Abstract
Recurrent spontaneous abortion (RSA) has various causes, including chromosomal abnormalities, prethrombotic state, and abnormal uterine anatomical factors. However, the pathogenesis of RSA is still unclear. Surprisingly, non-coding RNA can stably express at the maternal-fetal interface and regulate immune cells’ proliferation, apoptosis, invasion, metastasis, and angiogenesis. Accumulating evidence suggests that the competing endogenous RNA (ceRNA) regulatory network between non-coding RNAs complicates RSA’s pathological process and maybe a new starting point for exploring RSA. In this review, we mainly discuss the regulatory network and potential significance of non-coding RNA in the immune microenvironment of RSA patients. In addition, the cellular interactions of non-coding RNA transported through vesicles were introduced from aspects of trophoblast function and immune regulation. Finally, we analyze previous studies and further discuss that the stable expression of non-coding RNA may be used as a biomarker of some disease states and a prediction target of RSA.
Keywords: non-coding RNA, recurrent spontaneous abortion, maternal-fetal immune, trophoblast cells, ceRNA
Introduction
RSA is defined as two or more consecutive spontaneous abortions before 20 weeks of pregnancy, which is a common clinical pregnancy complication [1]. The incidence rate among women of childbearing age has reached 5%, which has seriously affected the quality of life of families. As a semi-allogeneic graft, half of the MHC molecules are of maternal, and the other half are of paternal origin. Hence, the fetus acts as a paternal antigen protected from rejection in the maternal immune system, which presents an immune response [2]. Disruption of the immune tolerance microenvironment of pregnancy can lead to complications such as recurrent miscarriage (RM), recurrent pregnancy loss [3], and preeclampsia [4]. In recent years, advances in the field of immunology and insights into RSA have revealed that macrophages, NK cells, trophoblast cells, Th17 cells, Treg cells, and cytokines are essential for normal pregnancy, providing new insights into the possible causes of the disease [5]. Teng et al. [6] reported that overexpression of lncRNA NEAT1 inhibited proliferation, migration, and invasion of trophoblast cells and promoted its apoptosis. Furthermore, Tang et al. [7] also found that miR-125b regulates the invasion and migration of extravillous trophoblast cells. Therefore, revealing the mechanism that the immune cells regulated by non-coding RNAs in RSA will provide potential targets for RSA therapy.
In recent years, the expression profile of ncRNAs can be stably expressed at the maternal-fetal interface and show differential expression in RSA and average pregnant women. Therefore, non-coding RNAs are a potential marker for the prediction, clinical diagnosis, and prognosis of disease. Non-coding RNA (ncRNA) is a type of RNA that does not code for proteins, and they can be classified according to their length, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). MiRNAs have about 19–25 nucleotides. However, lncRNAs are composed of at least 200 nucleotides in length and circular endogenous non-coding RNA molecules (circRNAs) formed by reverse splicing. While the pathogenesis of RSA is still unclear, non-coding RNA may be one of the mechanisms that explain the pathogenesis of RSA, as it is an essential regulator of transcription and protein expression [8]. For example, although ncRNA does not encode proteins, it participates in many physiological activities, such as chromosome remodeling, cell localization of proteins, gene transcription, and post-translational modification [9]. Not all RNA has functions, but a large number of functional ncRNAs are important for cells to maintain normal biological functions and participate in the occurrence and development of pathological processes. In recent years, our research on non-coding RNA has mainly focused on miRNA, lncRNA, and CircRNA. Therefore, our review focuses on the mechanism of these three RNAs in the immune microenvironment. Since the competitive endogenous RNA (ceRNA) network hypothesis was put forward in 2011 [10], ncRNA has ushered in further discussion. This article demonstrates, at the laboratory level, that endogenous RNAs regulate each other’s expression by binding to competing miRNAs. For example, circRNAs contain multiple miRNA binding sites that enable them to act as miRNA molecular sponges as competing endogenous RNA (ceRNA) that improve miRNA-induced inhibition of downstream target genes, thereby increasing their expression levels [11]. Several articles [12, 13] have confirmed the universality of ceRNA regulation. Researchers are increasingly aware of the potential significance of ceRNA network regulation in predicting and diagnosing some diseases.
It is generally believed that trophoblast dysfunction mediates pregnancy failure. Previous studies have shown that ncRNA is closely related to the proliferation, invasion, and angiogenesis of trophoblast cells [14]. In addition, many literatures [15, 16] reported that maternal-fetal immune tolerance and immune rejection may participate in the pathogenesis of RSA. Therefore, the abnormal expression of some ncRNA may provide new insights into the potential mechanism of RSA. In this review, we mainly focus on the role of ncRNA in the immune microenvironment of RSA patients and the potential application of the ceRNA network.
Pathogenesis of RSA
Current research on RSA has found that there are genetic factors [17], immune factors [18], environmental factors [19], and psychosocial factors [20] that affect its pathogenesis. Genetic factors are considered to be the leading cause of its onset, with fetal chromosomal abnormalities accounting for 50–60% [21]. During the early stages of pregnancy, the invasion of the placental trophoblast and normal physiological functions directly affect the embryo’s survival. After implantation of the placenta into the endometrium, the cytotrophoblast (CTB) differentiates into syncytiotrophoblast cells (STB) and extravillous trophoblastic tissue (EVT) [22]. Subsequently, EVT cells play a crucial role in placental anchoring, maternal spiral artery modeling, angiogenesis, cytokine secretion, and interactions with maternal immune cells [23]. The dysfunction of trophoblast cells may lead to the occurrence of RSA. Previous studies have also shown that non-coding RNA can lead to RSA by influencing multiple phenotypes such as invasion, proliferation, apoptosis, migration, and angiogenesis of trophoblast cells [24]. Therefore, the related mechanisms of trophoblast cell dysfunction may contribute to a deeper understanding of the pathogenesis of RSA. Moreover, IL-17 secreted by Th17 cells may promote appropriate responses during early pregnancy to protect mothers from extracellular pathogens. More importantly, IL-17 produced by helper T cells contributes to pregnancy by promoting the proliferation and invasion of trophoblast cells and inhibiting trophoblast cell apoptosis. This physiological function of Th17 cells suggests that it can generate an appropriate immune tolerance microenvironment during normal early pregnancy to prevent the mother from developing an excessive inflammatory response and reduce the incidence of RSA. Abnormal activation of uterine natural killer cells, macrophages [25], and unbalanced differentiation of helper T cell subsets can disrupt maternal-fetal immune tolerance. For example, researchers have found that in many pregnancy complications, miRNA can affect the balance of TH17/Treg cells [26], suggesting that miRNA is involved in establishing immune tolerance during pregnancy (Figure 1). The miRNA spectrum related to immune cells can be used as biomarkers and provide potential targets for the clinical treatment of RSA patients.
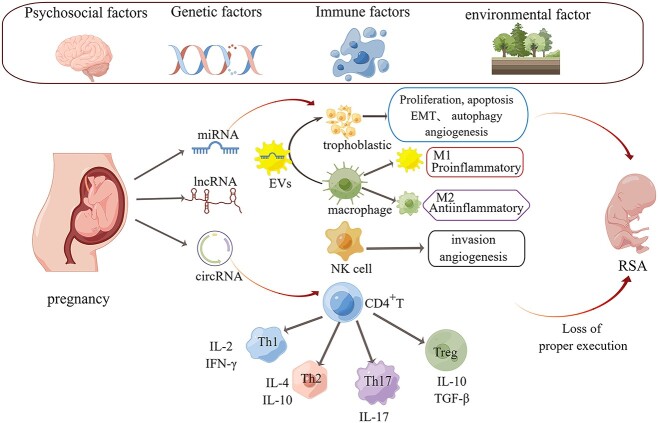
Figure 1.
The role of non-coding RNA in the immune microenvironment of the maternal-fetal interface. Normal pregnancy is influenced by sociopsychological, genetic, and immune memory environmental factors. Non-coding RNA can act on trophoblast cells and regulate their phenotypes such as proliferation, apoptosis, EMT, autophagy, and angiogenesis. While, M1-type macrophages can transport miRNA to trophoblastic cells through vesicular transport to play a role. Non-coding RNA can also act on macrophages, NK cells, and CD4 T cells to perform physiological functions. When the immune cells of the body lose their correct executive power, RSA can occur.
In recent years, with the increasing research on non-coding RNA in RSA, researchers have gained a further understanding of the mechanism of ncRNA regulating RSA. In fact, most studies on the pathogenesis of RSA focus on miRNA, lncRNA, and circRNA. However, the specific mechanism of how non-coding RNA participates in regulating RSA is unclear, leading to significant limitations in the clinical diagnosis and application of non-coding RNA. Next, our review will also analyze the regulatory relationship between non-coding RNA and RSA around these three molecules.
miRNA and RSA
Overview of miRNA
miRNAs are single-stranded RNA fragments with uniform length and generally contain 21–25 nucleotide sequences. Meanwhile, miRNAs are highly evolutionarily conserved, and more than 50% exist in clusters. The miRNA originally transcribed has hundreds or even thousands of bases long, called primary miRNA (pri-miRNA). Pri-miRNA is processed by the endonuclease Drosha to produce about 70–90 base miRNA precursors (pre-miRNA) (Figure 2). Subsequently, pre-miRNA is digested by Dicer to produce mature miRNA during its transport to the cytoplasm [27]. Next, miRNA molecules can downregulate gene expression by binding with 3-UTR of the target gene [28], which depends on the role of RNA-induced silencing complex (RISC). Lin-4, the first miRNA identified in Caenorhabditis elegans [29], negatively regulates the expression of Lin-14 mRNA and proteins by binding to the 3′UTR of Lin-14 mRNA, leading to impaired worm development. Since then, research on miRNAs has continued to deepen, and more than 1000 miRNAs are involved in regulating complex processes in the body, such as immune research, tissue remodeling, epigenetic modification, and cell development [30]. Interestingly, one miRNA can regulate the expression of multiple mRNA, and multiple miRNAs can also regulate one mRNA simultaneously that a small amount of miRNA in the body can regulate numerous physiological functions. For example, miR185 and miR133b are usually highly expressed in colorectal cancer patients, and the level of these two miRNAs can be used as prognostic markers of colorectal cancer. In addition, miRNAs have a more comprehensive application: a. for early screening of diseases, such as preeclampsia, b. for molecular typing of diseases, c. as potential drug targets. Therefore, some stable miRNAs (e.g. miR-146a-5p, miR-146b-5p [44], and miR-103 [31], etc.) in the maternal circulatory system can be used as biomarkers to monitor the normal pregnancy process and RSA patients, providing potential therapeutic targets.
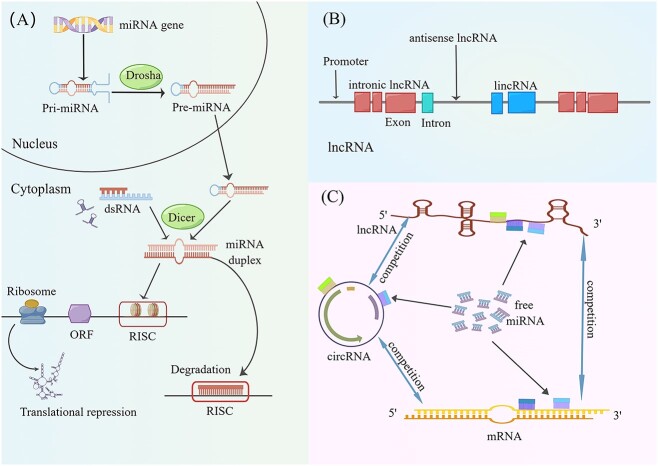
Figure 2.
The formation of miRNA and lncRNA and the effects of ceRNA regulatory networks. (A) The synthesis process of miRNA. The miRNA gene first synthesized pri-miRNA, followed by pre-miRNA synthesis under the action of Drosha. Pre-miRNA forms mature miRNA under the action of Dicer enzyme after enucleation. Mature miRNAs rely on RISC to perform transcriptional inhibition or degradation functions. (B) Gene structure of lncRNA. (C) The ceRNA regulatory network. LncRNA, CircRNA, and mRNA can compete with each other. Both lncRNA and CircRNA compete with mRNA for the opportunity to bind to miRNA to regulate downstream mRNA gene expression.
miRNA and RSA
Wang et al. [32] determined the miRNA expression profile in decidua or villi through high-throughput sequencing analysis, suggesting that the pathogenic process of RSA may be related to changes in miRNA expression profiles in decidua and villi. Studies have further confirmed that miRNA expression can affect physiological processes such as proliferation, apoptosis, invasion, metastasis, epithelial to mesenchyme transition (EMT), and angiogenesis of placental trophoblasts, thereby affecting the pathogenesis of RSA. Research on miRNAs in trophoblast cells found that they mainly come from the two most significant clusters (C14MC and C19MC) on chromosomes 14 and 19 [33]. The miRNA from C19MC primarily exists in embryonic stem cells and plays a role in cell proliferation, invasion, metastasis, and differentiation. Specifically, placental-derived miR519c (derived from C19MC) is produced by trophoblast cells and can be released or packaged as free miRNA into extracellular vesicles (EVs). It has been proven to inhibit TNF-αgene expression in placental transplant models, which involves the expression of endotoxin tolerance models [34]. In recent years, the significance and possible regulatory role of miRNA in normal pregnancy and adverse pregnancy outcomes (such as miscarriage) have been partially revealed. For example, downregulation of the expression of USP25 [35] in placental villus tissue of patients with RM inhibits the invasion and metastasis of trophoblast cells. At the same time, mimetics of miR27a-3p can upregulate the expression of USP25 to inhibit the occurrence of RM. In addition, miR146a-5p [36] directly regulates the expression of downstream Wnt2, affecting HTR-8/SVneo cell proliferation and EMT. As an upstream regulator of PLGF, miR-124-3p [37] directly acts on the downstream PLGF-ROS pathway to reduce the migration and invasion of trophoblast cells and promote the occurrence of apoptosis. Another study has shown that miR520 [38] promotes DNA damage-induced trophoblast apoptosis through targeting poly (ADP-ribose) polymerase 1 (PARP1), thereby enabling the occurrence and development of RSA. These data suggest that the expression of miRNAs is strictly spatiotemporal and tissue-specific. Moreover, vascular endothelial growth factor (VEGF) plays a crucial role in regulating the invasion and proliferation of trophoblast cells, and miR-16 [39] can inhibit the expression of VEGF and participate in the pathogenesis of RSA. In many pregnancy complications, miRNA may also serve as a biomarker by influencing the expression of Th17/Treg. Through a large number of high-throughput sequencing technologies and the mining of bioinformatics tools, we have conducted an in-depth understanding of miRNA [40]. For example, high expressions of miR-4497, miR135a-5p, miR93, and miR19b were found in the blood or villi tissues of RSA patients, while low expressions of miR16, miR494, miR219a, and miR155-5p were found. Based on the testing of clinical samples from RSA patients, the upregulation or downregulation of these differential genes facilitates us to select the differential genes with the highest specificity, which guides us to further excavate through which regulatory pathway of the differential genes leads to the occurrence of RSA, and provides a theoretical basis for the study of the detailed mechanism of RSA.
In summary, the potential mechanism of miRNAs regulating RSA may involve different target genes and binding sites (Table 1). However, in recent years, some researchers have found that the circulation contains many non-coding RNA, which is passively released as acellular circulating RNA or through tissue cell damage [41]. However, the non-coding RNA in these cycles is unstable and often encapsulated in cell vesicles (especially miRNA). The vast majority of miRNAs are directed for transcription by RNA pol II, and a small number of miRNAs (containing Alu sequences) are transcribed by RNA pol III. Therefore, when extracellular vesicles regulate two intercellular mechanisms of action by transporting miRNAs, inhibiting the expression of RNA pol II of miRNAs in the recipient cells and observing the expression of miRNAs can help us to further explore whether extracellular vesicles are involved in the study of the mechanism of non-coding RNA regulation of recurrent spontaneous abortion. This non-coding RNA molecule has been a hot topic in recent years to generate cell interaction through vesicular transport, indicating that the mechanism of ncRNA acting on RSA is very extensive and needs to be discovered. EVs are membrane-bound vesicles communicating between cells and participating in many biological processes. More and more evidence shows [42] that EVs are related to normal pregnancy or pregnancy complications. It can transfer proteins, various nucleic acids, lipids, and bioactive enzymes, to receptor cells to regulate their physiological behavior. Importantly, EVs are an integral part of fetal-maternal communication during implantation and placental processes, regulating maternal immune responses, maintaining normal physiological functions of trophoblast cells, and promoting fetal angiogenesis [43]. For example, TRAF6 can promote EMT, migration, and invasion of trophoblast cells. As upstream regulators of TRAF6, miR-146a-5p and miR-146b-5p downregulate its expression and promote the occurrence of RSA. M1-type macrophages function by transporting miR-146a-5p and miR-146b-5p to trophoblast cells through EVs [44]. Vilella et al. [45] found that when miR-30d of EVs in mouse endometrial fluid was transferred to mouse embryos, cell adhesion molecules such as Itgb3, Itga7, and Cdh5 were upregulated. In addition, the migration and invasion of vascular smooth muscle cells (VSMCs) play a crucial role in the reconstruction of placental spiral arteries, which has been proven to be promoted by the release of EVs from EVTs through the EVT-VSMC exosome pathway [46]. Moreover, various exosome miRNAs, including miR486-1-5p, miR150, and miR486-2-5p, are involved in trophoblast cell migration, placental development, and angiogenesis. These results suggest that EVs play an important role in embryo attachment. MiRNA is often unstable in plasma, and the “packaging” of vesicles will improve its stability and further play a role between cells. However, the research on this vesicular transport (microbubbles, exosomes, and apoptotic bodies) in RSA is incomplete. Studying the mechanisms of cellular interactions may be an important way for us to reconceptualize RSA.
Table 1.
MiRNA related to the pathogenesis of RSA
| ncRNA | Expression in RSA | Tissue | Function | Reference |
|---|---|---|---|---|
| miR27a-3p | Upregulation | Trophoblast | Promotes EMT, migration, and invasion of trophoblast cells | [20] |
| miR146a-5p | Upregulation | M1 macrophages | Inhibits EMT and maintains cellular interaction | [21] |
| miR-124-3p | Upregulation | Trophoblast | Inhibits invasion and metastasis, promotes apoptosis | [23] |
| miR520 | Upregulation | Trophoblast | Promotes DNA damage-induced apoptosis | [24] |
| miR-16 | Upregulation | Placenta | Regulates angiogenesis and development | [58] |
| miR-494 | Downregulation | Placental villi | Regulates abnormal cellular invasion and apoptosis | [59] |
| miR-219a | Downregulation | Trophoblast | Inhibits regulation of cellular proliferation and invasion | [60] |
| miR-4497 | Upregulation | Trophoblast | Downregulates SP1 mRNA and induces trophoblast cell apoptosis | [61] |
| miR-135a-5p | Upregulation | Trophoblast | Targets PTPN1 and inhibits proliferation, invasion, and migration of trophoblast cells | [62] |
| miR-365 | Upregulation | Decidua | Targets SGK1 and regulates cellular apoptosis | [63] |
lncRNA and RSA
Overview of lncRNA
LncRNAs are characterized by three features: (a) They are more than 200 nucleotides in length, (b) They do not encode proteins [47], (c) They are low in abundance, and they are also spatiotemporally specific. In terms of structure, lncRNA has 5 “cap structure and 3” PolyA tail structure and has a complex secondary or tertiary structure without high sequence conservation [47]. Interestingly, lncRNAs can be widespread in organisms and regulate many biological behaviors [47], such as handling the role of transcription factors and other transcriptional regulatory proteins, epigenetic modification, regulating cell signal transduction, initial chromatin remodeling, epigenetic silencing of gene clusters, oligomerization of activated proteins, and regulating gene expression. They are classified into five types according to their location in the genome: intergenic lncRNA, intronic lncRNA, sense lncRNA, antisense lncRNA, and bidirectional lncRNA [48]. LncRNAs also form four mechanistic patterns based on their molecular characteristics [49]: (a) Signal, lncRNA when after being transcribed, can directly combine with DNA at adjacent positions in chromosomes, regulate gene expression, and have the function of signal transduction, itself as a biological event marker. Examples include allele specificity, induction of DNA damage with synergistic activity (represented by enhancer RNAs (eRNAs)), and cold induction, which is mainly present in plants, (b) decoy, lncRNA can combine with downstream transcription factors or RNA to form a competitive binding mode. For example, in the ceRNA regulatory network (described in detail below), p21-associated non-coding RNA (PANDA) directly binds to NF-YA to inhibit apoptotic gene expression, and glucocorticoid receptor lncRNA Gas5 functions to induce cellular glucocorticoid resistance, among others, (c) Guide is a regulation model with positive and negative orienting effects. lncRNAs recruit chromatin modifiers through cis-acting and site-specific recruitment, e.g. RepA RNA recruits the PRC2 complex, or Air ncRNA interacts with Slc22a3 promoter chromatin and H3K9 histone methyltransferase G9a in the placenta. Conversely, lncRNA HOTAIR can also exert transcriptional regulation to affect tumor metastasis, among other things. (d) Scaffold refers to the fact that lncRNA acts as the backbone of a protein complex, connecting multiple proteins and related transcription factors. For example, HOX motif transcriptional antisense RNA (HOTAIR) plays a splicing and bridging role between PRC2 and the LSD1/CoREST/REST complex. In recent years, the field of RNA research is undergoing rapid development. As a regulatory molecule, lncRNA, which accounts for most non-coding RNA, has great potential for exploration. For example, researchers have identified 1449 differentially expressed lncRNA in the chorionic villi of patients with recurrent abortion (RM) [50], which provides a pathway for lncRNA to participate in the pathogenesis of human abortion.
lncRNA and RSA
Like miRNA, lncRNA can also directly act on target gene mRNA to regulate cell proliferation, invasion, migration, and apoptosis. Alternatively, gene expression can be regulated through sponge miRNA using a ceRNA regulatory network [51]. The first lncRNA discovered by humans was H19, which promoted the rapid development of the history of lncRNA research. In recent years, researchers have identified several types of lncRNA, which are differentially expressed in the placenta of patients with abortion compared to healthy patients (Table 2). For example, lncRNA H19, SNHG7-1, and MEG8. Through sequencing technology, it was found that the expression of lncRNA SNHG7-1 was reduced in the villi of RSA abortion patients. It directly affected the downstream miR-34a to inhibit the proliferation and invasion of trophoblast cells. The WNT/β-Catenin signaling pathway regulates the cell cycle and proliferation and invasion ability of trophoblast cells, and miR-34a inhibits the expression of WNT1 by binding to 3′-UTR. Based on the above research, this SNHG7-1-miR-34a-Wnt/β-Catenin has provided potential therapeutic targets in the development of RSA [52]. Some transcription factors can act as upstream factors of lncRNA, guiding the binding of lncRNA to downstream target genes, and influencing the occurrence of RSA. YY1 is a transcription factor that alters the function of trophoblast cells by regulating the expression of cytoskeleton-related proteins. Compared with healthy patients, the level of YY1 mRNA in trophoblasts in RSA patients was significantly lower. In addition, YY1 can directly bind to the PVT1 promoter, affecting the invasion and adhesion of trophoblasts through the downstream mTOR pathway [53]. Similarly, HOX transcriptional antisense RNA HOTAIR is also a downstream target gene of YY1. Promote the expression of HOTAIR, thereby causing the production of matrix metalloproteinase 2 (MMP2), and enhancing the invasion of trophoblast cells [54]. The expression changes of related lncRNA are shown in Table 2.
Table 2.
LncRNA and CircRNA related to the pathogenesis of RSA
| ncRNA | Expression in RSA | Tissue | Function | Reference |
|---|---|---|---|---|
| lncRNA H19 | Downregulation | Placental villi | Inhibits let-7and upregulates ITGB3 expression, and regulation of invasion and adhesion | [64] |
| lncRNA SNHG7-1 | Downregulation | Trophoblast | Regulates the proliferation and invasion of trophoblast cells by targeting miR-34a | [36] |
| lncRNA MEG8 | Upregulation | Trophoblast | Inhibits cell proliferation and invasion | [39] |
| lncRNA PVT1 | Downregulation | Trophoblast | Regulation of invasion and adhesion | [37] |
| lncRNA HOTAIR | Downregulation | Trophoblast | Metalloproteinase 2 and enhances cell invasion | [38] |
| Circ-ZUFSP | Upregulation | Trophoblast | Regulates migration and invasion of trophoblast cells via the CIRC-ZUFSP/miR203/STOX1 pathway | [48] |
| CircPUM1 | Downregulation | Placenta | Promotes trophoblast cell processes and anti-inflammatory effects via the miR-30a-5p/JunB axis | [49] |
| CircFOXP1 | Downregulation | Trophoblast | Regulates the function of trophoblast cells via the miR-143-3p/S100A11 pathway | [49] |
Moreover, RSA is also affected by imprinted lncRNA. Xiang et al. [52] used microarrays to analyze the differential expression of lncRNA during placental development and found that imprinted lncRNA Rian may play an important role in placental development. Its homologous sequence, lncRNA-MEG8 (RIAN), is highly expressed in human spontaneous abortion villi, inhibiting trophoblast proliferation and invasion. These results indicate that imprinted lncRNA exhibits dynamic spatiotemporal expression during placental development. In addition, DNA methylation and some enhancers can regulate gene expression. For example, DNA methylation monitoring in the villi of patients with miscarriage revealed an increase in methylation in the MEG8 promoter region. In addition, Lnc-SLC4A1-1 [55] is transcribed from H3K27ac and H3K4me1 labeled active enhancers. It induces TNF-αand IL-1 β elevated by activating CXCL8-α, leading to trophoblastic inflammation. In summary, lncRNA can participate in the pathological process of RSA, and understanding the regulatory mechanism of lncRNA can provide potential targets for our clinical treatment of RSA.
CircRNA and RSA
Overview of CircRNA
CircRNA is an endogenous RNA produced by special variable splicing, mainly in the cytoplasm [56]. Moreover, CircRNA has no 5 “cap and 3” PolyA tail structure and presents a closed circular structure [57], which is not easily degraded by exonuclease and is more stable than linear RNA. In general, the expression levels of circRNAs are relatively low. Still, the products of certain circRNAs are expressed at higher levels than homologous linearly transcribed RNAs, and their expression is not directly correlated with that of the corresponding linearly transcribed RNAs. CircRNA was first discovered in 1979 [58], but it was not until 2012 that it was discovered in large quantities through high-throughput sequencing, and it quickly became a hot spot in the following years. Studies have found that circRNA expression levels are higher than regular linear RNA regarding tissue, species, time specificity, and sequence conservation. Meanwhile, circRNAs are expressed in diverse forms, with expression specificity in different mammalian cells and tissues as well as in some biological processes. The expression of the same circRNA in different species is also heterogeneous; for example, only 10–20% of the circRNAs in humans are expressed in parallel tissues in mice. Currently, circRNA has been found to be an important participant in normal cell function and the occurrence and development of diseases, for example, as an miRNA sponge to regulate the operation of miRNA [59], as a transcription regulator or protein translation vector [60], interacting with proteins to regulate gene expression [61]. In many cases, circRNA has a homologous expression gene, which forms a circular RNA due to its particular splicing form, but its expression is usually not related to the expression of this homologous gene. This indicates that circRNA is not only a byproduct of mRNA splicing, but also a specially regulated molecular type. In addition, promoting the production and cyclization of circular RNA mainly involves several ways: (a) By utilizing complementary base pairing (such as enrichment of sites formed by Alu repeat sequences [62] in human exon circular RNA), reverse splicing is promoted due to reduced space at splicing sites, (b) The RBP binds [63] to the flanking introns and undergoes cyclization, shearing and bridging them. For example, the placenta can be described as a “controllable cancer” with some similar characteristics to the occurrence of cancer. Epithelial mesenchymal transition is a common process in placental development and many cancers. Interestingly, highly expressed RBP Quaking (QKI) can be found in the placenta to be involved in the occurrence of EMT-related circular RNAs. (c) Lariat introns that escape debranching. (d) The lariat-driven model of circularization. The role of circRNA in the study of pregnancy complications is crucial, and it may also provide new insights into cancer progression. Therefore, conducting in-depth research on the structure and function of circRNA is necessary. With the in-depth study of circRNA, it has a broader range of physiological and pathological processes. For example, many circRNAs are upregulated in neurogenesis and synaptogenesis, while Cdr1as knockout mice exhibit excitatory synaptic dysfunction and neuropsychiatric disorders. Furthermore, some circRNAs are derived from genome-specific loci with specific functions; for example, circRNAs derived from the ANRIL locus are positively associated with atherosclerosis risk. However, in patients with recurrent spontaneous abortion, circRNAs are associated with immune response and trophoblast function, e.g. CircPUM1 targets miRNA expression and thus regulates trophoblast function. Therefore, understanding the molecular functions of circRNAs and their regulatory mechanisms in vivo provides us with new perspectives to further explore the pathogenesis of RSA.
CircRNA and RSA
Studies have shown that CircRNA plays a key role in tumor regulation [64]. Although the physiological processes of embryo implantation and tumor are similar, there are relatively few studies on the role of circRNA in abortion. Qian et al. [65] found that RSA women have nearly 600 differentially expressed circular RNAs in the placenta compared to normal pregnancy. Li et al. [66] further confirmed that 123 differentially expressed cyclic RNAs were found in patients with early RSA compared to women with normal pregnancy, of which 78 were upregulated and 45 were downregulated. CircRNA, as a ceRNA molecule, can stimulate miRNA-induced target gene expression and interact with disease-related miRNAs, reducing miRNA-induced target gene inhibition (Table 2). For example, CircPUM1 promotes the function of trophoblast cells and fights inflammation by targeting the expression of microRNA-30a-5p/JUNB [67], thereby reducing the occurrence of RSA. CircPUM1 and JUNB can simultaneously bind to miR-30a-5p, influencing the expression of regulatory molecules. Furthermore, similar mechanisms of action include CDR1as and miR7, circRNA BIRC6 and miR34a, miR45, circRNA HIPK2, and miR124-2HG. Due to the lower expression of circRNA in vivo, its impact as an miRNA molecule is relatively limited compared to lncRNA. Li et al. [68] found that CIRC-ZUFSP affects the downstream STOX1 pathway by acting on miR-203, leading to trophoblast dysfunction. Therefore, inhibition of trophoblast migration and invasion through the CIRC-ZUFSP/miR-203/STOX1 pathway may lead to RSA. In addition, circFOXP1 [69] regulates the expression of S100 calcium-binding protein A11 (S100A11) by competitively binding to miR-143-3p, thereby regulating the function of trophoblast cells. These studies have shown that CircRNA and lncRNA have similarities and can bind to downstream corresponding miRNAs to regulate their gene expression, thereby controlling the progress of RSA. Significantly, circRNA can promote genomic instability through translocation. Rapid proliferation and hypoxia stress exist in cancer and placenta, which may lead to DNA damage. CircRNA forms RNA–DNA hybridization with its homologous DNA sites, leading to transcriptional arrest. If so, may the imbalance of cyclic RNA in early pregnancy affect genomic changes, leading to placental and fetal development? From the expression profiles of human-expressed circRNAs, we can speculate on the potential function of circRNAs that may serve as biomarkers. circRNAs’ intrinsic loop structure makes them relatively stable inside and outside the cell (intracellular stability is better than extracellular stability). Similarly, some circRNAs can be transported by exosomes into the extracellular fluid. Still, the authors believe that circRNAs as biomarkers need to first clarify its effect on gene expression in disease. In summary, the research on circRNA in RSA needs further exploration.
ceRNA network
Studies have confirmed that multiple miRNAs may regulate a single mRNA, and a single miRNA can also regulate multiple mRNA, forming a network-like structure. If the UTR regions of two RNAs can bind the same miRNA, will there be a regulatory relationship between them? In 2011, Salmena et al. [69] proposed the concept of a ceRNA regulatory network based on this. They demonstrated the regulatory phenomenon that endogenous RNA interacts with each other by competing for miRNA binding. The concept has been widely studied and it has been found that ceRNA networks are ubiquitous in organisms. The role of ceRNA networks in pregnancy diseases such as cancer progression [70] and recurrent implantation failure (RIF) [71] has been discovered. For example, the tumor suppressor gene PTEN uses the ceRNA network mechanism to play a role in cancer. Huang et al. [72] found the existence of lncRNA-miRNA-mRNA regulatory networks in RSA patients and elaborated on the potential mechanisms that may lead to RSA. Zhang et al. [73] used high-throughput sequencing to construct a ceRNA network related to SAB in the endometrium of pigs, further explaining the complex regulatory mechanisms between non-coding RNAs. However, the role of ceRNA networks in the pathogenesis of RSA still requires a lot of in-depth research.
For miRNA, lncRNA, and circRNA, there is a ceRNA network. LncRNA can act as a molecular sponge to competitively bind to miRNA, resulting in reduced binding to downstream target mRNA and restoration of mRNA activity [73]. For example, Huang et al. found that lncRNA-related ceRNA network contains 31 lncRNAs, one miRNA (hsa-miR-210-5p), and three mRNAs (NTNG2, GRIA1, and AQP1). Based on this, lncRNA DANCR can act on downstream miR-214-5p to promote invasion and migration of trophoblast cells in patients with preeclampsia [74]. In addition, the content of lncRNA H19 in the placental villi of SA patients decreased compared to healthy pregnant women. However, lncRNA H19 can inhibit the function of miRNA let-7 to prevent mRNA degradation, leading to the expression of ITGB3 increasing [75]. It indicates it participates in SA through the H19/let7/ITGB3 axis. Similarly, circRNA can act as a sponge for miRNA and bind to miRNA (Figure 2). Unlike histone, the circRNA DURSA reduces T cell apoptosis through the miR-760-HIST1H2BE axis. In contrast, histone H2B, the protein encoding the target gene Hist1h2b, whose deletion prevents DNA replication, leads to aberrant apoptosis of TCs and ultimately, to RSA. Tang et al. used microarray technology to compare placental villi from URSA patients with placental villi from healthy pregnancies, clarifying the expression of cyclic RNA and different competitive endogenous RNA (ceRNA) networks [76]. Some researchers also have found ceRNA networks of CircRNA-miRNA-mRNA in pregnancy disorders, such as preeclampsia [77] and recurrent implantation failure (RIF) [78]. These studies indicate that the ceRNA regulatory network of non-coding RNA has a broad impact on organisms. However, the research data on ceRNA in the field of RSA are insufficient, indicating that a lot of work should be shifted to the ceRNA regulatory network to reveal the potential mechanism for the occurrence of RSA in the future.
Discussion
In this review, we mainly describe the latest research on non-coding RNA and its potential molecular pathways in RSA pathology, highlighting the ceRNA regulatory network between non-coding RNA. Based on previous studies, we have found that non-coding RNA regulates physiological processes of cells, such as proliferation, apoptosis, invasion, metastasis, and angiogenesis. The emergence of ceRNA complicates the regulatory mechanism. Interestingly, with the progress of the new generation of biological information technology and the maturity of flux technology, it is beneficial for us to discover differentially further expressed genes in diseases, thereby guiding the entire process of disease discovery. In addition, epigenetic modification of genes, DNA methylation, and variable splicing may all play critical roles in RSA (not detailed in this article). Moreover, we are surprised to find an interaction between two types of cells in the maternal-fetal interface. For example, M1-type macrophages can transport miRNA to trophoblast cells through cell vesicles to play a physiological role. In this regard, recent research has focused on transmitting signaling molecules through vesicular transport to affect cell interactions. It is worth considering whether cell interaction can exert physiological effects through direct contact with proteins on the cell membrane or by secreting proteins. Although the study of non-coding RNA has become a hot topic, there are still some shortcomings in its clinical application. For example, non-coding is typically dynamic in the circulatory system, and continuous dynamic monitoring is crucial if ncRNA is to be used as a biomarker for predicting RSA. In addition, recombinant viruses are commonly used as vectors for transporting non-coding RNA, and the invasion of viruses may aggravate the immune response of the body. Therefore, non-coding RNA plays a crucial role in the pathological process of RSA and may be a potential target for the treatment of RSA. However, there is still a long way to go before it can be incorporated into clinical applications. We should invest more energy in gene research to demonstrate the clinical applicability of ncRNA in the diagnosis, treatment, and prognosis of pregnancy diseases, such as RSA and PE.
Acknowledgment
We are grateful to the participants for their contribution in accomplishing this study.
Footnotes
† Grant Support: This work has been supported by the National Natural Science Foundation of China (No. 82060294).
Contributor Information
Cen Tang, Obstetrics Department, Kunming Medical University Second Affiliated Hospital, Kunming, Yunnan, China.
Wanqin Hu, Obstetrics Department, Kunming Medical University Second Affiliated Hospital, Kunming, Yunnan, China.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. On behalf of all authors, the corresponding author states that there is no conflict of interest.
Author Contributions
Cen Tang conducted research and wrote articles, Wanqin Hu provided direction and revised the first draft. All authors have read and approved the final manuscript. Cen Tang is the First Author and Wanqin Hu is the Correspondent Author.
References
- 1. Naseema G, Ayalew T. Myeloproliferative neoplasms and pregnancy: overview and practice recommendations. Am J Hematol 2020; 96:354–366. [DOI] [PubMed] [Google Scholar]
- 2. Heitmann RJ, Weitzel RP, Feng Y, Segars JH, Tisdale JF, Wolff EF. Maternal T regulatory cell depletion impairs embryo implantation which can be corrected with adoptive t regulatory cell transfer. Reprod Sci 2017; 24:1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carp H. Immunotherapy for recurrent pregnancy loss. Best Pract Res Clin Obstet Gynaecol 2019; 60:77–86. [DOI] [PubMed] [Google Scholar]
- 4. Eunjung J, Roberto R, Lami Y, Nardhy G-L, Piya C, Adithep J, Francesca G, Offer E. The etiology of preeclampsia. Am J Obstet Gynecol 2022; 226:S844–S866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hajipour H, Nejabati HR, Latifi Z, Hamdi K, Bahrami-Asl Z, Fattahi A, Nouri M. Lymphocytes immunotherapy for preserving pregnancy: mechanisms and Challenges. Am J Reprod Immunol (New York, N.Y.: 1989) 2018; 80:80. [DOI] [PubMed] [Google Scholar]
- 6. Teng L, Liu P, Song X, Wang H, Sun J, Yin Z. Long non-coding RNA nuclear-enriched abundant transcript 1 (NEAT1) represses proliferation of trophoblast cells in rats with preeclampsia via the MicroRNA-373/FLT1 Axis. Med Sci Monit 2020; 26:e927305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tang J, Wang D, Lu J, Zhou X. MiR-125b participates in the occurrence of preeclampsia by regulating the migration and invasion of extravillous trophoblastic cells through STAT3 signaling pathway. J Recept Signal Transduct Res 2021; 41:202–208. [DOI] [PubMed] [Google Scholar]
- 8. Houshmand-Oeregaard A, Hjort L, Kelstrup L, Hansen NS, Broholm C, Gillberg L, Clausen TD, Mathiesen ER, Damm P, Vaag A. DNA methylation and gene expression of TXNIP in adult offspring of women with diabetes in pregnancy. PloS One 2017; 12:article e0187038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu L, Li N, Sun L, Zheng D, Shao G. Non-coding RNAs: The key detectors and regulators in cardiovascular disease. Genomics 2021; 113:1233–1246. [DOI] [PubMed] [Google Scholar]
- 10. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011; 146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang C, Changliang W, Huiyan S, Juexin W, Yanchun L, Yan W, Garry W. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 2020; 22:1706–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu Y, Xiaomin W, Xintong Z, Lin H, Jibiao W, Wenfeng Z, Huayao L, Chundi G, Changgang S. NcRNA-mediated ceRNA regulatory network: transcriptomic insights into breast cancer progression and treatment strategies. Biomed Pharmacother 2023; 162:114698. [DOI] [PubMed] [Google Scholar]
- 13.Shen J, Liang C, Su X, Wang Q, Ke Y, Fang J, Zhang D, Duan S. Dysfunction and ceRNA network of the tumor suppressor mi R-637 in cancer development and prognosis. Biomarker Res 2022, 10(1):72–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang C, Lim W, Park J, Park S, You S, Song G. Anti-inflammatory effects of mesenchymal stem cell-derived exosomal microRNA-146a-5p and microRNA-548e-5p on human trophoblast cells. Mol Hum Reprod 2019; 25:755–771. [DOI] [PubMed] [Google Scholar]
- 15.Yan ZQ, Hui LQ, Yao FY, Ren Chun E, Fang JA, Han MY. Decidual macrophages in recurrent spontaneous abortion. Front Immunol 2022; 13:994888–994888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L, Li Y, Sang Y, Li D-J, Du M. Crosstalk between trophoblasts and decidual immune cells: the cornerstone of maternal-fetal immunotolerance. Front Immunol 2021; 12:642392–642392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yongbo Z, Jiani S, Liping J. The N6-methyladenosine regulator ALKBH5 mediated stromal cell–macrophage interaction via VEGF signaling to promote recurrent spontaneous abortion: a bioinformatic and in vitro study. Int J Mol Sci 2022; 23:15819–15819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiuxiu X, Yonggang Z, Haiming W. Roles of HLA-G in the maternal-fetal immune microenvironment. Front Immunol 2020; 11:592010–592010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiqiang Z, Yan G, Min L, Zhaofeng Z, Junwei L, Yanyan M, Qianxi Z, Lin Z, Yupei S, Fujia C, Lingjin X, Lin H, Jing D. Integrated single-cell RNA-seq and DNA methylation reveal the effects of air pollution in patients with recurrent spontaneous abortion. Clin Epigenetics 2022; 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lisa M, Jennifer W. A meta-ethnography on the experience and psychosocial implications of providing abortion care. Soc Sci Med 1982; 2023:328. [DOI] [PubMed] [Google Scholar]
- 21. Tur-Torres MH, Garrido-Gimenez C, Alijotas-Reig J. Genetics of recurrent miscarriage and fetal loss. Best Pract Res Clin Obstet Gynaecol 2017; 42:11–25. [DOI] [PubMed] [Google Scholar]
- 22. Pollheimer J, Vondra S, Baltayeva J, Beristain AG, Knofler M. Regulation of placental extravillous trophoblasts by the maternal uterine environment. Front Immunol 2018; 9:2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y, Zhou J, Li MQ, Xu J, Zhang JP, Jin LP. MicroRNA-184 promotes apoptosis of trophoblast cells via targeting WIG1 and induces early spontaneous abortion. Cell Death Dis 2019; 10:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ojosnegros S, Seriola A, Godeau AL, Veiga A. Embryo implantation in the laboratory: an update on current techniques. Hum Reprod Update 2021; 27:501–530. [DOI] [PubMed] [Google Scholar]
- 25. Faas MM, De Vos P. Innate immune cells in the placental bed in healthy pregnancy and preeclampsia. Placenta 2018; 69:125–133. [DOI] [PubMed] [Google Scholar]
- 26. Abdolmohammadi Vahid S, Ghaebi M, Ahmadi M, Nouri M, Danaei S, Aghebati-Maleki L, Mousavi Ardehaie R, Yousefi B, Hakimi P, Hojjat-Farsangi M, Rikhtegar R, Yousefi M. Altered T-cell subpopulations in recurrent pregnancy loss patients with cellular immune abnormalities. J Cell Physiol 2019; 234:4924–4933. [DOI] [PubMed] [Google Scholar]
- 27. Volk N, Shomron N. Versatility of MicroRNA biogenesis. PloS One 2011; 6:e19391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ambros V. The functions of animal microRNAs. Nature 2004; 431:350–355. [DOI] [PubMed] [Google Scholar]
- 29. Lee RC, Feinbaum RL, Ambros V, The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 30. Chen X, Guo DY, Yin TL, Yang J. Non-coding RNAs regulate placental trophoblast function and participate in recurrent abortion. Front Pharmacol 2021; 12:646521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiaoxiao Z, Haiping L, Zhen Z, Ran W, Xianbin Z, Zhaoxia W, Lin Z, Qiang G, Yunhong Z, Chu C, Li W, Xia L. MiR-103 protects from recurrent spontaneous abortion via inhibiting STAT1 mediated M1 macrophage polarization. Int J Biol Sci 2020; 16:2248–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jian-Mei W, Yan G, Yao Z, Qian Y, Xuan Z, Lirong Y, Jian W. Deep sequencing identification of differentially expressed miRNAs in decidua and villus of recurrent miscarriage patients. Arch Gynecol Obstet 2016; 293:1125–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Donker RB, Mouillet JF, Chu T, Hubel CA, Stolz DB, Morelli AE, Sadovsky Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol Hum Reprod 2012; 18:417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 2018; 75:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ding J, Cheng Y, Zhang Y, Liao S, Yin T, Yang J. The mi R-27a-3p/USP25 axis participates in the pathogenesis of recurrent miscarriage by inhibiting trophoblast migration and invasion. J Cell Physiol 2019; 234:19951–19963. [DOI] [PubMed] [Google Scholar]
- 36. Pingping P, Huamei S, Chenghong X, Wenfei Z, Huigai M, Dandan X, Jingqiong Z, Xiaoqing Y, Aihua C, Jing T, Jufang Q. mi R-146a-5p-mediated suppression on trophoblast cell progression and epithelial-mesenchymal transition in preeclampsia. Biol Res 2021;5454(1):30–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tao J, Xia L-Z, Liang L, Chen Y, Wei D, Meng J, Wu SY, Wang Z. MiR-124-3p promotes trophoblast cell HTR-8/SVneo pyroptosis by targeting placental growth factor. Placenta 2020; 101:176–184. [DOI] [PubMed] [Google Scholar]
- 38. Dong X, Yang L, Wang H. mi R-520 promotes DNA-damage-induced trophoblast cell apoptosis by targeting PARP1 in recurrent spontaneous abortion (RSA). Gynecol Endocrinol 2017; 33:274–278. [DOI] [PubMed] [Google Scholar]
- 39. Zhu Y, Lu H, Huo Z, Ma Z, Dang J, Dang W, Pan L, Chen J, Zhong H. MicroRNA-16 inhibits feto-maternal angiogenesis and causes recurrent spontaneous abortion by targeting vascular endothelial growth factor. Sci Rep 2016; 6:35536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qin W, Tang Y, Yang N, Wei X, Wu J. Potential role of circulating microRNAs as a biomarker for unexplained recurrent spontaneous abortion. Fertil Steril 2016; 105:1247–1254.e3. [DOI] [PubMed] [Google Scholar]
- 41. Barth DA, Drula R, Ott L, Fabris L, Slaby O, Calin GA, Pichler M. Circulating non-coding RNAs in renal cell carcinoma-pathogenesis and potential implications as clinical biomarkers. Front Cell Dev Biol 2020; 8:828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zha Y, Li Y, Lin T, Chen J, Zhang S, Wang J. Progenitor cell-derived exosomes endowed with VEGF plasmids enhance osteogenic induction and vascular remodeling in large segmental bone defects. Theranostics 2021; 11:397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gaynullina DK, Schubert R, Tarasova OS. Changes in endothelial nitric oxide production in systemic vessels during early ontogenesis—a key mechanism for the perinatal adaptation of the circulatory system. Int J Mol Sci 2019; 20:1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jinli D, Yan Z, Xiaopeng C, Yi Z, Sisi Y, Jiayu W, Sainan Z, Tailang Y, Chaogang Y, Jing Y. Extracellular vesicles derived from M1 macrophages deliver mi R-146a-5p and mi R-146b-5p to suppress trophoblast migration and invasion by targeting TRAF6 in recurrent spontaneous abortion. Theranostics 2021; 11:5813–5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vilella F, Moreno-Moya JM, Balaguer N, Grasso A, Herrero M, Martínez S, Marcilla A, Simón C. Hsa-mi R-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 2015; 142:3210–3221. [DOI] [PubMed] [Google Scholar]
- 46. Gillet V, Ouellet A, Stepanov Y, Rodosthenous RS, Croft EK, Brennan K, Abdelouahab N, Baccarelli A, Takser L. miRNA profiles in extracellular vesicles from serum early in pregnancies complicated by gestational diabetes mellitus. J Clin Endocrinol Metab 2019; 104:5157–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pan L, Xiao X, Zhao Y, Yin L, Fu M, Zhang X, Jiang P. The functional roles of long noncoding RNA DANCR in Human Cancers. J Cancer 2020; 11:6970–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Richard JLC, Eichhorn PJA. Deciphering the roles of lncRNAs in breast development and disease. Oncotarget 2018; 9:20179–20212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinn Jeffrey J, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016; 17:47–62. [DOI] [PubMed] [Google Scholar]
- 50. Wang L, Tang H, Xiong Y, Tang L. Differential expression profile of long noncoding RNAs in human chorionic villi of early recurrent miscarriage. Clinica Chim Acta 2017; 464:17–23. [DOI] [PubMed] [Google Scholar]
- 51. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xiang H, Yan H, Sun B, Feng F, Chen P. Decreased expression of long non-coding RNA SNHG7 cause recurrent spontaneous abortion through suppression proliferation and invasion of trophoblast cells via mi R-34a. Am J Transl Res 2019; 11:463–472. [PMC free article] [PubMed] [Google Scholar]
- 53. Yang D, Ding J, Wang Y, Yuan M, Xian S, Zhang L, Liu S, Dai F, Wang F, Zheng Y, Zhao X, Liao S, et al. YY1-PVT1 affects trophoblast invasion and adhesion by regulating mTOR pathway-mediated autophagy. J Cell Physiol 2020; 235:6637–6646. [DOI] [PubMed] [Google Scholar]
- 54. Zhang Y, Jin F, Li XC, Shen FJ, Ma XL, Wu F, Zhang SM, Zeng WH, Liu XR, Fan JX, Lin Y, Tian FJ. The YY1-HOTAIR-MMP2 signaling axis controls trophoblast invasion at the maternal-fetal interface. Mol Ther 2017; 25:2394–2403. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55. Huang Z, Du G, Huang X, Han L, Han X, Xu B, Zhang Y, Yu M, Qin Y, Xia Y, et al. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine 2018; 38:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bauer-Negrini G, Cordenonsi da Fonseca G, Gottfried C, Herbert J. Usability evaluation of circRNA identification tools: Development of a heuristic-based framework and analysis. Comput Biol Med 2022; 147:105785. [DOI] [PubMed] [Google Scholar]
- 57. Chen L, Huang C, Wang X, Shan G. Circular RNAs in Eukaryotic Cells. Curr Genomics 2015; 16:312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979; 280:339–340. [DOI] [PubMed] [Google Scholar]
- 59.Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara AC, Fenske P, Trimbuch T, Zywitza V. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017; 357:eaam 8526. [DOI] [PubMed] [Google Scholar]
- 60. Li CH, Xiao Z, Tong JH, To KF, Fang X, Cheng AS, Chen Y. EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int J Cancer 2017; 140:120–129. [DOI] [PubMed] [Google Scholar]
- 61. Dong W, Dai ZH, Liu FC, Guo XG, Ge CM, Ding J, Liu H, Yang F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine 2019; 45:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jeck W, Sorrentino J, Wang K, Slevin M, Burd C, Liu J. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan SS, Andreas W, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125–1134. [DOI] [PubMed] [Google Scholar]
- 64. Xin Z, Ma Q, Ren S, Wang G, Li F. The understanding of circular RNAs as special triggers in carcinogenesis. Brief Funct Genom 2017; 16:80–86. [DOI] [PubMed] [Google Scholar]
- 65. Qian Y, Wang X, Ruan H, Rui C, Mao P, Cheng Q, Jia R. Circular RNAs expressed in chorionic villi are probably involved in the occurrence of recurrent spontaneous abortion. Biomed Pharmacother 2017; 88:1154–1162. [Google Scholar]
- 66.Qinghua L, Yangyang H, Peng X, Lingxuan Y, Yanru S, Cuijuan Z, Yuhan M, Weiguo F, Zhifang P, Zhiqin G, Jie L, Weiwei Y. Elevated microRNA-125b inhibits cytotrophoblast invasion and impairs endothelial cell function in preeclampsia. Cell Death Discov 2020; 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lihua Z, Lijuan S, Wenfeng Y, Shuping L, Xinmei L, Zonghao Z. Circular RNA PUM1 (Circ PUM1) attenuates trophoblast cell dysfunction and inflammation in recurrent spontaneous abortion via the microRNA-30a-5p (mi R-30a-5p)/JUNB axis. J Bioeng 2021; 12:6878–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Z, Zhou G, Tao F, Cao Y, Han W, Li Q. circ-ZUFSP regulates trophoblasts migration and invasion through sponging mi R-203 to regulate STOX1 expression. Biochem Biophys Res Commun 2020; 531:472–479. [DOI] [PubMed] [Google Scholar]
- 69. Gao Y, Tang Y, Sun Q, Guan G, Wu X, Shi F, Zhou Z, Yang W. Circular RNA FOXP1 relieves trophoblastic cell dysfunction in recurrent pregnancy loss via the mi R-143-3p/S100A11 cascade. Bioengineered 2021; 12:9081–9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wu X, Sui Z, Zhang H, Wang Y, Yu Z. Integrated analysis of lncRNA-mediated ceRNA network in lung adenocarcinoma. Front Oncol 2020; 10:554759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Xu H, Zhou M, Cao Y, Zhang D, Han M, Gao X, Xu B, Zhang A. Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene 2019; 720:144056. [DOI] [PubMed] [Google Scholar]
- 72. Huang Y, Hao J, Liao Y, Zhou L, Wang K, Zou H, Hu Y, Li J. Transcriptome sequencing identified the ceRNA network associated with recurrent spontaneous abortion. BMC Med Genomics 2021; 14:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xupeng Z, Ting G, Wenjing W, Chen Z, Yue D, Shengchen G, Zhiqian X, Yanshe X, Zicong L, Gengyuan C, Bin H, Linjun H, Zhenfang W. Integrated insight into the molecular mechanisms of spontaneous abortion during early pregnancy in pigs. Int J Mol Sci 2021; 22:6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qian Z, Zhenzhen W, Xianghong C, Haiyin W. lncRNA DANCR promotes the migration an invasion and of trophoblast cells through microRNA-214-5p in preeclampsia. Bioengineered 2021; 12:9424–9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sheng F, Sun N, Ji Y, Ma Y, Ding H, Zhang Q, Yang F, Li W. Aberrant expression of imprinted lncRNA MEG8 causes trophoblast dysfunction and abortion. J Cell Biochem 2019; 120:17378–17390. [DOI] [PubMed] [Google Scholar]
- 76. Tang M, Bai L, Wan Z, Wan S, Xiang Y, Qian Y, Cui L, You J, Hu X, Qu F, Zhu Y. circRNA-DURSA regulates trophoblast apoptosis via mi R-760-HIST1H2BE axis in unexplained recurrent spontaneous abortion. Mol Ther Nucleic Acids 2021; 26:1433–1445 production 2019, 157, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xiaoxiao X, Sha L, Ziwen X. Analysis of a circRNA-, miRNA-, and mRNA-associated ceRNA network reveals potential biomarkers in preeclampsia a ceRNA network in preeclampsia. Ann Med 2021;53(1):2354–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peigen C, Tingting L, Yingchun G, Lei J, Yanfang W, Cong F. Construction of circulating microRNAs-based non-invasive prediction models of recurrent implantation failure by network analysis. Front Genet 2021; 12:712150–712150. [DOI] [PMC free article] [PubMed] [Google Scholar]