Abstract
There is growing evidence that a number of oral Treponema species, in particular Treponema denticola, are associated with the progression of human periodontal disease. The major sheath (or surface) protein (Msp) of T. denticola is implicated in adhesion of bacteria to host cells and tissue proteins and is likely to be an important virulence factor. However, the binding regions of the Msp are not known. We have purified from Escherichia coli recombinant Msp (rMsp) polypeptides corresponding to the following: full-length Msp (rMsp) minus 13 N-terminal amino acid (aa) residues, an amino-terminal fragment (rN-Msp, 189 aa residues), a 57-aa residue segment from the central region (rV-Msp), and a C-terminal fragment (rC-Msp, 272 aa residues). rMsp (530 aa residues) bound to immobilized fibronectin, keratin, laminin, collagen type I, fibrinogen, hyaluronic acid, and heparin. The N- and V-region polypeptides, but not rC-Msp, also bound to these substrates. Binding of rMsp to fibronectin was targeted to the N-terminal heparin I/fibrin I domain. Antibodies to the N-region or V-region polypeptides, but not antibodies to the rC-Msp fragment, blocked adhesion of T. denticola ATCC 35405 cells to a range of host protein molecules. These results suggest that the N-terminal half of Msp carries epitopes that are surface exposed and that are involved in mediating adhesion. Binding of rMsp onto the cell surface of low-level fibronectin-binding Treponema isolates conferred a 10-fold increase in fibronectin binding. This confirms that Msp functions autonomously as an adhesin and raises the possibility that phenotypic complementation of virulence functions might occur within mixed populations of Treponema species.
Human periodontal disease is a chronic inflammatory condition of the gums and gingival tissues that can lead to bone resorption and tooth loss (30). The disease is associated with polymicrobial infections containing principally anaerobic gram-negative bacteria (38), and spirochetes of the genus Treponema are implicated in disease progression (36). Recently it has become apparent that ulcerative conditions of the feet or hooves of ruminants also contain complex microbial communities within which are present Treponema species closely related to human oral Treponema (11, 12). Interactions of these organisms with host tissues involve adhesion to epithelial or endothelial cells and to extracellular matrix components, penetration of tissue layers by chemotaxis, and tissue destruction through a combination of proteolysis and direct cytopathic effects (11, 36).
There are numerous designated species of oral spirochetes, including Treponema denticola, Treponema vincentii, Treponema pectinovorum, Treponema socranskii, and Treponema maltophilum (11). Detailed studies of subgingival plaque bacteria have identified 49 novel Treponema phylotypes, organisms that have yet to be cultivated (9, 32), suggesting high diversity within oral spirochete populations. However, T. denticola is perhaps the best characterized of oral species and is frequently found in association with Porphyromonas gingivalis and Tannerella forsythensis at diseased sites (22).
T. denticola adheres avidly to human epithelial cells, migrates through epithelial tissues, and binds a wide range of host tissue proteins. The bacteria express a number of putative virulence factors, including a chymotrypsin-like protease (CTLP [or dentilisin]) (16, 27), a trypsin-like protease (OpdB) (17), cystalysin (6), hyaluronidase (35), and a phospholipase C (37). An important adhesin and virulence factor is the major sheath protein (Msp) that forms an oligomeric cell surface complex with CTLP (33). Msp is a dominant antigen associated with the cell outer layers and is believed to be responsible for mediating many of the adhesive properties and cytopathic effects of T. denticola (16, 31).
Msps from different strains of T. denticola range in molecular mass between 53 kDa and 62 kDa (18, 19, 20, 24) and have been shown to bind fibronectin, fibrinogen, and laminin (18, 20), as well as collagen (39, 40). The msp gene from T. denticola ATCC 35405 has been expressed in Escherichia coli, and purified recombinant Msp binds fibronectin (18) and causes cytopathic effects on epithelial cells (16). The cytotoxicity is due to, at least in part, the ability of Msp to form ion channels within lipid bilayers, leading to depolarization and cell volume dysregulation (31). Msp also disrupts calcium signaling in fibroblasts (41) and induces release of MMP-8, MMP-9, cathepsin G, and elastase from neutrophils (10). Such a plethora of properties related to bacterial virulence underscores the importance of Msp in T. denticola-associated disease pathogenesis.
Despite the crucial role played by Msp in a wide range of interactions of T. denticola with host cells and tissue proteins, it is not known which regions of Msp are important for Treponema-host interactions. There is also some dispute as to whether or not Msp is exposed on the Treponema cell surface. Although electron microscopic studies have indicated that Msp contributes to the regular hexagonal arrays visible in the outer sheath of T. denticola (13), it has recently been proposed that Msp may be localized predominantly within the periplasmic region (4). In this article, we demonstrate that sequences within the N-terminal half of the 543-amino-acid (aa) residue Msp are cell surface exposed in T. denticola ATCC 35405 and mediate adhesion to a range of host protein molecules. In addition, we show that recombinant Msp binds onto the cells of heterologous Treponema strains, promoting their adhesion to fibronectin and thus confirming the ability of Msp to function autonomously as an adhesin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The Treponema strains used in this study were T. denticola ATCC 35405 and GM-1 (from R. J. Lamont, University of Florida, Gainesville); T. vincentii ATCC 35580 and D2A-2 (from P. E. Greenberg, University of Iowa); and Treponema sp. strain UB1090 isolated from a sheep with contagious ovine digital dermatitis (7, 12). Treponemes were maintained in New Oral Spirochaete (NOS) medium (21) in an anaerobic atmosphere of N2-CO2-H2 (8:1:1). Late-exponential-phase cultures, corresponding to an optical density at 600 nm (OD600) of between 0.4 and 0.6, were obtained following incubation for 3 to 4 days at 37°C. Escherichia coli XL-1 Blue was used as a host for preparing plasmid DNA for constructing expression vectors. For production of recombinant polypeptides, pQE30 plasmids (QIAGEN GmbH, Hilden) were maintained in E. coli M15 cultured on LB agar (34) or in LB broth containing appropriate antibiotics (ampicillin, 100 μg/ml; kanamycin, 25 μg/ml).
Expression and purification of recombinant Msp and Msp fragments.
DNA corresponding to the complete Msp coding sequence, minus a 13-aa-residue segment of the 20-aa-residue N-terminal leader peptide sequence that is otherwise toxic to E. coli (see also reference 18), was amplified by PCR with primers Ntermfor (5′CGGGATCCGTGCTCGTGGGCGGA) and Ctermrev (5′CGACGTCGACGGTAGATAACTTTAACACCGAT). Primer pairs utilized to PCR amplify the coding regions for recombinant Msp (rMsp) fragments (see Fig. 1) were as follows: rN-Msp, Ntermfor and Ntermrev (5′ AGTGGTACCCTTAGCTTTCCATG); rV-Msp, Vregfor (5′CGGATCCGCTCAAGGATCAACAGCT) and Vregrev (5′ GTCGACTTTCCGTCTTCACCAGCACCT); rC-Msp, Ctermfor (5′CGGATCCGCAGCAAACAAATATGC) and Ctermrev (as above). Each forward primer contained a BamHI site (underlined), and each reverse primer a SalI or KpnI site (underlined) in order that the correct in-frame fusion was obtained following cloning into pQE30. PCR products were initially cloned into pGEM-T (Promega Corp., Madison, Wis.) and maintained in E. coli XL-1 Blue. Plasmids were recovered using the GibcoBRL Concert Rapid Plasmid Miniprep system (Life Technologies Ltd., Paisley, United Kingdom) and were digested with a combination of BamHI and SalI or KpnI. Digested PCR products were gel purified using a QIAGEN gel extraction kit and ligated into similarly digested His6 tag vector pQE30 (QIAGEN). Ligation mixes were transformed into E. coli XL-1 Blue, recombinant plasmids were screened by restriction enzyme digest analysis, and inserts were sequenced to confirm authenticity. Suitable constructs were then transformed into the expression host E. coli M15 (QIAGEN). The reason that, in addition to the His6 tag, the rMsp and rN-Msp polypeptides each contained 7 aa residues from the putative Msp leader peptide was because of difficulties first encountered with in-frame cloning of msp closer to the leader peptide cleavage site. Recombinant His6-tagged proteins were expressed and purified according to the manufacturer's recommendations with amendments as follows. Briefly, E. coli M15 containing a plasmid construct was incubated with shaking in 100 ml LB medium containing ampicillin (100 μg/ml) and kanamycin (25 μg/ml) to OD600 = 0.5 to 0.7. Prewarmed LB medium (100 ml) containing 2 mM isopropyl-β-d-thiogalactopyranoside was then added, and the culture was incubated for a further 3 to 4 h at 37°C. Bacteria were harvested by centrifugation (10,000 × g for 10 min), and proteins were solubilized in 8 M urea containing 2% (vol/vol) Tween 20 (30 ml). Cellular debris was removed by centrifugation (15,000 × g for 15 min), and the supernatant containing His6-tagged protein was incubated with nickel-nitrilotriacetic acid resin (QIAGEN) with gentle agitation for 20 min at 20°C. Recombinant proteins were purified by column elution, dialyzed against phosphate-buffered saline (PBS; 0.01 M phosphate, 2.7 mM KCl, 0.137 M NaCl, pH 7.4) containing 0.4 mM phenylmethylsulfonyl fluoride to remove urea and then against water, and freeze-dried.
FIG. 1.
Diagrammatic representation of the Msp sequences from T. denticola ATCC 35520 (A), strain ATCC 35405 (B), and of recombinant Msp polypeptides rMsp (530 aa residues), rN-Msp (189 aa residues), rV-Msp (57 aa residues), and rN-Msp (272 aa residues), with N-terminal His6 tags derived from the msp sequence of strain ATCC 35405 (C). Designated regions of Msp, based on amino acid sequence conservation between ATCC 35520 (GenBank accession no. U66255) and ATCC 35405 (no. U29399) were as follows: LP, leader peptide (20 aa residues, 100% identical aa sequences); N, amino-terminal region (100% identity); V, variable region (32% identity); and C, carboxy-terminal region (99.6% identity [one aa residue change]).
Protein extraction, electrophoresis, and immunoblot detection.
Treponema surface proteins were extracted into Triton X-114 (16), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and electroblotted onto nitrocellulose membrane as previously described (12). Blots were incubated with primary antibodies, diluted as appropriate, and binding was detected with horseradish peroxidase (HRP)-conjugated secondary antibody in conjunction with ECL detection (Amersham Biosciences, Little Chalfont, United Kingdom). Antibodies to rMsp and rMsp fragments were raised in rabbits, which had been pre-bled and screened negative for antibodies reactive with T. denticola, following subcutaneous inoculations of proteins (0.1 mg) at two-weekly intervals.
ELISA.
The reactivity of antiserum with T. denticola ATCC 35405 cells was determined by enzyme-linked immunosorbent assay (ELISA). Portions of PBS-washed treponemes (50 μl, OD600 = 0.25) were immobilized onto Immulon 2HB plates (Dynex Technologies, Ashford, United Kingdom) in the presence of 0.25% (vol/vol) glutaraldehyde as described elsewhere (25). Wells were blocked with 1% (wt/vol) bovine serum albumin (BSA) incubation with primary antiserum for 1 h at 37°C, followed by three washes with PBS containing 0.1% (vol/vol) Tween 20 and 0.1% (wt/vol) BSA (PBSTB buffer). Antibody binding was detected with HRP-conjugated anti-rabbit immunoglobulin G (IgG; diluted 1:2,000 in PBSTB) and o-phenylenediamine (25).
Blot overlay assay.
Trypsin fragments of human plasma fibronectin were generated and subjected to SDS-PAGE as described previously (12), transferred onto nitrocellulose by electroblotting, and overlaid with either biotinylated Treponema cells (12) for 1 h at 20°C or purified rMsp (0.5 μg/ml in PBS) for 2 h at 20°C. Bound cells were detected with HRP-conjugated streptavidin (diluted 1:2,000), while bound rMsp was detected with antibodies to tetrahistidine (His4) (QIAGEN; diluted 1:2000) followed by HRP-conjugated anti-mouse IgG and ECL reagent.
Bacterial adhesion assays.
Late-exponential-phase Treponema cells were biotinylated as previously described (12). Proteins, including human plasma fibronectin (Roche Diagnostics Ltd., Lewes, United Kingdom), the 30-kDa heparin binding fragment of fibronectin (Sigma), human serum albumin (Sigma), or BSA, were immobilized onto the surface of Immulon 2HB 96-well plastic plates (Dynex Technologies; 0.1 μg/well) in carbonate buffer (0.02 M NaHCO3, 0.02 M Na2CO3, pH 9.3) for 16 h at 4°C, and the remaining protein binding sites were blocked with 1% (wt/vol) BSA in PBS for 1 h at 22°C. Biotinylated Treponema cell suspensions containing between 1 × 107 and 2 × 108 cells were applied in triplicate wells and incubated for 2 h at 20°C. Numbers of bacteria bound were calculated from absorbance values at 490 nm (A490) following detection with HRP-linked streptavidin, as previously described (12).
To determine the effects of rMsp on bacterial cell adhesion to immobilized fibronectin, rMsp (0 to 4 μg) was added to wells in 50 μl PBS and incubated for 30 min at 37°C prior to the addition of biotinylated cells (2.4 × 107) and measuring numbers of cells bound as described above. To test inhibition of bacterial adhesion by antibodies, biotinylated T. denticola cells were incubated with rMsp antisera, or preimmune sera, diluted in the range 1:10 to 1:200 in PBS for 1 h at 37°C. Cells were then washed twice by alternate centrifugation and suspension in PBS and assayed for adhesion as before.
Msp adhesion assays.
Recombinant Msp polypeptides (2.5 μg) in PBS were added to BSA-blocked wells containing 0.1 μg immobilized protein and incubated for 1 h at 37°C. Wells were washed twice with PBS, His4 antiserum (diluted 1:1,000 in PBSTB) was added, and plates were incubated for 1 h at 37°C. Bound antibodies were detected with HRP-conjugated anti-mouse IgG and o-phenylenediamine, and A490 values converted to equivalent μg rMsp from a standard plot constructed for each rMsp preparation. In control experiments, we could detect no binding of an irrelevant recombinant protein (PMA1 from yeast), carrying N-terminal His6 tag, to immobilized fibronectin, showing that the His6 tag does not bind fibronectin.
Statistical analyses.
Statistical significance was determined using Student's t test on paired samples.
RESULTS
Binding of Msp to fibronectin.
To investigate in detail the binding properties of Msp, we cloned and expressed various coding regions of the T. denticola ATCC 35405 msp gene in E. coli. Our initial studies focused on the recombinant rMsp, which comprised full-length Msp (543 aa residues) minus the N-terminal 13 aa residues (Fig. 1). It was found necessary to remove this portion of the coding region in order for the cloned msp gene to be tolerated by E. coli (see also reference 18). Previous studies have demonstrated that Msp binds fibronectin (18), but it is not known which regions of fibronectin are bound. Accordingly, we separated tryptic-digest fragments of fibronectin by SDS-PAGE, blotted them onto nitrocellulose, and determined to which of these fragments rMsp bound. The results in Fig. 2A show that rMsp bound exclusively to the 30-kDa N-terminal fragment of fibronectin, which contains heparin and fibrin binding sequences. This is the identical tryptic digest fragment of fibronectin to which T. denticola ATCC 35405 cells bound (Fig. 2A, lane 2) (12). In quantitative assays, rMsp bound to plastic wells coated with fibronectin, up to a maximum of approximately 1 μg protein bound to ∼0.1 μg fibronectin, and to wells coated with the 30-kDa fibronectin fragment (Fig. 2B), but not to wells coated with BSA (Fig. 2B). Binding of rMsp (2.5 μg) to immobilized fibronectin was 50% inhibited by preincubation of fibronectin with heparin (10 μg), but not with gelatin (results not shown). Binding of rMsp to surface-immobilized fibronectin was unaffected by the addition of 0.25 mM Arg-Gly-Asp-Ser (RGDS) peptide, which corresponds to the integrin recognition motif within the cell binding domain of fibronectin (29). Preincubation of rMsp (5 μg) with fluid-phase fibronectin (10 μg) or the 30-kDa fragment did not affect subsequent binding levels to immobilized fibronectin (data not shown). These results suggested that Msp had a higher affinity for immobilized fibronectin than for fluid-phase fibronectin and that the N-terminal heparin I/fibrin I binding region of fibronectin was preferentially recognized.
FIG. 2.
Binding of rMsp to human fibronectin and to 30-kDa N-terminal fragment of human fibronectin. (Panel A) Western blot overlay of trypsin-derived fragments of human plasma fibronectin reacted with rMsp (lane 1) or with biotinylated T. denticola ATCC 35405 cells (lane 2). Bound rMsp was detected with antibodies to tetrahistidine and HRP-conjugated secondary antibody, while T. denticola cells were detected with HRP-conjugated streptavidin. The pattern of Coomassie blue-stained fibronectin fragments and their associated binding regions is shown in lane 3 (23). Molecular mass markers are indicated (kDa). (Panel B) Binding of rMsp to plastic microtiter plate wells coated with 0.1 μg human fibronectin (filled circles), 30-kDa fragment (open circles), or BSA (open triangles). Binding was determined as described in Materials and Methods. Error bars indicate ± standard deviation of triplicates from three individual experiments.
rMsp inhibits T. denticola cell adhesion to fibronectin.
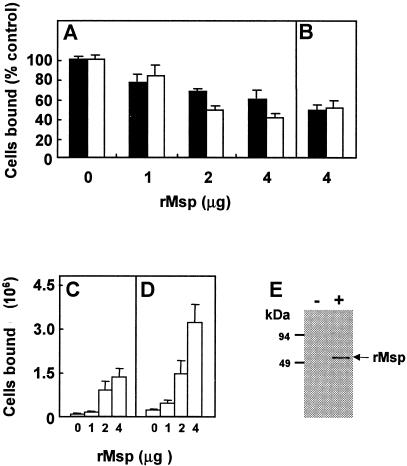
Since rMsp and T. denticola cells both bound to the 30-kDa fragment of fibronectin, we tested the ability of rMsp to inhibit cell adhesion to fibronectin. As anticipated, rMsp was found to be an effective inhibitor of T. denticola ATCC 35405 cell adhesion to both fibronectin and to the 30-kDa tryptic fragment. Inhibition by rMsp was dose dependent up to a maximum of 40% for binding to fibronectin and to 60% inhibition of cell binding to the 30-kDa fragment (Fig. 3A). rMsp from T. denticola ATCC 35405 was also effective in inhibiting adhesion of T. denticola GM-1 cells to immobilized fibronectin and the 30-kDa fragment (Fig. 3B).
FIG. 3.
Effect of exogenously added rMsp on adhesion of Treponema strains to immobilized human fibronectin or 30-kDa fibronectin fragment. (Panel A) Adhesion levels of T. denticola ATCC 35405 cells to 0.1 μg fibronectin (filled column) or 30-kDa fibronectin fragment (open column) preincubated with 0 to 4 μg rMsp. (Panel B) Adhesion levels of T. denticola GM-1 cells to fibronectin or 30-kDa fragment preincubated with 4 μg rMsp (relative to T. denticola GM-1 control). (Panels C and D) Adhesion of T. vincentii ATCC 35580 cells (C) or ovine Treponema strain UB1090 cells (D) to fibronectin preincubated with 0 to 4 μg rMsp. (Panel E) Western immunoblot of T. vincentii ATCC 35580 outer membrane proteins extracted from 1.2 × 108 control (untreated) cells (−) or cells incubated with 4 μg purified rMsp (+) and reacted with antibodies raised to rV-Msp from T. denticola ATCC 35405. Error bars are ± standard deviation of the mean of three experiments performed in triplicate.
rMsp promotes heterologous Treponema adhesion to fibronectin.
T. vincentii ATCC 35580 and a closely-related ovine isolate, Treponema strain UB1090, both adhere to fibronectin but show considerably lower binding levels than does T. denticola (12). In addition, unlike T. denticola, neither of these strains expresses CTLP. We therefore tested the effects of adding exogenous rMsp on adhesion of these heterologous Treponema species to fibronectin. Unexpectedly, we found that binding levels of both T. vincentii and Treponema strain UB1090 cells to fibronectin were enhanced by rMsp and that this was dose dependent (Fig. 3C and 3D). An approximately 10-fold increase in numbers of Treponema cells bound, for both strains, was observed at the maximum rMsp input (4 μg) (Fig. 3C and 3D). Adhesion levels of T. vincentii D2A-2 were also similarly increased in the presence of rMsp (data not shown). We considered the most likely explanation for these results was that rMsp acted as a bridging adhesin by becoming bound to the cell surface of the heterologous strains. To test this, T. vincentii ATCC 35580 cells (1.2 × 108) were incubated with rMsp (4 μg) for 2 h at 20°C and washed extensively. Outer membrane proteins were then solubilized from rMsp-treated or control (untreated) cells with Triton X-114, and Western blots were reacted with polyclonal antibodies to a central region fragment (rV-Msp) of Msp (Fig. 1), that do not react with T. vincentii proteins. The results in Fig. 3E demonstrate that exogenously added Msp associated tightly with the outer surface layers of heterologous cells of T. vincentii. These Msp-treated cells showed 10-fold-increased binding levels to immobilized fibronectin (as seen in Fig. 3C) and to the immobilized 30-kDa fibronectin fragment (not shown).
Binding regions of Msp to fibronectin and other host proteins.
The msp gene of T. denticola ATCC 35405 may be divided into three coding regions on the basis of sequence conservation with msp of T. denticola ATCC 33520 (Fig. 1). These comprise an N-terminal region of 609 bp; a central, sequence variable (V) region of approximately 200 bp; and a C-terminal region of about 800 bp (Fig. 1). To determine if these regions encoded Msp fragments with substrate binding properties, recombinant polypeptides corresponding to sequences within the N-terminal (rN-Msp, 14 to 202 aa residues), variable (rV-Msp, 203 to 259 aa residues), and C-terminal (rC-Msp, 272 to 543 aa residues) regions were purified. We were unable to express a stable recombinant polypeptide comprising the entire V region (203 to 271 aa residues). rMsp and rC-Msp migrated on SDS-PAGE according to their predicted molecular masses of 53 kDa and 31.7 kDa, respectively (Fig. 4A, lanes 1 and 4). However, rN-Msp and rV-Msp bands (predicted molecular masses, 22.9 kDa and 8.1 kDa, respectively) migrated more slowly than predicted in SDS-PAGE, with apparent molecular masses of 26 kDa and 12 kDa (Fig. 4A).
FIG. 4.
Recombinant Msp derivatives and rMsp antiserum specificity. (Panel A) SDS-PAGE gel of purified rMsp and rMsp fragments stained with Coomassie blue. Lane 1, rMsp; lane 2, rN-Msp; lane 3, rV-Msp; lane 4, rC-Msp (see Fig. 1 for details). (Panel B) Corresponding Western immunoblot of rMsp (lanes 1, 3, and 5), rN-Msp (lane 2), rV-Msp (lane 4), and rC-Msp (lane 6) reacted with antisera to rMsp fragments as shown. Molecular mass markers are indicated (kDa).
The binding properties of rMsp and rMsp fragments to a range of purified host tissue molecules are shown in Table 1. rMsp bound to immobilized fibronectin, 30-kDa fibronectin fragment, laminin, keratin, collagen I, and fibrinogen. rMsp also bound to heparin and hyaluronic acid, but at only 15 to 18% of the binding to fibronectin, and did not bind to BSA (Table 1) or to gelatin, fetuin, and human serum albumin (data not shown). The substrate binding profile of rV-Msp was similar to that of rMsp, with highest binding observed to keratin, fibronectin, and the 30-kDa fibronectin fragment (Table 1). The rN-Msp fragment also bound to fibronectin, the 30-kDa fragment, keratin, and fibrinogen, although at lower levels than rMsp. On the other hand, the rC-Msp fragment did not exhibit significant levels of binding to any of the substrates tested (Table 1). These data suggested that the N-terminal half of Msp, and in particular the V region, determined the binding properties of Msp.
TABLE 1.
Binding of rMsp or rMsp fragments to immobilized host moleculesa
| Substrate | rMsp (μg) bound to substrate ± SDb
|
|||
|---|---|---|---|---|
| rMsp | rN-Msp | rV-Msp | rC-Msp | |
| Fibronectin | 0.20 ± 0.02 | 0.05 ± 0.01 | 0.23 ± 0.01 | 0.02 ± 0.01 |
| 30-kDa fragment | 0.22 ± 0.02 | 0.10 ± 0.01 | 0.50 ± 0.02 | <0.01 |
| Laminin | 0.16 ± 0.01 | 0.03 ± 0.01 | 0.16 ± 0.01 | 0.03 ± 0.01 |
| Keratin | 0.23 ± 0.02 | 0.08 ± 0.02 | 0.57 ± 0.01 | 0.02 ± 0.01 |
| Collagen | 0.16 ± 0.01 | 0.02 ± 0 | 0.07 ± 0.01 | 0.02 ± 0 |
| Fibrinogen | 0.08 ± 0.01 | 0.05 ± 0 | 0.12 ± 0 | <0.01 |
| Hyaluronic acid | 0.04 ± 0.01 | 0.02 ± 0 | 0.05 ± 0.01 | <0.01 |
| Heparin | 0.03 ± 0 | 0.03 ± 0 | 0.05 ± 0.01 | <0.01 |
| Bovine serum albumin | <0.01 | <0.01 | <0.01 | <0.01 |
His6-tagged rMsp fragments (2.5 μg) were applied to plastic wells coated with host molecule substrate (0.1 μg), and amounts of rMsp bound were determined by ELISA with tetra-His antibody (see Materials and Methods).
Means of three experiments performed in triplicate.
Antibodies to rMsp inhibit T. denticola cell adhesion.
Antibodies to rN-Msp, rV-Msp, and rC-Msp fragments were raised in rabbits (see Materials and Methods). Each antiserum reacted monospecifically with the homologous antigen (Fig. 4B), and no cross-reactivities of the antisera with other recombinant Msp fragments could be demonstrated. The antisera showed similar titers in ELISA with immobilized rMsp (data not shown), and all reacted to similar extents with rMsp on Western immunoblots (Fig. 4B). The three antisera all reacted with Msp in outer membrane protein extracts of T. denticola ATCC 35405 cells (Fig. 5). The rN-Msp and rC-Msp antibodies reacted with some additional minor bands, probably partial Msp degradation products, while the rV-Msp antibodies reacted more or less exclusively with the Msp band (Fig. 5B).
FIG. 5.
Reactivities of Msp antisera with T. denticola ATCC 35405 outer membrane protein extracts. (Panel A) SDS-PAGE gel of outer membrane proteins. (Panel B) Western blots reacted with antisera to rN-Msp (lane 1), rV-Msp (lane 2), or rC-Msp (lane 3). Molecular mass markers are indicated (kDa).
To determine the effects of these region-specific antibodies on bacterial adhesion, biotinylated cells of T. denticola ATCC 35405 were preincubated with a range of dilutions of antiserum or preimmune serum (control). Adhesion levels of antiserum-treated cells to immobilized host tissue proteins were then compared with adhesion levels of cells incubated with an identical dilution of preimmune serum. Maximum inhibition of adhesion was obtained with antisera diluted 1:10. Antibodies to rV-Msp inhibited cell binding to fibronectin, keratin, and fibrinogen by 60% or greater and to laminin and collagen by 30 to 40% (Table 2). Antibodies to rN-Msp also inhibited cell binding to keratin, collagen, and laminin but were less effective than rV-Msp antibodies in inhibiting adhesion to fibronectin and fibrinogen (Table 2). Antibodies to rC-Msp demonstrated no adhesion inhibition properties (Table 2).
TABLE 2.
Effect of rMsp antibodies on adhesion of T. denticola ATCC 35405 cells to host moleculesa
| Substrate | No. of cells (106) bound to substrate ± SDb following incubation with serum:
|
|||
|---|---|---|---|---|
| Preimmune | rN-Msp | rV-Msp | rC-Msp | |
| Fibronectin | 2.9 ± 0.4 | 2.0 ± 0.4 | 0.8 ± 0.2 | 3.2 ± 0.5 |
| Laminin | 4.7 ± 0.7 | 2.5 ± 0.4 | 3.0 ± 0.6 | 4.6 ± 0.6 |
| Keratin | 2.0 ± 0.1 | 1.0 ± 0.2 | 0.8 ± 0.4 | 1.8 ± 0.2 |
| Collagen | 1.0 ± 0.1 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.9 ± 0.1 |
| Fibrinogen | 1.8 ± 0.2 | 1.3 ± 0.3 | 0.5 ± 0.3 | 1.7 ± 0.4 |
Biotinylated Treponema cells were preincubated with serum (diluted 1:10) for 30 min at 20°C and washed, and then the numbers of cells binding to immobilized substrates (0.1 μg) were determined with HRP-conjugated streptavidin as described in Materials and Methods.
Means of two independent experiments performed in triplicate.
Cell surface accessibility of Msp.
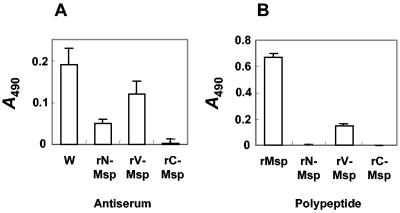
To determine if Msp antibodies reacted with immobilized T. denticola ATCC 35405 cells, spirochetes were fixed with glutaraldehyde, adsorbed onto plastic microwell plates, and reacted with antibodies to each of the rMsp fragments. Polyclonal antibodies raised to glutaraldehyde-fixed T. denticola ATCC 35405 cells served as a control. The whole-cell antiserum showed the highest ELISA value (Fig. 6A). Antibodies raised to rV-Msp and rN-Msp both reacted with fixed cells, while antibodies to rC-Msp were nonreactive (Fig. 6A). These results suggested that the N and V regions of Msp were accessible to antibodies under these conditions and thus might be surface exposed on viable cells. We then tested the polyclonal antiserum to T. denticola ATCC 35405 cells for reactivity with rMsp and rMsp fragments. The antiserum reacted strongly with rMsp and with the rV-Msp fragment (Fig. 6B), but not with the rN-Msp or rC-Msp fragments (Fig. 6B). Taken collectively, these results confirm Msp as a major antigenic determinant on T. denticola cells and suggest that prominent B-cell epitopes reside within the V region.
FIG. 6.
Interactions of rMsp antibodies with T. denticola ATCC 35405 cells and of whole-cell antibodies with rMsp fragments. (Panel A) Reactivity in ELISA of antiserum (1:250 diluted) to T. denticola ATCC 35405 cells (W) or antisera (1:250 diluted) raised to rMsp fragments (as indicated), with T. denticola ATCC 35405 cells (2 × 108) immobilized onto plastic microwells. (Panel B) Reactivity of polyclonal antiserum (W) (1:1,000 diluted) raised to T. denticola ATCC 35405 cells with rMsp or rMsp fragments (0.1 μg) immobilized onto plastic microwells. Error bars indicate ± standard deviation of triplicates from three independent experiments.
DISCUSSION
Treponema outer membrane components have been implicated as adhesion factors that mediate colonization of host tissues (14) and as virulence factors that promote cytopathic effects in the host (15, 16, 41). The Msp of T. denticola is an abundant outer sheath protein that exhibits pore-forming activity (13), binds extracellular matrix components (18), and induces cytopathic effects in cultured cells (31). However, relatively little is known about the binding properties of Msp to host-associated proteins other than fibronectin and laminin or about the regions of the polypeptide that confer binding properties. In this study, recombinant Msp and defined fragments were successfully purified from E. coli, with no deleterious effects on the expression host and with minimal degradation of recombinant polypeptides. Initial experiments investigating the adhesion properties of Msp to immobilized fibronectin demonstrated that rMsp bound to the 30-kDa N-terminal tryptic fragment of fibronectin that carries a heparin I/fibrin I binding domain. However, rMsp showed low affinity for binding fluid-phase fibronectin. These fibronectin interaction properties of Msp are similar to those of T. denticola cells and to those of T. vincentii and related Treponema species (12). rMsp was found to block, by up to 50%, binding of T. denticola ATCC 35405 cells to fibronectin, an observation that conflicts with a previous report showing that full-length recombinant Msp did not significantly affect adhesion to fibronectin (18). Different rMsp preparations or assay conditions could account for this discrepancy.
Receptor blocking experiments using rMsp from T. denticola ATCC 35405 confirmed that the N-terminal region of fibronectin is the only region targeted in vitro by T. denticola ATCC 35405. The same region of fibronectin was bound by T. denticola GM-1. These results show that Msp has the ability to function autonomously as an adhesin. It is relevant to consider, though, that Msp forms part of an oligomeric complex associated with the outer sheath of T. denticola. One of the other components of this complex is CTLP (33). Strains of Treponema species that are deficient in CTLP activity generally show lower levels of binding to fibronectin (12), even though they appear to express Msp-like proteins. Thus, it is likely that CTLP (26) or other components of the Msp complex may modulate fibronectin binding functions in vivo.
While msp genes have been identified in a number of strains of T. denticola and T. vincentii, it is not entirely clear at present whether all oral Treponema species express these proteins or if their distribution is restricted. Strains of T. socranskii and T. pectinovorum have been shown to produce 43-kDa Msp-like proteins (19), but their binding properties and their relationship to other Msp proteins from T. denticola are unknown. The Msp proteins from T. denticola ATCC 35405 and ATCC 35520 show high sequence identity (except for the central V region; Fig. 1), whereas the Msp polypeptide from T. denticola OTK is significantly different and antigenically distinct (19). The predicted sequence of Msp from T. denticola GM-1 is >95% identical to that of Msp from strain OTK (A. M. Edwards, unpublished data). All of these T. denticola strains show high binding levels to fibronectin (8, 12). In contrast, T. vincentii and some related Treponema species isolated from animal infections show lower levels of binding to fibronectin than T. denticola (12). In assays of T. vincentii ATCC 35580 and ovine Treponema strain UB1090 binding to fibronectin, exogenously added rMsp did not block adhesion of these strains but enhanced their cell adhesion levels. This might be explained by suggesting that vacant sites were available on the surface of these strains to acquire exogenously supplied Msp molecules, whereas these could not be incorporated at elevated levels onto the surface of T. denticola ATCC 35405 or GM-1. The ability of Msp to enhance fibronectin binding by low-level-adhering strains further shows that Msp can act as an autonomous adhesin. Recently it has been reported that Borrelia burgdorferi cells are able to incorporate exogenous proteins into the outer membrane (3). Spirochetes produce vesicles and release outer membrane fragments during growth, and so Msp molecules could potentially serve to enhance adhesion if they are bound back onto the cell surface. However, we have not tested the ability of outer membrane vesicles prepared from T. denticola to enhance binding of other Treponema species to fibronectin. Since it appears that Msp can enhance adhesion of heterologous Treponema species to fibronectin, the Msp could play a potentially significant role in subgingival colonization by multiple Treponema species. The ability of Treponema to acquire new phenotypic traits by complementation with exogenous protein factors would foster cooperative interactions between species and could partly account for the multispecies diversity of Treponema isolates at periodontal sites (5, 32).
The potential for Msp to modulate Treponema colonization under a variety of host environmental conditions is indicated by the results showing that rMsp binds keratin, collagen, and fibrinogen, in addition to fibronectin and laminin. Msp also bound weakly to hyaluronic acid and heparin. Comparison of binding levels of rMsp fragments to these molecules demonstrated that the Msp central variable sequence region (rV-Msp) had a binding spectrum similar to that of rMsp. In contrast, the N-terminal region of Msp (rN-Msp) bound only weakly to fibronectin, keratin, and fibrinogen and the C-terminal region (rC-Msp) did not bind significantly to any of the substrates. The implication from these in vitro binding assays is that the V region is very important for adhesion. This was supported by results obtained for region-specific antibody inhibition of T. denticola cell adhesion. Antiserum directed to the V region of Msp was a most effective inhibitor of T. denticola adhesion, and antiserum to the N-terminal region was generally less effective in inhibiting adhesion (except to laminin), while antibodies to the C-terminal region were completely without inhibitory effect. These results suggest that the N-terminal half of Msp presents major adhesion epitopes and extend previous work (16) showing that antibodies against Msp were effective inhibitors of T. denticola cell binding to fibronectin, laminin, and periodontal ligament epithelial cells. Although the isolated C-terminal region does not bind host molecules in vitro, a requirement for the C-terminal region in mediating cell adhesion is not ruled out by our studies. The presence of the C-terminal region could be crucial for correct incorporation of Msp into the outer sheath and for presentation of adhesion-mediating sequences at the cell surface. In future studies, it should be possible to investigate further the functional roles of the various Msp regions by constructing defined mutants with deletions or substitutions within the Msp protein.
On the basis of primary sequence analyses, it is proposed that Msp is a porin-like protein with β barrel secondary structure (18) that spans the cytoplasmic membrane. Since porins contain regions, usually peptide sequence loops, that are exposed on the outer face of the membrane, the porin-like topology of Msp would be consistent with the notion that Msp functions as a surface factor that mediates interactions of Treponema cells with host molecules. In accordance with this model, we have shown that antibodies raised to the V region of Msp are particularly effective in blocking adhesion of T. denticola to fibronectin and to several other host proteins. Antibodies raised to the N region were also inhibitory, although less so. Thus it is suggested that adhesion epitopes are formed from sequences within the N and V regions of Msp polypeptide. The accessibility of the V and N regions to antibodies was clearly demonstrated for cells of T. denticola that were fixed to microtiter plate wells. However, in immunofluorescence studies (not shown) it was found to be not possible to obtain any more than rather localized binding of V region antibodies to a relatively small proportion of the total cell population. Somewhat similar observations have been made by Caimano et al. (4), who demonstrated that Msp antibodies reacted only weakly with T. denticola cells encapsulated in gel microdroplets (to retain cell surface integrity) but strongly and uniformly with detergent-permeabilized cells. These authors suggested that only minor portions of Msp are normally surface exposed and that the protein may be predominantly periplasmic (4). It is possible that the presence of other surface proteins, closely associated with Msp, limits the access of antibody to the surface-exposed regions of Msp. Lipoproteins present in the outer membrane of B. burgdorferi have been shown to hinder reaction of antibodies with the exposed loop of outer membrane protein P66 (1). The ability to detect high levels of N-region and V-region antibody binding to T. denticola cells fixed to microtiter wells may be related to an effect on the cell surface as a result of deposition of cells onto the plastic. This could result in partial loss in integrity of the outer membrane or sheath, rendering the Msp molecules more accessible to antibodies. Interestingly though, the C-terminal region remained inaccessible to antibodies, indicating that Msp conformation or topography was at least partially preserved.
In summary, the results in this article provide evidence that Msp protein mediates, at least in part, binding of T. denticola cells to fibronectin, laminin, collagen, keratin, and fibrinogen. The central V region of Msp has three features that could be considered critical to the function of the polypeptide. The first is that the V region appears to carry major adhesion-mediating sequences. The second feature demonstrated is antigenic variation. This is a strategy frequently utilized by pathogenic bacteria to evade host immune defenses (2). The V region contains the dominant B-cell epitopes for animals immunized with T. denticola cells. The V region is thus highly immunogenic and would provide a major target for the host immune system. This provides an explanation for why antibodies raised to closely related Msp proteins, which differ only in their V-region sequences, have been shown to not cross-react (19). On the other hand, the invariable N and C regions, which may be important for maintaining structure and function, are not significantly immunogenic. A third feature is that, on intact cells, there may be a mechanism by which accessibility of Msp is regulated. In vivo this would potentially avoid immune responses that might be harmful to the organism. However, in vitro, N- and V-region antibodies were very effective in blocking cell adhesion. This could be explained by the kinetic and dynamic properties of T. denticola cell surface protein antigens. Surface antigens have been shown in several studies to become redistributed in accordance with environmental conditions. In particular, there is evidence for clustering of adhesins in response to cell contact with host proteins (8), so it is possible that Msp becomes more accessible to antibodies in the presence of adhesion substrate. Further understanding of the topology and important functional domains of Msp will be gained by developing techniques that preserve the molecular composition and integrity of the T. denticola cell surface (28) and allow real-time analyses of Msp interactions with host components, including antibodies. Meanwhile, the work presented here suggests that the central V region of T. denticola Msp is critical for function. This region is thus potentially an excellent target for development of strategies aimed at controlling oral colonization and tissue destruction by oral Treponema.
Acknowledgments
We thank Jane Brittan and Caroline Bamford for technical assistance; Mark Jepson for microscopy; Chris Fenno for helpful discussions; and R. J. Lamont, P. E. Greenberg, and I. Demirkan for providing bacterial strains.
A Seedcorn grant from the Veterinary Laboratories Agency and DEFRA funding through Project OD0213 are gratefully acknowledged.
Editor: V. J. DiRita
REFERENCES
- 1.Bunikis, J., and A. G. Barbour. 1999. Access of antibody or trypsin to an integral outer membrane protein (P66) of Borrelia burgdorferi is hindered by Osp lipoproteins. Infect. Immun. 67:2874-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bunikis, J., C. J. Luke, E. Bunikiene, S. Bergstrom, and A. G. Barbour. 1998. A surface-exposed region of a novel outer membrane protein (P66) of Borrelia spp. is variable in size and sequence. J. Bacteriol. 180:1618-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bunikis, J., H. Mirian, E. Bunikiene, and A. G. Barbour. 2001. Non-heritable change of a spirochaete's phenotype by decoration of the cell surface with exogenous lipoproteins. Mol. Microbiol. 40:387-396. [DOI] [PubMed] [Google Scholar]
- 4.Caimano, M. J., K. W. Bourell, T. D. Bannister, D. L. Cox, and J. D. Radolf. 1999. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect. Immun. 67:4072-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Gobel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu, L., J. L. Ebersole, and S. C. Holt. 1999. Hemoxidation and binding of the 46-kDa cystalysin of Treponema denticola leads to a cysteine-dependent hemolysis of human erythrocytes. Oral Microbiol. Immunol. 14:293-303. [DOI] [PubMed] [Google Scholar]
- 7.Collighan, R. J., R. D. Naylor, P. K. Martin, B. A. Cooley, N. Buller, and M. J. Woodward. 2000. A spirochete isolated from a case of severe virulent ovine foot disease is closely related to a treponeme isolated from human periodontitis and bovine digital dermatitis. Vet. Microbiol. 74:249-257. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, J. R., and R. P. Ellen. 1994. Clustering of fibronectin adhesins toward Treponema denticola tips upon contact with immobilized fibronectin. Infect. Immun. 62:2214-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewhirst, F. E., M. A. Tamer, P. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 10.Ding, Y., V. J. Uitto, M. Haapasalo, K. Lounatmaa, Y. T. Konttinen, T. Salo, D. Grenier, and T. Sorsa. 1996. Membrane components of Treponema denticola trigger proteinase release from human polymorphonuclear leukocytes. J. Dent. Res. 75:1986-1993. [DOI] [PubMed] [Google Scholar]
- 11.Edwards, A. M., D. Dymock, and H. F. Jenkinson. 2003. From tooth to hoof: treponemes in tissue-destructive diseases. J. Appl. Microbiol. 94:767-780. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, A. M., D. Dymock, M. J. Woodward, and H. F. Jenkinson. 2003. Genetic relatedness and phenotypic characteristics of Treponema associated with human periodontal tissues and ruminant foot disease. Microbiology 149:1083-1093. [DOI] [PubMed] [Google Scholar]
- 13.Egli, C., W. K. Leung, K.-H. Müller, R. E. W. Hancock, and B. C. McBride. 1993. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect. Immun. 61:1694-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellen, R. P. 2002. Adhesion of oral spirochaetes to host cells and its cytopathogenic consequences, p. 247-276. In M. Wilson (ed.), Bacterial adhesion to host tissues: mechanisms and consequences. Cambridge University Press, Cambridge, United Kingdom.
- 15.Ellen, R. P., J. R. Dawson, and P. F. Yang. 1994. Treponema denticola as a model for polar adhesion and cytopathogenicity of spirochetes. Trends Microbiol. 2:114-119. [DOI] [PubMed] [Google Scholar]
- 16.Fenno, J. C., P. M. Hannam, W. K. Leung, M. Tamura, V.-J. Uitto, and B. C. McBride. 1998. Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect. Immun. 66:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenno, J. C., S. Y. Lee, C. H. Bayer, and Y. Ning. 2001. The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect. Immun. 69:6193-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenno, J. C., K.-H. Müller, and B. C. McBride. 1996. Sequence analysis, expression, and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J. Bacteriol. 178:2489-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenno, J. C., G. W. K. Wong, P. M. Hannam, K.-H. Müller, W. K. Leung, and B. C. McBride. 1997. Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J. Bacteriol. 179:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haapasalo, M., K. H. Muller, V. J. Uitto, W. K. Leung, and B. C. McBride. 1992. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect. Immun. 60:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haapasalo, M., U. Singh, B. C. McBride, and V.-J. Uitto. 1991. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect. Immun. 59:4230-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haffajee, A. D., M. A. Cugini, A. Tanner, R. P. Pollack, C. Smith, R. L. Kent, Jr., and S. S. Socransky. 1998. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J. Clin. Periodontol. 25:346-353. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi, M., and K. M. Yamada. 1983. Domain structure of the carboxyl-terminal half of human plasma fibronectin. J. Biol. Chem. 258:3332-3340. [PubMed] [Google Scholar]
- 24.Heuner, K., B.-K. Choi, R. Schade, A. Moter, A. Otto, and U. B. Gobel. 1999. Cloning and characterization of a gene (mspA) encoding the major sheath protein of Treponema maltophilum ATCC 51939T. J. Bacteriol. 181:1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes, A. R., R. McNab, and H. F. Jenkinson. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect. Immun. 64:4680-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishihara, K., H. K. Kuramitsu, T. Miura, and K. Okuda. 1998. Dentilisin activity affects the organization of the outer sheath of Treponema denticola. J. Bacteriol. 180:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishihara, K., T. Miura, H. K. Kuramitsu, and K. Okuda. 1996. Characterization of the Treponema denticola prtP gene encoding a prolyl-phenylalanine-specific protease (dentilisin). Infect. Immun. 64:5178-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izard, J., B. F. McEwen, R. M. Barnard, T. Portuese, W. A. Samsonoff, and R. J. Limberger. 2004. Tomographic reconstruction of treponemal cytoplasmic filaments reveals novel bridging and anchoring components. Mol. Microbiol. 51:609-618. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn, K., and J. Eble. 1994. The structural bases of integrin-ligand interactions. Trends Cell Biol. 4:256-261. [DOI] [PubMed] [Google Scholar]
- 30.Loesche, W. J., and N. S. Grossman. 2001. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin. Microbiol. Rev. 14:727-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathers, D. A., W. K. Leung, J. C. Fenno, Y. Hong, and B. C. McBride. 1996. The major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect. Immun. 64:2904-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen, G., R. Naor, and M. N. Sela. 1999. Multiple forms of the major phenylalanine specific protease in Treponema denticola. J. Periodontal Res. 34:269-276. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Scott, D., I. R. Siboo, E. C. S. Chan, and R. Siboo. 1996. An extracellular enzyme with hyaluronidase and chondroitinase activities from some oral anaerobic spirochaetes. Microbiology 142:2567-2576. [DOI] [PubMed] [Google Scholar]
- 36.Sela, M. N. 2001. Role of Treponema denticola in periodontal diseases. Crit. Rev. Oral Biol. Med. 12:399-413. [DOI] [PubMed] [Google Scholar]
- 37.Siboo, R., W. Al-Joburi, M. Gornitsky, and E. C. S. Chan. 1989. Synthesis and secretion of phospholipase C by oral spirochetes. J. Clin. Microbiol. 27:568-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 39.Umemoto, T., and I. Namikawa. 1994. Binding of host-associated treponeme proteins to collagens and laminin—a possible mechanism of spirochetal adherence to host tissues. Microbiol. Immunol. 38:655-663. [DOI] [PubMed] [Google Scholar]
- 40.Umemoto, T., M. Y. Li, and I. Namikawa. 1997. Adherence of human oral spirochetes by collagen-binding proteins. Microbiol. Immunol. 41:917-923. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q., K. S. Ko, A. Kapus, C. A. McCulloch, and R. P. Ellen. 2001. A spirochete surface protein uncouples store-operated calcium channels in fibroblasts: a novel cytotoxic mechanism. J. Biol. Chem. 276:23056-23064. [DOI] [PubMed] [Google Scholar]