Abstract
Background
Pulmonary function (PF) progressively declines with aging. Forced expiratory volume in the first second (FEV1) and forced vital capacity (FVC) are predictors of morbidity of pulmonary and cardiovascular diseases and all-cause mortality. In addition, reduced PF is associated with elevated chronic low-grade systemic inflammation, glucose metabolism, body fatness, and low muscle strength. It may suggest pleiotropic genetic effects between PF with these age-related factors.
Methods
We evaluated whether FEV1 and FVC share common pleiotropic genetic effects with interleukin-6, high-sensitivity C-reactive protein, body mass index, muscle (grip) strength, plasma glucose, and glycosylated hemoglobin in 3 888 individuals (age range: 26–106). We employed sex-combined and sex-specific correlated meta-analyses to test whether combining genome-wide association p values from 2 or more traits enhances the ability to detect variants sharing effects on these correlated traits.
Results
We identified 32 loci for PF, including 29 novel pleiotropic loci associated with PF and (i) body fatness (CYP2U1/SGMS2), (ii) glucose metabolism (CBWD1/DOCK8 and MMUT/CENPQ), (iii) inflammatory markers (GLRA3/HPGD, TRIM9, CALN1, CTNNB1/ZNF621, GATA5/SLCO4A1/NTSR1, and NPVF/C7orf31/CYCS), and (iv) muscle strength (MAL2, AC008825.1/LINC02103, AL136418.1).
Conclusions
The identified genes/loci for PF and age-related traits suggest their underlying shared genetic effects, which can explain part of their phenotypic correlations. Integration of gene expression and genomic annotation data shows enrichment of our genetic variants in lung, blood, adipose, pancreas, and muscles, among others. Our findings highlight the critical roles of identified gene/locus in systemic inflammation, glucose metabolism, strength performance, PF, and pulmonary disease, which are involved in accelerated biological aging.
Keywords: Functional genome annotations, Inflammation, Longevity, Muscle, Obesity
Pulmonary function (PF), specifically forced expiratory volume in the first second (FEV1) and forced vital capacity (FVC), is often used to evaluate lung abnormalities, including asthma, fibrosis, and chronic obstructive pulmonary disease (COPD) (1–4). Reduced PF has shown associations with elevated inflammatory markers (interleukin-6 [IL-6] and high-sensitivity C-reactive protein [hsCRP] levels) (3,5), glucose metabolism (plasma glucose and glycosylated hemoglobin [HbA1c] levels) (6), body fatness (body mass index, BMI) (7), and with low muscle (grip) strength (8). PF and these correlated traits progressively become impaired with aging, which are biomarkers for a range of morbidities, such as pulmonary diseases, systemic inflammation, type 2 diabetes (T2D), and physical limitations that predict cardiovascular diseases (CVDs) and all-cause mortality (5,9).
The discovery of genetic variants associated with PF can provide etiological insights into pulmonary diseases and systemic inflammation, which may decrease the risk of T2D and CVD, contributing to the understanding of age-related diseases. Genome-wide association studies (GWASs) have identified over 300 genetic variants for PF (2,4,10,11). Additional variants have been discovered by whole-genome sequencing (12) and via functional genome annotations (13).
Correlated meta-analysis (CMA) (14,15) can enhance the ability to detect pleiotropic genetic variant effects on correlated traits that share common biological and etiological pathways while protecting against false-positive signals arising from nonindependent data. Our study aimed to identify pleiotropic genetic variants between PF measures with inflammatory markers, body fatness, muscle strength, and glucose metabolism through CMA in the Long Life Family Study (LLFS), which recruited participants for exceptional survival in the upper generation. Further, we performed bioinformatics analyses and reviewed the literature to determine whether our identified variants in the sex-specific and sex-combined CMAs might be tagging potentially functional genome elements and genes implicated in biologic pathways.
Method
Study Data
The LLFS (phs000397.v1.p1) was designed to study genetic, behavioral, and environmental factors in families exhibiting exceptional longevity. Families were sampled from 4 clinical centers: Boston University Medical Center in Boston, Massachusetts, Columbia College of Physicians and Surgeons in New York City, New York, the University of Pittsburgh in Pittsburgh, Pennsylvania, United States, and the University of Southern Denmark, Odense, Denmark. The characteristics, recruitment, eligibility, and enrollment were previously described (16). The first clinical exam started in 2006 and recruited 4 953 individuals in 539 two-generational families that demonstrated clustering for exceptional survival in the upper generation. The second clinical exam (2014–2017) revised 2 933 European descent individuals from 528 families. The current analyses included 3 888 European ancestry individuals with information on age, sex, PF, and genotype data recruited from the first clinical exam.
Age-Related Traits
FEV1 and FVC were measured with a portable spirometer (EasyOne, NDD Medical Technologies, Andover, MA) using American Thoracic Society guidelines, as previously reported (1,16). Participants with non-European ancestry, poor quality spirometry readings, and pulmonary fibrosis were excluded. BMI was calculated as weight (kg)/height (m)2. Muscle grip strength was measured twice in a seated position to the nearest 2 kg in the dominant hand and averaged using an isometric dynamometer (Jamar Hydraulic Hand Dynamometer, Lafayette, IN). Fasting plasma glucose, HbA1c, IL-6, and hsCRP were measured in blood at the University of Minnesota. Plasma glucose was assessed at ≥6-hour fasting. T2D was defined as fasting glucose ≥ 126 mg/dL (≥7.0 mmol/L), HbA1c ≥ 6.5%, taking diabetes medications, self-report of T2D, or having a doctor diagnosis of T2D. Current cigarette smoking (yes/no) was obtained by self-report. Inverse normal transformation on FEV1, FVC, IL-6, muscle strength, plasma glucose, and HbA1c was used to counteract departures from normality.
Genotype and Imputation
All participants were genotyped using the Illumina Human Omni chip 2.5 v1. Genotype quality control filters applied before imputation were call rate < 98%, minor allele frequency (MAF) < 1%, p value Hardy–Weinberg equilibrium < 1 × 10−6, and mismatched alleles between LLFS with 1000 Genomes Project (1000Gp3v5), which provided 1 421 289 SNPs. Genotype imputation was performed with the SHAPEIT and IMPUTE2 software based on the 1000Gp3v5 ALL reference panel, resulting in 81.2 million imputed variants. For CMA analyses, ~9.5 M single nucleotide polymorphisms (SNPs) with MAF > 0.01 and imputation quality (r2) > 0.5 were coded as allelic dosages and annotated on the NCBI-Genome Reference Consortium Human 38 (GRCh38).
LLFS data are available in the Database of Genotypes and Phenotypes (dbGaP, Study Accession: phs000397.v1.p1). All participants signed informed consent. The Institutional Review Boards approved all study procedures of participating institutions.
Association and Statistical Analyses
We performed GWAS implemented by lmekin function in the coxme R package (version 2.2-16), which accounts for dependency among family members as a function of their kinship coefficient estimated with the kinship2 (version 1.8.5) algorithm. Age, age2, sex, field centers, and 10 principal components were added as covariates in the linear regression model with an additive effect of the genetic variants for each trait. In addition, height and smoking also entered the model for PF traits.
We conducted the CMA to test whether pleiotropic genetic variants were shared between PF and other age-related traits (14,15). The method empirically estimates the covariance among GWAS to be included in a meta-analysis and corrects for the signal inference in the combined GWAS p value. In brief, CMA integrates GWAS p values of multiple traits, either correlated or independent, that might come from the same or different studies and environmental exposures. The CMA method prevents Type I error by accounting for all sources of dependencies between multiple genome scans under the null, including overlapping individuals, cryptic relatedness, and population structure. The p values are first converted to corresponding Z-scores, which are assumed to have a multivariate normal distribution. Because most SNPs in a GWAS are under the null for any given trait, the global degree of correlation between the GWAS can be calculated to assess the degree of nonindependence. Then, a tetrachoric correlation estimator, based on dichotomized p values (p ≤ .5; p > .5), is used to adjust the variance estimate of the combined Z-scores. Finally, the method combines p values for each variant and estimates the combined CMA p values for the number of tests/GWAS (k), assuming that the sum of −2ln(Pk) has a chi-squared distribution with 2k degrees of freedom.
We reported a genetic variant correlated with 2 or more traits if p < 5 × 10−2 for individual GWAS, CMA p < 5 × 10−8, and CMA p value < GWAS p value. We considered a novel locus if our lead variant (ie, the most significant SNP) presented a distance >500 kb from any lead variant previously reported in a GWAS (p < 5 × 10−8). We employed GWAS and CMA analyses in sex-combined and sex-specific samples.
A chi-squared test was calculated to determine differences in proportions between groups, and a t test was utilized to compare means between groups. The statistical significance threshold in these analyses was set at the nominal p < 5 × 10−2. The analyses were executed using SAS version 9.4 (SAS Institute, Inc., Cary, NC).
Bioinformatics Analyses
We selected all variants within 1 Mb and in high linkage disequilibrium (LD, r2 ≥ 0.8) with CMA lead variants to examine whether the lead variants might be tagging regulatory variants. The potentially functional implications of regulatory CMA variants were assessed using the ENCODE Consortium (https://www.encodeproject.org/) and the Roadmap Epigenome Mapping Consortium (http://www.roadmapepigenomics.org/) initiatives via HaploReg (v.4, https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php), and RegulomeDB (v.1.1, https://regulomedb.org/regulome-search/). HaploReg provides (i) the sequence conservation by the genetic evolutionary rate profiling (GERP) and SiPhy, (ii) the chromatin state segmentation for enhancer or promoter elements, (iii) histone modification ChIP-seq peaks, and (iv) DNase hypersensitivity data peaks from the Roadmap. In addition, HaploReg reports (i) the transcription factor (TF) binding and motif data from the ENCODE data, and (ii) the gene annotations from the UCSC Genome Browser and GENCODE. RegulomeDB presents regulatory gene expression information via ChIP factors, DNase sensitivity, and TF binding sites from ENCODE, and it reports Chromatin States from the Roadmap. We searched for the expression quantitative trait locus (eQTL) from the Genotype-Tissue Expression (GTEx Portal-V8, https://gtexportal.org/home/).
To identify the presence of physical interactions (Hi-C), we applied GWAS4D (from January 3, 2021) (17) using the CMA lead variants. GWAS4D utilizes context-dependent epigenomic weighting to prioritize regulatory variants and disease-associated genes to score each SNP after filtering and LD expansion steps. GWAS4D incorporates tissue/cell type-specific epigenome data, integrates and refines TF motifs from 8 public resources, processes Hi-C data at significant interactions at 5-kb resolution, links variants to its target regions, and annotates noncoding variants with comprehensive functional annotations. GWAS4D reports 3 probabilities: (i) ‘composite p’ shows the likelihood of the variant to be functional in gene regulation by context-free ensemble method, (ii) ‘cell p’ represents the condition-dependent regulatory potential in the current tissue/cell type, and (iii) ‘combined p’ is the final regulatory probability that jointly considers both context-free and context-dependent models. We reported GWAS4D findings for CMA variants that reached ‘combined p’ > .10 in fetal lung fibroblasts (IMR90), adipose mesenchymal stem cell, liver, hepatocellular-carcinoma cell (HepG2), T cells and B cells from peripheral blood, and skeletal muscle myoblasts (HSMM).
We interrogated publicly available GWAS for PF, IL-6, hsCRP, BMI, muscle strength, plasma glucose, HbA1c, and longevity via the NHGRI-EBI catalog (https://www.ebi.ac.uk/gwas, accessed on April 20, 2021). In addition, relevant biological insights for genes residing within a 1-Mb interval of CMA lead variants were sourced from NCBI (https://www.ncbi.nlm.nih.gov/) and GeneCards (https://www.genecards.org/).
Results
Descriptive Analysis
Table 1 shows the characteristics of study participants for covariates and age-related traits. The age mean and mean levels of FEV1, FVC, IL-6, height, BMI, muscle strength, plasma glucose, HbA1c, and T2D were significantly higher in men than women.
Table 1.
Study Population Characteristics
| Sex-Combined | Men | Women | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables | N | Mean (SD) or Frequency | Range Values | N | Mean (SD) or Frequency | Range Values | N | Mean (SD) or Frequency | Range Values |
| Age (years) | 3 888 | 68.8 (15.1) | 24–106 | 1 744 | 69.4 (14.8) | 32–101 | 2 144 | 68.3 (15.4)* | 24–106 |
| Sex (% women) | 3 888 | 2 138 (55.1%) | |||||||
| Smoking (%) | 3 888 | 279 (7.2%) | 1 740 | 125 (7.2%) | 2 138 | 154 (7.2%) | |||
| Height (cm) | 3 888 | 166.2 (10.4) | 128.5–205.2 | 1 744 | 173.9 (7.8) | 143.2–205.2 | 2 144 | 160.0 (7.7)† | 128.5–187.1 |
| FEV1 (mL) | 3 888 | 2 460.0 (868.2) | 305–5 253 | 1 744 | 2 906.8 (888.0) | 543–5 253 | 2 144 | 2 096.6 (656.7)† | 305–4 476 |
| FVC (mL) | 3 888 | 3 211.6 (1 054.9) | 552–6 690 | 1 744 | 3 817.6 (1 025.3) | 1 180–6 690 | 2 144 | 2 718.6 (788.4)† | 552–6 007 |
| IL-6 (pg/mL) | 3 780 | 2.1 (6.1) | 0.1–132 | 1 701 | 2.3 (6.7) | 0.1–132 | 2 079 | 1.9 (5.6)§ | 0.1–116.4 |
| hsCRP (mg/L) | 3 634 | 3.2 (7.4) | 0.1–202.8 | 1 628 | 3.2 (8.2) | 0.1–202.8 | 2 006 | 3.3 (6.7) | 0.1–147.8 |
| BMI (kg/m2) | 3 858 | 27.1 (4.8) | 15.3–57.4 | 1 734 | 27.6 (4.0) | 17.2–49.7 | 2 124 | 26.8 (5.3)‡ | 15.3–57.4 |
| Grip (kg) | 3 876 | 29.9 (12.2) | 2–88 | 1 739 | 38.2 (11.8) | 8–88 | 2 137 | 23.2 (7.4)† | 2–70 |
| Glucose (mg/dL) | 3 210 | 91.4 (11.8) | 34–183 | 1 408 | 93.7 (10.9) | 44–142 | 1 802 | 90.1 (10.7)† | 34–123 |
| Hb1Ac (%) | 3 782 | 5.6 (0.6) | 4–11.1 | 1 699 | 5.7 (0.6) | 4.1–11.1 | 2 083 | 5.6 (0.5)‡ | 4.0–9.7 |
| T2D (%) | 3 888 | 322 (8.3) | 1 744 | 174 (10.0%) | 2 144 | 148 (6.9%)‖ | |||
Notes: Total number of individuals, BMI = body mass index; FEV1 = forced expiratory volume in 1 second; FVC = forced vital capacity; Glucose = fasting plasma glucose; Grip = muscle (grip) strength; HbA1c = glycosylated hemoglobin; hsCRP = high-sensitivity C-reactive protein; IL-6 = interleukin-6; mean (SD) = mean levels (standard deviation); T2D = type 2 diabetes.
Significance levels for differences in proportions (chi-squared tests) or means (t test) between men and women: *p = .024. †p < 1 × 10−8. ‡p < 3 × 10−7. §p ≤ .05. ‖p = .0005.
Genetic Association Analyses
Firstly, we employed GWAS analysis for each of the age-related traits. The observed versus expected GWAS −log10 (p value) distributions (Q–Q plots; Supplementary Figure 1) and the estimated GWAS inflation factors (λ GC, from 0.996 [FVC] to 1.055 [muscle strength]; Supplementary Table 1) indicate an absence of genomic inflation, systematic technical bias, or population stratification for all traits. The Manhattan plots (Supplementary Figure 2) depict the GWAS −log10 (p value) on genomic scales for FEV1, FEVC, IL-6, hsCRP, BMI, muscle strength, plasma glucose, and HbA1c.
To identify whether pleiotropic genetic variants shared effects on age-related traits, we performed CMA analyses between PF (FEV1 or FEVC) with inflammatory markers (IL-6 and hsCRP), body fatness (BMI), muscle (grip) strength, or glucose metabolism (plasma glucose or HbA1c). The Manhattan plots (Supplementary Figure 3) represent the CMA −log10 (p value) on genomic scales. Supplementary Table 1 shows the estimated genomic inflation factors (λ GC < 1.1) for CMA analyses, which were corrected with tetrachoric correlations for nonindependence among GWAS (Supplementary Table 2).
Table 2 displays the p values for CMA and each univariate GWAS for the lead CMA variants. For all significant variants, the GWAS summary statistics and CMA p values are shown in Supplementary Table 3. We identified 6 pleiotropic loci with on PF measures and inflammatory markers (Supplementary Figure 4), including the NPVF/C7orf31/CYCS (7p15.3-p15.2; Figure 1), CALN1 (7q11.22), TRIM9 (14q22.1; Figure 1), AXIN2/CEP112 (17q23-q24.1), MICAL3 (22q11.21), and CMIP (16q23.2-q23.3). Also, 3 loci are associated with PF measures and body fatness (Table 2; Supplementary Figure 4), including the CYP2U1 (4q25; Figure 1), MDGA2/RPL10L (14q21.3), and LOC105372472 (19q13.43). In addition, the XXYLT1 (3q29) and MAL2 (8q24.12) loci show shared associations between PF with muscle strength, while the CBWD1/DOCK8 locus (9p24.3; Figure 1) is associated with PF and HbA1c variations. However, we did not find any correlated associations (CMA p < 5 × 10−8) for PF and plasma glucose.
Table 2.
Summary of SNP/Locus Sex-Combined Associations From Correlated Meta-analysis
| Trait_1_2_3 | SNP | Chr | Loci | Trait_1 p Value* | Trait_2 p Value* | Trait_3 p Value* | CMA p Value* |
|---|---|---|---|---|---|---|---|
| FVC_IL-6_hsCRP | rs67112407 | 7 | NPVF/CYCS | 6.14 × 10−06 | 4.47 × 10−03 | 9.17 × 10−04 | 1.91 × 10−08 |
| FEV1_IL-6_hsCRP | rs17349645 | 7 | CALN1 | 2.34 × 10−05 | 2.06 × 10−04 | 4.89 × 10−04 | 1.20 × 10−09 |
| FVC_IL-6_hsCRP | 2.44 × 10−06 | 2.06 × 10−04 | 4.89 × 10−04 | 2.75 × 10−10 | |||
| FEV1_IL-6_hsCRP | rs34171875 | 14 | TRIM9 | 3.30 × 10−03 | 2.90 × 10−04 | 1.12 × 10−04 | 3.49 × 10−08 |
| FVC_IL-6_hsCRP | 3.85 × 10−03 | 2.90 × 10−04 | 1.12 × 10−04 | 4.86 × 10−08 | |||
| FEV1_IL-6_hsCRP | rs11079574 | 17 | AXIN2/CEP112 | 1.56 × 10−04 | 3.57 × 10−04 | 5.04 × 10−04 | 9.17 × 10−09 |
| FVC_IL-6_hsCRP | rs2896013 | 22 | MICAL3 | 3.77 × 10−06 | 9.38 × 10−05 | 3.17 × 10−03 | 2.12 × 10−08 |
| FEV1_hsCRP | rs8044418 | 16 | CMIP | 6.81 × 10−06 | 1.18 × 10−04 | 9.57 × 10−09 | |
| FEV1_BMI | rs1493126 | 4 | CYP2U1 | 1.54 × 10−05 | 6.15 × 10−05 | 8.70 × 10−09 | |
| FVC_BMI | 6.72 × 10−05 | 6.76 × 10−05 | 4.63 × 10−08 | ||||
| FEV1_BMI | rs144725076 | 14 | MDGA2/RPL10L | 2.41 × 10−05 | 2.51 × 10−04 | 5.47 × 10−08 | |
| FVC_BMI | 1.15 × 10−05 | 2.51 × 10−04 | 3.64 × 10−08 | ||||
| FEV1_BMI | rs7256661 | 19 | LOC105372472 | 1.50 × 10−03 | 1.55 × 10−06 | 3.90 × 10−08 | |
| FVC_BMI | 1.20 × 10−03 | 1.55 × 10−06 | 3.84 × 10−08 | ||||
| FEV1_GRIP | rs9870813 | 3 | XXYLT1 | 1.38 × 10−04 | 4.57 × 10−06 | 7.10 × 10−09 | |
| FVC_GRIP | rs77980484 | 8 | MAL2 | 9.55 × 10−05 | 5.56 × 10−05 | 4.95 × 10−08 | |
| FVC_HbA1c | rs201917736 | 9 | CBWD1/DOCK8 | 4.35 × 10−04 | 1.28 × 10−06 | 8.44 × 10−09 |
Notes: The most significantly associated SNPs per locus within a 1-Mb region; BMI = body mass index; Chr = chromosome; FEV1 = forced expiratory volume in 1 second; FVC = forced vital capacity; GWAS = genome-wide association study; HbA1c = glycosylated hemoglobin; hsCRP = high-sensitivity C-reactive protein; IL-6 = interleukin-6.
*Trait_1 p value = GWAS p value for Trait_1 (FEV1 or FVC); Trait_2 p value = GWAS p value for Trait_2 (IL-6, BMI, Muscle (grip) strength or HbA1c); Trait_3 p value = GWAS p value for Trait_3 (hsCRP); CMA p value = p value for correlated meta-analysis.
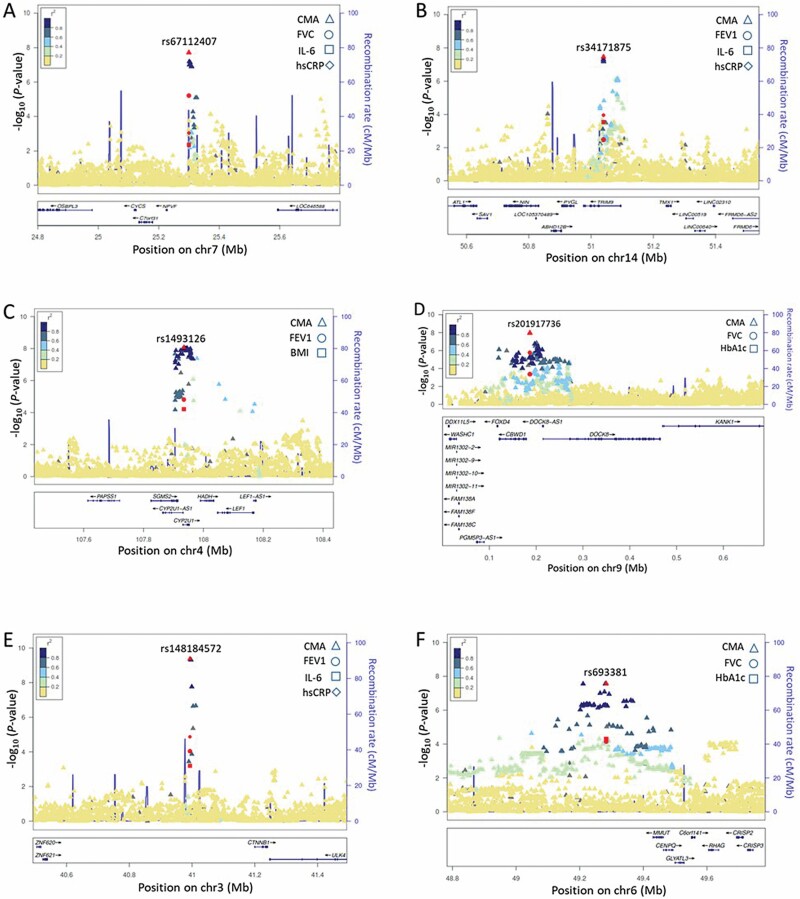
Figure 1.
Locuszoom plots of some novel CMA loci. (A) NPVF/C7orf31/CYCS for FVC_IL-6_hsCRP in a sex-combined analysis; (B) TRIM9 for FEV1_IL-6_hsCRP in a sex-combined analysis; (C) CYP2U1 for FEV1_IL-6_hsCRP in a sex-combined analysis; (D) CBWD1/DOCK8 for FVC_HbA1c in a sex-combined analysis; (E) CTNNB1 for FEV1_IL-6_hsCRP in women; (F) MMUT for FVC_HbA1c in women. All plots of novel CMA loci are shown in Supplementary Figures 4 and 5. CMA = correlated meta-analysis; FEV1 = forced expiratory volume in the first second; FVC = forced vital capacity; HbA1c = glycosylated hemoglobin; hsCRP = high-sensitivity C-reactive protein; IL-6 = interleukin-6.
Ten out of 12 pleiotropic identified loci are novel discoveries for PF (Supplementary Table 4). According to the NHGRI-EBI catalog, the MICAL3 locus was identified for FEV1 (10,18), FVC, and FEV1/FVC (10) in European ancestry and multiancestry GWAS meta-analyses. The MDGA2/RPL10L locus was associated with FEV1/FVC ratio in African ancestry (2), but it was not present in European ancestry (10). Also, genome-wide significant results were reported between MICAL3 and CMIP with hsCRP in European ancestry (19), MDGA2/RPL10L and LOC105372472-ZNF264 with BMI in multiancestry (19), and DOCK8 with plasma glucose in Asian ancestry (20). However, none of the variants within these reported loci reached a genome-wide significance level for the individual age-related GWAS traits in LLFS (Table 2). The CMA approach enhanced the power of joint association of traits in LLFS, which allowed replicating some of the previously reported GWAS findings.
Given the sex differences on the mean levels in the age-related traits (Table 1), we evaluated whether pleiotropic genetic effects contribute to sex-specific trait level variations by conducting GWAS and CMA analyses separately in men and women. Table 3 shows p values for the sex-specific univariate GWAS and CMA for the lead CMA variants. For all significant variants, the sex-specific GWAS summary statistics and CMA p values are provided in Supplementary Table 5. We identified 20 sex-specific pleiotropic loci for age-related traits. In men, we detected 9 pleiotropic locus effects on PF and inflammatory marker traits (Supplementary Figure 5), residing in AL138927.1/LINC01720 (1q31.1-q31.2), OTOL1/AC131211.1 (3q26.1), AC004053.1/CXXC4/AS1 (4q24), IYD/PPP1R14C/PLEKHG1 (6q25.1), C2CD3 (11q13.4), GRIP1 (12q14.3), LINC00645 (14q12), and MUC16/ZNF317 (19p13.2). NRXN1 (2p16.3) shares associations with PF and inflammatory markers and PF and body fatness (BMI). In addition, TLN2 (15q22.2) presents correlated genetic effects on PF and BMI.
Table 3.
Summary of SNP/Locus Sex-Specific Associations From Correlated Meta-analysis
| Trait_1_2_3 | SNP | Chr | Loci | Trait_1 p Value* | Trait_2 p Value* | Trait_3 p Value* | CMA p Value* |
|---|---|---|---|---|---|---|---|
| Men | |||||||
| FVC_IL-6_hsCRP | rs76268042 | 1 | AL138927.1/LINC01720 | 4.93 × 10−04 | 2.10 × 10−04 | 1.28 × 10−03 | 4.82 × 10−08 |
| FEV1_IL-6_hsCRP | rs539460509 | 2 | NRXN1 | 7.18 × 10−04 | 4.04 × 10−04 | 2.89 × 10−05 | 4.66 × 10−09 |
| FVC_IL-6_hsCRP | 5.52 × 10−05 | 4.04 × 10−04 | 2.89 × 10−05 | 5.55 × 10−10 | |||
| FVC_IL-6_hsCRP | rs73170189 | 3 | OTOL1/AC131211.1 | 5.22 × 10−04 | 1.31 × 10−03 | 1.45 × 10−04 | 3.85 × 10−08 |
| FVC_IL-6_hsCRP | rs62329654 | 4 | AC004053.1/CXXC4-AS1 | 2.74 × 10−04 | 6.52 × 10−03 | 3.71 × 10−05 | 3.98 × 10−08 |
| FVC_IL-6_hsCRP | rs594747 | 6 | IYD/PPP1R14C/PLEKHG1 | 1.24 × 10−04 | 4.03 × 10−04 | 6.45 × 10−04 | 1.45 × 10−08 |
| FEV1_IL-6_hsCRP | rs12788025 | 11 | C2CD3 | 3.28 × 10−04 | 1.71 × 10−06 | 1.21 × 10−04 | 9.17 × 10−11 |
| FVC_ IL-6_hsCRP | 5.51 × 10−04 | 1.71 × 10−06 | 1.21 × 10−04 | 1.73 × 10−10 | |||
| FVC_IL-6_hsCRP | rs58387290 | 12 | GRIP1 | 1.30 × 10−02 | 2.93 × 10−05 | 1.41 × 10−04 | 4.31 × 10−08 |
| FEV1_IL-6_hsCRP | rs78382897 | 14 | LINC00645 | 5.49 × 10−03 | 1.07 × 10−04 | 3.24 × 10−05 | 1.28 × 10−08 |
| FEV1_IL-6_hsCRP | rs202046252 | 19 | MUC16/ZNF317 | 3.89 × 10−03 | 2.07 × 10−04 | 6.76 × 10−05 | 2.61 × 10−08 |
| FVC_IL-6_hsCRP | 8.71 × 10−04 | 2.07 × 10−04 | 6.76 × 10−05 | 6.69 × 10−09 | |||
| FVC_BMI | rs539460509 | 2 | NRXN1 | 5.52 × 10−05 | 9.49 × 10−06 | 8.50 × 10−09 | |
| FVC_BMI | rs28704520 | 15 | TLN2 | 1.28 × 10−03 | 1.04 × 10−06 | 3.80 × 10−08 | |
| Women | |||||||
| FEV1_IL-6_hsCRP | rs148184572 | 3 | CTNNB1/ZNF621 | 9.12 × 10−05 | 6.35 × 10−04 | 1.35 × 10−05 | 4.31 × 10−10 |
| FVC_IL-6_hsCRP | 1.04 × 10−04 | 6.35 × 10−04 | 1.35 × 10−05 | 5.93 × 10−10 | |||
| FEV1_IL-6_hsCRP | rs1426949 | 4 | GLRA3/HPGD | 3.92 × 10−02 | 3.03 × 10−04 | 2.93 × 10−06 | 4.62 × 10−08 |
| FVC_IL-6_hsCRP | rs11734163 | 4 | LINC01060/LINC02508 | 3.77 × 10−03 | 5.62 × 10−04 | 2.79 × 10−05 | 2.61 × 10−08 |
| FEV1_IL-6_hsCRP | rs149824870 | 7 | DGKI | 5.32 × 10−03 | 3.57 × 10−04 | 8.89 × 10−07 | 1.96 × 10−09 |
| FVC_IL-6_hsCRP | 2.65 × 10−03 | 3.57 × 10−04 | 8.89 × 10−07 | 1.09 × 10−09 | |||
| FEV1_IL-6_hsCRP | rs74940149 | 13 | FLT3/URAD | 1.23 × 10−04 | 4.47 × 10−05 | 1.63 × 10−03 | 3.65 × 10−09 |
| FVC_IL-6_hsCRP | 5.07 × 10−04 | 4.47 × 10−05 | 1.63 × 10−03 | 1.46 × 10−08 | |||
| FVC_IL-6_hsCRP | rs61982030 | 14 | LOC105370611 | 7.12 × 10−04 | 9.38 × 10−05 | 1.34 × 10−03 | 2.89 × 10−08 |
| FEV1_IL-6_hsCRP | rs71350436 | 18 | GNAL/SLC35G4 | 7.84 × 10−04 | 1.65 × 10−03 | 1.35 × 10−04 | 4.32 × 10−08 |
| FVC_IL-6_hsCRP | rs62196356 | 20 | GATA5/SLCO4A1/NTSR1 | 1.78 × 10−03 | 3.81 × 10−04 | 1.16 × 10−04 | 2.58 × 10−08 |
| FEV1_Grip | rs80148245 | 5 | AC008825.1/LINC02103 | 5.11 × 10−06 | 7.44 × 10−04 | 4.30 × 10−08 | |
| FVC_Grip | 7.43 × 10−07 | 7.44 × 10−04 | 9.17 × 10−09 | ||||
| FVC_Grip | rs117500377 | 14 | AL136418.1 | 1.64 × 10−03 | 1.56 × 10−06 | 4.32 × 10−08 | |
| FVC_HbA1c | rs693381 | 6 | MMUT-CENPQ | 7.08 × 10−05 | 4.90 × 10−05 | 2.69 × 10−08 |
Notes: The most significantly associated SNPs per locus within a 1-Mb region separately by men and women; BMI = body mass index; Chr = chromosome; FEV1 = forced expiratory volume in 1 second; FVC = forced vital capacity; GWAS = genome-wide association study; HbA1c = glycosylated hemoglobin; hsCRP = high-sensitivity C-reactive protein; IL-6 = interleukin-6.
*Trait_1 p value = GWAS p value for Trait_1 (FEV1 or FVC); Trait_2 p value = GWAS p value for Trait_2 (IL-6, BMI, muscle (grip) strength, or HbA1c); Trait_3 p value = GWAS p value for Trait_3 (hsCRP); CMA p value = p value for correlated meta-analysis.
In women, we identified 8 pleiotropic loci sharing genetic associations on PF and inflammatory markers (Supplementary Figure 5), located on CTNNB1/ZNF621 (3p22.1; Figure 1), GLRA3/HPGD (4q34.1), LINC01060/LINC02508 (4q35.2), DGKI (7q33), FLT3/URAD (13q12.2), LOC105370611 (14q31.3), GNAL/SLC35G4 (18p11.21), and GATA5/SLCO4A1/NTSR1 (20q13.33). In addition, we found correlated associations between AC008825.1/LINC02103 (5p14.1) and AL136418.1 (14q12) with PF and muscle strength (Supplementary Figure 5), and MMUT/CENPQ with PF and HbA1c (6p12.3; Figure 1).
We computed p values for differences between men-specific and women-specific associations (21) for each GWAS trait (Supplementary Table 6) to test whether the identified sex-specific associations occurred on both (or multiple) traits or if the heterogeneity by sex was due to one trait but not the other. Fourteen out of 20 sex-specific associations for PF were confirmed with the multiple-comparison test (Bonferroni correct: p < 2.5 × 10−3). However, there was no statistically significant heterogeneity between sexes for PF with GLRA3/HPGD (4q34.1, FEV1), LINC01060/LINC02508 (4q35.2, FVC), GRIP1 (12q14.3, FVC), FLT3/URAD (13q12.2, FEV1), LINC00645 (14q12, FEV1), and GNAL/SLC35G4 (18p11.21, FEV1). The sex-specific significant CMA p values (p < 5.0 × 10−8) could be due to the β-coefficient mean differences between men and women for inflammatory markers but attenuated associations or no sex-specific associations for PF traits (eg, LINC00645 rs78382897: IL-6: β [SD] = −0.402 [0.104] in men and β [SD] = 0.079 [0.104] in women, t test p = 3.52 × 10−6, hsCRP: β [SD] = −0.458 [0.110] in men and β [SD] = −0.024 [0.108] in women, t test p = 6.52 × 10−5, and FEV1: β [SD] = 0.291 [0.105] in men and β [SD] = −0.011 [0.103] in women, t test p = 3.66 × 10−3).
Out of the 20 pleiotropic loci from sex-specific CMA analyses, 19 loci represent novel discoveries for PF. According to the NHGRI-EBI catalog (Supplementary Table 7), GRIP1 was already reported with FEV1/FVC in African ancestry (2) but not in European ancestry or multiancestry (10). Genome-wide associations were also previously described between GATA5/SLCO4A1/NTSR1 with hsCRP (19) and NRXN1 with BMI (13,22).
We further searched the NHGRI-EBI GWAS catalog to assess whether any of the novel CMA loci were identified at a suggestive significance level (p < 1 × 10−5 and p > 5 × 10−8) in prior GWASs with our age-related traits (Supplementary Tables 4 and 7). GWASs for PF reported suggestive associations in 13 loci out of 29 CMA loci, including CALN1, CEP112, GAN/CMIP, CYP2U1/RPSAP34, MAL2/AC021733.2, CBWD1/DOCK8/KANK1, NRXN1, CTNNB1/ZNF621/AC099560.1, AC131211.1/TOMM22P6, LOC101929468/AC004053.1, IYD/PPP1R14C/PLEKHG1, LINC00645, and GATA5/AL121832.1. Some of our CMA loci have also previously reached suggestive associations from GWAS for hsCRP (DGKI/AC083867.3), pulmonary artery enlargement in COPD (LOC105370611/GPR65), and longevity (AL138927.1/LINC01720, NRXN1, and DGKI/AKR1D1), among others.
Functional Annotations and Biological Insights
NCBI-dbSNP, HaploReg, RegulomeDB, GTEx, and GWAS4D were accessed to annotate variants concerning their functional consequence and regulatory potential. Supplementary Tables 8 and 9 provide the detailed functional regulatory features for lead CMA variants and their variants in high LD(r2 ≥ 0.8) for sex-combined and sex-specific CMA analyses. The summary of these findings (Supplementary Table 10) shows evidence of regulatory features for several of our CMA variants.
HaploReg indicated a total of 7 SNPs (5 loci) mapped in conserved syntenic regions by GERP and SiPhy, 18 SNPs (7 loci) located in promoter histone marks, 76 SNPs (20 loci) in enhancer histone marks, 42 SNPs (15 loci) at DNAse hypersensitive sites, 5 SNPs (4 loci) at protein regulatory binding sites, and 193 SNPs (29 loci) in TF binding motifs.
In addition, 95 variants (5 loci) have cis-eQTLs for target genes in several tissues from GTEx. For PF and adiposity located on CYP2U1/SGMS2 locus (4q25; Supplementary Tables 8 and 10), 7 variants of SGMS2 (including the rs17564275 in the 3-prime UTR region of the gene) and 45 variants on CYP2U1 are cis-eQTLs for CYP2U1 in the lung, adipose-visceral, artery-aorta, heart-atrial, fibroblasts, and thyroid. Forty-five variants on CYP2U1/SGMS2 locus are also cis-eQTLs for RP11-286E11.2 in fibroblasts.
For PF and glucose metabolism located on CBWD1/DOCK8 locus (9p24.3), 5 variants indicate evidence of cis-eQTLs (eg, rs1932751; Supplementary Table 8) for CBWD1 in 20 tissues, including lung, pancreas, liver, adipose-subcutaneous, adipose-visceral, muscle-skeletal, artery-aorta, artery-coronary, artery-tibial, heart-left ventricle, heart-atrial appendage, fibroblasts, whole blood, and others. Five variants on the CBWD1/DOCK8 locus are cis-eQTLs for the lncRNA-C9orf66 (DOCK8 antisense RNA) and DOCK8 in the pancreas, adipose-subcutaneous, liver, and brain. Twenty-nine associated variants on the MMUT/CENPQ locus (6p12.3) for PF and glucose metabolism are cis-eQTLs (eg, rs525559; Supplementary Tables 9 and 10) for CENPQ in 14 tissues, including lung, adipose-subcutaneous, adipose-visceral, muscle-skeletal, artery-aorta, artery-coronary, artery-tibial, heart-left ventricle, heart-atrial appendage, fibroblasts, whole blood, thyroid, and brain. The 29 variants are also suggested as cis-eQTLs for MMUT in the artery-tibial, nerve-tibial, fibroblasts, and adrenal gland.
For PF and inflammatory markers, 10 variants on the GLRA3/HPGD locus (4q34.1, eg, rs2332838; Supplementary Table 9) are cis-eQTLs for HPGD in whole blood and lung, and 3 variants on the TRIM9 locus (14q22.1, eg, rs55801251; Supplementary Table 9) are cis-eQTLs for TRIM9 expression in nerve-tibial.
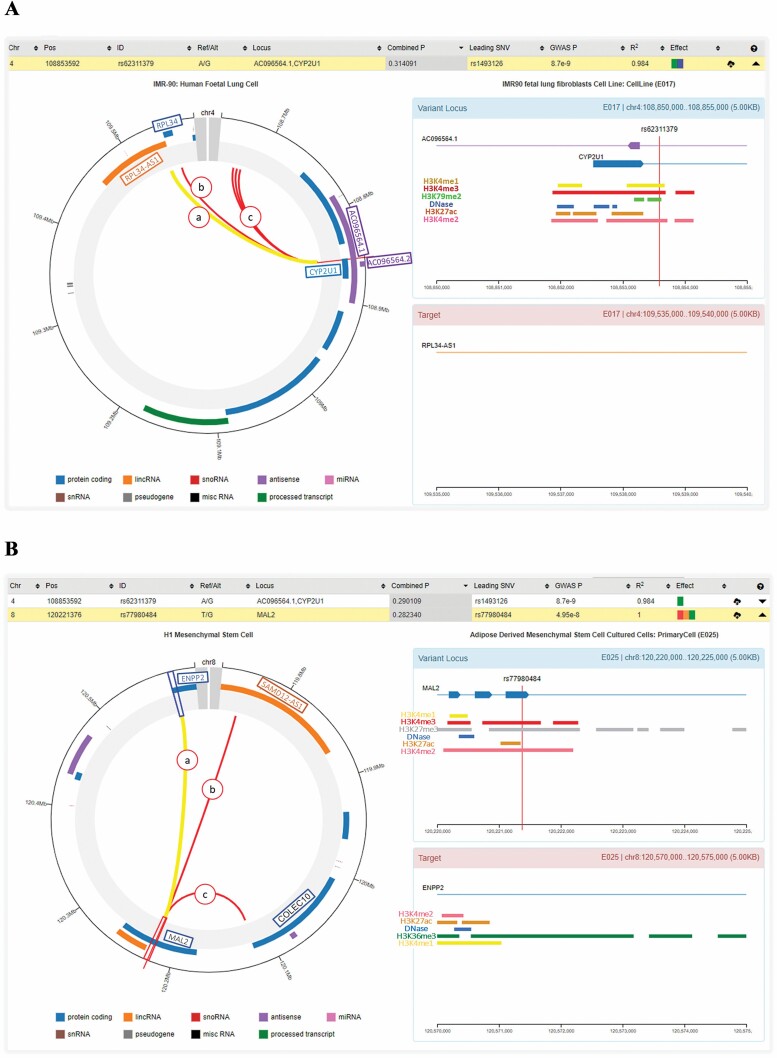
GWAS4D results indicate that several CMA variants (CMA p < 5 × 10−8; Supplementary Table 11) have effects of transcription regulatory binding affinity change, conserved syntenic regions, Hi-C interaction, and cell type-specific regulation. For PF and adipose in sex-combined analyses, for instance, Hi-C interaction links the rs62311379/CYP2U1 to LincRNA-RPL34 and RPL34 in fetal lung fibroblasts (Figure 2). Hi-C interactions are suggested between rs62311379/CYP2U1 with a genomic region at 5 kb in adipose, HepG2, T cells, and B cells. For PF and muscle strength, the rs77980484/MAL2 interacts with ENPP2, COLEC10, LincRNA-SAMD12 in adipose stem cells (Figure 2) and SAMD12 and LincRNA-SAMD12 in T cells (Supplementary Figure 6). Also, in T cells, the rs11079574/AC004805.1 (21-kb 5′ of AXIN2) interacts with CEP112 for PF with inflammatory markers (Supplementary Figure 7). Significant Hi-C interactions for PF and muscle strength in sex-specific analyses are between rs117500377/AL136418.1 with HEATR5A in adipose stem cells (Supplementary Figure 8) and with HECTD1, AKAP6, and HEATR5A in the liver (Supplementary Figure 9). For PF and inflammatory markers, the rs62196354/20q13.33 interacts with C20orf166/MIR1-1HG and RPL7P3 in fetal lung fibroblasts (Supplementary Figure 10) and with C20orf166/MIR1-1HG, RPL7P3, LINC00659, OGFR, COL9A3, and GATA5 in HepG2 (Supplementary Figure 11), among others. Hi-C results suggest regulatory functions, such as promoter-enhancer interactions, occurring between several of our CMA SNPs/loci with target genes/regions at 5-kb resolution.
Figure 2.
GWAS4D regulatory features for rs62311379 (4q25) on fetal lung cell lines (A) and rs77980484 (8q24.12) on adipose mesenchymal stem cells (B). Notes: Chr = chromosome; Pos = position (GRCh37, hg19); ID = prioritized regulatory variant; Ref/Alt = reference and alternative alleles; Combined p = combined regulatory probability; Leading SNP = the most significant SNP; GWAS p = correlated meta-analysis p value; r2 = square correlation coefficient between ID SNP and leading SNP (see Supplementary Table 11). (A) The prioritized SNP rs62311379, residing on the AC096564.1/CYP2U1 locus. The uniform processes of Hi-C data at 5-kb resolution are represented at spatiotemporal level (left graphic). Significant HI-C interactions (a–f internal lines) were detected between CYP2U1 intronic–rs62311379 with (a) RPL34-AS1 (score = 1.83), (b) RPL34 (score = 1.70), and (c) 108.65 Mb (scores = 1.33–1.38). rs62311379 is also located in the histone acetyltransferase activity (H3) regions and the DNase hypersensitivity site (right-top graphic). RPL34-AS1 (RPL34 antisense RNA) is the target gene (for “a,” right-bottom graphic). (B) The prioritized SNP rs77980484, residing on the MAL locus, indicated regulatory features on adipose mesenchymal stem cells. The uniform processes of Hi-C data at 5-kb resolution are represented at spatiotemporal level (left graphic). Significant HI-C interactions (a–c internal lines) were detected between MAL intronic–rs77980484 with (a) ENPP2 (score = 1.84), (b) SAMD12-AS1 (score = 1.39), (c) COLEC10 (score = 1.19). rs77980484 is also located in H3 regions and the DNase hypersensitivity site (right-top graphic). ENPP2 represents the target gene (for “a,” right-bottom graphic). GWAS = genome-wide association study.
In addition, we provide biological insights for several TF binding (eg, P300, HDAC2, IRF, POU2F, STAT, MYC, AP-1, MAF, PAX5, GATA, SOX, GR/NR3C1, NRSF/REST, ZEB1, and FOX) with evidence for regulating the expression of at least 3 of our identified CMA genes/loci and with prominent roles in PF and pulmonary diseases (eg, CALN1, CTNNB1, CYP2U1, SGMS2, CBWD1, MMUT, HPGD, GATA5, NPVF, C2CD3, and MUC16; Supplementary Table 12).
Discussion
The power gained by accounting for the GWAS correlations between PF with inflammatory markers, body fatness, muscle strength, or glucose metabolism via the CMA approach enabled the discovery of 32 loci (12 for sex-combined and 20 for sex-specific samples). Twenty-nine out of 32 are novel loci for FEV1 and FVC. Thirteen of the 29 CMA genome-wide significant loci were previously reported with only suggestive associations (p < 1 × 10−5 and p > 5 × 10−8) from GWAS for PF. Ten among the 13 loci suggestive associations were found in African ancestry only. In addition, the CMA approach allowed us to validate 10 loci previously reported in the literature from GWAS for PF or age-related traits, which were not significant for the LLFS GWAS traits.
We found potentially regulatory functional implications for several identified CMA genes/loci from genomic annotation and gene expression data. Variants on the CYP2U1/SGMS2 locus (4q25) for PF and body fatness show enhancer and promoter histone marks, DNAse hypersensitive sites, TF binding motifs, and cis-eQTLs for CYP2U1 in the lung, adipose, artery, heart, thyroid, and fibroblasts, and RP11-286E11.2 expression in fibroblasts. The Hi-C interaction analyses also indicate that rs62311379 of CYP2U1 links to LincRNA-RPL34 and RPL34 in fetal lung fibroblasts and has other interactions at 5-kb genomic region in adipose, liver (HepG2), and white blood cells of the immune system. These findings show evidence of the role of the CYP2U1/SGMS2 locus with regulatory effects on lung, adipose, artery, heart, and fibroblasts. Dysfunction in these tissues is related to reduced PF, impaired metabolic diseases associated with obesity, chronic low-grade inflammation, and aging, which are underlying causes of T2D, CVD, and mortality (5,9,23). Fibroblast migration is also involved in pathways relating to the cytoskeleton and plays an essential role in age-related idiopathic pulmonary fibrosis (18,24). CYP2U1 encodes a member of the cytochrome P450 superfamily of enzymes that catalyze many reactions in drug metabolism and synthesis of cholesterol, steroids, and other lipids (25). The enzyme encoded by SGMS2 contributes to sphingomyelin (SGM) synthesis and homeostasis at the plasma membrane. SGM regulates many physiological cellular responses inducing proliferation, apoptosis, membrane mobility, and airway smooth muscle functions. The SGM activity and Sgms2 gene expression in mouse lung was reduced in response to cigarette smoke (26).
Variants on the CBWD1/DOCK8 locus (9p24.3) for PF and glucose metabolism have enhancer histone marks, promoter histone marks, TF binding motifs, and cis-eQTLs for CBWD1 in lung, pancreas, liver, adipose, muscle-skeletal, artery, heart, fibroblasts, blood, and others. Also, variants on CBWD1/DOCK8 are cis-eQTLs for lncRNA-C9orf66 (DOCK8 antisense RNA) and DOCK8 in the pancreas, adipose, liver, or brain. The function of CBWD1 (cobalamin synthetase W domain-containing protein-1) remains elusive. A recent study has reported that high cobalamin levels (ie, vitamin B12) increase lung cancer risk (27). DOCK8 (dedicator of cytokinesis-8) encodes a member of the DOCK-C subfamily of guanine nucleotide exchange factors. DOCK8 is essential for T-cell survival, maintaining CD8+ T-cell memory (28), and regulating signal transduction events to control immunity (29). An experimental study demonstrated that mutation in the Dock8 gene led to an immune attack on mouse islet cells, resulting in insulin-dependent T2D in 20% of the animals (30). Deletion at 9p24.3, where DOCK8 is located, was associated with squamous cell lung carcinoma (31). In addition, for PF and glucose metabolism, variants on MMUT/CENPQ locus (6p12.3) are cis-eQTLs for CENPQ in the lung, adipose, muscle-skeletal, artery, heart, fibroblasts, blood, brain, and others. Variants on the MMUT/CENPQ locus are also cis-eQTLs for MMUT in the artery, fibroblasts, and adrenal gland. MMUT (alias MUT) encodes the mitochondrial methylmalonyl coenzyme-A mutase (MCM). In humans, the product of MMUT is a vitamin B12-dependent enzyme that catalyzes the isomerization of MCM to succinyl-CoA. A fine-scale haplotype mapping study reported the association of MMUT with insulin resistance (32). CENPQ is involved in the centromeric complex and plays a central function in coordinating chromosome congression mechanisms (33).
For PF and inflammatory markers, variants on the GLRA3/HPGD locus (4q34.1) have promoter histone marks, DNAse hypersensitive sites, TF binding motifs, and cis-eQTLs for HPGD in whole blood and lung. HPGD encodes 15-hydroxyprostaglandin dehydrogenase (15-PGDH) responsible for the metabolism of prostaglandins and has a function in various physiologic and cellular processes such as inflammation. Inhibition of 15-PGDH decreased alveolar epithelial cell apoptosis, fibroblast proliferation, and fibrocyte differentiation, and reduced collagen production in idiopathic pulmonary fibrosis precision-cut lung slices (34). In the bleomycin-induced pulmonary fibrosis murine model, the inhibition of the 15-PGDH led to an increase of anti-inflammatory actions of prostaglandin E2 known to antagonize TGFβ-mediated fibrotic signaling (35). TRIM9 variants (14q22.1), associated with PF and inflammatory markers, show promoter histone marks, DNAse hypersensitive sites, TF binding motifs, and cis-eQTLs for TRIM9 expression in the nerve-tibial. TRIM9 encodes E3 ubiquitin-protein ligase, which ubiquitinates itself in cooperation with an E2 enzyme UBE2D2/UBC4. E3 ubiquitin-protein ligase plays an essential role in various cellular processes, including cell proliferation and apoptosis (36). E3 ubiquitin-protein ligase knockdown in human lung cancer tissues and cell lines reduced expression of Bcl-2 (β-cell lymphoma-2), known to be involved in the apoptosis pathway (36).
In addition, several variants of CTNNB1/ZNF621 (3p22.1), GATA5/SLCO4A1/NTSR1 (20q13.33), NPVF/C7orf31/CYCS (7p15.3-p15.2), and CALN1 (7q11.22), associated for PF and inflammatory markers, show TF binding motifs, enhancer or promoter histone marks, and DNAse hypersensitive sites in blood, lung, brain, muscle, among others (Supplementary Tables 8 and 9). CTNNB1 encodes catenin β-1, which is part of a complex of proteins that constitute adherens junctions necessary to create and maintain epithelial cell layers by regulating cell growth and adhesion between cells. CTNNB1 plays a critical function in cancer regulation, including lung cancer (37,38). It is worth mentioning that our CMA association with CTNNB1 was identified in women, which is consistent with a previous study where 70% of primary lung adenocarcinoma patients presenting mutated CTNNB1 were women (38). GATA5 has been associated with many cardiovascular development and diseases (39), lung adenocarcinoma, and squamous cell lung carcinoma (40). NTSR1 is a neurotensin receptor-1 mediating several neurotensin functions such as hypotension, hyperglycemia, hypothermia, and antinociception. Intense and chronic neurotensin exposure leads to the activation of NTSR1 signaling, which enhances cell proliferation, survival, mobility, and invasion in several cancers, including lung cancer (41). The protein encoded by SLCO4A1 transports many structurally unrelated compounds, including hormones, bile acids, and prostaglandins (42). The cytochrome-C protein encoded by CYCS functions as a central component of the electron transport chain in mitochondria and participates in intrinsic apoptotic pathways, including lung (43). Patients in advanced stages of nonsmall cell lung cancer showed significantly lower levels of serum cytochrome-C than healthy individuals, and the lower levels were associated with worse survival outcomes (44).
For PF and muscle strength, the rs77980484 of MAL2 (8q24.12) has protein regulatory binding sites, TF binding motif, enhancer histone markers, promoter histone marks, and DNAse hypersensitive sites in lung, muscle, fat, brain, among others. The GWAS4D results also show that the rs77980484 interacts with ENPP2, COLEC10, LincRNA-SAMD12 in adipose stem cells and with SAMD12 and LincRNA-SAMD12 in T cells. MAL2 encodes a component of lipid rafts involved in the machinery of polarized transport. Although MAL2 has been associated with several cancer types, its association with lung cancer or its role in PF remains poorly understood. ENPP2 (ectonucleotide pyrophosphatase/phosphodiesterase-2) encodes autotaxin (ATX) enzyme that catalyzes the extracellular production of lysophosphatidic acid (LPA). LPA binds to plasma membrane G-protein-coupled receptors, regulating essential cellular functions including proliferation, migration, and apoptosis. ATX acts as an angiogenic factor by stimulating smooth muscle cells migration and microtubule formation (45). ATX/LPA signaling has been linked to inflammatory diseases, such as allergic asthma, pulmonary fibrosis, and lung carcinogenesis (46). MAL2/ENPP2 resides on 8q24, a chromosomal region of instability due to somatic copy number alterations, where the proto-oncogene MYC is also located (47). LPA was suggested to stimulate MYC expression and cooperate with it in cell transformation (46). MYC participates in numerous human cancers, including lung cancer (48).
It is worth noting that some of our identified genes/loci are part of the WNT signaling pathway, including CTNNB1, LEF1 (CYP2U1 locus), AXIN2, SLCO4A1-AS1, and NTSR1. The WNT pathway is one of the main signaling pathways involved in normal lung development and homeostasis. In contrast, WNT signaling deregulation contributes to tumor vascularization, drug response, and lung carcinogenesis progression (37,38). β-catenin encoded by CTNNB1 is an integral part of the canonical WNT signaling, which interacts with signaling regulators (eg, AXIN2, 17q23-q24), transcriptional regulators (eg, TCF [T-cell factor]/LEF1 [lymphoid enhancer-binding factor-1]), among other elements. The TCF/LEF1 initiates a program of transcription of downstream WNT target genes, while AXIN plays an essential role in regulating the stability of β-catenin in the WNT signaling. AXIN is a negative regulator of WNT signaling that promotes β-catenin phosphorylation leading to β-catenin degradation (37). The lncRNA SLCO4A1-AS1 facilitates cell growth and enhances the resistance of lung adenocarcinoma cells to chemotherapy via activating WNT/β-catenin signaling (49). The neurotensin protein, encoded by NTSR1, causes overexpression of EGFR (epidermal growth factor receptor). Inhibition of β-catenin can enhance the efficacy of irreversible EGFR tyrosine kinase inhibitor therapy in EGFR-mutant nonsmall cell lung cancer (50). However, the exact mechanism involving Wnt/β-catenin signaling in nonsmall cell lung cancer progression remains undetermined.
Our study has several important strengths but also some limitations. CMA is a powerful approach that detects associations at genome-wide significance level integrating correlated GWAS scans, corrected for dependencies due to overlapping individuals, cryptic relatedness, and population structure. However, CMA does not differentiate if the observed genetic variant is a single causal variant that affects 2 (or more) traits or if the variant is in a strong LD with causal variants colocalized in the same chromosome region that affects different traits. Some of our SNPs had previously shown evidence of association for age-related traits from published individual GWAS. However, replication in larger studies is relevant to validate our pleiotropic loci for PF with age-related traits. In addition, the generalization of our findings to other populations and ancestry groups needs confirmation because the allele frequency of the associated SNPs and the LD in European ancestry can differ in other ethnic groups. Several CMA variants/genes have supportive evidence by gene expression, genomic annotation data, and biologic pathways impacting pulmonary diseases, systemic inflammation, glucose metabolism, and muscle strength. However, further functional experimental studies are needed to confirm our findings.
In summary, our study identified variants sharing genetic effects on PF with IL-6 and hsCRP, HbA1c, BMI, and muscle strength. Several variants colocalized with gene expression (cis-eQTL) in lung, blood, adipose, pancreas, muscle, artery, heart, fibroblasts, and liver. Several variants also show evidence of 3 or more functional regulatory genomic elements. In addition, several genes are implicated in biological pathways. Together, these insights provide new avenues for investigating the underlying biology for pulmonary diseases, systemic inflammation, T2D, frailty, and the potential therapeutics for pulmonary disease.
Supplementary Material
Acknowledgments
We thank the Long Life Family Study participants and its members.
Funding
This work was supported by the National Institute on Aging (U01AG023746, U01AG023712, U01AG023749, U01AG023755, U01AG023744, and U19AG063893). A.J.S. was supported by a career development award from the National Institute on Aging (K01 AG057726).
Conflict of Interest
None declared.
Author Contributions
M.F.F. and M.A.P. participated in the conceptualization of the study, interpretation, writing, and revising the manuscript. M.A.P. conceptualized the correlated meta-analysis method. M.F.F. and L.W. performed the statistical and bioinformatics analyses. M.F.F., M.A.P., M.K.W., J.A.A., E.W.D., L.W., A.J.S., and M.N. revised and approved the manuscript.
References
- 1. Thyagarajan B, Wojczynski M, Minster RL, et al. Genetic variants associated with lung function: the Long Life Family Study. Respir Res. 2014;15:134. doi: 10.1186/s12931-014-0134-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lutz SM, Cho MH, Young K, et al. A genome-wide association study identifies risk loci for spirometric measures among smokers of European and African ancestry. BMC Genet. 2015;16:138. doi: 10.1186/s12863-015-0299-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang R, Burke GL, Enright PL, et al. Inflammatory markers and longitudinal lung function decline in the elderly. Am J Epidemiol. 2008;168(6):602–610. doi: 10.1093/aje/kwn174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Silverman EK. Genetics of COPD. Annu Rev Physiol. 2020;82:413–431. doi: 10.1146/annurev-physiol-021317-121224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McNeill JN, Lau ES, Zern EK, et al. Association of obesity-related inflammatory pathways with lung function and exercise capacity. Respir Med. 2021;183:106434. doi: 10.1016/j.rmed.2021.106434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKeever TM, Weston PJ, Hubbard R, Fogarty A. Lung function and glucose metabolism: an analysis of data from the Third National Health and Nutrition Examination Survey. Am J Epidemiol. 2005;161(6):546–556. doi: 10.1093/aje/kwi076 [DOI] [PubMed] [Google Scholar]
- 7. Peralta GP, Marcon A, Carsin AE, et al. Body mass index and weight change are associated with adult lung function trajectories: the prospective ECRHS study. Thorax 2020;75(4):313–320. doi: 10.1136/thoraxjnl-2019-213880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen L, Liu X, Wang Q, et al. Better pulmonary function is associated with greater handgrip strength in a healthy Chinese Han population. BMC Pulm Med. 2020;20(1):114. doi: 10.1186/s12890-020-1155-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duong M, Islam S, Rangarajan S, et al. Mortality and cardiovascular and respiratory morbidity in individuals with impaired FEV1 (PURE): an international, community-based cohort study. Lancet Glob Health 2019;7(5):e613–e623. doi: 10.1016/S2214-109X(19)30070-1 [DOI] [PubMed] [Google Scholar]
- 10. Shrine N, Guyatt AL, Erzurumluoglu AM, et al. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet. 2019;51(3):481–493. doi: 10.1038/s41588-018-0321-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sakornsakolpat P, Prokopenko D, Lamontagne M, et al. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet. 2019;51(3):494–505. doi: 10.1038/s41588-018-0342-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao X, Qiao D, Yang C, et al. Whole genome sequence analysis of pulmonary function and COPD in 19,996 multi-ethnic participants. Nat Commun. 2020;11(1):5182. doi: 10.1038/s41467-020-18334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kichaev G, Bhatia G, Loh PR, et al. Leveraging polygenic functional enrichment to improve GWAS power. Am J Hum Genet. 2019;104(1):65–75. doi: 10.1016/j.ajhg.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Province MA, Borecki IB. A correlated meta-analysis strategy for data mining “OMIC” scans. Pac Symp Biocomput. 2013:236–246. [PMC free article] [PubMed] [Google Scholar]
- 15. Feitosa MF, Kraja AT, Chasman DI, et al. Novel genetic associations for blood pressure identified via gene-alcohol interaction in up to 570K individuals across multiple ancestries. PLoS One. 2018;13(6):e0198166. doi: 10.1371/journal.pone.0198166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Newman AB, Glynn NW, Taylor CA, et al. Health and function of participants in the Long Life Family Study: a comparison with other cohorts. Aging (Albany NY) 2011;3(1):63–76. doi: 10.18632/aging.100242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang D, Yi X, Zhang S, et al. GWAS4D: multidimensional analysis of context-specific regulatory variant for human complex diseases and traits. Nucleic Acids Res. 2018;46(W1):W114–W120. doi: 10.1093/nar/gky407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wain LV, Shrine N, Artigas MS, et al. Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet. 2017;49(3):416–425. doi: 10.1038/ng.3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han X, Ong JS, An J, Hewitt AW, Gharahkhani P, MacGregor S. Using Mendelian randomization to evaluate the causal relationship between serum C-reactive protein levels and age-related macular degeneration. Eur J Epidemiol. 2020;35(2):139–146. doi: 10.1007/s10654-019-00598-z [DOI] [PubMed] [Google Scholar]
- 20. Hwang JY, Sim X, Wu Y, et al. Genome-wide association meta-analysis identifies novel variants associated with fasting plasma glucose in East Asians. Diabetes. 2015;64(1):291–298. doi: 10.2337/db14-0563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Randall JC, Winkler TW, Kutalik Z, et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013;9(6):e1003500. doi: 10.1371/journal.pgen.1003500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu Z, Guo Y, Shi H, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol. 2020;145(2):537–549. doi: 10.1016/j.jaci.2019.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weinmayr G, Schulz H, Klenk J, et al. Association of lung function with overall mortality is independent of inflammatory, cardiac, and functional biomarkers in older adults: the ActiFE-study. Sci Rep. 2020;10(1):11862. doi: 10.1038/s41598-020-68372-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. King TE Jr, Pardo A, Selman M. Idiopathic pulmonary fibrosis. Lancet 2011;378(9807):1949–1961. doi: 10.1016/S0140-6736(11)60052-4 [DOI] [PubMed] [Google Scholar]
- 25. Dhers L, Ducassou L, Boucher JL, Mansuy D. Cytochrome P450 2U1, a very peculiar member of the human P450s family. Cell Mol Life Sci. 2017;74(10):1859–1869. doi: 10.1007/s00018-016-2443-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gupta G, Baumlin N, Poon J, et al. Airway resistance caused by sphingomyelin synthase 2 insufficiency in response to cigarette smoke. Am J Respir Cell Mol Biol. 2020;62(3):342–353. doi: 10.1165/rcmb.2019-0133OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fanidi A, Carreras-Torres R, Larose TL, et al. Is high vitamin B12 status a cause of lung cancer? Int J Cancer. 2019;145(6):1499–1503. doi: 10.1002/ijc.32033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lambe T, Crawford G, Johnson AL, et al. DOCK8 is essential for T-cell survival and the maintenance of CD8+ T-cell memory. Eur J Immunol. 2011; 41(12):3423–3435. doi: 10.1002/eji.201141759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kearney CJ, Randall KL, Oliaro J. DOCK8 regulates signal transduction events to control immunity. Cell Mol Immunol. 2017; 14(5):406–411. doi: 10.1038/cmi.2017.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horwitz E, Krogvold L, Zhitomirsky S, et al. Beta-cell DNA damage response promotes islet inflammation in type 1 diabetes. Diabetes. 2018;67(11):2305–2318. doi: 10.2337/db17-1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang JU, Koo SH, Kwon KC, Park JW. Frequent silence of chromosome 9p, homozygous DOCK8, DMRT1 and DMRT3 deletion at 9p24.3 in squamous cell carcinoma of the lung. Int J Oncol. 2010;37(2):327–335. [PubMed] [Google Scholar]
- 32. Haydar S, Grigorescu F, Vintila M, et al. Fine-scale haplotype mapping of MUT, AACS, SLC6A15 and PRKCA genes indicates association with insulin resistance of metabolic syndrome and relationship with branched chain amino acid metabolism or regulation. PLoS One. 2019;14(3):e0214122. doi: 10.1371/journal.pone.0214122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bancroft J, Auckland P, Samora CP, McAinsh AD. Chromosome congression is promoted by CENP-Q- and CENP-E-dependent pathways. J Cell Sci. 2015;128(1):171–184. doi: 10.1242/jcs.163659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnthaler T, Theiler A, Zabini D, et al. Inhibiting eicosanoid degradation exerts antifibrotic effects in a pulmonary fibrosis mouse model and human tissue. J Allergy Clin Immunol. 2020;145(3):818–833 e811. doi: 10.1016/j.jaci.2019.11.032 [DOI] [PubMed] [Google Scholar]
- 35. Smith JNP, Witkin MD, Jogasuria AP, et al. Therapeutic targeting of 15-PGDH in murine pulmonary fibrosis. Sci Rep. 2020;10(1):11657. doi: 10.1038/s41598-020-68336-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gupta I, Singh K, Varshney NK, Khan S. Delineating crosstalk mechanisms of the ubiquitin proteasome system that regulate apoptosis. Front Cell Dev Biol. 2018;6:11. doi: 10.3389/fcell.2018.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rapp J, Jaromi L, Kvell K, Miskei G, Pongracz JE. WNT signaling—lung cancer is no exception. Respir Res. 2017;18(1):167. doi: 10.1186/s12931-017-0650-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou C, Li W, Shao J, Zhao J, Chen C. Analysis of the clinicopathologic characteristics of lung adenocarcinoma with CTNNB1 mutation. Front Genet. 2019;10:1367. doi: 10.3389/fgene.2019.01367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Song ZP, Yan B. Potential roles of GATA binding protein 5 in cardiovascular diseases. Rev Cardiovasc Med. 2020;21(2):253–261. doi: 10.31083/j.rcm.2020.02.5104 [DOI] [PubMed] [Google Scholar]
- 40. Gong C, Fan Y, Zhou X, Lai S, Wang L, Liu J. Comprehensive analysis of expression and prognostic value of GATAs in lung cancer. J Cancer. 2021;12(13):3862–3876. doi: 10.7150/jca.52623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Younes M, Wu Z, Dupouy S, et al. Neurotensin (NTS) and its receptor (NTSR1) causes EGFR, HER2 and HER3 over-expression and their autocrine/paracrine activation in lung tumors, confirming responsiveness to erlotinib. Oncotarget 2014;5(18):8252–8269. doi: 10.18632/oncotarget.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aregger M, Lawson KA, Billmann M, et al. Systematic mapping of genetic interactions for de novo fatty acid synthesis identifies C12orf49 as a regulator of lipid metabolism. Nat Metab. 2020;2(6):499–513. doi: 10.1038/s42255-020-0211-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matapurkar A, Lazebnik Y. Requirement of cytochrome c for apoptosis in human cells. Cell Death Differ. 2006;13(12):2062–2067. doi: 10.1038/sj.cdd.4401968 [DOI] [PubMed] [Google Scholar]
- 44. Javid J, Mir R, Julka PK, Ray PC, Saxena A. Extracellular cytochrome c as a biomarker for monitoring therapeutic efficacy and prognosis of non-small cell lung cancer patients. Tumour Biol. 2015;36(6):4253–4260. doi: 10.1007/s13277-015-3062-6 [DOI] [PubMed] [Google Scholar]
- 45. Nam SW, Clair T, Kim YS, et al. Autotaxin (NPP-2), a metastasis-enhancing motogen, is an angiogenic factor. Cancer Res. 2001;61(18):6938–6944. [PubMed] [Google Scholar]
- 46. Magkrioti C, Oikonomou N, Kaffe E, et al. The autotaxin-lysophosphatidic acid axis promotes lung carcinogenesis. Cancer Res. 2018;78(13):3634–3644. doi: 10.1158/0008-5472.CAN-17-3797 [DOI] [PubMed] [Google Scholar]
- 47. Watkins TBK, Lim EL, Petkovic M, et al. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature 2020;587(7832):126–132. doi: 10.1038/s41586-020-2698-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu J, Huang X, Liu D, et al. Demethyleneberberine induces cell cycle arrest and cellular senescence of NSCLC cells via c-Myc/HIF-1alpha pathway. Phytomedicine. 2021;91:153678. doi: 10.1016/j.phymed.2021.153678 [DOI] [PubMed] [Google Scholar]
- 49. Wei Y, Wei L, Li J, et al. SLCO4A1-AS1 promotes cell growth and induces resistance in lung adenocarcinoma by modulating miR-4701-5p/NFE2L1 axis to activate WNT pathway. Cancer Med 2020;9(19):7205–7217. doi: 10.1002/cam4.3270 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet. 2017;49(12):1693–1704. doi: 10.1038/ng.3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.