Abstract
The type III secretion (TTS) system is used by several animal and plant pathogens to deliver effector proteins into the cytosol of the eukaryotic target cell as a strategy to evade the defense reactions elicited by the infected organism. The fact that these systems are highly homologous implies that novel antibacterial agents that chemically attenuate the pathogens via a specific interaction with the type III secretion mechanism can be identified. A number of small organic molecules having this potential have recently been identified (A. M. Kauppi, R. Nordfelth, H. Uvell, H. Wolf-Watz, and M. Elofsson, Chem. Biol. 10:241-249, 2003). Using different reporter gene constructs, we showed that compounds that belong to a class of acylated hydrazones of different salicylaldehydes target the TTS system of Yersinia pseudotuberculosis. One of these compounds, compound 1, was studied in detail and was found to specifically block Yop effector secretion under in vitro conditions by targeting the TTS system. In this respect the drug mimics the well-known effect of calcium on Yop secretion. In addition, compound 1 inhibits Yop effector translocation after infection of HeLa cells without affecting the eukaryotic cells or the bacteria. A HeLa cell model that mimics in vivo conditions showed that compound 1 chemically attenuates the pathogen to the advantage of the eukaryotic cell. Thus, our results show proof of concept, i.e., that small compounds targeting the TTS system can be identified, and they point to the possible use of TTS inhibitors as a novel class of antibacterial agents.
Type III secretion (TTS) constitutes a common virulence system present in many gram-negative species, including Yersinia spp., Salmonella spp., Shigella spp., Pseudomonas aeruginosa, entheropathogenic Escherichia coli, enterohemoragic E. coli, and Chlamydia spp. (11, 24). The bacteria depend on their respective TTS system to invade the host, resist phagocytosis, grow in deep tissues, and cause disease. Furthermore, studies have revealed that several components of the TTS systems are conserved between different species (11, 42). These findings offer a possibility to develop novel antibacterial agents that target TTS-based virulence (32, 50). Moreover, small molecules that interfere with TTS can be utilized as tools in efforts aiming at increasing our understanding of complex bacterial virulence systems by using a chemical genetics approach (29, 50). The strategy of identifying and using small molecules in functional studies of microbial virulence is attractive and complements current methods in the field, as illustrated by some recent publications (7, 26, 27, 47).
The well-studied, 70-kb-plasmid-encoded Ysc (for Yersinia secretion) TTS system of Yersinia (51) represents a suitable target for both drug development (32) and a small-molecule approach to address protein function (50). Of the 11 known species of Yersinia, Y. pestis, Y. enterocolitica, and Y. pseudotuberculosis are pathogenic to mammals (51). The Ysc TTS apparatus is essential for the bacteria to evade the host immune defense, and compounds targeting this mechanism will result in attenuation without affecting bacterial growth. Interestingly 10 of the Ysc proteins have counterparts in almost all TTS systems, and it has been shown that some components of the secretion systems are interchangeable among different species (20), demonstrating evolutionary conservation. Since the TTS systems are conserved among the gram-negative bacteria utilizing this virulence mechanism it is likely that compounds targeting TTS machinery in Yersinia will also affect the TTS system in other species and that data generated with one species would also be valid for others. The importance of TTS studies is further stressed by the fact that the number of multiresistant strains in different species that utilize this virulence system is rising (38). Moreover, multiresistant strains of Y. pestis, a potential weapon in biological warfare and bioterrorism (25), have been isolated (18).
During the progress of an infection the Yersinia bacterium adheres to eukaryotic cells, e.g., macrophages, and injects a set of effector proteins, called Yops (for Yersinia outer proteins), through the Ysc machinery into the cytoplasm of the eukaryotic cell (10, 40, 42). The injected Yops cause inhibition of innate immunity, and the bacteria will avoid phagocytosis, proliferate, and eventually spread to new hosts (10). When the bacterium enters the host and senses the temperature shift to 37°C, 29 Ysc proteins that form the secretion apparatus spanning the inner and outer membranes of the bacterium are produced (12). The temperature-induced activator LcrF regulates expression of the Ysc, Yop, and specific Yop chaperon (Syc) proteins (8, 12). Before the bacterium experiences close contact with the eukaryotic cell, the expression of Yops is suppressed by the negative element LcrQ. When the bacterium adheres to the eukaryotic cell, LcrQ is secreted, resulting in elevated production of Yops, which are delivered to the Ysc apparatus by the cognate Sycs (39). In parallel, a poorly understood chain of events results in formation of a pore in the eukaryotic cell membrane. The Yops are secreted through the Ysc machinery and translocated through the pore into the cytoplasm of the eukaryotic cell, presumably in one step (21, 23, 33). In the eukaryotic cell six different Yops, i.e., YopE, YopH, YpkA (YopO), YopJ, YopM, and YopT, specifically inactivate the innate immune response by disturbing and disrupting events such as cytoskeleton dynamics and inflammatory responses, including production of proinflammatory cytokines (10).
In order to allow identification of novel agents that target TTS, we developed and utilized a whole-cell bacterial reporter gene assay in Y. pseudotuberculosis to identify a number of promising inhibitors from a 9,400-compound collection (27). In this study we characterized one class of inhibitors in detail and showed that this class of molecules directly targets the TTS and that the inhibition prevents protein translocation and thus inhibits virulence.
MATERIALS AND METHODS
Compounds.
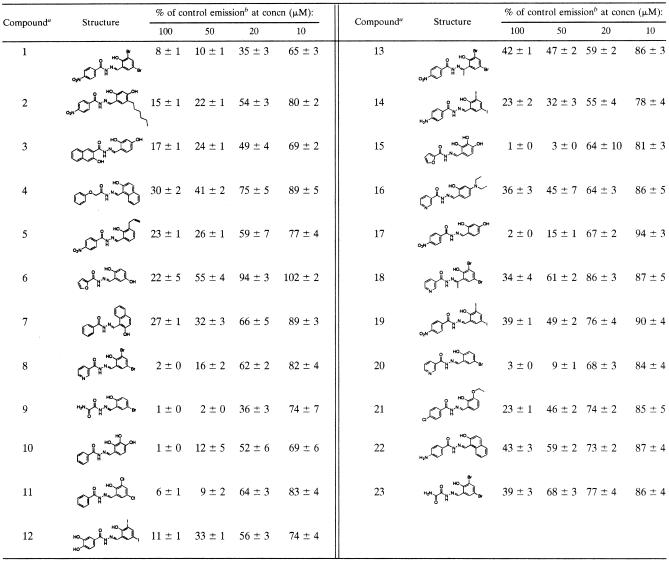
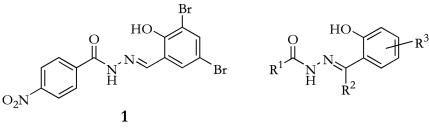
The type III secretion inhibitors 1 to 23 (Table 1) were synthesized according to literature procedures (1) from commercially available hydrazides and salicylaldehydes or acetophenones, with the exceptions of 3-allyl-salicylaldehyde (13) and 5-hexyl-4-hydroxy-salicylaldehyde (34), which have been described previously, and 2-phenoxyacethydrazide, which was prepared from methyl-2-phenoxyacetate and hydrazine (1). Compounds were characterized by 1H nuclear magnetic resonance spectroscopy and liquid chromatography-mass spectrometry. Dimethyl sulfoxide (DMSO) stock solutions were prepared and stored at −20°C.
TABLE 1.
Effect on luciferase light emission for strain YPIII(pIB29) in the presence of compounds 1 to 23 at different concentrations
The compounds were prepared as described in Materials and Methods.
Means and standard deviations (calculated with the Gauss approximation formula) are from triplicates, and experiments were reproduced at least twice as described previously (27).
Strains and growth conditions.
Y. pseudotuberculosis serotype III (YPIII) strains with wild-type (wt), ΔlcrQ, yscT::Tn5, ΔyopB, ΔlcrG, or ΔyopH mutant forms of the virulence plasmid pIB (pIB102, pIB26, pIB61, pIB604, pIB701, and pIB29, respectively) were grown at 26°C on LB plates (with 15 μg/ml of chloramphenicol or 50 μg/ml of kanamycin, depending on the strain) and then in liquid brain heart infusion (BHI) broth (Oxoid, Unipath Ltd., Basington, United Kingdom) supplemented with 2.5 mM CaCl2 (referred to as cultures with Ca2+) or supplemented with 20 mM MgCl2 and 5 mM EGTA for Ca2+ depletion (referred to as cultures without Ca2+) as described before (27).
HeLa cells were routinely cultured in Eagle's minimal essential medium containing 10% heat-inactivated fetal calf serum and 100 IU/ml of penicillin at 37°C in a humidified atmosphere as described before (44).
Plasmids and mutant constructions.
Constructions of transcriptional fusions pIB102 yopE-luxAB, pIB102 lcrF-luxAB, pIB102 lcrQ-luxAB, and pIB29 yopE-luxAB have been described elsewhere (14, 27). Constructions of pIB61 yopE-luxAB and pIB61 lcrF-luxAB were achieved using conjugations as described earlier (27), with the exception that YPIII(pIB61) was used as recipient in the conjugation instead of YPIII(pIB102). Construction of a translational fusion between LcrF and the FLAG epitope (28) followed a previously described strategy (14) for the transcriptional fusions except that the sequence coding for the FLAG epitope was introduced upstream of the LcrF stop codon. A BamHI/ClaI subfragment from BamHI fragment 7 of the pIB1 virulence plasmid was cloned into the BamHI site of pMMB66HE (17), resulting in the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible pLcrF.
Analysis of protein secretion.
Overnight cultures of the different Y. pseudotuberculosis strains were diluted 1/20 in BHI broth supplemented with 20 mM Mg2+ and 5 mM EGTA for Ca2+ depletion. Compound 1 (20 mM in DMSO) was added to a final concentration of 50 μM, and control cultures received only DMSO. Cultures were incubated at 26°C for 30 min and then shifted to 37°C for induction of Yop expression/export as described in Results. At the times indicated, total samples and supernatants were mixed 1:1 with sodium dodecyl sulfate (SDS) sample buffer. Material corresponding to 5 to 7 μl of bacterial culture was separated on a 12% SDS-polyacrylamide gel. Western blotting was performed with a polyclonal rabbit antiserum recognizing all Yops followed by a secondary horseradish peroxidase-coupled goat anti-rabbit antibody. For the analysis of the LcrF-FLAG fusion protein, bacteria corresponding to 15 μl of bacterial culture were loaded in each lane, and a monoclonal mouse anti-FLAG H2 antibody (Sigma Aldrich) followed by a secondary horseradish peroxidase-coupled goat anti-rabbit antibody was used for the Western blot. The signals on the nitrocellulose filters were visualized with ECL Western blot detection reagents (Amersham Biosciences Inc.). Western blot analysis of the effect of selected compounds on protein secretion (see Fig. 8) was carried out as described previously (27).
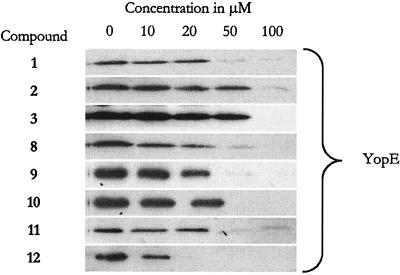
FIG. 8.
Western analysis of Yop secretion from YPIII(pIB29EL) in the presence of eight of the most potent compounds. The analysis was carried out as described previously (27), and only the YopE results are shown. All other Yops were similarly affected.
To analyze Yop secretion, wt YPIII(pIB102) and ΔlcrG YPIII(pIB701) cultures first were grown at 26°C for 30 min with or without 50 μM compound 1 in BHI broth containing 2.5 mM CaCl2 or depleted of Ca2+ and then were shifted to 37°C for 3 h of continued incubation. Secreted Yop proteins from filtered culture supernatants were, after concentration by trichloroacetic acid precipitation, separated by 12% SDS-polyacrylamide gel electrophoresis, and proteins were visualized by Coomassie blue staining.
Luciferase analysis.
Overnight cultures of wt or yscT::Tn5 insertion mutant Y. pseudotuberculosis strains containing transcriptional fusions of yopE, lcrQ, or lcrF with luxAB from Vibrio harveyi [YPIII(pIB102) yopE-luxAB, YPIII(pIB102) lcrQ-luxAB, YPIII(pIB102) lcrF-luxAB, YPIII(pIB61) yopE-luxAB, and YPIII(pIB61) lcrF-luxAB] were diluted 1/20 in 20 ml of BHI broth supplemented either with 20 mM Mg2+ and 5 mM EGTA for Ca2+ depletion or with 2.5 mM CaCl2. The flasks received compound 1 (50 μM final concentration) or DMSO alone at start of the 26°C incubation (−30 min) or at time points given in Fig. 2 and 3. Cultures were incubated at 26°C for 30 min and then (at t = 0 min) shifted to 37°C, and incubation was continued for 120 min. During the incubation, samples were removed and analyzed for absorbance at 600 nm and for luciferase activity using a luminometer (Aureon PhL; Aureon Biosystems GmbH, Austria) after the addition of 0.01% N-decyl aldehyde (Sigma Aldrich) to induce light production. Dose-response analysis of compounds 1 to 23 for inhibition of the reporter gene signal from YPIII(pIB29) yopE-luxAB (Table 1) was carried out as described previously (27).
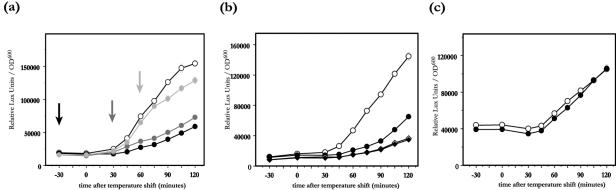
FIG. 2.
Induction of luciferase activity from yopE-luxAB (a, b) and lcrQ-luxAB (c) transcriptional fusions after temperature shift from 26°C to 37°C. Compound 1 (filled symbols) or DMSO (open symbols) was added to wt YPIII(pIB102) (circles) or to an yscT YPIII(pIB61) secretion mutant (diamonds). In panel a compound 1 was added to bacteria 30 min prior to the temperature shift (black symbols and arrow), 30 min after the temperature shift (medium grey symbols and arrow), or 60 min after the temperature shift (light grey symbols and arrow). In panels b and c compound 1 was added to bacteria at the start of the experiment (black symbols). OD600, optical density at 600 nm.
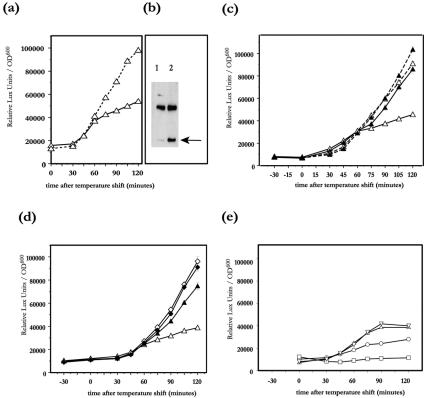
FIG. 3.
Induction of luciferase activity from lcrF-luxAB (a, c-e) transcriptional fusions grown in inducing BHI broth without Ca2+ (continuous lines) or in noninducing BHI broth with Ca2+ (dashed lines) after temperature shift from 26°C to 37°C. Compound 1 (filled symbols) or DMSO (open symbols) was added to the wt YPIII(pIB102) (triangles) or to a yscT YPIII(pIB61) secretion mutant (diamonds) 30 min prior to the temperature shift. Western blotting (b) for LcrF-FLAG (arrow) from wt YPIII(pIB102) grown in noninducing BHI broth with Ca2+ (lane 1) or in inducing BHI broth without Ca2+ (lane 2) 2 h after the temperature shift from 26°C to 37°C. The upper band in the Western blot is the result of a cross-reaction between the anti-FLAG antibody and a chromosomal Yersinia pseudotuberculosis protein; the band is included to demonstrate equal loading between the lanes. Induction of luciferase activity from the lcrF-luxAB transcriptional fusions in wt YPIII(pIB102) (e) in the absence of pLcrF (control), without (triangles) and with (inverted triangles) 1.0 mM IPTG and in the presence of pLcrF, and without (circles) and with (squares) induction with 1.0 mM IPTG. Note that pLcrF in trans with IPTG induction totally abolishes transcription from the endogenous lcrF promoter. OD600, optical density at 600 nm.
Virulence inhibition.
Wild-type Y. pseudotuberculosis, YPIII(pIB102), and ΔyopB YPIII(pIB604) were grown in LB overnight with shaking at 26°C. For analysis of virulence inhibition the overnight cultures were diluted 106 with tissue culture medium containing 10% fetal calf serum without antibiotics. The cultures were supplemented with either DMSO or compound 1 to a final concentration of 40 μM. The final DMSO concentration was kept below 1%. The inoculated cultures with or without compound 1 were grown at 26°C for an initial 30 min, followed by incubation at 37°C for 30 min. A volume of 400 μl of the preincubated bacterial cultures was added to a 24-well tissue culture plates containing 2 × 105 HeLa cells per well, and the plates (multiplicity of infection [MOI] = 0.01) were centrifuged at 400 × g for 5 min. The plates were incubated for 30 min at 37°C in a 5% CO2 humidified atmosphere, and thereafter the overlying medium was removed and replaced with 250 μl of antibiotic-free tissue culture medium without any compound and incubated for further 0 or 7 h at 37°C in a 5% CO2 humidified atmosphere. The HeLa cells were harvested by removal of the overlying medium, and cells were lysed by the addition of 400 μl of lysis buffer (0.1% NP-40, 15 mM NaCl, 1 mM Tris, pH 8). The resulting lysates were assayed for the number of viable bacteria by viable count at 0 and 7 h after the initial 30-min incubation (5). The experiment was run in duplicate in independent wells and reproduced three times.
Immunofluorescence.
Immunofluorescence staining was performed as described previously (36). Briefly, 0.5 × 105 HeLa cells were seeded on 1.2-cm-diameter round glass coverslips in 24-well microtiter plates 24 h prior to infection. Wild-type Y. pseudotuberculosis YPIII(pIB102) was pregrown for 30 min at 26°C in cell culture medium and then shifted to 37°C for 60 min. Bacteria at an MOI of 20 and 40 μM compound 1 or DMSO were added to the HeLa cells. The plates were spun at 400 × g for 5 min, and infection was continued for 90 min at 37°C. At the end of the infection the coverslips were washed twice with phosphate-buffered saline (PBS) and then incubated with 10 μg/ml of rhodamine-conjugated wheat germ agglutinin for 10 min at 24°C to visualize membranes. The coverslips were again washed twice with PBS, and the cells were fixed with 2% paraformaldehyde in PBS for 10 min and then permeabilized for 5 min with 0.5% Triton X-100, 1.0 mM EGTA, 4% polyethylene glycol 6000, and 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8. Permeabilized cells were then incubated with a goat anti-YopH antibody for 1 h at 37°C, washed, treated with a secondary Alexa-conjugated donkey anti-goat antibody in PBS, again washed, and then stained with the DNA-chelating fluorescent dye DAPI (4′,6′-diamidino-2-phenylindole). The coverslips were washed and mounted on slides with UMM mounting medium (6). Slides were analyzed in a microscope (Zeiss Axioskop) mounted with a digital camera (Hamamatsu C4742-95 with a Neofluor 40×/0.75 objective).
RESULTS
Compound 1 specifically blocks Yop secretion via the TTS system.
We have previously described a screening system based on viable Y. pseudotuberculosis for the identification of potential TTS inhibitors from chemical libraries in a high-throughput mode (27). The screening resulted in a number of hits, of which compounds 1 to 7 (Fig. 1; Table 1) belong to a class of acylated hydrazones of different salicylaldehydes. Compound 1 proved to be most potent, and it can readily be prepared on a multigram scale from inexpensive starting materials. Thus, compound 1 is a suitable candidate for continued efforts aimed at determination of the compound's mode of action. The screening strategy (27) exploited the tight regulatory coupling between Yop expression and LcrQ secretion in Yersinia (37) and therefore failed to determine if tested compounds acted directly or indirectly on the TTS system; e.g., they might affect transcription/translation per se and thereby indirectly affect the amount of protein exported. To discriminate between the two possibilities, we designed a number of experiments to answer these questions. First, we followed the kinetics of yopE transcriptional activation during induction of Yop expression/export by using the yopE-luxAB transcriptional fusion. Luciferase activity from the LuxAB protein produced by a yopE-luxAB transcriptional fusion was monitored in a virulent wt YPIII(pIB102) (yopE-luxAB) strain of Y. pseudotuberculosis. Cultures were grown in BHI broth lacking Ca2+, conditions allowing for induction of Yop secretion when cultures are shifted from 26°C to 37°C. Compound 1 (50 μM final concentration) was added to the cultures at different time points (Fig. 2a). In the control culture receiving only DMSO 30 min before the shift in temperature from 26°C to 37°C, an increase in luciferase activity could be monitored 45 min after the temperature shift (Fig. 2a). This corresponds to the time needed to assemble a functional TTS system after a temperature shift (16). As expected, the luciferase activity increased throughout the monitoring time (120 min). In the culture receiving compound 1 30 min prior to temperature shift, only a minor increase in luciferase activity could be seen during the 120-min induction period (Fig. 2a), showing that compound 1 interferes with the Yop secretion-coupled regulation of yopE transcription. Addition of compound 1 at 30 min after temperature shift, resulted in a small increase in luciferase activity compared to that in the culture receiving the drug 60 min earlier (Fig. 2a). When compound 1 was added to the culture 60 min after temperature shift, there was a 30-min delay before the rate of transcription from the yopE promoter was reduced (Fig. 2a). The fact that the effect on transcription cannot be seen earlier than 30 min after addition of the drug argues for the hypothesis that the drug does not directly block yop transcription or Yop translation.
FIG. 1.
Structure of compound 1 and general structure for acylated hydrazones of salicylic aldehydes.
To further test this hypothesis, we monitored the luciferase activity generated by the yopE-luxAB transcriptional fusion in the polar yscT::Tn5 mutant [YPIII(pIB61)], which is blocked for Yop secretion under conditions favorable for yop induction (16). In contrast to that of the wt strain, the luciferase activity of the yscT mutant increased only marginally after the temperature shift to 37°C (Fig. 2b). Addition of compound 1 (50 μM) 30 min prior to the temperature shift did not affect the luciferase activity of the yscT mutant, in contrast to the case for the wt strain, where compound 1 addition greatly reduced the induction (Fig. 2b). This further supports the notion that compound 1 does not directly affect Yop expression. Interestingly, the drug is almost as efficient in lowering yopE transcription as the ysc secretion mutant (Fig. 2b).
Compounds that lower the transcriptional activity from a promoter such as the yopE promoter either could act by specific mechanisms or could lower the expression of many activated promoters via some general (toxic) effect. To test if the effect of compound 1 on yop transcription was specific for LcrF-regulated promoters, we followed the transcriptional induction from the lcrQ promoter, which also is up-regulated at 37°C but is regulated independently of LcrF, employing an lcrQ-luxAB transcriptional fusion. The strain was grown under conditions favorable for Yop secretion (BHI broth without Ca2+), and the luciferase activity was measured in presence or absence of 50 μM compound 1 (Fig. 2c) The drug did not affect the induction of luciferase activity (Fig. 2c), indicating that the drug has no general effect on temperature-inducible promoters.
LcrF, the transcriptional activator of the Yersinia TTS system, is a member of the large AraC/XylS family of transcriptional activators (8, 19). One of the features of the well-studied AraC prototype of this family is that AraC is autoregulated and can control transcription from its own promoter both positively and negatively (22). When the transcription from the lcrF promoter using an lcrF-luxAB transcriptional fusion was studied, we found that bacteria grown under conditions favoring high Yop production and export of Yop proteins (37°C without Ca2+) exhibited an increased rate of transcription from the lcrF promoter starting 30 to 45 min after the temperature shift. Surprisingly, the rate of transcription was greatly reduced about 60 min after the temperature shift (Fig. 3a). When bacteria were grown under noninducible conditions (blocked Yop export with Ca2+), the rate of transcription initiated from the lcrF promoter also increased 30 to 45 min after the shift (Fig. 3a). However, in contrast to the situation under inducible conditions, the high rate of transcription was maintained during the whole incubation period (Fig. 3a). This is a paradox since the rate of lcrF transcription under noninducible conditions was elevated compared to that under inducible conditions, given that yop expression is higher under inductive conditions. If, however, LcrF is autoregulated, then the lower transcription from the lcrF promoter under inducing conditions could reflect a more efficient translation of the lcrF transcript, resulting in high LcrF levels and a subsequent down-regulation of lcrF transcription. This prompted us to analyze the levels of LcrF under inducing and noninducing conditions (with Ca2+ or without Ca2+). LcrF levels were monitored by introducing a FLAG epitope in the C-terminal end of LcrF, using the same strategy as for the luxAB fusions except that the design in this case resulted in a translational fusion between LcrF and the FLAG epitope. The hybrid protein was still active as a transcriptional activator. When the LcrF-FLAG fusion protein levels were analyzed by Western blotting using antibodies directed against the FLAG epitope, we found that despite the higher transcriptional activity from the lcrF promoter under noninducing conditions at 120 min (Fig. 3a), only small amounts of LcrF protein were found 120 min after the temperature shift (Fig. 3b, lane 1). In contrast, the cultures grown under inducing conditions contained large amounts of LcrF protein (Fig. 3b, lane 2) in spite of the lower rate of transcription seen from the lcrF promoter (Fig. 3a). Overexpression of LcrF in trans from the IPTG-inducible promoter of the pLcrF present in strain YPIII(pIB102) lcrF-luxAB resulted in a dose-dependent down-regulation of transcription from the endogenous lcrF promoter (Fig. 3e), indicating that LcrF, when expressed at high levels, suppress its own transcription. Together these results demonstrate that LcrF, in analogy with other regulators of the AraC family, is autoregulated.
The effect of compound 1 on lcrF transcription was studied by employing the YPIII(pIB102) lcrF-luxAB transcriptional fusion strain to follow the luciferase activity after the temperature shift. The bacteria were grown in BHI medium depleted of Ca2+ or in BHI medium with 2.5 mM Ca2+, and compound 1 (50 μM) was added 30 min prior to the temperature shift. Addition of compound 1 to induced cultures changed the transcription of lcrF to resemble that of the noninduced culture (Fig. 3c). Addition of compound 1 to cultures grown with Ca2+ did not further change the luciferase signal (Fig. 3c). We then employed the yscT::Tn5 YPIII(pIB61) (lcrF-luxAB) Yop secretion mutant strain and analyzed the rate of transcription from the lcrF promoter in presence and absence of compound 1 under inducing conditions (without Ca2+). As expected, the yscT secretion mutant failed to down-regulate lcrF transcription levels, in contrast to the wt control (Fig. 3d). Addition of compound 1 to the yscT mutant did not further change lcrF transcription (Fig. 3d). Based on these results, we conclude that compound 1 does not act directly on LcrF, since the autoregulatory loop of LcrF, which depends on a functional LcrF protein, is unaffected by the drug. However, compound 1 gives the same phenotype as that of a yscT mutant, further supporting a direct effect of compound 1 on TTS.
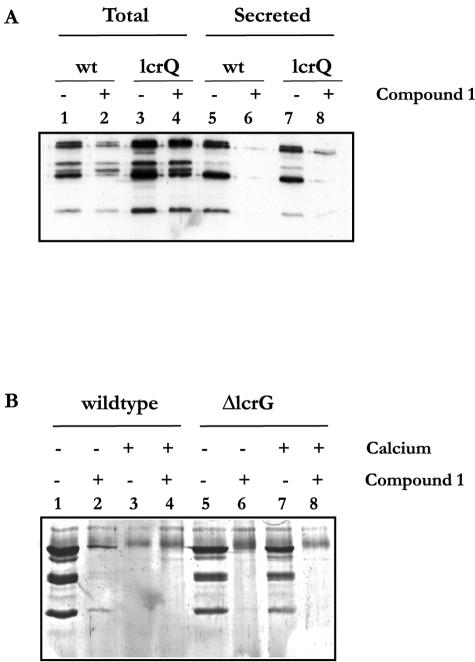
We have earlier demonstrated that a ΔlcrQ null mutant is derepressed for Yop expression even under conditions when Yop export is blocked (37). Furthermore, an lcrQ null mutant secretes Yop proteins only when grown in a medium depleted of Ca2+, and in this respect the mutant shows a wt phenotype (39). We tested if compound 1 could also block secretion of Yops in the lcrQ null mutant, which constitutively expresses high intracellular concentrations of Yop proteins. The lcrQ mutant and the wt strain were incubated in BHI broth lacking Ca2+ in the absence or presence of compound 1 (50 μm). The cultures were first incubated at 26°C for 30 min and then shifted to 37°C. The effect of the drug was monitored 2 h after the shift (Fig. 4). Total culture samples and supernatants were analyzed by Western blotting using anti-total-Yop antiserum. We found that the total Yop expression (Fig. 4A, lanes 1 to 4) in the wt cultures was lowered in the presence of compound 1 compared to the DMSO-treated control cultures (Fig. 4A, lanes 2 and 1, respectively). These results are in full agreement with the earlier observed lowered expression from the yopE promoter after addition of compound 1 (Fig. 2a). When total Yop levels in the lcrQ mutant were analyzed, elevated amounts were found relative to the wt culture, as expected given that the lcrQ mutant is derepressed for Yop expression (39) (Fig. 4A, lanes 3 and 1, respectively). Addition of compound 1 to the culture harboring the ΔlcrQ mutant did not affect the amount of Yop effector proteins expressed (Fig. 4A, lanes 3 and 4). When the levels of Yop proteins exported in presence or absence of compound 1 were analyzed (Fig. 4A, lanes 5 to 8), we found that addition of compound 1 to both wt and ΔlcrQ mutant strains efficiently blocked the export of Yops into the growth medium (Fig. 4A, lanes 5 and 7 compared to lanes 6 and 8, respectively). In agreement with the data presented in Fig. 2, this analysis using a ΔlcrQ mutant lacking the tight regulatory coupling between active export and yop transcription shows that compound 1 specifically blocks the export of Yop proteins.
FIG. 4.
Western blot (A) using anti-total-Yop antiserum on total culture samples (lanes 1-4) and on culture supernatants (lanes 5-6) from wt YPIII(pIB102) cultures (lanes 1-2 and 5-6) and from a lcrQ YPIII(pIB26 regulatory mutant (lanes 3-4 and 7-8). Bacteria were cultivated under Yop-inducing conditions for 2 h in BHI broth without Ca2+, supplemented with either DMSO (lanes 1, 3, 5, and 7) or compound 1 (lanes 2, 4, 6, and 8). Material corresponding to 7 μl of bacterial culture was loaded in each lane. Coomassie blue-stained SDS-polyacrylamide gel (B) with concentrated culture supernatants from wt YPIII(pIB102) cultures (lanes 1-4) or an lcrG YPIII(pIB701) secretion control mutant (lanes 5-8). Bacteria were cultivated under Yop-inducing conditions for 3 h in BHI broth without Ca2+ (lanes 1-2 and 5-6) or BHI broth with Ca2+ (lanes 3-4 and 7-8), supplemented with either DMSO (lanes 1, 3, 5, and 7) or compound 1 (lanes 2, 4, 6, and 8). Material corresponding to 1 ml of bacterial culture was loaded in each lane.
Compound 1 blocks Yop secretion in calcium-blind mutants.
To evaluate the effect of compound 1 on “calcium-blind” mutants, which secrete Yop proteins at 37°C regardless of the Ca2+ concentration, we analyzed the effect of compound 1 on a Y. pseudotuberculosis YPIII(pIB701) strain containing an in-frame deletion of lcrG, the inner gate of the translocation apparatus (46). Wild-type YPIII(pIB102) and ΔlcrG YPIII(pIB701) cultures were induced for Yop expression in BHI broth, either containing 2.5 mM CaCl2 or depleted of Ca2+, with or without addition of 50 μM compound 1. Supernatants from induced cultures were concentrated and checked for the presence of Yops by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining. As expected, wt Yersinia exported Yops only in BHI broth depleted of Ca2+ and only in absence of compound 1 (Fig. 4B, lanes 1 to 4). In contrast, the ΔlcrG mutant secreted Yop proteins both in the presence and in the absence of Ca2+ (Fig. 4B, lanes 5 and 7), but addition of 50 μM compound 1 blocked the Yop secretion both in the presence and in the absence of Ca2+, confirming that the drug also effectively blocks Yop secretion in a calcium-blind mutant (Fig. 4B, lanes 6 and 8). When testing another mutant exhibiting the same Ca2+-blind phenotype, the ΔyopN YPIII(pIB82) mutant (15), we found also for this mutant that compound 1 was equally effective in blocking Yop export in both the presence and absence of Ca2+ (data not shown). YopN has been suggested to act as the outer gate, regulating Yop secretion from the outside. These results show that the secretion-blocking effect of compound 1 is independent of the Ca2+ concentration in the growth medium and also imply that neither LcrG nor YopN is a molecular target for compound 1.
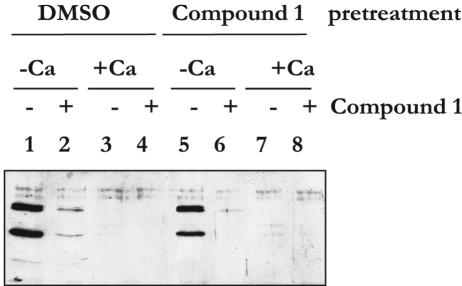
To study if the effect of compound 1 was reversible, we split a culture of YPIII(pIB102) wt bacteria grown in BHI broth supplemented with 2.5 mM Ca2+ into two cultures. One culture received only DMSO, the other received compound 1 (40 μM), and both cultures were incubated at 37°C for 120 min to allow full induction of the TTS machinery but without any secretion of Yop proteins into the growth medium. The bacteria were then aliquoted into eight tubes and washed once with 37°C BHI broth containing DMSO (0.5%, four tubes) or compound 1 (40 μM, four tubes) to remove Ca2+ from the medium, and then the bacteria were resuspended in BHI broth either containing or lacking compound 1 (40 μM) and/or Ca2+. The cultures were incubated at 37°C for an additional 45 min. Culture supernatants obtained after centrifugation were analyzed by Western blotting using a polyclonal rabbit anti-total-Yop protein antiserum (Fig. 5). Bacteria resuspended in medium containing Ca2+ were unable to export proteins both in the absence and in the presence of compound 1 and independent of the pretreatment conditions (Fig. 5, lanes 3, 4, 7, and 8). If, on the other hand, the bacteria were resuspended in medium depleted of Ca2+ and lacking compound 1, large amounts of Yops were secreted into the surrounding medium without influence of the pretreatment conditions (Fig. 5, lanes 1 and 5). Compound 1 effectively inhibited this secretion, and, importantly, this inhibition was also regardless of the pretreatment conditions (Fig. 5, lanes 2 and 6). These results show that the effect of compound 1 is fast and reversible and that the drug has no general toxic effect on the TTS system.
FIG. 5.
Western blot using anti-total-Yop protein antiserum on culture supernatants from wt YPIII(pIB102). Cultures were first grown at 37°C for 2 h in noninducing BHI broth with Ca2+ supplemented with DMSO (lanes 1-4) or compound 1 (lanes 5-6). The noninducing BHI broth with Ca2+ was then removed by centrifugation and bacteria washed in BHI broth with DMSO (lanes 1-4) or compound 1 (lanes 5-6). Bacteria were resuspended in inducing BHI broth without Ca2+ (lanes 1-2 and 5-6) or in noninducing BHI broth with Ca2+ (lanes 3-4 and 7-8), containing DMSO (lanes 1, 3, 5, and 7) or compound 1 (lanes 2, 4, 6, and 8). Incubation was continued for 45 min, and then culture supernatants were prepared and analyzed.
Compound 1 inhibits virulence in vivo.
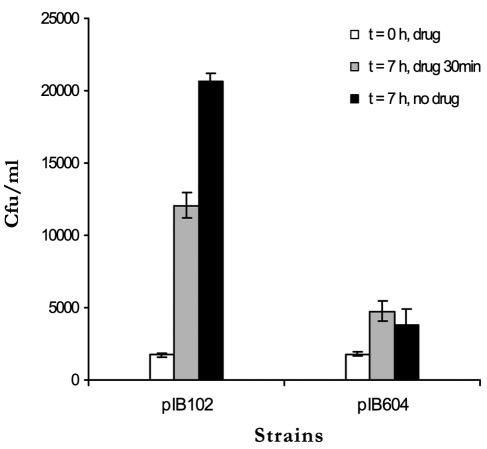
Our results clearly demonstrate that compound 1 inhibits the type III secretion in Y. pseudotuberculosis in vitro without affecting transcription/translation of Yops. With these results in hand, we were interested to investigate if compound 1 could inhibit virulence when eukaryotic cells were infected with wild-type Y. pseudotuberculosis. We employed a method developed by Bartra et al. to follow the success of infection using a HeLa cell model (5). This model allowed us to discriminate a functional TTS system from a nonfunctional one under semi-in vivo conditions (5). HeLa cells were infected with wt strain YPIII(pIB102) or the translocation-defective mutant YPIII(pIB604) (ΔyopB) at a multiplicity of infection of 0.01 in the presence of compound 1 (40 μM) or DMSO alone. The drug or DMSO was washed away 30 min after onset of infection, and the cells were incubated for an additional 7 h. The overlying medium was removed, and the HeLa cells were lysed by the addition of lysis buffer. When the lysed fraction was analyzed with respect to the number of viable bacteria, a 12-fold increase in bacterial CFU could be observed for wt bacteria, compared to a twofold increase for the translocation-deficient strain [YPIII(pIB604)], when monitored 7 h after onset of infection (Fig. 6). The wild-type bacteria can evade uptake by the eukaryotic cells by employing TTS, and extracellularly proliferating bacteria can be observed. The mutant strain YPIII(pIB604) adheres to the eukaryotic cells, but it cannot translocate effector proteins into the eukaryotic cell since it lacks YopB (21, 35). As a result of the malfunction of the translocation system, HeLa cells will in this case take up the bacteria. Internalized bacteria do not proliferate, and compared to cell cultures infected with wild-type bacteria, lower numbers of proliferating bacteria are observed (5). When HeLa cells were infected in the presence of compound 1 (40 μM), a 7-fold increase of wild-type YPIII(pIB102) bacteria could be observed, compared to a 2.5-fold increase for the mutant strain YPIII(pIB604) (Fig. 6). Thus, compared to control experiments in the absence of compound 1, a 40% inhibition of virulence was observed when HeLa cells were infected with wild-type YPIII(pIB102) bacteria in the presence of compound 1 at 40 μM (Fig. 6). The growth of bacteria in cell culture medium alone was examined by measuring absorbance and the viable count at different time points, and it was found that the compound did not affect bacterial growth under the experimental conditions used (data not shown). Thus, we essentially confirm the results obtained by Bartra et al. (5), and our results show that addition of compound 1 mimics a translocation-deficient mutant without any adverse effects on bacterial uptake by the HeLa cells.
FIG. 6.
HeLa cells were infected with pIB102 (wt)- and pIB604-carrying strains of Y. pseudotuberculosis in the presence of compound 1 at 40 μM (white and grey bars) or DMSO as a control (black bars). Thirty minutes after initial infection, unattached bacteria were removed and fresh medium was added. The number of cell-associated bacteria was determined by viable count at 0 h (white bars) and 7 h (grey and black bars) after the initial infection. The graph shows the average number of CFU for two independent wells. Error bars indicate standard deviations.
Compound 1 prevents protein secretion and translocation.
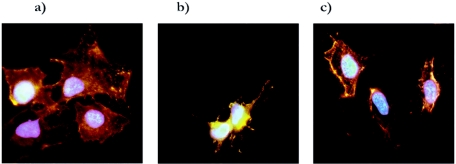
The in vitro method in use to induce Yop export is to deplete the culture of Ca2+. However, when Yersinia infects mammalian cells under cell culture conditions or infects animals, it encounters a high concentration of Ca2+ that is around 1 mM. Under in vivo conditions, it is direct contact between the bacterium and the mammalian cell that triggers the Yop secretion and the subsequent polarized transfer of Yop effectors across both the two bacterial membranes and the eukaryotic plasma membrane (10, 43). To test if the reduced virulence caused by compound 1 in the HeLa cell semi-in vivo model was a result of inhibition of Yop translocation, we infected cultured HeLa cells with the wt YPIII(pIB102) strain at a MOI of 20 for 90 min. At the start of infection either compound 1 (50 μM) or DMSO was added to the HeLa cells. One culture received compound 1 (50 μM) but was kept uninfected as a control for the direct effect of compound 1 on the HeLa cells. After infection the cells were immunostained using wheat germ agglutinin coupled to the fluorescent dye rhodamine to stain HeLa cell membranes red. Cells were fixed, permeabilized, and immunostained using a polyclonal goat antibody recognizing the YopH protein, followed by a secondary rabbit anti-goat antibody coupled to the fluorescent dye Alexa to stain the translocated YopH green. As a last step, the HeLa cell nucleus was visualized by staining with a DNA binding fluorescent drug, resulting in blue fluorescence from the nuclei. When the different samples were analyzed by fluorescence microscopy, it was found that compound 1 blocked translocation of YopH, while translocation of YopH was observed in the untreated infected control (Fig. 7). In addition, in the absence of compound 1, infected HeLa cells were also cytotoxically affected, with a rounded-up morphology caused by the effector protein YopE (41). Addition of compound 1 strongly reduced the YopE-mediated cytotoxic response of the HeLa cells (Fig. 7), indicating that the drug inhibits translocation of several Yop effectors. The effect on Yop translocation is likely due to the fact that the drug inhibits Yop secretion in general.
FIG. 7.
Immunostaining of noninfected HeLa cells (a) and HeLa cells infected for 90 min with wt YPIII(pIB102) Yersinia (b, c). At the start of infection 50 μM compound 1 in DMSO (a, c) or only DMSO (b) was added to the cultures. Cells were fixed and stained for membrane structures (red), DNA (blue), and translocated YopH (green).
Additional type III secretion inhibitors.
Compounds 1 to 7 are inhibitors that were identified in the primary screen (27), and in parallel with the detailed evaluation of compound 1, approximately 60 new compounds were prepared according to literature procedures (see Materials and Methods). All compounds share the common structural motif shown in Fig. 1, and from this set of compounds, compounds 8 to 23 represent additional inhibitors that show significant inhibition of the reporter gene signal in YPIII(pIB29) yopE-luxAB at 50 μM (Table 1). Inhibition of protein secretion was investigated for eight of the most potent compounds (i.e., compounds 1 to 3 and 8 to 12) in Table 1, and it was found that the level of reporter gene inhibition roughly matched inhibition of actual protein secretion (Table 1; Fig. 8), as previously found for compound 1 (27). These results show that second-generation TTS inhibitors within the same structural family as compounds 1 to 7 and with efficiency similar to that of compound 1 can be prepared.
DISCUSSION
Treatment of infections with broad-spectrum antibiotics stimulates the rapid emergence of multiply resistant bacterial strains, making restricted usage of these antibiotics an urgent requirement. Although a more restrictive use of current drugs might have an effect (31), there is an obvious need for new antimicrobial drugs with novel targets to combat resistant bacteria (31, 48). Preferably such drugs should have a mode of action that avoids emergence of resistance. However, the immediate need for new drugs does not correlate to the number of antimicrobial drugs in clinical trials (48). Moreover, with the exception of the oxazolidinones, the antibacterial drugs that have been developed during the last decades were obtained by structural variation of existing drugs (48). A strategy that relies on existing drugs and targets will most likely not be sufficient to meet the need for novel therapeutic regimens. Therefore, this requires the development of new antibacterial agents by engaging strategies that specifically counteract the development of resistance. It has been suggested that drugs that target bacterial virulence could have this effect (2, 29). It could be argued that the selection pressure on the total microflora is dramatically reduced if drugs are employed that specifically attenuate the pathogen without affecting growth, in contrast to classical antibiotics, which without discrimination attack the growth of all bacteria, including the commensal intestinal flora (45). One could also argue that wild-type strains are inherently more fit than mutants that have acquired resistance towards a drug. It is obvious that mutations leading to resistance produce changes of the TTS system per se, which as a consequence could result in reduced fitness. Antibacterial therapy, which involves chemical attenuation of the pathogen during infection without targeting bacterial growth, requires that the host's immune defense clears the infectious agent. To date, this is an unproven, novel strategy to cure infections. Antibacterial chemotherapy targeting gene products directly associated with virulence will likely have this characteristic. Type III secretion systems constitute one important virulence-associated mechanism that is commonly spread among gram-negative bacterial pathogens. With few exceptions, mutations affecting the TTS system lead to avirulent phenotypes (10, 12). Thus, if secretion of TTS system substrates can be blocked by the addition of small molecules without affecting bacterial viability, it is possible that novel antibacterial drugs can be developed that chemically attenuate the pathogen during infection. A small number of TTS inhibitors have been described in the literature, but no details on their mode of action have been described (26, 27, 30, 49). We have shown here that compound 1 specifically blocks the TTS system of Y. pseudotuberculosis. Compound 1 was identified in a chemical screen based on the strong coupling between Yop secretion and regulation of yop gene expression (27). This was defined by Allaoui et al. as feedback inhibition (3); i.e., if the TTS is blocked, which is achieved in vitro by the addition of millimolar concentrations of Ca2+, the transcription of yop genes is down-regulated. Thus, by employing a reporter system based on the luxAB genes regulated by the yopE promoter, we screened a chemical library and identified a number of compounds that inhibited the light signal emitted by LuxAB (27). These compounds can theoretically block the light signal at different levels: they could inhibit the luciferase directly, be toxic for the bacteria, mimic the effect of Ca2+, interfere directly with LuxAB expression, or actually block the TTS system and thereby indirectly reduce the level of transcription from the yopE promoter. We have earlier shown that compound 1 most likely blocks Yop secretion by targeting the TTS (27), but a direct effect on expression could not be excluded. We therefore initiated here a number of experiments that were aimed to discriminate between these two possibilities. We found that compound 1 specifically blocks the TTS system of Yersinia.
This is confirmed by experiments based on the following information. There are two regulatory loops that control Yop expression during infection (16). One positive loop responds to the environmental temperature during infection, and the activator LcrF is the master regulator of this loop (9, 16, 52). Importantly, LcrF is not directly involved in the feedback inhibition via the TTS system. In contrast, the negative control loop is directly coupled to the feedback inhibition of Yop expression. Mutational analysis (16) has defined three proteins, LcrH, YopD, and LcrQ, that are key elements in the negative regulation, and recent results have indicated that they work in concert to block expression of Yops (4). However, LcrQ is the only protein that inhibits Yop expression when overexpressed in a wild-type background (39). Moreover, LcrQ is rapidly secreted via the TTS system after activation of Yop secretion (shift to low Ca2+ in vitro) (37). These results have enabled us to suggest a model in which LcrQ is a negative element that is rapidly secreted when the TTS system is activated, resulting in a lowered intracellular concentration of LcrQ and consequently to relief of the inhibitory effect of LcrQ on Yop expression (37). Although an lcrQ null mutant constitutively expresses the Yops at 37°C, Yop secretion is still controlled in vitro by the Ca2+ concentration, and in this respect the mutant is behaving like the corresponding wild-type strain (39). We therefore used the lcrQ mutant to test if compound 1 had an effect on yop expression that was uncoupled from the TTS system-mediated feedback control mechanism. Our results showed that compound 1 did not influence either the level of Yop effector expression or the expression of LcrQ per se. Instead, our data suggest that the drug prevents secretion and that this is the cause of the inhibition of Yop expression. This idea is strengthen by the finding that compound 1 had no effect on either yopE or lcrF transcription in a yscT secretion mutant. The effect of compound 1 was faster (<15 min) if the drug was added prior to the time point where secretion via the TTS system was fully induced than if it was added after full induction of the system (∼30 min) (Fig. 2a). This difference can be explained in terms of intracellular LcrQ concentrations at the time point of drug addition. We argue that the level of LcrQ is lower due to secretion at the later time point and consequently a longer time is needed to build up repressive concentration of LcrQ. This further supports the role of compound 1 as a TTS inhibitor.
Compound 1 mimicked the effect of addition of Ca2+ to the bacteria, since addition of the drug down-regulated transcription from the yopE promoter and up-regulated lcrF transcription. The fact that lcrF transcription is up-regulated by addition of compound 1 shows that the drug is not generally toxic for the bacteria or that the drug negatively affects expression and/or enzymatic activity of the luciferase. Identical conclusions can also be drawn from our work with lcrQ-luxAB transcriptional fusions. Addition of compound 1 inhibited Yop secretion in the wt as well as in an lcrQ mutant, as did addition of Ca2+. The fact that compound 1 inhibited Yop secretion in an lcrQ mutant although Yop expression was derepressed argues strongly for a direct effect of compound 1 on Yop secretion. Moreover, Yop secretion was also blocked in the calcium-blind lcrG and yopN mutants, which are able to express and secrete Yops in the presence of Ca2+. Thus, the effect of the drug is independent of the calcium concentration. Collectively, all of our in vitro results are in favor of the idea that compound 1 specifically inhibits TTS, and we suggest that compound 1 directly targets one or several components of the TTS machinery of Yersinia, excluding YopN and LcrG.
We anticipated from these results that treatment of bacteria with compound 1 under in vivo conditions would block translocation of effector proteins into the eukaryotic target cell. No obvious effect of the drug on HeLa cells was noticed. Infection of the cells with wt Yersinia resulted in translocation of YopH and YopE, while treatment with compound 1, added at the time of infection, inhibited translocation of YopH and reduced the cytoxic effect of YopE on the HeLa cells. Importantly, it is known that only a small amount of translocated YopE is required to induce cytotoxicity. We conclude that compound 1 interferes with the TTS system also under in vivo conditions and thereby block the microinjection of effector proteins into the target cell. In our assay system the drug did not affect the HeLa cells, since they efficiently removed the translocation-defective yopB mutant from the culture medium via a phagocytic-like process (5) and this process was equally efficient in absence or presence of the drug. The mutant bacteria did not increase in numbers during incubation, since intracellularly located Y. pseudotuberculosis cannot replicate in this environment (44). In contrast, wt Yersinia blocked uptake, and consequently, during 7 h of incubation, bacteria increased in numbers in the infected culture. Addition of compound 1 to cells infected with wt Yersinia resulted in a reduction in bacterial numbers, suggesting that addition of the drug lowered translocation of the effector proteins, resulting in an increased uptake of the bacteria. It should be noted in this context that the drug does not affect the growth rate of the bacteria (27). Treatments with the drug in these experiments led to only a partial uptake of the wild-type strain, whereas the immunostaining experiments indicated a total blockage of Yop translocation. However, these results are not necessarily in conflict, since the quantitative uptake data in Fig. 6 may reflect the qualitative observation of protein translocation in Fig. 7. Nevertheless, both experiments support the notion that compound 1 inhibits virulence to the advantage of the eukaryotic cell.
In parallel with the detailed investigation of compound 1, around 60 compounds from the same class (Fig. 1) were prepared and evaluated in the luciferase assay. In total, this resulted in 23 compounds that exhibited significant inhibition of the luciferase signal from YPIII(pIB29) yopE-luxAB at 50 μM (Table 1). Compounds 1 to 7 were identified in the original screening campaign (27), and compounds 8 to 23 represent additional inhibitors. Western analysis of inhibition of protein secretion was carried out for eight of the most potent of these compounds (Fig. 8). In agreement with the observations made with compound 1 (reference 27 and the present study), the level of reporter gene inhibition roughly matched inhibition of actual protein secretion (Table 1; Fig. 8). Several of the compounds were as effective as compound 1 in terms of inhibition of the reporter gene signal and protein secretion. Although all inhibitors in Table 1 belong to the same compound class (Fig. 1), the data do not provide obvious structure-activity relationships except the basic information gained with compounds 1 to 7 (Fig. 1; Table 1). This is most likely because bacterium-based assays are used to assess biological activity. It cannot be unequivocally concluded what mechanism is actually affected by a certain change in chemical structure; e.g., a stronger inhibition can be a result of active uptake or reduced metabolism rather than improved interactions with the actual protein target. However, the compounds in Table 1 suggest that the chemical structure can be varied in order to improve TTS inhibition and physiochemical properties such as solubility. Furthermore, since screening and subsequent characterization are carried out with viable bacteria, all active compounds act on the bacterium, i.e., the exact agent that infects the host.
In conclusion, we have prepared and characterized a number of TTS inhibitors and shown that compound 1 specifically targets the TTS system of Yersinia and that compound 1 blocks translocation of Yop effectors, leading to a chemical attenuation resembling mutants that are defective in secretion and translocation. Such mutants are nonvirulent, and thus it is possible that virulence-blocking agents specifically targeting the TTS system can be developed as novel antibacterial agents. We are currently exploring this path.
Acknowledgments
We are grateful for financial support from the Swedish National Research Council, The Foundation of Strategic Research, The Foundation for Technology Transfer in Umeå, and Innate Pharmaceuticals AB.
We are grateful for chemical syntheses by undergraduate students in the medicinal chemistry course at the Department of Chemistry, Umeå University. We thank Fredrik Nordfelth for excellent technical assistance with the luciferase assays.
Editor: J. B. Bliska
REFERENCES
- 1.Ainscough, E. W., A. M. Brodie, W. A. Denny, G. J. Finlay, S. A. Gothe, and J. D. Ranford. 1999. Cytotoxicity of salicylaldehyde benzoylhydrazone analogs and their transition metal complexes: quantitative structure-activity relationships. J. Inorg. Biochem. 77:125-133. [DOI] [PubMed] [Google Scholar]
- 2.Alksne, L. E., and S. J. Projan. 2000. Bacterial virulence as a target for antimicrobial chemotherapy. Curr. Opin. Biotechnol. 11:625-636. [DOI] [PubMed] [Google Scholar]
- 3.Allaoui, A., R. Schulte, and G. R. Cornelis. 1995. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol. Microbiol. 18:343-355. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. M., K. S. Ramamurthi, C. Tam, and O. Schneewind. 2002. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J. Bacteriol. 184:1287-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartra, S., P. Cherepanov, A. Forsberg, and K. Schesser. 2001. The Yersinia YopE and YopH type III effector proteins enhance bacterial proliferation following contact with eukaryotic cells. BMC Microbiol. 1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, P. B., Jr., I. Rundquist, I. Svensson, and V. P. Collins. 1987. Use of cytofluorometry to evaluate binding of antibodies to the cytoskeleton of cultured cells. J. Histochem. Cytochem. 35:1381-1388. [DOI] [PubMed] [Google Scholar]
- 7.Carey, K. L., N. J. Westwood, T. J. Mitchison, and G. E. Ward. 2004. A small-molecule approach to studying invasive mechanisms of Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 101:7433-7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G., C. Sluiters, C. L. de Rouvroit, and T. Michiels. 1989. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G., J. C. Vanootegem, and C. Sluiters. 1987. Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb. Pathog. 2:367-379. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis, G. R., and H. Wolf-Watz. 1997. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23:861-867. [DOI] [PubMed] [Google Scholar]
- 13.Dauzonne, D., and L. Martinez. 1995. Synthesis of the 3-aminoflavone-8-acetic acid. Terahedron Lett. 36:1845-1848. [Google Scholar]
- 14.Forsberg, A., and R. Rosqvist. 1993. In vivo expression of virulence genes of Yersinia pseudotuberculosis. Infect. Agents Dis. 2:275-278. [PubMed] [Google Scholar]
- 15.Forsberg, A., A. M. Viitanen, M. Skurnik, and H. Wolf-Watz. 1991. The surface-located YopN protein is involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol. Microbiol. 5:977-986. [DOI] [PubMed] [Google Scholar]
- 16.Forsberg, A., and H. Wolf-Watz. 1988. The virulence protein Yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-plB1-encoded trans-acting elements controlled by temperature and calcium. Mol. Microbiol. 2:121-133. [DOI] [PubMed] [Google Scholar]
- 17.Furste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 18.Galimand, M., A. Guiyoule, G. Gerbaud, B. Rasoamanana, S. Chanteau, E. Carniel, and P. Courvalin. 1997. Multidrug resistance in Yersinia pestis mediated by a transferable plasmid. N. Engl. J. Med. 337:677-680. [DOI] [PubMed] [Google Scholar]
- 19.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ginocchio, C. C., and J. E. Galan. 1995. Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect. Immun. 63:729-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 22.Hamilton, E. P., and N. Lee. 1988. Three binding sites for AraC protein are required for autoregulation of araC in Escherichia coli. Proc. Natl. Acad. Sci. USA 85:1749-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmstrom, A., J. Petterson, R. Rosqvist, S. Hakansson, F. Tafazoli, M. Fallman, K. E. Magnusson, H. Wolf-Watz, and A. Forsberg. 1997. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol. Microbiol. 24:73-91. [DOI] [PubMed] [Google Scholar]
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inglesby, T. V., D. T. Dennis, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, J. F. Koerner, M. Layton, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, M. Schoch-Spana, and K. Tonat. 2000. Plague as a biological weapon—medical and public health management. JAMA 283:2281-2290. [DOI] [PubMed] [Google Scholar]
- 26.Kauppi, A. M., R. Nordfelth, U. Hagglund, H. Wolf-Watz, and M. Elofsson. 2003. Salicylanilides are potent inhibitors of type III secretion in Yersinia. Adv. Exp. Med. Biol. 529:97-100. [DOI] [PubMed] [Google Scholar]
- 27.Kauppi, A. M., R. Nordfelth, H. Uvell, H. Wolf-Watz, and M. Elofsson. 2003. Targeting bacterial virulence. Inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241-249. [DOI] [PubMed] [Google Scholar]
- 28.Knappik, A., and A. Pluckthun. 1994. An improved affinity tag based on the FLAG peptide for the detection and purification of recombinant antibody fragments. BioTechniques 17:754-761. [PubMed] [Google Scholar]
- 29.Lee, Y. M., F. Almqvist, and S. J. Hultgren. 2003. Targeting virulence for antimicrobial chemotherapy. Curr. Opin. Pharmacol. 3:513-519. [DOI] [PubMed] [Google Scholar]
- 30.Linington, R. G., M. Robertson, A. Gauthier, B. B. Finlay, R. van Soest, and R. J. Andersen. 2002. Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org. Lett. 4:4089-4092. [DOI] [PubMed] [Google Scholar]
- 31.Livermore, D. 2004. Can better prescribing turn the tide of resistance? Nat. Rev. Microbiol. 2:73-78. [DOI] [PubMed] [Google Scholar]
- 32.Muller, S., M. F. Feldman, and G. R. Cornelis. 2001. The type III secretion system of Gram-negative bacteria: a potential therapeutic target? Expert Opin. Ther. Targets 5:327-339. [DOI] [PubMed] [Google Scholar]
- 33.Neyt, C., and G. R. Cornelis. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 33:971-981. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen, S. F., S. B. Christensen, G. Cruciani, A. Kharazmi, and T. Liljefors. 1998. Antileishmanial chalcones: statistical design, synthesis, and three-dimensional quantitative structure-activity relationship analysis. J. Med. Chem. 41:4819-4832. [DOI] [PubMed] [Google Scholar]
- 35.Nordfelth, R., and H. Wolf-Watz. 2001. YopB of Yersinia enterocolitica is essential for YopE translocation. Infect. Immun. 69:3516-3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Persson, C., R. Nordfelth, A. Holmstrom, S. Hakansson, R. Rosqvist, and H. Wolf-Watz. 1995. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol. Microbiol. 18:135-150. [DOI] [PubMed] [Google Scholar]
- 37.Pettersson, J., R. Nordfelth, E. Dubinina, T. Bergman, M. Gustafsson, K. E. Magnusson, and H. Wolf-Watz. 1996. Modulation of virulence factor expression by pathogen target cell contact. Science 273:1231-1233. [DOI] [PubMed] [Google Scholar]
- 38.Poole, K. 2001. Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 39.Rimpilainen, M., A. Forsberg, and H. Wolf-Watz. 1992. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotuberculosis shows extensive homology to YopH. J. Bacteriol. 174:3355-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosqvist, R., I. Bolin, and H. Wolf-Watz. 1988. Inhibition of phagocytosis in Yersinia pseudotuberculosis: a virulence plasmid-encoded ability involving the Yop2b protein. Infect. Immun. 56:2139-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosqvist, R., A. Forsberg, M. Rimpilainen, T. Bergman, and H. Wolf-Watz. 1990. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol. Microbiol. 4:657-667. [DOI] [PubMed] [Google Scholar]
- 42.Rosqvist, R., S. Hakansson, A. Forsberg, and H. Wolf-Watz. 1995. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae and shigellae. EMBO J. 14:4187-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosqvist, R., K. E. Magnusson, and H. Wolf-Watz. 1994. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 13:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosqvist, R., and H. Wolf-Watz. 1986. Virulence plasmid-associated HeLa cell induced cytotoxicity of Yersinia pseudotuberculosis. Microb. Pathog. 1:229-240. [DOI] [PubMed] [Google Scholar]
- 45.Salyers, A. A., Gupta, A., and Y. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 46.Skryzpek, E., and S. C. Straley. 1993. LcrG, a secreted protein involved in negative regulation of the low-calcium response in Yersinia pestis. J. Bacteriol. 175:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsukahara, K., K. Hata, K. Nakamoto, K. Sagane, N. A. Watanabe, J. Kuromitsu, J. Kai, M. Tsuchiya, F. Ohba, Y. Jigami, K. Yoshimatsu, and T. Nagasu. 2003. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol. Microbiol. 48:1029-1042. [DOI] [PubMed] [Google Scholar]
- 48.Walsh, C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65-70. [DOI] [PubMed] [Google Scholar]
- 49.Warabi, K., K. Zimmerman, J. Shen, A. Gauthier, M. Robertson, B. Finlay, R. vanSoest, and R. Andersen. 2004. Pachymoside A—a novel glycolipid isolated from the marine sponge Pachymatisma johnstonia. Can. J. Chem. 82:102-112. [Google Scholar]
- 50.Ward, G. E., K. L. Carey, and N. J. Westwood. 2002. Using small molecules to study big questions in cellular microbiology. Cell Microbiol. 4:471-482. [DOI] [PubMed] [Google Scholar]
- 51.Wren, B. W. 2003. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55-64. [DOI] [PubMed] [Google Scholar]
- 52.Yother, J., T. W. Chamness, and J. D. Goguen. 1986. Temperature-controlled plasmid regulon associated with low calcium response in Yersinia pestis. J. Bacteriol. 165:443-447. [DOI] [PMC free article] [PubMed] [Google Scholar]