Abstract
Deformable image registration (DIR) between daily and reference images is fundamentally important for adaptive radiotherapy. In the last decade, deep learning-based image registration methods have been developed with faster computation time and improved robustness compared to traditional methods. However, the registration performance is often degraded in extra-cranial sites with large volume containing multiple anatomic regions, such as Computed Tomography (CT)/Magnetic Resonance (MR) images used in head and neck (HN) radiotherapy. In this study, we developed a hierarchical deformable image registration (DIR) framework, Patch-based Registration Network (Patch-RegNet), to improve the accuracy and speed of CT-MR and MR-MR registration for head-and-neck MR-Linac treatments. Patch-RegNet includes three steps: a whole volume global registration, a patch-based local registration, and a patch-based deformable registration. Following a whole-volume rigid registration, the input images were divided into overlapping patches. Then a patch-based rigid registration was applied to achieve accurate local alignment for subsequent DIR. We developed a ViT-Morph model, a combination of a convolutional neural network (CNN) and the Vision Transformer (ViT), for the patch-based DIR. A modality independent neighborhood descriptor was adopted in our model as the similarity metric to account for both inter-modality and intra-modality registration. The CT-MR and MR-MR DIR models were trained with 242 CT-MR and 213 MR-MR image pairs from 36 patients, respectively, and both tested with 24 image pairs (CT-MR and MR-MR) from 6 other patients. The registration performance was evaluated with 7 manually contoured organs (brainstem, spinal cord, mandible, left/right parotids, left/right submandibular glands) by comparing with the traditional registration methods in Monaco treatment planning system and the popular deep learning-based DIR framework, Voxelmorph. Evaluation results show that our method outperformed VoxelMorph by 6 % for CT-MR registration, and 4 % for MR-MR registration based on DSC measurements. Our hierarchical registration framework has been demonstrated achieving significantly improved DIR accuracy of both CT-MR and MR-MR registration for head-and-neck MR-guided adaptive radiotherapy.
Keywords: CT/MR deformable registration, Vision transformer, Patch-based registration, Multi-modality registration
1. Introduction
Recent technological advancement in radiotherapy (RT) has enabled online adaptive radiotherapy (ART) to optimize radiation treatment plans on-the-fly based on daily anatomy changes to improve treatment accuracy (Wu et al., 2011). ART is attractive in the treatment of head and neck cancer (HNC) because potential significant tumor shrinkage, large anatomical deformation of organs at risk (OARs), and substantial weight loss are commonly observed for HNC patients during treatment (Castelli et al., 2018; Bahl et al., 2019; Burela et al., 2019). Without ART, the anatomical changes can lead to a compromised plan for dose delivery, with either unnecessary damage to critical organs or under-treatment near the tumor boundary. Cone beam computed tomography (CBCT) has been in use for decades for patient alignment, it enables the observation of anatomical changes and assessing the necessity of conducting offline ART (Belshaw et al., 2019). However, the inferior image quality and poor soft tissue contrast of CBCT can limit its performance in clinical implementation. The advent of magnetic resonance (MR) imaging-guided linear accelerators (MR-Linacs) has provided an excellent platform for online ART, enabling MR-guided radiotherapy (MRgRT) (McDonald et al., 2021; Kupelian and Sonke, 2014; Raaymakers et al., 2009). Integrated MR-Linacs allow the acquisition of on-board MR images just before radiation delivery, thereby providing superior visualization of the soft tissue for daily online plan re-optimization.
In MRgRT, daily MR images are used for daily set up verification and plan adaptation. The key enabling technology for online ART is deformable image registration (DIR), which establishes spatial relationship between two images by transforming a moving image to a fixed image space. DIR establishes the relationship between the planning computed tomography (CT) and MRI images so that the original treatment plan on CT can be adapted and re-optimized based on daily anatomy on MRI to achieve optimal treatment delivery (Raaymakers et al., 2009; Owrangi et al., 2018; Schmidt and Payne, 2015). The current ART workflow relies on the DIR process to deform contours delineated on planning CT onto the daily MR image, followed by a full plan re-optimization using the established spatial relationship between CT and MR images. However, DIR between CT and MR images has long been a difficult task due to significant contrast differences and anatomical changes over time, particularly for extracranial anatomical sites. For example, the neck flexion is often unavoidable from fraction to fraction no matter how carefully the patient is set up for HNC treatment. In addition, the treatment volume of HNC often spans a large region from top of nasopharynx to lower neck or upper lung, further complicating the DIR task because of locoregional anatomical variations in the HN region, as shown in Fig. 1. Existing registration tools in clinic could not ensure accurate and robust contour propagation, especially for flexible structures such as the neck and spine (McDonald et al., 2021). They always require the physician to review and revise the deformed contours on daily MR images, which is tedious and time-consuming. Therefore, to facilitate the online ART workflow of MRgRT of HNC, it is crucial to develop a fast and efficient DIR method for CT-MR registration.
Fig. 1.
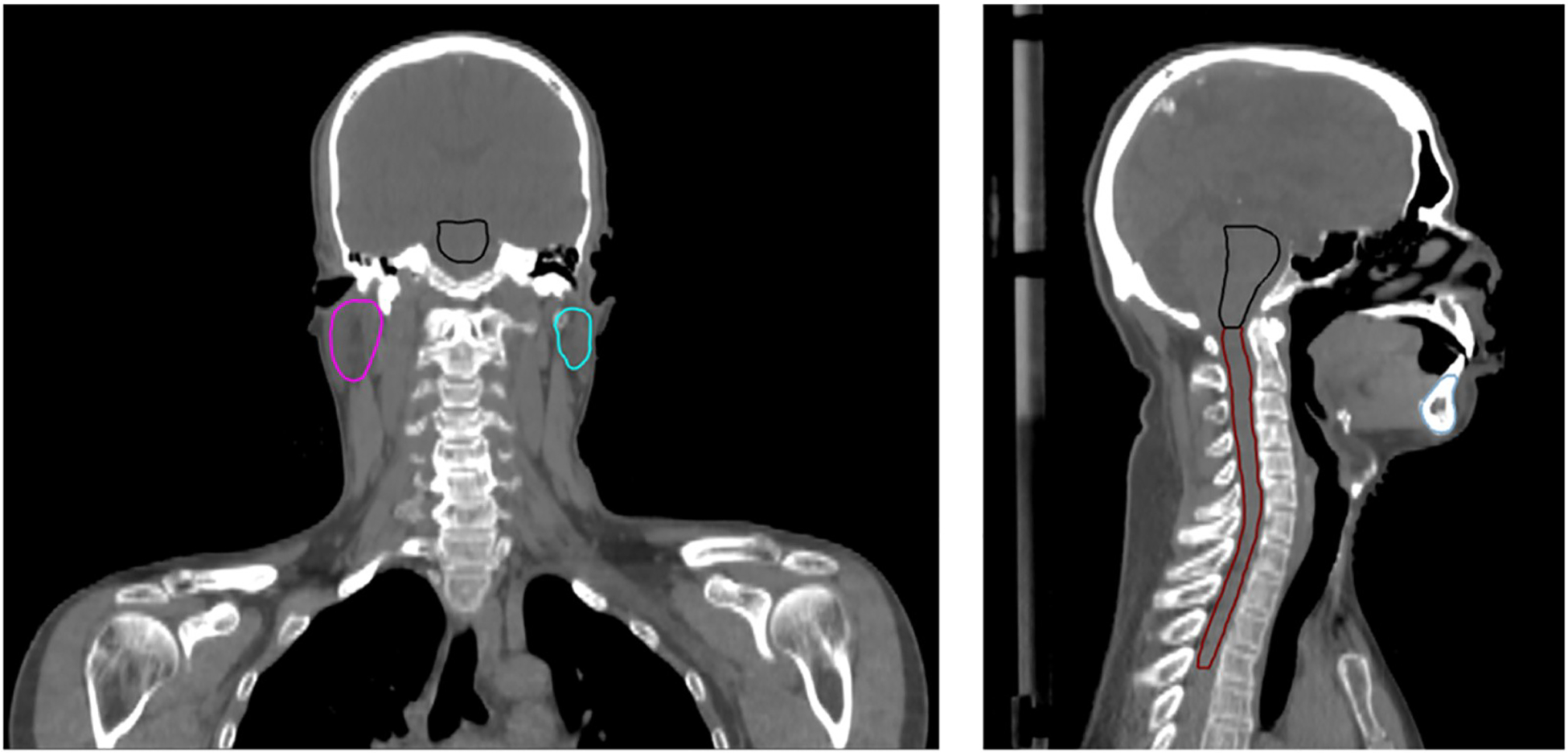
Illustration of the anatomical coverage of HNC patient.
Many approaches have been developed to address the challenges in inter-/intra modality image registration. For decades, traditional registration algorithms based on maximizing image similarity between two images have been successively implemented in many applications (Brock et al., 2017). However, these iterative optimization-based methods usually require high computational cost and are not robust or accurate for cases with large deformation. Recently, deep learning (DL)-based approaches have been demonstrated to have superior performance and speed compared to the traditional methods (Chen et al., 2021c, 2022; Fu et al., 2020; Haskins et al., 2020). The DL-based DIR methods are broadly categorized into (i) supervised and (ii) unsupervised learning methods (Chen et al., 2021c; Fu et al., 2020; Haskins et al., 2020). In supervised learning methods, the networks are trained with ground-truth deformation vector fields (DVFs) that are usually generated from traditional methods or synthetic data. The registration performance is limited by the quality of ground-truth DVFs which may be different from the actual anatomical deformation. On the other hand, unsupervised learning methods have been developed to overcome these limitations by training the networks to optimize similarity metrics between deformed and fixed images like traditional registration methods. VoxelMorph, proposed by Balakrishnan et al. (2019), was an example of unsupervised learning methods that utilized a spatial transformer network (STN) (Jaderberg et al., 2015) to generate the deformed image during training process. However, the application of the unsupervised registration methods for CT-MR comes with many challenges due to the inherent limitation of available similarity metrics. While mutual information (MI) (Maes et al., 1997; Viola and Wells, 1995) has been commonly used in CT-MR registration, its performance for DIR may be diminished due to its intrinsic global measurement and limited capacity in distinguishing tissue types with similar intensities (Heinrich et al., 2012a). To overcome this problem, translation methods using Generative Adversarial Network (GAN) (Goodfellow et al., 2020) have been proposed to generate synthetic CT (sCT) from MR images, which are used as a bridge for CT-MR registration. McKenzie et al. (2020) reduced the multi-modal registration problem to a mono-modal one by using a cycle-consistency GAN model to generate sCT from MR images for HNC patients. Xu et al. (2020) proposed to further combine the sCT-CT registration together with MR-CT registration, which could leverage the deformation predictions from both multi-modal and mono-modal registrations. However, the performance of this method is restricted by the reliability of the anatomical features in synthetic images (Xu et al., 2020). Furthermore, the convolutional neural network (CNN)-based registration methods with a limited size effective receptive field may suffer from loss of long-range spatial relations in moving and fixed images during registration. Recently, there has been an increase of applying transformer to computer vision tasks, such as segmentation (Chen et al., 2021b; Zhang et al., 2021a; Cao et al., 2023), image reconstruction (Güngör et al., 2022; Guo et al., 2022), and image recognition (Dosovitskiy et al., 2020; Wu et al., 2021). This technique has been demonstrated to have superior performance in image registration because of its self-attention mechanism and ability to build associations between distant parts of images (Chen et al., 2021a; Zhang et al., 2021b; Liu et al., 2022). However, to the best our knowledge, no studies have been conducted to address the challenges of DIR between CT and MR images specifically for HN sites.
In this study, we attempt to achieve accurate and rapid DIR for CT-MR and MR-MR in online MR-guided ART for HNC. Although our focus is precise DIR for inter-modality (CT-MR) images, we also demonstrate the usability of our proposed approach for intra-modality (MR-MR) images in the context of MR-guided ART. We propose a novel hierarchical registration framework named Patch-RegNet, where the patch-based registration is introduced to improve the local alignment. One difficulty in training a DL-based DIR network for HNC is to achieve a good initial position between moving and fixed images owing to their extensive coverage over multiple anatomic sites, including the head, neck, shoulders, and upper lungs. Our Patch-RegNet addresses this issue using a three-stage workflow: a whole volume rigid registration, a patch-based rigid registration, and a patch-based deformable registration. The patch-based rigid registration ensures an improved local pre-alignment between two images for subsequent DIR. Our patch-based DIR network, ViT-Morph, is built upon the combination of Vision Transformer (ViT) (Dosovitskiy et al., 2020) and VoxelMorph (Balakrishnan et al., 2019) to take advantage of both CNN features and the long-range spatial relationships from the transformer. In addition, the modality independent neighborhood descriptor (MIND) (Heinrich et al., 2012a) is used as the similarity metric in Patch-RegNet to account for both inter-(CT-MR) and intra- (MR-MR) modality DIR. Our proposed Patch-RegNet is evaluated on the clinical HNC CT and MR images acquired for radiotherapy both qualitatively and quantitatively.
2. Materials and methods
2.1. Overview
Patch-RegNet has a hierarchical registration framework to complete registration tasks robustly and automatically in a fully unsupervised manner. The overall framework is shown in Fig. 2.
Fig. 2.
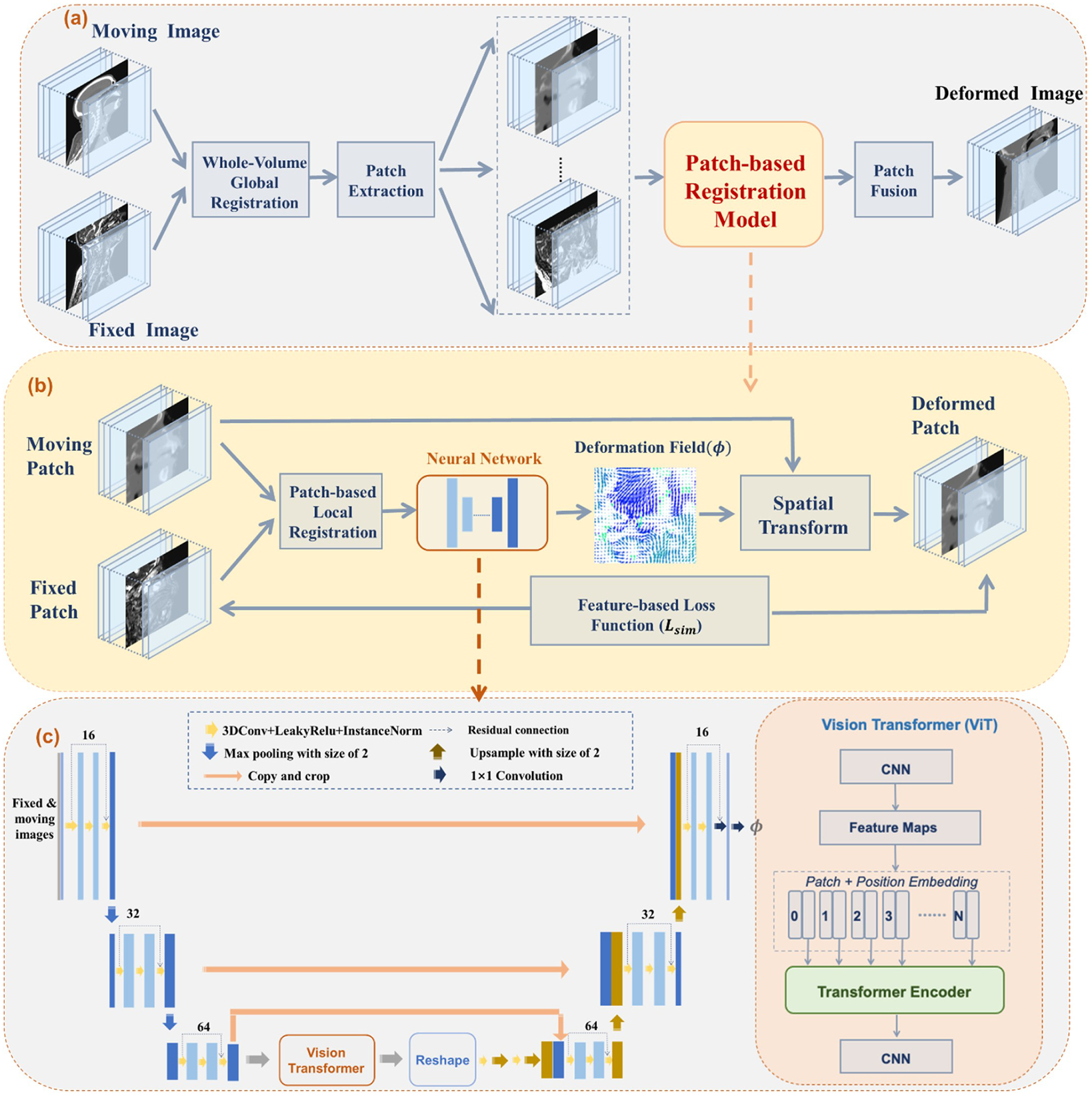
Overall framework of the proposed Patch-RegNet. (a) The hierarchical registration framework consists of three-stage registrations: a whole volume global registration, a patch-based local registration, and a patch-based deformable registration. The patch-based registration model that includes stages 2 and 3 is shown in (b) and (c). (b) The schematic illustration of ViT-Morph: a hybrid network of vision transformer (ViT) and VoxelMorph; (c) the details of the convolutional neural network (CNN): a combination of a modified residual-UNet and ViT. The transformer encoder consists of 12 alternating layers of Multihead Self-Attention (MSA) and Multi-Layer Perceptron (MLP) blocks.
The hierarchical registration framework starts with a whole-volume rigid registration. The moving and fixed images are first rigidly registered based on whole-image volumes. Overlapping patches are then extracted from the pre-aligned images for the subsequent patch-based registration model. Within patch-based registration, a rigid registration is applied to further refine the local alignment of patch pairs for following DIR, which is achieved by training a patch-based DIR network, ViT-Morph, for registration. The ViT-Morph model is trained to capture the deformation field between moving and fixed patches. A deformed patch is generated by warping the moving patch with the deformation field using a spatial transformer network (STN). Finally, the deformed patches are fused together to obtain the entire deformed image.
2.2. Whole volume global registration
Let VM, VF ϵ RD×H×W denote the whole volumes of moving and fixed images. VM and VF are first rigidly aligned to account for different anatomical scanning range as an initial step to facilitate the subsequent DL-based DIR process. In Patch-RegNet, we use SimpleITK (Beare et al., 2018) to implement a rigid registration method that minimizes the Mattes MI (Mattes et al., 2001) between moving and fixed images with a gradient optimization algorithm. Instead of using an affine transform, a similarity transform is employed for the linear registration. We observe that a similarity transform could often give more robust results than an affine transform for the sake of slightly inferior local alignment accuracy. After the whole volume linear registration, VF and pre-aligned VM are cropped to the overlapped region, represented by and for the following registration.
2.3. Patch-based local registration
The whole volume registration techniques generally do not provide sufficient accuracy in some local regions for subsequent DIR, especially for images covering a large area with multiple anatomical sites like HN images.
To address this problem, we propose the patch-based rigid registration to provide the DL-based registration with improved regional guidance. Let Pm, Pf ϵ Rd×h×w represent the corresponding patches extracted from the fixed and pre-aligned moving images. The parameters d, h, w are chosen to enable the patch to cover the complete anatomic site/sites and keep it with the flexibility of locoregional alignment. Body masks are generated for to assist the patch extraction. Based on the body mask, the patch centers are limited to be within the patient body to minimize the inclusion of air outside the body for each patch. This patch extraction strategy ensures that the extracted patches cover the entire patient body while leaving sufficient space outside the body for deformable registration of corresponding patches. As shown in Fig. 3, two tunable parameters, stride s and cropping c, allow the flexibility of patch extraction and border exclusion during patch fusion. As registration for each image voxel needs the surrounding spatial information, the patches are extracted with large overlapping regions which are controlled by the user specified strides s. However, registration in the peripheral regions of patches might be inaccurate due to the lack of sufficient surrounding spatial information. Thus, the patches are cropped to abandon the peripheral regions in the patch-fusion process.
Fig. 3.
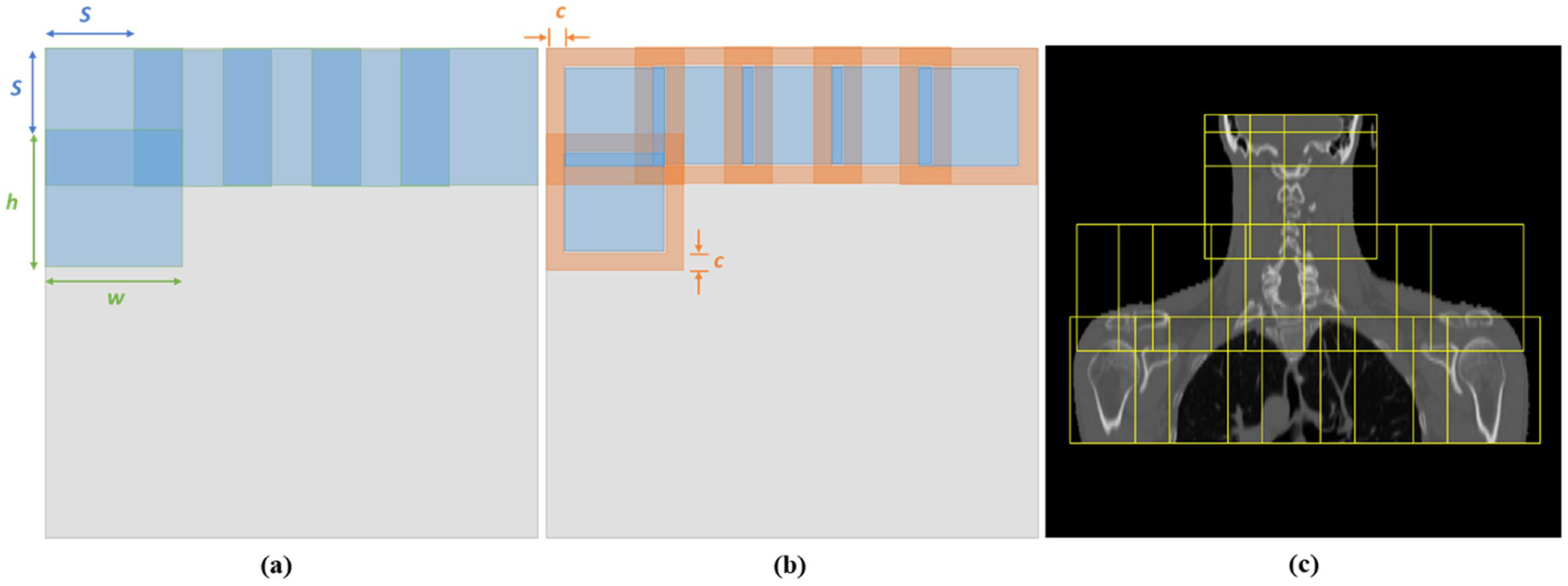
Illustration of patch processing with adjustable parameters stride s and cropping c. (a) Patch extraction. (b) Patch fusion. (c) Patch extraction is restricted within the patient body. h × w is the patch size. The diagram is shown in 2D, but the actual implementation is in 3D.
After patch extraction, Pm and Pf are registered using the same algorithm used in whole volume linear registration for further local alignment. The pre-aligned patch generated from Pm, is then deformably registered to Pf using the patch-based ViT-Morph network. During patch fusion, each patch is cropped by cropping parameter c to keep the central part of the patch for the final deformed image.
2.4. Patch-based deformable registration: ViT-Morph
The objective of the final stage is to predict a deformation vector field ϕ ϵ R3×d×h×w that can warp the moving patch to match the fixed patch Pf. Based on the framework of VoxelMorph, we adapted the UNet architecture and combine it with Vision Transformer (ViT) to construct the ViT-Morph network. The framework is shown in Fig. 2(b) and (c).
Specifically, a residual-UNet is adopted in the network. It can take advantage of long skip connections like UNet and alleviate the gradient vanishing issue with residual connections. The overview of our architecture is described in Fig. 2(b). The implementation of ViT has been detailed described in previous work (Chen et al., 2021a, 2021b; Dosovitskiy et al., 2020). ViT is integrated as an encoder with the residual-UNet in feature map level. To be more specific, after the images are encoded into feature maps through residual-UNet decoders, the high-level feature maps are fed into ViT and extracted into vectorized patches , where denotes the ith vectorized patch, P represents the patch size, is the number of patches, and C denotes the channel size. The patches are then mapped into a latent D-dimensional space using a trainable linear projection. The patch positional information is preserved by adding specific position embeddings to the patch embeddings:
where is the patch embedding projection and Epos ϵ RN×D is the position embedding.
Following the patch and position embeddings, the embedded patches are fed into the Transformer encoder, which consists of 12 layers of Multihead Self-Attention (MSA) and Multi-Layer Perceptron (MLP) blocks (Vaswani et al., 2017). The output of thel th Transformer encoder can be expressed as following:
where zl represents encoded image representation and denotes the layer normalization, which is added before each MSA and MLP block.
Finally, the resulting sequence of hidden feature from ViT is reshaped and decoded with residual-UNet decoders to output the final deformation field ϕ. STN is implemented to warp the moving patch P′m with ϕ, and the dissimilarity between Pf and is then calculated as the similarity loss function for network training.
2.5. Loss function and training strategy
The simple intensity-based similarity loss in VoxelMorph could not be used effectively for inter-modality (CT-MR) registration. MI was introduced for the rigid alignment of multi-modal images. However, its application to deformable CT-MR registration comes with many difficulties (Heinrich et al., 2012a). Because MI is intrinsically a global measurement, it would result in inaccurate image registration with the loss of local accuracy. In our network, we employed MIND (Heinrich et al., 2012a), a feature of 12 channels extracting the distinctive structure similarity from a local neighborhood, as the similarity metric for DIR, described further below. It has been demonstrated to have superior performance in MR-CT DIR tasks using conventional registration algorithms (Heinrich et al., 2012b). Following the similar implementation, the MIND features are calculated for fixed Pf and deformed patches during the training process. The MIND-based unsupervised loss ℒ MIND penalizes the differences between their MIND features and is defined as follows:
where represent the similarity loss between fixed Pf and deformed patches at voxel x, and MIND(⋅, ⋅, ⋅) denotes the MIND feature. Specifically, for voxel x in image patch I, its MIND feature in a local search region R is defined as:
where M is a normalization constant to ensure the maximum value of is 1, denotes the patch distance, V (I, x) represents the local variance estimation, and the spatial search region is set to be . To be more specific, D p(I, x, x + s) is the L2 distance between two patches P with the size of (2p+ 1) (Bahl et al., 2019) centered at voxel x and voxel x + s, defined as:
And the variance estimate V(I, x) is calculated based on the mean of the patch distances within a six-neighborhood n ϵ N:
In order to ensure the generation of a reasonable deformation vector field ϕ, a smoothness constraint is needed for regularization. In our study, the l2-norm of first order gradient of ϕ was applied as the diffusion regularizer as:
where ω is the set of all voxels and u denotes the displacement field. Thus, the final loss function used for training the network is:
where λ is a tunable regularization parameter.
2.6. Data acquisition and preprocessing
MR and CT images of 42 head and neck patients who were treated on a 1.5 T MR-Linac (Unity; Elekta AB; Stockholm, Sweden) at The University of Texas MD Anderson Cancer Center were included in this study. Each patient had a pre-treatment CT simulation scan and daily T2-weighted MR scans. The number of treatment fractions ranged from 2 to 35, giving a total of 266 pairs of CT-MR and 237 pairs of MR-MR scans. The voxel intensities of MR images can vary significantly among different patients, scans, and machines. To reduce the intensity variations in MR images, the Z-score normalization (Wahid et al., 2021) within the patient body was applied. The body contour for each MR image was generated in RayStation (version 11B; RaySearch Laboratories; Stockholm, Sweden), and the Z-score normalization was conducted only within the body contours.
After the whole volume registration in the first stage, all fixed and moving images were preprocessed to have an isotropic resolution of 1mm3 and spatial dimensions (420×420×300). In the following patch-based registration step, the stride was chosen to be s = 96, 96, 80, and the overlapping patches were extracted with a size of (160×160×128), which can sufficiently cover one complete anatomical structure in HN region. 40–80 patches were extracted from each scan, depending on the patient’s body size. In the patch fusion, we used the cropping parameter c = 20, 20, 16 to exclude the boundary region in the fusion process.
2.7. Evaluations and implementation
We selected 48 pairs of images (24 CT-MR and 24 MR-MR) from 6 patients as a testing dataset, and the remaining data were partitioned randomly into a training set (80 %) and a validation set (20 %). Seven manually labelled organs at risk (OARs) were used for evaluation, including brain stem, spinal cord, mandible, right and left parotid glands, right and left submandibular glands. These OARs were contoured separately on CT and T2-weigthed MR images by experienced radiation oncologists. After the DIR by our model, the deformed contours of the moving images were compared with those on the fixed images by calculating Dice similarity coefficient (DSC) and mean surface distance (MSD). Those labelled structures were only used for quantitative evaluation and not used for model training. Visual assessment was also performed by overlaying the deformed contours on those ground-truth contours in the fixed images.
To demonstrate the effectiveness of our method, we compared our Patch-RegNet with VoxelMorph-MIND and a clinical DIR tool in Monaco Treatment Planning System (version 5.4 Elekta AB; Stockholm, Sweden). VoxelMorph-MIND is the VoxelMorph network using MIND as the similarity metric and whole-volume global registration for pre-alignment. Input images were cropped and resized to 256 × 256 × 288 to fit the network. The paired two-tailed t-test was included in our study to evaluate the statistical difference between our proposed method and other methods at a significance level of 0.05 (defined by a p < 0.05). This comparison was performed for both inter-modality (CT-MR) and intra-modality (MR-MR) registrations.
In addition, we also trained a ViT-Morph-MIND model without using hierarchical registration framework for comparison. The primary purpose of training this model for comparison was to evaluate the effectiveness of the proposed network (a combination of visual transformer with a UNet) and the hierarchical framework. The ViT-Morph-MIND model was trained with whole-volume images, which have the same image size of 256 × 256 × 288 as that in VoxelMorph-MIND. This performance was performed for CT-MR registration only because we expect that the ViT-Morph-MIND performs consistently for CT-MR and MR-MR registrations when compared with the Patch-RegNet.
Our Patch-RegNet was implemented with the TensorFlow and trained on an NVIDIA QUADRO RTX 8000 for 120 epochs. We applied Adam optimizer (Kingma and Ba, 2014) to train our model with a learning rate of 0.0001. The regularization parameter λ is set to 1.0 to achieve the best network performance based on our investigation. To improve the model performance, we employed data augmentation techniques to the input image pairs during training, including rotation, random flipping, and non-linear transformations. Specifically, a horizontal flipping operation was randomly performed with a probability of 50 %. Furthermore, the image pairs underwent rotation around the longitudinal axis, with two random angles selected in the range of [− 30°, 30 °]. Non-linear transformations using random B-spline mapping were also applied to further enhance the variations of the training data.
3. Results
3.1. Registration accuracy: inter-modality (CT-MR) registration
The DIR results between CT and daily MR images were quantitatively and qualitatively analyzed to evaluate the performance of our Patch-RegNet for inter-modality registration.
Examples of the CT-MR registration results are shown in Fig. 4. The contours on the simulation CT images were propagated onto the daily MR images using different registration methods, including the traditional DIR tool in Monaco (a1, c1), VoxelMorph-MIND (a2, c2), and Patch-RegNet (a3, c3). The deformed contours were overlaid on the MR images and compared to the ground-truth contours shown in red color-wash for the evaluation of DIR accuracy. For better visualization, the insets (b1–b3, d1–d3) show the zoomed-in images of the deformed contours of specific structures (a1–a3, c1–c3) indicated by the dashed-line boxes.
Fig. 4.
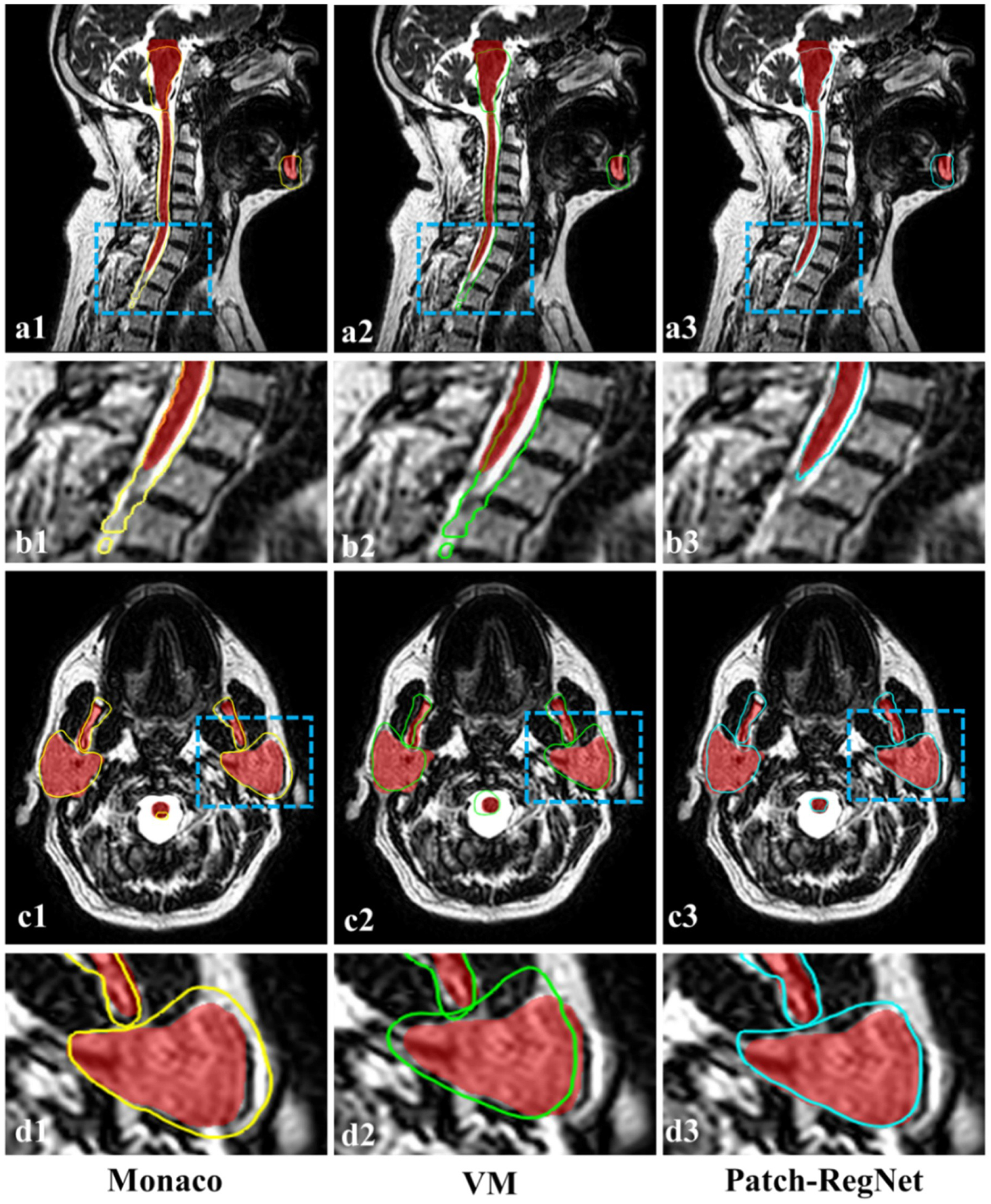
Qualitative evaluation results of different registration methods. The manual contours (red color-wash) are compared with the deformed contours from Monaco (yellow), VoxelMorph-MIND (green), and our Patch-RegNet (blue) methods. (VM: VoxelMorph-MIND).
As can be seen in Fig. 4, the deformed contours by Monaco have a considerable amount of disagreement with the manual contours, while our Patch-RegNet achieves the most accurate registration results among the four approaches. As can be seen from the zoomed-in images (a1–a3, c1–c3), there are clear distinctions between our Patch-RegNet and other methods, especially for the alignment of spinal cord and parotid glands.
Table 1 reports the average DSC and MSD over all organs calculated for different registration methods. Among them, our Path-RegNet achieved the best performance with the highest average DSC of 0.76 and lowest average MSD of 1.9 mm. Based on the DSC measurements, Our Patch-RegNet outperforms the VoxelMorph-MIND and the traditional DIR in Monaco by 6 %, and 10 %, respectively. The results demonstrate that our proposed Patch-RegNet achieves a statistically significant (p < 0.05 using a two-tailed t-test) improvement over other methods for inter-modality (CT-MR) registration.
Table 1.
Quantitative comparison of different methods for inter-modality (CT-MR) registration. The average Dice similarity coefficient (DSC) (%) and mean surface distance (MSD) (mm) and their standard deviations are calculated over all 7 organs for all test patients. The bolded numbers denote the highest scores.
| Methods | Rigid | Monaco | VoxelMorph | Patch-RegNet |
|---|---|---|---|---|
| DSC | 0.61 ± 0.16 | 0.69 ± 0.11 | 0.72 ± 0.10 | 0.76 ± 0.05 |
| MSD (mm) | 3.1 ± 1.8 | 2.6 ± 1.1 | 2.2 ± 0.7 | 1.9 ± 0.5 |
| Times (s) | - | 60 | 0.03 | 5.6 |
DSC: Dice similarity coefficient; MSD: mean surface distance.
The boxplots of organ-specific results are shown in Fig. 5 for detailed comparison. These results further demonstrate that our Patch-RegNet achieves improved registration performance for each organ under consideration. The improvement is more noticeable in organs/structures that are more likely to move from day to day due to setup, e.g. the mandible, because the local alignment in the hierarchical registration framework of Patch-RegNet can effectively correct this local displacement. In addition, Fig. 5 shows that our Patch-RegNet has smaller variance and improved median performance compared to other methods, indicating improved robustness and reliability of Patch-RegNet. The run-time for DIR process of each method is listed in Table 1 as well. The run-time included in the table is the average time for the DIR registration of one single case. The deep-learning methods show much faster registration than the traditional method in Monaco.
Fig. 5.
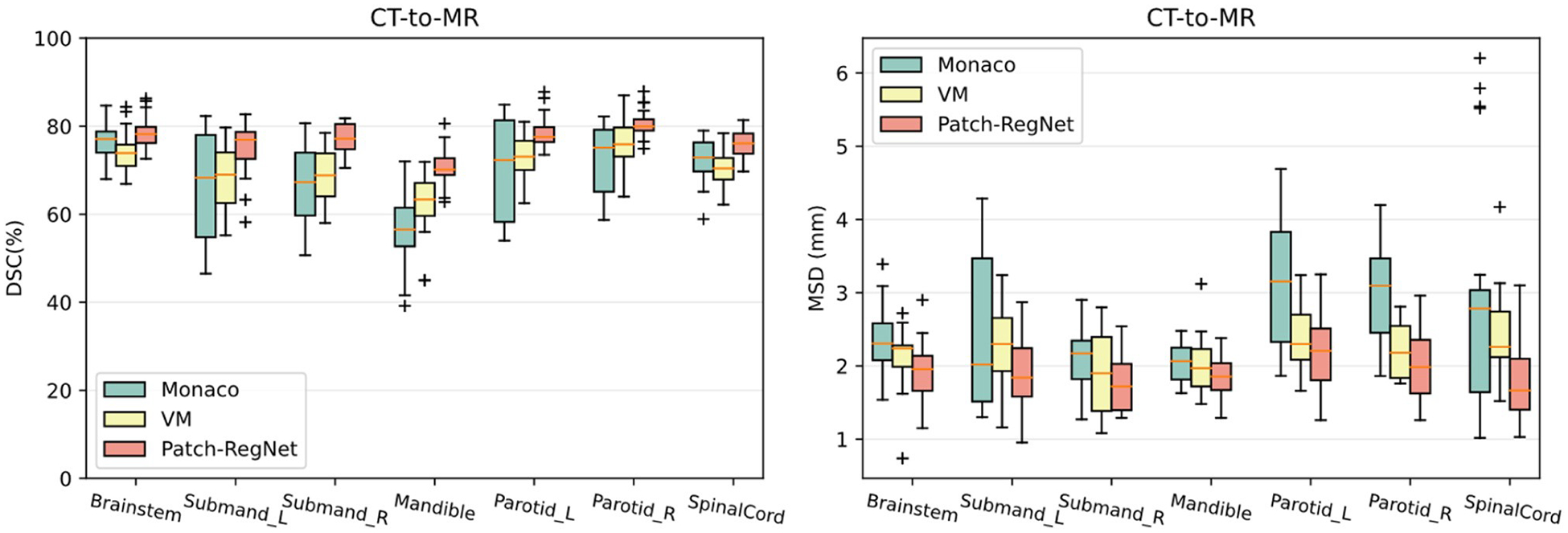
Quantitative evaluation results of different methods for inter-modality (CT-MR) registration. Boxplots showing DSC and MSD results for various anatomical structures using Monaco, VM (VoxelMorph-MIND), ViT-Morph (ViT-Morph-MIND), and our Patch-RegNet methods. DSC: Dice similarity coefficient. MSD: mean surface distance.
3.2. Registration accuracy: intra-modality (MR-MR) registration
The performance of our Patch-RegeNet for intra-modality (MR-MR) registration was also evaluated and compared with the traditional DIR tool in Monaco and VoxelMorph-MIND.
Table 2 summarizes the quantitative results of the average DSC and MSD over all organs calculated for different registration methods. Similar to the results of inter-modality registration, our Patch-RegNet achieves the best average DSC of 0.86 and average MSD of 0.9 mm. Compared with VoxelMorph-MIND and the traditional DIR tool in Monaco, Patch-RegNet improves the DSC accuracy by 4 % and 7 %, respectively. As shown in the quantitative results, our proposed Patch-RegNet achieves a statistically significant (p < 0.05 using a two-tailed t-test) improvement over all other methods for intra-modality (MR-MR) registration. It indicates our Patch-RegNet method can work effectively not only for inter-modality (CT-MR) but also for intra-modality (MR-MR) registration. The detailed DSC and MSD results for each structure are presented as boxplots in Fig. 6. Although Monaco is observed to achieve better registration results for some specific patients, its registration performance is unstable among different patients. On the contrary, our Patch-RegNet is much more robust for different cases, demonstrated by the smaller variance and improved median values in the boxplots. Therefore, overall performance of our Patch-RegNet is better than both the traditional DIR tool in Monaco and VoxelMorph-MIND. In addition, as shown in Tables 1 and 2, the average run-times for DIR process of CT-MR and MR-MR are comparable, which are more than 10 times faster than the traditional method.
Table 2.
Quantitative comparison of different methods for intra-modality (MR-MR) registration. The average Dice similarity coefficient (DSC) (%) and mean surface distance (MSD) (mm)) and their standard deviations are calculated over all 7 organs for all test patients. The bolded numbers denote the best scores.
| Methods | Rigid | Monaco | VoxelMorph | Patch-RegNet |
|---|---|---|---|---|
| DSC (%) | 0.76 ± 0.16 | 0.80 ± 0.11 | 0.83 ± 0.7 | 0.86 ± 0.06 |
| MSD (mm) | 2.0 ± 1.5 | 1.5 ± 1.5 | 1.1 ± 0.6 | 0.9 ± 0.3 |
| Times (s) | - | 60 | 0.03 | 5.2 |
DSC: Dice similarity coefficient; MSD: mean surface distance.
Fig. 6.
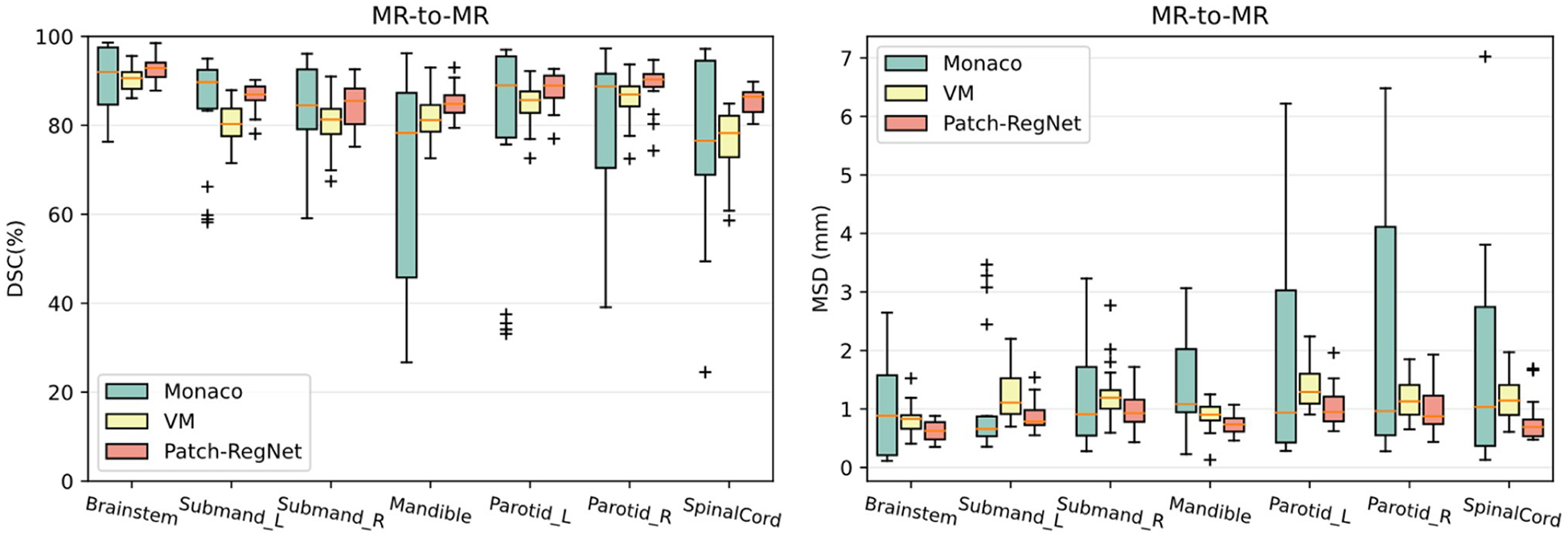
Quantitative evaluation results of different methods for intra-modality (MR-MR) registration. Boxplots showing DSC and MSD results for various anatomical structures using Monaco, VM (VoxelMorph-MIND), and our Patch-RegNet methods. DSC: Dice similarity coefficient. MSD: mean surface distance.
3.3. Effectiveness of ViT-Morph and hierarchical framework
To assess the effectiveness of our proposed network architecture, ViT-Morph, and the hierarchical registration framework, we trained a ViT-Morph-MIND model for CT-MR image registration without using the hierarchical registration framework for a comparative analysis. The overall quantitative results are presented in Table 3 and the organ-specific results are illustrated in Fig. 7 with boxplots. By comparing the ViT-Morph-MIND with the VoxelMorph-MIND, the results in Table 3 and Fig. 7 indicate that ViT-Morph-MIND achieves higher DSC scores and lower MSD values for both overall performance and organ-specific results. The superior performance of VIT-Morph-MIND suggests that the integration of Vision Transformer and UNet in the proposed ViT-Morph model leads to improved accuracy in image registration. This finding highlights the effectiveness of the proposed ViT-Morph as an effective network for image registration.
Table 3.
Quantitative comparison of different methods for inter-modality (CT-MR) registration. The average Dice similarity coefficient (DSC) (%) and mean surface distance (MSD) (mm) and their standard deviations are calculated over all 7 organs for all test patients. The bolded numbers denote the highest scores.
| Methods | VoxelMorph | ViT-Morph | Patch-RegNet |
|---|---|---|---|
| DSC | 0.72 ± 0.10 | 0.73 ± 0.09 | 0.76 ± 0.05 |
| MSD (mm) | 2.2 ± 0.7 | 2.1 ± 0.7 | 1.9 ± 0.5 |
| Times (s) | 0.03 | 0.03 | 5.6 |
DSC: Dice similarity coefficient; MSD: mean surface distance.
Fig. 7.
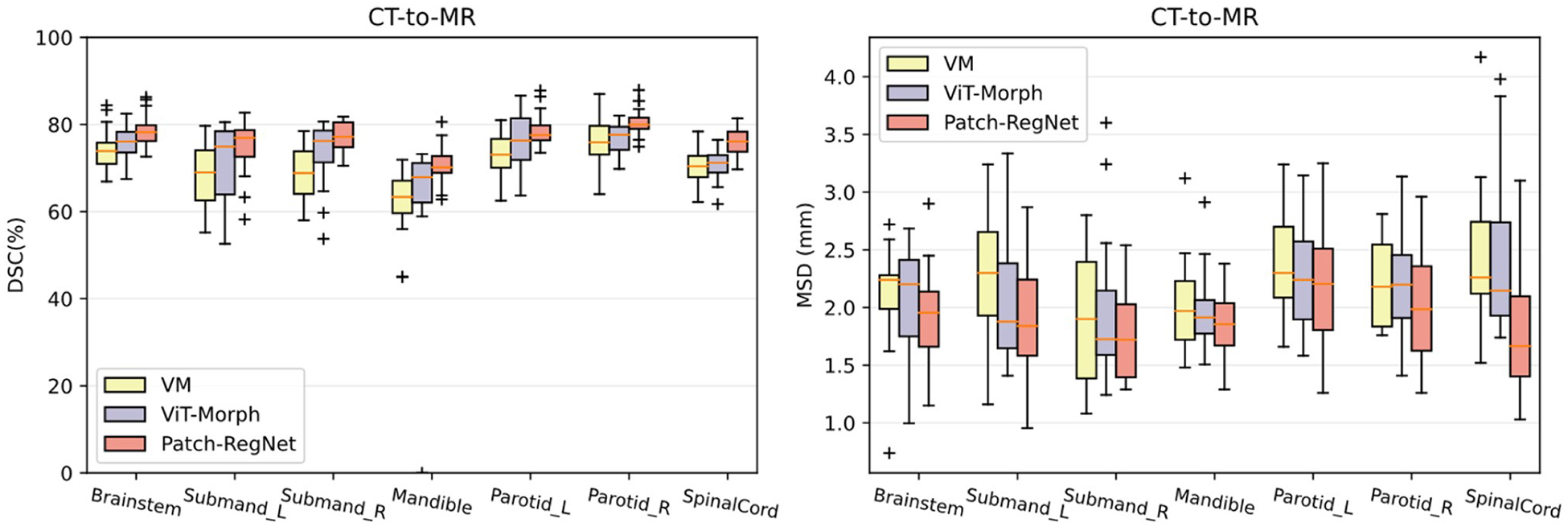
Quantitative evaluation results of different methods for inter-modality (CT-MR) registration. Boxplots showing DSC and MSD results for various anatomical structures using VM (VoxelMorph-MIND), ViT-Morph (ViT-Morph-MIND), and our Patch-RegNet methods. DSC: Dice similarity coefficient. MSD: mean surface distance.
On the other hand, the comparison between Patch-RegNet and ViT-Morph-MIND demonstrates that the Patch-RegNet outperforms the ViT-Morph-MIND 3 % on DSC measurement and 0.2 mm on MSD measurement overall. This improvement demonstrates the effectiveness of our hierarchical framework in improving the performance of the registration. The hierarchical framework in Patch-RegNet, which utilizes patch-based local registration, provides improved regional guidance for deep learning-based DIR. By focusing the registration on local regions and specific anatomical sites, it effectively addresses the challenge of inadequate alignment accuracy in some local regions that cannot be achieved using a global rigid registration. This comparison demonstrated how a combination of novel network architecture design and hierarchical registration framework in our Patch-RegNet improves the accuracy of deformable registration.
3.4. Regularization analysis
We investigated the influence of the hyperparameter λ, which regularizes the smoothness of the generated DVFs, to our Patch-RegNet. The average DSC results for the test dataset for different values of the regularization parameter λ are plotted in Fig. 8. The results for both CT-MR and MR-MR registration show that our method is relatively robust to the choice of λ values. The optimal value for the regularization parameter is demonstrated to be λ = 1.0 for both inter- and intra-modality registration.
Fig. 8.
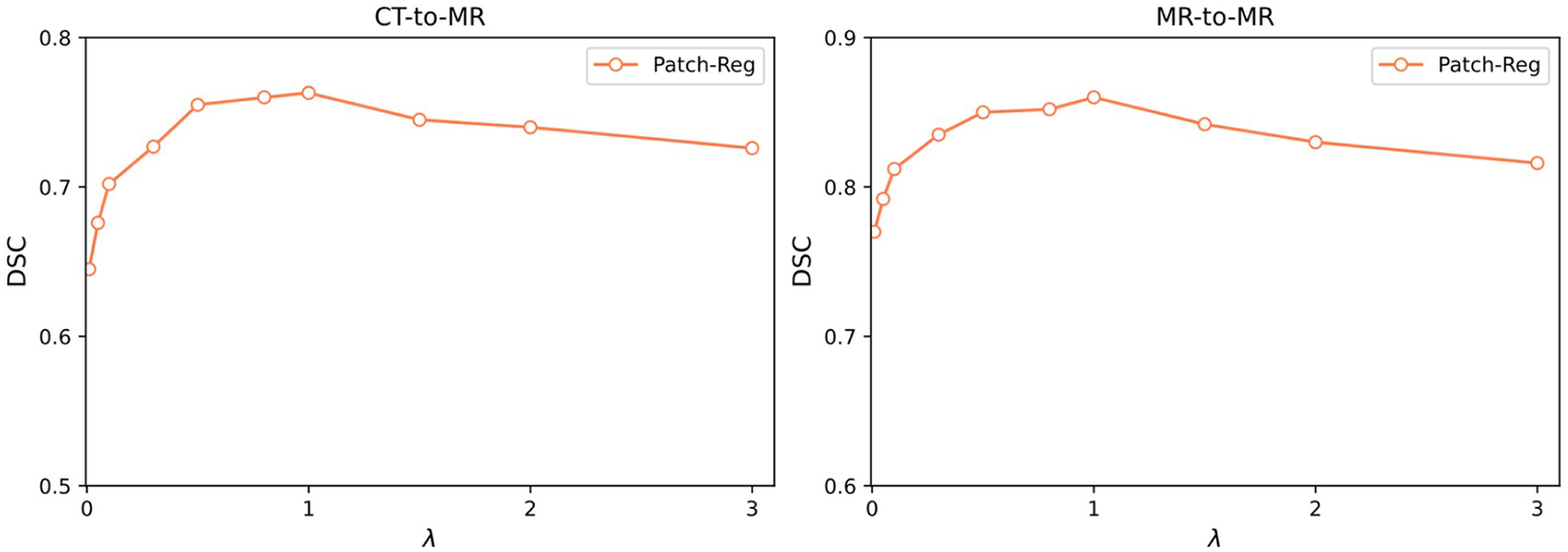
Average DSC results of test data for Patch-RegNet with varied regularization parameter λ.
4. Discussion
In last two decades, ART has gained widespread popularity due to its ability to deliver more personalized and accurate treatment. Meanwhile, the advent of MR-Linacs has significantly advanced the field of ART by enabling the integration of MR imaging into radiotherapy with online plan adaptation. DIR plays a crucial role in ART to ensure accurate contour propagation with daily anatomical changes so that treatment plan can be reoptimized in real time. In this study, we introduced a novel hierarchical framework for the implementation of efficient and precise CT-MR and MR-MR DIR. This framework features the utilization of patch-based registration, which can improve the local registration over a large anatomical region. The results of this study indicate that our proposed method significantly outperforms current DIR techniques in CT-MR and MR-MR registration accuracy. Compared to VoxelMorph, an average DSC improvement of 0.04 for 7 structures is not trivial and has significant impact in clinic, which can be demonstrated by statistical testing (p < 0.05 for all tests), subjective evaluations (Fig. 4), and quantitative evaluation for all individual structures (Figs. 5–7). Moreover, the study demonstrates that the proposed Patch-RegNet achieves faster registration speed compared to the traditional method in Monaco, as presented in Tables 1 and 2. Although our Patch-RegNet is slower than VoxelMorph-MIND due to multiple patch-based registrations, a total run time of 5–6 s for each case is acceptable for most clinical applications and the improved registration accuracy is worth the cost in registration time. On the other hand, a small improvement in contour propagation may save a lot of time for contour editing in online ART. This is particularly important in the HN region, where a lot of critical organs needs to be contoured for treatment planning. Our Patch-RegNet could greatly benefit the critical organs that are particularly susceptible to daily setup uncertainty during treatment. Despite the increased computation time compared to VoxelMorph, Patch-RegNet offers enhanced accuracy that can justify the slightly longer processing time, especially considering the applications in ART.
In this work, we introduced a novel deep learning-based model, named ViT-Morph, for DIR by incorporating Vision Transformer (ViT) into conventional Convolutional Neural Network (CNN) networks. ViT-Morph leverages both the local features provided by CNNs and the long-range image relationships obtained through the self-attention mechanism in ViT. We expected the combination of these two models would provide a more comprehensive representation of the medical images, which will provide superior image registration performance. The result of comparison between the performance of ViT-Morph-MIND and VoxelMorph-MIND in CT-MR DIR demonstrated a marked improvement in performance of ViT-Morph-MIND compared to the CNN-based network, VoxelMorph-MIND. These results proved that the combination of ViT and CNN indeed enhanced the performance of DIR.
One of the main challenges for deep learning-based registration methods is the pre-alignment of the moving and fixed images prior to the DIR process. It is common practice to apply a global rigid or affine registration to the moving and fixed images as an initial step for the following more complicated DIR process. However, this approach, which involves the application of rigid or affine registration to the entire image volumes alone, may not provide adequate alignment accuracy in some local regions for subsequent DIR, particularly for images that cover a large area and contain more than one anatomical site, such as HN images. Our study demonstrated that the hierarchical framework in Patch-RegNet can fundamentally address the local pre-alignment issue. This framework involves the extraction of patches from the whole image volumes and the application of patch-based local registration, which provides improved regional guidance for DL-based DIR by focusing the registration on local regions and specific anatomical sites. The results presented in Sections 3.1 and 3.2 indicate a substantial improvement in CT-MR and MR-MR DIR through the use of our hierarchical framework. This framework can greatly benefit organs that are particularly susceptible to daily setup uncertainty during treatment, such as the mandible and spinal cord. Our Patch-RegNet has been compared to the same DL network ViT-Morph-MIND that did not use a hierarchical framework in Section 3.3. The comparison results highlighted the effectiveness of the hierarchical framework, demonstrating that the improved performance of Patch-RegNet is not solely attributed to the introduction of a new DL network, but also to the combination of the new network and the hierarchical framework. Table 3 provides further evidence, showing the impact of our hierarchical framework on the DIR performance. Additionally, the flexibility of Patch-RegNet allows for the replacement of ViT-Morph-MIND with a more advanced networks, thus providing the potential for enhanced registration performance.
We used the HN patients to demonstrate the effectiveness of our Patch-RegNet for both inter-modality (CT-MR) and intra-modality (MR-MR) DIR. However, this registration method can be extended for the registration tasks of other body sites and other image modalities. To facilitate this expansion, a continuous process of data accumulation and thorough curation of large image datasets will be necessary. Additionally, the selection of appropriate patch size is crucial when extending the application of Patch-RegNet to other body sites. In this study, we employed overlapping patches with dimensions of 160× 160 × 128, ensuring adequate coverage for complete anatomic structures in the HN region. It is possible that varying patch sizes may be required for applications to other body sites.
Besides the development of the framework, the MIND descriptor was introduced as the similarity metric in Patch-RegNet for inter- and intra-modality DIR. The MIND extracts the distinctive features within a local neighborhood to create descriptor vectors, allowing the transformation of images from different modalities into a common domain. This enables straightforward similarity measurement through the use of metrics such as the sum of squared differences. Consequently, the incorporation of the MIND descriptor in Patch-RegNet enhances its robustness against image noise and non-linear intensity variations, making it suitable for the registration of images acquired from various modalities. Furthermore, the similarity metric used in Patch-RegNet can be easily changed to alternative metrics to accommodate other specific registration tasks.
In the results we presented, MR images were used as fixed images for CT-MR DIR. This choice was primarily driven by the clinical application, where contours delineated by physicians on simulation CT images are deformably mapped to daily MR images. Our contour deformation was performed using the binary mask deformation approach so that the MR image needed to be fixed to obtain a deformation vector field for the contour deformation. It is worth mentioning that our Patch-RegNet can be applied directly for MR -CT registration with a fixed CT image as well. In our initial investigation, we conducted a comparative analysis of the registration performances for both CT-MR and MR-CT registrations. Although the inverse consistency is not enforced explicitly in our algorithm, the registration performance in two directions did not show any significant difference. This observation can be attributed to accurate and consistent correspondence detection from both directions (CT-MR or MR-CT). It is known that some anatomical structures may be more visible on one image than the other. However, the structures exist on both images, and the modality independence loss function, i.e. MIND, used in our algorithm enables effective correspondence findings regardless of the choice of fixed images. Consequently, the selection of fixed images does not significantly impact the registration accuracy.
In this study, we evaluated the registration accuracy of our proposed deep learning-based image registration model, Patch-RegNet, through both qualitative and quantitative methods. Despite the promising results of our study, there are some limitations that should be noted. First, we only evaluated the performance of our proposed method for CT-MR and MR-MR registration. Thus, the applicability of Patch-RegNet to other modalities, such as PET and SPECT, remains to be investigated. Second, the study only focuses on head and neck cancer patients, and the extension of this work to other anatomical sites will need to be further assessed. Furthermore, the limited data used in our study may affect the generalizability of the results to other patient populations. Therefore, future studies with larger and more diverse datasets are necessary to validate the performance of our proposed method. Lastly, the performance of Patch-RegNet was only evaluated using a limited number of metrics, and further investigations are necessary to assess its clinical impact.
5. Conclusions
We developed the Patch-RegNet, a fully automated hierarchical registration framework, for inter-modality (CT-MR) and intra-modality (MR-MR) DIR. The hierarchical framework enables our Patch-RegNet to achieve markedly improved registration for large-volume images containing multiple anatomic sites. The patch-based ViT-Morph in our Patch-RegNet takes advantage of both CNN and ViT features of long-range spatial relationships. Additionally, MIND is incorporated as similarity metric to effectively train the network for multi-modality registrations. The Patch-RegNet is validated using HN cancer patient images and demonstrated superior results compared to the traditional DIR and other DL-based DIR methods.
Acknowledgements
This work was supported in part by a start-up fund from MD Anderson Cancer Center (YZ and JY), the Image Guided Cancer Therapy T32 Training Program T32CA261856 (BAM and CDF), and the National Institutes of Health through Cancer Center Support Grant P30CA016672. A part of the data was acquired under the MOMENTUM observational registry trial funded by Elekta AB.
Footnotes
CRediT authorship contribution statement
Yao Zhao: Conceptualization, Methodology, Software, Validation, Data curation, Writing – original draft. Xinru Chen: Methodology, Software, Validation Writing – review & editing. Brigid McDonald: Data curation, Writing – review & editing. Cenji Yu: Software, Writing – review & editing. Abdalah S R Mohamed: Data curation, Writing – review & editing. Clifton D Fuller: Writing – review & editing Laurence E Court: Conceptualization, Writing – review & editing. Tinsu Pan: Writing – review & editing. He Wang: Methodology, Writing – review & editing. Xin Wang: Methodology, Writing – review & editing. Jack Phan: Validation, Writing – review & editing. Jinzhong Yang: Conceptualization, Methodology, Validation, Supervision, Writing – review & editing.
Declaration of Competing Interest
CDF has received direct industry grant support, speaking honoraria, and travel funding from Elekta AB. The other authors have no conflicts of interest to disclose.
Data availability
The authors do not have permission to share data.
References
- Bahl A, Elangovan A, Dracham CB, et al. , 2019. Analysis of volumetric and dosimetric changes in mid treatment CT scan in carcinoma nasopharynx: implications for adaptive radiotherapy. J. Exp. Ther. Oncol 13 (1), 33–39. [PubMed] [Google Scholar]
- Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV, 2019. VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 38 (8), 1788–1800. [DOI] [PubMed] [Google Scholar]
- Beare R, Lowekamp B, Yaniv Z, 2018. Image segmentation, registration and characterization in R with SimpleITK. J. Stat. Softw 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw L, Agnew CE, Irvine DM, Rooney KP, McGarry CK, 2019. Adaptive radiotherapy for head and neck cancer reduces the requirement for rescans during treatment due to spinal cord dose. Radiat. Oncol 14 (1), 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock KK, Mutic S, McNutt TR, Li H, Kessler ML, 2017. Use of image registration and fusion algorithms and techniques in radiotherapy: report of the AAPM Radiation Therapy Committee Task Group No. 132. Med Phys. 44 (7), e43–e76. [DOI] [PubMed] [Google Scholar]
- Burela N, Soni TP, Patni N, Natarajan T, 2019. Adaptive intensity-modulated radiotherapy in head-and-neck cancer: a volumetric and dosimetric study. J. Cancer Res Ther 15 (3), 533–538. 10.4103/jcrt.JCRT_594_17. [DOI] [PubMed] [Google Scholar]
- Cao H, Wang Y, Chen J, et al. Swin-unet: unet-like pure transformer for medical image segmentation. In: Proceedings of the Computer Vision–ECCV 2022 Workshops: Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part III. Springer; 2023, 205–218. [Google Scholar]
- Castelli J, Simon A, Lafond C, et al. , 2018. Adaptive radiotherapy for head and neck cancer. Acta Oncol. 57 (10), 1284–1292. 10.1080/0284186X.2018.1505053. [DOI] [PubMed] [Google Scholar]
- Chen J, He Y, Frey EC, Li Y, Du Y, 2021a. Vit-v-net: Vision transformer for unsupervised volumetric medical image registration. ArXiv Prepr ArXiv210406468. Published online 2021. [Google Scholar]
- Chen J, Lu Y, Yu Q, et al. , 2021b. Transunet: Transformers make strong encoders for medical image segmentation. ArXiv Prepr ArXiv210204306. Published online 2021. [Google Scholar]
- Chen J, Frey EC, He Y, Segars WP, Li Y, Du Y, 2022. Transmorph: Transformer for unsupervised medical image registration. Med Image Anal. 82, 102615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Diaz-Pinto A, Ravikumar N, Frangi AF, 2021a. Deep learning in medical image registration. Prog. Biomed. Eng 3 (1), 012003. [Google Scholar]
- Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16×16 words: Transformers for image recognition at scale. ArXiv Prepr ArXiv201011929. Published online 2020. [Google Scholar]
- Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X, 2020. Deep learning in medical image registration: a review. Phys. Med Biol 65 (20), 20TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Pouget-Abadie J, Mirza M, et al. , 2020. Generative adversarial networks. Commun. ACM 63 (11), 139–144. [Google Scholar]
- Güngör A, Askin B, Soydan DA, Saritas EU, Top CB, Çukur T, 2022. TranSMS: transformers for super-resolution calibration in magnetic particle imaging. IEEE Trans. Med Imaging 41 (12), 3562–3574. [DOI] [PubMed] [Google Scholar]
- Guo P, Mei Y, Zhou J, Jiang S, Patel VM ReconFormer: accelerated MRI reconstruction using recurrent transformer. ArXiv Prepr ArXiv220109376. Published online 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskins G, Kruger U, Yan P, 2020. Deep learning in medical image registration: a survey. Mach. Vis. Appl 31, 1–18. [Google Scholar]
- Heinrich MP, Jenkinson M, Bhushan M, et al. , 2012a. MIND: modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal 16 (7), 1423–1435. 10.1016/j.media.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Heinrich MP, Jenkinson M, Bhushan M, et al. , 2012b. MIND: modality independent neighbourhood descriptor for multi-modal deformable registration. Med. Image Anal 16 (7), 1423–1435. [DOI] [PubMed] [Google Scholar]
- Jaderberg M, Simonyan K, Zisserman A, 2015. Spatial transformer networks. Adv. Neural Inf. Process Syst 28. [Google Scholar]
- Kingma DP, Ba J Adam: a method for stochastic optimization. ArXiv Prepr ArXiv14126980. Published online 2014. [Google Scholar]
- Kupelian P, Sonke JJ, 2014. Magnetic resonance–guided adaptive radiotherapy: a solution to the future. In: In: Seminars in Radiation Oncology, 24. Elsevier,, pp. 227–232. [DOI] [PubMed] [Google Scholar]
- Liu L, Huang Z, Li ò P, Schönlieb CB, Aviles-Rivero AI Pc-swinmorph: patch representation for unsupervised medical image registration and segmentation. ArXiv Prepr ArXiv220305684. Published online 2022. [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P, 1997. Multimodality image registration by maximization of mutual information. IEEE Trans. Med Imaging 16 (2), 187–198. [DOI] [PubMed] [Google Scholar]
- Mattes D, Haynor DR, Vesselle H, Lewellyn TK, Eubank W Nonrigid multimodality image registration. In: Medical Imaging 2001: Image Processing. Vol 4322. Spie; 2001:1609–1620. [Google Scholar]
- McDonald BA, Vedam S, Yang J, et al. , 2021. Initial feasibility and clinical implementation of daily mr-guided adaptive head and neck cancer radiation therapy on a 1.5 t mr-linac system: Prospective r-ideal 2a/2b systematic clinical evaluation of technical innovation. Int J. Radiat. Oncol. Biol. Phys 109 (5), 1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie EM, Santhanam A, Ruan D, O’Connor D, Cao M, Sheng K, 2020. Multimodality image registration in the head-and-neck using a deep learning-derived synthetic CT as a bridge. Med Phys. 47 (3), 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owrangi AM, Greer PB, Glide-Hurst CK, 2018. MRI-only treatment planning: benefits and challenges. Phys. Med Biol 63 (5), 05TR01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaymakers BW, Lagendijk JJW, Overweg J, et al. , 2009. Integrating a 1.5 T MRI scanner with a 6 MV accelerator: proof of concept. Phys. Med Biol 54 (12), N229. [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Payne GS, 2015. Radiotherapy planning using MRI. Phys. Med Biol 60 (22), R323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaswani A, Shazeer N, Parmar N, et al. , 2017. Attention is all you need. Adv. Neural Inf. Process Syst 30. [Google Scholar]
- Viola P, Wells WM, 1995. Alignment by maximization of mutual information. In: Proceedings of the IEEE International Conference on Computer Vision. IEEE,, pp. 16–23. [Google Scholar]
- Wahid KA, He R, McDonald BA, et al. , 2021. Intensity standardization methods in magnetic resonance imaging of head and neck cancer. Phys. Imaging Radiat. Oncol 20, 88–93. 10.1016/j.phro.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Hu R, Xiao Z, Chen J, Liu J, 2021. Vision Transformer-based recognition of diabetic retinopathy grade. Med Phys. 48 (12), 7850–7863. [DOI] [PubMed] [Google Scholar]
- Wu QJ, Li T, Wu Q, Yin FF, 2011. Adaptive radiation therapy: technical components and clinical applications. Cancer J. 17 (3), 182–189. 10.1097/PPO.0b013e31821da9d8. [DOI] [PubMed] [Google Scholar]
- Xu Z, Luo J, Yan J, et al. , 2020. Adversarial uni- and multi-modal stream networks for multimodal image registration. In: Martel AL, Abolmaesumi P, Stoyanov D, et al. (Eds.), Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. Springer International Publishing, pp. 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu H, Hu Q 2021a, Transfuse: Fusing transformers and cnns for medical image segmentation. In: Proceedings of the Twenty Fourth International Conference, Strasbourg, France, September 27–October 1, 2021, Medical Image Computing and Computer Assisted Intervention–MICCAI 2021:, Part I 24. Springer; 2021, 14–24. [Google Scholar]
- Zhang Y, Pei Y, Zha H, 2021b. Learning dual transformer network for diffeomorphic registration. In: Proceedings of the Twenty Fourth International Conference, Strasbourg, France, September 27–October 1, 2021, Medical Image Computing and Computer Assisted Intervention–MICCAI 2021, Part IV 24. Springer; 2021, 129–138. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.


