Abstract
Toxigenic Vibrio cholerae strains are lysogens of CTXΦ, a filamentous phage which encodes cholera toxin. The receptor for CTXΦ for invading V. cholerae cells is the toxin-coregulated pilus (TCP), the genes for which reside in a larger genetic element, the TCP pathogenicity island. We analyzed 146 CTX-negative strains of V. cholerae O1 or non-O1 isolated from patients or surface waters in five different countries for the presence of the TCP pathogenicity island, the regulatory gene toxR, and the CTXΦ attachment sequence attRS, as well as for susceptibility of the strains to CTXΦ, to investigate the molecular basis for the emergence of new clones of toxigenic V. cholerae. DNA probe or PCR assays for tcpA, tcpI, acfB, toxR, and attRS revealed that 6.85% of the strains, all of which belonged to the O1 serogroup, carried the TCP pathogenicity island, toxR, and multiple copies of attRS, whereas the remaining 93.15% of the strains were negative for TCP but positive for either one or both or neither of toxR and attRS. An analysis of the strains for susceptibility to CTXΦ, using a genetically marked derivative of the phage CTX-KmΦ, showed that all TCP-positive CTX-negative strains and 1 of 136 TCP-negative strains were infected by the phage either in vitro or in the intestines of infant mice. The phage genome integrated into the chromosome of infected V. cholerae O1 cells forming stable lysogens. Comparative analysis of rRNA gene restriction patterns revealed that the lysogens derived from nontoxigenic progenitors were either closely related to or distinctly different from previously described clones of toxigenic V. cholerae. To our knowledge, this is the first demonstration of lysogenic conversion of naturally occurring nontoxigenic V. cholerae strains by CTXΦ. The results of this study further indicated that strains belonging to the O1 serogroup of V. cholerae are more likely to possess the TCP pathogenicity island and hence to be infected by CTXΦ, leading to the origination of potential new epidemic clones.
Cholera caused by toxigenic Vibrio cholerae is a major public health problem in developing countries. Epidemiological surveillance of cholera and comparative molecular analysis of strains collected during outbreaks have demonstrated clonal diversity among epidemic strains and a continual emergence of new clones of toxigenic V. cholerae (6–8). The genetic basis of and the mechanisms involved in the origination of the new epidemic clones have not been adequately explained.
Cholera pathogenesis relies on the synergistic effect of a number of pathogenic factors produced by toxigenic V. cholerae. Profuse, watery diarrhea, which is characteristic of cholera, is caused by an enterotoxin, cholera toxin (CT), produced by V. cholerae when it colonizes the small intestine (27). Molecular analysis has revealed that in addition to genes encoding CT, all strains capable of causing cholera invariably carry genes for a colonization factor known as toxin-coregulated pilus (TCP) and a regulatory protein, ToxR, which coregulates the expression of both CT and TCP (15, 22, 34). Although the major subunit of TCP is encoded by the tcpA gene, the formation and function of the pilus assembly require the products of a number of other genes located on a large DNA region referred to as the TCP pathogenicity island, which includes the tcp and acf gene clusters (19). The ctxAB operon which encodes the A and B subunits of CT is part of a larger genetic element originally termed the CTX genetic element. Recent studies have shown that the CTX genetic element corresponds to the genome of CTXΦ, a lysogenic filamentous bacteriophage (35). The propagation of the phage has been studied in recipient laboratory strains of V. cholerae. CTXΦ uses TCP as its receptor for invading V. cholerae cells, in which the phage genome either integrates chromosomally at a specific attachment site (attRS), forming stable lysogens, or is maintained extrachromosomally as a replicative form (RF) of the phage genome (35). We previously studied the induction of CTXΦ prophage in naturally occurring strains of toxigenic V. cholerae and demonstrated that wild-type phage particles isolated from different native strains infected an attenuated CT-negative V. cholerae strain (10). This indicated that in the natural habitat, propagation of CTXΦ may be associated with horizontal gene transfer, leading to the origination of novel toxigenic strains of V. cholerae. To further validate this concept, the present study was undertaken to analyze a large number of nontoxigenic V. cholerae strains for the presence of major virulence-associated genes and for susceptibility of the strains to a genetically marked derivative of CTXΦ to investigate the molecular basis for the origination of novel toxigenic strains with epidemic potential.
MATERIALS AND METHODS
Bacterial strains, plasmids, and phages.
A total of 146 CTX-negative V. cholerae isolates from five different countries, obtained either from patients or from surface water samples, were included in the study. Clinical isolates from Bangladesh were obtained from patients who attended the treatment center of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B), located in Dhaka. The environmental isolates were from surface water samples in Dhaka isolated between 1992 and 1996. Other isolates consisted of four El Tor and three non-O1 strains from patients in Saudi Arabia (courtesy of M. H. Al-Jefrri, Parasitic and Infectious Diseases Unit, Department of Preventive Medicine, Ministry of Health, Riyadh, Kingdom of Saudi Arabia), three El Tor strains from Estonia and one El Tor strain from Latvia isolated from seawater (courtesy of Anja Siitonen, Laboratory of Enteric Pathogens, National Public Health Institute, Helsinki, Finland), and six clinical El Tor strains (28) from southern India (courtesy of G. B. Nair, National Institute for Cholera and Enteric Diseases, Calcutta, India). The strains were stored either in lyophilized form or in sealed deep nutrient agar at room temperature, in the culture collection of ICDDR,B. Before use, the identities of the cultures were confirmed by biochemical and serological methods (37), and the presence or absence of the CTX element was tested with specific DNA probes. The strains analyzed in this study are described in Table 1. The relevant characteristics of reference bacterial strains and properties of phages and plasmids used in this study are listed in Table 2.
TABLE 1.
Distribution of major virulence-associated genes among nontoxigenic Vibrio cholerae O1 and non-O1 strains isolated from cholera patients or environmental surface water and susceptibility of the strains to a genetically marked derivative of CTXΦ
| V. cholerae serogroup | Country | Yr of isolation | Source | No. of strains | Presence ofa:
|
No. of strains
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | zot | ace | tcpA | tcpI | acfB | toxR | attRS | Infected with CTX-KmΦb | Lysogenizedc | |||||
| O1, El Tor biotype | ||||||||||||||
| India | 1993 | Patient | 6 | − | − | − | + | + | + | + | + | 6 | 6 | |
| Estonia | 1995 | SWd | 3 | − | − | − | − | − | − | + | − | 0 | 0 | |
| Latvia | 1995 | SW | 1 | − | − | − | − | − | − | + | − | 0 | 0 | |
| Saudi Arabia | 1997 | Patient | 4 | − | − | − | + | + | + | + | + | 4 | 4 | |
| Non-O1, non-O139 | ||||||||||||||
| Saudi Arabia | 1997 | Patient | 1 | − | − | − | − | − | − | + | − | 0 | 0 | |
| Saudi Arabia | 1997 | Patient | 2 | − | − | − | − | − | − | + | + | 0 | 0 | |
| Bangladesh | 1992–1995 | SW | 3 | − | − | − | − | − | − | − | + | 0 | 0 | |
| Bangladesh | 1992–1995 | SW | 19 | − | − | − | − | − | − | − | − | 0 | 0 | |
| Bangladesh | 1994–1996 | SW | 23 | − | − | − | − | − | − | + | + | 0 | 0 | |
| Bangladesh | 1993–1996 | SW | 84 | − | − | − | − | − | − | + | − | 1 | 0 | |
The presence of different genes was detected with DNA probes and PCR assays (see the text for details). The size of the PCR-generated amplicon corresponding to the tcpA gene was characteristic of that of the El Tor biotype.
Susceptibility to the phage was determined with a genetically marked phage, CTX-KmΦ (see the text for details).
Integration of the phage genome into the host chromosome was determined by Southern blot analysis of genomic and plasmid DNA isolated from native and infected strains.
SW, surface water.
TABLE 2.
Characteristics of the V. cholerae reference strains used in the study
| Strain | Relevant characteristic | Reference or source |
|---|---|---|
| SM44 | Derivative of El Tor strain P27459 in which the CTX genetic element was marked with a kanamycin resistance cartridge by marker exchange disrupting the ctxAB operon | 14 |
| 569B | V. cholerae O1, classical Inaba strain | Laboratory collection |
| RV508 | Derivative of classical biotype strain 569B that constitutively expresses CT, TCP, and other ToxR-regulated gene products | 35 |
| O395 | Classical Ogawa streptomycin-resistant strain | Laboratory collection |
| O395(pCTX-Km) | Strain O395 carrying the RF of CTX-KmΦ derived from strain SM44 | This study |
The genetically marked phage CTX-KmΦ was initially derived from strain SM44, in which a kanamycin resistance (Kmr) determinant was introduced by marker exchange disrupting the ctxAB operon (14, 35). CTX-KmΦ isolated from a mitomycin C-induced culture of SM44 was used to infect the classical V. cholerae O1 strain O395. CTX-KmΦ was prepared for the present study from a culture of strain O395 carrying an RF of the phage, O395(pCTX-Km). Aliquots of the culture supernatants were sterilized by filtration through 0.22-μm-pore-size filters (Millipore Corp., Bedford, Mass.). To confirm that the filtrate did not contain any bacterial cells, aliquots of the filtrate were streaked on Luria agar plates and incubated overnight at 37°C. The filtrate was titrated for infectious phage particles by incubating aliquots of the supernatants for 30 min at 30°C with the classical strain RV508, which constitutively expresses TCP, and then selecting for colonies resistant to kanamycin.
Probes and hybridization.
The presence of virulence-associated genes was determined by using specific DNA probes or PCR assays. The gene probes used in this study to detect the CTX genetic element were a 0.5-kb EcoRI fragment of pCVD27 (16) containing part of the ctxA gene, an 840-bp region internal to the zonula occludens toxin gene (zot) amplified by PCR from the recombinant plasmid pBB241 as described previously (9), and a 2.1-kb SphI-XbaI fragment of pCTX-Km (35) containing the entire zot and ace genes and part of orfU. The toxR gene probe was a 2.4-kb BamHI fragment of pVM7 (22). The 18-bp attRS sequence was identified by using a synthetic oligonucleotide corresponding to the attRS sequence (25). The rRNA gene probe has been described by us previously and consisted of a 7.5-kb BamHI fragment of the Escherichia coli rRNA clone pKK3535 (3, 6). Colony blots or Southern blots were prepared with nylon filters (Hybond; Amersham International plc., Aylesbury, United Kingdom) and processed by standard methods (21, 31). The polynucleotide probes were labeled by random priming (11) with a random-primer DNA labeling kit (BRL) and [α-32P]dCTP (3,000 Ci/mmol; Amersham), and oligonucleotide probes were labeled by 3′ tailing with terminal deoxynucleotide transferase and [α-32P]dCTP. Southern blots and colony blots were hybridized with the labeled probes, and autoradiographs were developed as described by us previously (6–9).
PCR assays.
The presence of the TCP pathogenicity island was determined by PCR assays specific for the tcpA, tcpI, and acfB genes. All oligonucleotides used either as probes or as PCR primers were synthesized commercially by Oswel DNA Service (University of Edinburgh, Edinburgh, United Kingdom), and PCR reagents and kits were purchased from Perkin-Elmer Corp. (Norwalk, Conn.). The presence of tcpA genes specific for the classical and El Tor biotypes was determined by a multiplex PCR assay (18), and the tcpI gene was detected by a PCR assay described by us previously (7). The acfB gene was detected by a PCR assay based on the published sequence of acfB (5) in which two primers with the following sequences were used: 5′GGACCAAGCATTATTATCTCT and 5′AATGATAAACTTACTGATTAA. Thermocycle parameters for the PCR assay consisted of denaturation at 94°C for 2 min, annealing of primers at 50°C for 2 min, and primer extension at 72°C for 3 min. Amplification was performed for 25 cycles, and the size of the amplicon (1.9 kb, which is the expected size) was ascertained by electrophoresis in 1.5% agarose gels. The identities of all PCR products were further verified with specific oligonucleotide probes. The absence of the relevant genes in the PCR-negative strains was further confirmed by colony blot hybridization with the corresponding PCR-generated amplicons from a positive control strain, 569B, as specific probes.
Infection of V. cholerae strains with CTX-KmΦ.
The susceptibility of V. cholerae strains to the genetically marked derivative of CTXΦ was assayed under laboratory conditions and in the intestines of infant mice by modifications of previously described methods (10, 35). All strains were initially tested for susceptibility to the phage in vitro. The TCP-positive strains and a single TCP-negative strain which were found to be susceptible to the phage in vitro were subjected to further tests both in vitro and in infant mice. The recipient strains were grown in Luria broth (LB) at 37°C; the cells were precipitated by centrifugation and washed in fresh LB. The recipient cells and phage particles were mixed in LB to make an approximate final concentration of 106 bacterial cells and 106 phage particles per ml. For the in vitro assay, the mixture was incubated for 16 h at 30°C, and aliquots of the culture were diluted and plated on Luria agar plates containing kanamycin (50 μg/ml) to select for kanamycin-resistant colonies and on plates devoid of kanamycin to determine the total number of colonies.
For the in vivo assay, the same mixtures of phage and recipient cells were used to gastrointestinally inoculate groups of 5-day-old Swiss Albino mice obtained from the breeding facilities of the Animal Resources Branch of ICDDR,B. For each strain and phage combination, at least five mice were inoculated. Animals were sacrificed after 16 h, and their intestines were removed and homogenized in 10 mM phosphate-buffered saline, pH 7.2. The homogenate was centrifuged at low speed to precipitate debris, the supernatant was then centrifuged to precipitate bacterial cells, and the pellet was resuspended in phosphate-buffered saline. The suspension was then screened for the presence of Kmr V. cholerae colonies. The ratio of Kmr colonies to total colonies was calculated and expressed as the percentage of recipient cells carrying the phage genome.
Analysis of infected cells.
Representative infected colonies were grown overnight in LB containing kanamycin (50 μg/ml), and cells were precipitated by centrifugation. The supernatant fluids of the cultures were titrated for the presence of CTX-KmΦ particles by using strain RV508 as the recipient. Total DNA or plasmids were extracted from bacterial pellets by standard methods (21) and purified with microcentrifuge filter units (Ultrafree-Probind; Sigma). Integration of the phage genome into the chromosomes of the recipient cells was studied by comparative Southern blot analysis of total DNA and plasmid preparations from the phage-infected and the corresponding native strains by using the zot probe.
Analysis of rRNA gene restriction patterns.
All the V. cholerae O1 strains included in the study were subjected to comparative analysis of their rRNA gene restriction patterns (ribotype) as described by us previously (6, 7) to study clonal relationships among the strains. Briefly, 5-μg aliquots of total DNA were digested with the appropriate restriction enzyme and electrophoresed in 0.8% agarose gel, and the DNA fragments were blotted onto nylon membranes (Hybond; Amersham). The genomic blots were hybridized with the rRNA gene probe, and autoradiographs were developed as described previously (6–8). rRNA gene restriction patterns produced by the newly formed lysogens derived from nontoxigenic progenitors were compared with previously reported ribotype patterns derived from toxigenic V. cholerae strains isolated from outbreaks of cholera in different countries (4, 6–8, 26).
RESULTS AND DISCUSSION
Distribution of virulence genes among nontoxigenic V. cholerae.
Most toxigenic V. cholerae strains isolated from cholera patients simultaneously carry genes for CT, TCP, and ToxR (7, 8). In order to identify CTX-negative strains which are possible progenitors or intermediates in the origination of new toxigenic strains, we studied the distribution of genes encoding TCP and ToxR and the CTXΦ attachment sequence (attRS) among nontoxigenic strains of V. cholerae with either DNA probes or PCR assays. Since the genes encoding TCP have been suggested to be part of a large genetic element consisting of clusters of genes, we looked for the simultaneous presence of the tcpA, tcpI, and acfB genes to screen for the presence of the TCP pathogenicity island (17, 19). DNA probe and PCR analysis of a total of 146 CTX-negative strains belonging to the O1 and non-O1 serogroups (Table 1) showed that 6.85% of the strains were positive for the tcpA, tcpI, and acfB genes and presumably the entire TCP pathogenicity island in addition to toxR and attRS; 17.12% were positive for toxR and attRS but negative for the TCP pathogenicity island; 60.95% were positive for toxR but negative for TCP and attRS; and 2.04% were positive for attRS alone but negative for toxR and TCP. All strains simultaneously positive for TCP, toxR, and attRS belonged to the O1 serogroup and were of clinical origin, whereas none of the non-O1, non-O139 strains analyzed in the present study carried the genes for TCP. In addition, three V. cholerae O1 strains isolated from seawater in Estonia and Latvia were also negative for TCP. Previous studies (29, 30, 33) have reported the absence of TCP in V. cholerae O1 or non-O1 strains which do not produce CT. The present study demonstrated the existence of TCP-positive, CT-negative V. cholerae strains, although such strains were less prevalent than TCP-negative strains. Although in the present study we were able to identify 10 isolates of V. cholerae O1 from Saudi Arabia and India which were TCP positive and CT negative, these results did not reflect the real prevalence of TCP-positive, CT-negative strains, since in this study we did not analyze a statistically defined proportion of strains from patients or environments in different countries. We assume that the real prevalence of TCP-positive, CT-negative strains should be considerably lower, taking into consideration previous studies in which no such strain was isolated from the environment or from patients (13, 29, 33). Nevertheless, the demonstration of the existence of TCP-positive, CT-negative strains in the present study is significant for understanding possible mechanisms involved in the origination of novel toxigenic strains of V. cholerae. The reason for the observed low prevalence of CT-negative but TCP-positive strains may be that these strains do not cause full-blown cholera and hence are not adequately enriched through interactions with the mammalian host. Previous studies have shown that CT-positive V. cholerae O1 strains are more enriched in the intestinal environment than the corresponding CT-negative mutants (1). An alternative explanation for the low prevalence of CT-negative but TCP-positive strains may be that such strains are rapidly converted to toxigenic strains by CTXΦ either inside the host intestine or in the environment. The toxR gene was found to be widely distributed among nontoxigenic V. cholerae strains and was present in 84.93% of the total strains tested. This was probably because ToxR is involved in the regulation of a number of other genes in addition to those encoding TCP and CT (23, 24) and may be part of a common regulatory mechanism possessed by different serogroups of V. cholerae. Although toxR and attRS sequences were shared by both O1 and non-O1 strains, the possession of genes encoding TCP was confined to strains of the O1 serogroup. This also agreed with previous studies (13, 29, 30) in which V. cholerae non-O1, non-O139 strains have been very rarely found to possess the genes for TCP.
Infection and analysis of recipient strains.
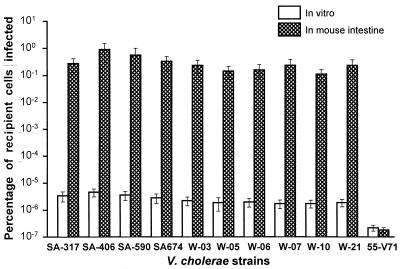
We used a genetically marked derivative of CTXΦ designated CTX-KmΦ (35) to study the susceptibility of the strains because the Kmr marker allowed us to conveniently detect and analyze infected cells. Two different methods were used to test the susceptibility of the nontoxigenic strains to CTX-KmΦ. Initially, all the strains were exposed to cell-free phage particles under in vitro laboratory conditions. Of the 146 nontoxigenic strains exposed to the phage, 10 strains belonging to the O1 serogroup and 1 non-O1, non-O139 strain were infected by the phage. However, the O1 strains which carried the TCP genes were infected more efficiently than the TCP-negative, non-O1 strain (Fig. 1).
FIG. 1.
Susceptibility of TCP-positive and TCP-negative nontoxigenic V. cholerae O1 and non-O1 strains to CTX-KmΦ under in vitro laboratory conditions and in the intestines of infant mice. Strains SA-317, SA-406, SA-590, and SA-674 are TCP-positive V. cholerae O1 clinical strains isolated in Saudi Arabia; strains W-03, W-05, W-06, W-07, W-10, and W-21 are TCP-positive V. cholerae O1 clinical strains isolated in India; strain 55-V71 is a TCP-negative non-O1, non-O139 environmental strain isolated in Bangladesh. Values represent the averages of five independent observations.
Previous reports suggested that TCP is the receptor for CTXΦ infection and the expression of TCP is a prerequisite for susceptibility to the phage (10, 35). Since TCP is known to be expressed more efficiently in vivo, we decided to use the infant mouse model to further test the susceptibility of the 11 strains which were infected in the initial screening. When the TCP-positive strains were tested in the mouse intestine, the proportion of infected cells recovered was higher than in the in vitro assay. However, in the case of the single non-O1 strain which was devoid of TCP, the infant mouse assay did not produce a higher level of infection than the in vitro assay (Fig. 1). In the present study, the proportion of infected cells recovered from both in vitro and in vivo studies possibly included progeny of infected cells as well as fresh infection of uninfected cells with phage particles derived from infected cells during the prolonged incubation period. Nevertheless, for the TCP-positive strains, the proportion of total cells carrying the phage genome was found to be significantly higher when the strains were grown inside the mouse intestine than after an equal length of incubation under laboratory conditions. Although these results supported previous reports (10, 35) that recipient strains were more efficiently infected under conditions conducive to the expression of TCP, this study also demonstrated that at least one strain which did not carry the genes for TCP was infected by the phage, albeit at a low efficiency (Fig. 1). The TCP-negative non-O1, non-O139 strain was distinctly susceptible to the phage in repeated in vitro assays, producing a small number of infected colonies, while the rest of the 135 TCP-negative strains were completely resistant to infection by the phage when assayed under identical conditions. It was not clear what determined the different phage sensitivities of these TCP-negative strains. These results indicated that in addition to the TCP-mediated mechanism, there may be a second mechanism for CTXΦ infection. In agreement with this observation, a study in India has reported the presence of CT-positive but TCP-negative V. cholerae strains belonging to non-O1 serogroups from both patients and the environment (13).
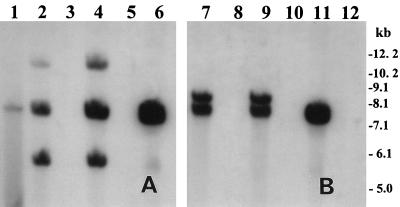
In all V. cholerae O1 strains which were infected by CTX-KmΦ, the phage genome integrated into the chromosome of the host, forming lysogens (Fig. 2). Hybridization of genomic DNA digested with BglI or BglII from infected and corresponding wild-type progenitor strains with the zot probe produced three bands for BglI and two bands for BglII digests corresponding to the number of fragments carrying the zot sequence. Since there is a single BglI site and there are no BglII sites within the zot gene (2), interpretation of the restriction patterns confirmed that the phage genome integrated into the chromosome of the infected V. cholerae cells, and two copies of the phage genome were present in tandem repeats in the host chromosome (Fig. 2). Although plasmid preparations from the freshly infected cells showed the presence of pCTX-Km, the lysogens spontaneously lost the RF of the phage genome, as confirmed by subsequent plasmid preparations (data not shown). The integrated form of the phage, however, was stable, and the lysogens did not lose the prophage when grown or stored in the absence of kanamycin. In the single non-O1, non-O139 strain, which was infected by the phage, the phage genome was maintained as the RF and produced a band corresponding to the linearized pCTX-Km (Fig. 2). This strain produced high titers of infectious phage particles in the supernatant fluids of the culture (mean, 2.7 × 106 particles/ml), as determined by titration with strain RV508 as the recipient. Previous studies (20, 35) described the induction and transmission of CTXΦ in the intestines of infant mice based on observed transduction of recipient strains. In the present study, the presence of at least one non-O1, non-O139 strain in the environment, which was capable of harboring the RF of CTXΦ and producing infectious phage particles, indicated that some environmental non-O1 strains may have a role in supporting the replication and propagation of CTXΦ in the environmental habitat. A recent demonstration of the presence of CT-positive but TCP-negative non-O1, non-O139 strains in the environment (13) supports this assumption. It is also interesting that a previous study in India reported an increased incidence of toxigenicity among freshwater isolates of V. cholerae non-O1, non-O139 strains during a cholera outbreak caused by V. cholerae O139 (12). This emphasizes the importance of further studies to understand the propagation of CTXΦ and its role in the ecology of V. cholerae.
FIG. 2.
Southern hybridization analysis of genomic DNA or plasmids isolated from V. cholerae strains infected with CTX-KmΦ and the corresponding native strains. Total DNA or plasmids were digested with BglI (A) or BglII (B) and probed with an 850-bp PCR-generated probe for the zot gene. Lane 1, pCTX-Km linearized with BglI; lanes 2 and 7, total DNA from SA-317(pCTX-Km); lanes 3 and 8, total DNA from SA-317 (native); lanes 4 and 9, total DNA from SA-406(pCTX-Km); lanes 5 and 10, total DNA from SA-406 (native); lanes 6 and 11, total DNA and plasmid, respectively, from 55V-71(pCTX-Km); lane 12, total DNA from 55V-71 (native). Integration of CTX-KmΦ DNA into the chromosome of recipient V. cholerae O1 strains SA-317 and SA-406 is shown, whereas in non-O1 strain 55V-71, the phage genome is shown to be present in the RF. Numbers indicating the molecular sizes of bands correspond to a 1-kb DNA ladder (Bethesda Research Laboratories).
Analysis of rRNA gene restriction patterns.
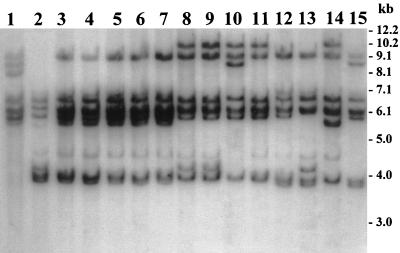
In regions where cholera is endemic, molecular analyses of V. cholerae strains collected during different epidemics have shown clonal diversity among strains and a continual emergence of new epidemic clones (6–8). In these previous studies, clonal diversity was demonstrated based on restriction fragment length polymorphisms in conserved rRNA genes. We performed rRNA gene restriction pattern analysis (ribotyping) of the new lysogens derived from nontoxigenic progenitors or the native nontoxigenic strains and compared these patterns with those of previous epidemic isolates as well as with published data on the ribotype patterns of toxigenic V. cholerae strains (4, 6–8, 26). Ribotyping was done with the restriction enzyme BglI, which was previously used by us and others for comparative analysis of ribotypes. Nontoxigenic V. cholerae O1 strains analyzed in the present study revealed four different BglI restriction patterns of their rRNA genes (Fig. 3). The four different patterns corresponded to strains collected from four countries—Estonia, Latvia, India, and Saudi Arabia—whereas all the O1 strains collected from each of these countries were clonal and produced identical restriction patterns (Fig. 3). Strains from Saudi Arabia and from India were positive for the TCP pathogenicity island and were lysogenized by the CTX-KmΦ phage, whereas those from Estonia and Latvia were negative for TCP and were resistant to the phage. The rRNA gene restriction patterns of the V. cholerae O1 strains from Estonia, Latvia, and Saudi Arabia were distinctly different from the restriction patterns of V. cholerae previously reported by us and other investigators who analyzed a large number of toxigenic V. cholerae strains isolated in different countries (4, 6–8, 26). This data provided evidence that the observed clonal diversity among epidemic strains of V. cholerae may have arisen, at least partly, due to the emergence of new toxigenic strains from nontoxigenic V. cholerae which are possibly undergoing a continuous genetic reassortment. This study also suggested that in an area where cholera is endemic, clinical and environmental surveillance for the presence of nontoxigenic V. cholerae carrying the genetic determinants for TCP and ToxR and the attRS sequence may be a means for predicting the origination of new toxigenic V. cholerae strains. The ribotype pattern of the new CTX-KmΦ lysogens derived from the nontoxigenic southern India strains in this study, however, was very similar to that of a clone of toxigenic V. cholerae strains isolated from cholera outbreaks in 1987 in Guinea-Bissau (4), situated along the West African coast. This suggested that the lysogenic conversion by CTXΦ demonstrated in the present study under laboratory conditions may indeed have been occurring efficiently in the ecological habitat.
FIG. 3.
BglI restriction patterns of rRNA genes of nontoxigenic V. cholerae O1 strains compared to those of selected toxigenic strains of V. cholerae O1 isolated from epidemic outbreaks of cholera. A Southern blot of BglI-digested genomic DNA was hybridized with the 7.5-kb BamHI fragment of the E. coli rRNA clone pKK3535. Lanes 1 and 2, TCP-negative CTX-KmΦ-resistant strains Fin-41642 and Fin-41643, isolated from seawater in Estonia and Latvia, respectively, in 1995; lanes 3 through 7, TCP-positive CTX-KmΦ-susceptible strains W-03, W-05, W-07, W-10, and W-21 respectively, isolated from patients in India in 1993; lanes 8 and 9, TCP-positive CTX-KmΦ-susceptible strains SA-317 and SA-674, respectively, isolated from patients in Saudi Arabia in 1997; lanes 10 through 15, epidemic strains of toxigenic V. cholerae isolated in different countries (the strains and their countries and years of isolation are as follows: AK-21445, Bangladesh, 1995; Syria-5, Syria, 1992; AF-9710, Bangladesh, 1990; 62-6-91, Tanzania, 1991; AG-19438, Bangladesh, 1991; and AH-806, Bangladesh, 1992). Numbers indicating molecular sizes of bands correspond to a 1-kb DNA ladder (Bethesda Research Laboratories).
Molecular basis for the emergence of potential epidemic strains.
Previous analysis of V. cholerae strains isolated from cholera epidemics has shown that in addition to genes encoding CT, these strains invariably possessed genes for the pilus colonization factor, TCP, and the regulatory gene toxR (7, 8). Hence, nontoxigenic strains capable of causing cholera outbreaks following conversion by CTXΦ should carry genes for TCP and ToxR, not only because TCP facilitates colonization inside the host intestine but also because TCP is the phage receptor in recipient cells. The presence of attRS as well as toxR in 25 of 132 non-O1 strains (18.93%) in the present study suggested that such strains were probably capable of integrating the CTXΦ genome and support the expression of the ctxAB operon, although in most cases CTXΦ would not infect such strains due to lack of the receptor, TCP. Hence, the presence of the TCP pathogenicity island remains the limiting factor for infection and conversion of nontoxigenic environmental strains by CTXΦ. The proposed TCP pathogenicity island shares several characteristics with those of other species of pathogenic bacteria. These include the presence of groups of virulence genes, a regulator of virulence genes, a transposase gene, specific (att-like) attachment sites flanking each end of the island, and an integrase with homology to a phage integrase gene (17, 19). The structure of the pathogenicity island is suggestive of horizontal transfer of the gene clusters as a possible mechanism for the origination of new pathogenic clones of V. cholerae. In the present study, all strains positive for tcpA, which encodes the major subunit of TCP, were simultaneously positive for tcpI and acfB genes, providing further evidence in favor of the pathogenicity island concept for the transfer of genes encoding TCP.
It has been suggested that the acquisition of the TCP pathogenicity island and the CTX element has allowed specific strains of V. cholerae to become adapted to the human intestinal environment (17). In the present study, the identification of V. cholerae O1 strain carrying the TCP pathogenicity island but devoid of the CTX element led us to speculate on the mechanism for the possible origin of such strains. These strains could have originated by acquisition of the TCP pathogenicity island by nontoxigenic TCP-negative O1 strains or alternatively by deletion of the complete CTX element from toxigenic TCP-positive O1 strains. Since these strains produced TCP and were likely to be infected rapidly by CTXΦ, as shown in the present study, it is highly improbable that the CT-negative, TCP-positive strains emerged from toxigenic strains by excision of the entire CTX element (the CTXΦ genome). Lazar and Waldor (20) have recently proposed that the CTXΦ genome is maintained in TCP-positive V. cholerae cells by a continuous reinfection of cells by resulting phage particles. The demonstration of the existence of V. cholerae O1 strains which were negative for both TCP and CT, and strains which were positive for TCP but negative for CT, was more consistent with the concept of origination of the later group of strains by acquisition of the TCP pathogenicity island by TCP-negative, CT-negative progenitors. However, the transmission of the TCP pathogenicity island to a recipient strain under laboratory conditions is yet to be demonstrated. The recent discovery of CTXΦ has provided an impetus to investigate the role of CTXΦ in the emergence of new toxigenic strains. In the present study, we have demonstrated for the first time lysogenic conversion of naturally occurring nontoxigenic V. cholerae strains by CTXΦ. Since TCP is the receptor for CTXΦ infection, the sequence of events that leads to the origination of new toxigenic strains should include acquisition of TCP, followed by infection and lysogenic conversion with CTXΦ. The different combinations of virulence-associated genes carried by different groups of V. cholerae strains analyzed in the present study and the demonstration of lysogenic conversion support this assumption.
Epidemic outbreaks of cholera are known to be caused by V. cholerae belonging to the O1 serogroup or the recently emerged O139 serogroup, which possibly evolved from the O1 El Tor strains by serotype-specific genetic changes (32, 36). In the present study, all the nontoxigenic strains which carried the genes for TCP and were subsequently converted by the CTX phage also belonged to the O1 serogroup. That none of the 132 non-O1, non-O139 strains analyzed carried the genes for TCP suggested that possession of the TCP pathogenicity island may be characteristic of the O1 vibrios. Since the TCP pathogenicity island is the initial genetic element required for the origination of epidemic strains, this explains why V. cholerae O1 and O139 (possibly derived from a parental O1 strain) are the only serogroups capable of causing epidemic cholera. It is not clear, however, what determines the efficient acquisition of the TCP pathogenicity island by the O1 serogroup of V. cholerae.
In the present study, most of the nontoxigenic V. cholerae O1 strains expressed TCP and hence ToxR, as evidenced by the fact that the strains were efficiently infected by CTX-KmΦ. It was not possible to study the expression of CT in the corresponding lysogens of CTX-KmΦ, since the ctxAB operon in the phage had been disrupted by the insertion of the kanamycin resistance gene. However, the native strains were isolated from outbreaks of diarrhea, and since these strains carried functional genes for TCP and ToxR, it is understandable that such strains should produce CT when infected with the wild-type CTXΦ and thus emerge as toxigenic strains capable of expressing all three major virulence determinants—ToxR, TCP, and CT—normally found in epidemic V. cholerae isolates. The present study thus demonstrated the genetic basis for the origination of new V. cholerae strains with epidemic potential from nontoxigenic strains. However, the predominance of the new strains over existing epidemic strains may involve environmental as well as host factors associated with the natural selection and enrichment of particular toxigenic clones. Further studies are required to understand the interactions of these factors in the evolution of V. cholerae strains with epidemic potential.
ACKNOWLEDGMENTS
This research was funded by the U.S. Agency for International Development (USAID) under grant HRN-5986-A-00-6005-00 with the ICDDR,B. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries.
REFERENCES
- 1.Baselski V S, Medina R A, Parker C D. In vivo and in vitro characterization of virulence-deficient mutants of Vibrio cholerae. Infect Immun. 1979;24:111–116. doi: 10.1128/iai.24.1.111-116.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudry B, Fasano A, Ketley J, Kaper J B. Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae. Infect Immun. 1992;60:428–434. doi: 10.1128/iai.60.2.428-434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brosius J, Ullrich A, Raker M A, Gray A, Dull T J, Gutell R R, Noller H F. Construction and fine mapping of recombinant plasmids containing the rRNB ribosomal RNA operon of E. coli. Plasmid. 1981;6:112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- 4.Dalsgaard A, Mortensen H F, Mølbak K, Dias F, Serichantalergs O, Echeverria P. Molecular characterization of Vibrio cholerae O1 strains isolated during cholera outbreaks in Guinea-Bissau. J Clin Microbiol. 1996;34:1189–1192. doi: 10.1128/jcm.34.5.1189-1192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everiss K D, Hughes K J, Kovach M E, Peterson K M. The Vibrio cholerae acfB colonization determinant encodes an inner membrane protein that is related to a family of signal-transducing proteins. Infect Immun. 1994;62:3289–3298. doi: 10.1128/iai.62.8.3289-3298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruque S M, Roy S K, Alim A R M A, Siddique A K, Albert M J. Molecular epidemiology of toxigenic Vibrio cholerae in Bangladesh studied by numerical analysis of rRNA gene restriction patterns. J Clin Microbiol. 1995;33:2833–2838. doi: 10.1128/jcm.33.11.2833-2838.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque S M, Ahmed K M, Alim A R M A, Qadri F, Siddique A K, Albert M J. Emergence of a new clone of toxigenic Vibrio cholerae O1 biotype El Tor displacing V. cholerae O139 Bengal in Bangladesh. J Clin Microbiol. 1997;35:624–630. doi: 10.1128/jcm.35.3.624-630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque S M, Ahmed K M, Siddique A K, Zaman K, Alim A R M A, Albert M J. Molecular analysis of toxigenic Vibrio cholerae O139 Bengal strains isolated in Bangladesh between 1993 and 1996: evidence for emergence of a new clone of the Bengal vibrios. J Clin Microbiol. 1997;35:2299–2306. doi: 10.1128/jcm.35.9.2299-2306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque S M, Comstock L, Kaper J B, Albert M J. Distribution of zonula occludens toxin (zot) gene among clinical isolates of Vibrio cholerae O1 from Bangladesh and Africa. J Diarrhoeal Dis Res. 1994;12:222–224. [PubMed] [Google Scholar]
- 10.Faruque S, Asadulghani M, Alim A R M A, Albert M J, Islam K M N, Mekalanos J J. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feinberg A, Volgelstein B. A technique for radio labelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh A R, Koley H, De D, Garg S, Bhattacharya M K, Bhattacharya S K, Manna B, Nair G B, Shimada T, Takeda T, Takeda Y. Incidence and toxigenicity of Vibrio cholerae in a freshwater lake during the epidemic of cholera caused by serogroup O139 Bengal in Calcutta, India. FEMS Microbiol Ecol. 1994;14:285–291. [Google Scholar]
- 13.Ghosh C, Nandi R K, Dasgupta S K, Nair G B, Hall R H, Ghose A C. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb Pathog. 1997;22:199–208. doi: 10.1006/mpat.1996.0105. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg I, Mekalanos J J. Effect of a recA mutation on cholera toxin gene amplification and deletion events. J Bacteriol. 1986;165:723–731. doi: 10.1128/jb.165.3.723-731.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili and ToxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaper J B, Morris J G, Jr, Nishibuchi M. DNA probes for pathogenic Vibrio species. In: Tenover F C, editor. DNA probes for infectious disease. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 65–77. [Google Scholar]
- 17.Karaolis D K, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keasler S P, Hall R H. Detection and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 19.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 20.Lazar S, Waldor M K. ToxR-independent expression of cholera toxin from the replicative form of CTXΦ. Infect Immun. 1998;66:394–397. doi: 10.1128/iai.66.1.394-397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 22.Miller V L, Mekalanos J J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci USA. 1984;81:3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsot C, Mekalanos J J. Expression of the Vibrio cholerae gene encoding aldehyde dehydrogenase is under control of ToxR, the cholera toxin transcriptional activator. J Bacteriol. 1991;173:2842–2851. doi: 10.1128/jb.173.9.2842-2851.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popovic T, Bopp C, Olsvic Ø, Wachsmuth K. Epidemiological application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabbani G H, Greenough W B. Cholera. In: Lebenthal E, Duffy M, editors. Text book of secretory diarrhea. New York, N.Y: Raven Press; 1990. pp. 233–253. [Google Scholar]
- 28.Saha P K, Koley H, Mukhopadhyay A K, Bhattacharya S K, Nair G B, Ramakrishnan B S, Krishnan S, Takeda T, Takeda Y. Nontoxigenic Vibrio cholerae O1 serotype Inaba biotype El Tor associated with a cluster of cases of cholera in southern India. J Clin Microbiol. 1996;34:1114–1117. doi: 10.1128/jcm.34.5.1114-1117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Said B, Smith H R, Scotland S M, Rowe B. Detection and differentiation of the gene for toxin co-regulated pili (tcpA) in Vibrio cholerae non-O1 using the polymerase chain reaction. FEMS Microbiol Lett. 1995;125:205–210. doi: 10.1111/j.1574-6968.1995.tb07359.x. [DOI] [PubMed] [Google Scholar]
- 30.Sharma C, Thungapathra M, Ghosh A, Mukhopadhyay A K, Basu A, Mitra R, Basu I, Bhattacharya S K, Shimada T, Ramamurthy T, Takeda T, Yamasaki S, Takeda Y, Nair G B. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 32.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor R, Shaw C, Peterson K, Spears P, Mekalanos J. Safe, live Vibrio cholerae vaccines? Vaccine. 1988;6:151–154. doi: 10.1016/s0264-410x(88)80019-7. [DOI] [PubMed] [Google Scholar]
- 34.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 36.Waldor M K, Mekalanos J J. ToxR regulates virulent gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.World Health Organization. World Health Organization guidelines for the laboratory diagnosis of cholera. Geneva, Switzerland: Bacterial Disease Unit, World Health Organization; 1974. [Google Scholar]