Abstract
Mammalian β-defensins are small cationic peptides possessing broad antimicrobial and physiological activities. Because dogs are particularly resilient to sexually transmitted diseases, it has been proposed that their antimicrobial peptide repertoire might provide insight into novel antimicrobial therapeutics and treatment regimens. To investigate this proposal, we cloned the full-length cDNA of three canine β-defensin isoforms (cBD-1, -2, and -3) from canine testicular tissues. Their predicted peptides share identical N-terminal 65-amino-acid residues, including the β-defensin consensus six-cysteine motif. The two longer isoforms, cBD-2 and -3, possess 4 and 34 additional amino acids, respectively, at the C terminus. To evaluate the antimicrobial activity of cBD, a 34-amino-acid peptide derived from the shared mature peptide region was synthesized. Canine β-defensin displayed broad antimicrobial activity against gram-positive bacteria (Listeria monocytogenes and Staphylococcus aureus; MICs of 6 and 100 μg/ml, respectively), gram-negative bacteria (Escherichia coli, Klebsiella pneumoniae, and Neisseria gonorrhoeae; MICs of 20 to 50, 20, and 50 μg/ml, respectively), and yeast (Candida albicans; MIC of 5 to 50 μg/ml) and lower activity against Ureaplasma urealyticum and U. canigenitalium (MIC of 200 μg/ml). Antimicrobial potency was significantly reduced at salt concentrations higher than 140 mM. All three canine β-defensins were highly expressed in testis. In situ hybridization indicated that cBD-1 was expressed primarily in Sertoli cells within the seminiferous tubules. In contrast, cBD-2 was located primarily within Leydig cells. The longest isoform, cBD-3, was detected in Sertoli cells and to a lesser extent in the interstitium. The tissue-specific expression and broad antimicrobial activity suggest that canine β-defensins play an important role in host defense and other physiological functions of the male reproductive system.
The mucosal physical barrier is an important component protecting luminal surfaces from bacteria. Discovery of epithelium-derived antimicrobial peptides with activity at mucosal surfaces extends luminal protection from the traditional barrier effect to innate immune capabilities. These endogenous peptides are natural antibiotics that are widely distributed in nature and represent an important mechanism of innate defenses against microbial infection (1, 5). Among these naturally occurring antimicrobial peptides, defensins form a unique family of cysteine-rich, small cationic peptides (17).
The defensin family is large and has been documented in mammals, plants, insects, and mollusks (6, 27, 29, 39, 47). In mammals, defensins are subdivided into α-, β-, and θ-defensins based on the organization of conserved six-cysteine motifs (43, 50). With the exception of bovine neutrophils, β-defensins are synthesized primarily in epithelial tissues (18, 27). Because epithelia of the male reproductive tract are essentially isolated from adaptive immune capabilities and because the luminal contents of the testis and epididymis are of vital importance for the preservation of the species, it follows that these epithelia have developed immediate and rapid mechanisms to counteract urogenital pathogens.
Sexually transmitted diseases and infections of the urinary and reproductive tracts remain important public health problems worldwide. Although β-defensins have been identified in the urogenital tract of several species, including humans (4, 12, 19, 27, 33, 38, 42, 52), mice (12, 32), rats (3, 9, 15, 36, 52), and nonhuman primates (2, 4), their existence in dogs has not been documented. Because of the clinical similarities in urinary tract infections between humans and dogs, this is a significant omission. For example, urinary tract infections are the most common infections in human hospitals and the second most common infections observed in the general population (13). Escherichia coli and Klebsiella pneumoniae represent the causative agents of 70 to 90% of all urinary tract infections in humans (16, 35, 40). This clinical feature is almost identical in dogs, where these two bacteria cause more than 80% of urinary tract infection cases reported in this species (26, 30, 44). Information on the antimicrobial peptide capabilities of the canine urogenital system may provide basic comparative knowledge for the development of novel therapeutic agents and treatments for urogenital infections.
Here we report the molecular cloning and identification of three β-defensins from canine testicular tissues. These newly identified antimicrobial peptides effectively kill gram-positive and -negative bacteria and yeast in a salt-dependent manner. The presence of these antimicrobial peptides in the testicular tissue of a species that is remarkably resilient to sexually transmitted diseases strongly suggests that β-defensins play a pivotal role in host defense mechanisms of the canine urogenital system.
MATERIALS AND METHODS
Cloning of cBD cDNA.
To begin our identification of cBDs, the conserved cysteine-motifs of hBD-5 and hBD-6 were used as a query sequence to search the National Center for Biotechnology Information GenBank expressed sequence tag (EST) database. This search resulted in a partial sequence of one cDNA (accession number BM537999) from the Canis Testicular cDNA Library (Cold Spring Harbor Laboratory, NY). Based on this EST sequence, two pairs of primers were designed (Table 1). To obtain full-length cDNA sequences, a modified rapid amplification of 5′ cDNA ends (5′-RACE), which selects for nontruncated 5′-capped mRNAs, was used, (FirstChoice RLM-RACE kit; Ambion, Inc., Austin, TX). Briefly, 10 μg of total RNA from canine testes were treated with calf intestinal phosphatase, followed by the addition of tobacco acid pyrophosphatase to remove the cap structure. T4 RNA ligase was used to ligate the 45-bp RNA adapter, and the RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase and random decamers. cDNAs were amplified with nested primers to the RNA adapter and an antisense gene-specific outer primer. The first PCR product was reamplified by using a nested inner RNA adapter primer (RLM-RACE kit; Ambion) and a gene-specific inner primer. A similar nested PCR strategy was also used for 3′-RACE to obtain the 3′-terminal cDNA sequences. In this case, the 3′-adapter was directly annealed to mRNA by its poly (T) tail and used as the primer for nonspecific reverse transcription of mRNA. Two rounds of nested PCR were then conducted with outer- and inner-adapter primers paired with the outer and inner sense gene-specific primers, respectively.
TABLE 1.
RACE and RT-PCR primers for canine β-defensins
| Primer type and name | Primer sequence | Location in cDNA (nt)a | Product size (bp) | GenBank accession no. |
|---|---|---|---|---|
| RACE primers | ||||
| Sense outer primer | 5′-GCTTGTCACTTCGTCAAGACCAGA-3′ | 90-113 | BM537999 | |
| Antisense outer primer | 5′-AATGCTTCCAGCTTTGTTCTTC-3′ | 428-407 | 339 | |
| Sense inner primer | 5′-CCTGAAGACATGAAGGCTTT-3′ | 131-150 | ||
| Antisense inner primer | 5′-TTCCAGCTTTGTTCTTCTTTATCA-3′ | 423-400 | 293 | |
| RT-PCR primers | ||||
| cBD-1 (sense) | 5′-CCTGAAGACATGAAGGCTTT-3′ | 46-65 | AY169790 | |
| cBD-1 (antisense) | 5′-TGAGATCAGACTTGGGACAGG-3′ | 313-293 | 268 | |
| cBD-2 (sense) | 5′-CCTGAAGACATGAAGGCTTT-3′ | 47-66 | AY169791 | |
| cBD-2 (antisense) | 5′-TTCCAGCTTTGTTCTTCTTTATCA-3′ | 409-386 | 363 | |
| cBD-3 (sense) | 5′-CCTGAAGACATGAAGGCTTT-3′ | 46-65 | AY169792 | |
| cBD-3 (antisense) | 5′-AATGCTTCCAGCTTTGTTCTTC-3′ | 343-322 | 298 | |
| GAPDH (sense) | 5′-TGGYATCGTGGAAGGRCTCAT-3′ | 561-581 | BC025925 | |
| GAPDH (antisense) | 5′-RTGGGWGTYGCTGTTGAAGTC-3′ | 930-910 | 370 |
nt, nucleotides.
The nested PCR products were purified with a column-based PCR purification kit (Qiagen Inc, Valencia, CA) and cloned into plasmids with pGEM-T Easy Vector Systems (Promega Co., Madison, WI), followed by transformation into E. coli (JM109; Stratagene Co., La Jolla, CA) and colony screening with PCR. The primers used for PCR clone screening were the sense and antisense gene-specific primers. Plasmids were extracted from bacterial culture derived from identified single colonies, and the inserts were sequenced with SP6/T7 vector primers (Promega) on an automated ABI 3700 DNA analyzer at the Kansas State University Sequencing and Genotyping Facility (Manhattan, KS). Each cDNA sequence was generated from the sequencing results of at least five identical plasmid extracts from individual colonies. The full-length cDNA sequence was then generated by ligation of the 5′ and 3′ sequences and deletion of the adaptor and overlapped region.
Synthesis and preparation of a cBD peptide.
A 34-amino acid peptide (KCWNLRGSCREKCIKNEKLYIFCTSGKLCCLKPK) spanning the cBD cysteine motif was chemically synthesized (AC Scientific, Inc., Duluth, GA). The material eluted as a single peak on reverse-phase high-pressure liquid chromatography, and the peptide identity was confirmed by mass spectroscopy. The final purity of the peptide was 93% with a mass of 3,994.95 Da. The peptide was lyophilized and dissolved in 0.01% acetic acid at 2 mg/ml (∼0.5 mM) as a stock solution and stored at −135°C until use. Formation of the three disulfide bonds was induced by air oxidation catalyzed by Cu2+ (52). Briefly, the peptide (2 mg/ml) was dialyzed in a 500-μl dialysis tube (molecular mass cutoff, 3,500 Da; Spectrum Laboratories, Inc., Rancho Dominguez, CA) at 4°C in 1 liter of 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM dithiothreitol overnight. Dialysis bags were then transferred to 1 liter of 0.1 M NaHCO3, containing10 mM cysteine and 10 μM CuCl2. Oxidized peptides were dialyzed against 1 liter of 0.01% acetic acid for 4 h at 4°C and passed through a 0.2-μm-pore-size filter. The peptide concentration was determined with a BCA protein assay (Pierce, Inc., Rockford, IL). Human serum albumin (Sigma Aldrich, St. Louis, MO) was added to the peptide solution to achieve a final concentration of 0.1% immediately before the peptide was used.
Three-dimensional structure analysis of cBD.
Structural models were generated for cBD via homology modeling. The target structure was predicted via alignment to a set of three human β-defensin templates (isoforms 1, 2, and 3, respectively) for which experimental structural characterization was available (24, 42). Canine β-defensin sequences were aligned to the human templates via the CLUSTALW program (48), using the Blosum 30 substitution matrix, a gap-opening penalty of 10, and a gap-extension penalty of 0.1. These resulted in multiple alignments, and the corresponding three-dimensional human β-defensin peptide structures were then processed via the Modeller program (41) to yield structural predictions for the canine β-defensin target. Modeller's default simulated annealing cycles were used for structural refinement. Analysis of the peptide secondary structure and surface characteristics was carried out on the resulting structures via SYBYL (SYBYL6.9.2,;TRIPOS, Inc., St. Louis, MO).
Antimicrobial activity.
Candida albicans ATCC 11006 and 14053, Escherichia coli ATCC 25922 and 700336, Klebsiella pneumoniae ATCC 10031, Neisseria gonorrhoeae ATCC 10150, Listeria monocytogenes ATCC 19115, Staphylococcus aureus ATCC 10832, Ureaplasma canigenitalium ATCC 51252, and U. urealyticum ATCC 27619 were used for the microbicidal assays. C. albicans was grown overnight (35°C) to stationary phase on potato dextrose agar and managed as described by the NCCSL standards (33). U. canigenitalium and U. urealyticum were stored frozen (−20°C) in urea broth 10B (per American Type Culture Collection specifications) and thawed at 5°C for 2 h before assay. N. gonorrhoeae was regularly maintained on chocolate agar, single strength BBL GC II agar base (Becton Dickinson, Sparks, MD) with 2% hemoglobin (BBL) and incubated at 37°C in 5% CO2. All other bacteria were maintained on Trypticase soy agar (Difco, Detroit, MI) plates at 37°C. N. gonorrhoeae and other bacteria were cultured to mid-logarithmic phase by transferring single cell colonies grown overnight into GC II agar base broth (36) or Trypticase soy broth (Difco), followed by incubation and shaking for 3 h at 37°C. Subcultures were centrifuged for 10 min at 4°C at 900 × g, and the bacterial pellets were washed once and suspended in 10 mM sodium phosphate buffer (PB [pH 7.4], [Na+] = 17.83 mM).
Initial bacterial concentrations were determined as previously described (12). N. gonorrhoeae at 108 CFU/ml was obtained at an optical density of 0.1 at 620 nm; the same initial population was estimated for the other bacterial cultures at an optical density of 0.1 at 600 nm. C. albicans were suspended in 10 mM PB to a McFarland standard of 0.5 for an initial 108 CFU/ml according to NCCLS recommendations (33), except that the recommended 0.85% saline solution was replaced with 10 mM PB. All microbial working dilutions were in 10 mM PB thereafter.
To determine the antimicrobial activity of cBD, a broth microdilution method adapted from the NCCLS methods (12, 33) was used as previously described (46). Briefly, 50 μl of the CBD working stocks were added to the wells of a microtiter plate in which 25 μl of 30 mM sodium PB (pH 7.4, [Na+] = 53.49 mM), 25 μl of H2O, and 50 μl of the microbial suspension had been previously combined. Microtiter plates were incubated at 37°C for 2 h in a shaking incubator (100 rpm), followed by the addition of 150 μl of the corresponding nutritive broth. Growth inhibition was determined by measuring MICs, defined as the lowest concentration in which microbial growth was prevented, as indicated by the lack of turbidity or color change (urea broth 10B) after 24 h of incubation at 37°C. Because N. gonorrhoeae does not show turbidity as evidence of growth in GC broth and because of its high autolytic activity in liquid medium, 50 μl was spotted onto chocolate agar and the MIC was defined as the lowest concentration at which no colonies developed after 24 h of incubation at 37°C in 5% CO2.
To determine the salt sensitivity of cBD bactericidal activity, two representative bacteria (one gram-negative and one gram-positive bacteria) were used; E. coli ATCC 25922 and L. monocytogenes ATCC 19115, respectively. The final sodium concentrations of the bactericidal mixture were adjusted to 15, 30, 50, 140, and 300 mM with NaCl. All assays were performed in triplicate.
Tissue expression of cBD mRNA.
To determine the tissue expression of cBD genes, samples were obtained from healthy dogs according to procedures approved by the Kansas State University Institutional Animal Care and Use Committee, placed immediately in liquid nitrogen, and stored at −135°C until use. Testicular tissues were collected from three clinically healthy dogs. All dogs had a thorough clinical examination and preoperative screening, including hematology, blood chemistry, and urinalysis. All clinical laboratory results were within normal limits. No clinical signs of urogenital disease (i.e., urethral secretion, orchitis, balanitis, and cystitis) were present at time of the elective surgical procedure. Total RNA was extracted with TRI reagent (Sigma-Aldrich) after frozen tissues were ground in liquid nitrogen. A one-step reverse transcription-PCR (RT-PCR) was used to detect expression of cBD mRNA. Briefly, total RNA was treated with RQ1 RNase-free DNase I (Promega), and RNA samples (250 ng) were then used in a 25-μl RT-PCR mixture with 0.1 μM concentrations of each sense and antisense primer (Table 1). Conventional one-step RT-PCR was performed by using an AccessQuick RT-PCR System kit (Promega). Complementary DNA synthesis and predenaturation were performed for 1 cycle at 48°C for 45 min and 95°C for 3 min; amplification was carried out for 40 (cBD) or 25 (glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and the final extension was at 72°C for 10 min in a 25-μl reaction mixture. After amplification, 10 μl of each reaction mixture was subjected to electrophoresis on 2% agarose gels, and bands were visualized by ethidium bromide staining. The efficiency of the RT procedure was standardized by assurance of comparable levels of the house-keeping gene GAPDH, amplified with PCR (51). For Southern analysis, RT-PCR products of cBDs were subjected to electrophoresis on 1.5% agarose gels and then capillary transferred overnight onto nylon membranes. Hybridization was performed with 32P-labeled cDNA probes generated from related fragments in the sequence-confirmed cBD cDNA clones. Probes were labeled with Prime-a-Gene Labeling System (Promega), and visualization of the signal was by exposure of X-ray films to the hybridized membrane.
To validate the RT-PCR and Southern analysis data, a two-step real-time RT-PCR assay was conducted. Briefly, RT was performed in 25-μl reactions by using optimal MgCl2 (∼4 mM) in the first-strand buffer containing 50 mM Tris-HCl (pH 8.3) and 75 mM KCl. The priming oligonucleotides (40-pmol gene-specific primers) were annealed to 2.5 μg of total RNA in a total volume of 13 μl, incubated at 65°C for 5 min, and then cooled to 4°C. RT was performed by adding 7 μl of the RT mix such that the final concentration was 1× first-strand buffer, 0.5 mM deoxynucleoside triphosphate, 5 mM dithiothreitol, 40 U RNase inhibitor, and 200 U of superscript III reverse transcriptase (Invitrogen, Carlsbad, CA), and the mixture was incubated at 50°C for 60 min, heated to 70°C for 15 min, and cooled to 4°C. The reaction mixture of the subsequent PCR consisted of cDNA (250 ng), 0.5 μM concentrations of each primer, 1.5 μl of LightCycler Fast Start DNA Master SYBR Green 1 mix (Roche Diagnostics), and 4 mM MgCl2 in a total volume of 25 μl. Quantitative PCR was performed by using a SmartCycler system (Cepheid, Inc., Sunnyville, CA) and the following profile: stage 1, 95°C for 10 min; and stage 2, 95°C for 30 s and 58°C for 30 s, followed by an extension at 72°C for 40 s with the optic on at the primer annealing step. Stage 2 was repeated for 40 cycles. Stage 3 involved a melt curve assay that determined the specificity of the amplified PCR product. Analysis of amplification utilized increased fluorescence intensity of the reporter dye, SYBR Green, which was plotted against the cycle number. Threshold cycle (CT) was determined by exponential product amplification and determined from subsequent increased fluorescence intensity above background. The data were processed by using a standard curve generated with a series of doses of identical targets amplified at the same time.
In situ hybridization.
To localize cBD expression in canine testes, in situ hybridization was performed on tissues collected at the Kansas State University Veterinary Medical Teaching Hospital by using procedures approved by the Institutional Animal Care and Use Committee. Testicles were collected from clinically healthy dogs undergoing elective surgery (castration). All dogs were between 6 and 12 months of age. Briefly, testes were cut into ∼0.5-cm3 blocks, placed in 4% paraformaldehyde diethyl pyrocarbonate (DEPC)-treated phosphate-buffered saline (PBS; pH 7.5), and incubated at 4°C for 2 to 4 h with two changes of the same fixative solution. Fixed tissues were transferred to 30% sucrose-buffered, DEPC-treated PBS at 4°C overnight. Tissues were then stored in a freezing compound at −80°C until sectioning. Sections (15 μm) were prepared with a cryostat and attached to precoated slides (Fisher Scientific, Inc.). Cryosections were immediately postfixed with 400 μl of fixative on the slide for 5 min at 22°C. Sections were washed twice for 5 min each with DEPC-PBS and then permeabilized with 0.5 mg/ml of proteinase K (Roche Diagnostics) at 22°C for 10 min. Sections were then washed twice as described above and incubated in 2 mg/ml of glycine and PBS for 5 min. Tissues were treated with 0.1 M triethanolamine-HCl-0.25% acetic anhydride (vol/vol) for 10 min and equilibrated for 15 min in 5× SSC (0.75 M NaCl, 0.075 M sodium citrate). Prehybridization was conducted for 2 h at 55°C in prehybridization mixture (50% formamide, 5× SSC, 1 mg/ml of yeast tRNA, 200 μl/section).
To generate probe templates, 3′-RACE products of cBDs in the pGEM-T vector (Promega) were treated with HindIII and SpeI (Promega) resulting in the T7 polymerase promoter sequence linked to the isoform-specific 3′-terminal sequences at 180, 322 and 314 bp for cBD-1, cBD-2, and cBD-3, respectively. The remaining ∼80-bp shared regions after HindIII restriction were then removed by exonuclease III by using an Erase-a-Base System (Promega). The linearized plasmids were purified from agarose gel with a QIAquick gel extraction kit (Qiagen). Isoform-specific antisense probes were generated by using T7-polymerase (Roche Diagnostics) and sense probes were generated by using SP6-polymerase (Roche Diagnostics). Hybridization was carried out with the addition of 0.5 μg of labeled probe/μl in prehybridization solution at 55°C in a humidified chamber for 24 h. Slides were subsequently rinsed in 2× SSCT (2× SSC containing 0.1% Tween 20) twice at 50°C for 30 min each and then washed in 0.2× SSCT at 50°C for 30 min. Sections were blocked in 1% blocking reagent (wt/vol; Roche Diagnostics) in malate buffer (100 mM maleic acid, 150 mM NaCl [pH 7.5]) for 10 min, followed by incubation for 2 h with an anti-digoxigenin antibody conjugated to alkaline phosphatase (Roche Diagnostics) diluted 1:1,000 in blocking solution at room temperature. After a wash with 0.02% Tween-PBS (vol/vol) twice for 30 min, sections were equilibrated in color development buffer (50 mM Tris-HCl [pH 9.5], 100 mM NaCl, 50 mM MgCl2) for 5 min. The location of cBD transcripts was visualized by using antibody-conjugated alkaline phosphatase to catalyze the Fast Red TR-aphthol substrate (Sigma Fast AS-MX tablets; Sigma-Aldrich) at 22°C for 2 h. Slides were washed three times with DEPC-H2O for 10 min each, counterstained with probe-hematoxylin (Biomeda, Foster City, CA) for 2 min, and mounted with crystal-mount (Biomeda).
Nucleotide accession numbers.
The nucleotide sequences reported here have been registered in GenBank under accession numbers AY169790, AY169791, and AY169792.
RESULTS
Identification of three cBDs.
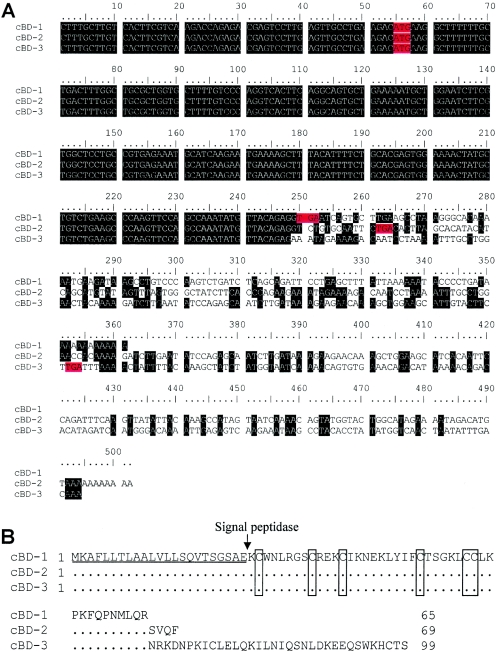
After a canine EST was located by using query sequences from hBD-5 and hBD-6, 5′- and 3′-RACE were used to identify three cBDs from testes (Fig. 1A). Full-length cDNA sequences for cBD-1 (361 bp), cBD-2 (503 bp), and cBD-3 (494 bp) have been deposited under GenBank accession numbers, AY169790, AY169791, and AY169792, respectively. All three isoforms have an identical 249-bp 5′-terminal sequence, including 5′-terminal untranslated regions (UTR) and the entire open reading frame (ORF) of the shortest isoform, cBD-1. However, the isoforms are diverse at the 3′-terminal sequences including the 3′-UTR and part of the ORF of cBD-2 and cBD-3 (Fig. 1A). The predicted pre-proproteins of cBD-1, -2, and -3 are composed of 65, 69, and 99 amino acid residues, respectively. All of the cBD isoforms contain the typical β-defensin consensus with a cysteine spacing pattern in their cysteine motif of 6-3-9-5-0 (Fig. 1B). The putative mature peptides of the cBDs contain nine positively charged residues (arginine, lysine, and histidine), which is an average number of all identified β-defensins (43). The cBD-1 precursor, which contains 65 amino acid residues, provides a putative backbone for the three cBD isoforms. In addition to this N-terminal 65-amino acid segment, cBD-2 and cBD-3 possess 4 and 34 additional amino acids at their C-terminal (Fig. 1B), respectively. It is noteworthy that the 34-amino-acid C terminus of cBD-3 contains seven positively charged residues, a ratio that is comparable to the mature peptide region (Fig. 1B). Prediction by the SignLP program in ExPASy (http://us.expasy.org/tools/) indicates that the cBD precursors have a signal peptide cleavage site between amino acid residues 22 and 23 from their N terminus, suggesting that they are secretory peptides. The cBDs share a common feature found in all previously identified β-defensins, i.e., the adjacent carboxyl-terminal amino acid residue of the second cysteine is lysine (Fig. 1B).
FIG. 1.
cDNA and predicted amino acid sequences for canine β-defensins. (A) Canine β-defensin cDNA sequences showing their identical 249 bp 5′-terminal regions (shaded black) and diversity at their 3′-terminal ends. ORFs are indicated with start (ATG) and stop (TGA) codons highlighted in red in each sequence. (B) Alignment of predicted pre-proprotein sequences of three canine β-defensins. Identical amino acids are indicated as periods in cBD-2 and cBD-3. The cleavage site for a signal peptidase is indicated by the arrow. Conserved cysteine residues are boxed.
Comparison of cBDs with β-defensins from other species.
The primary peptide sequence of the three isoforms of cBD show some similarity to β-defensins isolated from other species. Figure 2 shows the putative mature peptide regions of cBD aligned with several identified β-defensins that share the 6-3-9-5-0 cysteine spacing pattern in their cysteine motifs. In the aligned regions, cBD is 71% identical to a predicted human DEFB-22, and 51% identical to a putative human DEFB-23 (43). All other aligned β-defensin peptides represent a group of testis- or epididymal-specific β-defensins such as hBD-4, -5, -6, and -18, murine Defb-12, and testis defensin-like peptide, all showing identities to cBD of 20 to 41% (52). Several other epididymus-specific β-defensins, including human EP2 gene encoded β-defensin variants C, D, and E and Bin1-b, a rat epididymis-specific β-defensin, show ca. 20% identity to cBD (15, 57). Using the basic local alignment search tool against cBD peptides identified three human β-defensins, DEFB-25, -27, and -28, which are encoded by three of five novel β-defensin genes (DEFB25 to DEFB29) clustered on chromosome 20p13 and functional within different segments of epididymis (38). cBD has ca. 40% identity to DEFB-25, -27, and -28 in their putative mature peptide region; however, these three novel human β-defensins have a cysteine spacing pattern different than cBD peptides (43).
FIG. 2.
Comparison of deduced peptide sequences of canine β-defensin mature peptide with congeners from humans and mice. Conserved cysteine residues are boxed. GenBank accession numbers are as follows: cBD1, -2, and -3, AY169790, AY169791, and AY16979, respectively; hBD-4, AJ314834; hBD-5, AB089180; hBD-6, AB089181; hBD-18, AY122471; hBD-22, AY122474; hBD-23, AY122475; Tdl (testis defensin-like), NP660139; and mBD-12, AB089182.
Three-dimensional structural analysis of synthetic cBD peptide.
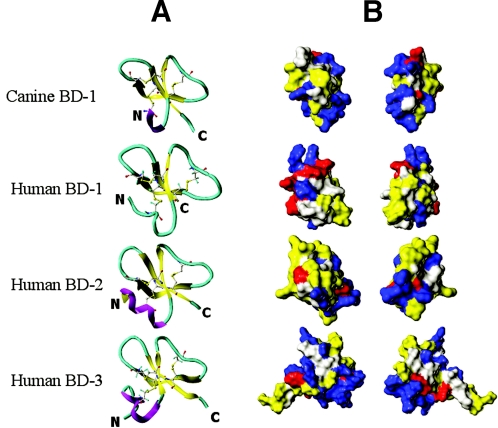
The predicted three-dimensional structure of cBD displaying 3-disulfide bonds (Cys2-Cys29, Cys9-Cys23, and Cys13-Cys30) was modeled based on the three human β-defensins hBD1, -2, and -3 for which experimental structural characterization was available (4). cBD forms a short N-terminal amphipathic alpha-helix (Trp3-Arg6) and three-stranded twisted antiparallel beta-sheets formed by Cys9-Glu11, Ile21-Cys23, and Leu28-Cys30, respectively (Fig. 3A). The three-dimensional structure model revealed that the cBD peptide possesses three-stranded antiparallel beta-sheets with a short helical segment near the N-terminal segment. This finding was in agreement with other previously reported human beta-defensin structures (42). The electrostatic analysis of cBD (Fig. 3B) demonstrated that the distribution of the basic and acidic amino acids seen on the solvent accessible surface model was in agreement with the amphipathic properties of other previously reported defensins. This regional concentration of basic and hydrophobic amino acids displayed by all mammalian β-defensins has been suggested to play an important role in their antimicrobial activity (43).
FIG. 3.
Secondary structure assignment and surface analysis for human and canine β-defensins. (A) The leftmost column depicts ribbon diagrams showing α-helix (purple), β-sheet (yellow), and coil (turquoise) secondary structure elements for the four peptides. (B) The two rightmost columns depict front and back views of peptide solvent accessible surfaces. Acidic (anionic) residues are red, basic (cationic) residues are blue, and hydrophobic residues are yellow.
Antimicrobial activity of synthetic cBD peptide.
To evaluate the antimicrobial activity of cBD, a 34-amino-acid sequence corresponding to the putative mature peptide region conserved among all three cBD isoforms was used as a template to chemically synthesize a cBD peptide. As shown in Table 2, synthetic cBD displayed bactericidal activity against nonuropathogenic E. coli, uropathogenic E. coli (ATCC 25922), and K. pneumoniae with MICs of 20, 50, and 20 μg/ml, respectively. cBD also possessed bactericidal activity against L. monocytogenes, N. gonorrhoeae, and S. aureus with MICs of 10, 50, and 100 μg/ml, respectively. The cBD peptide had broad antimicrobial activity as shown by inhibition of growth of C. albicans (ATCC 11006 and 14053; MICs of 5 and 50 μg/ml, respectively) and U. canigenitalium and U. urealyticum (MIC of 200 μg/ml for both bacterial species).
TABLE 2.
Antimicrobial activity of canine β-defensin
| Microorganisma | ATCC no. | MIC (μg/ml)b |
|---|---|---|
| Candida albicans (hematogenous) | 14053 | 50 |
| Candida albicans (vaginal epithelium) | 11006 | 5 |
| Escherichia coli* (generic) | 25922 | 20 |
| Escherichia coli* (uropathogenic) | 700336 | 50 |
| Klebsiella pneumoniae* | 10031 | 20 |
| Listeria monocytogenes† | 19115 | 10 |
| Staphylococcus aureus | 10832 | 100 |
| Neisseria gonorrhoeae | 10150 | 50 |
| Ureaplasma canigenitalium | 51252 | 200 |
| Ureaplasma urealyticum | 27619 | 200 |
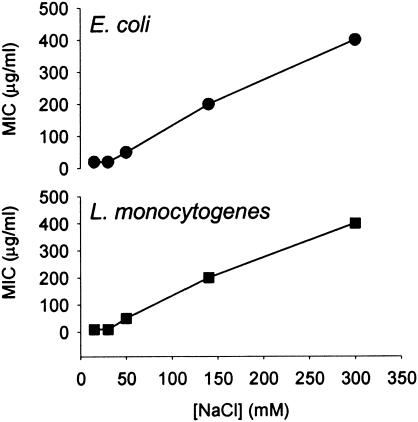
Similar to β-defensins in other species, the antimicrobial activity of cBD was highly dependent on salt concentration (Fig. 4). The bactericidal activity observed at a low salt concentration (≤15 mM NaCl), with MICs of 20 and 10 μg/ml for E. coil and L. monocytogenes, respectively, was inhibited at higher salt concentrations. At 140 mM [Na+] in the assay mixture, the MIC of cBD (for both bacteria) increased 10- to 40-fold. These findings are in agreement with those of the salt sensitivity evaluations of other β-defensins (3, 19, 32, 52).
FIG. 4.
Effect of sodium chloride on antimicrobial activity of canine β-defensin. A broth microdilution method was used to determine the MIC of cBD against two strains of bacteria, gram-negative E. coli (ATCC 25922) and gram-positive L. monocytogenes (ATCC 19115) in various salt concentrations. The final sodium concentration of the bactericidal mixture was adjusted to 15, 30, 50, 140, and 300 mM with NaCl. All assays were performed in triplicate.
Gene expression profiles of cBDs.
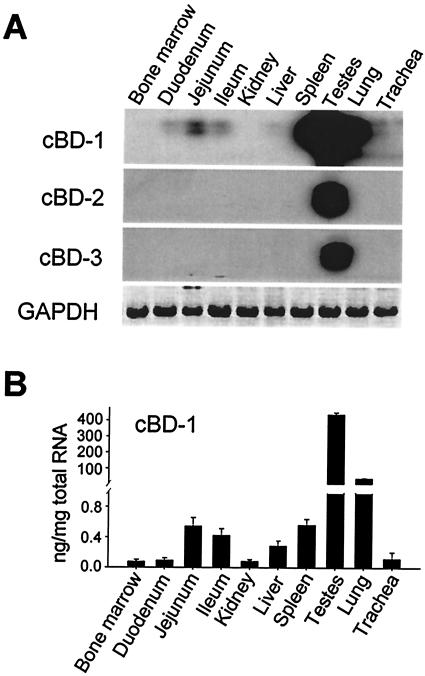
cBD-1 was predominantly expressed in testis, with lower expression in lung and intestinal tissues such as jejunum, ileum, and duodenum (Fig. 5A). Expression of the two longer isoforms, cBD-2 and cBD-3, were testis specific. Measurable expression of cBDs was not observed in any of the other tissues evaluated, including kidney, liver, spleen, lung, and bone marrow. To confirm our Southern analysis data, real-time RT-PCR was conducted on the same tissues. As shown in Fig. 5B, cBD-1 is highly expressed in testis, with an absolute value of 0.1 ng/250 ng of RNA, which is ca. 10-fold higher than that present in lung and 1,000- to 6,000-fold higher than in other tissues.
FIG. 5.
Tissue expression of canine β-defensins. (A) Fragments of cDNA were amplified from total RNA (250 ng) with one-step RT-PCR (40 cycles) with the primers listed in Table 1 and hybridized with 32P-labeled probes generated from cloned cDNA with the same primers. cDNA fragments of GAPDH were amplified (25 cycles) with the same RNA samples as a control. (B) Real-time RT-PCR of cBD-1 mRNA. Total RNA (250 ng) was reverse transcribed into cDNA and quantified with a SYBR Green-based PCR on a SmartCycler. Specific amplicon concentrations were normalized by using threshold cycle values against standard curves generated by using the same PCR conditions with serial dilutions of the cloned cBD-1 cDNA fragments.
In situ localization of cBD mRNA in testes.
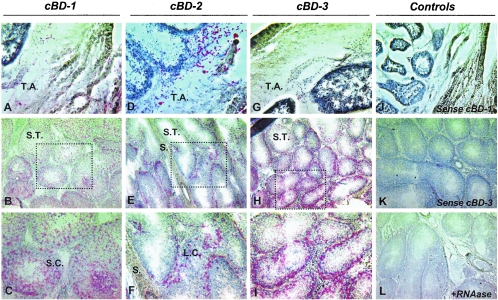
To localize cBD mRNA in testes, in situ hybridization was conducted. cBD-1 was highly expressed in Sertoli cells within the seminiferous tubules extending from the basement membrane to the tubular lumen (Fig. 6B and C). In contrast, cBD-2 was primarily detected in interstitial tissue within Leydig cells, with no overt expression in the seminiferous tubules (Fig. 6E and F). cBD-3 expression was detected in Sertoli cells primarily adjacent to the basement membrane (Fig. 6H and I).
FIG. 6.
Localization of canine β-defensin mRNA in normal canine testes. Frozen sections of canine testes were analyzed by in situ hybridization with digoxigenin-labeled antisense (A to I) and sense (J and K) probes. Panels C, F, and I are higher-magnification images of the framed regions in panels B, E, and H, respectively. Red precipitates are the products of anti-digoxigenin antibodies conjugated with alkaline phosphatase catalyzing Fast Red (Sigma-Aldrich) substrates and indicate locations of hybridized mRNA by the cBD probes. (L) RNase-treated control. The sections were counterstained with hematoxylin. T.A., tunica albuginea; S.T., seminiferous tubules; S., septa; S.C., Sertoli cells; L.C., Leydig cells.
DISCUSSION
Our understanding of the cellular and molecular mechanisms that comprise innate defense mechanisms of the mammalian male reproductive tract is rudimentary. In the present study we describe for the first time the identification and characterization of three novel β-defensins in the canine testis.
Typically, β-defensin genes are comprised of two exons and an intermediate intron. The first exon encodes the prepro-region, and the second exon encodes the mature peptide (43). The pre-proproteins are processed into the mature peptides, which are generally composed of 36 to 47 amino acids. However, two groups of β-defensins do not fit this pattern. One group is comprised of the three alternate splicing variants of the human HE2/EP2 gene, EP2C, EP2D, and EP2E, which all share the conserved cysteine motif of β-defensins. Their peptide precursors are composed of 113, 133, and 80 amino acid residues, respectively (9, 14, 15, 43). The second group resides among the five newly identified human β-defensin genes (DEFB25 to DEFB29) clustered on chromosome 20p13; the longest, DEFB-29, is 183 amino acids, and DEFB-25 to −28 are at 156, 111, 99, and 93 amino acids, respectively (38). The additional length of these β-defensin isoforms is generated mostly through extensions in the carboxy terminus following the consensus cysteine motifs. The three cBDs reported here exhibit similar C-terminal extensions. In comparison to other β-defensins, cBD-1 and cBD-2 are within the average length of most; however, cBD-3 is longer due to its 34 additional amino acids at the C terminus. The function of these additional C-terminal amino acids is unknown. One possibility is that they serve as a regulatory domain to promote the attachment of the antimicrobial peptide to the membrane of bacteria. Alternatively, the two longer isoforms, i.e., cBD-2 and cBD-3, might be testis-specific storage antimicrobial peptides. We theorize that they may act as endogenous antimicrobial molecules by providing immediate access to a gene product that could be rapidly processed into a shorter active isoform, i.e., cBD-1, when it is required. In this case, the C-terminal extension may, in fact, protect the stored peptides from degradation or unnecessary antimicrobial activity.
The structural analysis revealed that the global folding of cBD assumes all of the common features of the β-defensin family, such as three disulfide bonds, three antiparallel β-sheets and an α-helix usually located near the N-terminal of the peptide (Fig. 3A). cBD also displayed another essential feature common in all β-defensins which was the high concentration of cationic residues (Fig. 2). The number of positively charged residues (Arg, Lys, and His) reported in the mature peptides of other mammalian β-defensins (range, 6 to 14; average, 9) (43) was in good agreement with the number of cationic amino acid residues (6 Lys, 2 Arg; n = 8) found in the cBD sequence (Fig. 2 and 3B). It has been proposed that this high cationic charge density enables β-defensins to bind and insert into the cellular membrane, leading to the killing of the microorganism by forming multiple membrane pores (55). Our findings on the surface analysis of the cBD (Fig. 3B) established three well-defined cationic areas that may allow potent electrostatic interactions with bacterial membranes, furnishing cBD with the broad antimicrobial spectrum demonstrated in the in vitro assays.
Bacteria used in the present study were selected based on their ability to produce urinary tract infections (uropathogenic E. coli and K. pneumonia) and sexually transmitted diseases (N. gonorrhoeae, C. albicans, U. urealyticum, and U. canigenitalium). The antimicrobial effect of cBD was also evaluated against L. monocytogenes, S. aureus, and E. coli according to previous reports that have used these bacteria in assays of antibacterial activity for novel antimicrobial peptides (25, 34, 52). cBD effectively killed gram-positive and -negative bacteria, yeast, and Ureaplasma spp. The antimicrobial activity of cBD was comparable to that reported for other synthetic or natural β-defensin peptides, such as mBD-12, hBD-1, and hBD-3 (22, 49, 52). More importantly, synthetic cBD was more effective in killing C. albicans isolated from vaginal epithelium than one isolated from hematogenous origin, strongly suggesting that cBD plays an essential role as an epithelium-derived antibiotic that functions as an immediate line of defense against genital pathogens. The MIC for cBD for C. albicans is in agreement with previous reports (7, 23). Similar to other β-defensins (17), the bactericidal activity of cBD is dependent on the salt concentration used in the in vitro assay (52). Furthermore, our data suggest that the C termini of cBD-2 and cBD-3 may not be necessary for antimicrobial activity. Future studies will be required to determine differences in antimicrobial activities of the cBD isoforms and to investigate their individual role in relation to urogenital infections.
The biological activity of β-defensins is not limited to direct killing of microorganisms (1, 5, 6, 9, 43). These epithelium-derived molecules also participate actively in other aspects of adaptive immunity, including chemotactic effects on immature dendritic cells and memory T cells exerted through the CCR6 receptor pathway (53, 54). Despite the lack of information about the RNA expression pattern of cBDs in other epithelial surfaces of the canine urogenital tract, the results from the present study allow us to propose a speculative scenario. Considering that the expression of the shortest isoform, cBD-1, was not restricted to the testis, it is likely that this β-defensin may work in conjunction with other epithelial components of the mucosal barrier of the collecting ducts, ureters, bladder, and urethra to counter bacterial invasion of the urogenital tract. Conversely, the two longer cBD isoforms (cBD-2 and cBD-3) may diffuse to the site of infection, actively orchestrating chemotactic signals that direct the migration of circulating cells needed to initiate an adaptive immune response at the site of inflammation. The fact that uropathogenic E. coli and K. pneumoniae (the two bacterial strains that account for up to 90% of the urinary tract infections in humans and dogs) were sensitive to cBD supports the hypothesis that this epithelium-derived defensin may be also expressed in urinary tract epithelia.
The existence of multiple β-defensin isoforms in the male reproductive tracts of humans, rats, and mice (10, 20, 28, 52) suggests that these antimicrobial cationic peptides might specifically and cooperatively contribute to protect the reproductive system against pathogenic microbes. The presence of three different testis-specific cBDs isoforms suggests synergistic peptide function might also occur in the male dog. cBD-1, which displayed a more ubiquitous tissue expression pattern, could be the primary endogenous antibiotic line of defense against bacterial invasion in testis and other epithelial surfaces (i.e., lung, and small intestine). The two testis-specific β-defensins (cBD-2 and cBD-3) could possibly interact with cBD-1, providing a synergistic antimicrobial effect, thus rendering enhanced local protection to the canine reproductive system.
It is worth noting that only a few bacterial sexually transmitted diseases (Mycoplasma, Ureaplasma, and Brucella spp.) occur with any frequency in male dogs. Moreover, a direct relationship between these pathogens and clinical signs has not been fully documented and remains controversial (8, 11, 21). Our results clearly showed that cBD possesses broad antibacterial effects against the sexually transmitted disease pathogens used in the present study. cBD also effectively killed two strains of C. albicans; however, Ureaplasma spp. appeared to be resistant to cBD. This fact may be explained by the cell membrane composition of Ureaplasma spp. Ureaplasma spp. are among the few bacteria that possess a higher concentration of cholesterol within their cell membranes, protecting them from the antibacterial activity of defensins (21, 37). It has been suggested that eukaryotic cells are more resistant to the pore-forming effects of defensins due to the presence of cholesterol in the cellular membrane (56). The lack of cBD killing effect may explain why Ureaplasma is commonly isolated from the canine genital tract, where it is present as normal microflora (21).
Previous studies have reported that N. gonorrhoeae is very sensitive to protregrins (antimicrobial peptides derived from porcine neutrophils) but resistant to several human α-defensins (45). Our results conclusively showed that N. gonorrhoeae is killed by cBD. This inconsistency could reflect species-specific differences or more likely the site of defensin production; cBD is of epithelial origin (the first site of contact for a sexually transmitted disease pathogen), whereas most α-defensins are produced by circulating blood cells, which most likely are not the primary contact tissue for genital pathogens.
Several microbial pathogens are able to invade and colonize reproductive tract tissues and semen. If successful, they may cause serious clinical consequences to the reproductive status of the individual. Therefore, innate immunity should play a crucial role against bacterial invasion of the urethra, epididymis, and testes to protect and preserve the spermatozoa. Anatomically, the epididymis is the continuation of the urethra, exposing it to a permanent risk for ascending microbial infections. Our in situ hybridization results clearly showed that cBD-1 and cBD-3 are expressed in Sertoli cells (supporting cells for the developing spermatozoa), suggesting that these epithelium-derived peptides may actively participate as local antimicrobial molecules keeping the environment sterile for adequate sperm development and maturation. Moreover, these results are in agreement with previous reports showing the expression of β-defensins in the epididymis and seminiferous tubules, chiefly Sertoli cells, of the reproductive tract of rats (10, 31).
In summary, we have cloned the full-length cDNA of three canine β-defensins—cBD-1, cBD-2, and cBD-3—from testicular tissues. Canine β-defensin has broad antimicrobial activity, particularly against pathogens of the urogenital tract. The expression pattern of cBDs in Sertoli and Leydig cells of the testis suggests that they play a role in host defense of the male reproductive tract. In light of the pattern of expression and activity of canine β-defensins, further studies are warranted to investigate the role of β-defensins in host defense and the physiological function of the male reproductive system.
Acknowledgments
We thank Julie Hix for excellent technical assistance in preparation of frozen sections; Samir El-Zarkouny, Danielle Goodband, and Shannon Walsh for excellent technical support; Gerald H. Lushington, Molecular Graphics and Modeling Laboratory, University of Kansas, for invaluable input with the three-dimensional structure analysis; and Annika Linde, Thomas Schermerhorn, and Danielle Goodband for valuable discussions and critical reading of the manuscript.
This study was supported by National Institutes of Health Grant P20 RR017686 (T.M.) from the Institutional Development Award Program of the National Center for Research Resources.
Editor: J. D. Clements
REFERENCES
- 1.Bals, R. 2000. Epithelial antimicrobial peptides in host defense against infection. Respir. Res. 1:141-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bals, R., C. Lang, D. J. Weiner, C. Vogelmeier, U. Welsch, and J. M. Wilson. 2001. Rhesus monkey (Macaca mulatta) mucosal antimicrobial peptides are close homologues of human molecules. Clin. Diagn. Lab. Immunol. 8:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bals, R., X. Wang, M. Zasloff, and J. M. Wilson. 1998. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. USA 95:9541-9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, F., K. Schweimer, E. Kluver, J. R. Conejo-Garcia, W. G. Forssmann, P. Rosch, K. Adermann, and H. Sticht. 2001. Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity. Protein Sci. 10:2470-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boman, H. G. 1995. Peptide antibiotics and their role in innate immunity. Annu. Rev. Immunol. 13:61-92. [DOI] [PubMed] [Google Scholar]
- 6.Boman, H. G. 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254:197-215. [DOI] [PubMed] [Google Scholar]
- 7.Burd, R. S., J. L. Furrer, J. Sullivan, and A. L. Smith. 2002. Murine beta-defensin-3 is an inducible peptide with limited tissue expression and broad-spectrum antimicrobial activity. Shock 18:461-464. [DOI] [PubMed] [Google Scholar]
- 8.Carmichael, L. E., and J. C. Joubert. 1988. Transmission of Brucella canis by contact exposure. Cornell Vet. 78:63-73. [PubMed] [Google Scholar]
- 9.Cole, A. M., and T. Ganz. 2000. Human antimicrobial peptides: analysis and application. BioTechniques 29:821-822. [DOI] [PubMed] [Google Scholar]
- 10.Com, E., F. Bourgeon, B. Evrard, T. Ganz, D. Colleu, B. Jegou, and C. Pineau. 2003. Expression of antimicrobial defensins in the male reproductive tract of rats, mice, and humans. Biol. Reprod. 68:95-104. [DOI] [PubMed] [Google Scholar]
- 11.Doig, P. A. 1981. Bovine genital mycoplasmosis. Can. Vet. J. 22:339-343. [PMC free article] [PubMed] [Google Scholar]
- 12.Ferraro, M. J., M. A. Wikler, W. A. Craig, M. N. Dudley, G. M. Eliopoulos, D. W. Hecht, J. Hindler, L. B. Reller, A. T. Sheldon, J. M. Swenson, F. C. Tenover, R. T. Testa, and M. P. Weinstein. 2003. Methods for Dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard, 6th ed. NCCLS, Wayne, Pa.
- 13.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S-13S. [DOI] [PubMed] [Google Scholar]
- 14.Frohlich, O., N. M. Ibrahim, and L. G. Young. 2003. EP2 splicing variants in rhesus monkey (Macaca mulatta) epididymis. Biol. Reprod. 69:294-300. [DOI] [PubMed] [Google Scholar]
- 15.Frohlich, C. O., Po, T. Murphy, and L. G. Young. 2000. Multiple promoter and splicing mRNA variants of the epididymis-specific gene EP2. J. Androl. 21:421-430. [PubMed] [Google Scholar]
- 16.Gales, A. C., H. S. Sader, and R. N. Jones. 2002. Urinary tract infection trends in Latin American hospitals: report from the SENTRY antimicrobial surveillance program (1997-2000). Diagn. Microbiol. Infect. Dis. 44:289-299. [DOI] [PubMed] [Google Scholar]
- 17.Ganz, T. 2002. The role of hepcidin in iron sequestration during infections and in the pathogenesis of anemia of chronic disease. Isr. Med. Assoc. J. 4:1043-1045. [PubMed] [Google Scholar]
- 18.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 19.Goldman, M. J., G. M. Anderson, E. D. Stolzenberg, U. P. Kari, M. Zasloff, and J. M. Wilson. 1997. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 88:553-560. [DOI] [PubMed] [Google Scholar]
- 20.Hamil, K. G., P. Sivashanmugam, R. T. Richardson, G. Grossman, S. M. Ruben, J. L. Mohler, P. Petrusz, M. G. O'Rand, F. S. French, and S. H. Hall. 2000. HE2β and HE2γ, new members of an epididymis-specific family of androgen-regulated proteins in the human. Endocrinology 141:1245-1253. [DOI] [PubMed] [Google Scholar]
- 21.Harasawa, R., Y. Imada, H. Kotani, K. Koshimizu, and M. F. Barile. 1993. Ureaplasma canigenitalium sp. nov., isolated from dogs. Int. J. Syst. Bacteriol. 43:640-644. [DOI] [PubMed] [Google Scholar]
- 22.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 23.Harris, R. H., D. Wilk, C. L. Bevins, R. S. Munson, Jr., and L. O. Bakaletz. 2004. Identification and characterization of a mucosal antimicrobial peptide expressed by the chinchilla (Chinchilla lanigera) airway. J. Biol. Chem. 279:20250-20256. [DOI] [PubMed] [Google Scholar]
- 24.Hoover, D. M., K. R. Rajashankar, R. Blumenthal, A. Puri, J. J. Oppenheim, O. Chertov, and J. Lubkowski. 2000. The structure of human beta-defensin-2 shows evidence of higher order oligomerization. J. Biol. Chem. 275:32911-32918. [DOI] [PubMed] [Google Scholar]
- 25.Hoover, D. M., Z. Wu, K. Tucker, W. Lu, and J. Lubkowski. 2003. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob. Agents Chemother. 47:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, J. R., N. Kaster, M. A. Kuskowski, and G. V. Ling. 2003. Identification of urovirulence traits in Escherichia coli by comparison of urinary and rectal E. coli isolates from dogs with urinary tract infection. J. Clin. Microbiol. 41:337-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehrer, R. I., and T. Ganz. 2002. Cathelicidins: a family of endogenous antimicrobial peptides. Curr. Opin. Hematol. 9:18-22. [DOI] [PubMed] [Google Scholar]
- 28.Li, P., H. C. Chan, B. He, S. C. So, Y. W. Chung, Q. Shang, Y. D. Zhang, and Y. L. Zhang. 2001. An antimicrobial peptide gene found in the male reproductive system of rats. Science 291:1783-1785. [DOI] [PubMed] [Google Scholar]
- 29.Lopez, L., G. Morales, R. Ursic, M. Wolff, and C. Lowenberger. 2003. Isolation and characterization of a novel insect defensin from Rhodnius prolixus, a vector of Chagas disease. Insect Biochem. Mol. Biol. 33:439-447. [DOI] [PubMed] [Google Scholar]
- 30.Low, D. A., B. A. Braaten, G. V. Ling, D. L. Johnson, and A. L. Ruby. 1988. Isolation and comparison of Escherichia coli strains from canine and human patients with urinary tract infections. Infect. Immun. 56:2601-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palladino, M. A., T. A. Mallonga, and M. S. Mishra. 2003. Messenger RNA (mRNA) expression for the antimicrobial peptides beta-defensin-1 and beta-defensin-2 in the male rat reproductive tract: beta-defensin-1 mRNA in initial segment and caput epididymidis is regulated by androgens and not bacterial lipopolysaccharides. Biol. Reprod. 68:509-515. [DOI] [PubMed] [Google Scholar]
- 32.Park, I. Y., J. H. Cho, K. S. Kim, M. S. Y. B. Kim, Kim, and S. C. Kim. 2004. Helix stability confers salt resistance upon helical antimicrobial peptides. J. Biol. Chem. 279:13896-13901. [DOI] [PubMed] [Google Scholar]
- 33.Pfaller, M. A., V. Chaturvedi, A. Espinel-Ingroff, M. A. Ghannoum, L. L. Gosey, F. C. Odds, J. H. Rex, M. G. Rinaldi, D. J. Sheehan, T. J. Walsh, and D. W. Warnock. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts, approved standard, 2nd ed. NCCLS, Wayne, Pa.
- 34.Porter, E. E. M., van Dam, E. V. Valore, and T. Ganz. 1997. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect. Immun. 65:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prais, D., R. Straussberg, Y. Avitzur, M. Nussinovitch, L. Harel, and J. Amir. 2003. Bacterial susceptibility to oral antibiotics in community acquired urinary tract infection. Arch. Dis. Child. 88:215-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu, X. D., K. C. Lloyd, J. H. Walsh, and R. I. Lehrer. 1996. Secretion of type II phospholipase A2 and cryptdin by rat small intestinal Paneth cells. Infect. Immun. 64:5161-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razin, S., G. K. Masover, M. Palant, and L. Hayflick. 1977. Morphology of Ureaplasma urealyticum (T-mycoplasma) organisms and colonies. J. Bacteriol. 130:464-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez-Jimenez, F. J., A. Krause, S. Schulz, W. G. Forssmann, J. R. Conejo-Garcia, R. Schreeb, and D. Motzkus. 2003. Distribution of new human beta-defensin genes clustered on chromosome 20 in functionally different segments of epididymis. Genomics 81:175-183. [DOI] [PubMed] [Google Scholar]
- 39.Romestand, B., F. Molina, V. Richard, P. Roch, and C. Granier. 2003. Key role of the loop connecting the two beta strands of mussel defensin in its antimicrobial activity. Eur. J. Biochem. 270:2805-2813. [DOI] [PubMed] [Google Scholar]
- 40.Ronald, A. 2002. The etiology of urinary tract infection: traditional and emerging pathogens. Am. J. Med. 113(Suppl. 1A):14S-19S. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez, R., and A. Sali. 2000. Comparative protein structure modeling. Introduction and practical examples with modeler. Methods Mol. Biol. 143:97-129. [DOI] [PubMed] [Google Scholar]
- 42.Schibli, D. J., H. N. Hunter, V. Aseyev, T. D. Starner, J. M. Wiencek, P. B. McCray, Jr., B. F. Tack, and H. J. Vogel. 2002. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 277:8279-8289. [DOI] [PubMed] [Google Scholar]
- 43.Schutte, B. C., and P. B. McCray, Jr. 2002. βdefensins in lung host defense. Annu. Rev. Physiol. 64:709-748. [DOI] [PubMed] [Google Scholar]
- 44.Seguin, M. A., S. L. Vaden, C. Altier, E. Stone, and J. F. Levine. 2003. Persistent urinary tract infections and reinfections in 100 dogs (1989-1999). J. Vet. Intern. Med. 17:622-631. [DOI] [PubMed] [Google Scholar]
- 45.Shafer, W. M., X. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, J., C. R. Ross, M. M. Chengappa, M. J. Sylte, D. S. McVey, and F. Blecha. 1996. Antibacterial activity of a synthetic peptide (PR-26) derived from PR-39, a proline-arginine-rich neutrophil antimicrobial peptide. Antimicrob. Agents Chemother. 40:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomma, B. P., B. P. Cammue, and K. Thevissen. 2003. Mode of action of plant defensins suggests therapeutic potential. Curr. Drug Targets Infect. Disord. 3:1-8. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valore, E. V., C. H. Park, A. J. Quayle, K. R. Wiles, P. B. McCray, Jr., and T. Ganz. 1998. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J. Clin. Investig. 101:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, W., A. M. Cole, T. Hong, A. J. Waring, and R. I. Lehrer. 2003. Retrocyclin, an antiretroviral theta-defensin, is a lectin. J. Immunol. 170:4708-4716. [DOI] [PubMed] [Google Scholar]
- 51.Whelan, J. A., N. B. Russell, and M. A. Whelan. 2003. A method for the absolute quantification of cDNA using real-time PCR. J. Immunol. Methods 278:261-269. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi, Y., T. Nagase, R. Makita, S. Fukuhara, T. Tomita, T. Tominaga, H. Kurihara, and Y. Ouchi. 2002. Identification of multiple novel epididymis-specific beta-defensin isoforms in humans and mice. J. Immunol. 169:2516-2523. [DOI] [PubMed] [Google Scholar]
- 53.Yang, D., A. Biragyn, L. W. Kwak, and J. J. Oppenheim. 2002. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 23:291-296. [DOI] [PubMed] [Google Scholar]
- 54.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schroder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
- 55.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 56.Zasloff, M. 2002. Innate immunity, antimicrobial peptides, and protection of the oral cavity. Lancet 360:1116-1117. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, C. X., Y. L. Zhang, L. Xiao, M. Zheng, K. M. Leung, M. Y. Chan, P. S. Lo, L. L. Tsang, H. Y. Wong, L. S. Ho, Y. W. Chung, and H. C. Chan. 2004. An epididymis-specific beta-defensin is important for the initiation of sperm maturation. Nat. Cell Biol. 6:458-464. [DOI] [PubMed] [Google Scholar]