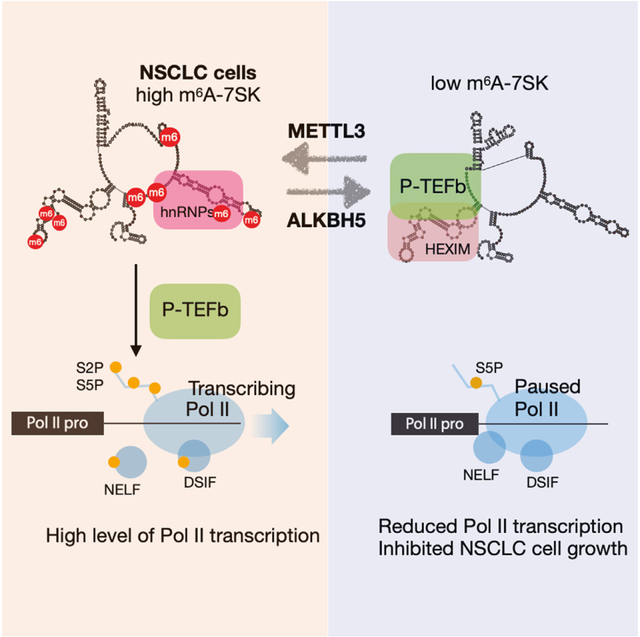
SUMMARY
N6-methyladenosine (m6A) modifications play crucial roles in RNA metabolism. How m6A regulates RNA polymerase II (RNA Pol II) transcription remains unclear. We find that 7SK small nuclear RNA (snRNA), a regulator of RNA Pol II promoter-proximal pausing, is highly m6A-modified in non-small cell lung cancer (NSCLC) cells. In A549 cells, we identified eight m6A sites on 7SK and discovered methyltransferase-like 3 (METTL3) and alkB homolog 5 (ALKBH5) as the responsible writer and eraser. When the m6A-7SK is specifically erased by a dCasRx-ALKBH5 fusion protein, A549 cell growth is attenuated due to reduction of RNA Pol II transcription. Mechanistically, removal of m6A leads to 7SK structural rearrangements that facilitate sequestration of the positive transcription elongation factor b (P-TEFb) complex, which results in reduction of serine 2 phosphorylation (Ser2P) in the RNA Pol II C-terminal domain and accumulation of RNA Pol II in the promoter-proximal region. Taken together, we uncover that m6A modifications of a non-coding RNA regulate RNA Pol II transcription and NSCLC tumorigenesis.
Graphical Abstract
In brief
Wang et al. discovered that 7SK RNA contains high levels of m6A in lung cancer cells. METTL3 and ALKBH5 modify 7SK at eight different m6A sites. Removal of m6A induces 7SK conformational change and sequesters P-TEFb, leading to RNA Pol II promoter-proximal pausing, inhibited transcription, and cancer cell growth.
INTRODUCTION
Higher organisms have evolved sophisticated mechanisms for transcriptional control, and proper regulation of transcription is fundamental to normal gene expression and cellular function. Transcription initiation by RNA polymerase II (RNA Pol II) is recognized as a key regulatory step in eukaryotic gene expression.1–3 In eukaryotic cells, RNA Pol II is paused soon after transcription initiation, about 20–60 nucleotides downstream from the transcription start site (TSS).4–6 Positive transcription elongation factor b (P-TEFb), composed of cyclin-dependent kinase 9 (CDK9) and cyclin T1, is required for the release of RNA Pol II promoter-proximal pausing. P-TEFb phosphorylates the second serine residue of the heptapeptide (YSPTSPS) repeats at the carboxyl-terminal domain of the largest subunit of RNA Pol II (RNA Pol II CTD serine 2 phosphorylation [Ser2P]), as well as negative elongation factors, such as negative elongation factor (NELF) and DRB-sensitivity-inducing factor (DSIF).7–9 Therefore, RNA Pol II CTD Ser2P serves as a hallmark for the transition from transcription initiation to productive elongation.
Human 7SK is an abundant and conserved 332-nucleotide small nuclear RNA (snRNA) that inhibits transcription elongation through negatively regulating P-TEFb kinase activity.10–12 Specifically, 7SK forms small nuclear ribonucleoprotein (snRNP) complexes with several RNA-binding proteins to modulate RNA Pol II promoter-proximal pausing. The mature 7SK folds into four major stem-loop structures (SLI, SLII, SLIII, and SLIV), providing docking sites for 7SK snRNP core proteins. The capping enzyme methylphosphate capping enzyme (MePCE) and the La-related protein 7 (Larp7) constitutively associate with 7SK for RNA stability, whereas other proteins dynamically associate with 7SK.13–15 In the transcription inactive state, the canonical 7SK core snRNP complex consists of hexamethylene bisacetamide-induced protein1/2 (HEXIM1/2) and P-TEFb, thus sequestering P-TEFb from phosphorylating and activating paused RNA Pol II.16,17 When P-TEFb and HEXIM are released, 7SK associates with heterogeneous nuclear ribonucleoproteins (hnRNPs), including hnRNP A1, hnRNP A2/B1, hnRNP R, and hnRNP Q.18–20 Interestingly, a recent study detected putative N6-methyladenosine (m6A) within the HEXIM1/2 and hnRNPs binding sites in 7SK (on SLI and SLIII) from human MOLM13 cells by nanopore sequencing.21 However, the possible function and underlying mechanism of these m6A sites remain unknown.
m6A is the most abundant internal modification on messenger RNAs (mRNAs) and is also widespread in long non-coding RNAs in eukaryotic cells. On mRNAs, m6A is deposited by the methyltransferase complex (MTC), consisting of METTL3, METTL14, Wilms tumor 1-associated protein (WTAP), and other adaptor proteins, including VIRMA (KIAA1429), RBM15/15B, and ZC3H13, and can be reversed by m6A erasers, such as ALKBH5 and fat mass- and obesity-associated protein (FTO).22–29 m6A affects transcription state and various cellular functions through m6A readers, including YT521-B homology (YTH) domain family proteins (YTHDF1/2/3 and YTHDC1/2) and hnRNP proteins, which directly or indirectly recognize m6A-marked transcripts, thereby affecting RNA metabolism.30–34 Accumulating evidence suggests that m6A modification controls multiple fundamental biological processes by regulating m6A-marked mRNA stability and translation.22,35,36 However, the molecular roles of m6A remain largely confined to more abundant mRNAs and in the process of post-transcriptional regulation.37,38 Despite recent studies suggesting that m6A modification regulates global transcription through chromosome-associated regulatory RNAs, including promoter-associated RNAs (paRNAs), RNA transcribed from transposable elements (repeat RNA), and enhancer RNA (eRNA),22,39,40 its importance in transcriptional control via long non-coding RNA remains unknown. Interestingly, the m6A MTCs are recruited to the promoter regions to regulate the pausing of RNA Pol II in Drosophila S2R+ cells.41 Given that 7SK plays a central role in RNA Pol II transcriptional regulation and its enrichment at the promoter region, we set out to test whether m6A involves in 7SK-mediated RNA Pol II promoter-proximal pausing.
Here, we found that 7SK is highly modified with m6A in non-small cell lung cancer (NSCLC) cells. We identified eight m6A sites on 7SK and found that METTL3 and ALKBH5 are responsible for writing and erasing the m6A modifications. When the m6A modification was erased, 7SK adopts a structure facilitating the sequestration of P-TEFb, resulting in a decrease in Ser2P-RNA Pol II and an accumulation of Ser2P-RNA Pol II at the promoter-proximal region. Consequently, the removal of m6A led to a significant decrease in RNA Pol II transcription and inhibition of NSCLC cell growth. Our findings suggest a previously unrecognized role of m6A modification on a non-coding RNA in regulating RNA Pol II transcription and NSCLC.
RESULTS
7SK snRNA contains m6A modifications in NSCLC cells
Previous studies suggest that aberrantly elevated m6A levels are closely associated with the progression of lung cancer, making it an ideal model to detect putative m6A modifications on different RNAs.42 We first examined whether 7SK harbors m6A in NSCLC A549 cells using m6A RNA immunoprecipitation (RIP) (MeRIP).43 MeRIP-enriched RNAs were then analyzed with reverse transcription followed by quantitative PCR (RT-qPCR) and northern blots. As shown in Figures 1A and 1B, 7SK, along with U6 snRNA, which is known to be m6A-modified,44 can be isolated by a m6A-specific antibody but not by the control immunoglobulin G (IgG) (Figure 1B, compare lane 3 with 6). This result suggests that 7SK harbors m6A modifications, which is consistent with a recent study that used nanopore sequencing to probe 7SK methylation in MOLM13 cells.21 By contrast, there was no enrichment of the negative controls RCN2 mRNA or tRNA-lysine in MeRIP over IgG control42 (Figures 1A and 1B).
Figure 1. m6A modification of 7SK is dynamically controlled by METTL3 and ALKBH5 in NSCLC cells.
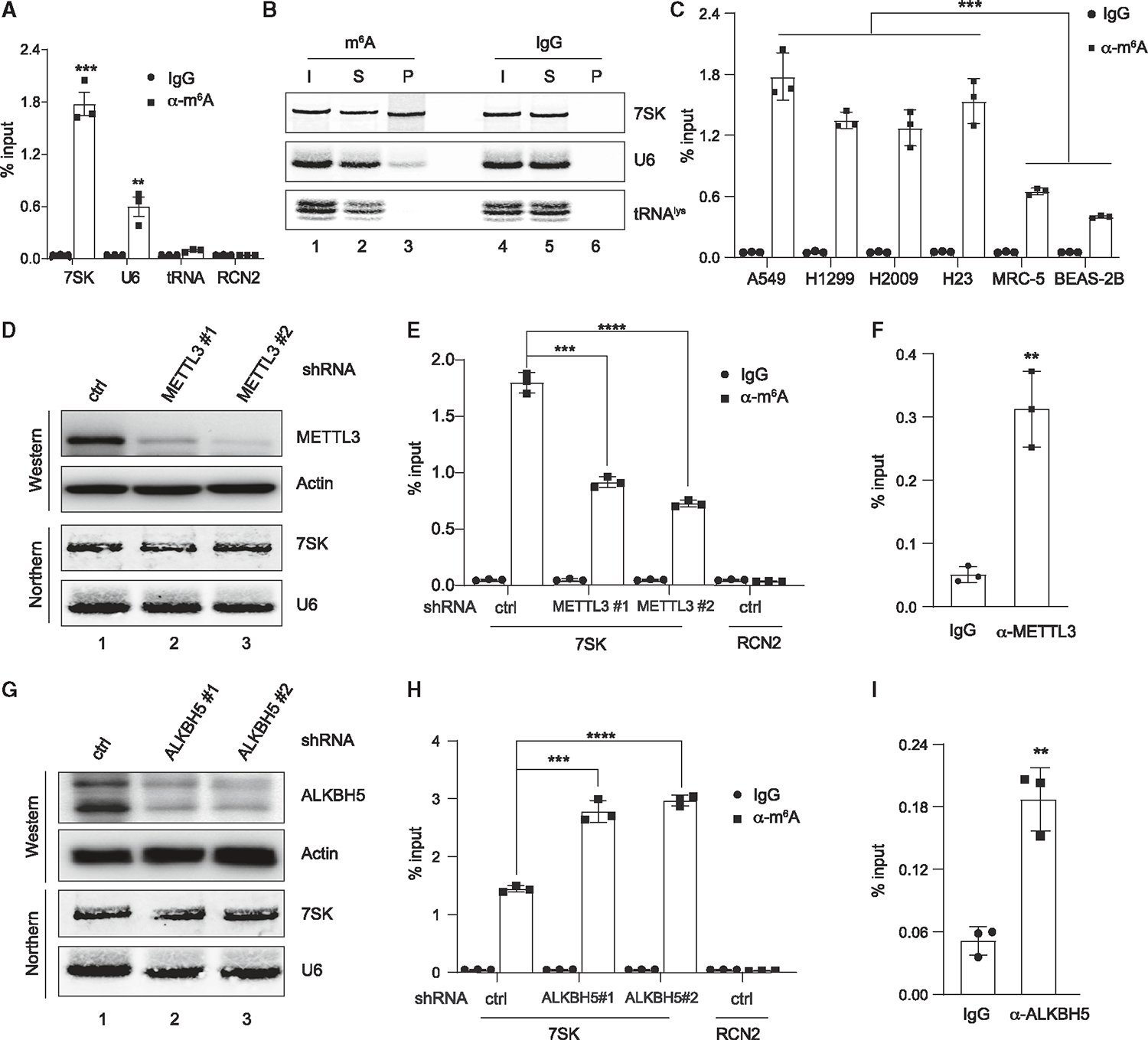
(A) Total RNA from A549 cells was subjected to immunoprecipitation using a m6A-specific antibody or IgG control, followed by RT-qPCR analyses of 7SK, U6, tRNAlys, and RCN2 mRNA.
(B) Same MeRIP experiment as in (A), except that the immunoprecipitated RNAs were analyzed via northern blot to detect 7SK, U6, and tRNAlys. Input (I) and supernatant (S) are 1.2% of the pellet (P).
(C) MeRIP analyses were performed in four different NSCLC cell lines, including A549, H1299, H2009, and H23, as well as the control cell lines, MRC-5 and BEAS-2B, followed by RT-qPCR analyses of 7SK.
(D and G) Western blot analyses show the knockdown of METTL3 (D) or ALKBH5 (G) by two different stably expressed shRNAs in A549 cells, with actin as a control. Northern blots detect the abundance of 7SK and U6 in A549 cells with METTL3/ALKBH5 depletion.
(E and H) MeRIP experiments were performed in METTL3 (E) or ALKBH5 (H) knockdown cells, followed by RT-qPCR detection of 7SK or RCN2.
(F and I) RIP analyses were performed in A549 cell lysates using α-METTL3 (F) or α-ALKBH5 (I) antibodies, followed by RT-qPCR detection of 7SK. Error bars represent standard deviation (SD). A t test calculated the p values, **p < 0.01, ***p < 0.001, ****p < 0.0001. See also Figure S1.
Next, we compared the abundance of m6A-modified 7SK (or m6A-7SK) in four different NSCLC cell lines (A549, NCI-H1299, NCI-H2009, and NCI-H23) and non-malignant lung epithelial (MRC-5) and fibroblast (BEAS-2B) cells. As shown in Figure 1C, 7SK exhibits significantly higher levels of m6A in NSCLC cell lines compared with the two control cell lines. The four NSCLC cell lines harbor different mutations in oncogenes such as KRAS and CDKN2A and tumor suppressors, including LKB1 and TP53 (A549: mutations in LKB1, CDKN2A, and KRAS; NCI-H2009: mutations in KRAS and TP53; and NCI-H23: mutations in LKB1, KRAS, and TP53). Therefore, our study uncovers that 7SK has higher m6A modifications in NSCLC cells with diverse genetic backgrounds.
m6A modification of 7SK is dynamically controlled by METTL3 and ALKBH5 in NSCLC cells
The above observations prompted us to identify specific m6A methyltransferase (writer) and demethylase (eraser) for 7SK. Therefore, we knocked down METTL3 and METLL16, two major m6A writers,38 in A549 cells by short hairpin RNAs (shRNAs). The knockdown efficiency of METTL3 and METTL16 was estimated by western blot analyses and RT-qPCR (Figures 1D and S1A–S1C). We observed that m6A-7SK can be significantly decreased by METTL3 depletion but not METTL16 (Figures 1E and S1D). Consistently, using RIP assays, we found that METTL3 but not METTL16 directly binds to 7SK (Figures 1F, S1E, and S1F). By contrast, U6, a known substrate of METTL16,44 was successfully enriched by METTL16-RIP (Figure S1G). Our results are consistent with the report that METTL16 does not interact with 7SK,44 and suggest that METTL3 can methylate 7SK.
Next, we tried to identify the m6A eraser for 7SK. ALKBH5 and FTO are two well-characterized RNA m6A erasers, although there are some disputes on FTO demethylase activity toward m6A and N6, 2′-O-dimethyladenosine (m6Am).27,28,45 Using the same approach as the m6A-writer identification for 7SK, we knocked down endogenous ALKBH5 and FTO in A549 cells using shRNAs (Figures 1G and S1H–S1J). We observed that ALKBH5, but not FTO, ablation led to an increase in m6A-7SK (Figures 1H and S1K). Consistently, we found that ALKBH5, but not FTO, can significantly enrich 7SK in RIP experiments (Figures 1I and S1L). As a control, 7SK abundance is not altered by knockdown of either m6A writers or erasers (Figures 1D, 1G, S1C, S1J, S1M, and S1N). Collectively, our data suggest that METTL3 and ALKBH5 are responsible for 7SK methylation and demethylation in NSCLC cells.
Identification of m6A sites on 7SK
Previous transcriptome-wide m6A mapping studies reveal that this modification generally occurs at the “RRACH” (R = G or A; H = A, C, or U) consensus motif, where the central A can be converted to m6A.30,46 Accordingly, we observe five potential m6A sites, namely, A172, A220, A228, A281, and A288 in 7SK, within the RRACH motif (Figure 2A). Interestingly, a recent study using nanopore sequencing analysis suggests that 7SK is potentially m6A modified at sites A43, A56, A65, A186, A230, A231, A238, A239, and A245, which are not within the traditional RRACH motif.21 To identify the exact m6A sites in 7SK, we screened all the above-mentioned m6A candidate sites (in the canonical RRACH motif and predicted by the nanopore sequencing study) using a single-base elongation- and ligation-based qPCR amplification method (SELECT-qPCR)47 (Figure 2B). In the SELECT assay, a pair of primers anneal to sequences flanking the putative m6A site. The primers are joined by polymerase elongation and DNA ligation, which are inhibited by a m6A modification. Therefore, compared with primers designed for an adjacent non-A site (control), primers targeting a genuine m6A site provide fewer ligated products to be detected by qPCR (with higher quantification cycle [Cq] value). As shown in Figures 2C, 2D, and S2A, SELECT assays suggest that 7SK can be methylated at A43, A56, A65, A186, A238, A245, A281, and A288. Together, we identified eight 7SK m6A modification sites, many of which locate in stem loops SLI and SLIII that interact with HEXIM1/2 and hnRNPs, respectively.18,21,48,49
Figure 2. Identification of m6A modification sites on 7SK.
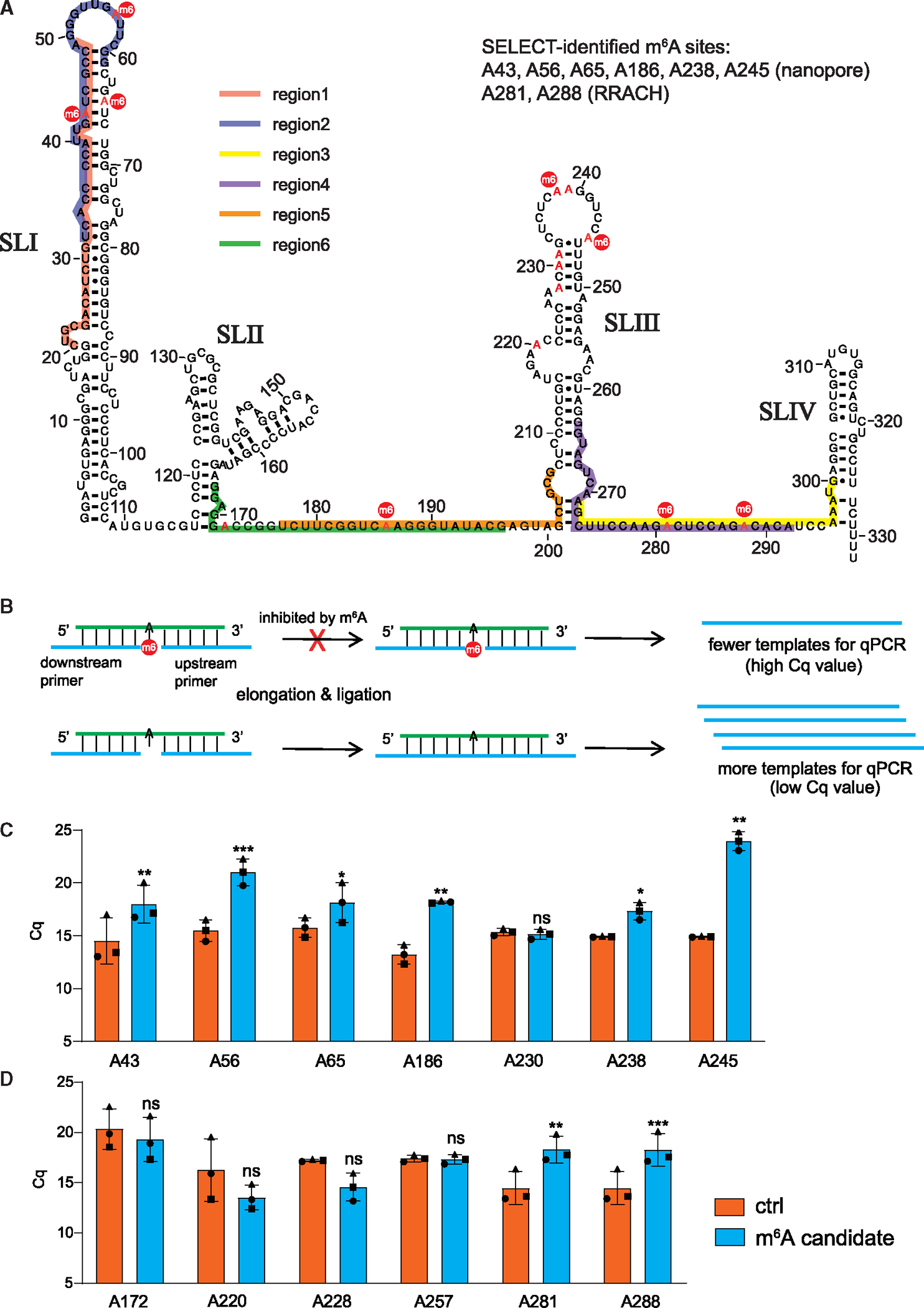
(A) Secondary structure of 7SK contains four stem-loops (SLI–SLIV) with putative m6A sites highlighted in red. The sites confirmed by the SELECT assay are marked with a red circle (m6). Colored lines labeled “regions 1–6” indicate the sequences targeted by DNA oligonucleotides in the RNase H cleavage assays.
(B) Illustration of the SELECT assay for m6A detection, detailed in STAR Methods.
(C and D) SELECT analyses of candidate m6A sites indicated by the nanopore sequencing (C) or within the RRACH consensus motif, as well as A257 (D). The y axis shows the quantification cycle (Cq) value of the SELECT-qPCR measurement. Error bars represent SD from 3 experiments (paired t test statistics were used, *p < 0.05, **p < 0.01, ***p < 0.001, and ns, not significant). See also Figure S2.
m6A modifiers of 7SK, METTL3 and ALKBH5, influence RNA Pol II Ser2P and transcription
Because we observed that m6A modifications of 7SK have no effect on 7SK abundance, and some of these sites appear at regions where HEXIM/P-TEFb interact with 7SK, we next determined whether m6A modifications affect 7SK-mediated transcriptional repression. To this end, we first changed the levels of the 7SK m6A writer (METTL3) and eraser (ALKBH5). We transfected A549 cells with plasmids expressing FLAG-tagged METTL3 wild-type (WT) and enzymatic mutant (D395A).50 Western blot analyses confirm that both WT and mutant METTL3 are expressed at comparable levels after plasmid transfection (Figure 3A). We observed that overexpression of WT METTL3 led to modest but significantly increased levels of m6A-7SK (Figure S2C). The modest increase is probably due to low transfection efficiency in A549 cells as shown in the immunofluorescent labeling experiment (Figure 3B). However, at the individual level, the cells with overexpression of WT METTL3 show much higher levels of Ser2P-RNA Pol II (Figure 3B, white arrows; quantification in Figure 3C). Conversely, shRNA knockdown of METTL3, which leads to reduction of m6A-7SK (Figure 1E), led to a reduction in Ser2P-RNA Pol II (Figure 3D). We further confirmed that the reduction of Ser2P-RNA Pol II is not due to change in the levels of P-TEFb (CDK9/cyclin T1) (Figure S2D).
Figure 3. m6A modifiers of 7SK, METTL3, and ALKBH5 influence RNA Pol II serine 2 phosphorylation and transcription.
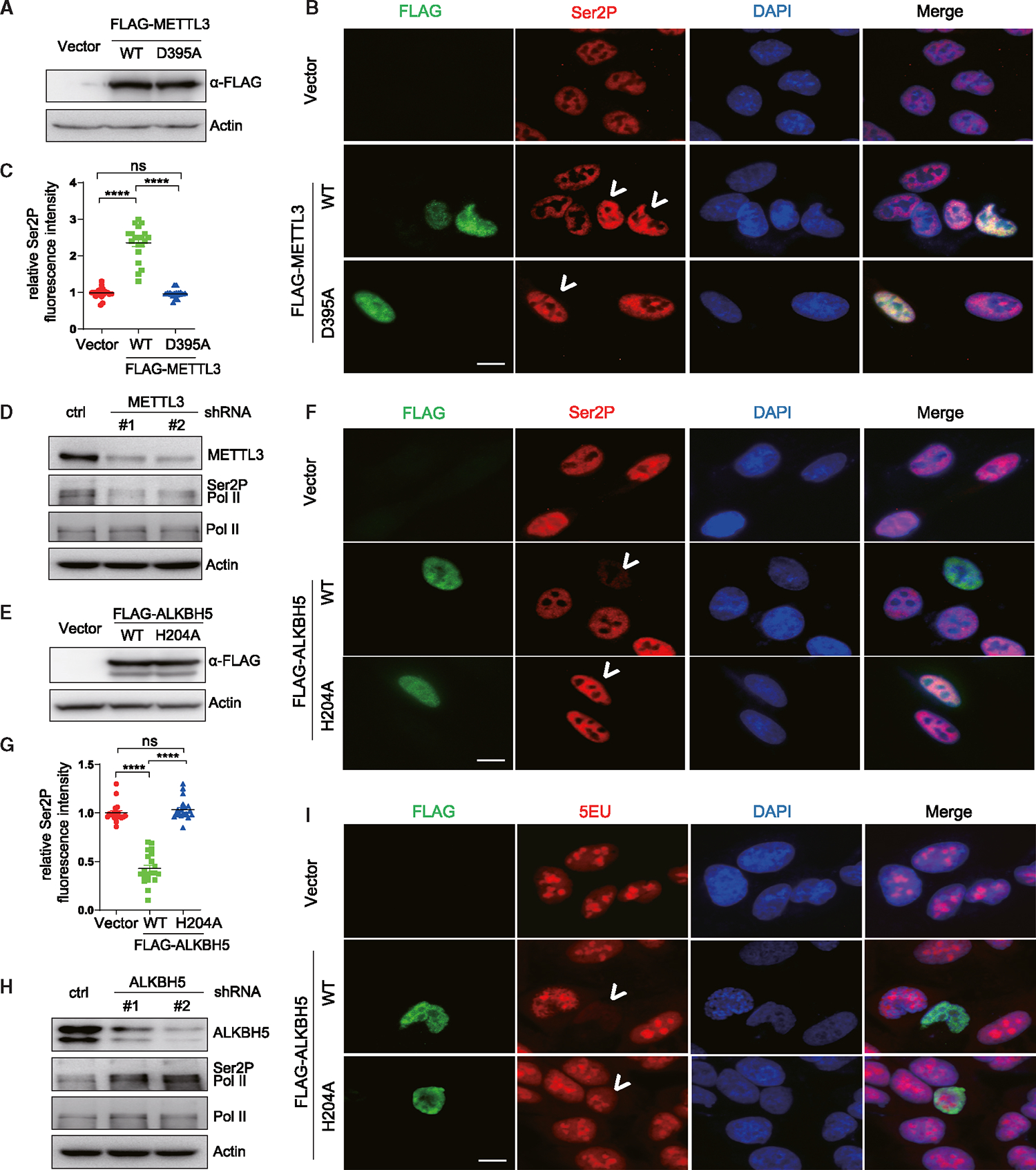
(A and E) Western blots using anti-FLAG antibody demonstrate the overexpression of FLAG-METTL3 (WT and D395A) (A) and FLAG-ALKBH5 (WT and H204A) (E) in A549 cells, with actin as a loading control.
(B and F) Representative immunofluorescence images of A549 cells transfected with plasmids encoding FLAG-METTL3 (WT and D395A) (B) and FLAG-ALKBH5 (WT and H204A) (F).
(C and G) The cells were dual stained using RNA Pol II Ser2P (red) and FLAG (green) antibodies, with quantification of RNA Pol II Ser2P levels presented in (C) and (G), respectively. White arrows point to cells transfected with the plasmids.
(D and H) Western blots of METTL3 (D) or ALKBH5 (H) in A549 cells stably expressing shRNA against these proteins. Total RNA Pol II and RNA Pol II Ser2P levels are also detected, with actin as a loading control.
(I) EU incorporation and staining in A549 cells transfected with plasmids encoding FLAG-ALKBH5 (WT and H204A). White arrows point to cells transfected with the plasmids. Scale bars, 10 μm. See also Figure S2.
To further confirm that METTL3 but not METTL16 is responsible for m6A modifications on 7SK, we used SELECT assay to measure several m6A sites in METTL3 and METTL16 knockdown cells (A43, A56, A186, A245, A281, and A288; see Figure 2). Knockdown of METTL3, but not METTL16, shows decreased m6A modification of examined sites (Figures S2E–S2J), corroborating with our earlier experiments that show METTL3 affects overall abundance of m6A-7SK.
Additionally, we observed that WT ALKBH5 but not ALKBH5 enzymatic mutant (H205A)28 overexpression (Figure 3E) significantly reduced m6A-7SK (Figure S2K), as well as Ser2P-RNA Pol II (Figure 3F, white arrows; quantification in Figure 3G). Conversely, ALKBH5 depletion markedly increased levels of Ser2P-RNA Pol II (Figure 3H) without affecting the levels of CDK9 or cyclin T1 (Figure S2L).
The observation that manipulating m6A-7SK, either by METTL3 or ALKBH5 knockdown/overexpression, control levels of Ser2P-RNA Pol II prompted us to examine whether m6A-7SK regulates RNA Pol II transcription. Indeed, reducing m6A-7SK by overexpression of WT ALKBH5 markedly represses global transcription level, as evidenced by 5-ethynyl-uridine (5EU) incorporation in nascent RNAs followed by immunostaining analysis (Figure 3I, white arrows). Taken together, these observations are consistent with recent findings in Drosophila S2R+ cells that modulation of m6A modifiers changes RNA Pol II transcription.41 We note that modulation of METTL3 and ALKBH5 would affect m6A levels on a broad spectrum of RNAs in addition to 7SK. Although our results suggest that m6A positively regulates RNA Pol II transcription, the specific involvement of m6A-7SK needs to be established.
Establishment of a CRISPR-CasRx system to specifically modulate m6A-7SK
Having identified potential m6A modification sites of 7SK by SELECT analyses, we next wanted to determine whether removing these m6A modifications is able to alter levels of Ser2P-RNA Pol II thereby affecting transcription. To achieve this goal, we aimed to establish an A549 cell line stably expressing m6A eraser ALKBH5 fused to a deactivated CasRx protein (dCasRx, an RNA-targeting CRISPR protein) together with a sgRNA against 7SK (Figure 4A).43 To determine the accessible regions on 7SK that are suitable for sgRNA targeting, we carried out an RNase H assay in A549 cell lysate. DNA oligonucleotides were designed against potential single-stranded regions on 7SK (Figure 2A). After forming the DNA/RNA duplexes, the endogenous RNase H cleaves the 7SK into smaller pieces, which can be detected by northern blots. As shown in Figure S2B, full-length 7SK was cleaved to different extents by endogenous RNase H after hybridization with DNA oligonucleotides targeting regions shown in Figure 2A, revealing accessible regions for sgRNA targeting.
Figure 4. Establishment of a CRISPR-CasRx system to specifically modulate m6A-7SK.
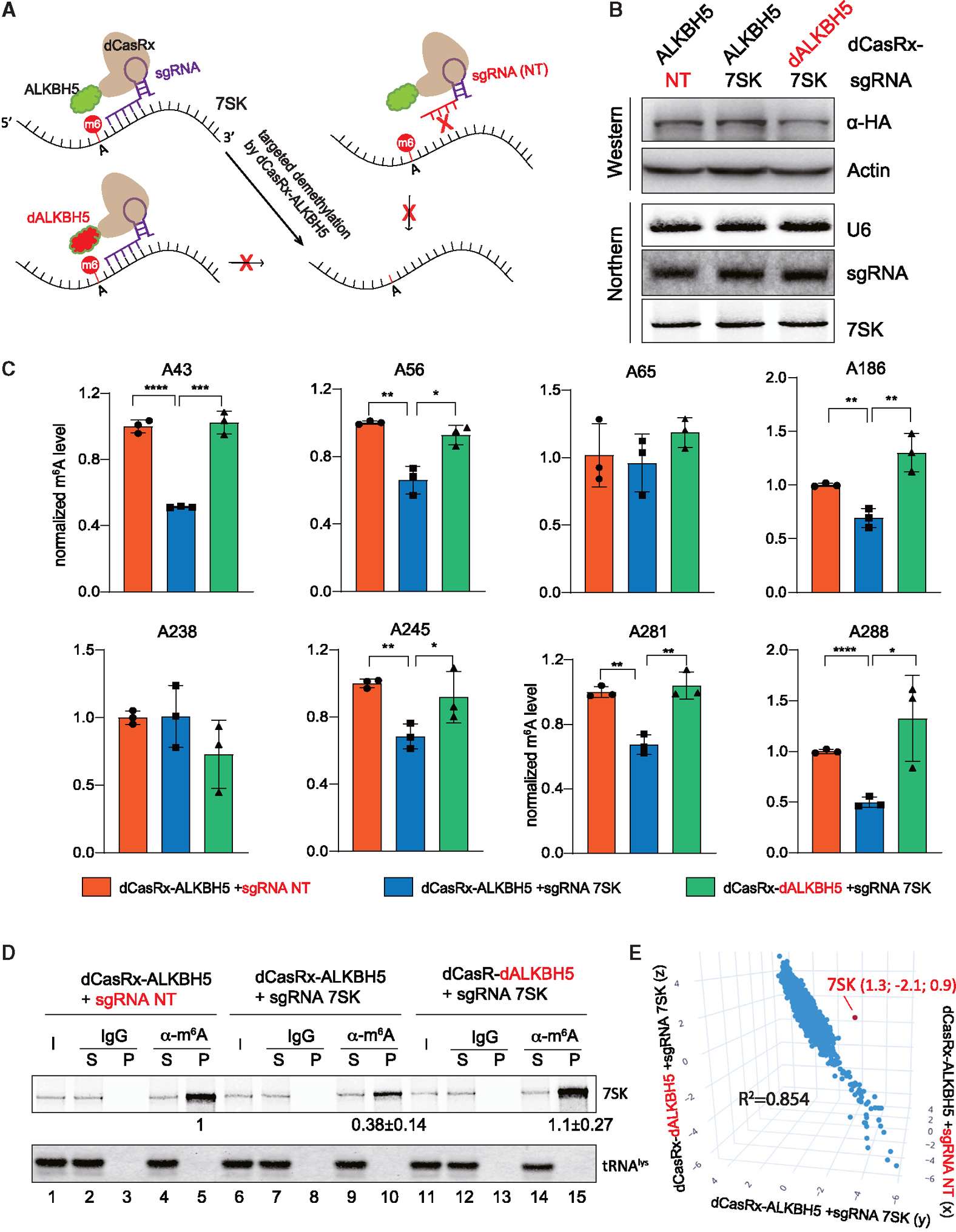
(A) Overview of the fusion dCasRx-ALKBH5 (WT or H204A) proteins for removing m6A modification on 7SK.
(B) Western blot and northern blot detected the dCasRx-ALKBH5 fusion protein expression level and the abundance of sgRNA and 7SK in the 3 engineered A549 cell lines stably expressing fusion dCasRx-ALKBH5.
(C) Normalized m6A levels at A43, A56, A65, A186, A238, A245, A281, and A288 detected by SELECT in 3 A549 cell lines as in (B) (error bars represent SD, t test calculated the p values, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
(D) Northern blot analyses of 7SK and tRNAlys after immunoprecipitation using a m6A-specific antibody or IgG control in 3 A549 cell lines as in (B). Input (I) and supernatant (S) are 1.2% of the pellet (P). Quantitation of relative m6A-7SK levels (mean ± SD) in the pellet was derived from 3 independent experiments.
(E) 3D scatter plot of transcriptome-wide relative m6A modifications (log2[m6A-IP/input]) in 3 different A549 cell lines as in (B), detected by triplicate MeRIP-seq experiments. The red dot indicates 7SK. See also Figures S2–S4.
With a sgRNA targeting the most accessible region on 7SK (nt 271–300, region 3 in Figures 2A and S2B), we generated an A549 cell line expressing dCasRx-ALKBH5 (Figure 4A). We also generated two isogenic control cell lines: one cell line expressing dCasRx fused with a deactivated ALKBH5 (H204A) together with a sgRNA against 7SK and the other cell line expressing dCasRx-ALKBH5 with a non-targeting (NT) sgRNA. In these three engineered A549 cell lines, dCasRx-ALKBH5 fusion proteins and the sgRNAs are expressed at similar levels (Figure 4B). The dCasRx-ALKBH5 and sgRNA interactions are comparable as shown in the RIP experiments (Figure S3A). The abundance of 7SK in these stable cell lines is also comparable (Figure 4B) and the dCasRx-ALKBH5 fusion proteins have similar subcellular localization in the nuclei (Figures S3B and S3C).
Next, using SELECT assays, we confirmed that 7SK-targeting dCasRx-ALKBH5 is able to reduce m6A modification at A43, A56, A186, A245, A281, and A288 in A549 cells (Figure 4C, compare blue bars with orange and green bars). However, A65 and A238 were not affected. Accordingly, cells expressing 7SK-targeting dCasRx-ALKBH5 have lower levels of 7SK enriched by MeRIP (Figure 4D, compare lane 10 with lanes 5 and 15). These data further confirm the m6A sites on 7SK and suggest that our 7SK-targeting dCasRx-ALKBH5 can reduce m6A modifications on certain sites.
A previous study reported that ALKBH5 can demethylate m6A of the MYC mRNA at A5553, which we successfully detected with SELECT assay (Figure S3D).43 Confirming the specificity of 7SK-targeting dCasRx-ALKBH5, the MYC A5553 m6A modification is not affected in our three engineered A549 cell lines (Figure S3E). To further confirm the specificity of 7SK-targeting dCasRx-ALKBH5, we performed MeRIP followed by RNA sequencing (RNA-seq) (MeRIP-seq) in triplicate to map m6A methylomes in the three engineered A549 cell lines. As intended, 7SK is the only RNA that has significant m6A change in the cells expressing 7SK-targeting dCasRx-ALKBH5 compared with the two control cell lines (Figures 4E and S4). Comparison between each of the two A549 cell lines revealed highly consistent m6A status in the transcriptome (Figure S4, R2 > 0.85).
Reducing m6A-7SK inhibits NSCLC cell growth and RNA Pol II transcription
Higher m6A modification is an important factor affecting the growth, survival, and invasion in cancer cells.42 We questioned whether higher levels of m6A-7SK contribute to lung cancer cell growth. To this end, we measured the growth of the three A549 cell lines expressing dCasRx-ALKBH5 in colony formation assays. The cells with specific m6A-7SK reduction exhibit the lowest numbers of colonies, suggesting that m6A modification of 7SK contributes to NSCLC cell growth (Figures 5A and 5B).
Figure 5. Reducing m6A-7SK inhibits NSCLC cell colony formation and RNA Pol II transcription.
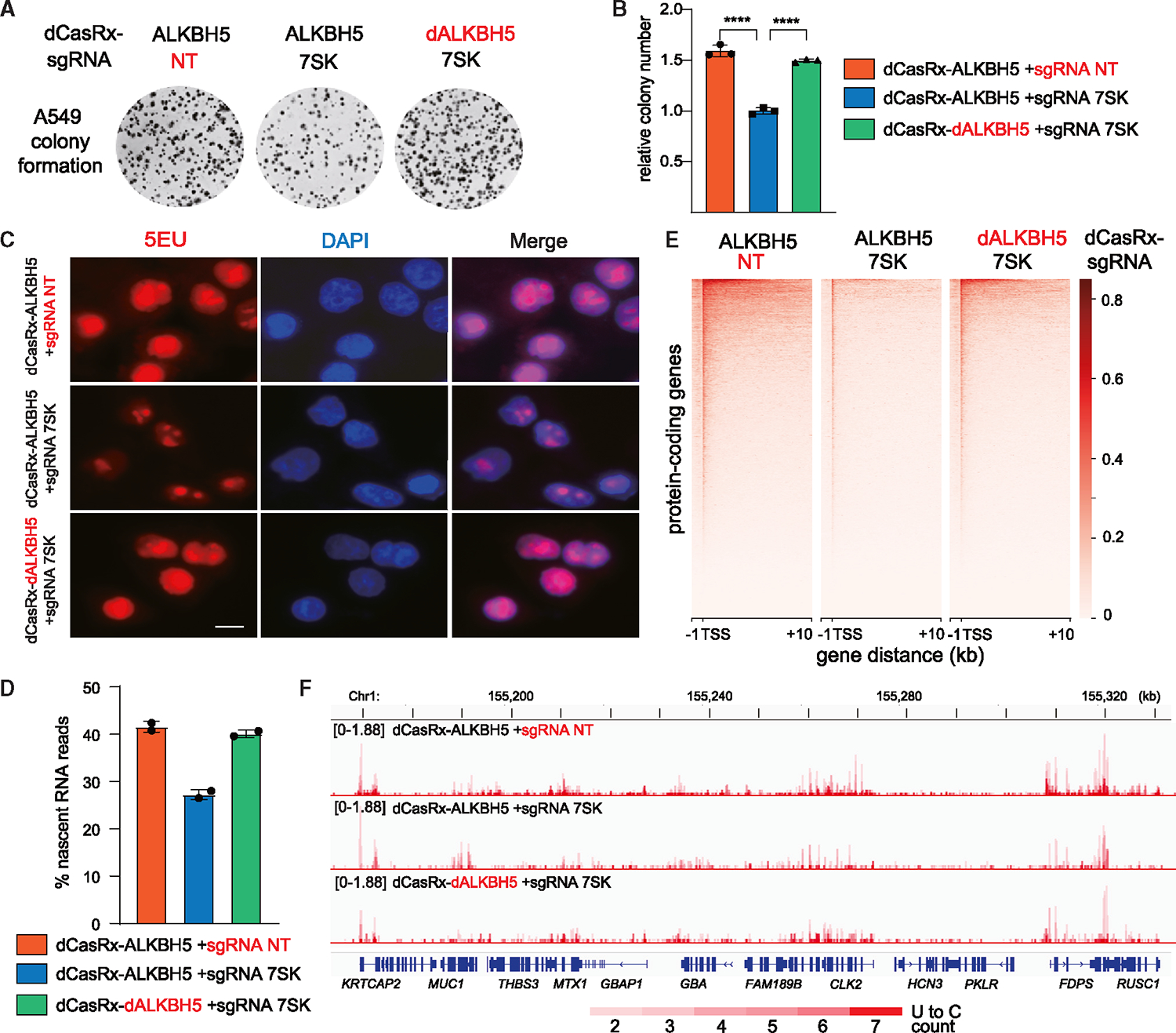
Experiments were performed in the engineered A549 cell lines stably expressing fusion dCasRx-ALKBH5 against 7SK, and two control cell lines as in Figure 4. (A and B) Representative image (A) and relative colony number (B) in colony formation assays (error bars represent SD; t test calculated the p value, ****p < 0.0001).
(C) EU incorporation and staining of nascent RNAs. Scale bars, 10 μm.
(D) Histogram profile showing the percentage of mapped nascent reads that contain 2–7 U-C mutations from duplicate nascent RNA sequencing (RNA-seq) experiments.
(E) Heatmap distribution of nascent RNA reads mapped to protein-coding genes, centered at the TSS (−1 kb to +10 kb).
(F) Genome browser shots of nascent RNA-seq tracks, spanning 155,160–155,330 kb of chromosome 1. See also Figure S5.
The reduction in A549 cell colony formation due to 7SK demethylation is most likely connected with 7SK’s role in regulating RNA Pol II transcription. To explore this possibility, we first measured the global transcription in these cells by nascent RNA 5EU staining. Indeed, removing m6A modifications of 7SK led to a reduction of nascent RNAs, reflecting a transcriptional defect (Figure 5C).
To further define the transcriptional defect related to lower m6A-7SK levels, we employed nascent RNA-seq with 4-thiouridine (4SU) metabolic labeling. 4SU-labeled nascent RNAs were enriched through conjugation with methanethiosulfonate (MTS)-biotin and reacted with iodoacetamide (IAA) to induce U to C mutation during sequencing (detailed in STAR Methods).51,52 Within the nascent RNA-seq data, about 99% of the mapped reads contain 0–7 U-C mutations (Figure S5A). Inspection of the reads containing 0 or 1 U-C mutation revealed that close to 80% of these reads are mapped to exons (Figure S5B), representing mature RNA rather than nascent RNAs (Figures S5C and S5D, blue tracks). Reads containing 8+ U-C mutations cannot be reliably mapped (data not shown). By contrast, reads containing 2–7 U-C mutations include ~40%–60% intronic reads (Figure S5B) and are therefore grouped as nascent RNA reads in subsequent analyses (Figures S5C and S5D, red tracks).
When we examined the nascent RNA-seq data from duplicate experiments in three engineered A549 cell lines, the percentage of nascent reads was reduced in the cells containing lower m6A-7SK (Figure 5D, compare blue bar with orange and green bars). Consistently, metagene analyses of the nascent RNA reads showed that reducing m6A-7SK results in the reduction of transcription in protein-coding genes (Figure 5E). We observed widespread instances of dampened nascent RNA synthesis across the genome in cells with reduced m6A-7SK (Figure 5F, compare the middle track with the top and bottom tracks), whereas the steady-state RNA levels are comparable (Figure S5E). Therefore, RNA Pol II transcription appears to be inhibited when 7SK contains lower levels of m6A modifications.
m6A modification affects the equilibrium of 7SK snRNP
During RNA Pol II transcription, 7SK enhances RNA Pol II promoter-proximal pause by sequestering P-TEFb.53 Given the global transcription inhibition in A549 cells with reduced m6A-7SK, we asked whether 7SK with lower m6A modifications facilitates P-TEFb sequestration. To this end, we immunoprecipitated CDK9, the catalytic subunit of P-TEFb complex, from lysate prepared from the three A549 cell lines described above (Figure 4) and measured associating 7SK by northern blots. As expected, reducing m6A modification of 7SK significantly strengthened the sequestration of CDK9 (Figure 6A, compare lane 10 with lanes 5 and 15). Because CDK9 is responsible for the phosphorylation of RNA Pol II Ser2, the mark of productively elongating RNA Pol II, we observed a reduction of Ser2P-RNA Pol II in A549 cells where CDK9 is more sequestered by 7SK with fewer m6A modifications (Figure 6B).
Figure 6. Reducing m6A modification of 7SK attenuates productive RNA Pol II elongation.
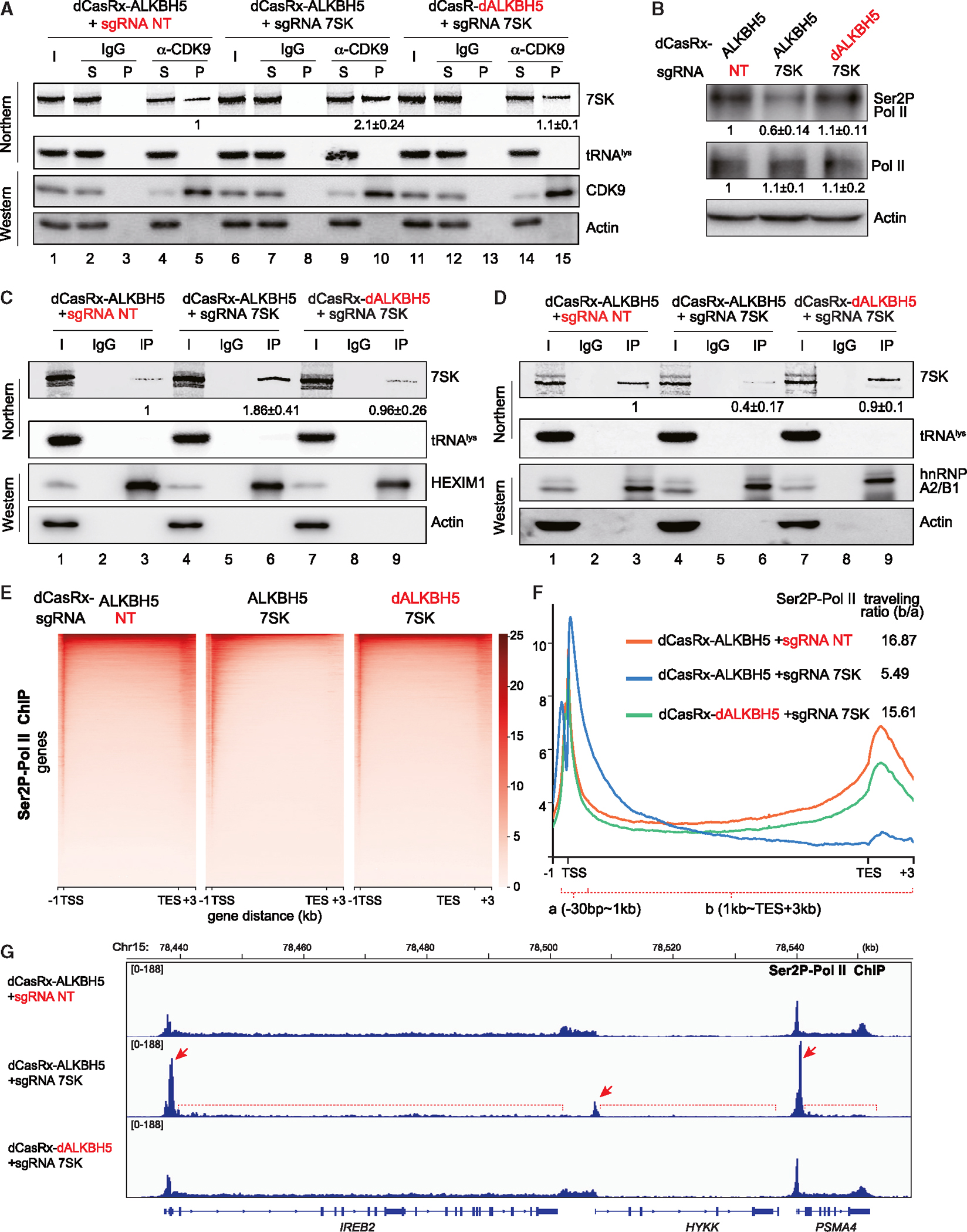
Experiments were performed in the engineered A549 cell lines stably expressing fusion dCasRx-ALKBH5 against 7SK and two control cell lines as in Figure 4. (A, C, and D) Northern blot analyses of 7SK and tRNAlys from RIP experiments performed using a CDK9 (A), HEXIM1 (C), and hnRNP A2B1 (D) antibodies or IgG. Western blots demonstrate IP efficiency. Inputs (I) are 5% of the pellet (P). Quantitation of relative m6A-7SK levels (mean ± SD) was derived from 3 independent experiments.
(B) Western blot of total RNA Pol II and Ser2P-RNA Pol II, with actin as a control. Quantitation of relative Ser2P-RNA Pol II and RNA Pol II levels (mean ± SD) were derived from 3 independent experiments.
(E) Heatmaps of Ser2P-RNA Pol II occupancy (ChIP relative to input), centered at the TSS (−1 to +3 kb).
(F) Metagene profiles of Ser2P-RNA Pol II ChIP, showing the distribution of Ser2P-RNA Pol II and the traveling ratio.
(G) Track examples of RNA Pol II Ser2P ChIP-seq in the region spanning genes IREB2, HYKK, and PSMA4. See also Figure S6.
We next examined the association of m6A-7SK with other 7SK snRNP components by RIP assays. As expected, HEXIM1, a protein that associates with 7SK together with P-TEFb, also interacts more with the demethylated 7SK (Figure 6C, compare lane 6 with lanes 3 and 9). By contrast, hnRNP A2/B1, which associates with P-TEFb-free 7SK, interacts less with the demethylated 7SK (Figure 6D, compare lane 6 with lanes 3 and 9). The constitutive 7SK-binding proteins MePCE and LARP7 interact with 7SK equally well regardless of its methylation status (Figures S6A and S6B).
To further confirm that the reduction of P-TEFb and m6A-7SK interaction reflects an equilibrium of transcriptionally active/inactive P-TEFb rather than an overall reduction of P-TEFb level, we performed a nuclear fractionation experiment.54 We found that the ratio between chromatin-bound P-TEFb (in high-salt fraction or HSF, active transcription) and chromatin-free P-TEFb (in low-salt fraction or LSF, inactive transcription) is significantly lower in A549 cells with lower m6A-7SK (Figure S6C). Similarly, the non-malignant MRC-5 cells, which have lower m6A-7SK levels compared with A549 cells (Figure 1C), have a lower ratio of active/inactive P-TEFb (Figure S6D).
Finally, we asked whether stimuli that were previously shown to promote P-TEFb releasing from 7SK, such as hexamethylene bisacetamide (HMBA), affect m6A modifications of 7SK.55 Indeed, MeRIP experiments showed that HMBA-treated A549 cells contain higher levels of m6A-7SK (Figure S6E). Taken together, these data suggest that m6A modifications of 7SK actively modulate the equilibrium of 7SK-released P-TEFb (active transcription) and 7SK-associated P-TEFb (inactive transcription).
Reducing m6A modification of 7SK attenuates productive RNA Pol II elongation
To gain mechanistic insights on m6A-7SK-related transcription regulation, chromatin immunoprecipitation sequencing (ChIP-seq) was performed for Ser2P-RNA Pol II and RNA Pol II to determine possible polymerase elongation defects. Metagene analysis of the ChIP-seq showed that reducing m6A-7SK results in the accumulation of Ser2P-RNA Pol II around the TSS and reduction in the gene body and downstream of transcription termination site (TES) (Figure 6E). We further estimated the RNA Pol II elongation defect by measuring the traveling ratio of the Ser2P-RNA Pol II, which is calculated by the ratio between ChIP-seq reads in gene body region (from 1 kb after TSS till 3 kb after TES) and reads in the promoter-proximal region (from 30 bp upstream to 1 kb downstream of TSS) (Figure 6F). Ser2P-RNA Pol II traveling ratio in the A549 cell with lower m6A-7SK is almost 3-fold lower than the control cells (Figure 6F, compare blue line with the orange and green lines). Finally, a gene-level analysis of genomic regions with high levels of ChIP-seq reads revealed Ser2P-RNA Pol II elongation defect that is consistent with the metagene analyses results (Figure 6G). For the three representative neighboring genes (IREB2, HYKK, and PSMA4), in cells with lower m6A-7SK, Ser2P-RNA Pol II accumulates at the promoter-proximal region (Figure 6G, red arrows) and fails to travel into the gene bodies (Figure 6G, red brackets with dotted lines). In the RNA Pol II ChIP-seq data, RNA Pol II is similarly more concentrated in the promoter-proximal region when 7SK is demethylated (Figures S6F–S6H). Taken together, our data provide compelling evidence that erasing m6A modifications of 7SK not only reduces the overall level of Ser2P-RNA Pol II but also attenuates them at the promoter-proximal region through 7SK-mediated sequestration of P-TEFb.
Reducing m6A modification of 7SK alters its RNA structure
The structure of 7SK RNA has been previously studied in vitro and in vivo, in a variety of systems.56 It has become clear that the 7SK snRNP is dynamic and that the RNA structure and protein interactions change in response to cellular conditions to control transcription.57,58 Specifically, 7SK adopts distinct structures to sequester or release P-TEFb, although the detailed structural differences between these two states are still being debated.57–61 Furthermore, the molecular mechanism that induces such 7SK conformational changes is unknown.
To examine the effect of m6A perturbation on 7SK structure, we performed in-cell selective 2′-hydroxyl acylation analyzed by primer extension, combined with mutational profiling (SHAPE-MaP) on 7SK in A549 cells expressing our CRISPR dCasRx-ALKBH5 systems (Figure 4). SHAPE-MaP is a chemical probing method that uses a hydroxyl-selective electrophile (such as 2′-methylnicotinic acid imidazolide or NAI) to react with the RNA 2′-hydroxyl position when it is accessible.62,63 Subsequently, the 2′-O-modified bases are converted into sequence mutations during RT and deep sequenced (Figure 7A). Cells were treated with NAI or dimethyl sulfoxide (DMSO) only and analyses were performed using the ShapeMapper v2 pipeline64,65 (detailed in STAR Methods).
Figure 7. Structural switch of 7SK RNA upon modulation of m6A-7SK.
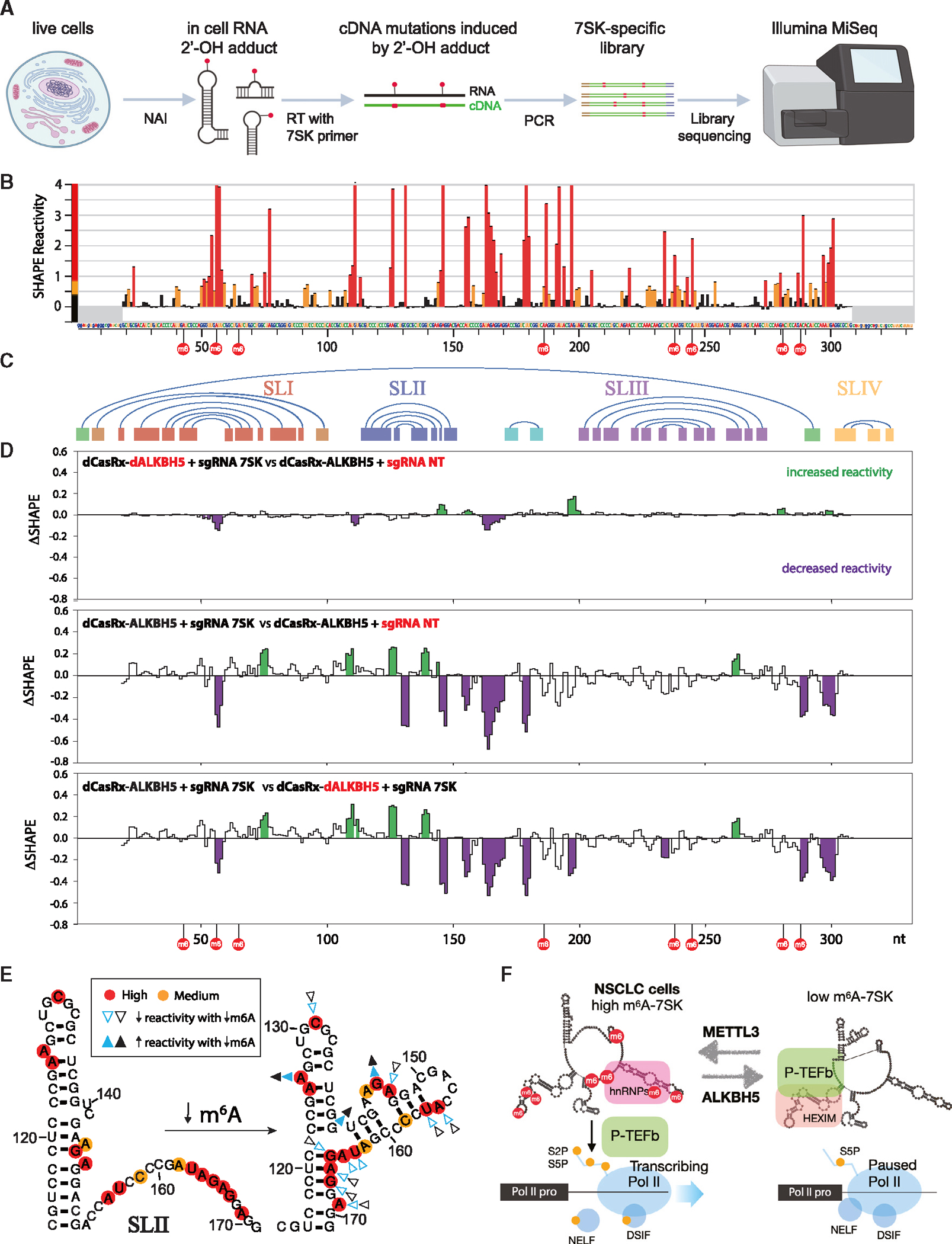
Experiments were performed in the engineered A549 cell lines stably expressing fusion dCasRx-ALKBH5 against 7SK and two control cell lines as in Figure 4. (A) Overview of SHAPE-MaP.
(B) Histogram profile depicting SHAPE-MaP reactivity of 7SK in A549 cells. The nucleotides and numbering across the x axis correspond to the reference sequence NR_001445. m6A sites are indicated by red circles. Highly reactive nucleotides are colored in red, medium in orange, and low in black. Regions that do not contain structural information due to primer binding (nt 1–18, 319–332) are shaded in gray. Error bars indicate SD from duplicate experiments.
(C) Graphic representation of interacting helices in a consensus model of 7SK. The region corresponding to SLI is shown in red, SLII in blue, SLIII in purple, SLIV inyellow, and the circularization helix in green.
(D) Pairwise ΔSHAPE analyses highlighting the differences between 3 SHAPE-MaP conditions. Analyses were performed with a sliding window of 3 nt and regions with increased reactivity are shaded in green, whereas decreased reactivity is shown in purple (p < 0.05).
(E) Comparison of 7SK SLII (nt 114–171) structures modeled with SHAPE-MaP activity and DSHAPE. High-reactivity positions are shown in red, and medium are in orange. The arrowheads show changes in reactivity upon reduction of m6A levels in 7SK. The colors (blue and black) represent 2 replicate experiments. Filled arrowheads indicate positions that become more reactive when m6A is decreased, whereas open arrowheads mark positions that are less reactive.
(F) Model of m6A-7SK-mediated RNA Pol II transcription activation. See also Figure S7.
The SHAPE-MaP reactivity profiles (Figures 7B and S7A–S7F) show that reactive regions on 7SK fall predominantly within single-stranded regions based on previous structure models (Figures 7C and S7G).66,67 Regions in 7SK that were affected by different m6A levels were identified by performing ΔSHAPE analyses to compare reactivity across different conditions.68 Comparison of the two control conditions showed very little difference between the profiles (Figure 7D, top). By contrast, comparison of the m6A reduced condition to either control sample showed large, highly overlapping differences (Figure 7D, middle and bottom), suggesting that modulation of m6A is sufficient to alter the 7SK RNA structure.
As seen in Figures 7D and S7G, alterations in SHAPE reactivity were present in the terminal loop of SLI (A56, which is an m6A site, and U57), as well as the internal loop (G74 and C75), which is of interest as this is the primary location of HEXIM binding.48,49 Two other m6A sites, A43 and A65, are also located in SLI. To test if these m6A sites directly affect binding of 7SK with HEXIM, we performed an in vitro binding assay with 5′-biotinylated 7SK SLI (nt 30–80, Figure S7H) with or without m6A modifications in A549 cell lysates. This experiment revealed that m6A on SLI inhibits the interaction between 7SK and HEXIM1 (Figure S7I).
The largest cluster of changes was distributed across SLII and the region immediately downstream (A155, U156, U163-G167, and A169), with a loss of reactivity upon m6A depletion. These changes agree well with the formation of an extended SLII as in Figure 2A, one of the original 7SK models determined by chemical probing69 and thus become less accessible (Figure 7E). SLIII was unperturbed except for an unusual increase in reactivity of G262, which is modeled in the midst of a helix, and no concurrent increase in the reactivity of its putative partner nucleotide was detected. Finally, there is a decrease in reactivity in nucleotides C289 and A297–A301, adjacent to the helix responsible for 5′ to 3′ circularization. The placement of the amplification primers prevents SHAPE analysis on one side of the helix, but no changes were detected on the 3′ side of the circularization helix (A290–A295). Biological replicates of the SHAPE-MaP experiment show good reproducibility (Figures S7A–S7F), and mapping of these changes onto a circular model of 7SK suggests that erasing the m6A modification shifts 7SK toward a conformation favoring P-TEFb sequestration (Figure S7G).
DISCUSSION
Despite numerous studies suggesting that m6A modification on mRNAs post-transcriptionally modulates various crucial biological processes,35,36,70–73 the exact role of m6A in transcriptional regulation via non-coding RNAs is yet to be defined. In this study, we uncovered m6A modifications on 7SK snRNA, an RNA Pol III transcribed non-coding RNA.12 Of note, during the revision of this manuscript, recent publications similarly suggest m6A-7SK interacts with specific reader proteins and activates transcription in HeLa cells.74,75
Among the major m6A writers (METTL3 and METTL16) and erasers (FTO and ALKBH5), we determined METTL3 and ALKBH5 to be responsible for 7SK methylation (Figure 1). A previous study detected association between METTL16 and 7SK by RIP-seq,76 suggesting that METTL16 is a potential m6A writer for 7SK. However, another study found that METTL16 neither binds to 7SK snRNA nor regulates m6A modification of 7SK.44 Consistent with the latter study, we did not detect METTL16-7SK association by RIP, and knockdown or overexpression of METTL16 did not change levels of m6A-7SK (Figure S1) nor the m6A sites detected by SELECT assays (Figures S2E–S2J). Instead, we observed that METTL3 is responsible for m6A installation on 7SK, which is consistent with recent publications.74,75
Global epitranscriptomic analyses suggest that RNA m6A modifications predominantly occur at the RRACH consensus motif.30,46 However, among the eight m6A sites we identified on 7SK, only two sites (A281 and A288) are within the RRACH motif (Figure 2A). The other six sites are in GAU (A43, A56, and A65), CAA (A186 and A238), or CAU (A245) motifs. Four of these non-RRACH m6A sites can be specifically demethylated by the 7SK-targeting dCasRx-ALKBH5, providing further evidence that these are bona fide m6A sites (Figure 4C). Nanopore sequencing analyses suggested many lower-confidence 7SK candidate m6A sites, which require further validation.21 Nonetheless, many of these candidate sites are similarly outside of a RRACH motif. The predominant non-RRACH distribution of 7SK m6A sites is unexpected, suggesting that a distinct mechanism for m6A modification may exist.
A common function of the m6A-mark is altering the stability of mRNAs.35,77,78 Unexpectedly, our study suggests that m6A modification of 7SK does not affect its stability (Figure 4), consistent with the recent findings.74 Instead, m6A modification induces marked structural rearrangement of 7SK, thereby facilitating the release of P-TEFb complex to activate RNA Pol II transcriptional elongation (Figures 5, 6, and 7). m6A modification can alter the RNA structure by directly affecting RNA base pairing and by influencing RNA-protein interactions.36,79 In the case of m6A-7SK, both mechanisms may play a role in determining the 7SK structure: several validated m6A sites reside in duplexes where 7SK would interact with HEXIM and hnRNPs (Figure 2) and SHAPE activity data suggest changes in RNA base pairing across 7SK. In SLI, A43 and A65 in the two GAUC motifs critical for HEXIM binding are both methylated.48 Our data suggest that these two m6A modifications may directly inhibit HEXIM binding to 7SK (Figure S7I). In low m6A-7SK cells, decrease in SHAPE activity on A56-U59 perhaps results from HEXIM binding (Figure S7G). The most prominent structural change occurs in stem-loop II (Figures 7D and 7E), suggesting that an extended SLII structure may favor HEXIM/P-TEFb association. SLIII for hnRNP binding also contains two m6A sites in A238 and A245, and m6A modifications on 7SK increase its interaction with hnRNPs (A1, A2/B1, R, and Q).74 Follow-up studies are needed to determine whether the m6A nucleotides or the structure of m6A-7SK are recognized by hnRNPs. Furthermore, given the wide spectrum of m6A reader proteins, additional proteins other than hnRNPs may specifically recognize m6A-modified 7SK.31–33,74,80
Most previous studies focus on how the fates of m6A-marked transcripts are determined by m6A modification, confined to the regulation at post-transcriptional level. More recently, several studies suggest that m6A modification affects global cellular transcription through chromosome-associated regulatory RNAs, including nascent mRNAs and eRNAs. However, the identity and the mode of action for such m6A-containing transcripts that regulate transcription are far from clear. Some studies claim that m6A modification on eRNAs suppresses transcription by promoting decay of m6A-marked eRNAs,22,39 whereas others suggest that m6A modification on eRNAs promotes transcription by facilitating transcriptional condensate formation.40 Nonetheless, it is clear that m6A MTC (METTL3METTL14) is recruited to chromatin at the RNA Pol II promoter proximal region and promotes gene expression by releasing paused RNA Pol II.41,81 Whether MTC exerts its function through methylating RNAs or interacting with the integrator complex is under debate.41,81 Our study provides compelling evidence that 7SK is a key substrate for MTC in the promoter region. m6A-7SK promotes RNA Pol II transcription by inhibiting promoter-proximal pausing, providing another layer of m6A-mediated gene regulation.
Having established that m6A modification facilitates RNA Pol II transcription by regulating 7SK-mediated promoter-proximal pausing, we further probed the physiological function of this regulation. Interestingly, we found that m6A modification of 7SK is much more abundant in NSCLC cells such as A549, NCI-H1299, NCI-H2009, and NCI-H23 than in non-malignant MRC-5 and BEAS-2B cells (Figure 1). Blocking m6A modification of 7SK markedly inhibits colony-forming ability of A549 cells, suggesting that m6A-7SK plays a crucial role in NSCLC tumorigenesis (Figure 5). Intriguingly, NSCLC patients have a high prevalence of epidermal growth factor (EGF) receptor (EGFR) mutations, which can lead to constitutive activation of EGF signaling that, ultimately, phosphorylates and activates METTL3.74,82 In addition, two studies suggest that m6A writer METLL3 is required for the growth, survival, and invasion of the lung cancer cells by facilitating translation of oncogenes.42,83 Aberrant m6A levels induced by overexpressed/hyperactivated METTL3 are involved in the progression of lung cancer, including cell proliferation, invasion, metastasis, drug resistance, and tumor environment.37,38 Because METTL3 potentially methylates 7SK to aberrantly activate RNA Pol II transcription in cancer cells, our study provides the molecular basis for potential therapeutics that modulate m6A-7SK for NSCLC treatment.
Limitations of the study
We demonstrated that modulation of m6A-7SK influences RNA Pol II transcription in A549 cells (Figures 4 and 5). We cannot, however, pinpoint which specific m6A site(s) contribute to 7SK’s transcription regulation function. Likewise, the two m6A sites (A65 and A238) that are not erased by dCasRx-ALKBH5 may also be important for 7SK’s function (Figure 4C). Future work using a different 7SK-targeting sgRNA may elucidate the functional importance of m6A sites on A65 and A238 (Figures 2A and S2B). In addition, the utility of other RNA-targeting CRISPR-Cas systems, such as Cas13b, can expand our strategy in site-specific m6A modulation on 7SK.84 Finally, direct RNA-seq, such as the nanopore technology, holds the potential to map m6A sites accurately and precisely on 7SK at the single-molecule level.
Our study focuses on the major 7SK RNP, which regulates RNA Pol II transcription through P-TEFb. There exist at least two other 7SK RNPs that can regulate transcription: 7SK-LEC (little elongation complex) and 7SK-BAF (chromatin remodeling complex).60,85 Future studies are required to determine the possibility that m6A also affects 7SK assembly in these two much less abundant RNPs.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Dr. Mingyi Xie (mingyi.xie@ufl.edu).
Materials availability
All the materials generated in this study are accessible upon request.
Data and code availability
All the sequencing data generated by this study have been deposited to NCBI Sequence Read Archive (SRA) database under SRA accession number PRJNA938517
All original code has been deposited at https://github.com/UF-Xie-Lab/7SK-python-tools, which is also available through: https://doi.org/10.5281/zenodo.8341420
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Cell culture
A549, H1299 and H23 cells were cultured in RPMI-1640 medium (Sigma, R8758) supplemented with 10% fetal bovine serum (FBS) (Gibco, A5256701), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, 15140-122). 293T, H2009 and MRC5 cells were cultured in DMEM medium (Sigma, D5796) supplemented with 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. BEAS-2B cells were cultured in BEGM BulletKit (Lonza, CC-3170) supplemented with 100 U/mL penicillin and 100 mg/mL streptomycin at 37°C, 5% CO2.
METHOD DETAILS
RNA isolation and RT-qPCR
Cells were washed with cold phosphate buffer-saline (PBS) and RNA was isolated from cells using TRIzol, following the manufacturer’s protocol. The cDNA synthesis was performed using reverse transcription kit. All RT-qPCR experiments used Actin mRNA or U6 snRNA for normalization. PCR primers used are listed in Table S1.
m6A RNA immunoprecipitation
MeRIP were modified according to previously published protocols.43,84 Briefly, 100 μg total RNA were incubated with 5 μg a-m6A antibody or control IgG in 1× reaction buffer (150 mM NaCl, 10 mM Tris–HCl, pH 7.5 and 0.1% NP-40) for four hours at 4°C. Then, 30 μL M-280 sheep anti-Rabbit IgG dynabeads were washed and added to the mixture with rotation at 4°C overnight. The enriched RNA was extracted with TRIzol and co-precipitated with 15 μg glycoblue. Finally, m6A-RNA enrichment was determined by RT-qPCR analysis or MeRIP-seq. The MeRIP-seq libraries were generated using the SMARTer Stranded Total RNA-Seq Kit v3 following the manufacturer’s protocol.
Data analysis for MeRIP-seq
All MeRIP-seq data sets were first checked for base call quality using the FastQC program (bioinformatics.babraham.ac.uk/projects/fastqc/). Sequencing reads were processed by Trimmomatic to remove adapter content and low-quality reads.86 PEAR was used to combine paired reads into one forward read.87 Unique molecular tags (UMTs) generated during library preparation were removed using cutadapt.88 Reads were mapped using HISAT2 to the human reference genome (GRCh38 release 106) using default parameters.89 The resulting alignment files were sorted and indexed using Samtools sort and Samtools index tools.90 Per gene read counts were calculated using the HTSeq htseq-count tool.91 Low abundance genes were characterized as genes with fewer than 10 reads in any of the three input samples or any of the three MeRIP samples and were removed from the analysis. Each condition was normalized to the input control by taking log2 (m6A IP/input). Both 3-dimensional (Figure 4E) and 2-dimensional (Figure S4) plots comparing the m6A IP from the 7SK-targeting dCasRx-ALKBH5 to the two controls were generated using Plotly.92
RNA immunoprecipitation assay
RIP analyses were modified according to a previously published protocol.44 10 million A549 cells were fixed with 0.1% formaldehyde in PBS at room temperature for 10 min. Glycine (final concentration 125 mM) was added to terminate formaldehyde fixation at room temperature. Cells were washed with cold PBS and scraped, followed by centrifuging at 500 ×g for 5 min at 4°C. Cell pellets were resuspended in 600 μL lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM KCl, 0.5 mM DTT, 1% Triton X-100 and 1 mM PMSF) on ice for 30 min. After centrifugation at 16000 ×g, at 4°C for 15 min, the supernatant was collected (5% saved as input) and incubated with 1× protease inhibitor cocktail, 2 μL RNAaseOUT (Invitrogen, 10777019), 40 μL M-280 sheep anti-Rabbit IgG dynabeads that are pre-coated with 5 μg antibody/IgG, then rotate at 4°C overnight. The beads were washed by lysis buffer twice and the bound RNAs were extracted by TRIzol reagent. Amount of target transcripts in both the input and RIP samples were analyzed with RT-qPCR.
Western blot and northern blot
Total protein was extracted with NP-40 lysis buffer (1% NP-40, 10% Glycerol, 150 mM NaCl, 50 mM Tris-HCl, 5 mM EDTA) and quantified using DC protein assay kit. Equal amount of protein samples was separated in 10% polyacrylamide-SDS gel and then transferred to PVDF membranes. The membranes were blocked in 5% non-fat milk, and then incubated in primary and secondary antibodies. The primary antibodies used in this study are listed in key resources table. Near-infrared northern blot was performed as previously described.96 Briefly, total RNA was isolated by TRIzol and separated on 10% denaturing polyacrylamide 8 M urea gel and subsequently transferred onto an Amersham Hybond-N+ membrane and UV cross-linked. Full-length antisense 7SK labeled with near-infrared (IR) dye was used as an RNA probe. Other probes are either IR-containing DNA oligonucleotides or azide-containing DNA oligonucleotides that are labeled with IR dye through click chemistry (Table S2).96
KEY RESOURCES TABLE.
| REAGENTor RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| m6A | Synaptic systerms | Cat#202003; RRID:AB_2279214 |
| METTL3 | ABClonal | Cat#A21572 |
| ALKBH5 | Sigma | Cat#HPA007196; RRID:AB_1850461 |
| FTO | Sigma | Cat#SAB2106776 |
| METTL16 | ABClonal | Cat#A15894; RRID:AB_2763325 |
| CDK9 | ABClonal | Cat#A11145; RRID:AB_2861506 |
| RNAP II | Abcam | Cat#ab300575 |
| Ser2P | Abcam | Cat#ab238146 |
| HA | Abcam | Cat#ab9110; RRID:AB_307019 |
| Flag | Sigma | Cat#F7425; RRID:AB_439687 |
| Actin | Proteintech | Cat#60008-1-Ig; RRID:AB_2289225 |
| Alexa Fluor 488 AffiniPure Goat Anti-Mouse IgG | Jackson ImmunoResearch | Cat#115-545-003; RRID: AB_2338840 |
| Alexa Fluor 594 AffiniPure Goat Anti-Rabbit IgG | Jackson ImmunoResearch | Cat#111-585-003; RRID: AB_2338059 |
| MePCE | Proteintech | Cat#14917-1-AP; RRID:AB_2250635 |
| Larp7 | Proteintech | Cat#17067-1-AP; RRID:AB_2132693 |
| HEXIM1 | Proteintech | Cat#15676-1-AP; RRID:AB_2248363 |
| CyclinT 1 | Cell Singnal | Cat#81464; RRID:AB_2799973 |
| H3 | Sigma | Cat# H0164; RRID: AB_532248 |
| hnRNPA2/B1 | Santa Cruz | Cat#sc-374053; RRID:AB_10947257 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Lipofectamine 3000 | Invitrogen | Cat# L3000015 |
| Streptavidin Dynabeads | Thermo Fisher | Cat# 65002 |
| TRIzol reagent | Thermo Fisher | Cat# 15596018 |
| Dynabeads Protein G | Thermo Fisher | Cat# 88848 |
| Proteinase K | VWR | Cat# 97062-238 |
| Protease Inhibitor Cocktail | Sigma | Cat# 5056489001 |
| MTSEA BIOTIN | Biotium | Cat# 90066 |
| 4-THIOURIDINE | Sigma | Cat# T4509 |
| NAI | Millipore | Cat# 03-310 |
| SsoAdvanced Universal SYBR Green Supermix | Biorad | Cat# 1725274 |
| Glycoblue Coprecipitant | Thermo Fisher | Cat# AM9516 |
| Potassium Chloride | Affymetrix | Cat# 75896 |
| Dynabeads M-280 Sheep anti-Rabbit IgG | Thermo Fisher | Cat# 11204D |
|
| ||
| Critical commercial assays | ||
|
| ||
| Qubit 1X dsDNA HS Assay Kit | Invitrogen | Cat# Q33230 |
| Qubit RNA HS assay Kit | Invitrogen | Cat# Q32852 |
| TURBO DNA-free Kit | Invitrogen | Cat# AM1907 |
| Smart Stranded Total RNA-Seq Kit v3 | Takara | Cat# 634488 |
| ThruPLEX Tag-seq 96D Kit | Takara | Cat# R400586 |
| SuperScript II Reverse Transcriptase | Invitrogen | Cat# 18064014 |
| iClick EU Andy Fluor 594 Imaging Kit | ABP Biosciences | Cat# A010 |
| QuantiTect Reverse Transcription Kit | QIAGEN | Cat# 205313 |
| Agencourt RNAclean XP beads | Beckman Coulter | Cat# A63881 |
| Protein Assay Reagent S | Bio-Rad | Cat# 5000115 |
| Protein Assay Reagent A | Bio-Rad | Cat# 5000113 |
|
| ||
| Deposited data | ||
|
| ||
| The raw and processed sequencing data | This paper | SRA: PRJNA938517 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| HEK293T | Laboratory of Joan A. Steitz | N/A |
| A549 | Laboratory of Lingtao Jin | N/A |
| BEAS-2B | Laboratory of Lizi Wu | N/A |
| MRC-5 | Laboratory of Lizi Wu | N/A |
| H2009 | Laboratory of Lizi Wu | N/A |
| H23 | Laboratory of Lizi Wu | N/A |
| H1299 | Laboratory of Lizi Wu | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| RT-qPCR primer sequences | This paper | See Table S1 |
| Northern blot probe sequences | This paper | See Table S2 |
| siRNA sequences | This paper | See Table S3 |
| SELECT primer sequences | This paper | See Table S4 |
| DNA oligonucleotides for RNase H | This paper | See Table S5 |
| 7SK SLI sequences | This paper | See Table S6 |
| Primer sequences for SHAPE-MaP library | This paper | See Table S7 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pMSCV-dCasRx-ALKBH5 | Addgene | 175582 |
| pMSCV-dCasRx-ALKBH5 H204A | This paper | N/A |
| pCDNA3.1-Flag-ALKBH5 | This paper | N/A |
| pCDNA3.1-Flag-ALKBH5 H204A | This paper | N/A |
| pCDNA3.1-Flag-METTL3 | This paper | N/A |
| pCDNA3.1-Flag-METTL3 D395A | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Prism 9 | Gaphpad Software | https://www.graphpad.com/scrientifificsofware/prism/ |
| Custom sequencing analysis scripts | This paper | https://doi.org/10.5281/zenodo.8341420 |
| FastQC program | Babraham Bioinformatics | bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trimmomatic | Bolger et al.86 | N/A |
| PEAR | Zhang et al.87 | N/A |
| cutadapt | Martin88 | N/A |
| HISAT2 | Kim et al.89 | N/A |
| Samtools | Li et al.90 | N/A |
| HTSeq | Anders et al.91 | N/A |
| Plotly | Inc., P.T.92 | N/A |
| DeepTools | Huang et al.93 | N/A |
| MACS2 callpeak | Rami´rez et al.94 | N/A |
| HISAT-3N | Zhang et al.95 | N/A |
| deltaSHAPE.py | Laboratory of Kevin M. Weeks | weekslab.com/software |
| StructureEditorv6.2 | Laboratory of David H. Mathews | rna.urmc.rochester.edu/GUI/html/StructureEditor.html |
Molecular cloning
The original pMSCV-dCasRx-ALKBH5-PURO plasmid (175582) and lenti-sgRNA-BSD (175583) were purchased from Addgene. dCasRx-dALKBH5-PURO containing a single mutation at H204A was constructed. sgRNAs targeting 7SK were designed and cloned into lenti-sgRNA-BSD. The guide sequences are listed in Table S5. For transient transfection experiments, coding sequences for FLAG-tagged ALKBH5 (WT or H204A) or METTL3 (WT or D395A) were cloned into the pCDNA3.1 vector. For knockdown experiments, shRNA sequences against METTL3, ALKBH5, METTL16 and FTO were cloned into PLKO.1 plasmid vector. shRNA sequences were listed in Table S3
Generation of stable cell lines with lentivirus
80% confluent 293T cells in 10 cm dishes were transfected with lentivirus packaging plasmids (6 μg of psPAX2 and 4 μg pMD2.G) and 10 μg cargo plasmids (encoding dCasRx-ALKBH5, sgRNA or shRNA) with lipofectamine 3000. After transfection, the cells were cultured in FreeStyle 293 Expression Medium (Gibco, 12338018) for 48 h and the supernatant containing the viruses was harvested. The viruses were concentrated with Lenti-X concentrator (Takara, 631232) following the manufacturer’s protocol. 2 mL concentrated lentivirus were used to infect A549 cells in 6 well plate for 48h and followed by 1 μg/mL puromycin selection to obtain polyclonal stable cell lines. For A549 cells expressing both dCasRx-ALKBH5 and sgRNA, dCasRX-ALKBH5-expressing cells were first established via puromycin selection. The cells were then infected with lentiviruses carrying sgRNA, followed by selection with blasticidin (10 μg/mL) and puromycin to obtain polyclonal stable cell lines. Due to growth defect in cells with low m6A-7SK, experiments were performed using stable cells with low passage numbers.
SELECT qPCR for m6A site detection
SELECT qPCR was conducted following a previously published protocol with minor changes.43,47 Briefly, 1 μg total RNA was mixed with 40 nM each of the upstream and downstream primers and 5 μM dNTP (NEB#N0446S) in 17.5 μL 1× CutSmart buffer (NEB, B7204S). The reaction mixtures were incubated with annealing cycle: 90°C, 80°C, 70°C, 60°C, 50°C for 1 min and 40°C for 6 min. 2.5 μL of enzyme mixture including 0.5 U SplintR ligase (NEB, M0375S), 0.01 U Bst 2.0 DNA polymerase (NEB, M0537S) and 10 nM ATP (NEB, P0756S) were added and the reaction was incubated at 40°C for 20 min and 80°C for 20 min. Subsequently, 2 μL of reaction mixture, 200 nM SELECT primers and 2× SYBR Green Master Mix were mixed for qPCR with the following program: 95°C, 1 min; 95°C, 20 s, 60°C, 60 s for 40 cycles; 95°C, 15 s; 60°C, 1 min; 95°C, 15 s; 4°C, hold. Cq values of samples were normalized to their corresponding Cq values of control. Primers are listed in Table S4.
Immunofluorescence
For immunofluorescence, 5×104 A549 cells were cultured on glass slides in 24 well plates for 12 h. The plasmids with HA or FLAG tagged ALKBH5/METTL3 (500 ng) were transfected into A549 cells with Lipofectamine 3000. After 36 h incubation, cells were fixed in 4% paraformaldehyde (in 1× PBS) for 15 min, permeabilized in 0.5% Triton X-100 for 15 min at room temperature. Then the cells were blocked in 5% bovine serum albumin for 1h at room temperature and incubated with HA (1:500), FLAG (1:500) or Ser 2P (1:1000) antibody overnight at 4 °C. Cells were stained with Alexa Fluor 488 (1:500) or Alexa Fluor 594-conjugated (1:500) secondary antibodies for 1h at room temperature. The nuclei were stained with DAPI. Representative images were captured using the LEICA DFC7000T. Using the Image J software, we quantified the fluorescence intensity of Ser-2P in 20 randomly selected transfected cells (with FLAG signal) and normalized to 20 cells from the vector transfection experiment. For quantification of the vector transfection experiment, 20 cells were randomly selected.
EU labeling assay
The EU incorporation assay was performed as previously described.97 Briefly, EU was added to the culture medium after 36 h of initial transfection with a final concentration of 250 μM, and cells were incubated at 37 °C for 1.5 h. After EU labeling, the cells were washed with cold PBS twice and fixed in 3.7% formaldehyde at room temperature for 15 min, permeabilized in 0.5% Triton-X 100 at room temperature for 15 min. Then the cells were washed with PBS, stained with Alexa 594–azide (1:500) and incubated with DAPI according to the manufacturer’s instructions.
Endogenous RNase H cleavage assay
10 million A549 cells were resuspended in 400 μL Sucrose Buffer I (0.32 M sucrose, 3 mM CaCl2, 2 mM Mg acetate, 0.1 mM EDTA, 10 mM Tris-HCl pH 8.0, 1 mM DTT, 0.5 mM PMSF and 0.5% (v/v) NP-40). After centrifugation at 500 ×g for 5 min at 4°C, the supernatant was collected as cytoplasmic extract. 250 μL low salt Buffer (20 mM HEPES, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF) was added to resuspend the nuclei pellet at 4°C. 1/5 volume of high salt Buffer (20 mM HEPES, pH 7.9, 25% glycerol, 1.5 mM MgCl2, 0.8 M KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 1% NP-40) was added to the nuclei suspension. The nuclear extract and cytoplasmic extract were then mixed, and centrifuged at 21,000 ×g for 5 min at 4 °C. The supernatant was taken out for RNase H reaction. For each reaction 10 μL of 100 μM DNA oligos were added to 90 μL cell extract and incubated for 30 min at 37°C. TRIzol was added to extract total RNA. Sequence of the DNA oligos is provided in Table S5.
Colony formation assay
Two thousand A549 cells per well were seeded in 6-well plates. 10 days later, cells were fixed with 4% paraformaldehyde and stained with crystal violet dye (0.1% w/v). The images were taken by a ChemiDoc Imaging system (Bio-Rad).
Nuclear fractionation
The nuclear fractionation experiment was prepared as previously described with minor changes.54 A549 cell pellets were resuspended in buffer A (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1mM DTT and 13 protease inhibitor cocktail) and incubated on ice for 15 min. After centrifuged at 250 ×g for 5 min, the pellets were resuspended with same volume of low-salt buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 150 mM KCl, 1% NP-40, 1mM DTT and 1× protease inhibitor cocktail) for 15 min and centrifuged at 5000 ×g for 5 min at 4°C. The supernatant was saved as low-salt fraction (LSF, containing chromatin-unbound P-TEFb). Subsequently, the pellets were resuspended with high-salt buffer (10 mM HEPES pH 7.9, 300 mM NaCl, 1.5 mM MgCl2, 20% Glycerol, 0.4 mM EDTA, 0.5% NP40, 1 mM DTT and 1× protease inhibitor cocktail) on ice for 30 min and centrifuged at 12,000 ×g at 4°C for 15 min. The supernatant was saved as the High-salt fraction (HSF, containing chromatin-bound P-TEFb).
In vitro m6A RNA binding assay
This experiment was performed following a previously published protocol with minor modifications.93 Biotinylated 7SK SLI RNA (nt 30–80) with or without m6A modifications were denatured at 95°C for 5 minutes and cooled down slowly to room temperature before use. 50 μl streptavidin magnetic beads was mixed with the RNA in binding buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% NP-40, 1 mM EDTA, 1 mM DTT, 1 μL RNase inhibitor) at 4°C for 5 hours. Then the cell lysate was mixed with streptavidin beads in a final volume of 300 μl overnight at 4°C. The beads were washed three times with washing buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% NP-40, 1 mM EDTA), before being denatured in the loading buffer for western blot analysis. 7SK SLI oligonucleotides are listed in Table S6.
ChIP-seq
The cells were crosslinked as described in RIP, except the formaldehyde concentration was 1%. Cell lysates were generated by addition of 1 mL sonication buffer (0.1% SDS, 1% Triton X-100, 10 mM Tris-HCl, 1 mM EDTA, 0.1% sodium deoxycholate, 0.25% Sarkosyl, 1 mM DTT and protease inhibitors) per 40 million cells. The DNA was sheared to an average fragment size of 100 – 300 bp using Biorupter Pico (Diagenode). The sonicated samples were centrifuged for 10 min at 12,000 ×g at 4°C. The supernatant was collected (5% saved as input) and incubated with 40 μL Protein G dynabeads that are pre-coated with 5 μg antibody/IgG, 1× protease inhibitor cocktail at 4°C overnight. The IP was washed twice with 1 mL of RIPA 0.3 buffer (0.1% SDS, 1% Triton X-100, 10 mM Tris-HCl, 1 mM EDTA, 0.1% sodium deoxycholate, 0.3 M NaCl), RIPA 0 buffer (0.1% SDS, 1% Triton X-100, 10 mM Tris-HCl, 1 mM EDTA, 0.1% sodium deoxycholate), LiCl buffer (250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl) and TE buffer (10 mM Tris-HCl, 1 mM EDTA). 300 μL SDS elution buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl) was added to the beads and incubated at 65°C for 6 hours with 1 μL Proteinase K (20 mg/mL) and 2 μL RNase A (0.6 mg/mL). DNA was purified by Phenol/Chloroform/Isoamyl alcohol (PCA) (25:24:1). The libraries were generated using the ThruPLEX Tag-seq Kit following the manufacturer’s protocol.
Data analysis for ChIP-seq
All ChIP-seq data sets (two repeats for Ser2P Pol II and three repeats for Pol II) were first processed as the MeRIP-seq data, as described above, until alignment files were sorted with Samtools sort tool. DeepTools bamCoverage tool was used to generate Bigwig files for visualization of read pileups in a genome browser with a smooth length of 60 and were normalized using reads per genomic coverage (RPGC) with an effective genome size of 2,913,022,398.94 Narrow peaks were called using MACS2 callpeak comparing the ChIP to input samples using default parameters.95 Bigwig files from replicate values were combined using the DeepTools bigwigAverage function. Evaluation of read coverage across all protein coding genes was completed using deepTools compute-matrix scale-regions with the parameters “-m 20000 -b 1000 -a 3000 –skipZeros –transcriptID gene –transcript_id_designator gene_id”.94 Read profiles and heatmaps were generated using deepTools plotProfile and plotHeatmap with the “–perGroup” parameter.94 The traveling ratio for this analysis is defined by the ratio of reads in the gene body region, as defined by 1kb downstream of the TSS to 3kb downstream of the TES, to reads from the TSS regions, as defined by 30 bp upstream of the TSS to 1kb downstream of the TSS. The coverage in the TSS and gene body regions was calculated using the DeepTools computeMatrix function, as before, but with the added –unscaled5prime 1000 condition. From this matrix, the traveling ratio was calculated using the python script traveling_ratio.py (https://github.com/UF-Xie-Lab/7SK-python-tools).
Nascent RNA-seq
Nascent RNA sequencing was performed as previously described with minor modifications.40,52 The cells were treated with 700 mM 4SU for 15 minutes at 37 °C and then the total RNA was extracted immediately via TRIzol. 50 μg total RNA was added in biotinylation reaction mixture (MTS-Biotin 0.2 mg/mL, 1 mM EDTA, 20 μM HEPES pH 7.4) and incubated at RT in the absence of light for 35 min, then the equal volume of PCA was added to each tube for RNA purification. Biotinylated RNA was captured by 25 μL Dynabeads MyOne Streptavidin C1 dynabeads according to the manufacturer’s protocol. 4SU RNA was eluted from Streptavidin C1 beads in the elution buffer (100 mM DTT, 20 mM HEPES, 1 mM EDTA, 100 mM NaCl, 0.05% (v/v) Tween-20). 8 mM fresh iodoacetamide (IAA), 50 mM NaPO4 and DMSO were mixed with 600 ng 4SU RNA in the final volume of 50 μL and incubated at 50°C for 15min. After incubation, 1 μL 1 M DTT was added to stop the reaction and RNA was precipitated with 20 μg glycogen, 300 mM NaOAc and 100% ethanol. The libraries were generated using the SMARTer Stranded Total RNA-Seq Kit v3 following the manufacturer’s protocol.
Data analysis for nascent RNA-seq
All nascent RNA-seq data sets were first processed as the MeRIP-seq data as described above through UMT removal. The reads were mapped using HISAT-3N to the human reference genome (GRCh38, release 106) with setting the “–base-change T,C”.98 The resulting alignment files were sorted and indexed using the Samtools sort and index tools.90 The percent of reads that contain a T to C mutation was calculated from HISAT-3N conversion tables using the python script conversion_rate.py (https://github.com/UF-Xie-Lab/7SK-python-tools). Alignment files were split by the number of T to C mutations found in the aligned sequence using Samtools view with the “-d Yf:#” flag option.90 deepTools bamCoverage tool was run with default settings to generate Bigwig files for visualization of read pileups in a genome browser with each alignment normalized to the total number of reads containing 0 mutations within that sample. The deepTools computeMatrix reference-point tool was run with the parameters “-b 1000 -a 3000 –skip-Zeros” and plotHeatmap tool was used to generate a heatmap of the first 10 kb downstream of the TSS.94
In vivo SHAPE modification
Aliquots of two million A549 cells per sample were pelleted. The cell pellets were resuspended to a volume of 465 μL in 13 PBS and 10 μL of SUPERase-In RNase Inhibitor. SHAPE samples were treated with 25 μL of 2 M NAI for a final concentration of 100 mM, and control cells were treated with 25 μL of anhydrous DMSO. Reactions were incubated at 37°C with rotation for 12 min. Reactions were stopped by centrifuging the cells, removing the supernatant and resuspending the samples in 1 mL of TRIzol. RNA was extracted using the DirectZol RNA Miniprep Plus kit (Zymo, R2071), including an on-column DNase treatment according to manufacturer’s instructions. Samples were eluted into 50 μL of nuclease-free water. To ensure no genomic DNA carry-over, samples were then further treated with TURBO DNase Enzyme for 30 min at 37°C. Reactions were stopped using a DNase inactivation reagent in slurry form as per manufacturer’s instructions. RNA was quantitated and stored at −80°C until use.
MaP reverse transcription and library construction
MaP RT was performed as previously described.63 1 μg total RNA was used as input for RT (25°C 12min, 42°C 3h, 70°C 15min, 10°C ∞) and the RT products were purified by 2× volume of AMPure XP beads. Libraries were constructed by two-step PCR. One-tenth of the purified RT reaction was used for PCR1 with the following program: 98°C for 30 s; 98°C for 10 s, 68°C for 20 s, 72°C for 20 s with 10 cycles; and 72°C for 2 min. PCR1 product was purified by AMPure XP beads (1× volume). 5 ng product was used for PCR2 with the following program: 98°C for 30 s; 98°C for 10 s, 65 °C for 30 s, 72°C for 20 s with 10 cycles; and 72°C for 2 min. PCR2 product was purified by AMPure XP beads (1× volume) and sequenced with an Illumina MiSeq instrument using 2 × 250 paired-end sequencing. Primers for the two-step PCR are listed in Table S7.
Data analysis for SHAPE-Map
All libraries were processed with the ShapeMapper2 (v2.1.5) pipeline using the human 7SK reference sequence based on NR_001445 using fastq files and the –amplicon flag. All comparisons were performed between NAI-treated cells (–modified) and DMSO only (–untreated) controls.
The ΔSHAPE analysis was performed using the python script deltaSHAPE.py (weekslab.com/software) with the –colorfill flag to highlight the significant differences. The structure in Figure S7G was rendered using StructureEditorv6.2 (rna.urmc.rochester.edu/GUI/html/StructureEditor.html).
All sequencing data associated with this study have been deposited in the NCBI Sequence Read Archive (SRA) database and are accessible under the accession number SRA:PRJNA938517.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical data performed using the Student’s t-test, as described in the figure legends. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 and ns, not significant.
Supplementary Material
Highlights.
7SK snRNA contains m6A modifications that are regulated by METTL3 and ALKBH5
m6A on 7SK does not always occur at the canonical “RRACH” motif
Reduced m6A-7SK in NSCLC cells suppresses proliferation and RNA Pol II transcription
Demethylated 7SK adopts a structure that sequesters P-TEFb, resulting in RNA Pol II pause
ACKNOWLEDGMENTS
We thank Drs. Qi Xie, Xin Zhou, Lizi Wu, Lingtao Jin, Erin E. Duffy, and Matthew D. Simon for reagents and protocols and UF Interdisciplinary Center for Biotechnology Research for sequencing services. This work was supported by grants from the National Institutes of Health (R35GM128753 to M.X., R01CA246418 to J.B., and P50NS048843 to M.S.S.), the American Cancer Society (Research Scholar Award RSG-21-118-01-RMC to M.X.), and the Florida Department of Health (Live Like Bella Pediatric Cancer Initiative 21L03 to M.X.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2023.09.020.
REFERENCE
- 1.Sainsbury S, Bernecky C, and Cramer P (2015). Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143. [DOI] [PubMed] [Google Scholar]
- 2.Roeder RG (2005). Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579, 909–915. [DOI] [PubMed] [Google Scholar]
- 3.Hochheimer A, and Tjian R (2003). Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17, 1309–1320. [DOI] [PubMed] [Google Scholar]
- 4.Guo J, and Price DH (2013). RNA polymerase II transcription elongation control. Chem. Rev. 113, 8583–8603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonkers I, and Lis JT (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adelman K, and Lis JT (2012). Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall NF, and Price DH (1995). Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270, 12335–12338. [DOI] [PubMed] [Google Scholar]
- 8.Cheng B, and Price DH (2007). Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J. Biol. Chem. 282, 21901–21912. [DOI] [PubMed] [Google Scholar]
- 9.Yu F, Shi G, Cheng S, Chen J, Wu S-Y, Wang Z, Xia N, Zhai Y, Wang Z, and Peng Y (2018). SUMO suppresses and MYC amplifies transcription globally by regulating CDK9 SUMOylation. Cell Res. 1, 670–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen VT, Kiss T, Michels AA, and Bensaude O (2001). 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414, 322–325. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z, Zhu Q, Luo K, and Zhou Q (2001). The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414, 317–322. [DOI] [PubMed] [Google Scholar]
- 12.Peterlin BM, Brogie JE, and Price DH (2012). 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 3, 92–103. 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C,Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. (2007). Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27, 262–274. 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, and Zhou Q (2008). A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell 29, 588–599. 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. (2008). LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36, 2219–2229. 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michels AA, Nguyen VT, Fraldi A, Labas V, Edwards M, Bonnet F, Lania L, and Bensaude O (2003). MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23, 4859–4869. 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, and Zhou Q (2003). Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12, 971–982. 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 18.Van Herreweghe E, Egloff S, Goiffon I, Jády BE, Froment C, Monsarrat B, and Kiss TJTEJ (2007). Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 26, 3570–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hogg JR, and Collins K (2007). RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 13, 868–880. 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrandon C, Bonnet F, Nguyen VT, Labas V, and Bensaude O (2007). The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 27, 6996–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leger A, Amaral PP, Pandolfini L, Capitanchik C, Capraro F, Miano V, Migliori V, Toolan-Kerr P, Sideri T, Enright AJ, et al. (2021). RNA modifications detection by comparative nanopore direct RNA sequencing. Nat. Commun. 12, 7198. 10.1038/s41467-021-27393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, Zhao S, Shen B, Gao Y, Han D, et al. (2020). N 6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, and Deng X (2014). A METTL3–METTL14 complex mediates mammalian nuclear RNA N 6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ping X-L, Sun B-F, Wang L, Xiao W, Yang X, Wang W-J, Adhikari S, Shi Y, Lv Y, and Chen Y-S (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, and Jaffrey SR (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, and Hess D (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl (2). Genes Dev. 32, 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, and Yang Y-G (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, Vågbø CB, Shi Y, Wang W-L, and Song S-H (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteller M, and Pandolfi PP (2017). The epitranscriptome of noncoding RNAs in cancer. Cancer Discov. 7, 359–368. 10.1158/2159-8290.CD-16-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, and Kupiec M (2012). Topology of the human and mouse m 6 A RNA methylomes revealed by m 6 A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- 31.Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, and Min J (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 10, 927–929. 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, and Tavazoie SF (2015). HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162, 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, and Ma J (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, and Jia G (2014). N 6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu N, Dai Q, Zheng G, He C, Parisien M, and Pan T (2015). N 6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 518, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, and Liu T (2019). The critical role of RNA m6A methylation in cancer. Cancer Res. 79, 1285–1292. [DOI] [PubMed] [Google Scholar]
- 38.Huang H, Weng H, and Chen J (2020). m6A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell 37, 270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei J, Yu X, Yang L, Liu X, Gao B, Huang B, Dou X, Liu J, Zou Z, and Cui X-L (2022). FTO mediates LINE1 m6A demethylation and chromatin regulation in mESCs and mouse development. Science 376, 968–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT, Moroz-Omori EV, Shao J, Zhu X, Bolt MJ, et al. (2021). Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 81, 3368–3385.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akhtar J, Renaud Y, Albrecht S, Ghavi-Helm Y, Roignant JY, Silies M, and Junion G (2021). m(6)A RNA methylation regulates promoterproximal pausing of RNA polymerase II. Mol. Cell 81, 3356–3367.e6. 10.1016/j.molcel.2021.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Lin S, Choe J, Du P, Triboulet R, and Gregory RI (2016). The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia Z, Tang M, Ma J, Zhang H, Gimple RC, Prager BC, Tang H, Sun C, Liu F, Lin P, et al. (2021). Epitranscriptomic editing of the RNA N6-methyladenosine modification by dCasRx conjugated methyltransferase and demethylase. Nucleic Acids Res. 49, 7361–7374. 10.1093/nar/gkab517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, and Conrad NK (2017). The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur J-J, and Chen Q (2017). Reversible methylation of m 6 A m in the 5′ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Y, Wang Y, Tang Q, Wei L, Zhang X, and Jia G (2018). An elongation-and ligation-based qPCR amplification method for the radiolabeling-free detection of locus-Specific N6-methyladenosine modification. Angew. Chem. Int. Ed. Engl. 57, 15995–16000. [DOI] [PubMed] [Google Scholar]
- 48.Lebars I, Martinez-Zapien D, Durand A, Coutant J, Kieffer B, and Dock-Bregeon AC (2010). HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 38, 7749–7763. 10.1093/nar/gkq660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muniz L, Egloff S, Ughy B, Jády BE, and Kiss T (2010). Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 6, e1001152. 10.1371/journal.ppat.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, and Tang CJN (2016). Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature 534, 575–578. [DOI] [PubMed] [Google Scholar]
- 51.Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, and Simon MD (2015). Tracking distinct RNA populations using efficient and reversible covalent chemistry. Mol. Cell 59, 858–866. 10.1016/j.molcel.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Herzog VA, Reichholf B, Neumann T, Rescheneder P, Bhat P, Burkard TR, Wlotzka W, von Haeseler A, Zuber J, and Ameres SL (2017). Thiol-linked alkylation of RNA to assess expression dynamics. Nat. Methods 14, 1198–1204. 10.1038/nmeth.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C Quaresma AJ, Bugai A, and Barboric M (2016). Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb. Nucleic Acids Res. 44, 7527–7539. 10.1093/nar/gkw585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou K, Zhuang S, Liu F, Chen Y, Li Y, Wang S, Li Y, Wen H, Lin X, Wang J, et al. (2022). Disrupting the Cdk9/cyclin T1 heterodimer of 7SK snRNP for the Brd4 and AFF1/4 guided reconstitution of active P-TEFb. Nucleic Acids Res. 50, 750–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Contreras X, Barboric M, Lenasi T, and Peterlin BM (2007). HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3, 1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Egloff S, Studniarek C, and Kiss T (2018). 7SK small nuclear RNA, a multifunctional transcriptional regulatory RNA with gene-specific features. Transcription 9, 95–101. 10.1080/21541264.2017.1344346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Y, Liu S, Egloff S, Eichhorn CD, Hadjian T, Zhen J, Kiss T, Zhou ZH, and Feigon J (2022). Structural basis of RNA conformational switching in the transcriptional regulator 7SK RNP. Mol. Cell 82, 1724–1736.e7. 10.1016/j.molcel.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olson SW, Turner AW, Arney JW, Saleem I, Weidmann CA, Margolis DM, Weeks KM, and Mustoe AM (2022). Discovery of a large-scale, cell-state-responsive allosteric switch in the 7SK RNA using DANCE-MaP. Mol. Cell 82, 1708–1723.e10. 10.1016/j.molcel.2022.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krueger BJ, Varzavand K, Cooper JJ, and Price DH (2010). Themechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One 5, e12335. 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Flynn RA, Do BT, Rubin AJ, Calo E, Lee B, Kuchelmeister H, Rale M, Chu C, Kool ET, Wysocka J, et al. (2016). 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 23, 231–238. 10.1038/nsmb.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brogie JE, and Price DH (2017). Reconstitution of a functional 7SKsnRNP. Nucleic Acids Res. 45, 6864–6880. 10.1093/nar/gkx262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siegfried NA, Busan S, Rice GM, Nelson JA, and Weeks KM (2014). RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965. 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smola MJ, Rice GM, Busan S, Siegfried NA, and Weeks K.M.J.N.p. (2015). Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat. Protoc. 10, 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, and Chang HY (2013). RNA SHAPE analysis in living cells. Nat. Chem. Biol. 9, 18–20. 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Busan S, and Weeks KM (2018). Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148. 10.1261/rna.061945.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, and Bensaude O (2009). Evolution of 7SK RNA and its protein partners in metazoa. Mol. Biol. Evol. 26, 2821–2830. 10.1093/molbev/msp198. [DOI] [PubMed] [Google Scholar]
- 67.Bubenik JL, Hale M, McConnell O, Wang ET, Swanson MS, Spitale RC, and Berglund JAJR (2020). RNA structure probing to characterize RNA–protein interactions on low abundance pre-mRNA in living cells. RNA 27, 343–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smola MJ, Calabrese JM, and Weeks KMJB (2015). Detection of RNA–protein interactions in living cells with SHAPE. Biochemistry 54, 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wassarman DA, and Steitz JA (1991). Structural-Analyses of the 7sk Ribonucleoprotein (Rnp), the Most Abundant Human Small Rnp of Unknown Function. Mol. Cell. Biol. 11, 3432–3445. 10.1128/mcb.11.7.3432-3445.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, and Manor YS (2015). m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006. [DOI] [PubMed] [Google Scholar]
- 71.Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, and Jaffrey SR (2015). 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, and Qian S-B (2015). Dynamic m 6 A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xiang Y, Laurent B, Hsu C-H, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, and Wang S (2017). RNA m 6 A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perez-Pepe M, Desotell AW, Li H, Li W, Han B, Lin Q, Klein DE, Liu Y, Goodarzi H, and Alarcón CR (2023). 7SK methylation by METTL3 promotes transcriptional activity. Sci. Adv. 9, eade7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu X, Ma L, Zhang X, Guo S, Guo W, Wang Y, Qiu S, Tian X, Miao Y, and Yu Y (2023). A positive feedback circuit between RN7SK snRNA and m6A readers is essential for tumorigenesis. Mol. Ther. 31, 1615–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, Sloan KE, and Bohnsack MT (2017). Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zaccara S, and Jaffrey SR (2020). A Unified Model for the function of YTHDF proteins in regulating m6A-modified mRNA. Cell 181, 1582–1595.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, Hanna JH, Black DL, Darnell JE, and Darnell RB (2017). m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 31, 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu B, Shi H, Rangadurai A, Nussbaumer F, Chu C-C, Erharter KA, Case DA, Kreutz C, and Al-Hashimi H.M.J.N.c. (2021). A quantitative model predicts how m6A reshapes the kinetic landscape of nucleic acid hybridization and conformational transitions. Nat. Commun. 12, 5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Patil DP, Pickering BF, and Jaffrey SR (2018). Reading m6A in the transcriptome: m6A-binding proteins. Trends Cell Biol. 28, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu W, He C, Kaye EG, Li J, Mu M, Nelson GM, Dong L, Wang J, Wu F, Shi YG, et al. (2022). Dynamic control of chromatin-associated m(6)A methylation regulates nascent RNA synthesis. Mol. Cell 82, 1156–1168.e7. 10.1016/j.molcel.2022.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. (2004). EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500. 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 83.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, and Sliz P (2018). mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J, Chen Z, Chen F, Xie G, Ling Y, Peng Y, Lin Y, Luo N, Chiang CM, and Wang H (2020). Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res. 48, 5684–5694. 10.1093/nar/gkaa269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Egloff S, Vitali P, Tellier M, Raffel R, Murphy S, and Kiss T (2017). The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. EMBO J. 36, 934–948. 10.15252/embj.201695740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J, Kobert K, Flouri T, and Stamatakis A (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martin M.J.E.j. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- 89.Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anders S, Pyl PT, and Huber W.J.b. (2015). HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Inc. PT (2015). Collaborative Data Science (Plotly Technologies Inc.). [Google Scholar]
- 93.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, and Yuan CL (2018). Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke TJNa.r. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller BR, Wei T, Fields CJ, Sheng P, and Xie M (2018). Near-infrared fluorescent northern blot. RNA 24, 1871–1877. 10.1261/rna.068213.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jao CY, and Salic A (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA 105, 15779–15784. 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Park C, Bennett C, Thornton M, and Kim D (2021). Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 31, 1290–1295. 10.1101/gr.275193.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the sequencing data generated by this study have been deposited to NCBI Sequence Read Archive (SRA) database under SRA accession number PRJNA938517
All original code has been deposited at https://github.com/UF-Xie-Lab/7SK-python-tools, which is also available through: https://doi.org/10.5281/zenodo.8341420
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


