Abstract
Two-component systems are widely distributed in prokaryotes where they control gene expression in response to diverse stimuli. To study the role of the sixteen putative two-component systems of Listeria monocytogenes systematically, in frame deletions were introduced into 15 out of the 16 response regulator genes and the resulting mutants were characterized. With one exception the deletion of the individual response regulator genes has only minor effects on in vitro and in vivo growth of the bacteria. The mutant carrying a deletion in the ortholog of the Bacillus subtilis response regulator gene degU showed a clearly reduced virulence in mice, indicating that DegU is involved in the regulation of virulence-associated genes.
All free-living bacteria have the ability to respond and adapt rapidly to changes in their environment by coordinate changes in the expression of sets of genes. The signal transduction mechanisms allowing bacteria to modulate gene expression in response to diverse stimuli often involve two-component systems (TCSs), which are composed of a sensor kinase (HK) and a cognate response regulator (RR) (16). The sensor histidine kinase is composed of an N-terminal input domain which is frequently exposed to the periplasmic or extracellular space and a highly conserved cytoplasmic kinase domain which, in the presence of the appropriate stimulus, autophosphorylates at a highly conserved histidine residue. The phosphate group is subsequently transferred to an aspartic acid residue in the receiver domain of the cognate response regulator. Phosphorylation of the RR triggers a conformational change of the protein activating its output domain which frequently has DNA binding capability.
Listeria monocytogenes, a facultative intracellular bacterial pathogen (43) is known to either live in the environment or to infect humans and other mammals, thereby encountering very different microenvironments. The expression of the listerial virulence genes is coordinately regulated to allow the bacteria to proceed through their intracellular life cycle. Central to virulence gene regulation is a protein called PrfA (25), which binds to palindromic DNA sequences in the upstream regions of most known virulence genes.
So far, TCSs have not been studied comprehensively in L. monocytogenes. The availability of the complete genomic sequence of the L. monocytogenes strain EGD-e and the related apathogenic species Listeria innocua, allowed the in silico identification of 16 TCSs (15) in L. monocytogenes of which only one was absent in L. innocua.
In order to study the role of the listerial TCSs for in vitro and in vivo survival and growth of L. monocytogenes more systematically, we constructed mutants with in-frame deletions in 15 out of the 16 response regulator genes. The characterization of the mutants demonstrated that only the deletion of the degU gene had a significant effect on the in vitro and in vivo growth of L. monocytogenes. Furthermore, we show that deletion of degU renders L. monocytogenes nonmotile due to the lack of flagellum expression.
In silico analysis of the TCS genes identified in the genome sequence of L. monocytogenes.
The genome sequence of L. monocytogenes EGD-e contains 15 and 16 open reading frames (ORFs) encoding two-component histidine kinases and response regulators, respectively, including the chemotaxis regulatory system CheYA (Lmo0691/Lmo0692) (12, 15) (Table 1). With the exception of lmo2515 all the response regulator genes are genetically linked to an ORF encoding a cognate histidine kinase. Out of these TCSs Lmo1377/Lmo1378 (LisRK) and Lmo2422/2421 (CesRK) have already been characterized in some detail and were shown to contribute to stress tolerance and antibiotic resistance and to affect virulence (7, 8, 23). Lmo0051/Lmo0050 (AgrAC) which was reported to be required for the full virulence of L. monocytogenes in mice (3) is homologous to the AgrAC TCS of Staphylococcus aureus which is involved in quorum sensing and virulence gene regulation (2). Several TCSs identified showed significant homology to well-characterized TCSs from B. subtilis: Lmo1948/Lmo1947 (ResDE) is homologous to the TCS ResDE (40) which participates in the regulation of both aerobic and anaerobic respiration, while Lmo2501/Lmo2500 (PhoPR) is homologous to the phosphate starvation regulatory system PhoPR. Lmo0287/Lmo0288 is orthologous to the essential TCS YycFG which is probably involved in the regulation of genes involved in cell wall metabolism and membrane protein synthesis (10, 21). The orphan response regulator Lmo2515 (DegU) is highly homologous to DegU. In B. subtilis DegUS is a pleiotropic regulatory system which is involved in the induction of extracellular degradative enzyme synthesis, the expression of late competence genes, and down-regulation of the σD regulon (18, 27). Lmo2678/Lmo2679 (KdpED) is homologous to the KdpED TCS of Escherichia coli which contributes to the adaptation to high-osmolarity conditions by regulating the expression of a high-affinity potassium uptake system encoded by the kdpABC genes (6, 34). In L. monocytogenes orthologs of kdpABC are located adjacent to lmo2678/lmo2679. From the remaining L. monocytogenes response regulators four contain OmpR-like DNA-binding output domains (Lmo1060, Lmo1507, Lmo1745, Lmo2583), one (Lmo1022) has an output domain of the NarL type, and response regulator Lmo1022 contains an exceptionally large output domain with an AraC-like helix-turn-helix motif located at the very C terminus. The output domain of the putative response regulator Lmo1172 does not contain a common DNA-binding motif but carries the RNA-binding ANTAR domain which is present in the AmiR protein of Pseudomonas aeruginosa and other transcription antitermination regulatory proteins (31, 36).
TABLE 1.
In silico analysis of the L. monocytogenes TCSsa
| Histidine kinase | HK group | TMD(s) (extracytoplasmic loop) | Additional sensing domain | Response regulator | Output domain |
|---|---|---|---|---|---|
| AgrB (Lmo0050) | 6 | AgrA (Lmo0051) | LytTr | ||
| YycG (Lmo0288) | IIIA | 2 (158) | PAS | YycF (Lmo0287) | OmpR |
| CheA (Lmo0692) | CheY (Lmo0691) | ||||
| Lmo1021 | II | 2 (22) | Lmo1022 | NarL | |
| Lmo1061 | IIIA | 2 (125) | Lmo1060 | OmpR | |
| Lmo1173 | Lmo1172 | ANTAR | |||
| LisK (Lmo1378) | IIIA | 2 (144) | LisR (Lmo1377) | OmpR | |
| Lmo1508 | IIIA | 2 (142) | Lmo1507 | OmpR | |
| Lmo1741 | IIIA | 2 (10) | Lmo1745 | OmpR | |
| ResE (Lmo1947) | IIIA | 2 (146) | ResD (Lmo1948) | OmpR | |
| Lmo2011 | I | 2 (252) | Lmo2010 | AraC-HTH | |
| CesK (Lmo2421) | IIIA | 2 (40) | CesR (Lmo2422) | OmpR | |
| PhoR (Lmo2500) | IIIA | 2 (147) | PAS | PhoP (Lmo2501) | OmpR |
| DegU (Lmo2515) | NarL | ||||
| Lmo2582 | IIIA | 2 (137) | Lmo2583 | OmpR | |
| KdpD (Lmo2679) | IIIA | 4 | KdpE (Lmo2678) | OmpR |
Classification of the histidine kinases is based on the sequence motifs surrounding the phosphorylated histidine residue according to an analysis of Fabret and Hoch (10), who compared the TCS proteins of B. subtilis. For the prediction of transmembrane domains (TMDs) the “DAS”-Transmembrane Prediction server (http://www.sbc.su.se/∼miklos/DAS/tmdas.cgi) was used. When two TMDs have been predicted in the N-terminal domain, the size of the extracytoplasmic loops (in amino acids) confined by the TMDs is given in parentheses. The output domains are classified as OmpR- or NarL-like according to structural similarities with the E. coli proteins (10). The prediction of PAS domains (41) in the sensor proteins and DNA-binding motifs in the response regulators differing from the OmpR and NarL types was performed using SMART—Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de).
Construction of L. monocytogenes mutants with in-frame deletions in 15 out of 16 two-component response regulator genes.
We constructed individual mutants with large in-frame deletions in the response regulator genes in the strain L. monocytogenes Sv1/2a EGD using a two step integration/excision procedure (5) which is based on the mutagenesis plasmid pLSV1 (45). The respective knock-out plasmids were constructed as follows: DNA fragments of approximately 400 bp derived from the upstream and downstream regions, respectively, of the respective regulator gene under study were amplified from chromosomal DNA using the appropriate primer pairs, called geneN1/geneN2 (upstream fragment; Table 2) and geneC1/geneC2, (downstream fragment; Table 2). Both fragments were cut at the KpnI site provided by the geneN2 and geneC1 primers and ligated. Then PCR amplifications were performed with the outmost primers using the ligation mixture as template. The obtained fragments were cloned via the BamHI and EcoRI sites introduced by primers geneN1 and geneC2, respectively, into pLSV1 giving rise to plasmids pLSVΔTCS which were transformed into L. monocytogenes EGD and chromosomal integration was then induced by growing the bacteria at 43°C. The integration mutants were subcultured at 30°C over several days and then screened for the loss of erythromycin resistance. Sensitive clones were screened by PCR to identify mutants with correct in-frame deletions. With one exception this strategy allowed the almost complete deletion of the coding region of the respective response regulator gene with only several base triplets encoding few amino acids from the N and C terminus still present in a short ORF.
TABLE 2.
Primers used for the construction of the L. monocytogenes ΔTCS-RR mutants
| Name of gene to be deleteda | Primer name | Sequence 5′-3′ |
|---|---|---|
| resD | resDN1 | GATC GGATCC AAGATAAAGCCATTGTAGAAATTAC |
| resDN2 | GATC GGTACC ACTCATAAGTACTCATCCCCTAAC | |
| resDC1 | GATC GGTACC GATTAATAAAACAGACTAAGATAACG | |
| resDC2 | GATC GAATTC AGTATCTGGTGACTGTGATTGATAG | |
| lmo1060 | 1060N1 | GATC GGATCC ACGTGAAACAATCAACAAGATTTA |
| 1060N2 | GATC GGTACC TTCCATATTTGCCTCCTACTGTTC | |
| 1060C1 | GATC GGTACC GAATAAAGTGAGAACCAGAATTAG | |
| 1060C2 | GATC GAATTC CTGTTCCACCTGAATAATCCG | |
| phoP | phoPN1 | GATC GGATCC GGCTTGTAGTTTTGGGTGCTCGTGC |
| phoPN2 | GATC GGTACC TACCAACGTACTTCCCATCCC | |
| phoPC1 | GATC GGTACC GGCTTCGGTTATAAAATGGAGAACG | |
| phoPC2 | GATC GAATTC GATTGAAATACCAACGCTTTCCCC | |
| lmo2422 (cesR) | cesRN1 | GATC GGATCC ATATCATGTATGAGGCTTATAAATTG |
| cesRN2 | GATC GGTACC TGTCATACTCATTGTCCTTTCCAG | |
| cesRC1 | GATC GGTACC ATCTAAACTGGAACTGTTGTTTACG | |
| cesRC2 | GATC GAATTC ACGTTCCAGTTCTTGAATTTCCGG | |
| lmo2678 (kdpE) | 2678N1 | GATC GGATCC AATTCCGGAAAATCGTTTGGCAG |
| 2678N2 | GATC GGTACC ATTACTGATGCCATCTTGATCTTC | |
| 2678C1 | GATC GGTACC TAATAATATGTTATGATAAAAATAAAC | |
| 2678C2 | GATC GAATTC ACAAATAAGCAAGACTCCGAGTAC | |
| cheY | cheYN1 | GATC GGATCC GATGCAGATATGGCTGCTGAAATG |
| cheYN2 | GATC GGTACC CAACATAAAATACTACTCCCATTC | |
| cheYC1 | GATC GGTACC AAGTAGAGAAATGGAGGTGGATAGC | |
| cheYC2 | GATC GAATTC TTGCTCTGGGTGTAAAAGTGCTTC | |
| lmo0287 | 0287N1 | GATC GGATCC CAAGTAGCGAATTTGCTGATTATC |
| 0287N2 | GATC GGTACC TGCCATTTTTTTACAGTCCTCTC | |
| 0287C1 | GATC GGTACC GAATAAATAACAAGAAGTGGTTTC | |
| 0287C2 | GATC GAATTC TTTCTTCTCTAGCATTATAATCC | |
| lisR | lisRN1 | GATC GGATCC AATTCCAGTACCAACATTCACTGC |
| lisRN2 | GATC GGTACC ATTCATTTGGCCTAACCCTCTC | |
| lisRC1 | GATC GGTACC ACATGACGACTAGCCCATTTTCC | |
| lisRC2 | GATC GAATTC GGTTTATTCATAATAAACTTATC | |
| lmo2515 (degU) | degUN1 | GATC GGATCC AATAATGATCCAACATAAGGAACC |
| degUN2 | GATC GGTACC TGCCATAATGACTACTCCTCCTTC | |
| degUC1 | GATC GGTACC GCGGTGGTAACGGCAATCAAGCAC | |
| degUC2 | GATC GAATTC AATTCTCCTGGGGCAGGTTGAGAG | |
| lmo0051 (agrA) | 51N1 | GATC GGATCC GCAATTTTGGAAGTAGTTGAACCG |
| 51N2 | GATC GGTACC CGGTAGCATAAATTCATCCCC | |
| 51C1 | GATC GGTACC TTATAAAAGTGGCCTTAAAGG | |
| 51C2 | GATC GAATTC CTTCTTCACTACGCTTAATCCG | |
| lmo1172 | 1172N1 | GATC GGATCC GCTCAAATCGCTGAAGGTGCGC |
| 1172N2 | GATC GGTACC CTTCCATTCATTCCTGTCACGTCC | |
| 1172C1 | GATC GGTACC ATGACGGATGACTAAAACGATTCG | |
| 1172C2 | GATC GAATTC GAATTAGGACGGCAATAACTCGC | |
| lmo1507 | 1507N1 | GATC GGATCC ATCACTACTTGAAGCCTCTTC |
| 1507N2 | GATC GGTACC TTTCATATTCTGTTCTCACTCC | |
| 1507C1 | GATC GGTACC ACCGTCTGGGGTGTTGGCTAC | |
| 1507C2 | GATC GAATTC TTCTTACTAACCGTTTCACCAC | |
| lmo1745 | 1745N1 | GATC GGATCC TTGTATTCTTTTTAGCAACAGG |
| 1745N2 | GATC GGTACC TACCATTTCTAGTGCCTCCC | |
| 1745C1 | GATC GGTACC GAATGACGAAAAAACAGGGAG | |
| 1745C2 | GATC GAATTC CTAAAGCTCGACCTTTTTGAG | |
| lmo1022 | 1022N1 | GATC GGATCC GTTTGCAAAAAGGCATTGAGG |
| 1022N2 | GATC GGTACC AACTACTTCCATGTCATCTTGC | |
| 1022C1 | GATC GGTACC AAGTAGCAAGGAGGTAACGTAC | |
| 1022C2Bam | GATC GGATCC ACGTCGTCCCATGTCTCGCTC | |
| lmo2010 | 2010N1 | GATC GGATCC GTACCATCATTCTGTTTCAAG |
| 2010N2 | GATC GGTACC CCTTGGTATCACACTGTCTGTC | |
| 2010C1 | GATC GGTACC TAAGATTATTTCTAATATTTGTC | |
| 2010C2Bam | GATC GGATCC ACCAAGAAACAGCCCTACG | |
| lmo2583 | 2583N1 | GATC GGATCC CAAAATTCTGCTACATTTCATC |
| 2583N2 | GATC GGTACC TTTCATTTTCACCGCTCACC | |
| 2583C1 | GATC GGTACC AGATGAAATCGTTATACAGTCG | |
| 2583C2 | GATC GAATTC GGTATTTCGCACATCATTATC |
According to Glaser et al. (15).
In accordance with previous observations of Kallipolitis and Ingmer (22) who inactivated several response regulator genes of L. monocytogenes by insertion mutagenesis we were unable to generate an in-frame deletion in the gene lmo0287. More than 50 colonies derived from three independent experiments were tested and proved to be revertants to the wild type. The gene lmo0287 is orthologous to the essential response regulator gene yycF of B. subtilis which has recently been shown to be involved in regulation of the cell division operon ftsAZ (13) and of several genes involved in cell wall metabolism (21). Orthologs of yycF and the gene encoding the cognate histidine kinase, yycG, are present in all the available genomes of low-G+C gram-positive bacteria, and it has been reported that yycF is also essential in Staphylococcus aureus, Streptococcus pneumoniae, and Enterococcus faecalis (26, 28, 42). Besides yycFG only few essential TCSs have been identified including the CtrA/CckA system of Caulobacter crescentus (19) which regulates the cell cycle by controlling DNA replication, DNA methylation, and flagellar biogenesis, as well as the essential response regulators MtrA of Mycobacterium tuberculosis (46) and HP166 and HP1043 of Helicobacter pylori, the functions of which remain to be identified (4).
In vitro growth characteristics and motility of the ΔTCS-RR mutants.
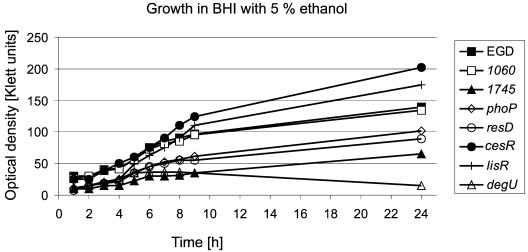
The 15 ΔTCS-RR mutants displayed normal growth in brain heart infusion (BHI) medium at 37°C, 20°C, and 43°C under aerobic conditions. Similarly, addition of 9% NaCl and 0.025% H2O2 to BHI broth had no effect on the growth of any of the mutants, demonstrating that none of the TCSs plays a significant role for the survival under these stress conditions. Interestingly, in-frame deletion of the L. monocytogenes ortholog of kdpE which in E. coli is involved in maintenance of cell turgor by regulating the expression of a high-affinity K+ transporter encoded by the kdpABC genes, did not affect growth at high osmolarity as was already observed by others (6). In B. subtilis, which does not contain orthologs of kdpED and kdpABC, the influx of potassium ions is mediated by the K+ transporters KtrAB and KtrBC (20), orthologs of which are present in L. monocytogenes, which might explain the unaltered phenotype of the kdpE mutant in response to high osmolarity. The addition of 5% ethanol to BHI markedly reduced the growth of the ΔresD, ΔphoP, and Δlmo1745 mutants and led to enhanced growth of the strains ΔcesR and ΔlisR (Fig. 1), according to earlier findings (7, 22). The ΔdegU mutant did not grow at all in the presence of 5% ethanol. In addition to contributing to tolerance to ethanol the TCSs LisRK and CesRK have been shown to influence the sensitivity of L. monocytogenes to various antibiotics and CesRK has been suggested to play a role in sensing and responding to changes in cell wall integrity (8, 23). Similarly, degU, lmo1745, resD, and phoP might directly or indirectly have an impact on the composition of the cell envelope resulting in an altered tolerance to ethanol stress. When cells were cultured anaerobically, no difference in growth was observed between the L. monocytogenes EGD wild type and the ΔTCS-RR mutants. In contrast to B. subtilis where nitrate respiration is controlled by ResDE, L. monocytogenes is unable to perform nitrate respiration since it lacks a dissimilative nitrate reductase (15). Our results indicate that neither ResDE nor the other TCSs of L. monocytogenes are involved in the regulation of the various fermentative pathways used by this organism.
FIG. 1.
Growth of the L. monocytogenes ΔTCS-RR mutants in BHI in the presence of 5% ethanol. Overnight cultures of all mutant strains were diluted 1:50 and cultured aerobically in BHI medium supplemented with 5% ethanol at 37°C. Optical density was measured every hour. The growth curves of a subset of mutants are shown.
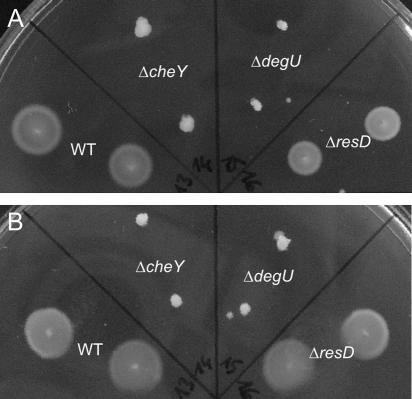
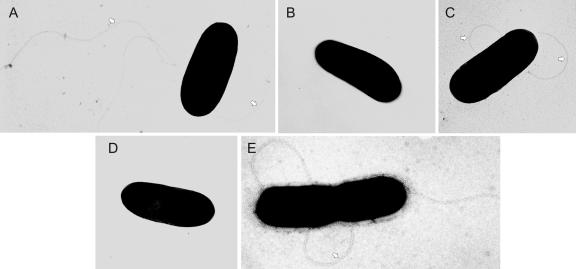
Motility of the L. monocytogenes ΔTCS-RR mutants was determined by swim-plate assays on semisolid TSB agar. Swimming of the bacteria on and within the agar resulted in halos around the point of inoculation. As already described (9), the L. monocytogenes ΔcheY mutant showed a defect in the chemotactic behavior since the mutant lacks the response regulator protein interacting with the flagellar motor. However, swimming was also completely abolished in the ΔdegU mutant, while the other ΔTCS-RR mutants behaved like the wild-type bacteria, irrespective of whether they were cultivated at 24°C or 37°C (Fig. 2). As was observed previously for laboratory-adapted strains (44), the L. monocytogenes EGD strain used here showed temperature-independent swimming on semisolid agar plates. Electron microscopy demonstrated that in contrast to the ΔcheY mutant which produces flagella, the ΔdegU mutant did not produce flagella indicating a role of degU in flagellar gene expression (Fig. 3). In B. subtilis the DegUS TCS is involved in the complex regulation of transition state-specific processes including the synthesis of degradative enzymes, the development of natural competence and motility (29) and also contributes to gene expression under high-osmolarity conditions (38). Motility is controlled by the phosphorylated form of DegU which negatively regulates the expression of the alternative sigma factor σD controlling the transcription of genes involved in flagellar biosynthesis and chemotaxis (1, 27). In L. monocytogenes which lacks a homolog of σD, the mechanisms of flagellar regulation have not yet been investigated thoroughly. It was shown recently that the repressor protein MogR is involved in the downregulation of expression of the flagellin gene flaA at low temperature (17). A role of DegU for the motility of L. monocytogenes was recently also presented by Knudsen et al., (24) by showing that DegU is necessary for the transcription of the flagellin-encoding flaA gene but the details of how DegU contributes to the process of flagellar gene regulation remain to be elucidated.
FIG. 2.
Swimming of L. monocytogenes mutants in soft agar. The L. monocytogenes wild-type (WT) strain EGD and the ΔTCS-RR mutants were stabbed into semisolid agar plates with 0.25% agar and incubated at 24°C (A) or 37°C (B) for 24 h. The results for L. monocytogenes EGD and the ΔdegU, ΔcheY, and ΔresD mutants are presented. Pictures were taken with a standard digital camera (Camedia C300; OLYMPUS).
FIG. 3.
Electron micrographs of the flagellated L. monocytogenes wild-type (WT) strain EGD (A and C) and the nonflagellated ΔdegU mutant (B and D). The flagellated but nonmotile ΔcheY mutant is included as a control (E). Bacteria were grown at 24°C (A, B, and E) or 37°C (C and D) and examined under a transmission electron microscope (TEM 100; Zeiss) after negative staining with 0.5% uranyl acetate for 1 min. The flagella present in the WT and ΔcheY strain are clearly visible (arrowheads). Original magnification: 20,000-fold.
Expression of virulence factors and interaction with mammalian cells.
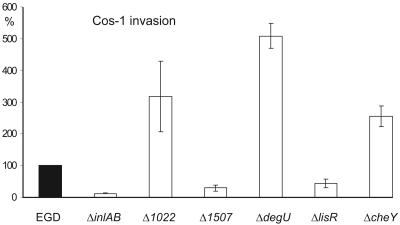
To test whether the inactivation of individual TCS in L. monocytogenes would affect the expression of known virulence factors, we checked culture supernatants and whole-cell extracts for the presence of the proteins listeriolysin O (LLO), phosphatidylcholine-specific phospholipase C (PlcB), and ActA by immunoblot analysis and found that the deletion of none of the response regulators had any obvious effect on the expression of three of the most important virulence factors of L. monocytogenes at least under in vitro growth (data not shown). The interaction of the ΔTCS mutants with mammalian cells and their ability to proceed through the well described L. monocytogenes intracellular life cycle were analyzed using four different in vitro cell culture assays. We tested (i) invasion into Cos-1 fibroblast cells, (ii) intracellular growth in J774 macrophages, (iii) F-actin tail formation. and (iv) cell-to-cell spread (both in Caco-2 cells). The invasiveness of most of the mutants for Cos-1 fibroblasts was essentially like that of the wild-type bacteria. Only two mutants (Δ1507 and ΔlisR) showed an about threefold reduction in invasion into Cos-1 fibroblasts, which was, however, not as prominent when compared to a ΔinlAB strain lacking internalin and InlB expression (5). The reasons for these changes in invasion capability are not clear at the moment. Interestingly, the nonmotile L. monocytogenes ΔcheY and ΔdegU mutants as well as the Δ1022 mutant showed a slightly increased ability to invade Cos-1 fibroblast cells than the wild-type strain (Fig. 4). In contrast, Dons et al. (9) reported that L. monocytogenes ΔcheY, ΔcheA, and ΔflaA mutants showed significantly reduced association with and invasion into Caco-2 cells when the bacteria were grown at 24°C. When the mutants were centrifuged onto the monolayer, the defect in cell association was less pronounced indicating a role for motility to ensure the initial contact of the bacteria with the host cell. Consequently, when bacteria were grown at 37°C which is the nonpermissive temperature for flagella expression, no differences in cell association between the wild-type strain and the flaA mutant were observed (9). In our experimental settings L. monocytogenes was grown at 37°C prior to invasion since temperature-dependent repression of flagellar expression was no longer observed in the wild-type strain used. While in case of the ΔdegU mutant other features in addition to the lack of flagellar-based motility might account for the increased invasion into Cos-1 cells, the different invasion capabilities into Caco-2 and Cos-1 cells, respectively, of cheY in frame deletion mutants in the L. monocytogenes strains 12067 and EGD remain unexplained.
FIG. 4.
Invasive capacity for Cos-1 fibroblast cells of the ΔTCS-RR mutants. Invasion assays were performed with all the L. monocytogenes ΔTCS-RR mutants, and the results for a subset of the mutants are shown. For invasion assays, the cells were inoculated with bacteria at a multiplicity of infection of about 10 bacteria per cell in RPMI 1640 medium without fetal calf serum. After centrifuging the bacteria onto the Cos-1 cells for 10 min, the cells were further incubated together with the bacteria for 50 min at 37°C to allow entry. The cells were then washed three times with phosphate buffered saline (PBS) and inoculated with fresh RPMI 1640 medium containing gentamicin (50 μg ml−1) for 1 h at 37°C and then lysed by adding a PBS/Triton X-100 solution (0.1%) and the titer of intracellular bacteria was determined by spreading onto BHI plates. The numbers of bacteria recovered (expressed as percentages of the number recovered for the control, taken as 100%) are shown. Values are means and standard deviations (error bars) of the results of one representative experiment run in triplicate. Open bars indicate that differences from the control are statistically significant.
All mutants replicated in J774 macrophages essentially like the wild-type strain (data not shown) indicating that neither the escape of the mutant bacteria out of the phagocytic vacuole was impaired, nor their cytosolic replication. Fluorescein isothiocyanate (FITC)-phalloidin staining of infected Caco-2 cells demonstrated that all mutants induced the formation of F-actin tails like the wild-type strain and all mutants displayed normal cell-to-cell spread which was demonstrated using gfp-expressing bacteria (data not shown). In summary, the ΔTCS-RR mutants behaved indistinguishably from the wild-type bacteria regarding escape from the phagocytic vacuole, intracellular multiplication, and intracellular movement by actin polymerization and cell-to-cell spread which is in line with our observation that the major virulence factors LLO, ActA, and PlcB are efficiently expressed in the mutants.
In vivo properties of the mutants.
To test the virulence of the ΔTCS mutants, groups of five 6- to 8-week-old female BALB/c mice (Harlan Winkelmann, Germany) were infected intravenously via the tail vein with a dose of 5 × 103 CFU of the bacteria in 100 μl of endotoxin-free phosphate-buffered saline (PBS; Invitrogen). Mice were sacrificed 3 days postinfection, the livers and spleens removed and homogenized, and CFU per organ were calculated. The Student t test was used for statistical analysis, and P values of <0.05 were considered as statistically significant. While several mice infected with the wild-type and mutant bacteria showed signs of disease, mice infected with L. monocytogenes ΔdegU had a normal, healthy appearance. Mice infected with the ΔdegU strain carried a bacterial load which is more than 1 log lower in the spleen (P < 0,05) and more than 1.5 logs lower in the liver (P < 0.05) compared to the wild-type strain at day 3 of infection. For all other mutants tested, the differences of bacterial counts in livers and spleens compared to the wild-type mice are small and in most cases not statistically significant. This virulence defect of the ΔdegU mutant probably cannot be attributed to the lack of flagella observed in this mutant since an aflagellate ΔflaA mutant of L. monocytogenes 12067 grown at 24°C exhibited an increased bacterial load in the spleen of experimentally infected mice (9). Furthermore, it was shown that the repressor protein MogR is required for the down-regulation of motility gene expression and that deletion of mogR resulted in a 250-fold decrease in virulence (17). In addition, others did not observe a significant virulence attenuation of an isogenic ΔflaA mutant of L. monocytogenes 10403S after both orogastric and intravenous application (44). In accordance with results obtained with a ΔcheAY mutant administered orally (9) we did not detect a virulence attenuation of our ΔcheY mutant when intravenously injected in mice. In contrast, we did not observe a significant decrease in virulence of the ΔlisR and ΔagrA mutants which had been reported when the bacteria were administered intraperitoneally and intravenously, respectively (3, 22). Similarly, ΔkdpE and ΔcesR mutants of strain L. monocytogenes LO28 reported to be attenuated in the mouse model upon oral administration (22) behaved like the wild-type bacteria in our analysis.
Protein pattern of the L. monocytogenes ΔdegU strain.
The reduced virulence of the ΔdegU strain prompted us to characterize the extracellular protein pattern of this mutant by performing two-dimensional gel electrophoresis. Extracellular proteins of logarithmically grown bacteria were isolated according to procedures described by the manufacturer of the isoelectric focusing equipment (Amersham Biosciences). The proteins were resuspended in urea buffer containing CHAPS (7 M urea, 2 M thiourea, 4% [wt/vol] 3-[{3-cholamidopropyl}-dimethylammonio]-1-propanesulfonate, 70 mM dithiothreitol) and then loaded onto Immobiline DryStrips (Amersham Biosciences) with a pH gradient ranging from 4 to 7. For the isoelectric focusing a total of approximately 180,000 Vh was applied with several stepwise increases in voltage up to 8,000 V (IPGphor IF Unit, Amersham Biosciences). The second dimension was a standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% polyacrylamide gels (ISODalt; Amersham Biosciences) after subsequent treatment of the strips with equilibration buffer (50 mM Tris [pH 8.8], 6 M urea, 30% [vol/vol] glycerol, 2% [vol/vol] SDS, some crystals bromphenol blue) supplemented with 64 mM dithiothreitol for 15 min and with equilibration buffer supplemented with 135 mM iodacetic acid for another 15 min. The gels were fixed and stained either with silver nitrate or with Coomassie blue G250 according to standard procedures.
When analyzing proteins from the supernatant, a number of protein spots were either less prominent or induced in the mutant (data not shown). Seven of the differentially expressed proteins were excised from Coomassie blue-stained gels and further analyzed by mass spectroscopy as described previously (37).
Five out of the seven spots (four up regulated and one down regulated in the mutant) gave clear results in the mass spectroscopy as shown in Table 3 and, surprisingly, turned out to be mostly typical intracellular proteins. Only one of the identified proteins (encoded by lmo0644) has a typical signal sequence; however, the function of this conserved protein which is repressed in the ΔdegU strain is not exactly known. It is most likely a membrane protein belonging to a family of cell wall biogenesis proteins (14). The four proteins whose expression was shown to be induced in the ΔdegU mutant are all typical cytoplasmic proteins; two of them, TufA, encoded by lmo2653, and Tsf, encoded by lmo1657, belong to the translational machinery (30). It is unclear at present why these proteins are differently expressed in the ΔdegU background and why they are found in the supernatant. The listerial superoxide dismutase (SOD) is necessary for the detoxification of reactive oxygen intermediates and may play a role in defending L. monocytogenes against antilisterial host mechanisms. However, a clear role for SOD in listerial pathogenesis has not yet been demonstrated (43). In a recent report SOD, TufA, and Tsf were also detected on the surface of L. monocytogenes (35) supporting our observation that these normally cytoplasmic proteins can indeed be present outside the cell.
TABLE 3.
Putative DegU-regulated extracellular proteins of L. monocytogenes EGD grown at 37°C identified by mass spectroscopya
| Putative DegU regulation | Protein | Gene | Mol. mass (kDa)b | pIb |
|---|---|---|---|---|
| Inductionc | EF-Tu | tufA (lmo2653) | 43 | 4.81 |
| Glyceraldehyde 3-phosphate dehydrogenase | gap (lmo2459) | 36 | 5.20 | |
| Translation elongation factor | tsf (lmo1657) | 33 | 5.11 | |
| Superoxide dismutase | sod (lmo1439) | 23 | 5.23 | |
| Repressiond | Conserved hypothetical protein (phosphoglycerol transferase?) | lmo0644 | 69 | 5.42 |
Supernatant proteins from bacteria grown to log phase were analyzed.
Theoretical molecular (mol.) masses and pI values of the L. monocytogenes proteins were calculated from the corresponding amino acid sequences deposited in the NCBI protein database (http://www.ncbi.nlm.nih.gov/Entrez) by using ExPASy proteomics tool Compute pl/Mw (http://www.expasy.ch). Putative signal peptide cleavage sites in the precursors of the identified proteins were predicted with ExPASy proteomics tool SignalP.
Proteins show higher expression levels in in L. monocytogenes EGD than the ΔdegU mutant.
Proteins show higher expression levels in the ΔdegU mutant than in L. monocytogenes EGD.
Finally, expression of the glyceraldehyde-3-phosphate dehydrogenase (GAP) was found to be induced in the ΔdegU strain. GAP is a key enzyme in glycolysis, and its encoding gene gap is present in an operon together with the transcriptional regulator gene cggR (15). Interestingly, GAP has been identified as a major surface protein of gram-positive group A streptococci where it was shown to have also ADP-ribosylating activity and binding activity to multiple cellular surface proteins (32, 33). GAP has also been found on the surface of L. monocytogenes recently (35), and it is therefore tempting to speculate that this enzyme may also play a role in the interaction of L. monocytogenes with host cells.
In summary, in the present study we have shown that one response regulator of L. monocytogenes is essential for growth under standard culture conditions and that all the other response regulators are not directly involved in the regulation of several important virulence genes in L. monocytogenes. The response regulator DegU is necessary for full virulence in mice and seems to act as a pleiotropic regulator whose function remains to be characterized in detail. Interestingly, degU is the only response regulator gene in L. monocytogenes which is not genetically linked to a cognate sensor kinase. No ortholog of the sensor kinase gene degS of B. subtilis is present in the genome of L. monocytogenes. It is therefore unclear how the response regulator DegU gets activated. Since a single histidine kinase may phosphorylate several response regulators, as has been shown for the NarX/NarL and NarQ/NarP systems of E. coli (39), it is conceivable that DegU interacts with one of the 15 histidine kinases of L. monocytogenes. Since DegU belongs to the subclass of NarL-like response regulators, Lmo1021 would be a good candidate histidine kinase since it is the only sensor protein with a group II kinase domain (Table 1) which is predicted to interact with NarL-like response regulators (11).
Acknowledgments
We thank S. Pilgrim for help with the animal experiments, A. Spory for advice concerning the 2D-SDS-PAGE, G. Krohne for help with the electron microscopy, and A. Bosserhoff for performing the mass spectrometry. Special thanks go to W. Goebel for constant support and encouragement.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB 479-B5 (M.K.), by the BMBF Competence Network PathoGenoMik (T.W. and D.B.), and by the Deutscher Frauenbund (T.W.).
Editor: D. L. Burns
REFERENCES
- 1.Amati, G., P. Bisicchia, and A. Galizzi. 2004. DegU-P represses expression of the motility fla-che operon in Bacillus subtilis. J. Bacteriol. 186:6003-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:65-188. [DOI] [PubMed] [Google Scholar]
- 3.Autret, N., C. Raynaud, I. Dubail, P. Berche, and A. Charbit. 2003. Identification of the agr locus of Listeria monocytogenes: role in bacterial virulence. Infect. Immun. 71:4463-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann, B., D. Raffelsbauer, M. Kuhn, M. Goetz, S. Hom, and W. Goebel. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol. Microbiol. 43:557-570. [DOI] [PubMed] [Google Scholar]
- 6.Brondsted, L., B. H. Kallipolitis, H. Ingmer, and S. Knöchel. 2003. kdpE and a putative RsbQ homologue contribute to growth of Listeria monocytogenes at high osmolarity and low temperature. FEMS Microbiol. Lett. 219:233-239. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, P. D., N. Emerson, C. G. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter, P. D., C. M. Guinane, and C. Hill. 2003. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dons, L., E. Eriksson, Y. Jin, M. E. Rottenberg, K. Kristensson, C. N. Larsen, J. Bresciani, and J. E. Olsen. 2004. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 72:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabret, C., and J. A. Hoch. 1998. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J. Bacteriol. 180:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fabret, C., V. A. Feher, and J. A. Hoch. 1999. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol. 181:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flanary, P. L., R. D. Allen, L. Dons, and S. Kathariou. 1999. Insertional inactivation of the Listeria monocytogenes cheYA operon abolishes response to oxygen gradients and reduces the number of flagella. Can. J. Microbiol. 45:646-652. [PubMed] [Google Scholar]
- 13.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 14.Galperin, M. Y., and M. J. Jedrzejas. 2001. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins 45:318-324. [DOI] [PubMed] [Google Scholar]
- 15.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chétouani, E. Couvé, A. de Daruvar, P. Dehoux, E. Domann, G. Domínguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K.-D. Entian, H. Fsihi, F. Garcia-Del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L.-M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. Mata Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J.-C. Pérez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J.-A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 16.Gross, R., B. Arico, and R. Rappuoli. 1989. Families of bacterial signal-transducing proteins. Mol. Microbiol. 3:1661-1667. [DOI] [PubMed] [Google Scholar]
- 17.Gründling, A., L. S. Burrack, H. G. A. Bouwer, and D. E. Higgins. 2004. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl. Acad. Sci. USA 101:12316-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamoen, L. W., A. F. Van Werkhoven, G. Venema, and D. Dubnau. 2000. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:9246-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecht, G. B., T. Lane, N. Ohta, J. M. Sommer, and A. Newton. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtmann, G., E. P. Bakker, N. Uozumi, and E. Bremer. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in the adaptation to hypertonicity. J. Bacteriol. 185:1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 22.Kallipolitis, B. H., and H. Ingmer. 2001. Listeria monocytogenes response regulators important for stress tolerance and pathogenesis. FEMS Microbiol. Lett. 204:111-115. [DOI] [PubMed] [Google Scholar]
- 23.Kallipolitis, B. H., H. Ingmer, C. G. Gahan, C. Hill, and L. Sogaard-Andersen. 2003. CesRK, a two-component signal transduction system in Listeria monocytogenes, responds to the presence of cell wall-acting antibiotics and affects beta-lactam resistance. Antimicrob. Agents Chemother. 47:3421-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knudsen, G. M., J. E. Olsen, and L. Dons. 2004. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 240:171-179. [DOI] [PubMed] [Google Scholar]
- 25.Kreft, J., and J.-A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 26.Le Breton, Y., G. Boel, A. Benachour, H. Prevost, Y. Auffray, and A. Rince. 2003. Molecular characterization of Enterococcus faecalis two-component signal transduction pathways related to environmental stresses. Environ. Microbiol. 5:329-337. [DOI] [PubMed] [Google Scholar]
- 27.Mäder, U., H. Antelmann, T. Buder, M. K. Dahl, M. Hecker, and G. Homuth. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genom. 268:455-467. [DOI] [PubMed] [Google Scholar]
- 28.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Msadek, T., F. Kunst, and G. Rapoport. 1995. A signal transduction network in Bacillus subtilis includes the DegS/degU and ComP/ComA two-component systems, p. 447-471. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 30.Nyborg, J., and M. Kjeldgaard. 1996. Elongation in bacterial protein biosynthesis. Curr Opin. Biotechnol. 7:369-375. [DOI] [PubMed] [Google Scholar]
- 31.O'Hara, B. P., R. A. Norman, P. T. C. Wan, S. M. Roe, T. E. Barrett, R. E. Drew, and L. H. Pearl. 1999. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 18:5175-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pancholi, V., and V. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pancholi, V., and V. Fischetti. 1993. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. USA 90:8154-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polarek, J. W., G. Williams, and W. Epstein. 1992. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J. Bacteriol. 174:2145-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaumburg, J., O. Dieckmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Kärst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 36.Shu, C. J., and I. B. Zhulin. 2002. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem. Sci. 27:3-5. [DOI] [PubMed] [Google Scholar]
- 37.Spory, A., A. Bosserhoff, C. von Rhein, W. Goebel, and A. Ludwig. 2002. Differential regulation of multiple proteins of Escherichia coli and Salmonella enterica serovar Typhimurium by the transcriptional regulator SlyA. J. Bacteriol. 184:3549-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steil, L., T. Hoffmann, I. Budde, U. Völker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart, V., and R. S. Rabin. 1995. Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli, p. 233-252. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 40.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 178:1374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. R. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 43.Vazquez-Boland, J.-A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Way, S. S., L. J. Thompson, J. E. Lopes, A. M. Hajjar, T. R. Kollmann, N. E. Freitag, and C. B. Wilson. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell. Microbiol. 6:235-242. [DOI] [PubMed] [Google Scholar]
- 45.Wuenscher, M. D., S. Köhler, W. Goebel, and T. Chakraborty. 1991. Gene disruption by plasmid integration in Listeria monocytogenes: insertional inactivation of the listeriolysin determinant lisA. Mol. Gen. Genet. 228:177-182. [DOI] [PubMed] [Google Scholar]
- 46.Zahrt, T. C., and V. Deretic. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]