Abstract
Infections with avian pathogenic Escherichia coli (APEC) cause colibacillosis, an acute and largely systemic disease resulting in significant economic losses in poultry industry worldwide. Although various virulence-associated genes have been identified in APEC, their actual role in pathogenesis is still not fully understood, and, furthermore, certain steps of the infection process have not been related to previously identified factors. Here we describe the application of a signature-tagged transposon mutagenesis (STM) approach to identify critical genes required for APEC infections in vivo. Twenty pools of about 1,800 IMT5155 (O2:H5) mutants were screened in an infection model using 5-week-old chickens, and potentially attenuated mutants were subjected to a secondary screen and in vivo competition assays to confirm their attenuation. A total of 28 genes required for E. coli septicemia in chickens were identified as candidates for further characterization. Among these disrupted genes, six encode proteins involved in biosynthesis of extracellular polysaccharides and lipopolysaccharides; two encode iron transporters that have not been previously characterized in APEC in in vivo studies, and four showed similarity to membrane or periplasmic proteins. In addition, several metabolic enzymes, putative proteins with unknown function, and open reading frames with no similarity to other database entries were identified. This genome-wide analysis has identified both novel and previously known factors potentially involved in pathogenesis of APEC infection.
Escherichia coli typically colonizes the avian gastrointestinal tract and other mucosal surfaces. While most strains are commensal, certain strains designated avian pathogenic E. coli (APEC) have the ability to cause severe disease. Predominant serotypes of APEC are O1:K1, O2:K1, and O78:K80 (7, 13, 18, 26). APECs most likely enter and colonize the avian respiratory tract by inhalation of fecal dust, leading to localized infections such as airsacculitis and pneumonia. In certain cases, they spread into various internal organs and typically cause pericarditis, perihepatitis, peritonitis, salpingitis, and other extraintestinal diseases. Colibacillosis of poultry is characterized in its acute form by septicemia, commonly resulting in sudden death (6).
Several bacterial factors have been associated with the virulence of APEC, including adhesins, toxins, iron acquisition systems, colicin V plasmid, serum resistance proteins, and capsule as well as lipopolysaccharide complexes (15, 21, 37). However, the mechanisms underlying pathogenicity are still not fully understood, and only certain steps of the infection process can be accounted for by these known virulence factors. In recent years, genome-wide analyses have led to a better understanding of the molecular mechanisms of pathogenicity. New molecular approaches have also aided in the identification of genes involved in pathogenesis, including in vivo expression technology, selective capture of transcribed sequences (SCOTS), differential fluorescence induction, and signature-tagged transposon mutagenesis (STM) (27, 33, 42, 64). Recently, suppression subtractive hybridization has been used successfully to identify genes present in the genome of two APEC strains but which are absent in E. coli K-12 MG1655. Dozois et al. (16) applied SCOTS to identify conserved genes in APEC strain χ7122 that are expressed in infected chicken tissues. More recently, a genomic subtraction was performed between the APEC strain MT512 and the nonpathogenic E. coli strain EC79 (58). Pathogen-specific DNA or cDNA such as putative adhesin, lipopolysaccharide core synthesis, iron-responsive metabolic enzymes, plasmid- and phage-encoded genes, and genes of unknown function were successfully enriched and isolated by these authors. However, neither genomic subtraction nor SCOTS gives direct information about the significance of the isolated genes in virulence. Here we report the application of STM in a chicken infection model using APEC wild-type strain IMT5155 (O2:H5), which was responsible for a severe outbreak of avian colisepticemia in Germany. We report the identification of both known and novel APEC genes involved in pathogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
E. coli strain IMT5155 (O2:H5) was used for infection studies, mutant construction and STM analyses. The strain was isolated from the internal organs of a 4-month-old laying hen in Germany with clinical symptoms of colisepticemia. Preliminary infection studies confirmed the virulence of the strain that caused severe symptoms of colibacillosis and high mortality rates in 6-month-old chickens (data not shown). Furthermore, IMT5155 has been included in large-scale phylogenetic analysis of 150 APEC strains and was grouped into one of the most representative clonal lineages in Germany. The strain harbors the virulence-associated genes fyuA, irp2, cva/cvi, tsh, fimC, iss, ompA, traT, vat, and iucD but is negative for papC, pic, sat, and hlyA, as shown by hybridization and PCR experiments (20).
A spontaneous nalidixic acid-resistant mutant was generated by growing IMT5155 in the presence of the antibiotic and plating 109 CFU to solid medium containing nalidixic acid and confirmed to retain full pathogenicity in the chicken model (data not shown). E. coli CC118 λpir was used for maintaining the tagged pUTmini-Tn5km2 plasmids, and E. coli S17-1 λpir was used as the donor strain for conjugation. Bacterial strains were routinely cultured at 37°C in Luria-Bertani (LB) broth or on LB agar plates containing the appropriate antibiotics at the following concentrations: ampicillin (Amp), 50 μg/ml; kanamycin (Kan), 50 μg/ml; nalidixic acid (Nal), 30 μg/ml. Strain IMT5104 (O8:H−) served as a negative control during animal tests. This strain was isolated from the feces of a healthy laying hen and harbors none of the previously identified virulence-associated genes common to APEC strains.
Selection of the tags and construction of the APEC mutant library.
A pool of tagged pUTmini-Tn5km2 plasmids (Ampr, Kmr) was kindly provided by David Holden (Imperial College, London, UK). Briefly, each tag contains a 40-bp random sequence flanked by two 20-bp invariable sequences. The invariable sequences serve as primers for amplification of the tags (33). E. coli CC118 λpir was transformed with pUTmini-Tn5km2 containing signature tags, and a total of 384 transformants were isolated for further screening by hybridization with their cognate labeled tags. After extensive cross-hybridization experiments, 90 clones that showed the strongest hybridization signals with single-labeled tags and that did not cross-react with each other were collected for further investigations (23). Finally, plasmids containing specific tags were purified from the clones and individually transformed into donor strain E. coli S17-1 λpir. Independent matings were set up by growing each E. coli S17-1 λpir (with tag) clone and the IMT5155 Nalr to late log phase. To construct tagged transposon mutants of APEC, 400 μl donor and 400 μl recipient cells were mixed and centrifuged. After discarding the supernatant the cells were resuspended in 10 μl of a 10 mM MgSO4 solution and plated onto LB agar plates and incubated at 37°C for 8 h. Colonies were collected and resuspended in phosphate-buffered saline (PBS) and plated onto LB agar containing kanamycin and nalidixic acid. Following selective overnight growth, single colonies were resuspended in 300 μl LB supplemented with nalidixic acid and kanamycin. After an overnight incubation, 53 μl glycerol (15%) was added and a total of 1,827 mutants were stored at −80°C in microtiter plates.
Animal model, screening of the STM transposon mutant library, and in vitro and in vivo competition assay.
A newly established chicken infection model for E. coli septicemia was used to perform STM analyses. Five-week-old SPF white leghorn specific pathogen-free chickens (Lohmann Selected Leghorn; Lohmann Tierzucht GmbH, Cuxhaven, Germany) were inoculated into the trachea with a 0.5 ml suspension containing 108 CFU of strain IMT5155 for preliminary studies. For preparation of mutant inoculants, frozen plates of pooled APEC mutants were taken out of −80°C storage and subcultured by transferring 10 μl from each well to a new 96 well round bottom plate containing 300 μl LB with kanamycin and nalidixic acid. Following overnight culture, each plate was pooled and 10 ml of pooled bacteria was added to 90 ml LB and then incubated at 37°C with aeration to an optical density at 600 nm of approximately 1. Cultures were diluted in PBS to approximately 2 × 108 bacteria per ml. A total of 0.5 ml of each mutant pool was used to infect at least four 5-week-old SPF chickens per mutant pool via intratracheal application. After 48 h and 72 h of infection, chickens were killed and bacteria were reisolated from spleens. The organs from at least two chickens were homogenized in sterile PBS and then plated onto LB agar supplemented with kanamycin and nalidixic acid. Following overnight incubation, at least 10,000 colonies (5,000 colonies from each chicken) were pooled in 10 ml PBS to form the “output pool.” A 5 ml aliquot of this pool was used to prepare genomic DNA. Mutants with reduced output hybridization signals were subsequently used individually to quantify their relative growth rates.
For both in vitro and in vivo competition assays, cultures of mutant and wild-type strain were mixed at a ratio of 1:1. For in vitro competition, the bacteria were incubated in LB broth for 4 h at 37°C and then plated onto media with or without kanamycin. For in vivo competition, four chickens were infected with 108 CFU of this mixture. After 48 h of infection, spleen, heart, liver, lung, and kidney were collected, weighed, and homogenized, and the dilutions were plated on media with or without kanamycin for selection of mutants or total bacteria, respectively. A competitive index (CI) was calculated for each mutant by dividing the output ratio (mutant/wild type) by the input ratio (mutant/wild type). Due to this CI, mutants outcompeted up to 10-fold were evaluated as slightly, up to 100-fold as moderately, and more than 100-fold as highly attenuated.
DNA manipulations and sequence analysis.
DNA manipulations and transformations were performed using standard methods (4). Restriction and DNA-modifying enzymes were obtained from Roche Molecular Biochemicals (Mannheim, Germany). Hybridization was performed with digoxigenin (DIG)-labeled probes using a Roche labeling and detection kit (Roche, Mannheim, Germany). A total of 500 to 1,000 ng of DNA was used for hybridization experiments. Blotting onto positively charged nylon membranes (Roche, Mannheim, Germany) was performed as recommended by the manufacturer.
Probe labeling and hybridizations for identification of mutants disappeared from recovered pools.
We modified the strategy described by Fuller et al. (23). To significantly reduce background of hybridization, PCR generated tag sequences were used as the target DNA for dot blots rather than using the entire plasmid containing the tagged transposon. In addition, we tested lower concentrations of DIG-dUTP to ensure more complete digestion by HindIII and removed the conserved sequences of tags by gel electrophoresis (data not shown).
For amplification of DNA tags, either 5 ng of plasmid DNA or 5 μg of genomic DNA from input and recovered pools was used as a template in first round PCRs using the conditions described by Hensel et al. (33). The amplified tags from both the inoculum and the recovered pools were compared by labeling with digoxigenin (DIG)-dUTP using a PCR DIG probe synthesis kit (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions. After purification, the PCR products were labeled in a volume of 50 μl using 50 pmol of each primer P2 and P4 (Table 1) and a 1:1 mix of DIG-dUTP and 2.5 mM deoxynucleoside triphosphate stock solution with the same cycle conditions as for the first round PCR (23). The labeled PCR products were digested with HindIII in a total volume of 400 μl, ethanol precipitated, and isolated from a 4% 3:1 NuSieve GTG:agarose gel (Cambrex Corporation, New Jersey). The region containing the labeled tag was excised, and the entire gel slice was directly used as probe in hybridizations. Plasmids containing the cognate tags from the original plate were used as templates for the amplification of these tags with primer pair P2 and P4 (Table 1). Identical dot blots were prepared by transferring these amplicons onto positively charged nylon membranes followed by hybridization. Mutants that showed hybridization signals with the probe from the input pool but not with that from the recovered pool were selected for further analysis.
TABLE 1.
Oligonucleotide primers used in this study and results of PCRs to detect genes of pathogenicity island
| Primer | Sequence (5′-3′) | Presence of genea |
|---|---|---|
| P2 | TACCTACAACCTCAAGCT | |
| P4 | TACCCATTCTAACCAAGC | |
| P6 | CCTAGGCGGCCAGATCTGAT | |
| P7 | GCACTTGTGTATAAGAGTCAG | |
| P9 | CGCAGGGCTTTATTGATTC | |
| Arbi1 | GGCCACGCGTCGACTAGTAC(N)10 GATAT | |
| Arbi2 | GGCCACGCGTCGACTAGTAC | |
| Arbi3 | GGCCACGCGTCGACTAGTAC(N)10 TGACG | |
| Arbi4 | GGCCACGCGTCGACTAGTAC(N)10 ACGCC | |
| Arbi5 | GGCCACGCGTCGACTAGTAC(N)10 TACNG | |
| P19 | ATTCAACGGGAAACGTCTTG | |
| P20 | ACTGAATCCGGTGAGAATGG | |
| PapG-F | TTTGCGAGTGGAGTGTATTT | |
| PapG-R | TACCTAACCCAACCGAAAAT | |
| PapC-F | TGATATCACGCAGTCAGTAGC | |
| PapC-R | CCGGCCATATTCACATAA | |
| R14-15-F | GCCAGTGACACATACTGAGAGC | + |
| R14-15-R | CAGATGTACAGTGGCGCG | |
| R2 plus R3-F | GCTGTCAGAATATTTCGCTCG | + |
| R2 plus R3-R | AGTCCTGTCACGCTGAACG | |
| R1 plus R2-F | AGCCTTTCTGTTTTGAGCAT | + |
| R1 plus R2-R | TCGCTACTATTGATTCTTGC | |
| R1 plus f447-F | CCGCAAGAATCAATAGTAGC | + |
| R1 plus f447-R | CTGGCGAGAAGGGGATAATG |
PCR elaborated for detection of metV genomic island.
Identification of the transposon insertion sites and sequence analyses.
All of transposon insertion sites were amplified by arbitrary PCR (48). For the first round, the arbitrary primers Arbi1, Arbi3, Arbi4, and Arbi5 were used in combination with transposon-specific primer P9 (Table 1). Subsequently, 1 μl of each PCR product was used in a second round of nested PCR with primers Arbi2 and P6 (Table 1). Arbi2 is homologous to the 5′ sequences of the above-mentioned arbitrary primers, and P6 is a transposon I terminus-specific primer. Genomic DNA from the wild-type strain served as negative control in all reactions. The second round PCR products were purified and sequenced. DNA sequencing was performed commercially (AGOWA GmbH, Berlin, Germany). Data analysis was done using public DNA and protein databases (http://www.ncbi.nlm.nih.gov/BLAST) and employing the BLASTX and BLASTN algorithms (1). All DNA sequences were compared with sequences in the TIGR comprehensive microbial resource.
Analysis of LPS expression.
Lipopolysaccharide (LPS) was obtained by proteinase K digestion of whole cells as previously described (34). LPS samples were separated on 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel according to standard methods (4) and silver staining as described by Tsai and Frasch (63a).
RESULTS AND DISCUSSION
Construction of a signature-tagged transposon mutant library of avian pathogenic E. coli.
A library of 1,827 signature-tagged mini-Tn5km2 mutants of strain IMT5155 was constructed according to the methods of Hensel et al. (33) using 90 prescreened non-cross-reactive tags that could be amplified and labeled efficiently as described in Materials and Methods. Nineteen of the mutants (about 1%) were ampicillin resistant, indicating the presence of the suicide plasmid containing the transposon within the cytoplasm or integrated into the chromosome. Eight mutants lost kanamycin resistance spontaneously and were eliminated from the library. To determine whether the transposon had any insertional hot spots, Southern blotting analysis was performed on 18 randomly chosen mutants, digested with PstI and probed with a fragment of the kanamycin resistance gene amplified from the pUT-mini-Tn5km2 by primer pair P19 and P20 (Table 1). The hybridization patterns from each PstI-restricted mutant differed from one another, and each mutant showed only one positive band (data not shown). Overall, these tests indicated a broad distribution of transposon insertion sites among the mutants and only one insertion in each mutant. Following this procedure the mutants were stored in 96 well microtiter plates at −80°C for further in vivo testing in a chicken infection model.
Screening of the APEC mutant library in a chicken model.
Preliminary experiments were performed to obtain an optimal protocol for screening in vivo attenuation of STM mutants. Critical parameters for STM studies are the selection of a highly pathogenic strain, determination of an optimal 50% infective dose value with regard to the complexity of tagged pools, appropriate infection duration, and determination of the optimal time for reisolation of bacteria from different internal organs. A range of experimental factors, including the infectious dose and the method of recovery, were investigated before we could reproducibly recover the majority of individual mutants. Using intratracheal challenge doses of 105, 106, and 107 CFU of strain IMT5155 Nalr, only a low percentage of animals developed typical signs of colibacillosis. In contrast, the use of 109 CFU for intratracheal inoculation resulted in the sudden death of several chickens before reisolation of bacteria took place. An inoculation dose of 108 CFU showed reproducible disease progression, and bacteria could be reisolated from about 85% of infected chickens. Irrespective of the bacterial concentration used for inoculation, the control strain, IMT5104, was not recovered from any internal organ and infected animals showed no clinical signs of colibacillosis or of macroscopic lesions typical of the disease, as shown by necropsy.
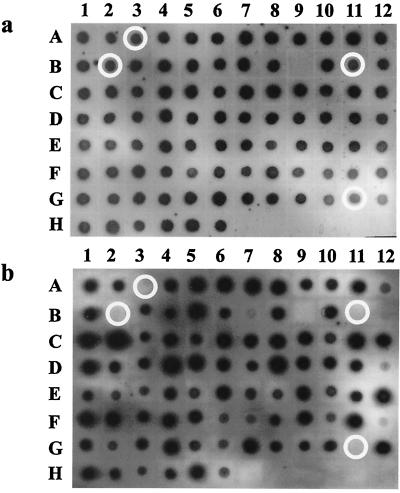
In a following experiment, three randomly selected pools of 90 mutants each were used to infect chickens with 108 CFU of IMT5155 Nalr and bacteria were recovered from spleens at 48 and 72 h after infection. Similar hybridization results were obtained when blots were probed with tags from bacteria recovered from at least two chickens after 48 h of infection. In most cases, three or four mutants were absent from each output pool (Fig. 1). In contrast, unusually high numbers of mutants were absent in the recovered pools when bacteria were reisolated after 72 h, suggesting that many mutants were simply outgrown by others. As these results were reproducible in several experiments, we therefore decided to test pools of 90 mutants with an inoculating dose of 108 CFU and recovered bacteria from spleens after 48 h of infection. A total of 1,800 mutants from 20 pools were used for screening, and 65 mutants absent from the recovered pools were selected for further studies. These 65 mutants were subjected to a second screen as described above, and each pool was tested twice to verify the results. After the second round of STM screening, 30 mutants showed a reproducible loss in hybridization signals. Of these mutants, 19 were used for in vivo competition assay based on their belonging to one of the eight classes listed in Table 2.
FIG. 1.
Example of STM results from a representative pool. Most mutants grew as well in vivo as in vitro. (a) Dot blot of input pool 16. (b) Dot blot of recovered pool 16. Signals of mutants A3, B2, B11, and G11 were absent and selected for further study.
TABLE 2.
Genetic loci disrupted in mutants of APEC strain IMT5155 Nalr identified by STM
| In vivo-identified mutant class | Mutant name | Similar E. coli genes (GenBank)
|
|||
|---|---|---|---|---|---|
| Genea | Accession no. | % identitya | Putative function | ||
| Capsule | M03A03 | kpsM | ECU59301 | 97 | Polysaccharide transport |
| M03E05 | kpsM | ECU59301 | Polysaccharide transport | ||
| M18H02 | kpsS | AAN82139 | 99 | Polysaccharide transport | |
| Colanic acid | M03G02 | wcaE | AF320069 | 97 | Putative glycosyltransferase |
| LPS | M13F03 | waaW | AAN82886 | 99 | LPS R1 core biosynthesis region |
| M12D10 | waaW | AAN82886 | 98 | LPS R1 core biosynthesis region | |
| M03D10 | waaL | NP_756310 | 97 | Polymer ligase | |
| M00G10 | None | None | 0 | O-antigen synthesisb | |
| Iron uptake system | M04E08 | sitB | AAN80284 | 98 | Iron transporter |
| M11B04 | chuA | AAN82744 | Heme utilization/transport protein | ||
| Membrane and periplasmic proteins | M17F12 | mppA | ECU88242 | 97 | Periplasmic murein peptide-binding protein precursor |
| M19A01 | narK | AAN80151 | 93 | Nitrite extrusion | |
| M19B12 | None | AAN80227 | 100 | Transport, drug/analog sensitivity | |
| M08E08 | sbmA | AAN78960 | 96 | Putative membrane, drug/analog sensitivity, possible envelope | |
| Metabolic enzymes | M00C03 | R3 | AAN81852 | 98 | Beta-cystathionase |
| M04C10 | tktA | AAN81968 | 91 | Transketolase 1 enzyme, central intermediary metabolism | |
| Regulators | M14G11 | carP | CAA60164 | 91 | Pyrimidine regulation |
| M18E10 | yjjQ | NP_757290 | 98 | Putative regulator | |
| Unknown | M03C05 | ykgC | NP_752357 | 100 | Putative oxidoreductase |
| M11E02 | malX | NP_755283 | 98 | Phosphotransferase system | |
| M17E09 | ygcW | AAC75816 | 100 | Hypothetical oxidoreductase | |
| M00C03 | ycjM | NP_753684 | 98 | Putative polysaccharide hydrolase | |
| M13H03 | None | AAN81851 | 96 | Phosphosugar isomerase | |
| M18E07 | ychF | NP_753566 | 100 | GTP-binding proteins | |
| M18B06 | None | AAB40738 | 99 | Inserted sequence | |
| M16D12 | yahA | AAC73418 | 100 | Unknown | |
| M00B02 | yagM | AAC73382 | 96 | Unknown | |
| M02E05 | None | NP_755082 | 98 | Unknown, hypothetical protein | |
| M00F07 | None | None | 0 | Unknown | |
| M18D02 | None | None | 0 | Unknown | |
Genetic locus with the closest match to the sequence interrupted by the transposon in each mutant.
As determined in this study.
Identification and characterization of the interrupted genes.
The insertion sites of mini-Tn5km2 in the selected 30 mutants were determined by amplifying their flanking DNA regions by arbitrarily primed PCR followed by sequencing of the amplified DNA products which ranged between 150 and 750 bp. For sequence analyses, BLASTX or BLASTN hits with the highest score and lowest e values were recorded. Of the 30 mutants, 28 were found to have transposon insertions in different genes (Table 2).
Quantification of virulence defects of selected mutants by in vivo competition assays.
In vivo competition assays were performed to validate the results of our STM screen and to quantify the degree of virulence attenuation of individual mutants. A total of 19 of the 30 mutants were selected, based on their belonging to one of the 8 classes listed in Table 2. These 19 mutants were mixed with the parental strain at a 1:1 ratio and inoculated into four chickens by intratracheal application. Total bacteria were recovered from lung, liver, heart, spleen, and kidney after 48 h, and CFU were enumerated on selective medium. A competitive index (CI) was calculated as described in Materials and Methods. Of the 19 mutants tested, 14 (73.7%) were outcompeted by the wild-type strain in all five internal organs examined, while the other five mutants (26.3%) showed reduced recovery only in certain organs. Furthermore, 15 (78.9%) of the mutants were outcompeted in certain organs more than 10-fold, and of these 6 (31.6%) were outcompeted more than 100-fold in certain organs, confirming the attenuation of the 19 selected mutants. The results of in vivo competition assays are shown in Table 3. These results suggested that most of the 30 mutants identified by the two rounds of STM screening were reproducibly attenuated.
TABLE 3.
Results of in vivo competition assay between IMT5155 NalR and its mutants
| Mutant | Gene/ homologue | CIa in vitro | CI of lung | CI of liver | CI of heart | CI of spleen | CI of kidney |
|---|---|---|---|---|---|---|---|
| M03A03 | kpsM | 1.56 | 3.5 × 10−1 | <1.0 × 10−3 | <1.0 × 10−3 | <1.0 × 10−3 | 1.5 × 10−1 |
| M18H02 | kpsS | 5.1 × 10−1 | 4.2 × 10−2 | <1.0 × 10−3 | <1.0 × 10−3 | 1.7 × 10−1 | <1.0 × 10−3 |
| M03G02 | wcaE | 8.0 × 10−1 | <1.0 × 10−3 | —b | 2.0 × 10−4 | 9.0 × 10−3 | 1.2 × 10−1 |
| M12D10 | waaW | 7.0 × 10−1 | 2.6 × 10−1 | 1.0 | 5.1 × 10−2 | 2.6 × 10−1 | 1.5 × 10−1 |
| M03D10 | waaL | 6.0 × 10−1 | 5.0 × 10−1 | 2.5 × 10−1 | 1.0 × 10−1 | 5.1 × 10−2 | 1.2 × 10−1 |
| M00G10 | None | 1.67 | 1.6 × 10−2 | 1.0 × 10−1 | 6.3 × 10−3 | —b | 2.7 × 10−1 |
| M04E08 | sitB | 3.2 × 10−1 | 8.3 × 10−2 | 2.1 × 10−1 | 4.8 × 10−2 | 1.3 × 10−2 | 8.8 × 10−2 |
| M11B04 | chuA | 9.6 × 10−1 | 6.45 × 10−2 | 1.8 × 10−1 | 4.0 × 10−2 | 4.12 × 10−2 | 1.5 × 10−1 |
| M17F12 | mppA | 4.6 × 10−1 | 6.6 × 10−1 | 3.3 × 10−1 | 1.0 | 2.0 × 10−1 | 7.0 × 10−1 |
| M19A01 | narK | 1.40 | 3.8 × 10−1 | 4.7 × 10−2 | 3.2 × 10−2 | 4.0 × 10−1 | 4.8 × 10−1 |
| M19B12 | None | 6.2 × 10−1 | 5.4 × 10−1 | 1.2 | 6.1 × 10−1 | 2.5 × 10−1 | 1.0 |
| M08E08 | sbmA | 6.4 × 10−1 | 2.0 × 10−1 | 1.4 × 10−1 | 2.2 × 10−1 | 2.5 × 10−1 | —b |
| M00C03 | r3 | 5.2 × 10−1 | 5.4 × 10−1 | 6.1 × 10−1 | 1.3 | 2.8 × 10−1 | 1.1 |
| M04C10 | tktA | 1.7 × 10−1 | 5.51 × 10−1 | 6.0 × 10−4 | 1.7 × 10−1 | 3.9 × 10−3 | 9.6 × 10−2 |
| M21C03 | ycjM | 1.08 | 5.3 × 10−1 | 1.7 × 10−1 | 6.12 × 10−2 | 4.6 × 10−1 | 3.6 × 10−1 |
| M11E02 | malX | 1.05 | 3.1 × 10−2 | 2.2 × 10−1 | 1.6 × 10−2 | 3.8 × 10−1 | 2.1 × 10−2 |
| M13H03 | None | 8.0 × 10−1 | 2.8 × 10−2 | 3.1 × 10−1 | —b | 2.3 × 10−1 | 2.9 × 10−1 |
| M18E07 | ychF | 5.8 × 10−1 | 3.1 × 10−2 | 1.42 × 10−2 | <1.0 × 10−4 | 1.5 × 10−2 | 1.8 × 10−2 |
| M18D02 | None | 2.07 | 9.0 × 10−2 | 2.9 × 10−1 | 2.2 × 10−1 | 6.4 × 10−2 | 8.5 × 10−1 |
CI, competitive index. Values > 1 indicate mutant outcompeted wild type, values <1 indicate wild type outcompeted mutant; mutants being outcompeted up to 10-fold were evaluated as slightly, up to 100-fold as moderately, and more than 100- fold as highly attenuated. In vivo competitive indices (CIs) are the average of results gained with two to four animals.
—, not tested.
Synthesis of different types of exopolysaccharides (EPS) is necessary for APEC virulence.
Several of the attenuated mutants had insertions in genes involved in the synthesis of extracellular polysaccharides. Two separate mutants were shown to have an insertion in the kpsM locus (M03A03, M03E05), while another was disrupted in the kpsS locus (M18H02). Both kpsM and kpsS encode proteins required for translocation of E. coli group II capsular polysaccharide across the inner membrane (11). The degree of attenuation for the mutants was high (more than 100-fold reduction in certain organs), but differed for both the mutant and target organs, with the most severe attenuation observed in liver and heart (Table 3). Previous epidemiological studies noted that the K1 (group II) antigen is frequently associated with APEC, particularly serotype O1 and O2 (29). More recently, a spontaneous K1 mutant of the APEC strain MT78 was found to show a decreased colonization potential in infected chickens and contributed to increased resistance to the bactericidal effects of chicken serum and phagocytosis by interaction with complement system (46, 47). The isolation of three independent attenuated mutants by STM in this study therefore provides further evidence for the importance of K1 capsule biosynthesis in APEC pathogenicity.
An additional STM mutant (M03G02) was found to harbor a disruption in a sequence related to wcaE, a putative colanic acid (CA) glycosyl transferase. This CA mutant was attenuated for survival in different organs, with the most severe defect in heart tissue (Table 3). Besides capsule biosynthesis, E. coli produces a variety of exopolysaccharides (EPS), e.g., colanic acid (CA). CA contains l-fucose, d-glucuronic acid, d-galactose, d-glucose, and pyruvate and forms a thick, mucoid matrix on the cell surface. Similar to the group IA capsular polysaccharides, CA is characterized by high molecular weight, specific sugar composition, and a chromosomal location near the his operon of the responsible biosynthetic genes (60). Hanna et al. (31) found that the capsular polysaccharide colanic acid of an uropathogenic E. coli did not contribute to bacterial adhesion to bladder epithelium as had been previously suggested but rather blocked the establishment of specific binding as well as time-dependent interactions between the uropathogenic strain and inert substrates. A CA-deficient mutant of E. coli O157:H7 demonstrated a reduction in heat and acid tolerance (43), suggesting that this exopolysaccharide confers a protective effect against host environmental stresses, such as body temperature and acid environment in macrophages (10). This might be advantageous for APEC which survive and replicate in an environment of 42°C, the normal body temperature of birds. APEC have also been demonstrated in macrophages in the air sac of chickens 24 h after infection, where they appear to be resistant to the acidic intracellular environment (53). Thus, for the first time, we provide direct evidence for an association of colanic acid with virulence and fitness of this pathogen. Furthermore, we established that mutants with insertions in genes involved in the synthesis of several distinct extracellular polysaccharide structures including group II capsule are attenuated in the ability to cause septicemia in a chicken model.
Lipopolysaccharide (LPS) contributes to viability of APEC in chickens.
LPS is one of the major components of the bacterial outer membrane. This amphipathic molecule is composed of lipid A, a core oligosaccharide, and a polysaccharide chain termed O-specific antigen. Four of the isolated STM mutants were shown to have transpositional insertions in genes involved in biosynthesis of LPS. M03D10 had a disruption in a gene encoding a surface polymer ligase termed waaL which joins newly synthesized O-polysaccharide to the lipid A core (14). In addition, two independent mutants (M13F03 and M12D10) had different transposon insertion sites in the same gene which showed similarity to waaW. This gene is involved in LPS R1 core biosynthesis and functions as a (galactosyl) LPS alpha-1,2 galactosyltransferase (32). The R1 core structure is the most prevalent among clinical isolates of E. coli (2, 25). The in vivo tested mutant M12D10 showed moderate attenuation in heart and slight attenuation in lung, spleen, and kidney but not in the liver. Clones corresponding to genes involved in the synthesis of the R1-type core LPS have recently been identified by selective capture of transcribed sequences (SCOTS) analysis (16). The O78-antigen of the APEC strain χ7122 used in this prior study was previously shown to be required for virulence by increasing the bacterial resistance to serum (9). We identified another mutant, M00G10, with a disruption in a gene locus of unknown function which does not share similarity with E. coli K12, CFT073, or EDL933 strains. Immediately downstream of this gene, there is a locus encoding a putative protein similar to glucose-1-phosphate thymidylyltransferase (RmlA), which is involved in O-antigen biosynthesis of Shigella boydii and E. coli (19, 65).
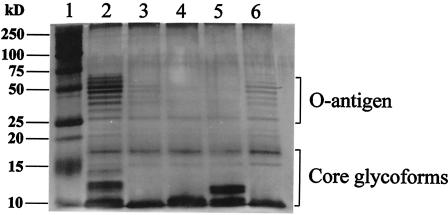
In order to confirm defects in LPS synthesis, LPS of the four mutant and wild-type strains was isolated and subjected to SDS-PAGE and silver staining. Compared to the parental strain, all four mutants showed reduced LPS production. As expected, M13F03 and M12D10, both being disrupted in waaW, also showed altered banding patterns for the LPS core region compared to wild-type strain. Mutant M03D10 (waaL) had alterations in both O-antigen and core region, whereas M00G10 (no similarity to any known gene) demonstrated altered banding patterns only in the O-antigen region (Fig. 2). These results indicated that the waa orthologues and the gene interrupted in M00G10 most likely function in different steps of LPS biosynthesis and are required for the expression of a complete LPS molecule, as was also shown in a previously STM study investigating the gastrointestinal colonization of infant rats by an E. coli K1 strain (49). Our findings of four attenuated LPS mutants by STM further confirm the importance of LPS, including O-antigen in the pathogenesis of APEC.
FIG. 2.
SDS-PAGE analysis of LPS from IMT5155 and its mutants. The gel was loaded with whole cell lysates of strains as follows: lane 1: ladder; lane 2, wild type; lane 3, waaW mutant (M13F03); lane 4, waaL mutant (M03D10); lane 5, mutant M00G10; lane 6, waaW mutant (M12D10). The positions of the repeating subunits of the O-antigen and the core region are indicated. All mutants yielded lower LPS production compared to wild type, and waaW mutants have altered LPS core structures; waaL mutant showed differences in both O-antigens and the core region and mutant M00G10 has altered O-antigen region.
Several iron uptake systems ensure bacterial survival in different host environments.
Two mutants with insertions in genes likely to be involved directly or indirectly in iron uptake were also isolated. Both mutants (sitB and chuA) showed moderate attenuation in different tissues. Sequestration of iron by host compounds results in an extremely low free iron concentration in vivo and has been suggested to limit bacterial growth in this environment. Consistent with this, iron acquisition systems have been associated with bacterial virulence, especially for bacteria causing septicemia. chuA encodes an outer membrane protein involved in heme uptake in enterohemorrhagic E. coli and uropathogenic E. coli (UPEC) strains (49, 61, 62) and has been implicated as a determinant of virulence in E. coli (28). chuA has previously been identified by suppression subtractive hybridization in APEC without any further functional characterization. Thus, the isolation of a chuA mutant and its attenuation in vivo is the most compelling evidence so far that this gene is important for the pathogenicity of APEC.
The second iron uptake protein identified in this study is highly similar to Salmonella enterica serovar Typhimurium and Shigella flexneri SitB, which is encoded in a four-member operon that mediates manganese and iron transport (56, 67). In Salmonella spp., the sit iron acquisition locus is encoded within a pathogenicity island and is required for full virulence of this pathogen (36, 67). A sitA mutant of Shigella flexneri showed reduced in vitro growth compared to the wild type, and sitA is likely located on a pathogenicity island in this species as well (56). More recently, the SitABCD system has been identified in an APEC strain MT512 by genomic subtraction (58). However, no direct evidence was shown that Sit is involved in the virulence of APEC by these authors. Here, again we provide the first striking evidence for a role of this factor in the pathogenesis of avian colibacillosis.
In addition to the chu and sit gene loci, IMT5155 possesses aerobactin and yersiniabactin as iron transport systems (20). The occurrence of multiple iron transport systems in APEC suggests the importance of this metal ion for bacterial survival, possibly helping the bacteria to adapt to various host environments during the infection process (63a).
Membrane and periplasmic proteins contribute to the virulence of APEC.
Several genes with high similarity (93 to 100%) to genes coding for diverse membrane and periplasmic proteins were also identified by STM. The mutant M08E08 showed a slight 4- to 6-fold attenuation in lung, liver, heart, and spleen and was shown to harbor a disruption in a gene similar to sbmA. SbmA is predicted to be an inner membrane transport protein and functions in the uptake of microcins B17 and J25 and bleomycin in E. coli (39). A sbmA orthologue mutant of Sinorhizobium (S.) meliloti exhibited increased sensitivity to agents such as hydrophobic dyes and detergents, indicating a role for SbmA in maintaining envelope integrity (22). In S. meliloti and Brucella abortus, orthologues of SbmA have also been shown to be important for long-term survival within host cells (40).
Mutant M17F12 was found to harbor a disruption in a gene with sequence similarity (97%) to mppA, encoding a periplasmic protein essential for import of bacterial cell wall peptide l-alanyl-γ-d-glutamyl-meso-diaminopimelate (51). The mutant showed a slight attenuation in lung, liver, spleen, and kidney. During growth, E. coli breaks down over one-third of its cell wall with each generation and reutilizes the tripeptide for synthesis of murein for the biosynthesis of the murein sacculus, the scaffolding structure of the bacterial cell wall. However, the principal pathway for uptake and reutilization of this tripeptide is indirect and the MppA pathway does not appear to play a key role in recycling (35, 50). This might suggest that a reduction in uptake/recycling through the MppA pathway would not have a significant effect on murein biosynthesis. However, as shown in other studies, MppA negatively regulates the expression of marA and hence MarA-dependent multiple antibiotic resistance genes in a signal transduction pathway (41). For the Salmonella enterica serovar Typhimurium, it has been demonstrated that the mar locus plays an important role in the interaction with porcine lung macrophages and is involved in adherence to human gut cells and invasion of and persistence in internal organs of chicken (54). Thus, presumably mppA mediates effects on APEC virulence indirectly, through signal transduction pathways affecting other virulence loci.
An additional STM mutant (M19A01) showed disrupted narK, a gene encoding a membrane protein involved in nitrite transport (12). Moderate attenuation was found in liver (∼20-fold) and heart (∼30-fold). The observed attenuation could be the result of loss of nitrate respiration and nitrate as an electron acceptor required for growth under anaerobic conditions (55), or the defect in NarK may limit utilization of nitrate as a nitrogen source. We also identified a gene encoding a putative protein 100% identical to an acriflavine resistance protein (M19B12). This mutant showed slight attenuation in lung, heart, and spleen but not in liver and kidney.
The ability to synthesize certain metabolic enzymes may be required for pathogenesis of colisepticemia.
The ability to adapt to the host environment is a key component of bacterial pathogenicity. We indeed identified two genes whose products are involved in metabolic pathways and nutrient uptake following our negative in vivo selection. Mutant M04C10 showed slight to high attenuation in liver, spleen, kidney, and heart and was slightly reduced in growth in vitro. The disrupted gene tktA encodes a transketolase catalyzing the reversible transfer of a ketol group between several donor and acceptor substrates. TktA has also recently been identified in APEC strain MT512 by genomic subtraction (58). This key enzyme is a link between glycolysis and the pentose phosphate pathway. E. coli contains two transketolase isozymes encoded by the tktA and tktB genes, with tktA encoding the major enzymatic activity in E. coli. Due to its wide substrate specificity the enzyme is involved in the catabolism of pentose sugars, in the formation of d-ribose 5-phosphate, and in the provision of d-erythrose 4-phosphate, a precursor of aromatic amino acids, aromatic vitamins, and pyridoxine (59). Since 4-(phosphohydroxy)-l-threonine and 1-deoxy-d-xylulose 5-phosphate are believed to be direct precursors of vitamin B6 in E. coli, and transketolase catalyses the formation of each precursor, vitamin B6 may be important for in vivo survival of APEC (57).
A further mutant M14G11 affecting the synthesis of pyrimidine and indirectly of vitamin B1 (vitamin B1 harbors a pyrimidine ring) is interrupted in the pyrimidine regulation gene carP (Table 2), a finding further pointing towards a crucial role of yet another vitamin, vitamin B1, for in vivo survival of APEC. Similar findings concerning the importance of vitamins were found in SCOTS analyses with a highly virulent APEC strain (16). A gene encoding a putative dethibiotin synthase was highly expressed in vivo during APEC infection. The authors suggested that biotin limitation may be an innate host response against the bacterium, as the biotin-binding protein avidin is generally induced in avian tissues after E. coli infection.
Identification of a putative new pathogenicity island.
One of the selected mutants, M00C03, is disrupted in a gene with similarity to r3 encoding a β-cystathionase. This enzyme converts cystathione into homocysteine (8). In a previous study it was reported that a cell-free β-cystathionase preparation of Bordetella avium was highly toxic against a variety of eukaryotic cell lines, including embryonic bovine tracheal cells and osteogenic cells (24). M00C03 was slightly outcompeted by its parental strain in lung, liver, and spleen but not in heart and kidney. Another mutant, M13H03, was shown to be disrupted in a gene encoding a putative phosphosugar isomerase. It demonstrated slight attenuation in liver, spleen, and kidney and moderate attenuation in the lung. In mutant M11E02, the transposon inserted into a gene with significant similarity to malX (synonym r2) which encodes the maltose- and glucose-specific component IIa of a phosphoenolpyruvate-dependent phosphotransferase system. This mutant was shown to be moderately attenuated in lung, heart, and kidney.
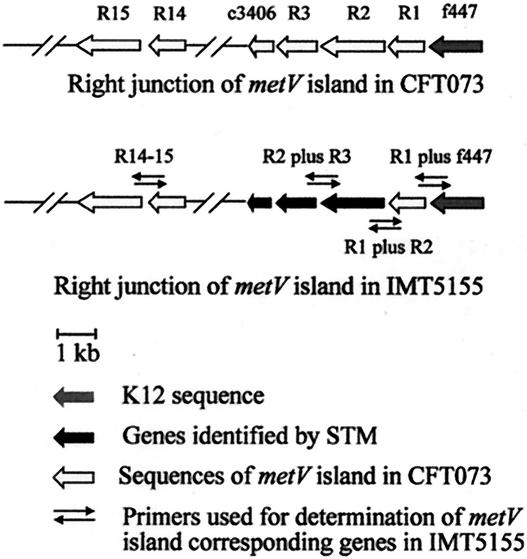
These genes have so far only been found in the uropathogenic E. coli (UPEC) strain CFT073 (30) but not in any other bacterial species. Interestingly, in CFT073 these genes are localized on a pathogenicity island inserted near the tRNA gene metV. It is well known that UPEC and APEC, both causing extraintestinal diseases, share identical virulence genes (66). In order to identify a similar pathogenicity island as described for CFT073, we utilized primer pairs R2 plus R3, R1 plus R2, and R1 plus f447 (Table 1), and indeed we were able to amplify the right junction of a metV genomic island in IMT5155 as well. This genomic island also harbored insertion sequences r14 and r15 identified in CFT073, and open reading frames c3406 to R3 were found in identical order as in CFT073 (Fig. 3). As all three newly identified STM mutants M00C03, M11E02, and M13H03 showed attenuation by in vivo competition assay, we predict a role of this metV genomic island in APEC pathogenesis. However, further studies are required to characterize this island and to determine whether the term “pathogenicity island” is justified.
FIG. 3.
Schematic representation of the metV island-associated sequences of APEC strain IMT5155. Genes r2 (syn. malX), r3, and c3406 (phosphosugar isomerase) were identified in APEC strain IMT5155 by STM. Their links with r1 and f447 were confirmed by PCR with primers outlined in Table 1.
Other mutants.
We further isolated two putative oxidoreductases, one GTP-binding protein, one putative polysaccharide hydrolase, and one putative regulator. In addition, three open reading frames are predicted to encode proteins with a high degree of similarity to proteins of unknown function, and a further two mutants had disruptions in genes which have no similarity to any known sequences in the databases (Table 2). Thus, it is a major future challenge to elucidate the functions of such genes in APEC pathogenesis.
Conclusion.
In this study, we have used STM, a large-scale simultaneous screen (33), to identify genes required for in vivo survival of the highly virulent APEC strain IMT5155 (O2:H5) in the chicken, the natural host organism. To our knowledge, this is the first time STM has been used for E. coli infections of the respiratory tract of chicken. Prior studies with E. coli strains have used STM analyses only for urinary tract and gastrointestinal tract infections (5, 45).
After two rounds of screening and individual infection studies, 1.8% of our total pool of mutants was confirmed to be attenuated for survival in internal organs. We identified 28 genes resulting in in vivo attenuation, including both previously known and novel factors critical for E. coli infections in chickens. Our results confirmed the importance of LPS, capsule, and iron acquisition systems as colisepticemia-associated virulence determinants in APEC. Two mutants were identified with interruptions in genes directly or indirectly involved in the synthesis of vitamins. We further identified many genes known to be common virulence determinants for bacteria in general, some of which are described for APEC for the first time in this study. We did not, however, target all virulence-associated factors described for APEC so far (15, 21, 38). One reason for this could be that our study does not represent an exhaustive search for virulence-associated genes in APEC.
Virulence genes critical for disease, especially toxins such as the APEC vacuolating autotransporter toxin (52), may not appreciably affect survival of the bacterium in the host and thus would not be regularly selected by STM. The method may also not identify mutants with insertions in genes encoding secreted products such as the temperature-sensitive hemagglutinin (Tsh) of APEC which could theoretically be trans-complemented by other bacteria in the mixed population of mutant pools (17). Perhaps not surprisingly, we did not identify genes of the fim cluster encoding F1 fimbriae in APEC strains. It remains controversial whether F1 fimbriae contribute to the adhesion of APEC to epithelial cells of trachea and lung, since fim mutants show conflicting results in in vivo tests (3, 44, 46). Our results contrast those from an STM study of E. coli urinary tract infections, where eight fim mutants were selected in the UPEC strain CFT073 (5). The authors confirmed the potential role of type 1 fimbriae for the pathogenesis of urinary infection by isolating six different fim genes, demonstrating the attenuation of these mutants in a murine infection model.
UPEC strains also express pyelonephritis-associated fimbriae that have been shown to be critical for urinary tract infections (66). Our strain does not possess the pap operon, as we could not target these genes by PCR and hybridization analyses. However, the high virulence of IMT5155 questions an essential role for P-fimbriae in the APEC infection process. Beyond LPS and capsule genes, we did not identify other genes encoding serum resistance factors in APEC such as outer membrane protein A, transfer protein T, or increased serum survival protein, although some of these factors have recently been shown to be expressed in vivo during APEC infection (16). The failure to detect the above mentioned factors for serum resistance could be due to limitations of STM described above. Another reason could be the differing inoculation route of bacteria in the cited work. While we used intratracheal infections, the intra-air sac inoculation is the predominant route of infection in experimental chicken infection studies with APEC. This suggests an influence of the route of infection and expression of virulence-associated genes in different host compartments. Future studies in our laboratory should help clarify these questions. In addition, functional analyses of the disrupted genes are needed to elucidate the mechanisms by which they exert their effects as well as complementation and construction of defined deletion mutations to confirm the observed attenuation. This and future studies will hopefully lead to a more comprehensive understanding of APEC virulence in the chicken.
Acknowledgments
We thank David Holden (Imperial College, London, UK) for the gift of E. coli strains CC118 λpir and S17-1 λpir as well as a pool of tagged pUTmini-Tn5km2 plasmids. Thanks are extended also to R. Mueller and U. Boettcher for indispensable help with the animal tests, to Ines Diehl for technical help, and to K. Tedin and J. Jores for helpful suggestions on the manuscript.
This work was supported by grant WI 1436/5-1 from the Deutsche Forschungsgemeinschaft (DFG) and by Lohmann Tierzucht GmbH, Cuxhaven, Germany.
Editor: A. D. O'Brien
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelmelk, B. J., Y. Q. An, T. A. Hekker, L. G. Thijs, D. M. MacLaren, and J. de Graaf. 1994. Frequencies of lipopolysaccharide core types in Escherichia coli strains from bacteraemic patients. Microbiology 140:1119-1124. [DOI] [PubMed] [Google Scholar]
- 3.Arne, P., D. Marc, A. Bree, C. Schouler, and M. Dho-Moulin. 2000. Increased tracheal colonization in chickens without impairing pathogenic properties of avian pathogenic Escherichia coli MT78 with a fimH deletion. Avian Dis. 44:343-355. [PubMed] [Google Scholar]
- 4.Ausubel, F. M. 1994. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, H. J., and W. B. Gross. 1997. Colibacillosis, p. 131-141. In W. B. Gross (ed.), Diseases of poultry. Iowa State University Press, Ames, Iowa.
- 7.Blanco, J. E., M. Blanco, A. Mora, W. H. Jansen, V. Garcia, M. L. Vazquez, and J. Blanco. 1998. Serotypes of Escherichia coli isolated from septicaemic chickens in Galicia (northwest Spain). Vet. Microbiol. 61:229-235. [DOI] [PubMed] [Google Scholar]
- 8.Brown, E. A., R. D'Ari, and E. B. Newman. 1990. A relationship between l-serine degradation and methionine biosynthesis in Escherichia coli K12. J. Gen. Microbiol. 136:1017-1023. [DOI] [PubMed] [Google Scholar]
- 9.Brown, P. K., and R. Curtiss III. 1996. Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 93:11149-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheville, N. F., J. Hostetter, B. V. Thomsen, F. Simutis, Y. Vanloubbeeck, and E. Steadham. 2001. Intracellular trafficking of Mycobacterium avium ssp. paratuberculosis in macrophages. Dtsch. Tieraerztl. Wochenschr. 108:236-243. [PubMed] [Google Scholar]
- 11.Clarke, B. R., R. Pearce, and I. S. Roberts. 1999. Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J. Bacteriol. 181:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clegg, S., F. Yu, L. Griffiths, and J. A. Cole. 2002. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Mol. Microbiol. 44:143-155. [DOI] [PubMed] [Google Scholar]
- 13.Cloud, S. S., J. K. Rosenberger, P. A. Fries, R. A. Wilson, and E. M. Odor. 1985. In vitro and in vivo characterization of avian Escherichia coli. I. Serotypes, metabolic activity, and antibiotic sensitivity. Avian Dis. 29:1084-1093. [PubMed] [Google Scholar]
- 14.David, E., J. Heinrichs, J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30:221-232. [DOI] [PubMed] [Google Scholar]
- 15.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 16.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dozois, C. M., M. Dho-Moulin, A. Bree, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dozois, C. M., J. M. Fairbrother, J. Harel, and M. Bosse. 1992. pap-and pil-related DNA sequences and other virulence determinants associated with Escherichia coli isolated from septicemic chickens and turkeys. Infect. Immun. 60:2648-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Souza, J. M., L. Wang, and P. Reeves. 2002. Sequence of the Escherichia coli O26 O-antigen gene cluster and identification of O26 specific genes. Gene 297:123-127. [DOI] [PubMed] [Google Scholar]
- 20.Ewers, C., T. Janβen, S. Kieβling, H. C. Philipp, and L. H. Wieler. 2004. Molecular epidemiology of avian pathogenic Escherichia coli (APEC) isolated from colisepticemia in poultry. Vet. Microbiol. 104:91-101. [DOI] [PubMed] [Google Scholar]
- 21.Ewers, C., T. Janβen, and L. H. Wieler. 2003. [Avian pathogenic Escherichia coli (APEC)]. Berl. Munch. Tieraerztl. Wochenschr. 116:381-395. [PubMed] [Google Scholar]
- 22.Ferguson, G. P., R. M. Roop, 2nd, and G. C. Walker. 2002. Deficiency of a Sinorhizobium meliloti BacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 184:5625-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 24.Gentry-Weeks, C. R., J. M. Keith, and J. Thompson. 1993. Toxicity of Bordetella avium beta-cystathionase toward MC3T3-E1 osteogenic cells. J. Biol. Chem. 268:7298-7314. [PubMed] [Google Scholar]
- 25.Gibb, A. P., G. R. Barclay, I. R. Poxton, and F. di Padova. 1992. Frequencies of lipopolysaccharide core types among clinical isolates of Escherichia coli defined with monoclonal antibodies. J. Infect. Dis. 166:1051-1057. [DOI] [PubMed] [Google Scholar]
- 26.Glantz, P. J., S. Narotzky, and G. Bubash. 1962. Escherichia coli serotypes isolated from salpingitis and chrinic respiratory disease of poultry. Avian Dis. 6:322-328. [Google Scholar]
- 27.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths, E. 1987. The iron-uptake systems of pathogenic bacteria, p. 69-137. In J. J. Bullen (ed.), Iron and infection. John Wiley, London, United Kingdom.
- 29.Gross, W. B. 1994. Diseases due to Escherichia coli in poultry, p. 237-259. In C. L. Gyles (ed.), Escherichia coli in domestic animals and humans. CAB International, Wallingford, UK.
- 30.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanna, A., M. Berg, V. Stout, and A. Razatos. 2003. Role of capsular colanic acid in adhesion of uropathogenic Escherichia coli. Appl. Environ. Microbiol. 69:4474-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrichs, D. E., M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hybrid of those found in Escherichia coli K-12 and Salmonella enterica. Structure and function of the R2 WaaK and WaaL homologs. J. Biol. Chem. 273:8849-8859. [DOI] [PubMed] [Google Scholar]
- 33.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 34.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for beta-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 37.Janβen, T., C. Schwarz, P. Preikschat, M. Voss, H. C. Philipp, and L. H. Wieler. 2001. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. Int. J. Med. Microbiol. 291:371-378. [DOI] [PubMed] [Google Scholar]
- 38.La Ragione, R. M., and M. J. Woodward. 2002. Virulence factors of Escherichia coli serotypes associated with avian colisepticaemia. Res. Vet. Sci. 73:27-35. [DOI] [PubMed] [Google Scholar]
- 39.Lavina, M., A. P. Pugsley, and F. Moreno. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J. Gen. Microbiol. 132:1685-1693. [DOI] [PubMed] [Google Scholar]
- 40.LeVier, K., R. W. Phillips, V. K. Grippe, R. M. Roop 2nd, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492-2493. [DOI] [PubMed] [Google Scholar]
- 41.Li, H., and J. T. Park. 1999. The periplasmic murein peptide-binding protein MppA is a negative regulator of multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 181:4842-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 43.Mao, Y., M. P. Doyle, and J. Chen. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marc, D., P. Arne, A. Bree, and M. Dho-Moulin. 1998. Colonization ability and pathogenic properties of a fim− mutant of an avian strain of Escherichia coli. Res. Microbiol. 149:473-485. [DOI] [PubMed] [Google Scholar]
- 45.Martindale, J., D. Stroud, E. R. Moxon, and C. M. Tang. 2000. Genetic analysis of Escherichia coli K1 gastrointestinal colonization. Mol. Microbiol. 37:1293-1305. [DOI] [PubMed] [Google Scholar]
- 46.Mellata, M., M. Dho-Moulin, C. M. Dozois, R. Curtiss III, P. K. Brown, P. Arne, A. Bree, C. Desautels, and J. M. Fairbrother. 2003. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mellata, M., M. Dho-Moulin, C. M. Dozois, R. Curtiss III, B. Lehoux, and J. M. Fairbrother. 2003. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect. Immun. 71:494-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merrell, D. S., D. L. Hava, and A. Camilli. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol. Microbiol. 43:1471-1491. [DOI] [PubMed] [Google Scholar]
- 49.Nagy, G., U. Dobrindt, M. Kupfer, L. Emody, H. Karch, and J. Hacker. 2001. Expression of hemin receptor molecule ChuA is influenced by RfaH in uropathogenic Escherichia coli strain 536. Infect. Immun. 69:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, J. T. 1993. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J. Bacteriol. 175:7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park, J. T., D. Raychaudhuri, H. Li, S. Normark, and D. Mengin-Lecreulx. 1998. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide l-alanyl-gamma-d-glutamyl-meso-diaminopimelate. J. Bacteriol. 180:1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parreira, V. R., and C. L. Gyles. 2003. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 71:5087-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pourbakhsh, S. A., M. Boulianne, B. Martineau-Doize, C. M. Dozois, C. Desautels, and J. M. Fairbrother. 1997. Dynamics of Escherichia coli infection in experimentally inoculated chickens. Avian Dis. 41:221-233. [PubMed] [Google Scholar]
- 54.Randall, L. P., and M. J. Woodward. 2001. Role of the mar locus in virulence of Salmonella enterica serovar Typhimurium DT104 in chickens. J. Med. Microbiol. 50:770-779. [DOI] [PubMed] [Google Scholar]
- 55.Rowe, J. J., T. Ubbink-Kok, D. Molenaar, W. N. Konings, and A. J. Driessen. 1994. NarK is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol. Microbiol. 12:579-586. [DOI] [PubMed] [Google Scholar]
- 56.Runyen-Janecky, L. J., S. A. Reeves, E. G. Gonzales, and S. M. Payne. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakai, A., N. Kinoshita, M. Kita, T. Katsuragi, and Y. Tani. 2003. Investigation of 1-deoxy-d-xylulose 5-phosphate synthase and transketolase of Bacillus subtilis in relation to vitamin B6 biosynthesis. J. Nutr. Sci. Vitaminol. (Tokyo) 49:73-75. [DOI] [PubMed] [Google Scholar]
- 58.Schouleur, C., F. Koffmann, C. Amory, S. Leroy-Setrin, and M. Moulin-Schouleur. 2004. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology 150:2973-2984. [DOI] [PubMed] [Google Scholar]
- 59.Sprenger, G. A. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164:324-330. [DOI] [PubMed] [Google Scholar]
- 60.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stocki, S. L., L. A. Babiuk, N. A. Rawlyk, A. A. Potter, and B. J. Allan. 2002. Identification of genomic differences between Escherichia coli strains pathogenic for poultry and E. coli K-12 MG1655 using suppression subtractive hybridization analysis. Microb. Pathog. 33:289-298. [DOI] [PubMed] [Google Scholar]
- 62.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 63.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63a.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 64.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 65.Wang, L., W. Qu, and P. R. Reeves. 2001. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect. Immun. 69:6923-6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou, D., W. D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]