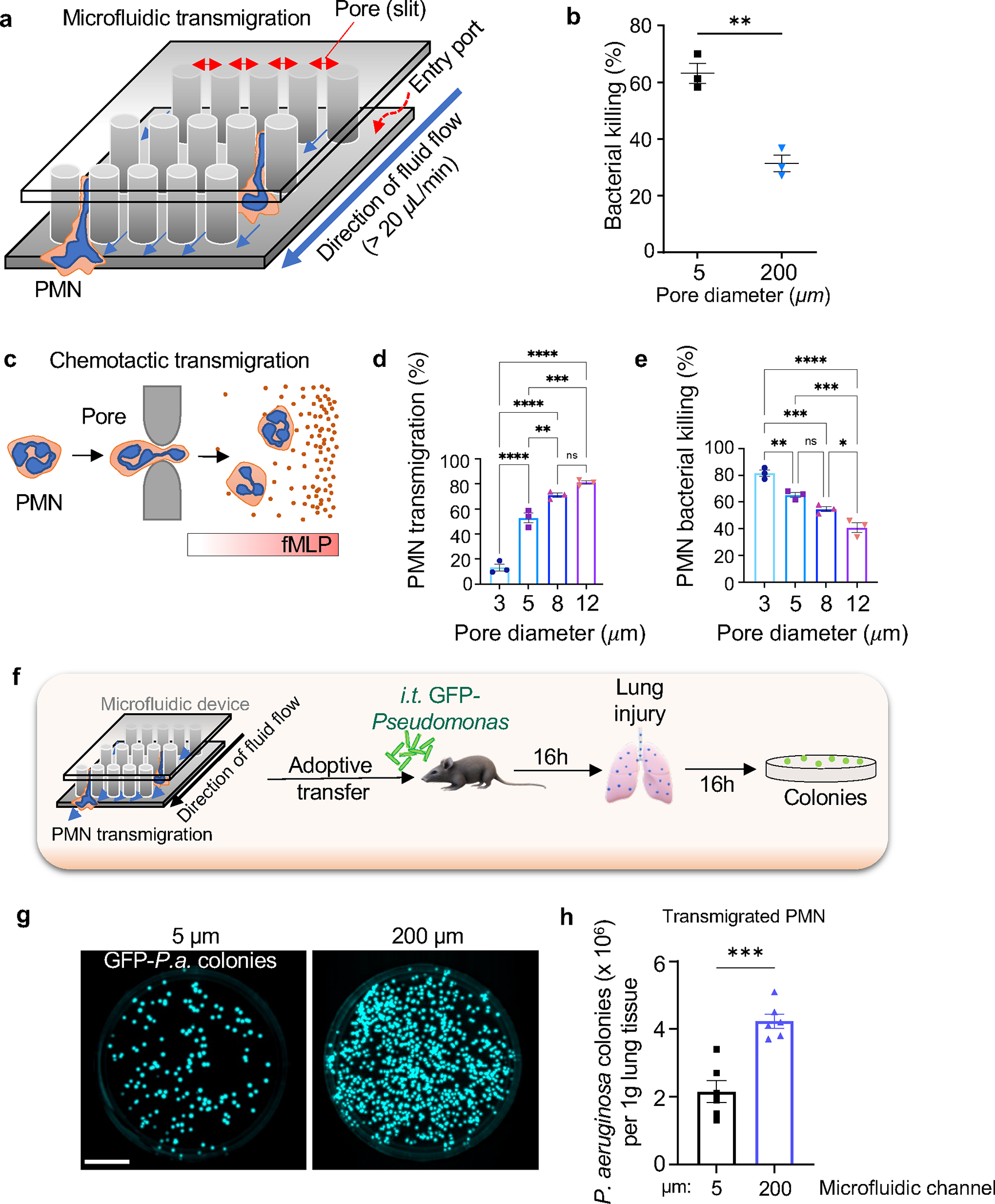
a. Design of the microfluidic system containing pores through which PMN transmigrated. b. Bone-marrow murine PMN transmigrated through 5μm diameter pores showed 2-fold greater E. coli killing ex vivo as compared to PMN transmigrating through 200μm diameter pores. Data are obtained from 3 independent experiments. c. Bone-marrow murine PMN transmigration through the Transwell system with pore diameters ranging from 3 to 12μm in response to a gradient of the chemoattractant fMLP. d. Percentage of transmigrated PMN through different pore sizes as shown in c. PMN transmigration is a direct function of pore diameter. e. Bactericidal activity of PMN transmigrated through different pore diameters as in c. PMN transmigrating through smaller pore augmented bacterial killing. Data are from 3 independent experiments. f. Schematic of the adoptive transfer experiment in which mice challenged with GFP-Pseudomonas aeruginosa (GFP-P.a.) received 1×106 PMN through i.t. route. g-h. Infected mice adoptively transferred with bone-marrow-derived murine PMN subjected to transmigration through 5μm diameter pores (as in a) showed increased clearance of Pseudomonas aeruginosa as compared to control PMN passed through 200 μm pores. Data are obtained from 6 mice per group from 6 independent experiments. Scale bar, 25 mm. Additional information is provided as Suppl. Figure 2a–b.
To further address the augmented bacterial killing phenotype of transmigrated PMN, we carried out an adoptive transfer experiment in which these PMN were instilled intratracheally (i.t.) (Figure 2f). Here we used murine bone-marrow PMN that had been perfused through either 50μm or 200μm diameter pores in the microfluidic system. We used a model of pneumonia induced by Pseudomonas aeruginosa instillation in lungs 42 and studied changes in bactericidal activity of PMN that had been pre-exposed to transmigration. We observed that PMN migrating across 5μm pores showed improved clearance of Pseudomonas aeruginosa from lungs of infected mice as compared to PMN passed through 200μm gaps (Figure 2g,h).
Piezo1 sensing of membrane tension induces calcium signaling in PMN during transendothelial migration
As PMN experience distortion forces during migration across restrictive AJs, we addressed the possible role of the mechanosensor Piezo1, a non-selective Ca2+ channel 43, expressed in PMN in mediating the PMN phenotype shift. To assess changes in Ca2+, we intravitally imaged Ca2+ influx in PMN during their transmigration phase using the Ca2+ indicator GCaMP6f constitutively expressed in PMN. Using dual-channel two photon live-cell imaging microscopy, we observed cyclic cytosolic Ca2+ transients in PMN occurred only during the transmigration phase (Figure 3a–c; supplementary Video S1). The response waned as the cells exited the junctions (Figure 3b–c). A similar pattern in cytosolic Ca2+ transients was observed in the PMN migrating across endothelial cells of cremaster venules (Suppl. Figure 2c–e, supplementary Video S4), indicating that Ca2+ spikes were a generalized response in all transmigrating PMN. To address whether Ca2+ influx observed during PMN transmigration was mediated by Piezo1 mechanosensing, we transfected PMN derived from HL-60 44 cells with control or Piezo1 shRNA constructs (Suppl. Figure 3a). Results showed that Piezo1 depletion in PMN prevented the Ca2+ influx seen in PMN transmigrating through 5μm pores (Figure 3d–f). To address the basis of Piezo1 mediated Ca2+ flux, we treated PMN with thapsigargin, a non-competitive inhibitor of endoplasmic reticulum (ER) Ca2+ ATPase (SERCA pump) that releases Ca2 from ER stores 45 or used the ionophore ionomycin to induce Ca2+ entry through the plasma membrane 46 by a Piezo1-independent mechanism. Treatment of PMN with ionomycin but not with thapsigargin enhanced bactericidal function of PMN (Suppl. Figure 3b). The latter experiments were made under nominally-free extracellular Ca2+ condition to prevent store-operated Ca2+ entry after ER Ca2+ depletion (Suppl. Figure 3b). These results support the central role of plasmalemma Ca2+ entry in PMN during transmigration via the Piezo1 channel as a central signaling mechanism of PMN bactericidal function.
Figure 3. PMN migration through endothelial junctions activates calcium signaling via Piezo1 in PMN.
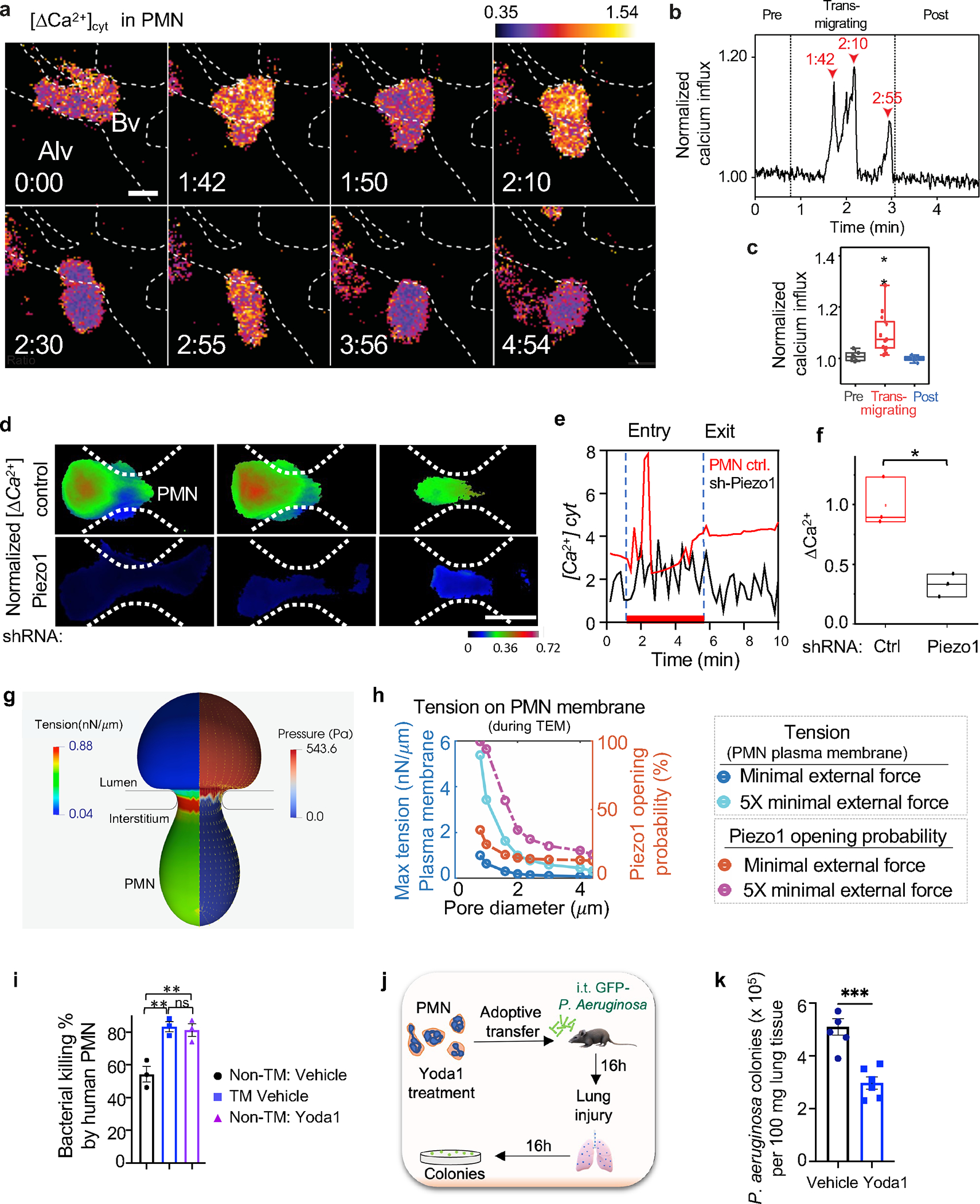
a. Ca2+ transients in PMN determined by motion-corrected 2-photon lung intra-vital imaging of the Ca2+ indicator GCaMP6f (green) before, during, and after PMN transmigration in endothelial AJs of lung micro-vessels. PMN reporter was td-Tomato expressed under Ly6G promoter (red); endothelial junction labeled by anti-PECAM1antibody (blue). Realtime data is supplied as supplementary Video S1. Scale, 0.3 mm/pixel; image capture rate, 2 seconds/frame. The color scheme shows blue as the lowest and yellow as highest increase in cytosolic [Ca2+]. The dotted line shows the lung capillary wall with a monolayer of endothelial cells. Bv, blood vessel; Alv, alveolus. Time is in min and sec. Scale bar, 5μm. b. Changes in cytosolic [Ca2+] during PMN transmigration in a. Arrowheads indicate Ca2+ spikes and corresponding times during PMN transmigration. c. Quantification of Ca2+ peaks normalized to baseline fluorescence prior to transmigration. Data are presented for 5 transmigration events observed in 5 mice. The boundaries of the box-plot indicate the 25th and 75th percentiles; whiskers show 1.5x interquartile range; median is shown by the straight line. d. Time-lapse images of Fluo4 in control and shRNA silenced Piezo1 PMN (HL-60 derived) migrating through 5μm pores of the microfluidic system. Scale bar, 8 μm. Individual Fluo4 tracings (e) and quantifications (f) demonstrating changes in cytosolic [Ca2+] in control and Piezo1 deleted PMN in d. The boundaries of the boxplot indicate the 25th and 75th percentiles. The median is shown by the straight line. Data are obtained from 3 independent experiments. g-h. Finite element simulations of PMN (8.5μm in diameter) transmigrating through the restrictive AJs. g. A cross-sectional view of coupled finite element and boundary element simulations of predicted tension and pressure on PMN plasma membrane during passage through a pore with diameter of 2.4μm under external force of 1.8nN, which is about 5 times the minimal external force required to push the PMN through the pore. The color maps show color-coded ranges for PMN membrane tension and pressure. h. Graph demonstrating maximum tension values at the PMN membrane (dark blue and cyan) and activation probability of the tension sensor Piezo1 (brown and pink) during PMN migration as a function of pore diameter under two different external forces, the minimal critical external force (dark blue and brown) and five times (5x) the minimal external force (cyan and pink) needed for PMN to pass through pores. i. Percent of E. coli killed by human PMN cultured under static condition and treated with vehicle control or Yoda1 to induce Piezo1 activation or post-transmigration through 5μm pores of the microfluidic system. Data are obtained from 3 independent experiments. The response was increased by Yoda1 treatment to the same degree as passing PMN through 5μm pores. j-k. Mice received i.t. adoptive transfer of 106 bone marrow derived murine PMN pretreated with Yoda1 showed marked improvement of Pseudomonas aeruginosa clearance at 16h post-i.t. instillation of 106 cfu bacteria per mouse as compared to vehicle control. Data were obtained from 5–6 mice per group from 3 independent experiments. Additional information is provided as Suppl. Figure 2c–j & 3a–e.
To assess next whether PMN traversing AJs could generate sufficient tension to activate Piezo1, we used a computational model to determine spatial changes in membrane tension of human PMN with the mean diameter of 8.5μm. Membrane tension was found to be increased during the transmigration phase at the sites of PMN contact with junctions (Figure 3g; supplementary Video S2). Maximum membrane tension experienced by PMN was inversely proportional to the pore diameter (Figure 3h) and increased with the total external force needed to push PMN through junctions (Figure 3h). The total pressure applied to PMN also increased proportionally to the pore diameter (Suppl. Figure 2f). The pressure consisted of intracellular force generated by the PMN actomyosin apparatus and the force generated by the hydrodynamic pressure difference between blood vessel and extravascular tissue (Figure 3h, Suppl. Figure 2f). PMN membrane tension generated by these forces from 0.7 nN/μm to 1.4 nN/μm was in the range required to gate the Piezo1 channel 47,48. As shear stress has also been shown to activate Piezo1 39,49, we determined whether PMN traversing through the 5μm pores in the microfluidic system (Suppl. Figure 2g) encountered sufficient mechanical fluid shear to affect Piezo1 channel opening during the transmigration phase (see Methods for details). We demonstrated migration and squeezing of PMN through 5μm pores of the system in real time at 30,000 frames sec−1 (supplementary Video S3). The flow field inside the microfluidic system was used to predict the velocity and shear stress in the channel under given flows (Suppl. Figure 2h). The maximum fluid shear in was estimated to be 191.924 Pa for flow of 20 μl/min and it was linearly proportional to the flow. Passage of PMN through this system generated sufficient plasma membrane tension in the range 43,44 required to gate Piezo1 (Suppl. Figure 2i–j, supplementary Video S5).
Piezo1 in transmigrating PMN activates bactericidal function
To address whether Piezo1 activation in the PMN plasma membrane was required for enhanced PMN bactericidal function, we treated freshly isolated human peripheral blood PMN with the activator of Piezo1, Yoda1 50 (Figure 3i). Both activation of Piezo1 with Yoda1 and mechanical forces of microfluidic system during PMN migration across 5μm pores increased bactericidal activity as compared to control PMN (Figure 3i). However, activation of Piezo1 did not alter phagocytosis of bacteria (Suppl. Figure 3c). Furthermore, adoptive i.t. transfer of bone-marrow murine PMN pre-treated with Yoda1 improved the clearance of Pseudomonas aeruginosa from lungs of i.t 106 GFP-expressing Pseudomonas aeruginosa infected mice (Figure 3j–k).
To genetically study the role of Piezo1 activation in transmigrating PMN in bacterial killing, we generated homozygous PMN-restricted Piezo1 mutant mice (Piezo1ΔPMN) by crossing Piezo1fl/fl with CatchupIVM-red mice containing Ly6G-Cre (Suppl. Figure 3d,e). Deletion of Piezo1 in PMN had no significant effect on the PMN number in peripheral blood (Suppl. Figure 3f) and did not alter maturation of bone marrow PMN (Suppl. Figure 3g,h). In Piezo1ΔPMN mice instilled with i.t 106 GFP-expressing Pseudomonas aeruginosa (Figure 4a–k), we observed greater number of Pseudomonas colonies in lungs (Figure 4a–d) as well as augmentation of inflammatory lung injury (Figure 4e–f) and increased mortality (Figure 4g) as compared to wild type. These results show the obligatory role of PMN-expressed Piezo1 in regulating the PMN bactericidal function.
Figure 4. Activation of Piezo1 in transmigrated PMN increases bactericidal activity.
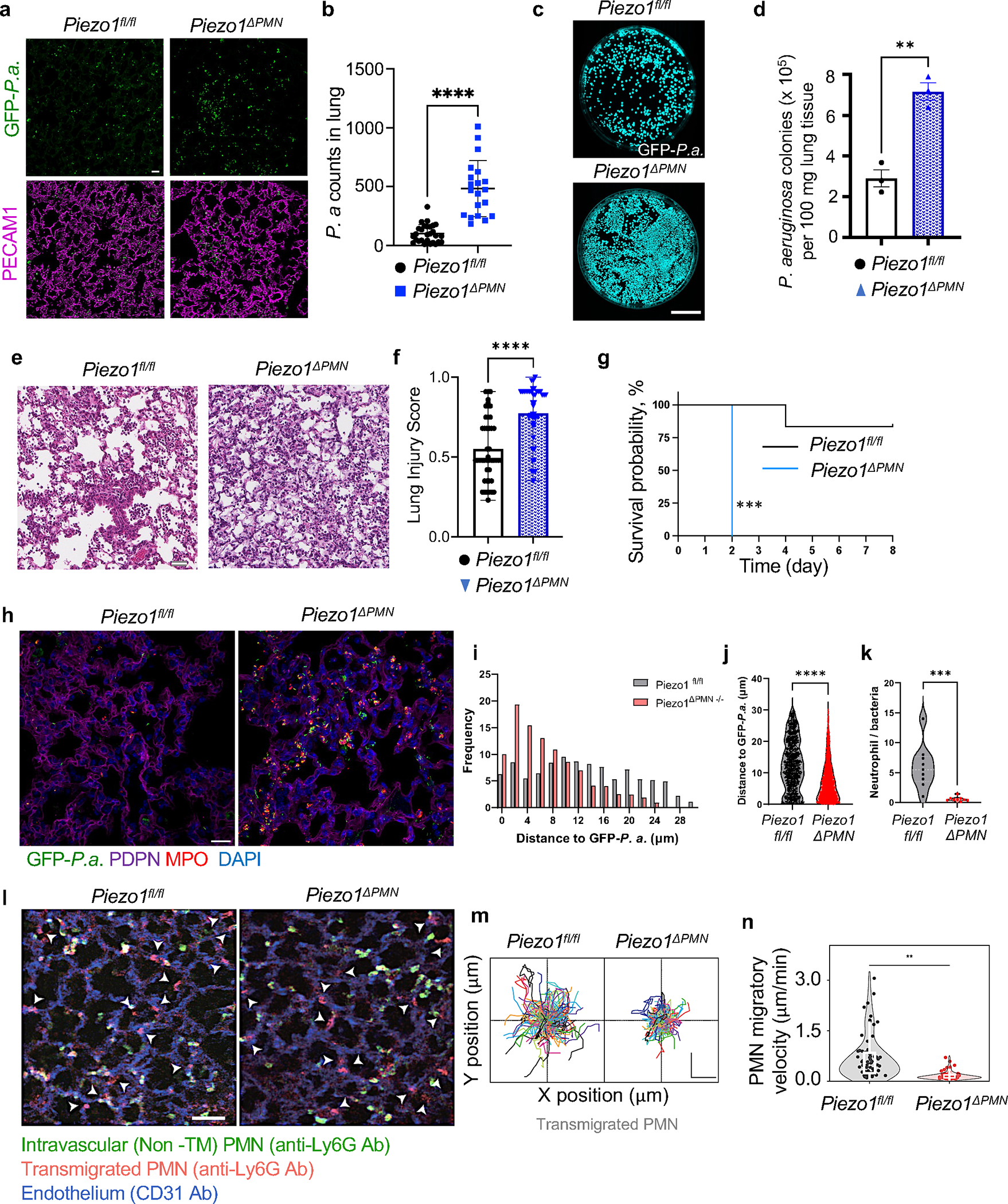
a-k. Genetic deletion of Piezo1 in PMN augmented lung infection due to diminished antimicrobial activity of PMN. a. Representative images of lung sections stained for GFP-Pseudomonas aeruginosa (green) and PECAM-1 (magenta) and b. quantification of GFP-Pseudomonas aeruginosa counts in lung tissue of control (Piezo1fl/fl) and PMN-specific Piezo1 deleted (Piezo1ΔPMN) mice challenged with 106 cfu bacteria per mouse for 12h. Scale bar, 20μm. c. Images of GFP-expressing Pseudomonas aeruginosa colonies grown on agar plates as in a-b. Scale bar, 25 mm. d. Quantification of data in c. Data were obtained from 3 mice per group from 3 independent experiments. e-f. Histopathological assessment of lung injury induced by Pseudomonas aeruginosa in Piezo1ΔPMN vs. control mice. Representative images of H&E-stained lung tissue from 3 independent experiments (e) and lung injury scores (f) of mice as in a-b. Scale bar, 20μm. g. Survival rates of mice challenged with i.t. 106 cfu Pseudomonas aeruginosa per mouse. Genetic deletion of Piezo1 in PMN markedly enhanced mortality. Data were obtained from 9 mice per group from 3 independent experiments. h-k. Genetic deletion of Piezo1 in PMN reduced PMN-to-bacteria ratio. h. Representative images of lung sections stained for GFP-Pseudomonas aeruginosa (green), podoplanin (PDPN, magenta) from 3 independent experiments were used for tissue architecture assessment, PMN marker myeloperoxidase (MPO, red), and 4′,6-diamidino-2-phenylindole (DAPI, blue). Scale bar, 20μm. i-k. histogram distribution (i) and mean distance (j) between PMN and GFP-Pseudomonas aeruginosa in lung tissue; and PMN-to-bacteria ratio (k) in lungs of control (Piezo1fl/fl) and PMN-specific Piezo1-deleted (Piezo1ΔPMN) mice as in a-b. l-n. Genetic deletion of Piezo1 in PMN reduced migratory velocity of PMN in lung tissue. l. 2 photon microscopic images of lung tissue PMN of control (Piezo1fl/fl) and PMN-specific Piezo1-deleted (Piezo1ΔPMN) mice 4h after challenge with insufflated LPS. Total lung intravascular and tissue PMN (red) were detected by i.v. staining of PMN with Alexa594-labeled α-Ly6G antibody before LPS insufflation. Intravascular PMN (green) were stained with BV421-labeled α-Ly6G antibody prior to intravital imaging. Endothelium (blue) was stained with α-CD31 antibody. Arrow heads show transmigrated PMN in extravascular tissue. Scale bar, 50μm. m-n. PMN migratory trajectory (m) and PMN migratory velocity (n) of transmigrated PMN. Piezo1ΔPMN PMN showed reduced migration velocity which was coupled to reduced bacteria killing as shown in a-d and k. Scale bar, 20μm. Data are obtained from 3 mice per group from 6 independent experiments. Additional information is provided as Suppl. Figure 3.
To address the basis of Piezo1-mediated increase in bactericidal activity, we analyzed factors such as the distance between Pseudomonas aeruginosa foci and PMN in infected lungs. We observed that deletion of Piezo1 in PMN did not alter their ability to phagocytize bacteria (Figure 4h–j; Suppl. Figure 3i) whereas Piezo1-deficient PMN localized closer to the bacteria as compared to control PMN (Figure 4h–j). Notably the number of PMN with phagocytized bacteria was similar between groups (Suppl. Figure 3i). Although lungs of Piezo1ΔPMN mice contained more PMN (Suppl. Figure 3j), their proportion to Pseudomonas aeruginosa was reduced as compared to lungs of control mice (Figure 4k). Furthermore, Piezo1 deletion in PMN had no effect on survival of transmigrated lung PMN cultured ex vivo (Suppl. Figure 3k). Another important phenotype of Piezo1ΔPMN mutant mice was their inability to form extracellular traps (NETs) in response to Pseudomonas aeruginosa challenge (Suppl. Figure 3l). We also analyzed the effects of Piezo1 deficiency on PMN migration in lung tissue following the onset of inflammatory lung injury induced by insufflated LPS (Figure 4l–n). Here we distinguished transmigrated and non-transmigrated PMN by chase labeling of lung tissue PMN with α-Ly6G-BV421 antibody before carrying out intravital imaging (see Methods). Analysis of migratory velocity of extravascular PMN (Ly6G-Alexa594+) (Figure 4l) showed that deletion Piezo1 in PMN reduced the velocity of PMN from 3 μm/min of control PMN to 0.8 μm/min (Figure 4m, n). Together these data showed Piezo1 expression in PMN is crucial in mediating bactericidal, NET forming, and migratory functions of PMN.
Piezo1 signaling in transmigrating PMN activates immune regulatory genes
We carried out a transcriptome analysis of transmigrated (TM) vs. non-transmigrated (Non-TM) PMN in mice to identify possible Piezo1 dependent genomic alterations that could explain their enhanced bactericidal function of TM PMN. TM and Non-TM PMN showed distinct transcriptomic profiles in wild-type and Piezo1ΔPMN mice (Figure 5a–f; Table S1 showing all differentially expressed genes (DEGs)). We found NOX4, the constitutively active isoform that generates hydrogen peroxide in the phago-lysosomal compartment 30,31,32, was upregulated in TM PMN and but reduced in TM PMN of Piezo1ΔPMN mice (Figure 5e,f) suggesting the relationship between Piezo1 activation and Nox4 expression. The role of NOX4 was thus specifically addressed as described below.
Figure 5. Genetic analysis of transmigrated and non-transmigrated PMN in lungs.
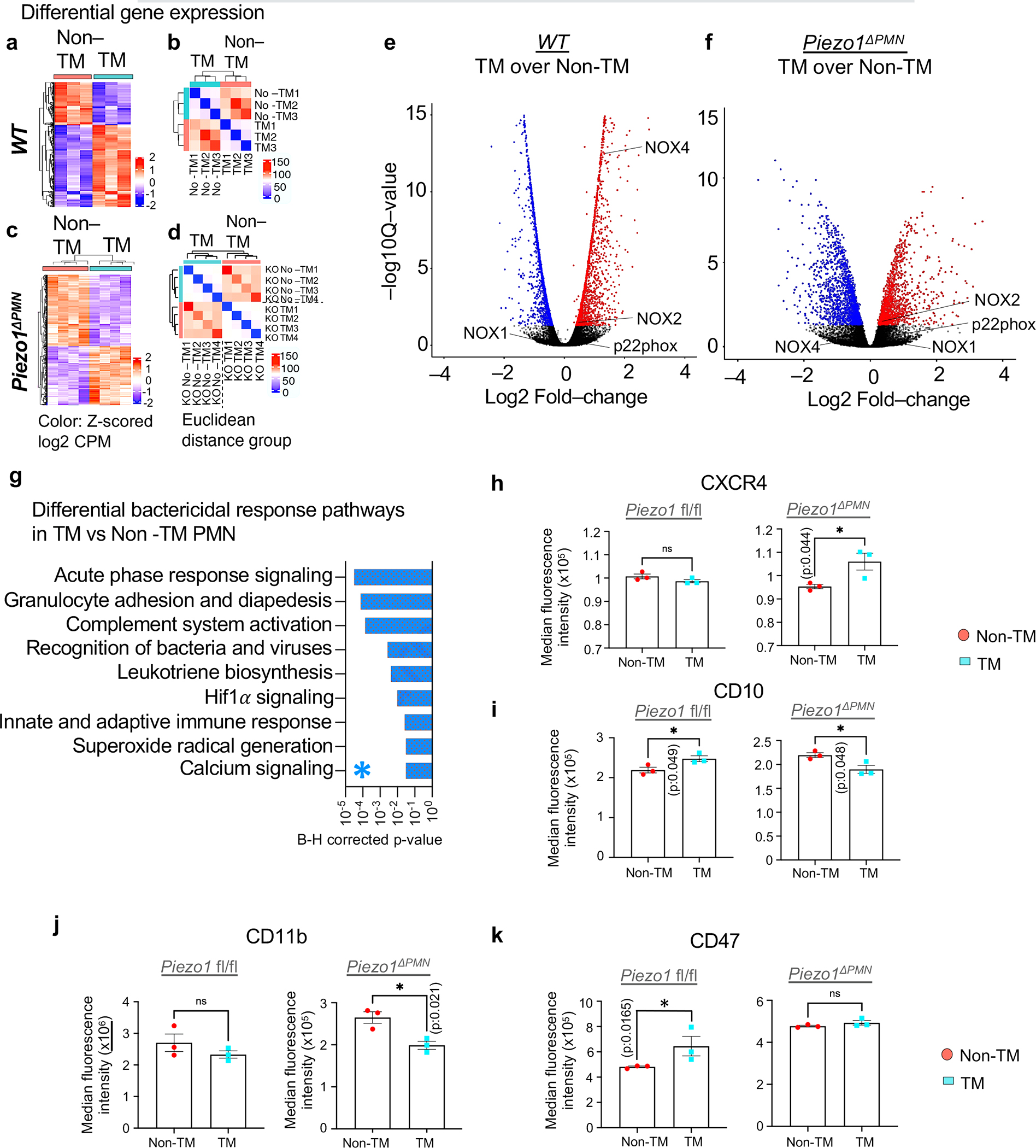
a-d. RNA-seq analysis of transmigrated vs non-transmigrated lung PMN of control (a-b) and PMN-specific Piezo1 mutant (Piezo1ΔPMN) mice (c-d). See supplementary Table S1 for details. a, c. Heat maps of gene expression profiles of non-transmigrated vs. transmigrated PMN in lungs of control mice and LPS insufflated mice as in Fig 1a–b, b, d Sample-to-sample distance matrix with color intensity representing Euclidean distance in the gene expression space. Data were obtained from 4 mice per group from 3 independent experiments. e-f. Volcano plots showing the overall changes in gene expression in wild-type and Piezo1ΔPMN PMN as a function of transmigration. NOX4 was upregulated in transmigrated wildtype PMN as compared to Piezo1ΔPMN PMN. g. The most significant bactericidal pathways derived from IPA analysis are shown (see supplementary Table S1 for further details). HIf1α and Ca2+ signaling pathways are among most altered pathways in Piezo1ΔPMN PMN. h-k. Changes in protein-level expression of cell surface markers CXCR4 (h), CD10 (i), CD11b (j), and CD47 (k) in control and Piezo1ΔPMN PMN upon transmigration as assessed by spectral flow-cytometry. Compared to control, the surface expression of CD47 and CD10 were reduced in transmigrated Piezo1ΔPMN PMN. Results are shown as mean fluorescence intensity. Data are obtained from 3 mice per group. Additional information is provided as Suppl. Figure 4 & 5.
To identify whether Piezo1 activation improved PMN host-defense function, we used ingenuity pathway analysis (Figure 5g; Suppl. Figure 4; Table S1). The most altered bactericidal response pathways included acute phase response signaling, pathogen recognition receptors, communications between innate and adaptive immunity, Hif1α, and Ca2+ signaling (Figure 5g). The predicted association between Piezo1-induced Ca2+ signaling, Hif1α signaling, and Nox4 expression in TM PMN suggests a role for Piezo1-mediated Ca2+ influx in Hif1α stabilization 51 which may transcriptionally upregulate the phago-lysosomal protein NOX4 51 causing enhanced bacterial killing. This prediction was experimentally addressed as described below.
We observed, the inflammatory and Ca2+ signaling pathways in TM PMN of Piezo1ΔPMN mice were not upregulated (Suppl. 4d, g, f). Analysis showed the downregulation of genes linked to Hif1α signaling in TM Piezo1ΔPMN PMN (Suppl. Figure 4d), suggesting that Piezo1 activation was required for the upregulation of Hif1α target genes. We also found that Hif1α was stabilized within minutes of Piezo1 activation (Suppl. Figure 4h) and that siRNA-mediated depletion of Hif1α abrogated the Piezo1 dependent enhancement of bacterial killing (Suppl. Figure 4i,j).
We next determined using multi-colored spectral flow cytometry whether differential expression of PMN surface markers was altered in TM vs. non-TM PMN (Figure 5h–k, Suppl. Figure 5). TM PMN of wild type mice showed upregulation of Mme (CD10) (Figure 5i; Suppl. Figure 5a), the gene encoding membrane metalloproteinase. TM PMN also showed upregulation CD47 a.k.a. integrin-associated protein (Figure 5k; Suppl. Fig 5a). These changes, however, were reduced in PMN of Piezo1ΔPMN mutant mice (Figure 5i,k). TM PMN of Piezo1ΔPMN mice also showed significant upregulation of transcript and surface protein expression of CXCR4, the C-X-C chemokine receptor type 4 (Figure 5h: Suppl. 5a, b), known to promote chemotaxis and suppress cell death 52. We did not observe significant change in surface expression of CD11b in TM PMN from wild-type mice; CD11b surface expression was reduced in TM PMN of Piezo1ΔPMN mice (Figure 5j). These data together showed that TM of PMN activated a range of transcriptional programs some of which were altered by deletion of Piezo1. The results support the important role of TM of PMN in activating multiple Piezo1 dependent genomic programs that altered the function of transmigrating cells.
Upregulation of Nox4 in transmigrating PMN is required for bactericidal function
As our transcriptome data above suggested that Piezo1 induced upregulation of Nox4 in TM PMN (Figure 5e,f), we addressed whether function of Piezo1-NOX4 signaling was a critical requirement for activation of the TM PMN and increased bactericidal function of these cells. Like other NADPH oxidases 20 NOX4 mediates bacterial killing via ROS generation. Analysis of Nox isoforms in TM vs. non-TM PMN challenged with LPS showed that mRNA expression of Nox2 and Nox4 was upregulated in transmigrated PMN whereas Nox1 was downregulated (Figure 6a). Critically, genetic ablation of Piezo1 in PMN interfered only with Nox4 upregulation in the TM PMN (Figure 6b). Furthermore, activation of Piezo1 with Yoda1 increased Nox4 mRNA in PMN as early as 30 min post-treatment (Figure 6c). We also observed increased NOX4 protein expression PMN stimulated with Yoda1 (Figure 6d). NOX4 protein expression was upregulated in HL-60 derived PMN that transmigrated in 5μm pores in response to fMLP chemotactic gradient (Figure 6e). Increase in NOX4 expression was significantly less in PMN transmigrating through larger 12 μm diameter pores and in Piezo1-depleted PMN (Figure 6e). In other control experiments, fMLP exposure of PMN grown in suspension or non-TM PMN showed no upregulation of NOX4 (Figure 6e).
Figure 6. Piezo1 upregulates Nox4 expression to activate bactericidal activity of transmigrated PMN.
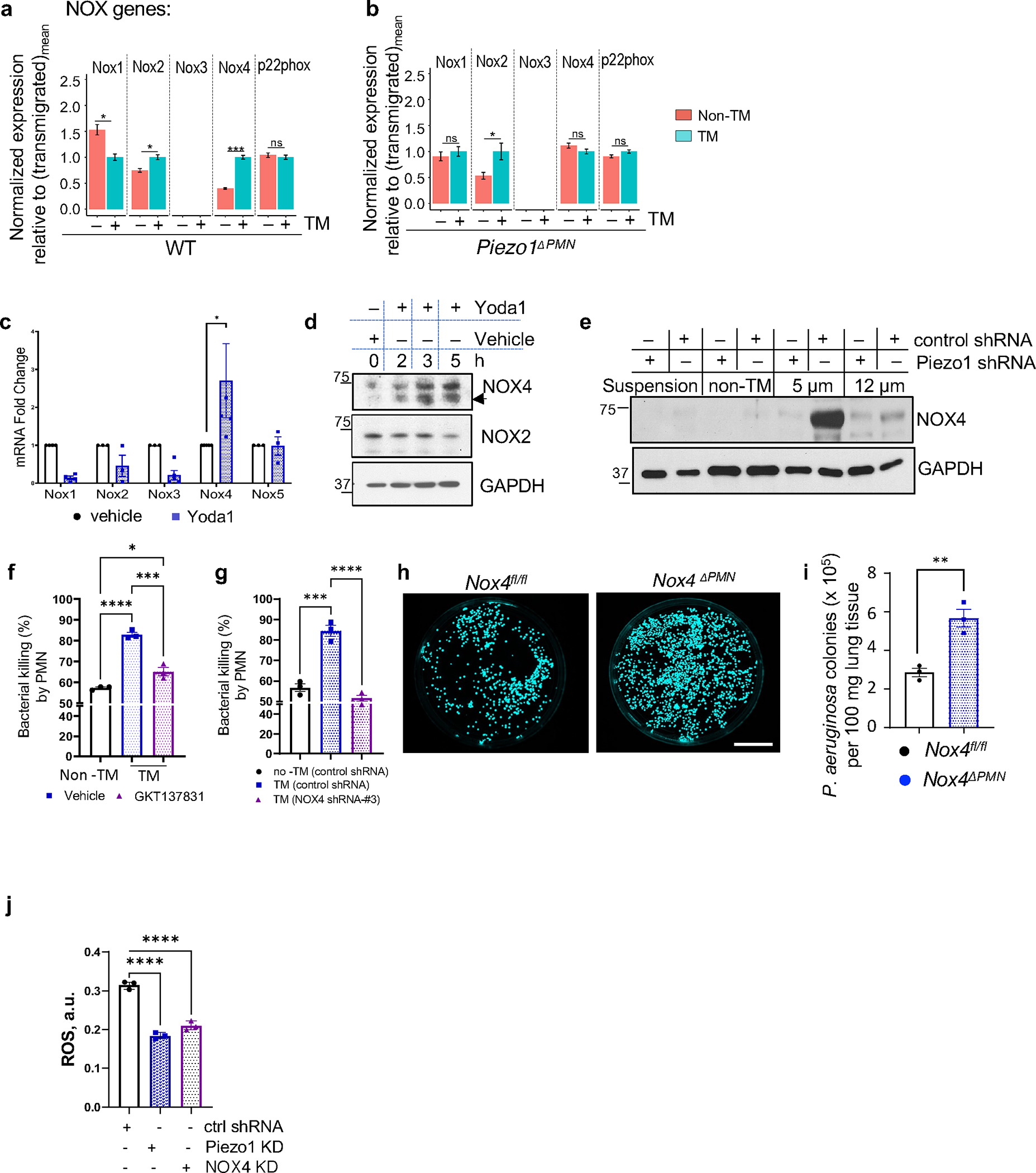
a-b. Changes in expression of NOX isoforms in transmigrated vs. non-transmigrated lung PMN from control (a) and Piezo1ΔPMN PMN (b), as in Fig. 5a, as assessed by RNA-seq analysis (details in supplementary Table S1). Increase in NOX4 expression was greater relative to NOX2 in the transmigrated PMN of control but not Piezo1ΔPMN mice. c. Yoda1 treatment of freshly isolated human PMN showed increased NOX4 mRNA expression as early as 30 min. Data were obtained from 5 healthy donors from 5 independent experiments. d. Western blot showing time-course of NOX2 and NOX4 expression in human PMN treated with Yoda1. Yoda1 increased only NOX4 protein expression. e. NOX4 protein expression is upregulated in control but not in Piezo1 depleted HL-60 derived PMN at 2 hr post-transmigration through the Transwell system with of 5μm pore diameter. NOX4 was not upregulated in fMLP-exposed PMN grown in suspension or in non-transmigrated PMN. Data are obtained of 3 independent experiments. f-g. Pharmacological inhibition (f) or genetic ablation (g) of NOX4 function in bone marrow and HL-60 derived PMN, respectively, reversed the augmented bacterial killing responses induced by PMN transmigration. Data are obtained from 3 independent experiments. h-i. Bacterial clearance was abrogated in lungs of Nox4ΔPMN mice. Results show Pseudomonas aeruginosa colonies in lungs of Nox4fl/fl and Nox4ΔPMN mice challenged with 106 cfu Pseudomonas aeruginosa per mouse for 16 hr. Lung tissue was lysed and plated on agar plates, and images of Pseudomonas aeruginosa colonies (h) and quantification (i) are shown. Data were obtained from 3 mice from 3 independent experiments. Scale bar, 25 mm. j. Piezo1 and downstream expression of NOX4 are required for ROS generation in transmigrated PMN. ROS production measured in HL-60 derived PMN stably transduced with control, Piezo1, or NOX4 shRNA lentiviral particles. ROS concentration was assessed at 1.5 h post-PMN transmigration through the microfluidic system. Data are obtained from 3 independent experiments. Additional information is provided as Suppl. Figure 6.
As NOX4 appeared to be important in bacterial killing by TM PMN, we next studied NOX4 by tagging it with green fluorescent protein (GFP). GFP-NOX4 localized to the cup-shaped invaginations of the plasma membrane of HL-60 derived PMN (Suppl. Figure 6a,b) and co-localized with F-actin cytoskeleton (Suppl. Fig 6c). Upon incubation with Pseudomonas aeruginosa, NOX4-GFP was seen within the early and late phagosomes as well as phagolysosomes (Suppl. Figure 6a,b,d). To address the specific role of NOX4 in Piezo1-activated bactericidal function in TM PMN, we treated PMN with a dual Inhibitor of NOX1 and NOX4, GKT137831 53 or depleted NOX4 using siRNA in HL-60 derived PMN (Figure 6f–g; Suppl. Figure 6e). Reduction in NOX4 expression and function reduced the bactericidal activity of PMN transmigrating through the 5μm pores (Figure 6f–g).
To compare in vivo bactericidal activity in lung tissue PMN of control (Nox4fl/fl) and homozygous Nox4-ablated (Nox4ΔPMN) mice (see Methods for details), we exposed mice to 106 GFP-expressing Pseudomonas aeruginosa, and observed crucially that deletion of Nox4 in PMN increased the number of Pseudomonas colonies in lungs (Figure 6h–i). Inhibition of NOX4 with GKT137831 in WT bone-marrow murine PMN perfused through 5μm pores of the microfluidic system also reduced the clearance of Pseudomonas aeruginosa from lungs of infected recipient mice similar to results seen in Piezo1 deficient PMN (Suppl. Figure 6f,g). Inhibition of NOX4 in Piezo1 deficient PMN had no additive effect (Suppl. Figure 6f,g). These data together show that Piezo1 enhanced PMN host-defense function through upregulation of NOX4. Furthermore, TM PMN were shown to generate augmented ROS via NOX4 as a function of mechanosensing by Piezo1 during the phase of PMN transmigration (Figure 6j).
Discussion
PMN function is driven by multiple factors including mechanical stimulation and chemoattractants that induce PMN activation and lead to killing of bacteria and other pathogens 54,55. Activation of PMN is thus essential for their fundamental host-defense function. Here, we assessed alterations in PMN function induced by mechanical stimuli during the transmigration phase of PMN through endothelial AJs. This transmigration event is the initial step in activating PMN host defense system as PMN breaches the vascular endothelial barrier and reaches the site of infection 7,12,56,57. The AJs form “pores” in the nanometer range, a defining characteristic of all continuous microvascular endothelia 58, through which PMN extravasate in response to chemotactic signals into infected tissue. We have described the fundamental role of AJs in re-programming the PMN bactericidal function during transendothelial migration as was evident by the inverse relationship between restrictiveness of the endothelial junctional barrier and the activation of the bactericidal function of the PMN entering tissue.
Our results raise the question of relevance of such a PMN transmigration event. Breakdown of AJs due to inflammatory injury and release of permeability-increasing mediators compromise the restrictiveness of AJs 58 which as we showed control the PMN activation state. This was evident in studies in which the restrictiveness of endothelial barrier was genetically increased by forcing the interaction of VE-cadherin, the primary adhesive protein of AJs, and its partner VE-PTP that normally binds VE-cadherin to stabilize VE-cadherin homotypic interaction to restrict the barrier 40,59. We observed decreased transmigration of PMN in these mice, but the transmigrated PMN mounted an intense bactericidal response as compared to PMN migrating across looser endothelial junctions. Our results are consistent with observations showing that influx of PMN into tissue via open or injured junctions did not effectively kill bacteria 60,61 as opposed to PMN transmigrating via normal junctions 41. These results help to explain the pathogenic role of PMN in mediating tissue injury during inflammatory disease such as ARDS associated with severe disruption of AJs resulting in augmented tissue PMN accumulation 62. Our findings also raise the conundrum that severity of leakiness of endothelial junctional barrier in inflammatory diseases reduces bacterial killing such that PMN become less effective bactericidal cells when their function is most required.
The present observations revealed the requisite role of the mechanosensor Piezo1 localized in the PMN membrane in mediating the effects of ‘squeezing’ of PMN during the transmigration phase, and in increasing bactericidal efficacy. Computational modeling showed that transmigrating PMN experienced plasma membrane tension in endothelial AJs from 0.7nN/μ to 1.4nN/μ, well in the range required to gate the Piezo1 channel 47,48. Our central conclusion is that PMN transmigration through restrictive AJs induced the PMN bactericidal phenotype through Piezo1. Piezo1 sensing of tension was required to reprogram the extravasating PMN and augment their bactericidal function.
Since Ca2+ signaling regulates multiple functions of PMN 63, we determined using 2-photon intravital microscopy in vivo whether Ca2+ changes occurring via Piezo1 gating were responsible for the shift in PMN phenotype. Data here showed tri-shoulder Ca2+ transients during PMN passage across endothelial junctions of lung capillaries. Ca2+ transients were not seen during the initial adhesion of PMN to the endothelium or the PMN exit phase. The site of highest Ca2+ signal intensity did not correlate with maximum membrane tension seen at the PMN bottleneck region. Although the mechanism is not clear, it is possible that Piezo1 was spatially restricted to the rear in order to establish rear-front polarity during PMN transmigration or Piezo1 was activated by intracellular pressure which also increased in the rear as PMN crosses AJs. This may be also the result of initial phase of Ca2+ entry occurring via Piezo1 which triggers other Ca2+ release pathways such as via Ca2+-induced-Ca2+-release (CICR) channel 64,65 or cAMP-potentiated release 66. To reinforce these fundings seen during transendothelial PMN migration, we also found similar Ca2+ waves in the microfluidic transmigration system, suggesting the restrictiveness of the junctional barrier was the central cause of Piezo1 dependent Ca2+ influx. Piezo1 activation was crucial in mediating the Ca2+ influx response since Piezo1 deletion in PMN prevented the Ca2+ transients seen in transmigrating PMN.
Piezo1 activation in lung macrophages unlike PMN has been shown to enhance their phagocytosis capacity 51. In contrast to lung macrophages, the primary effect of Piezo1 in PMN was enhancement of bacterial killing function. We showed this to be an obligatory mechanism for the PMN host-defense function since genetic ablation of Piezo1 in mouse PMN reduced bactericidal function, and the mice showed increased susceptibility to bacterial infection. Furthermore, the activation of Piezo1 signaling in PMN through adoptive transfer experiments provided therapeutic advantage even in the face of disrupted AJs seen in inflammatory lung injury. Therefore, the present results demonstrate an essential role of Piezo1 expression and activation in PMN as central to promoting the resolution of bacterial pneumonia.
The PMN nucleus is sufficiently deformable to enable migration through the endothelial junctional barrier but it cannot be deformed to the extent that it releases DNA and histones leading to wholesale formation of extracellular traps, NETs) 67,68. PMN nuclei are likely preserved during migration through AJs as the result of composition of the nuclear lamina, a flexible meshwork composed of intermediate filaments woven from type A and type B lamin proteins adjacent to the inner nuclear membrane 69. However, NET formation did occur in wild type mice but it was reduced in Piezo1ΔPMN suggesting the forces to which PMN are exposed during transmigration makes them susceptible to NETosis.
Transcriptomic analysis showed that transmigration activated multiple host-defense transcriptional programs in PMN, and crucially, that these were inhibited by deletion of Piezo1. We also observed impaired upregulation of genes comprising the Ca2+ signaling pathway in the transmigrated Piezo1ΔPMN PMN as compared to wild-type PMN. In addition, transmigrated PMN showed HIF1α dependent expression of the NADPH oxidase isoform NOX4 gene in Piezo1ΔPMN PMN, suggesting that NOX4 functioned as an effector of Piezo1 signaling downstream of HIF1α stabilization. Bacterial killing induced by transmigrated PMN required Piezo1 activation and NOX4 expression and was inhibited by HIF1α deletion. While Hif1α signaling has been shown to induce NOX4 transcription 70, the present study invokes the role of Piezo1 in signaling Hif1α dependent expression of NOX4. NOX4 is localized in the cup-shaped invaginations of the plasma membrane, specifically in phagosomes and phagolysosomes of PMN 30. In contrast, NOX2 is rapidly assembled an active protein complex that delivers superoxide into the phagosomal lumen 24,25. NOX4 generates hydrogen peroxide 30,31,32, and thus may have a specialized bactericidal role in phagosomal oxidation and acidification as compared to NOX2. These results provide insights into the obligatory Piezo1-dependent NOX4 expression and heightened ROS production in mediating the phenotypic switch in transmigrating PMN into more potent bactericidal cells in tissue.
Piezo1 as a channel permitting Ca2+ may itself induce stabilization of Hif1α by the Ca2−/calmodulin-dependent serine/threonine phosphatase, calcineurin 71, leading to NOX4 transcription in transmigrated PMN. We cannot rule out the alternate possibility that tissue hypoxia such as encountered during inflammation 70 may also activate NOX4 expression independent of Piezo1 through direct Hif1α-NOX4 signaling. Transcriptomic analysis showed that while Hif1α signaling was linked to NOX4 expression whereas no such association was seen between Hif1α and Piezo1 expression, suggesting that Hif1α was not involved in upregulation of Piezo1 by transcription. Nevertheless, it is possible that hypoxia amplifies NOX4 expression directly through Hif1α in addition to activation by Piezo1-induced calcium Ca2+ influx and thereby augments the bactericidal activity of transmigrated PMN.
In conclusion, the present studies show that Piezo1 is the essential mechanosensor mediating PMN activation during PMN passage through restrictive AJs into the infected tissue. Piezo1 activation in PMN is a key host-defense adaptation mechanism required for bacterial killing in tissue through the expression NOX4. These findings raise the possibility of cell-based therapy through adoptively transferring PMN subjected to Piezo1 activation as a means of normalizing PMN host-defense function and preventing bacterial-induced tissue injury in diseases such as ARDS.
Limitations of study
Although our study provides evidence for Piezo1–Hif1α–NOX4 axis mediated mechanical signal relay in transmigrated PMN, the RNAseq data also suggest involvement of other pathways and effectors as players in the mechano-transduction circuit. Future efforts in data mining from our open-source raw data sets (see STAR Methods and supplementary data for details) would expand the understanding of other mechano-transduction signaling pathways. Furthermore, the details of the mechanisms of real-time mechanical signal sensing, relay to the immediate early mediators, and specificities of mechanisms of mechano-chemical coupling are not well understood.
STAR Methods
Lead contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Asrar B. Malik (abmalik@uic.edu).
Materials Availability
Requests for reagents and resources generated in this study will be fulfilled by the lead contact, Asrar B. Malik (abmalik@uic.edu).
Data and code availability
Sequencing data is available on Gene Expression Omnibus. The accession number is GSE210111. The reviewer token is wlebcwqwdncrvir. Computational modeling and simulation data is uploaded using indigo: https://figshare.com/s/0eac4b5603eff08aba58.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human blood samples
All studies involving human subjects were performed in accordance with the Declaration of Helsinki and approved by the University of Illinois at Chicago’s Institutional Review Board (IRB). Human peripheral blood was collected by venipuncture from 11 random healthy volunteers of 18 years of age or older, recruited in an unbiased fashion irrespective of ethnicity and gender. Donors provided written informed consent that was documented. Information about gender was collected. Gender-based analysis was not performed due to low number of recruited donors per experiment (3–5 donor of both genders per experiment). Blood PMNs were isolated by immune-magnetic negative selection method (Easystep human neutrophil isolation kit, Stemcell technologies, #19666). 80–100 × 106 PMN yield were obtained from 20 ml of human blood.
Human cell lines
Human promyelocytic leukemia (HL-60) cells (ATCC #CCL-240) 44 were grown in suspension with RPMI1640 medium containing L-glutamine and 25 mM HEPES (Thermo Fisher #72400047), supplemented with 15% FBS (Thermo Fisher #A4766801) and penicillin-streptomycin-amphotericin B (Thermo Fisher #15240096), at 37°C, 5% CO2. Cells were passaged before the density reached 2×106/ml in 25 cm2 flask. HL-60 cell were differentiated into PMN-like cells with 1.3% DMSO for 6 days. Differentiated cells were routinely stained for DNA using 4′,6-diamidino-2-phenylindole (DAPI) or Hoechst 33342 and appeared to be free of cytosolic DNA.
Mouse models
All the mice were handled in accordance with NIH guidelines for the care and use of laboratory animals and UIC animal care and use committee (ACC)’s regulations. All procedures were approved by the UIC IACUC. Male mice of 8–10 weeks of age, body weight ranging from 18–22 g, were used for all experiments. C57BL/6 (IMSR_JAX:000664) mice were purchased from Jackson laboratory. C57BL/6-Ly6g(tm2621(Cre-tdTomato), known as “Catchup” mice were a gift from Dr. Matthias Gunzer 37. VE-cadherin-FKBP - VE-PTP-FRB (known as: Cdh5tm4(Cdh5/Fkbp, Ptprb/Mtor*)Dvst) mice were a gift from Dr. Dietmar Vestweber 38. In these mice, the two coding sequences of the fusion constructs VE-cadherin-FK506 and VE-PTP-FRB were separated by an internal ribosomal entry site (IRES). This cDNA construct was then inserted into the VE-cadherin locus of the target mice which led to the replacement of endogenous VE-cadherin. Mice containing this bi-cistronic construct express the two cDNAs under the endogenous VE-cadherin promotor, but as separate proteins. NADPH oxidase 4 (Nox4) floxed mice were a gift from Dr. Viswanathan Natarajan. Calcium indicator Salsa6f (Jackson cat. #IMSR_JAX:031968) and Piezo1 floxed B6.Cg-Piezo1tm2.1Apat/J (Jackson cat. #IMSR_JAX:029213) mice were purchased from vendor. PMN specific genetic deletion was generated by crossing indicated transgenic mice with Catchup mice. Deletion of specific genes were confirmed by Western blotting.
METHOD DETAILS
Isolation of transmigrated and non-transmigrated PMN
Segregation method of transmigrated and vascular PMNs have been adopted from Paul Frenette (see Results section: Adherens junctions (AJs) enhance bactericidal function of transmigrating PMN) 36. PMN were mobilized to lung of 8–10 weeks old male Catchup (heterozygous) or non-labelled C57BL/6-background mice by exposure to 1mg/ml or 5mg/ml (for short duration PMN transmigration kinetics) in nebulized LPS (Sigma, Escherichia coli O55:B5) inside the fumigation chamber. 20 μg of α-Ly6G-BV421 antibody (Biolegend, #127628) was i.v. injected 2 minutes before sacrifice. Lungs were perfused with 10 ml of PBS through right ventricle of heart and collected for PMN isolation, biochemical, and histopathological analyses. For PMN isolation, a single cell suspension was prepared by digestion of minced lung lobes with collagenase A (1 mg/ml; Millipore-Sigma # SCR103) at a 37°C shaking water bath for 45 min. Digested lung samples were passed through 16-17G needle and filtered through 40 μm cell strainer (Thermo Fisher, #08-771-1). Red blood cells were removed by using RBC lysis buffer (Thermo Fisher, #00-4333-57). A single cell suspension prepared from Catchup (heterozygous) mice were directly subjected to flow sorting for separation of td-Tomato-positive, or double td-Tomato and α-Ly6G-BV421-positive PMN. Suspension from non-labelled mice were further stained with α-Ly6G-AF594 antibody (Biolegend, #127636) before sorting for α-Ly6G-AF594, or dually labelled α-Ly6G-AF594 and α-Ly6G-BV421-positive cells.
Fluorescence-activated cell sorting (FACS) and flow cytometry
PMN, labelled with td-Tomato (or α-Ly6G-AF-594) and td-Tomato and α-Ly6G-BV-421 (or α-Ly6G-AF-594 and α-Ly6G-BV-421), were sorted in a sterile environment by MoFlo Astrios and MoFlo Astrios EQ (Beckman Coulter) cell sorter equipped with 405, 488, 561 and 635 nm lasers, and collected in RPMI-1640 medium (Thermo Fisher #72400047). Gates were initially established on forward and side scatter, based on granularity and relative size. Aggregates were gated out accordingly. Selective gates were drawn to sort the single positive and dual positive PMN. Software SUMMIT, 6.1 (Beckman Coulter) was used for sorting and analysis. CytoFLEX-S (Beckman) flow cytometer, equipped with 405, 488, 561 and 635 nm lasers, was used for data acquisitions of peripheral blood PMN. Data analyses were performed by Kaluza Analysis 2.1 (Beckman) software. The number of neutrophils in peripheral blood of control Piezo1fl/fl and Piezo1ΔPMN mice was quantified using flow cytometry. Blood analyzer, Siemens Advia 120, was used for absolute counts of PMN from blood.
As conventional flow cytometry is limited for multicolor analysis, we used spectral flow cytometry (Cytek Aurora, Cytek Biosciences’) and spectral unmixing to distinguish the differentially expressed surface markers in transmigrated and non-transmigrated PMNs. Briefly, samples were run through a 5-laser spectral flow cytometer (Aurora, Cytek Biosciences). Appropriate quality control was performed per the manufacturer recommendations. For single stained controls, beads (Invitrogen) were used and were acquired first, followed by samples. Cells were used as controls to determine autofluorescence and to determine live/dead staining. Unmixing matrix with autofluorescence extraction was calculated using SpectroFlo 2.2.1 software (Cytek Biosciences). Unmixed data was then analyzed using Kaluza 2.1 software (Beckman Coulter). Fluorescence minus one (FMO) controls were used to determine appropriate gating. Expression changes were calculated by measuring the Median Fluorescent Intensity (MFI). Gating strategy where cells were gated on scatter properties followed by singlets. Live cells negative for staining with Zombie-NIR live/dead stain were selected for further analyses. Gating strategy using Fluorescence minus one (FMO) controls are shown.
Bacterial culture
E. coli DH5α (ATCC #25922GFP) and Pseudomonas aeruginosa (PA-GFP-01 strain; gift from Dr. Terry Manchen, University of California, Berkeley, CA 72) constitutively expressing GFP, were grown in LB Broth (Sigma L3522) with carbenicillin (Goldbio, #C-103-5). Concentration of E. coli at OD 0.4–0.6 was calculate (https://www.agilent.com/store/biocalculators/calcODBacterial.jsp) using Agilent E. coli calculator. The titer of P. aeruginosa was calculated from colony forming units (c.f.u) obtained from serial dilution of over-night grown culture plated on LB-carbenicillin plates. Culture, storage, and handling were followed in BSL2 environment according to the University of Illinois at Chicago’s Institutional Biosafety Committee.
Phagocytosis and intracellular bacterial killing in vitro
PMN mediated bacterial phagocytosis and killing assay was adapted from 73,74. In brief, constant number of PMN were incubated with GFP-E. coli at 20:1 (cell: bacteria) ratio for 30–90 min for ingestion and non-phagocytosed bacteria were removed from incubation medium by gentle centrifugation at 200–300 rcf. Bacteria killing was assessed 30–210 min later. Internalized bacteria were released from PMN by 1% Triton-X. Percentage of bacterial killing was normalized to the ingestion sample.
Colocalization of Nox4 with phagosomal and phago-lysosomal markers
HL-60 cells were transduced with Nox4-mGFP-tagged lentiviral particles (OriGene, #RC208007L4V) and expressing cells were selected with puromycin. The Nox4-mGFP expressing HL-60 were differentiated as described above, plated on a glass bottom dish coated with fibrinogen, and were allowed to adhere for 1h. Pseudomonas aeruginosa at 1:50 (cell: bacteria) ratio were added to the dish on ice to allow for the bacteria to settle down as described elsewhere 75. The HL-60 derived PMN were then incubated at 37°C for 5, 20, and 60 min, fixed, and stained with phalloidin and immunofluorescent stained with antibodies for the early phagosome marker RAB5A (Proteintech, #11947-1-AP), the late phagosome marker RAB7 (Cell Signaling, #9367), and the phagolysosomal marker LAMP1 (Cell Signaling, #15665), respectively. Z-stacks images were captured on the Carl Zeiss LSM 710 BiG confocal microscope driven by the ZEN software. ImageJ (Version 1.53r) and the plugin JaCoP 76 were used to analyze he colocalization of Nox4 with the markers. 3D surface rendering of images was applied using the surface module in Imaris (Version 9.5). In brief, the images of Nox4, F-actin, or phagosome and lysosome markers were individually reconstructed with surface segmentation threshold before compositing into a single image.
Manipulation of restrictiveness of endothelial junction in vivo
The restrictiveness of endothelial junction in mice was decreased by IV injection of 20 μg functional blocking α-VE-cadherin antibody (α-CD144, clone 11D4.1; BD Biosciences #555289) 41. Isotype matched antibody (IgG2a κ, clone R35-95, BD Biosciences # 559073) was IV injected as control. 1h post administration of antibody, animals were subjected to LPS inhalation (1mg/ml in total volume of 5 ml) and used for the experiments at 3 h post-exposure. Transmigrated and non-transmigrated PMN were isolated as described above. Restrictiveness of EC junction was increased by IV injection of 125 μg (6.2 mg/kg) rapalog everolimus (Sigma SML2282) into Cdh5tm4(Cdh5/Fkbp, Ptprb/Mtor*)Dvst mice 38 at 8 h and 4 h before exposure of nebulized LPS. Vehicle was used as control. PMNs derived from bone marrow or lung were treated with 5 nM of everolimus in control experiments.
Fabrication of microfluidic device.
Microfluidic devices were fabricated using soft photolithography 77. Briefly, a glass photomask with the desired microfluidic channels was designed using AutoCAD and fabricated from company (Fineline-imaging Inc). The microchannels were then transferred from the photomask onto a 3” silicon wafer using dry film protocols 78. Microchannels were replicated into the Polydimethylsiloxane (PDMS, Sylgard™ 184, Dow Corning, USA) by casting base and curing agent (10:1 wt/wt) on the silicon wafer, which was cured on a hotplate of 80°C for 2h. PDMS slabs with the microchannels were then bonded onto 1” × 3” glass slides (#12-544-4, Fisher Scientific, USA) using oxygen plasma (PE-50, Plasma Etch Inc, USA). Inlet and outlet holes were punched using a biopsy punch before the bonding. The microchannels include a parallel-array of diamond-shaped with 4–5 μm gap size or circular posts with 5 and 200μm gap size, and the heights of the channels were either 20μm or 50μm, respectively.
Chemotactic PMN transmigration through trans-well pores
PMNs were chemotactically transmigrated through trans-well hanging cell inserts with various pore diameters (polyethylene terephthalate inserts were 3 μm (Millipore MCSP24H48), 5μm (Millipore MCMP24H48), 8 μm (Millipore MCEP24H48) in diameter; polycarbonate inserts were 12 μm (Millipore PIXP01250) in diameter). 1 μM fMLP (N-Formyl-Met-Leu-Phe; Sigma #F3506) chemo-attractant was added to 500 μL of RPMI-1640 medium in 24 well cell culture plate. 5×105 bone marrow PMNs were added on top of hanging cell insert and allowed to chemotactically transmigrate for 90 min. Transmigrated PMNs were collected from both the bottom layer of the insert and the medium. PMN numbers were calculated by automated cell counter (Countess 3, Thermo Fisher).
PMN isolation from bone marrow
Femur and tibia bones were removed from mice immediately after euthanasia by ketamine/xylazine/acepromazine. Bones were initially kept in RPMI medium containing heparin (10 U/ml), and then flushed with HBSS-prep buffer (Ca2+ and Mg2+–free HBSS with 20 mM HEPES, 0.1% BSA). Cell suspension was filtered through 40 μm cell strainer (Falcon 352340), centrifuged at 300g for 5 min at 4°C to pellet down. Cells were gently resuspended in 1 ml of HBSS buffer containing Ca2+ and Mg2+ and overlayed carefully over 5 ml of 63% Precoll (GE Healthcare, #17-0891-01) and centrifuged for 30 min at 500g at 4°C with medium break. PMNs were collected from the bottom of the gradient, treated with RBC lysis buffer (NH4Cl 8.03g/L, KHCO3 1g/L, EDTA-4Na 0.002%) for 1 min on ice. Cells were pelleted at 300g, 5 min, 4°C, and resuspended with 500–800 μL of RPMI or HBSS. Cell densities were kept less than 10×106 per ml.
Adoptive transfer
Bone marrow derived PMNs from donor control and Piezo1ΔPMN mice were transmigrated through microfluidic device, or sheared by rheometer, before transferring i.t. to syngeneic recipient mice of the same age. In the experiments with Nox4 inhibitor, bone marrow murine PMN were treated with 10μM GKT137831 or vehicle prior i.t. adoptive transfer of the cells into lungs of mice. Recipient mice were prior infected with 1×106 GFP-expressing Pseudomonas aeruginosa. After 16 hours, recipient mice were euthanatized at BSL2 facility.
Clearance of lung infection
Mice were infected with 1×106 GFP-expressing Pseudomonas aeruginosa through intra-tracheal route. 16 h post-challenge, lungs were perfused with HBSS, right lung lobes were excised for plating of bacteria into agar plates whereas left lung was inflated by 10% formaldehyde solution (Sigma HT501128) applied through trachea. 1 mg of left lung fragment was homogenized and plated upon serial dilution. Bacterial colonies formed per mg lung lobe were counted at multiple dilutions.
Immuno-histochemistry
Tissue processing, embedding, and sectioning of murine lung samples were performed by UIC’s Research Histology Core. Briefly, following the fixation in 10% formalin, murine lung samples were loaded into ASP 300s automated tissue processor (Leica Biosystems) and dehydrated by a series of ascending graded ethanol, cleared in xylene, and infused with paraffin following a preset protocol. Samples were then embedded in paraffin (Paraplast, Leica Microsystem), and five micrometer sections were cut at three levels with 200μm step. Sections were mounted on positively charged slides, dried, and baked at 60°C for an hour. The lung samples were stained with H&E was by UIC’s Research Histology Core and imaged by UIC’s Research Tissue Imaging Core. Briefly, adhered sections were deparaffinized, stained with H&E, and dehydrated on Leica Autostainer XL (Leica Biosystems) following a preset protocol. Sections were mounted with Micromount media (Leica Biosystems) on CV5030 automated cover-slipper (Leica Biosystems). Stained slides were scanned at 40X magnification on Aperio AT2 brightfield digital scanner (Leica Microsystems). Leica Aperio Imagescope software was used to analyze the images for determining lung injury index.
Assessment of Acute Lung Injury (ALI) Score
The score was calculated according to previously described method 79. Briefly, histology of the lung was used to score for: (A) PMNs in the alveolar space, (B) PMNs in the interstitial space, (C) hyaline membranes, (D) proteinaceous debris filling the airspace, and I alveolar septal thickening. Each item (a-e) was given a score between 0–2. The total score of the field was calculated by:
where is the variable described above and is the number fields analyzed. A minimum score of zero would represent a healthy lung and a maximum score of 1 represents a lung with severe acute lung injury.
Immunofluorescent staining
Immunofluorescent staining was performed to study the clearance of Pseudomonas aeruginosa in murine lung samples. Briefly, adhered sections of murine lung samples were deparaffinized and re-hydrated with descending graded ethanol. Heat antigen retrieval was then performed with a pH 6.0 sodium citrate buffer. The slides were then stained with α-GFP antibody (Santa Cruz Biotechnology, # sc-9996; 1:50) and α-PECAM1 antibody (Abcam, # ab28364; 1:50), or α-GFP antibody (Santa Cruz Biotechnology, # sc-9996; 1:50), α-PDPN antibody (DSHB, #8.1.1; 1:100), α-MPO antibody (abcam, #ab9535; 1:25) overnight at 4°C. The slides were then incubated with appropriate secondary antibodies. Images of stained samples were captured on the Carl Zeiss LSM 710 BiG confocal microscope driven by the ZEN software. ImageJ (Version 1.53r) was used to quantify the number of GFP-positive bacteria and PMN, the distance between bacteria and PMN, bacterial engulfment events, PMN to bacteria ratio.
Cell culture and lentiviral transgenesis
3 to 5 targeting shRNA containing lentiviral particles were purchased from Sigma. Lentivirus were added to HL-60 cells at a 1: 2 (cell: virus) ratio. Infected cells were selected by 2 μg/ml of puromycin (Sigma P8833). The most efficiently depleted cloned were selected for further analyses.
In vivo lung intravital imaging of transmigrated neutrophils.
Surgical methods for gaining access to lung was previously described 80. Briefly, mice anesthetized mice received intravenous injection via retro-orbital sinus of 10 μg per mouse Alexa594-labeled α-Ly6G antibody to stain total PMN before the induction of LPS injury. Mice inhaled 5mg of aerosolized LPS (Lipopolysaccharides from Escherichia coli O55: B5, L2880, Lot#028M4094V; Sigma-Aldrich) dissolved in PBS using nebulizer connected with inhalation box (Compressor nebulizer, MODEL: NE-C30; OMRON). Lungs were imaged after 4 hours post-LPS. Intravenous injection via retro-orbital sinus of SeTau647(SETA BioMedicals)-labeled α-CD31 antibody (25 μg/mice) (390, Biolegend) and BV421-labeled α-Ly6G (Biolegend) antibody (10 μg/mice) were used to stain intravascular PMNs and lung microvascular structures before surgery. The multi-color images were collected as described for calcium imaging. ImageJ and Origin (OriginLab) and customized LabVIEW programs (National instruments) were used to quantify PMN dynamics.
Lung intravital calcium imaging during PMN transendothelial migration
Lung intravital calcium imaging was performed based on Tsukasaki et al 81. Briefly, PMN calcium dynamics in mouse lungs was assessed at 24 hours post-endotoxin challenge induced by i.p. LPS. Intravenous injection via retro-orbital sinus of SeTau647(SETA BioMedicals)-labeled CD31 antibody (25 μg/mice) (390, Biolegend) were performed to stain PMNs and lung microvascular structures before surgery. A resonance-scanning two photon microscope (Ultima Multiphoton Microscopes, Bruker) with an Olympus XLUMPlanFL N 20x (NA 1.00) and Immersion oil (Immersol W (2010); Carl Zeiss) were employed to collect multi-color images (Dichroic mirror; 775 long pass filter (775 LP; Bruker), IR blocking filter; 770 short pass filter (770 SP; Bruker), Emission filter; 546/SP nm for GCaMP (FF01-546/SP; Semrock), 595/60 nm for tdTomato (Bruker) and 708/75 nm for SeTau647 (FF01-708/75-25; Semrock) with 940 nm excitation (Laser power: 18.8 mW/cm2 at back aperture of objective) at video rate. Lung imaging data containing motion artifacts were stabilized by using our own computer vision algorithms 8180. (https://github.com/YoshiTsukasaki/CoVSTii or https://uofi.box.com/v/StandaloneCoVSTii). ImageJ 1.53q and Origin 2022b (OriginLab) and customized LabVIEW programs (National instruments) were used to quantify PMN calcium dynamics. Supplementary Video S1 demonstrates temporal changes in cytosolic [Ca2+] in PMN before, during, and after transmigration through lung endothelial junction labeled by anti-PECAM1antibody (blue).
Cremaster muscle Intravital calcium imaging during PMN transendothelial migration.
PMN transmigration in cremaster muscle was induced by injection of 250 μl of MIP-2 (4 μg/ml; R&D Systems) solution intrascrotally. PMN transmigration in cremaster muscle was 1 h after MIP-2 application. Surgical methods for gaining access to cremaster muscle were previously described 82,83. An Olympus BX upright microscope with LUMPlanFLN60x/1.00W objective and Hamamatsu ORCA Flash 4.0 high-speed cameras (VIVO intravital imaging system, 3i) was used to collect bright field and dual-color images. The images of GCaMPf and tdTomato were collected using EGFP (FITC/Cy2) and DSRed (TRITC/Cy3) (Chrome) filter sets. Respectively. Videos, added as supplementary Video S4, were acquired, processed, and analyzed by using SlideBook (3i) and imageJ and was captured at rate, 2 seconds/frame.
Imaging calcium in PMNs migrating through microfluidic pores
HL-60-derived PMN stably expressing control and Piezo1 shRNA were used in this study. Prior to microfluidic transmigration PMNs were diluted in HBSS buffer (with Ca2+ and Mg2+; Thermo Fisher, # 14025092) to 104 – 5×104 cells per ml. Cells were stained with Fluor-4 AM dye (1 μg/ml; Invitrogen, #F14201) and cell mask membrane stain (Thermo Fisher, #C10046) in suspension for 5 min. After staining, 1ml of cell suspension was loaded into a 5 ml syringe (NORM-JECT ®, Air-Tite Products Co., Inc) and connected with microfluidic device by tubing. Cells were pumped (KD Scientific Inc, # Legato 180) into the device at a minimum volumetric flow rate of 20μl/min flow rate to squeeze through the gaps between the posts. Fluo4 and cell mask membrane stain were imaged using FITC (480/40Ex; 535/50Em) and mCherry (560/40Ex; 590lpEm) filters and LED light source (Xylist120). Time-laps images were captured using an Olympus IX-83 epi-fluorescence microscope equipped with a high-resolution sCMOS camera (Zyla 5.5, Andor Inc). Images were captured at 0.5fps for both channels. Cellsens software was used to record and analyze the data.
Computational modeling and 3D simulation
We coupled the finite element method (FEM) for the cell membrane with a boundary element method of Stokes flow 84 to simulate a PMN-like cell passing through a circular pore at the inter-endothelial gap. The PMN was modeled as an isotropic hyperelastic spherical shell with a typical diameter of 8.5 μm. The initial tension of the cortical cytoskeleton was set to 35 pN/μm 85. The cortical tension under deformation was modeled as:
| (1) |
where is the area change ratio and and are current and initial area. is the maximum allowed area change 85 is the initial cortical tension. In Eq. 1, the tension will reach infinity when area change approaches 2.0, while it gives infinitely large compression stress to prevent collapse when J approaches 0.0. When J=1 (no area deformation), it gives the initial tension . The interior of the PMN is filled with an incompressible viscous fluid. Its dynamics is solved using the boundary element approach 84 with a viscosity of 400 Pa, which is 5 orders higher than the viscosity of water (0.001 Pa s) 86. The detailed coupling algorithm was previously published 84. The contribution of the nucleus in resistance force was ignored due to its “three-lobed” structure, which has a high surface-area-to-volume ratio, leading to negligible resistance. Endothelial cells, which form the circular pore, are modeled as 1-μm thick rigid plate of smoothly rounded edge with radius of 0.5 μm. The diameter of the pore is varied from 0.5 μm to 5 μm based on experimental measurements of the inter-endothelial gap 1313.
We used COMSOL Multiphysics to simulate the flow field in the microfluidic device under given flow rate. The device geometry was imported from a CAD model. The finite element method was used to predict the velocity, pressure, and shear stress in the channel. The shear stress is calculated by multiplying the shear rate by the water viscosity (0.0012 Pa s). Realtime details of computational 3D modeling of membrane tension in a PMN passing through an endothelial junctional pore is shown as supplementary Video S2. The color map shows PMN membrane tension and pressure experienced by the plasma membrane. The simulation on provided supplementary Video S2 are based on the pore of 2 μm in diameter and PMN diameter of 8.5 μm.
Calculating activation probability of Piezo1 channel
Correlation between critical force on membrane to the deformability and activation of Piezo1 channel was obtained from 47,87–89. Mechanical spring constant values of Piezo1 was used from studies by Lin et al. 2019 47. The critical force on membrane was estimated as 18 pN to activate Piezo1 and the critical tension (half-activation tension) as 0.6 kBT nm−2, which is between the values of 0.35 kBT nm−2 87 and 1.2 kBT nm−2 88. These values can be translated to 1440 pN/μm, 2468 pN/μm, and 7820pN/μm, respectively. Recently, Yang et al. 2022 89 calculated a half maximal activation tension of about 1900 p/μm, based on the measured in-plane membrane area expansion and stiffness constant of Piezo1. Lewis and Grandl measured Piezo1 currents evoked by membrane stretch in three patch configurations by experiments 87. The current curve is fitted as where is current, is the plateau, is tension, is the tension of half-maximal activation, and is the slope factor. For +5 mmHg pre-pulse, values were obtained as . We consider the activation probability of PIEZO1 under tension as,
| (2) |
When is large, the probability approaches 1. When . When . Using this formula, we determined the activation probability of PIEZO1 in our simulations and plotted the right Y axis based on the membrane tension , obtained from simulations. Similarly, we also predicted the activation probability of PIEZO1 based on the pressure on the membrane by
| (3) |
Where is the pressure on the membrane, and and is estimated by and PMN diameter (8.5μm) based on the Young-Laplace equation.
For computational modeling of the transmigration process, we used our in-house code (Lu and Peng, Physics of Fluids, 31:031902, 2019), in which finite element modeling of cell membrane is coupled with the boundary integral method analysis of surrounding fluid. It is written in Fortran. We also used open-source code Paraview 5.7.0 for visualization.
Mouse survival experiments
PMN-specific Piezo1−/− and NOX4−/−and their respective floxed mice were infected with GFP-expressing Pseudomonas aeruginosa (PA-GFP-01 strain) at 1×106 concentration by intra tracheal route at the BSL2 facility. 9 animals per group were used for the survival studies.
RNA isolation
Total RNA from flow sorted lung PMNs were isolated by RNeasy Plus kit (Qiagen, #74034). PMNs sorted from 6 mice were pooled into 1 sample. Sorting was performed at different days to prepare three independent sample sets. Concentrations of isolated RNA samples were measured using Nanodrop 1000 (Thermo Fisher) at 260 nm, and purity was assessed by 260/280 nm optical density ratio. RNA samples were transcribed to complementary DNAs with random primers using “high-capacity cDNA reverse transcription kit” (Thermo Fischer, #4368814).
RT-qPCR
Quantitative gene expressions were determined by real-time PCR upon mixing the cDNAs with FastStart Universal SYBR Green Master (Rox: Millipore Sigma, #4913914001) with specific qPCR primers. Amplification of PCR reactions were carried out on ABI Prism 7000 system and real-time amplification data was analyzed on Quant studio7 Flex Real-Time PCR System (Thermo Fisher). Results were calculated using the comparative CT-method 90, and expressed relative to the expression of the housekeeping gene.
RNA sequencing
RNA quality and quantity was assessed using the Agilent bio-analyzer. All samples showed RNA integrity number > 8. Strand-specific RNA-SEQ libraries were prepared using a TruSEQ mRNA RNA-SEQ library protocol (Illumina provided). Library quality and quantity was assessed using the Agilent bio-analyzer and libraries were sequenced using an Illumina NovaSEQ6000 (Illumina provided reagents and protocols).
RNAseq analysis of transmigrated and non-transmigrated neutrophils.
Gene expression quantification.
Raw reads were trimmed to remove Truseq adapters and bases from the 3’ end with quality scores less than 20 using cutadapt (Martin, 2011); trimmed reads shorter than 40bp were discarded. Trimmed reads were aligned reference genome mm10 using STAR aligner 91. ENSEMBL gene expression was quantified using FeatureCounts 92.
Differential expression.
Differential expression statistics (fold-change and p-value) were computed using edgeR 93,94, using generalized linear models to correct for batch effect while testing for differences between LPS treatment groups. P-values were adjusted for multiple testing using the false discovery rate (FDR) correction of Benjamini and Hochberg 95. Significant genes were determined based on an FDR threshold of 5% (0.05).
Pathway analysis.
We ran the core analysis in Ingenuity Pathway Analysis on differentially expressed genes to obtain canonical pathways and upstream regulators that were associated with differentially expressed genes. Significantly enriched pathways were determined based on FDR-adjusted p-value < 0.05. Groups of significantly enriched pathways with similar function were grouped into super-pathways based on manual curation of the terms. Z-scored log-scaled normalized gene expression levels for all differentially expressed genes within these super-pathways were plotted in heatmaps.
Intracellular ratiometric Ca2+ measurements
Dual-color images were captured simultaneously for tdTomato and Fluo-4. Ratiometric calcium signal analysis was performed according to previously published method 96. For comparisons of relative changes in [Ca2+]cyt a cross-sectional view of the intensity values of Fluo-4 normalized to tdTomato intensity values was generated at each time point using Image J, and the relative changes in [Ca2+]cyt were calculated.
Western blotting
PMNs were lysed using RIPA buffer (Sigma-Aldrich Cat. R0278) and concentration was determined using Bicinchoninic acid assay/Pierce BCA Protein Assay Kit (Thermo Scientific, REF. 23225). Samples were resolved by SDS-PAGE, transferred to 0.45um Nitrocellulose Membrane (Bio-Rad Cat. #1620115) and incubated overnight with Piezo1 (Protein Tech, 15939-1-AP), NOX4 (Novus, NB110-58849), Vinculin (Abcam, ab129002) antibodies. After incubation with secondary antibody, protein was detected by chemiluminescence using PICO PLUS (Thermo scientific, REF 34580) and captured using Autoradiography films (LabForce, 1141J51).
ROS generation assay
Generation of reactive oxygen species by HL-60-derived PMN stably expressing control, Piezo-1 and NOX4 shRNA were measured by amplex red reagent (Invitrogen A22188). Amplex red (10-acetyl-3,7-dihydroxy-phenoxazine) detects H2O2 (a direct product of Nox4 activity, or by-product of free oxygen radicals) upon converting to resorufin (Exm: 571nm; Emm: 585nm; absorbance: 560nm). Phorbol-ester was used to enhance the signal.
NETosis assay
FACS-sorted transmigrated and non-transmigrated neutrophils were incubated with GFP Pseudomonas aeruginosa @ 1: 10 m.o.i. for 1h. 1×105 neutrophils were used per assay. Post incubation supernatant was used for elastase ELISA following manufacturers (mouse neutrophil elastase ELISA kit; ab252356) protocol. Frequency of NET formation is plotted as OD at 450 nm.
Neutrophil longevity assay
Transmigrated and non-transmigrated neutrophils sorted by FACS were cultured ex vivo in RPMI supplemented with a supernatant of mouse primary lung microvascular endothelial cell (7:3). Viability of neutrophils were tested at different time points by staining with trypan blue.
QUANTIFICATION, STATISTICAL ANALYSIS AND REPRODUCIBILITY OF EXPERIMENTS
Ratiometric calcium signal quantitation was plotted by Origin Pro, rest of the graphs were generated using GraphPad Prism. All data were expressed as mean ± standard error. One-way Anova was used for pairwise comparison with multiple groups, and two-tailed t-test was used to determine statistical significance. Number of samples or mice per group, replication in independent experiments and statistical tests are mentioned in the figure legends. For studies in mice, significance level of 0.05 and power of 0.9 were used to estimate the group sizes (JMP release 6 software: SAS, Cary NC). In RNA-seq analyses, the log2 fold-change was used to determine the effect size.
Supplementary Material
Table S1. RNAseq analyzed data; differentially expressed genes in WT and Piezo1ΔPMN PMN; pathway analysis, related to Figure 5a–g and Suppl. Figure 4a–e.
Video S1. Lung intra-vital imaging showing relative changes in cytosolic [Ca2+] in PMN during trans-endothelial migration, related to Figure 3a–c and STAR Methods.
Video S2. Computational 3D modeling of membrane tension in PMN passing through endothelial junctional pore, related to Figure 3g and STAR Methods.
Video S3. Squeezing event of a PMN passing through 5 μm pores of the microfluidic system, related to Suppl. Figure 2g and STAR Methods.
Video S4. Raw intravital video of PMN transmigration and calcium fluctuation in cremaster muscle, related to Suppl. Figure 2c–e and STAR Methods.
Video S5. Computational modeling of membrane tension in PMN passing through the microfluidic device, related to Suppl. Figure 2i–j and STAR Methods.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal α-mouse Ly-6G conjugated with Brilliant Violet 421™ | Biolegend | Cat #127628 |
| Rat monoclonal α-mouse Ly-6G conjugated with Alexa Fluor® 594 | Biolegend | Cat #127636 |
| Rat monoclonal α-VE-cadherin | BD Biosciences | Cat #555289 |
| Rat monoclonal isotype control | BD Biosciences | Cat # 559073 |
| Mouse monoclonal α-Green fluorescent protein | Santa Cruz Biotechnology | Cat # sc-9996 |
| Rabbit polyclonal α-CD31 (PECAM1) | Abcam | Cat # ab28364 |
| Rat monoclonal α-mouse CD31 conjugated with Brilliant Violet 421™ | Biolegend | Cat # 102424 |
| Rabbit polyclonal α-Piezo1 | Proteintech | Cat # 15939-1-AP |
| Rabbit polyclonal α-Nox4 | Novus | Cat # NB110-58849 |
| Rabbit polyclonal α-Vinculin | Abcam | Cat # ab129002 |
| Mouse monoclonal α-LAMP1 (D4O1S) | Cell Signaling | Cat # 15665 |
| Rabbit monoclonal α-RAB7 (D95F2) | Cell Signaling | Cat # 9367 |
| Syrian hamster monoclonal α-Podoplanin (PDPN) | DSHB | Cat # 8.1.1 |
| Rabbit polyclonal α-RAB5A | Proteintech | Cat # 11947-1-AP |
| APC anti-human CD62L Antibody | Biolegend | Cat # 304809 |
| Brilliant Violet 510™ anti-human CD184 (CXCR4) Antibody | Biolegend | Cat # 306535 |
| Brilliant Violet 711™ anti-human CD15 (SSEA-1) Antibody | Biolegend | Cat # 323049 |
| Alexa Fluor® 700 anti-human CD47 Antibody | Biolegend | Cat # 323125 |
| Brilliant Violet 785™ anti-human CD10 Antibody | Biolegend | Cat # 312237 |
| Brilliant Violet 650™ anti-human CD15 (SSEA-1) Antibody | Biolegend | Cat # 323033 |
| Alexa Fluor® 594 anti-mouse/human CD11b Antibody | Biolegend | Cat # 101254 |
| Zombie NIR live dead stain | Biolegend | Cat # 77184 |
| Bacterial and virus strains | ||
| Escherichia coli-GFP | ATCC | Cat #25922GFP |
| Pseudomonas aeruginosa (PA-GFP-01 strain) | Laboratory of Terry Manchen | Davey et. al., 2003 |
| Chemicals, peptides, and recombinant proteins | ||
| The collagenase type I from Clostridium histolyticum | Millipore-Sigma | Cat # SCR103 |
| Lipopolysaccharides from Escherichia coli O55:B5 | Millipore-Sigma | Cat # L2880 |
| Carbenicillin | Goldbio | Cat #C-103-5 |
| Everolimus | Millipore-Sigma | Cat # SML2282 |
| Formaldehyde solution | Millipore-Sigma | Cat # HT501128 |
| N-Formyl-Met-Leu-Phe | Millipore-Sigma | Cat # F3506 |
| Puromycin | Millipore-Sigma | Cat # P8833 |
| Fluor-4, AM | Invitrogen | Cat # F14201 |
| CellMask™ Deep Red Plasma membrane stain | Thermo Fisher | Cat #C10046 |
| E. coli-derived mouse CXCL2/GRO beta/MIP-2/CINC-3 protein | R&D Systems | Cat # 452-M2-050 |
| Yoda1 | Millipore-Sigma | Cat # SML1558 |
| Setanaxib (GKT137831) | Selleckchem | Cat # S7171 |
| Thapsigargin | Millipore-Sigma | Cat # T9033 |
| Ionomycin | Millipore-Sigma | Cat # I9657 |
| Amplex red reagent | Invitrogen | Cat # A22188 |
| FastStart Universal SYBR Green Master | Millipore-Sigma | Cat # 4913914001 |
| Pico Plus | Thermo Fischer | Cat # 34580 |
| Critical commercial assays | ||
| High-capacity cDNA reverse transcription kit” | Thermo Fisher | Cat #4368814 |
| RNeasy Plus kit | Qiagen | Cat # #74034 |
| Easystep human neutrophil isolation kit | Stemcell technologies | Cat #19666 |
| Pierce BCA Protein Assay Kit | Thermo Fischer | Cat # 23225 |
| neutrophil elastase ELISA kit | Abcam | Cat # ab252356 |
| Experimental models: Cell lines | ||
| Human promyelocytic leukemia (HL-60) cells | ATCC | Cat # CCL-240 |
| C57BL/6 Mouse Primary Lung Microvascular Endothelial Cells | Cell Biologics | Cat # C57-6011 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory | RRID: IMSR_JAX:000664 |
| Mouse: C57BL/6-Ly6g(tm2621(Cre-tdTomato) | from Dr. Matthias Gunzer’s lab | Hasenberg, A. et al., 2015 |
| Mouse: Cdh5tm4(Cdh5/Fkbp, Ptprb/Mtor*)Dvst | from Dr. Dietmar Vestweber’s lab | Broermann, A. et al., 2011 |
| Mouse: B6.Cg-Piezo1tm2.1Apat/J | The Jackson Laboratory | RRID: #IMSR_JAX:029213 |
| Mouse: B6(129S4)-Gt(ROSA)26Sortm1.1(CAG-tdTomato/GCaMP6f)Mdcah/J | The Jackson Laboratory | RRID: #IMSR_JAX:031968 |
| Mouse: Nox4 floxed | from Dr. Junichi Sadoshima | Ref.97 |
| Piezo1ΔPMN | this manuscript | N/A |
| NOX4ΔPMN | this manuscript | N/A |
| Oligonucleotides | ||
| MISSION® shRNA lentiviral particles Piezo1 shRNA#2: CTCACCAAGAAGTACAATCAT | Millipore-Sigma | TRCN0000121969 |
| MISSION® shRNA lentiviral particles Piezo1 shRNA#3: GTACTTCGTGAAGTGCATCTA | Millipore-Sigma | TRCN0000143190 |
| MISSION® shRNA lentiviral particles Nox4 shRNA#1: GAGCCTCAGCATCTGTTCTTA | Millipore-Sigma | TRCN0000046092 |
| MISSION® shRNA lentiviral particles Nox4 shRNA#2: CAGAGTTTACCCAGCACAAAT | Millipore-Sigma | TRCN0000046091 |
| MISSION® shRNA lentiviral particles Nox4 shRNA#3: CCCTCAACTTCTCAGTGAATT | Millipore-Sigma | TRCN0000046090 |
| MISSION® shRNA lentiviral particles Nox4 shRNA#4: GCTGTATATTGATGGTCCTTT | Millipore-Sigma | TRCN0000046089 |
| MISSION® shRNA lentiviral particles Nox4 shRNA#5: CCTGTGGTGTTACTATCTGTA | Millipore-Sigma | TRCN0000046088 |
| MISSION® shRNA lentiviral particles | Millipore-Sigma | TRCN0000003808 |
| Hif1α shRNA#1: CCGCTGGAGACACAATCATAT | ||
| MISSION® shRNA lentiviral particles Hif1α shRNA#2: CCAGTTATGATTGTGAAGTTA | Millipore-Sigma | TRCN0000003809 |
| MISSION® shRNA lentiviral particles Hif1α shRNA#3: GTGATGAAAGAATTACCGAAT | Millipore-Sigma | TRCN0000003810 |
| MISSION® shRNA lentiviral particles Hif1α shRNA#4: CGGCGAAGTAAAGAATCTGAA | Millipore-Sigma | TRCN0000003811 |
| MISSION® shRNA lentiviral particles Hif1α shRNA#5: TGCTCTTTGTGGTTGGATCTA | Millipore-Sigma | TRCN0000010819 |
| Primer: Nox1 Forward: CCGGTCATTCTTTATATCTGTG | This manuscript | N/A |
| Primer: Nox1 Reserved: CAACCTTGGTAATCACAACC | This manuscript | N/A |
| Primer: Nox3 Forward: CAAACACAACCACTGAATTG | This manuscript | N/A |
| Primer: Nox3 Reserved: CATGATCAAGACTAAAGCCAG | This manuscript | N/A |
| Primer: Nox4 Forward: AGTCCTTCCGTTGGTTTG | This manuscript | N/A |
| Primer: Nox4 Reserved: AAAGTTTCCACCGAGGAC | This manuscript | N/A |
| Primer: Nox5 Forward: AGGTGTTCTATTGGACTCAC | This manuscript | N/A |
| Primer: Nox5 Reserved: CTCCAGGAAAAACAAGATTCC | This manuscript | N/A |
| Primers: Nox2 | Santa Cruz Biotechnology | Cat # 35503-PR |
| Software and algorithms | ||
| SUMMIT, 6.1 | Beckman Coulter | https://www.beckmancoulter.com/wsrportal/techdocs?docname=cyan_adp_instructions_for_use.pdf |
| Kaluza Analysis 2.1 | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| E. coli calculator | Agilent | https://www.agilent.com/store/biocalculators/calcODBacterial.jsp |
| AutoCAD | AutoDESK | https://www.autodesk.com/products/autocad/overview?term=1-YEAR&tab=subscription |
| Countess™ 3 | Thermo Fisher | https://www.thermofisher.com/order/catalog/product/A51025 |
| Rspace | Netzsch | https://analyzing-testing.netzsch.com/en-US/products/software/rspace-software |
| Aperio ImageScope | Leica Biosystems | https://www.leicabiosystems.com/us/digital-pathology/manage/aperio-imagescope/ |
| Zen | Carl Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| Alpha Fold | DeepMind | https://alphafold.ebi.ac.uk/ |
| UCSF Chimera | UCSF | https://www.cgl.ucsf.edu/chimera/ |
| Image j | NIH | https://imagej.nih.gov/ij/notes.html |
| Slidebook (3i) | Intelligent Imaging | https://www.intelligent-imaging.com/slidebook |
| In-house computer vision algorithms 8180808080802 for lung 2-photon intravital imaging | Yoshikaju Tsukasaki (Author of this paper) | (https://github.com/YoshiTsukasaki/CoVSTii or https://uofi.box.com/v/StandaloneCoVSTii) |
| LabView | National instruments corp. | https://www.ni.com/en-us/shop/labview.html |
| Paraview 5.7.0 | Kitware | https://www.paraview.org/ |
| Cellsens | Olympus | https://www.olympus-lifescience.com/en/software/cellsens/ |
| Cutadapt v3.4 | Marcel Martin | https://cutadapt.readthedocs.io/en/v3.4/installation.html |
| STAR v2.7.0 aligner | Alexander Dobin | https://academic.oup.com/bioinformatics/article/29/1/15/272537Z |
| FeatureCounts v1.5.0 | Yang Liao | http://subread.sourceforge.net/ |
| R package edgeR v.3.36 | https://www.r-project.org/ | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Ingenuity Pathway Analysis | Qiagen | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-ipa/ |
| Gene Ontology Biological Process | MSigDB | https://www.gsea-msigdb.org/gsea/msigdb/ |
| Origin Pro | Origin lab | https://www.originlab.com/origin |
| Prism | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| Illustrator | Adobe | https://www.adobe.com/products/illustrator.html |
| Biorender | Biorender | https://biorender.com/ |
| Spectroflo 2.2.1 | Cytek Biosciences | https://cytekbio.com/pages/spectro-flo |
| Other | ||
| RBC lysis buffer | Thermo Fisher | Cat #00-4333-57 |
| RPMI 1640 Medium, HEPES | Thermo Fisher | Cat #22400089 |
| Endothelial Cell Medium/w Kit | Cell Biologics | Cat # M1168 |
| Cell strainer | Thermo Fisher | Cat #08-771-1 |
| polyethylene terephthalate insert 3μm diam. | Millipore-Sigma | Cat #MCSP24H48 |
| polyethylene terephthalate insert 5μm diam. | Millipore-Sigma | Cat #MCMP24H48 |
| polyethylene terephthalate insert 8μm diam. | Millipore-Sigma | Cat #MCEP24H48 |
| polycarbonate insert 12 μm diam. | Millipore-Sigma | Cat #PIXP01250 |
| Percoll® PLUS | GE Healthcare | Cat #17-0891-01 |
Highlights.
Trans-endothelial migration regulates transcription & antimicrobial function of PMN
Increased membrane tension during transmigration triggers Piezo1 gating & Ca2+ influx
Piezo1 signals transcriptional upregulation of Nox4 via Hif1α stabilization
Acknowledgements
We thank Dr. Matthias Gunzer from University Duisburg-Essen, Essen, Germany for providing the Catchup mouse; Dr. Dietmar Vestweber from Max Plank Institute, Germany, for VE-PTP-VE-cadherin transgenic mouse; Qiyue Luan from University of Illinois, Chicago, for assistance with live-cell imaging; Drs. Sreeparna Chakraborty, Abhalaxmi Singh and Abdul S. Qadir from University of Illinois, Chicago, for help with isolation of lung PMN; Flow Cytometry Core facility for sorting and flow analysis; Research Histology Core facility for preparing H&E sections; Research Informatics Core facility for RNA-seq data analysis; Genomics Facility of University of Chicago for RNA-sequencing; Guilan Liu and Dr. Shuangping Zhao for experiments with mice. This work was funded by R01 HL045638, P01 HL 151327, P01 HL 060678, R01 HL149300 and UL1TR002003.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of Interests
We declare that none of the authors have competing financial or non-financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Degrossoli A, Muller A, Xie K, Schneider JF, Bader V, Winklhofer KF, Meyer AJ, and Leichert LI (2018). Neutrophil-generated HOCl leads to non-specific thiol oxidation in phagocytized bacteria. Elife 7. 10.7554/eLife.32288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dziarski R, Platt KA, Gelius E, Steiner H, and Gupta D (2003). Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood 102, 689–697. 10.1182/blood-2002-12-3853. [DOI] [PubMed] [Google Scholar]
- 3.Balamayooran G, Batra S, Fessler MB, Happel KI, and Jeyaseelan S (2010). Mechanisms of neutrophil accumulation in the lungs against bacteria. Am J Respir Cell Mol Biol 43, 5–16. 10.1165/rcmb.2009-0047TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yipp BG, Kim JH, Lima R, Zbytnuik LD, Petri B, Swanlund N, Ho M, Szeto VG, Tak T, Koenderman L, et al. (2017). The Lung is a Host Defense Niche for Immediate Neutrophil-Mediated Vascular Protection. Sci Immunol 2. 10.1126/sciimmunol.aam8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekheri M, El Kebir D, Edner N, and Filep JG (2020). 15-Epi-LXA(4) and 17-epi-RvD1 restore TLR9-mediated impaired neutrophil phagocytosis and accelerate resolution of lung inflammation. Proc Natl Acad Sci U S A 117, 7971–7980. 10.1073/pnas.1920193117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nourshargh S, and Alon R (2014). Leukocyte migration into inflamed tissues. Immunity 41, 694–707. 10.1016/j.immuni.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 7.Mittal M, Nepal S, Tsukasaki Y, Hecquet CM, Soni D, Rehman J, Tiruppathi C, and Malik AB (2017). Neutrophil Activation of Endothelial Cell-Expressed TRPM2 Mediates Transendothelial Neutrophil Migration and Vascular Injury. Circ Res 121, 1081–1091. 10.1161/CIRCRESAHA.117.311747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prinyakupt J, and Pluempitiwiriyawej C (2015). Segmentation of white blood cells and comparison of cell morphology by linear and naive Bayes classifiers. Biomed Eng Online 14, 63. 10.1186/s12938-015-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etich J, Bergmeier V, Frie C, Kreft S, Bengestrate L, Eming S, Mauch C, Eckes B, Ulus H, Lund FE, et al. (2013). PECAM1(+)/Sca1(+)/CD38(+) vascular cells transform into myofibroblast-like cells in skin wound repair. PLoS One 8, e53262. 10.1371/journal.pone.0053262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemiec MJ, De Samber B, Garrevoet J, Vergucht E, Vekemans B, De Rycke R, Bjorn E, Sandblad L, Wellenreuther G, Falkenberg G, et al. (2015). Trace element landscape of resting and activated human neutrophils on the sub-micrometer level. Metallomics 7, 996–1010. 10.1039/c4mt00346b. [DOI] [PubMed] [Google Scholar]
- 11.Grinstein S, Furuya W, and Cragoe EJ Jr. (1986). Volume changes in activated human neutrophils: the role of Na+/H+ exchange. J Cell Physiol 128, 33–40. 10.1002/jcp.1041280107. [DOI] [PubMed] [Google Scholar]
- 12.Muller WA (2011). Mechanisms of leukocyte transendothelial migration. Annu Rev Pathol 6, 323–344. 10.1146/annurev-pathol-011110-130224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heemskerk N, Schimmel L, Oort C, van Rijssel J, Yin T, Ma B, van Unen J, Pitter B, Huveneers S, Goedhart J, et al. (2016). F-actin-rich contractile endothelial pores prevent vascular leakage during leukocyte diapedesis through local RhoA signalling. Nat Commun 7, 10493. 10.1038/ncomms10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hind LE, Ingram PN, Beebe DJ, and Huttenlocher A (2018). Interaction with an endothelial lumen increases neutrophil lifetime and motility in response to P aeruginosa. Blood 132, 1818–1828. 10.1182/blood-2018-05-848465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gronloh MLB, Arts JJG, and van Buul JD (2021). Neutrophil transendothelial migration hotspots - mechanisms and implications. J Cell Sci 134. 10.1242/jcs.255653. [DOI] [PubMed] [Google Scholar]
- 16.Stroka KM, and Aranda-Espinoza H (2011). Endothelial cell substrate stiffness influences neutrophil transmigration via myosin light chain kinase-dependent cell contraction. Blood 118, 1632–1640. 10.1182/blood-2010-11-321125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warnatsch A, Tsourouktsoglou TD, Branzk N, Wang Q, Reincke S, Herbst S, Gutierrez M, and Papayannopoulos V (2017). Reactive Oxygen Species Localization Programs Inflammation to Clear Microbes of Different Size. Immunity 46, 421–432. 10.1016/j.immuni.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee WL, Harrison RE, and Grinstein S (2003). Phagocytosis by neutrophils. Microbes Infect 5, 1299–1306. 10.1016/j.micinf.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Dale DC, Boxer L, and Liles WC (2008). The phagocytes: neutrophils and monocytes. Blood 112, 935–945. 10.1182/blood-2007-12-077917. [DOI] [PubMed] [Google Scholar]
- 20.Panday A, Sahoo MK, Osorio D, and Batra S (2015). NADPH oxidases: an overview from structure to innate immunity-associated pathologies. Cell Mol Immunol 12, 5–23. 10.1038/cmi.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Neill S, Brault J, Stasia MJ, and Knaus UG (2015). Genetic disorders coupled to ROS deficiency. Redox Biol 6, 135–156. 10.1016/j.redox.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deffert C, Cachat J, and Krause KH (2014). Phagocyte NADPH oxidase, chronic granulomatous disease and mycobacterial infections. Cell Microbiol 16, 1168–1178. 10.1111/cmi.12322. [DOI] [PubMed] [Google Scholar]
- 23.Buvelot H, Posfay-Barbe KM, Linder P, Schrenzel J, and Krause KH (2017). Staphylococcus aureus, phagocyte NADPH oxidase and chronic granulomatous disease. FEMS Microbiol Rev 41, 139–157. 10.1093/femsre/fuw042. [DOI] [PubMed] [Google Scholar]
- 24.Canton J, and Grinstein S (2014). Priming and activation of NADPH oxidases in plants and animals. Trends Immunol 35, 405–407. 10.1016/j.it.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Fairn GD, and Grinstein S (2012). How nascent phagosomes mature to become phagolysosomes. Trends Immunol 33, 397–405. 10.1016/j.it.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Moltyaner Y, Geerts WH, Chamberlain DW, Heyworth PG, Noack D, Rae J, Doyle JJ, and Downey GP (2003). Underlying chronic granulomatous disease in a patient with bronchocentric granulomatosis. Thorax 58, 1096–1098. 10.1136/thorax.58.12.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winterbourn CC, Kettle AJ, and Hampton MB (2016). Reactive Oxygen Species and Neutrophil Function. Annu Rev Biochem 85, 765–792. 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 28.Coste B, Mathur J, Schmidt M, Earley TJ, Ranade S, Petrus MJ, Dubin AE, and Patapoutian A (2010). Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60. 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kefauver JM, Ward AB, and Patapoutian A (2020). Discoveries in structure and physiology of mechanically activated ion channels. Nature 587, 567–576. 10.1038/s41586-020-2933-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nisimoto Y, Diebold BA, Cosentino-Gomes D, and Lambeth JD (2014). Nox4: a hydrogen peroxide-generating oxygen sensor. Biochemistry 53, 5111–5120. 10.1021/bi500331y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buck T, Hack CT, Berg D, Berg U, Kunz L, and Mayerhofer A (2019). The NADPH oxidase 4 is a major source of hydrogen peroxide in human granulosa-lutein and granulosa tumor cells. Sci Rep 9, 3585. 10.1038/s41598-019-40329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Lohneysen K, Noack D, Wood MR, Friedman JS, and Knaus UG (2010). Structural insights into Nox4 and Nox2: motifs involved in function and cellular localization. Mol Cell Biol 30, 961–975. 10.1128/MCB.01393-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yap B, and Kamm RD (2005). Mechanical deformation of neutrophils into narrow channels induces pseudopod projection and changes in biomechanical properties. J Appl Physiol (1985) 98, 1930–1939. 10.1152/japplphysiol.01226.2004. [DOI] [PubMed] [Google Scholar]
- 34.Makino A, Shin HY, Komai Y, Fukuda S, Coughlin M, Sugihara-Seki M, and Schmid-Schonbein GW (2007). Mechanotransduction in leukocyte activation: a review. Biorheology 44, 221–249. [PubMed] [Google Scholar]
- 35.Kumar S (2014). Cellular mechanotransduction: stiffness does matter. Nat Mater 13, 918–920. 10.1038/nmat4094. [DOI] [PubMed] [Google Scholar]
- 36.Chiang EY, Hidalgo A, Chang J, and Frenette PS (2007). Imaging receptor microdomains on leukocyte subsets in live mice. Nat Methods 4, 219–222. 10.1038/nmeth1018. [DOI] [PubMed] [Google Scholar]
- 37.Hasenberg A, Hasenberg M, Mann L, Neumann F, Borkenstein L, Stecher M, Kraus A, Engel DR, Klingberg A, Seddigh P, et al. (2015). Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat Methods 12, 445–452. 10.1038/nmeth.3322. [DOI] [PubMed] [Google Scholar]
- 38.Broermann A, Winderlich M, Block H, Frye M, Rossaint J, Zarbock A, Cagna G, Linnepe R, Schulte D, Nottebaum AF, and Vestweber D (2011). Dissociation of VE-PTP from VE-cadherin is required for leukocyte extravasation and for VEGF-induced vascular permeability in vivo. J Exp Med 208, 2393–2401. 10.1084/jem.20110525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedrich EE, Hong Z, Xiong S, Zhong M, Di A, Rehman J, Komarova YA, and Malik AB (2019). Endothelial cell Piezo1 mediates pressure-induced lung vascular hyperpermeability via disruption of adherens junctions. Proc Natl Acad Sci U S A 116, 12980–12985. 10.1073/pnas.1902165116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nawroth R, Poell G, Ranft A, Kloep S, Samulowitz U, Fachinger G, Golding M, Shima DT, Deutsch U, and Vestweber D (2002). VE-PTP and VE-cadherin ectodomains interact to facilitate regulation of phosphorylation and cell contacts. EMBO J 21, 4885–4895. 10.1093/emboj/cdf497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gotsch U, Borges E, Bosse R, Boggemeyer E, Simon M, Mossmann H, and Vestweber D (1997). VE-cadherin antibody accelerates neutrophil recruitment in vivo. J Cell Sci 110 (Pt 5), 583–588. 10.1242/jcs.110.5.583. [DOI] [PubMed] [Google Scholar]
- 42.Suresh Kumar V, Sadikot RT, Purcell JE, Malik AB, and Liu Y (2014). Pseudomonas aeruginosa induced lung injury model. J Vis Exp, e52044. 10.3791/52044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murthy SE, Dubin AE, and Patapoutian A (2017). Piezos thrive under pressure: mechanically activated ion channels in health and disease. Nat Rev Mol Cell Biol 18, 771–783. 10.1038/nrm.2017.92. [DOI] [PubMed] [Google Scholar]
- 44.Millius A, and Weiner OD (2010). Manipulation of neutrophil-like HL-60 cells for the study of directed cell migration. Methods Mol Biol 591, 147–158. 10.1007/978-1-60761-404-3_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thastrup O, Cullen PJ, Drøbak BK, Hanley MR, and Dawson AP (1990). Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc Natl Acad Sci U S A 87, 2466–2470. 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C, and Hermann TE (1978). Characterization of ionomycin as a calcium ionophore. J Biol Chem 253, 5892–5894. [PubMed] [Google Scholar]
- 47.Lin Y-C, Guo YR, Miyagi A, Levring J, MacKinnon R, and Scheuring S (2019). Force-induced conformational changes in PIEZO1. Nature 573, 230–234. 10.1038/s41586-019-1499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Z, Pak OS, Feng Z, Liu AP, and Young Y-N (2016). On the gating of mechanosensitive channels by fluid shear stress. Acta Mechanica Sinica 32, 1012–1022. 10.1007/s10409-016-0606-y. [DOI] [Google Scholar]
- 49.Ranade SS, Qiu Z, Woo SH, Hur SS, Murthy SE, Cahalan SM, Xu J, Mathur J, Bandell M, Coste B, et al. (2014). Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc Natl Acad Sci U S A 111, 10347–10352. 10.1073/pnas.1409233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Syeda R, Xu J, Dubin AE, Coste B, Mathur J, Huynh T, Matzen J, Lao J, Tully DC, Engels IH, et al. (2015). Chemical activation of the mechanotransduction channel Piezo1. Elife 4. 10.7554/eLife.07369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Solis AG, Bielecki P, Steach HR, Sharma L, Harman CCD, Yun S, de Zoete MR, Warnock JN, To SDF, York AG, et al. (2019). Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity. Nature 573, 69–74. 10.1038/s41586-019-1485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada M, Kubo H, Kobayashi S, Ishizawa K, He M, Suzuki T, Fujino N, Kunishima H, Hatta M, Nishimaki K, et al. (2011). The increase in surface CXCR4 expression on lung extravascular neutrophils and its effects on neutrophils during endotoxin-induced lung injury. Cell Mol Immunol 8, 305–314. 10.1038/cmi.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green DE, Murphy TC, Kang BY, Kleinhenz JM, Szyndralewiez C, Page P, Sutliff RL, and Hart CM (2012). The Nox4 inhibitor GKT137831 attenuates hypoxia-induced pulmonary vascular cell proliferation. Am J Respir Cell Mol Biol 47, 718–726. 10.1165/rcmb.2011-0418OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Artemenko Y, Axiotakis L Jr., Borleis J, Iglesias PA, and Devreotes PN (2016). Chemical and mechanical stimuli act on common signal transduction and cytoskeletal networks. Proc Natl Acad Sci U S A 113, E7500–E7509. 10.1073/pnas.1608767113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ekpenyong AE, Toepfner N, Chilvers ER, and Guck J (2015). Mechanotransduction in neutrophil activation and deactivation. Biochim Biophys Acta 1853, 3105–3116. 10.1016/j.bbamcr.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 56.Burns AR, Smith CW, and Walker DC (2003). Unique structural features that influence neutrophil emigration into the lung. Physiol Rev 83, 309–336. 10.1152/physrev.00023.2002. [DOI] [PubMed] [Google Scholar]
- 57.Nauseef WM, and Borregaard N (2014). Neutrophils at work. Nat Immunol 15, 602–611. 10.1038/ni.2921. [DOI] [PubMed] [Google Scholar]
- 58.Komarova YA, Kruse K, Mehta D, and Malik AB (2017). Protein Interactions at Endothelial Junctions and Signaling Mechanisms Regulating Endothelial Permeability. Circ Res 120, 179–206. 10.1161/CIRCRESAHA.116.306534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Juettner VV, Kruse K, Dan A, Vu VH, Khan Y, Le J, Leckband D, Komarova Y, and Malik AB (2019). VE-PTP stabilizes VE-cadherin junctions and the endothelial barrier via a phosphatase-independent mechanism. J Cell Biol 218, 1725–1742. 10.1083/jcb.201807210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tsuda Y, Takahashi H, Kobayashi M, Hanafusa T, Herndon DN, and Suzuki F (2004). Three different neutrophil subsets exhibited in mice with different susceptibilities to infection by methicillin-resistant Staphylococcus aureus. Immunity 21, 215–226. 10.1016/j.immuni.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Schauer C, Janko C, Munoz LE, Zhao Y, Kienhofer D, Frey B, Lell M, Manger B, Rech J, Naschberger E, et al. (2014). Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat Med 20, 511–517. 10.1038/nm.3547. [DOI] [PubMed] [Google Scholar]
- 62.Ngamsri KC, Fuhr A, Schindler K, Simelitidis M, Hagen M, Zhang Y, Gamper-Tsigaras J, and Konrad FM (2022). Sevoflurane Dampens Acute Pulmonary Inflammation via the Adenosine Receptor A2B and Heme Oxygenase-1. Cells 11. 10.3390/cells11071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hann J, Bueb JL, Tolle F, and Brechard S (2020). Calcium signaling and regulation of neutrophil functions: Still a long way to go. J Leukoc Biol 107, 285–297. 10.1002/JLB.3RU0719-241R. [DOI] [PubMed] [Google Scholar]
- 64.Vig M, and Kinet JP (2009). Calcium signaling in immune cells. Nat Immunol 10, 21–27. 10.1038/ni.f.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feske S (2007). Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol 7, 690–702. 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 66.Santana Nunez D, Malik AB, Lee Q, Ahn SJ, Coctecon-Murillo A, Lazarko D, Levitan I, Mehta D, and Komarova YA (2023). Piezo1 induces endothelial responses to shear stress via soluble adenylyl Cyclase-IP(3)R2 circuit. iScience 26, 106661. 10.1016/j.isci.2023.106661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoffmann K, Sperling K, Olins AL, and Olins DE (2007). The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma 116, 227–235. 10.1007/s00412-007-0094-8. [DOI] [PubMed] [Google Scholar]
- 68.Doerschuk CM, Beyers N, Coxson HO, Wiggs B, and Hogg JC (1993). Comparison of neutrophil and capillary diameters and their relation to neutrophil sequestration in the lung. J Appl Physiol (1985) 74, 3040–3045. 10.1152/jappl.1993.74.6.3040. [DOI] [PubMed] [Google Scholar]
- 69.Shin JW, Spinler KR, Swift J, Chasis JA, Mohandas N, and Discher DE (2013). Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc Natl Acad Sci U S A 110, 18892–18897. 10.1073/pnas.1304996110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diebold I, Petry A, Hess J, and Gorlach A (2010). The NADPH oxidase subunit NOX4 is a new target gene of the hypoxia-inducible factor-1. Mol Biol Cell 21, 2087–2096. 10.1091/mbc.e09-12-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu YV, Hubbi ME, Pan F, McDonald KR, Mansharamani M, Cole RN, Liu JO, and Semenza GL (2007). Calcineurin promotes hypoxia-inducible factor 1alpha expression by dephosphorylating RACK1 and blocking RACK1 dimerization. J Biol Chem 282, 37064–37073. 10.1074/jbc.M705015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davey ME, Caiazza NC, and O’Toole GA (2003). Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol 185, 1027–1036. 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gille C, Spring B, Tewes L, Poets CF, and Orlikowsky T (2006). A new method to quantify phagocytosis and intracellular degradation using green fluorescent protein-labeled Escherichia coli: comparison of cord blood macrophages and peripheral blood macrophages of healthy adults. Cytometry A 69, 152–154. 10.1002/cyto.a.20222. [DOI] [PubMed] [Google Scholar]
- 74.Isberg RR, and Falkow S (1985). A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317, 262–264. 10.1038/317262a0. [DOI] [PubMed] [Google Scholar]
- 75.Chow CW, Downey GP, and Grinstein S (2004). Measurements of phagocytosis and phagosomal maturation. Curr Protoc Cell Biol Chapter 15, Unit 15 17. 10.1002/0471143030.cb1507s22. [DOI] [PubMed] [Google Scholar]
- 76.Bolte S, and Cordelieres FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 77.Mukherjee P, Nebuloni F, Gao H, Zhou J, and Papautsky I (2019). Rapid Prototyping of Soft Lithography Masters for Microfluidic Devices Using Dry Film Photoresist in a Non-Cleanroom Setting. Micromachines (Basel) 10. 10.3390/mi10030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shelly M, Lee SI, Suarato G, Meng Y, and Pautot S (2017). Photolithography-Based Substrate Microfabrication for Patterning Semaphorin 3A to Study Neuronal Development. Methods Mol Biol 1493, 321–343. 10.1007/978-1-4939-6448-2_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matute-Bello G, Downey G, Moore BB, Groshong SD, Matthay MA, Slutsky AS, Kuebler WM, and Acute Lung Injury in Animals Study, G. (2011). An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol 44, 725–738. 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Looney MR, Thornton EE, Sen D, Lamm WJ, Glenny RW, and Krummel MF (2011). Stabilized imaging of immune surveillance in the mouse lung. Nat Methods 8, 91–96. 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsukasaki Y, Toth PT, Davoodi-Bojd E, Rehman J, and Malik AB (2022). Quantitative Pulmonary Neutrophil Dynamics Using Computer-Vision Stabilized Intravital Imaging. Am J Respir Cell Mol Biol 66, 12–22. 10.1165/rcmb.2021-0318MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li J, Kim K, Hahm E, Molokie R, Hay N, Gordeuk VR, Du X, and Cho J (2014). Neutrophil AKT2 regulates heterotypic cell-cell interactions during vascular inflammation. J Clin Invest 124, 1483–1496. 10.1172/JCI72305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu L, Cara DC, Kaur J, Raharjo E, Mullaly SC, Jongstra-Bilen J, Jongstra J, and Kubes P (2005). LSP1 is an endothelial gatekeeper of leukocyte transendothelial migration. J Exp Med 201, 409–418. 10.1084/jem.20040830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu H, and Peng Z (2019). Boundary integral simulations of a red blood cell squeezing through a submicron slit under prescribed inlet and outlet pressures. Physics of Fluids 31, 031902. 10.1063/1.5081057. [DOI] [Google Scholar]
- 85.Evans E, and Yeung A (1989). Apparent viscosity and cortical tension of blood granulocytes determined by micropipet aspiration. Biophysical journal 56, 151–160. 10.1016/S0006-3495(89)82660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai MA, Frank RS, and Waugh RE (1993). Passive mechanical behavior of human neutrophils: power-law fluid. Biophysical journal 65, 2078–2088. 10.1016/S0006-3495(93)81238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lewis AH, and Grandl J (2015). Mechanical sensitivity of Piezo1 ion channels can be tuned by cellular membrane tension. Elife 4, e12088. 10.7554/eLife.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cox CD, Bae C, Ziegler L, Hartley S, Nikolova-Krstevski V, Rohde PR, Ng C-A, Sachs F, Gottlieb PA, and Martinac B (2016). Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat Commun 7, 10366–10366. 10.1038/ncomms10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang X, Lin C, Chen X, Li S, Li X, and Xiao B (2022). Structure deformation and curvature sensing of PIEZO1 in lipid membranes. Nature. 10.1038/s41586-022-04574-8. [DOI] [PubMed] [Google Scholar]
- 90.Schmittgen TD, and Livak KJ (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108. 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 91.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 93.McCarthy DJ, Chen Y, and Smyth GK (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297. 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benjamini Y, and Yosef Hochberg(1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) vol. 57, 289–300. [Google Scholar]
- 96.Geyer M, Huang F, Sun Y, Vogel SM, Malik AB, Taylor CW, and Komarova YA (2015). Microtubule-Associated Protein EB3 Regulates IP3 Receptor Clustering and Ca(2+) Signaling in Endothelial Cells. Cell Rep 12, 79–89. 10.1016/j.celrep.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuroda J, Ago T, Matsushima S, Zhai P, Schneider MD, and Sadoshima J (2010). NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc Natl Acad Sci U S A 107, 15565–15570. 10.1073/pnas.1002178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. RNAseq analyzed data; differentially expressed genes in WT and Piezo1ΔPMN PMN; pathway analysis, related to Figure 5a–g and Suppl. Figure 4a–e.
Video S1. Lung intra-vital imaging showing relative changes in cytosolic [Ca2+] in PMN during trans-endothelial migration, related to Figure 3a–c and STAR Methods.
Video S2. Computational 3D modeling of membrane tension in PMN passing through endothelial junctional pore, related to Figure 3g and STAR Methods.
Video S3. Squeezing event of a PMN passing through 5 μm pores of the microfluidic system, related to Suppl. Figure 2g and STAR Methods.
Video S4. Raw intravital video of PMN transmigration and calcium fluctuation in cremaster muscle, related to Suppl. Figure 2c–e and STAR Methods.
Video S5. Computational modeling of membrane tension in PMN passing through the microfluidic device, related to Suppl. Figure 2i–j and STAR Methods.
Data Availability Statement
Sequencing data is available on Gene Expression Omnibus. The accession number is GSE210111. The reviewer token is wlebcwqwdncrvir. Computational modeling and simulation data is uploaded using indigo: https://figshare.com/s/0eac4b5603eff08aba58.


