Abstract
The Campylobacter genus of Gram-negative bacteria is characterized by expression of N-linked protein glycosylation (pgl) pathways. As Campylobacter concisus is an emerging human pathogen, a better understanding of the variation of the biosynthetic pathways across the genus is necessary to identify the relationships between protein glycosylation and disease. The pgl pathway of C. concisus strains have been reported to diverge from other Campylobacter in steps after the biosynthesis of UndPP-diNAcBac-GalNAc, which is catalyzed by PglC and PglA, a phosphoglycosyl transferase (PGT) and a glycosyltransferase (GT) respectively. Here we characterize the PglJ GTs from two strains of C. concisus. Chemical synthesis was employed to access the stereochemically-defined glycan donor substrates, UDP-GalNAcA and UDP-GlcNAcA, to allow biochemical investigation of PglJ. Evidence for the PglJ substrate specificity structural determinants for the C-6” carboxylate-containing sugar was obtained through variant-based biochemical assays. Additionally, characterization of a UDP-sugar dehydrogenase encoded in the pgl operon, that is similar to the Pseudomonas aeruginosa WbpO responsible for oxidization of a UDP-HexNAc to UDP-HexNAcA, supports the availability of a UDP-HexNAcA substrate for a GT that incorporates the modified sugar and provides evidence for the presence of a HexNAcA into the N-linked glycan. Utilizing sequence similarity network (SSN) analysis, we identified conserved sequence motifs among PglJ glycosyltransferases, shedding light on substrate preferences and offering predictive insights for enzyme functions across the Campylobacter genus. These studies now allow detailed characterization of the later steps in the pgl pathway in C. concisus strains and provide insights into enzyme substrate specificity determinants for glycan-assembly enzymes.
Introduction.
Campylobacter concisus (C. concisus) is an emerging human pathogen in the oral-gastrointestinal tract.1–4 Cell-surface glycans represent a primary point of contact between bacteria and their environments, underscoring the importance of understanding these glycoconjugate biosynthetic pathways.5 The asparagine (N)-linked protein glycosylation (pgl) pathway in Campylobacter jejuni (C. jejuni) has been extensively studied.6–9,10, 11 Glycan assembly pathways are initiated by a phosphoglycosyl transferase (PGT) that catalyzes the transfer of a phosphosugar onto a membrane-anchored undecaprenyl-phosphate acceptor from a soluble nucleotide-diphosphate sugar (NDP-sugar) donor (Figure 1A). The growing undecaprenyl-diphosphate-linked glycan is elaborated by sequential glycosyl transferases (GTs). GTs catalyze stereo- and regiospecific reactions between sugar acceptor and donor substrates. The GTs in the pgl pathways feature either GT-A or GT-B folds.12
Figure 1.
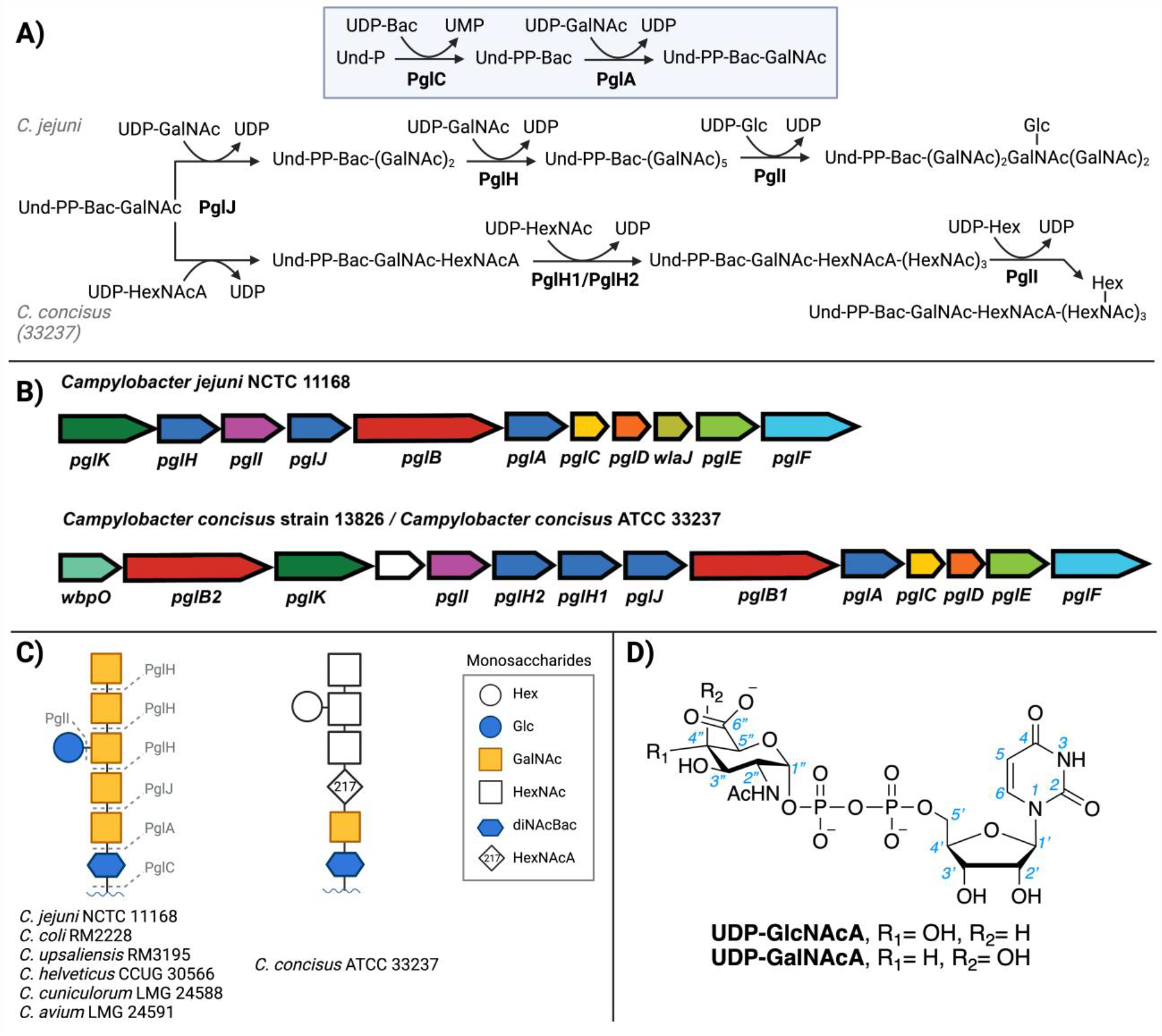
A) The protein glycosylation (pgl) pathways in C. jejuni and C. concisus, B) pgl loci, C) the diverse, isolated N-linked glycans from different Campylobacter bacteria14 and D) the possible UDP-HexNAcA sugar donors for the PglJ glycosyltransferase.
N-linked glycosylation is conserved across Campylobacter species, although the glycan structures are variable,13, 14 and result from the variation of genes in the Campylobacter pgl pathway operons (Figure 1B). Although the C. jejuni N-linked glycan has been fully characterized,6–9, 13 the glycans from the common C. concisus strains have only been partially annotated.13, 15, 16 Glycan structures and GT specificities are defined through multiple complementary approaches. The glycoconjugate can be isolated from the target organism and glycan composition can be ascertained through fragmentation-based mass spectrometry (MS). However, MS does not provide an unambiguous annotation because many sugars, like the C4 epimers GalNAc and GlcNAc are isobaric. Additionally, due to the fragmentation nature of the technique the anomeric stereochemistry (α or β) or the glycosidic linkage (e.g., 1,3 or 1,4) between monosaccharides cannot be defined. Nuclear magnetic resonance (NMR) is also a valuable characterization tool, but the resulting spectra can be difficult to assign, even at high field, because of overlapping signature sugar peaks and, also, analysis by NMR requires significant amounts of pure glycan. Genetic knockout strategies can also be employed, but the function of the biosynthetic pathway in vivo can be rescued or compensated for by other enzymes. In vitro biochemical assays are informative for substrate specificity, however, are often limited by the commercial or synthetic availability of unusual UDP-sugar substrates. It is of interest to biochemically validate additional GT/UDP-sugar partners to develop better predictive capabilities for connecting GT sequence information to substrate specificity and ultimately the cell-surface bacterial glycan structures and the related Campylobacter pgl pathways present an excellent opportunity for developing such approaches.
The N-linked glycans of Campylobacter species are structurally diverse.13, 15, 17, 18 The C. jejuni glycan is composed of N,N’-diacetylbacillosamine (diNAcBac), glucose (Glc), and five N-acetylgalactosamine (GalNAc) monosaccharides (Figure 1C). The glycan from C. concisus strain 13826 that was previously characterized is reported to closely resemble the C. jejuni heptasaccharide.13 In contrast, although C. concisus ATCC 33237 (Cc 33237) is reported to feature a similar glycan, there is variation of the third carbohydrate from the reducing end of the isolated C. concisus 33237 N-linked glycan. This glycoconjugate features a monosaccharide with a molecular weight of 217 (g/mol).15 The corresponding monosaccharide in the C. concisus 13826 N-glycan is annotated as GalNAc.13 For C. concisus 33237, glycan characterization was previously achieved through fragmentation-based MS, which affords molecular composition information, but does not define the stereochemistry of the sugar. The 217 monosaccharide has been annotated as a HexNAcA suggesting this fragment corresponds to either N-acetyl-D-glucuronic acid (GlcNAcA) or N-acetyl-D-galacturonic acid (GalNAcA) (Figure 1D). The goal of this study is to biochemically characterize the molecular basis for the substrate specificities of the C. concisus (Cc 33237 and Cc 13826) PglJ enzymes and define the role of the dehydrogenase (WbpO) in the pgl operon for the N-glycan biosynthesis.
Materials and Methods.
Chemical synthesis of candidate UDP-N-acetylhexuronic acid (UDP-HexNAcA) sugar epimers.
Oxidation of UDP-GlcNAc to UDP-GlcNAcA is achieved by employing a platinum catalyst in the presence of oxygen gas. 19, 20 However, a similar approach for the oxidation of UDP-GalNAcA produced significant side products, and we hypothesized that the GalNAc C4 stereochemistry might account for this unexpected reactivity. Specifically, once oxidation at C6 occurs, the acidity of the C5 hydrogen increases due to resonance delocalization into the neighboring carboxylic acid, and, unlike GlcNAc, the C5 hydrogen in GalNAc is anti-periplanar to the C4 hydroxyl (Figure S9), which results in undesired elimination products.20 With this in mind, we reduced the reaction time, which compromises conversion, but allows for clean product formation without undesired side product contaminants. Purification of both UDP-GlcNAcA and UDP-GalNAcA was accomplished by anion-exchange fast protein liquid chromatography (FPLC, Figures S2, S6–8). The FPLC-purified UDP-sugars were characterized by 1H, 13C, and 31P nuclear magnetic resonance (NMR, Figures S3–5, S10–12) and high-resolution mass spectrometry (HRMS).
Nano differential scanning fluorimetry (nanoDSF).
nanoDSF was utilized as a facile screen to rapidly investigate the interactions of glycosyl transferases with ligands by monitoring changes in thermal stability.21 Measurements were carried out using the Prometheus NT.48 (NanoTemper). Reactions contained 0.4 mg/mL (9.3 μM of C. concisus ATCC 33237 PglJ, 9.5 μM of C. concisus 13826 strain PglJ, 10 μM of C. jejuni PglJ) of enzyme, 1 mM UDP-sugar substrate, 10 mM HEPES, pH 7.5, 150 mM NaCl, 5% glycerol, 2 mM DTT, 0.03% DDM. For C. jejuni PglJ, we adjusted the buffer to 50 mM HEPES to prevent the protein from precipitating, while maintaining the same pH and other components. The reactions were incubated on ice for 20 minutes. Samples were loaded into glass capillaries, irradiated at 280 nm (100 % excitation power) and fluorescence emission was measured at 330 nm and 350 nm. The samples were heated to 70 °C at a slope of 1 °C/min, and the inflection point from the 1st derivative plot of F350/F330 was used to determine melting transitions. Measurements were carried out in triplicate for C. concisus ATCC 33237/13826 strain PglJ, and in duplicate for C. jejuni PglJ.
Glycosyltransferase activity assay
The glycosyltransferase reactions were monitored by the UDP-Glo™ glycosyltransferase assay from Promega (Cat # V6963). This bioluminescent assay determines the amount of UDP produced in a reaction by converting UDP product of the GT to ATP which is then assessed using a luciferase reaction to generate light. The resulting luminescence signal can be converted to micromolar (μM) concentrations of UDP using a standard curve. The quenching solution was prepared as described by Promega. For the single enzyme substrate screen, the assays included PglJ (varying strains and concentrations), 0.1% Triton X-100, 50 mM HEPES at pH 7.5, 100 mM NaCl, 5 mM MgCl2, 25 μM UDP-sugar, and 20 μM Und-PP-diNAcBac-GalNAc in a final volume of 11 μL and were quenched after 30 minutes. For the single enzyme kinetics, the assays included 0.1 nM PglJ, 0.1% Triton X-100, 50 mM HEPES at pH 7.5, 100 mM NaCl, 5 mM MgCl2, UDP-GalNAcA (varying concentrations), and 20 μM Und-PP-diNAcBac-GalNAc in a final volume of 48 μL. Then 11 μL aliquots were quenched into 11 μL of the UDP detection reagent at 1, 3, 5, and 10 min. For all reactions, Und-PP-diNAcBac-GalNAc was pre-incubated with PglJ in assay buffer for two minutes before the addition of the UDP-sugar to initiate the reaction. The samples were transferred to white, nonbinding surface 96-well plates (Corning), the plate was shaken at low speed for 30 s and incubated for 1 h at 25 °C, then the luminescence was measured on the plate reader. All luminescence values were background subtracted before conversion to UDP concentrations.
Enzymatic synthesis and isolation of the Und-PP-linked trisaccharide products of PglC, PglA, and PglJ enzymes followed by 2-aminobenzamide (2-AB) labeling and analysis
The Und-PP-linked trisaccharide composition after PglC, PglA, and PglJ in C. concisus (strains 33237 and 13826) and C. jejuni reactions were determined through a coupled enzymatic synthesis. The three reactions were set up in 7 mL scintillation vials. Each reaction contained a total volume of 1.5 mL and consisted of 265 μM UndP, 300 μM UDP-diNAcBac, 600 μM UDP-GalNAc, 300 μM UDP-GalNAcA, 600 nM PglC, 200 nM PglA, 100 nM PglJ, 50 mM HEPES pH 7.5, 100 mM NaCl, 0.1% Triton X-100, 5 mM MgCl2 and 10% DMSO. The C. jejuni experiment was performed in the absence of UDP-GalNAcA. The reaction was initiated by the addition of the UDP-sugars and allowed to proceed at ambient temperature for 2 h. The reaction was quenched with 2 mL of (2:1) CHCl3/MeOH. The organic layer was washed with 1 mL PSUP (Pure Solvent Upper Phase = 15 mL CHCl3, 240 mL MeOH, 1.83 g KCl, 235 mL H2O) and the aqueous layer was removed. The aqueous layer was then back extracted with 1 mL (2:1) CHCl3/MeOH. The combined organic fractions were washed three times with 1 mL PSUP and concentrated under a stream of N2. The 2-AB labeling was performed following a previously established procedure with slight modifications.7, 22 The Und-PP-trisaccharide products were hydrolyzed with 500 μL of n-propanol/2M trifluoroacetic acid (1:1) and heated at 50 °C for 15 min. The resulting solution was evaporated to dryness. The 2-AB labeling reagent was prepared by dissolving 5 mg of 2-aminobenzamide in 100 μL of acetic acid/DMSO (1:2.3). The entire solution was added to 6 mg of sodium cyanoborohydride to provide the 2-AB labeling reagent cocktail. This reagent (5 μL) was added to the dried, hydrolyzed saccharide and heated to 60 °C for 2–4 h. The resulting mixture was diluted with 100 μL of H2O and purified by HPLC with fluorescence monitoring. The product was separated from excess dye using a reverse-phase analytical HPLC column (Prozyme GlykoSepR, GKI4727). A gradient of 0–100% B over 40 min with a flow rate of 0.7 mL/min was used with solvent A (50 mM ammonium formate (pH 4.4)/10% MeOH (v/v)) and solvent B (50 mM ammonium formate (pH 4.4)/20% MeOH (v/v)) as mobile phases. The peaks were detected using a fluorescence detector with λex= 330 nm and λem= 420 nm, collected, lyophilized, and analyzed by ESI(−) MS (Figure S19).
Sequence Similarity Network (SSN) Generation
The SSN was generated using EFI-EST (https://efi.igb.illinois.edu/efi-est/)23 with C. concisus ATCC 33237 PglJ sequence as input. All-by-all Basic Local Alignment Search Tool (BLAST) calculations (BLAST E-value of 1×10−5) were run using the EFI-EST, while the SSN edge calculation was performed using BLAST E-value of 1×10−100. The sequences were filtered using an alignment score threshold of 35 and a sequence length range of 200 – 1,000 amino acids. A Python script was used to extract non-redundant UniRef90 sequences from the network and to identify sequences containing the ‘SER’ and ‘NEC’ motifs. The SSN was visualized in Cytoscape using the yFiles Organic layout algorithm. Cluster formation within the SSN was performed using the ClusterMaker application and the Markov Clustering Algorithm (MCL), with a granulation parameter set to 4 and an edge cutoff set to 43. The genome neighborhood diagrams were generated using the Enzyme Function Initiative-Genome Neighborhood Tool (EFI-GNT;https://efi.igb.illinois.edu/efi-gnt/)24, using the SSN as input. The reading frame was set to 10 frames and the minimal co-occurrence was set to 20%.
HPLC for UDP-sugar analysis
The C. concisus dehydrogenase assays were monitored with a Waters 600 HPLC at 260 nm (Waters 2487 Absorbance Detector) with a semi-preparative anion-exchange column (Dionex CarboPac PA1, cat #: 057178). The gradient consisted of 65–100% B over 20 minutes, then kept at 100% B for 10 minutes with a flow rate of 3 mL/min. Buffer A: 0.1 mM NaOH and Buffer B:1 M NaOAc in 0.1 mM NaOH were used as the mobile phases. Each injection contained 50 μL of 20 μM UDP-sugar from the dehydrogenase assay (Figure S21).
Results and Discussion.
Elucidation of UDP-sugar substrate specificity of the second glycosyltransferase, PglJ.
The first two membrane committed steps in the pgl pathway for C. jejuni have been previously characterized.7, 25, 26 These enzymes from C. concisus have high sequence identity to C. jejuni suggesting they have similar function (72% for PglC and 48–49% for PglA).27, 28 To provide initial insight into the candidate glycosyl donor for the subsequent enzymes, PglJ in Cc 33237 and the related Cc 13826, we turned to nanoDSF. This technique harnesses intrinsic tryptophan and tyrosine fluorescence to evaluate protein stability over a thermal temperature gradient. A melting temperature (Tm) can be extrapolated from the change in the fluorescence intensity upon protein unfolding as the local environment around the fluorescent residues changes. Small-molecule ligand binding can be evaluated by determining the stability of the enzyme by measuring changes in the Tm. To reveal binding preference, we screened Cc PglJ from both 33237 and 13826 strains with a panel of nucleotide ligands at 1 mM (Figure 2A–B). The panel included commercially available uridine diphosphate (UDP) and UDP-N-acetylhexosamines (UDP-GalNAc, UDP-GlcNAc). Additionally, we studied the two synthetic UDP-N-acetylhexuronic acid epimers, UDP-GalNAcA and UDP-GlcNAcA, prepared from UDP-GalNAc and UDP-GlcNAc, respectively, through the platinum-catalyzed oxidization.19, 20 Significant stabilization was observed with UDP, the nucleotide product of the glycosyltransferase reaction, compared to the wild-type apoenzyme. Minimal changes in the stability of the protein were observed with UDP-GalNAc and UDP-GlcNAc. In contrast, the C6”-carboxylate-containing UDP-sugars (UDP-GalNAcA and UDP-GlcNAcA) stabilized the PglJ enzymes from both strains of C. concisus studied. Of the epimeric UDP-sugars studied, UDP-GalNAcA preferentially stabilizes both C. concisus PglJ enzymes (7–13 °C) (Figure 2). In the same nanoDSF experiment conducted with C. jejuni PglJ, which is known to have a preference for UDP-GalNAc, the results were notably different. Specifically, there was very little stabilization observed in the presence of UDP-GalNAcA (2 °C). However, in the presence of UDP-GalNAc, significant stabilization was detected, evidenced by an increase of >5 °C in the melting temperature (Figure 2C). Such differences in stabilization patterns between C. jejuni and C. concisus PglJ further suggest distinct substrate preferences.
Figure 2.
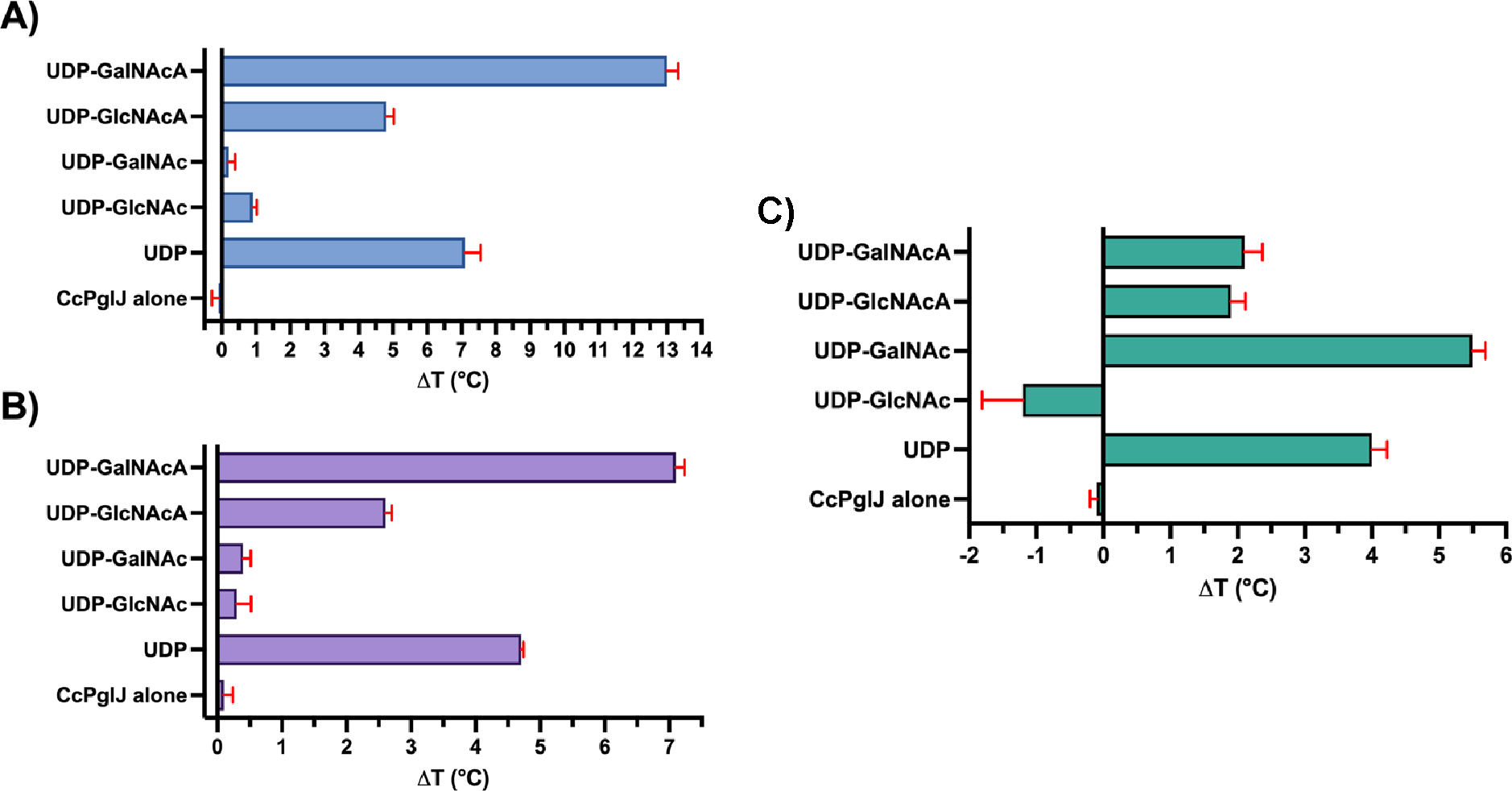
NanoDSF Tm determination experiments for (A) PglJ Cc 33237 (blue), (B) PglJ Cc 13826 (purple), and (C) PglJ Cj 11168 (green) tested against 1 mM UDP and a panel of UDP-sugars. Each data point represents the mean value derived from independent measurements (n = 2–3) with error bars indicating standard deviation (± SD). The unliganded PglJ Cc 33237 exhibited a melting point, Tm of 41.0 ± 0.2 °C, while PglJ Cc 13826 demonstrated a Tm of 47.5 ± 0.1 °C. Note that the melting transition for Cj PglJ, with UDP-GalNAc occurred at two points: 45.2 ± 0.1°C and at 51.4 ± 0.2°C.
As a significant stabilization was observed with both strains of Cc PglJ in the presence of UDP-GalNAcA by nanoDSF, this suggests that this UDP-sugar is the preferred donor substrate for PglJ enzymes. To further investigate this, PglJ was subjected to biochemical assays with a panel of UDP-sugars in the presence of the UndPP-diNAcBac-GalNAc acceptor to complement the nanoDSF experiments (Figure 3A). We included Cc PglJ from strains 33237 and 13826, as well as the PglJ from C. jejuni for comparison. At 10 nM, the two enzymes from C. concisus were selective for UDP-GalNAcA. At the same concentration, the PglJ from C. jejuni preferred UDP-GalNAc which is consistent with previous characterization.7 We note that when only one UDP-sugar is available in a non-competitive environment, we observed substrate promiscuity with the C. jejuni PglC. Although we observed stabilization with both oxidized sugars by nanoDSF, the enzyme did show preference for the transfer of C-4” axial hydroxyl-containing sugar, GalNAcA, onto the prenyldiphosphate-linked acceptor.
Figure 3.
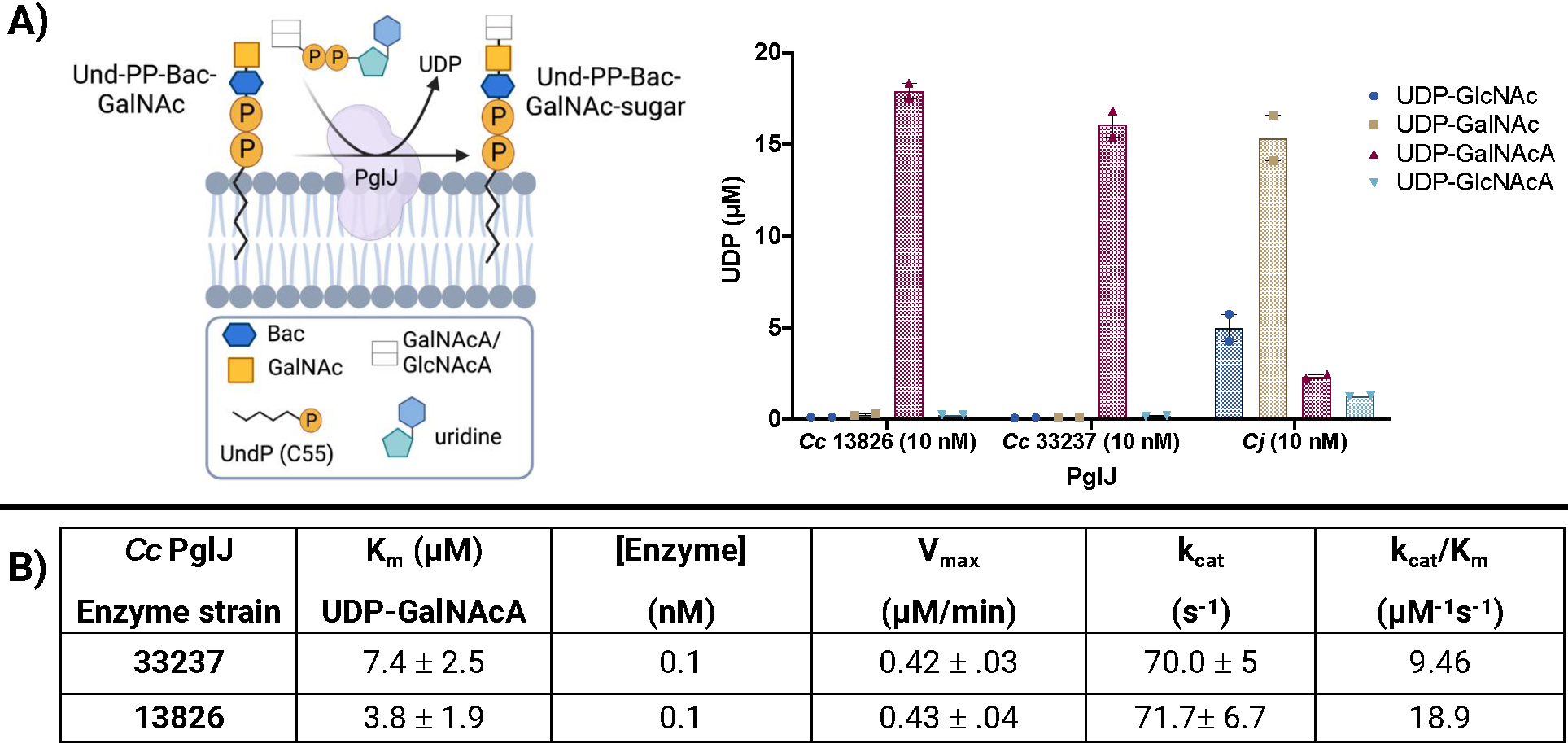
A) UDP-sugar substrate screen with PglJs from C. concisus and C. jejuni. Error bars are given for mean ± SEM, n = 2. B) Single enzyme kinetic parameters. Confidence intervals (95%) and parameters were calculated using nonlinear regression (curve fit, Michaelis-Mention) using GraphPad Prism 8, n = 2–3.
We then evaluated the kinetics of C. concisus PglJ enzymes from both strains (33237 and 13826) by screening with varying amounts of UDP-GalNAcA (2.5–100 μM) in the presence of 20 μM Und-PP-diNAcBac-GalNAc and monitored by UDP-Glo (Figure 3B). Under steady-state kinetic conditions, we derived the Michaelis constants (Km) of 7.4 ± 2.5 and 3.8 ± 1.9 μM for PglJ strains 33237 and 13826, respectively, from Michaelis Menten and Lineweaver-Burke plots (Figure S18). These experiments were performed with enzymes at the same concentration (0.1 nM) which provided similar turnover numbers (kcat) for both PglJ enzymes (70–71.7 s−1). With the culmination of this work, in addition to the high sequence identity (94.1%), we concluded that the enzymes from these two strains perform the same reaction and thus the previous annotation13 should be amended. To graphically designate the epimeric HexNAcA monosaccharides, GlcNAcA and GalNAcA, we propose combination of the square logo from the Symbol Nomenclature for Glycans (SNFG) database that represents a HexNAc monosaccharide with the horizontal line used to depict hexuronic acids (HexA)29 in blue for GlcNAcA, and yellow for GalNAcA.
Although the UDP Glo assay is a convenient high-throughput approach to monitor glycosyltransferase reactions, UDP release could also indicate nucleotide substrate hydrolysis. To verify incorporation of GalNAcA, we isolated the Und-PP-trisaccharide product of the three-component [PglC, PglA, and PglJ] reactions and performed 2-aminobenzamide (2-AB) fluorescent labeling (Figure 4), a well-established strategy to facilitate characterization of glycans.7, 30 Once extracted, the glycan was subsequently removed from the undecaprenyl diphosphate (Und-PP) through acid-catalyzed hydrolysis, followed by a reductive amination with 2-aminobenzamide with sodium cyanoborohydride (NaBH3CN) to furnish the fluorescently-labelled glycan, which was analyzed by fluorescence-based high performance liquid chromatography (F-HPLC). The desired peaks were isolated and further analyzed by negative-mode electrospray ionization-mass spectrometry (ESI-MS), which provided 2-AB-diNAcBac-GalNAc-GalNAcA for both strains of the C. concisus PglC, PglA, and PglJ tandem reactions. As a control, the corresponding C. jejuni PglC, PglA, and PglJ reaction produced the expected 2-AB-diNAcBac-(GalNAc)2 glycan. We note that there was no observable formation of 2-AB-diNAcBac-(GalNAc)2 in the Cc experiments. Moreover, the UDP-Glo coupled assays performed under analogous conditions with PglC, PglA, and PglJ (Figure S20) are consistent with the 2-AB labeled product findings; GalNAc is not installed by PglJ from C. concisus.
Figure 4.
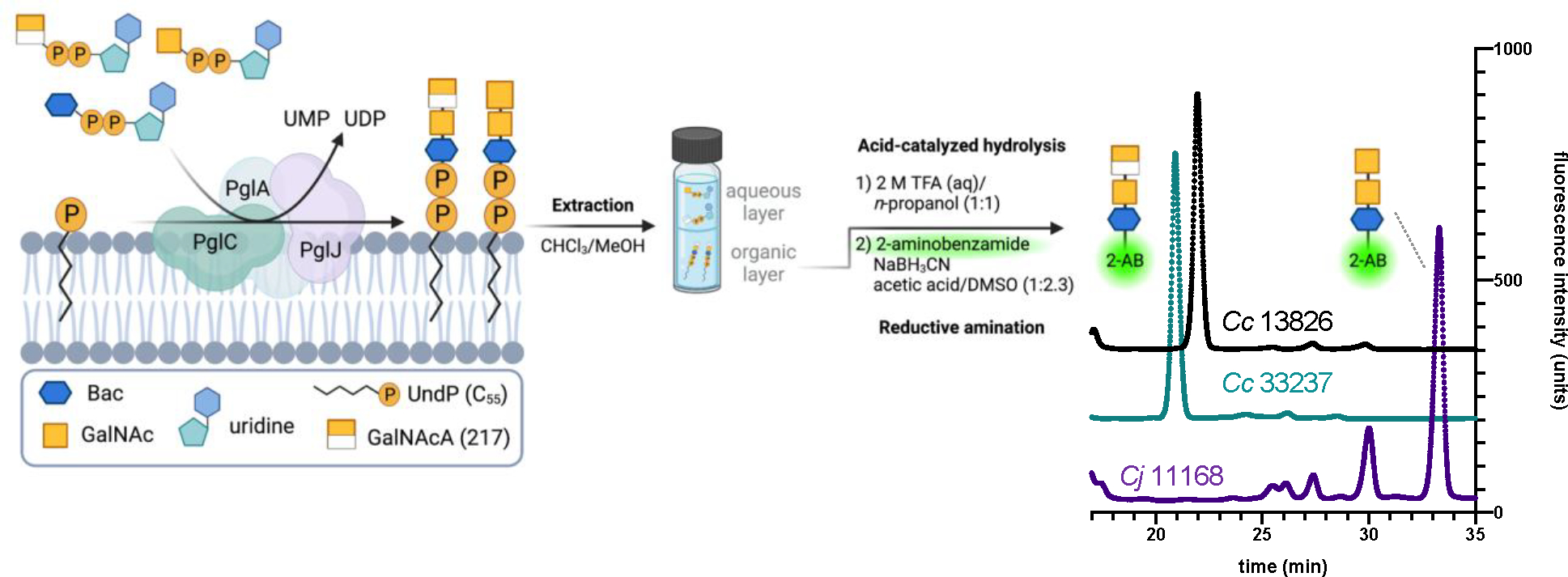
Extraction of the Und-PP-glycan produced from PglC, PglA, and PglJ, followed by hydrolysis and 2-aminobenzamide fluorescent labeling. The HPLC spectra were normalized to comparable fluorescence intensity units to better visualize retention time differences between the PglJ products. See the supporting information for full HPLC traces and the corresponding mass spectra.
We additionally aligned and categorized PglJ sequences from Campylobacter and Helicobacter31 to determine if substrate preference for UDP-GalNAcA by these enzymes could be correlated with specific sequence motifs (Figure 5A). Interestingly, PglJ from the Cc strain 13826, which has been previously annotated to use UDP-GalNAc, fell outside the UDP-GalNAc group in our alignment. To identify residues significant for substrate specificity, we analyzed sequence conservation within these two distinct groups. As “sequence signatures” - we observed that the amino acids at positions 120 (serine, S) and 122 (arginine, R) are preferentially conserved in the GalNAcA-group, suggesting their possible role in the C6-carboxylate substrate specificity. We experimentally tested this hypothesis by engineering the 33237 strain S120N/R122C PglJ variant, swapping the serine and arginine with the asparagine (N) and cysteine (C) residues commonly observed at comparable positions in GalNAc utilizing PglJs. We observe that this variant now transfers GalNAc from UDP-GalNAc onto the growing Und-PP-glycan (Figure 5B). The variant also exhibits activity towards a wider range of substrates than the wild-type counterpart. The observed expansion of substrate compatibility following the S120E121R122 to N120E121C122 mutation in PglJ supports our hypothesis on the importance of these motifs in determining substrate specificity. In light of these results, we propose that the NEC motif is related to an ‘ancestral’ or more ‘promiscuous’ state of the enzyme, allowing it to act on a wider range of substrates. Such findings underline the evolutionary trajectory where enzymes, over time, may transition from a promiscuous state to one of increased specificity, adapting to their cellular environments and metabolic demands. 32, 33 Based on the previous characterization of the Cc 13826 N-linked glycan,13 the PglJ sequence appeared “outside” the GalNAcA group, however this enzyme includes the residues we have now established to be critical for C6” carboxylate specificity. Cc PglJ 13826 was previously annotated to install UDP-GalNAc, but the trend observed in the sequence alignment, in combination with the biochemical assays supports that this enzyme prefers UDP-GalNAcA and should be included in the same grouping as C. concisus 33237.
Figure 5.
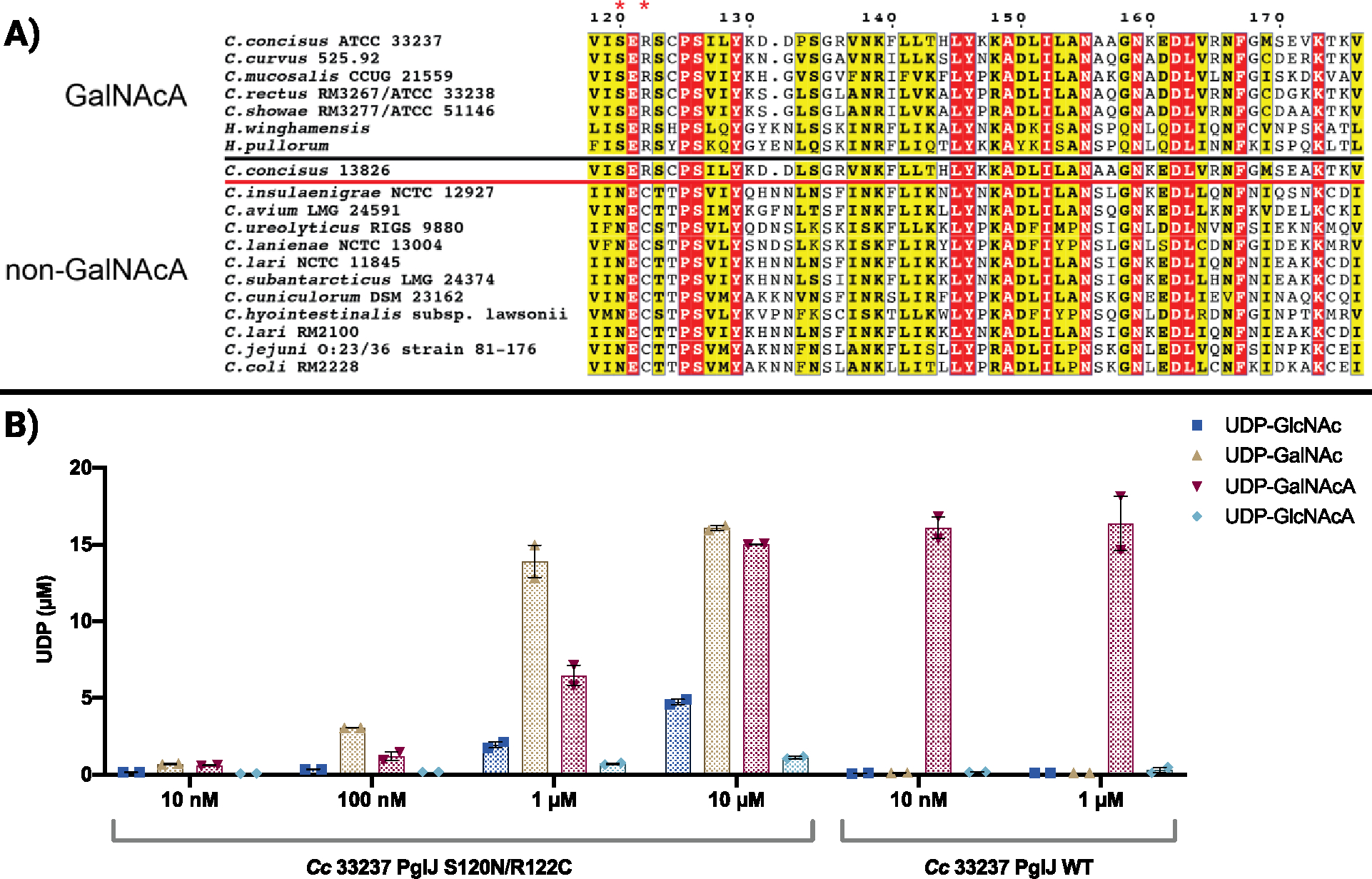
A) Displayed are PglJ sequence alignments. PglJs above the black line prefer GalNAcA according to established literature, while those below react with non-GalNAcA substrates. Our study determined that C. concisus PglJ 13826 has a preference for GalNAcA, suggesting it belongs to the group delineated by the red line. B) Glycosyltransferase activity assays containing a UDP-sugar substrate screen with varying concentrations of C. concisus 33237 S120N/R122C PglJ. Error bars are given for mean ± SEM, n = 2.
Prevalence of SER/NEC motifs across PglJ homologs
To evaluate the conservation of these motifs among PglJ glycosyltransferases, we generated a sequence similarity network (SSN) that comprised 1329 unique homologs of Cc PglJ 13826/33237 (Figure S15). The homologs have a sequence identity range of 23–99% relative to Cc PglJ 33237. As illustrated in Figure S15, the SER motif is widespread among PglJ homologs (320 sequences). About 20% (59 sequences), are orthologs identified in Campylobacter, Helicobacter, and Wolinella succinogenes. Notably, PglJ from W. succinogenes has been reported to react with N-acetylhexuronamide (HexNAcAN).14 Considering the fact that W. succinogenes PglJ only shares 38% sequence identity with Cc PglJ 13826, it is possible that the differences in the surrounding residues resulted in an altered substrate specificity, despite retaining this motif. The NEC motif is represented differently among bacteria, with all 136 instances belonging to Campylobacter members. A sequence alignment of all Campylobacter sequences from the SSN (percent sequence identity between 30%–99%) revealed that the SER/NEC motifs are highly conserved. Exceptions were found in only two PglJs, derived from C. hyointestinalis subsp. hyointestinalis and C. hominis, which exhibit NER and NEG motifs, respectively. The NEC motif emerges as the most common among Campylobacter sequences (Figure 6).
Figure 6.

Sequence logo showcasing the SER/NEC motifs among Campylobacter sequences within the sequence similarity network. The residues are color-coded based on their hydrophobicity characteristics. The sequence logo was generated using WebLogo.34
Characterization of a dehydrogenase in Campylobacter concisus.
The second glycosyltransferase in the C. concisus pgl pathway prefers UDP-GalNAcA in vitro. To establish if this GT utilizes UDP-GalNAcA in vivo, we analyzed if the pgl operon in C. concisus encodes for an enzyme to oxidize UDP-GalNAc. The previously characterized dehydrogenase, WbpO, is an NAD+-dependent enzyme responsible for oxidation of UDP-GalNAc to UDP-GalNAcA for incorporation into the O-antigen of Pseudomonas aeruginosa. This enzyme was used as a query sequence to identify an unannotated nucleotide sugar dehydrogenase in the C. concisus strain 13826 pgl operon based on sequence identity (62.5%).35, 36 Initial assays containing 1 mM UDP-GalNAc or UDP-GlcNAc with 2.5 mM NAD+ in buffer containing 10 mM Tris pH 8.2 and 5 mM MgCl2 only provided activity towards UDP-GlcNAc; no conversion of UDP-GalNAc to UDP-GalNAcA was observed (Figure S21). However, removal of MgCl2 and introduction of ammonium sulfate into the assay buffer (100 mM) provided complete conversion of both UDP-sugars to the corresponding C6”-oxidized products after 1 h at 37 °C (Figure 7). It is hypothesized that these dehydrogenases require potassium or ammonium cations for enzymatic activation,36 which is consistent with our findings. Additionally, during the chemical oxidation of UDP-GalNAc to UDP-GalNAcA we observed degradation products. This underscores the importance of enzyme catalysis providing the desired compound with stereo- and regiochemical precision.
Figure 7.
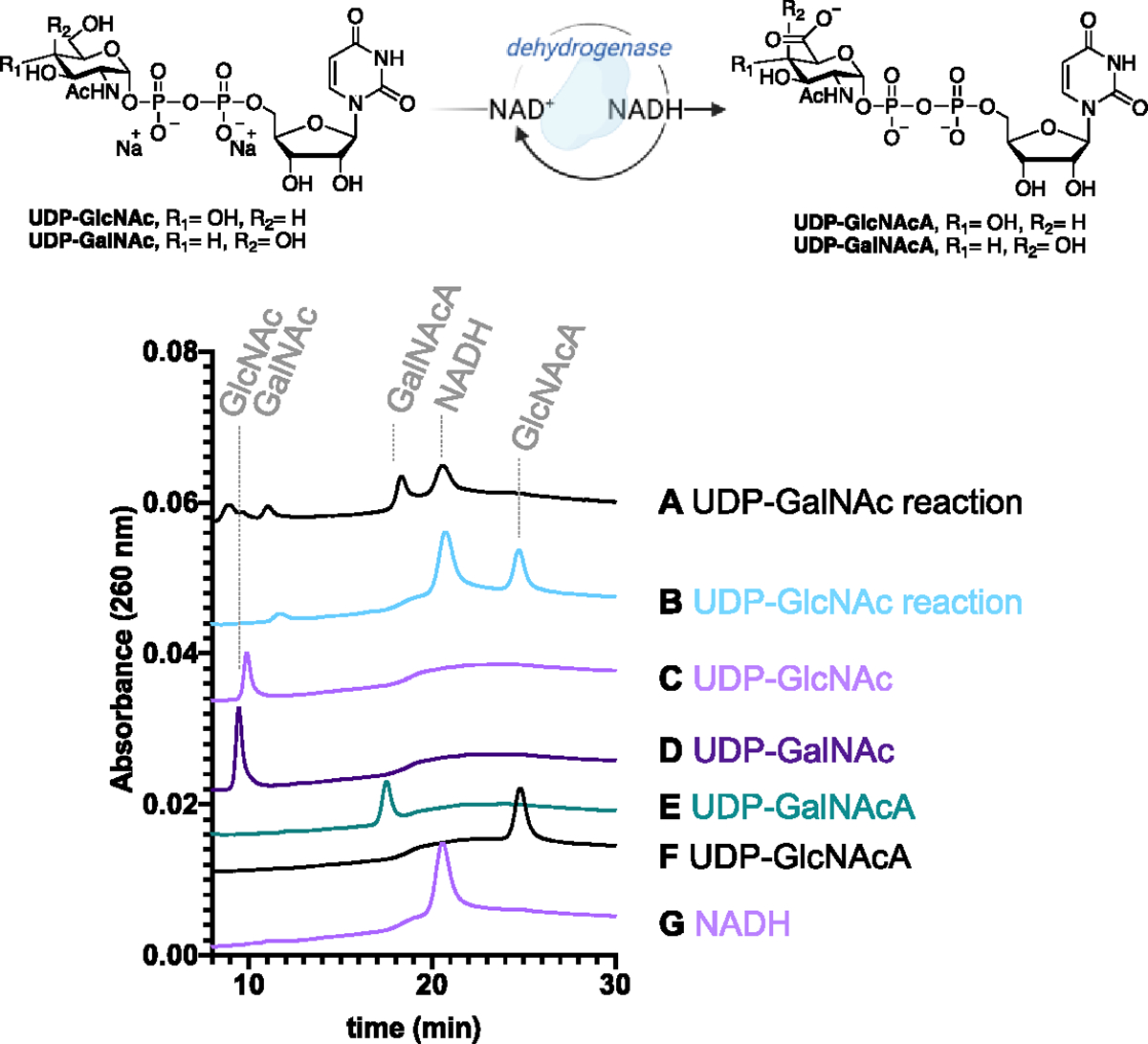
An HPLC-based assay to validate the activity of the dehydrogenase from C. concisus strain 13826. Lanes: A) UDP-GalNAc reaction, B) UDP-GlcNAc reaction, C) UDP-GlcNAc standard, D) UDP-GalNAc standard, E) UDP-GalNAcA standard, F) UDP-GlcNAcA standard, and G) NADH standard. The supporting information contains full HPLC traces.
Co-Occurrence of HexNAcA PglJs and dehydrogenases
Considering the role of dehydrogenase in UDP-HexNAcA biosynthesis, we proposed that UDP-HexNAcA PglJ enzymes would demonstrate a high frequency of co-localization with this enzyme within the pgl operon. In contrast, an absence of co-localization for those PglJs that do not use UDP-HexNAcA as a substrate is predicted. To assess this, the genome neighborhood diagrams of PglJs derived from annotated genomes of Campylobacter species in the SSN were analyzed. In all instances of Campylobacter PglJs with preference for UDP-HexNAcA, a dehydrogenase was co-localized within the pgl operon (Table S3). In contrast, for Campylobacter species predicted to utilize UDP-HexNAc substrates from the occurrence of the NEC sequence motif, 95% of them (130 out of 136) did not show co-localization of a dehydrogenase within the pgl operon. Four exceptions were found where a dehydrogenase was co-localized (Table S4). This could be a result of the functionality of the NEC motif which, as demonstrated in our study, allows for a broader range of substrates, possibly making the UDP-HexNAcA pathway a possible route when the primary substrate is unavailable. This could warrant the co-presence of a dehydrogenase in these cases, highlighting the potential for a higher degree of metabolic flexibility within these species.
Conclusions.
Biochemical confirmation of substrate specificity will lay the groundwork for elucidating the structural determinants of UDP-donor specificity of GT-Bs. In the case of C. concisus, PglJ demonstrated specificity toward GalNAcA over GlcNAcA, the C4 epimer, which is three bonds away from the location of the glycosylation reaction. Additionally, an engineered variant of the C. concisus PglJ eliminated the preference for the C-6” carboxylate of UDP-GalNAcA to mimic the C. jejuni PglJ specificity of UDP-GalNAc. This underscores the specificity of the GT donor binding site. The carboxylate-containing UDP-sugars can now be added to the rapidly growing toolkit for the biochemical characterization of UDP-sugar utilizing enzymes. Moreover, this addition will allow biochemical investigation of the H. pullorum pgl pathway,31, 37as well as other group II cluster Campylobacter bacteria which include C. rectus, C. showae, C. mucosalis, and C. curvus. All of which have been annotated to install HexNAcA at the third position from the reducing end of the glycan.13
By determining the kinetic parameters for PglJ from both strains of C. concisus, we ascertained that these enzymes catalyze the same reaction. This is consistent with the high sequence identity (94.1%) between the two enzymes. Additionally, these results are consistent with the Linton and co-workers isolation of glycans from both C. concisus strains 13826 and 33237 which contain HexNAcA.15, 38 Significantly, our analysis revealed a consistent co-occurrence of HexNAcA-installing PglJ with a specific dehydrogenase within the pgl operon. This co-occurrence offers a predictive framework, hinting at potential evolutionary and functional couplings between these enzymes. Such insights, derived from sequence similarity networks and co-occurrence patterns, will be invaluable for directing future efforts in enzyme substrate prediction and could streamline the elucidation of glycosylation pathways in uncharacterized bacterial strains
Future work will involve demonstrating that the acceptor bearing GalNAcA can be utilized by the subsequent enzyme in the pathway and determining acceptor substrate specificity determinants in general. In C. jejuni, PglH accepts the undecaprenyl-diphosphate-linked glycan produced from PglJ. In C. concisus, it is unclear which enzyme succeeds PglJ as the operons encode two PglH enzymes (Figure 1B). We hypothesize that the next enzymes in the pathway are PglH1 and PglH2, which install the remaining HexNAc monosaccharides, followed by hexose (Hex) addition by PglI to complete the glycan assembly in C. concisus (Figure 8). The presence of two PglH enzymes may be the result of variability in the acceptor substrates between C. concisus (Und-PP-diNAcBac-GalNAc-GalNAcA) and C. jejuni (Und-PP-diNAcBac-(GalNAc)2). In depth kinetic and structural characterization of the PglHs enzymes in C. concisus will be explored in future work.
Figure 8.
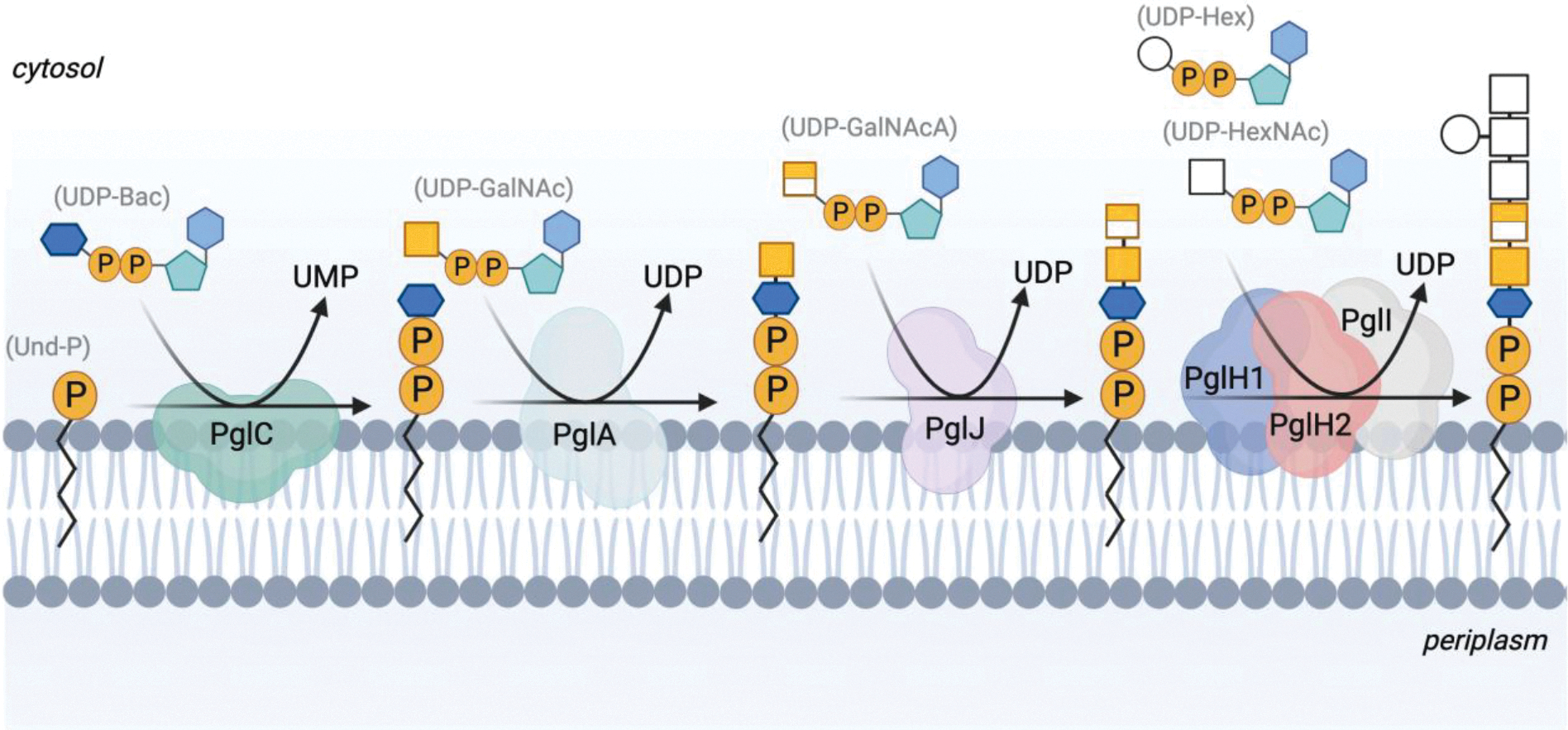
The proposed N-linked protein glycosylation pathway in C. concisus strain 33237.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Hayley Knox for providing purified C. jejuni PglJ used in activity assays and nanoDSF experiments. Figures 1, 3, 4, 7, and 8 were made with BioRender.com.
Funding Sources
Financial support from the National Institute of Health (GM039334 to B.I. and K.N.A., F32 GM136023 to C.A.A., and F32 GM146421 to N.V.) is gratefully acknowledged.
ABBREVIATIONS
- 2-AB
2-aminobenzamide
- TFA
trifluoroacetic acid
- HPLC
high-performance liquid chromatography
- Und-P
undecaprenyl phosphate
- LC-MS
liquid chromatography-mass spectrometry
Data Availability
The sequence similarity network (SSN) files have been deposited in Mendeley (DOI: 10.17632/24g8n465k9.1). All other data can be found either within the manuscript or in the Supplementary Information (SI).
REFERENCES
- (1).Liu F; Ma R; Wang Y; Zhang L The Clinical Importance of Campylobacter concisus and Other Human Hosted Campylobacter Species. Front. Cell. Infect. Microbiol. 2018, 8, Review. DOI: 10.3389/fcimb.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Man SM The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011, 8 (12), 669–685. DOI: 10.1038/nrgastro.2011.191. [DOI] [PubMed] [Google Scholar]
- (3).Kaakoush NO; Mitchell HM Campylobacter concisus - A new player in intestinal disease. Front Cell Infect Microbiol 2012, 2, 4. DOI: 10.3389/fcimb.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Lastovica AJ Clinical relevance of Campylobacter concisus isolated from pediatric patients. J. Clin. Microbiol. 2009, 47 (7), 2360. DOI: 10.1128/jcm.00568-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Varki A Biological roles of glycans. Glycobiology 2016, 27 (1), 3–49. DOI: 10.1093/glycob/cww086 (acccessed 6/29/2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Szymanski CM; Yao R; Ewing CP; Trust TJ; Guerry P Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 1999, 32 (5), 1022–1030. DOI: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- (7).Glover KJ; Weerapana E; Imperiali B In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 2005, 102 (40), 14255–14259. DOI: 10.1073/pnas.0507311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Karlyshev AV; Ketley JM; Wren BW The Campylobacter jejuni glycome. FEMS Microbiol. Rev. 2005, 29 (2), 377–390. DOI: 10.1016/j.fmrre.2005.01.003 (acccessed 6/29/2023). [DOI] [PubMed] [Google Scholar]
- (9).Wacker M; Linton D; Hitchen PG; Nita-Lazar M; Haslam SM; North SJ; Panico M; Morris HR; Dell A; Wren BW; et al. N-Linked Glycosylation in Campylobacter jejuni and Its Functional Transfer into E. coli. Science 2002, 298 (5599), 1790–1793. DOI: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- (10).Alemka A; Nothaft H; Zheng J; Szymanski CM N-Glycosylation of Campylobacter jejuni Surface Proteins Promotes Bacterial Fitness. Infect. Immun. 2013, 81 (5), 1674–1682. DOI: 10.1128/iai.01370-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Day C; Semchenko E; Korolik V Glycoconjugates Play a Key Role in Campylobacter jejuni Infection: Interactions between Host and Pathogen. Front. Cell. Infect. Microbiol. 2012, 2, Review. DOI: 10.3389/fcimb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lairson LL; Henrissat B; Davies GJ; Withers SG Glycosyltransferases: Structures, Functions, and Mechanisms. Annu. Rev. Biochem. 2008, 77 (1), 521–555. DOI: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- (13).Nothaft H; Scott NE; Vinogradov E; Liu X; Hu R; Beadle B; Fodor C; Miller WG; Li J; Cordwell SJ; et al. Diversity in the Protein N-Glycosylation Pathways Within the Campylobacter Genus. Mol. Cell. Proteom. 2012, 11 (11), 1203–1219. DOI: 10.1074/mcp.M112.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Nothaft H; Szymanski CM Bacterial Protein N-Glycosylation: New Perspectives and Applications*. Journal of Biological Chemistry 2013, 288 (10), 6912–6920. DOI: 10.1074/jbc.R112.417857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jervis AJ; Butler JA; Lawson AJ; Langdon R; Wren BW; Linton D Characterization of the Structurally Diverse N-Linked Glycans of Campylobacter Species. J. Bacteriol. 2012, 194 (9), 2355–2362. DOI: 10.1128/jb.00042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Chung HKL; Tay A; Octavia S; Chen J; Liu F; Ma R; Lan R; Riordan SM; Grimm MC; Zhang L Genome analysis of Campylobacter concisus strains from patients with inflammatory bowel disease and gastroenteritis provides new insights into pathogenicity. Scientific Reports 2016, 6 (1), 38442. DOI: 10.1038/srep38442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Young NM; Brisson J-R; Kelly J; Watson DC; Tessier L; Lanthier PH; Jarrell HC; Cadotte N St. Michael F; Aberg E.; et al. Structure of the N-Linked Glycan Present on Multiple Glycoproteins in the Gram-negative Bacterium, Campylobacter jejuni. J. Biol. Chem. 2002, 277 (45), 42530–42539. DOI: 10.1074/jbc.M206114200 (acccessed 2023/06/29). [DOI] [PubMed] [Google Scholar]
- (18).Szymanski CM; Michael FS; Jarrell HC; Li J; Gilbert M; Larocque S; Vinogradov E; Brisson J-R Detection of Conserved N-Linked Glycans and Phase-variable Lipooligosaccharides and Capsules from Campylobacter Cells by Mass Spectrometry and High Resolution Magic Angle Spinning NMR Spectroscopy J. Biol. Chem. 2003, 278 (27), 24509–24520. DOI: 10.1074/jbc.M301273200 (acccessed 2023/06/29). [DOI] [PubMed] [Google Scholar]
- (19).Larkin A; Imperiali B Biosynthesis of UDP-GlcNAc(3NAc)A by WbpB, WbpE, and WbpD: Enzymes in the Wbp Pathway Responsible for O-Antigen Assembly in Pseudomonas aeruginosa PAO1. Biochemistry 2009, 48 (23), 5446–5455. DOI: 10.1021/bi900186u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Rejzek M; Mukhopadhyay B; Wenzel CQ; Lam JS; Field RA Direct oxidation of sugar nucleotides to the corresponding uronic acids: TEMPO and platinum-based procedures. Carbohydr. Res. 2007, 342 (3), 460–466. DOI: 10.1016/j.carres.2006.10.016. [DOI] [PubMed] [Google Scholar]
- (21).Kotov V; Bartels K; Veith K; Josts I; Subhramanyam UKT; Gunther C; Labahn J; Marlovits TC; Moraes I; Tidow H; et al. High-throughput stability screening for detergent-solubilized membrane proteins. Sci Rep 2019, 9 (1), 10379. DOI: 10.1038/s41598-019-46686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Arbour CA; Nagar R; Bernstein HM; Ghosh S; Al-Sammarraie Y; Dorfmueller HC; Ferguson MAJ; Stanley-Wall NR; Imperiali B Defining early steps in Bacillus subtilis biofilm biosynthesis. mBio 0 (0), e00948–00923. DOI: 10.1128/mbio.00948-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Gerlt JA; Bouvier JT; Davidson DB; Imker HJ; Sadkhin B; Slater DR; Whalen KL Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochimica et biophysica acta 2015, 1854 (8), 1019–1037. DOI: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zallot R; Oberg NO; Gerlt JA ‘Democratized’ genomic enzymology web tools for functional assignment. Curr Opin Chem Biol 2018, 47, 77–85. DOI: 10.1016/j.cbpa.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Glover KJ; Weerapana E; Chen MM; Imperiali B Direct Biochemical Evidence for the Utilization of UDP-bacillosamine by PglC, an Essential Glycosyl-1-phosphate Transferase in the Campylobacter jejuni N-Linked Glycosylation Pathway. Biochemistry 2006, 45 (16), 5343–5350. DOI: 10.1021/bi0602056. [DOI] [PubMed] [Google Scholar]
- (26).Weerapana E; Glover KJ; Chen MM; Imperiali B Investigating Bacterial N-Linked Glycosylation: Synthesis and Glycosyl Acceptor Activity of the Undecaprenyl Pyrophosphate-Linked Bacillosamine. Journal of the American Chemical Society 2005, 127 (40), 13766–13767. DOI: 10.1021/ja054265v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ray LC; Das D; Entova S; Lukose V; Lynch AJ; Imperiali B; Allen KN Membrane association of monotopic phosphoglycosyl transferase underpins function. Nat. Chem. Biol. 2018, 14 (6), 538–541. DOI: 10.1038/s41589-018-0054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Das D; Kuzmic P; Imperiali B Analysis of a dual domain phosphoglycosyl transferase reveals a ping-pong mechanism with a covalent enzyme intermediate. Proc. Natl. Acad. Sci. U.S.A. 2017, 114 (27), 7019–7024. DOI: 10.1073/pnas.1703397114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Varki A; Cummings RD; Aebi M; Packer NH; Seeberger PH; Esko JD; Stanley P; Hart G; Darvill A; Kinoshita T; et al. Symbol Nomenclature for Graphical Representations of Glycans. Glycobiology 2015, 25 (12), 1323–1324. DOI: 10.1093/glycob/cwv091 (acccessed 7/1/2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Arbour CA; Nagar R; Bernstein HM; Ghosh S; Al-Sammarraie Y; Dorfmueller HC; Ferguson MAJ; Stanley-Wall NR; Imperiali B Defining Early Steps in B. subtilis Biofilm Biosynthesis. bioRxiv 2023, 2023.2002.2022.529487. DOI: 10.1101/2023.02.22.529487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Jervis AJ; Wood AG; Cain JA; Butler JA; Frost H; Lord E; Langdon R; Cordwell SJ; Wren BW; Linton D Functional analysis of the Helicobacter pullorum N-linked protein glycosylation system. Glycobiology 2018, 28 (4), 233–244. DOI: 10.1093/glycob/cwx110 (acccessed 6/29/2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Daughtry KD; Huang H; Malashkevich V; Patskovsky Y; Liu W; Ramagopal U; Sauder JM; Burley SK; Almo SC; Dunaway-Mariano D; et al. Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis. Biochemistry 2013, 52 (32), 5372–5386. DOI: 10.1021/bi400659k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Pandya C; Farelli JD; Dunaway-Mariano D; Allen KN Enzyme promiscuity: engine of evolutionary innovation. J Biol Chem 2014, 289 (44), 30229–30236. DOI: 10.1074/jbc.R114.572990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Crooks GE; Hon G; Chandonia JM; Brenner SE WebLogo: a sequence logo generator. Genome Res 2004, 14 (6), 1188–1190. DOI: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhao X; Creuzenet C; Bélanger M; Egbosimba E; Li J; Lam JS WbpO, a UDP-N-acetyl-galactosamine Dehydrogenase from Pseudomonas aeruginosa Serotype O6 *. J. Biol. Chem. 2000, 275 (43), 33252–33259. DOI: 10.1074/jbc.M004191200 (acccessed 2023/06/28). [DOI] [PubMed] [Google Scholar]
- (36).Miller WL; Matewish MJ; McNally DJ; Ishiyama N; Anderson EM; Brewer D; Brisson J-R; Berghuis AM; Lam JS Flagellin Glycosylation in Pseudomonas aeruginosa PAK Requires the O-antigen Biosynthesis Enzyme WbpO. J. Biol. Chem. 2008, 283 (6), 3507–3518. DOI: 10.1074/jbc.M708894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Jervis AJ; Langdon R; Hitchen P; Lawson AJ; Wood A; Fothergill JL; Morris HR; Dell A; Wren B; Linton D Characterization of N-Linked Protein Glycosylation in Helicobacter pullorum. J. Bacteriol. 2010, 192 (19), 5228–5236. DOI: 10.1128/jb.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Frost H N-linked protein glycosylation in Campylobacter and Helicobacter species. . Doctoral Thesis 2016, The University of Manchester; (Manchester, UK: ). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence similarity network (SSN) files have been deposited in Mendeley (DOI: 10.17632/24g8n465k9.1). All other data can be found either within the manuscript or in the Supplementary Information (SI).


