Significance
Lipid droplet (LD) accumulation is common in clear cell renal cell carcinoma (ccRCC), providing survival advantage and association with aggressiveness. However, the mechanisms controlling their biogenesis remain poorly known. Understanding their formation is crucial for developing targeted strategies. We show that MYC inhibition in ccRCC resulted in increased triglyceride accumulation in LDs in a glutamine-dependent process driven by enhanced HILPDA expression. Downregulation of HILPDA reduced tumor growth, and it was only highly expressed in ccRCC in comparison with other tumors or healthy renal tissue. Collectively, we highlight the role of MYC expression and glutamine metabolism in LD biogenesis and propose HILPDA as a specific target for precision medicine, providing an attractive therapeutic approach for this aggressive malignancy.
Keywords: clear cell renal cell carcinoma, lipid droplets, HILPDA, MYC, glutamine
Abstract
Metabolic reprogramming is critical during clear cell renal cell carcinoma (ccRCC) tumorigenesis, manifested by accumulation of lipid droplets (LDs), organelles that have emerged as new hallmarks of cancer. Yet, regulation of their biogenesis is still poorly understood. Here, we demonstrate that MYC inhibition in ccRCC cells lacking the von Hippel Lindau (VHL) gene leads to increased triglyceride content potentiating LD formation in a glutamine-dependent manner. Importantly, the concurrent inhibition of MYC signaling and glutamine metabolism prevented LD accumulation and reduced tumor burden in vivo. Furthermore, we identified the hypoxia-inducible lipid droplet–associated protein (HILPDA) as the key driver for induction of MYC-driven LD accumulation and demonstrated that conversely, proliferation, LD formation, and tumor growth are impaired upon its downregulation. Finally, analysis of ccRCC tissue as well as healthy renal control samples postulated HILPDA as a specific ccRCC biomarker. Together, these results provide an attractive approach for development of alternative therapeutic interventions for the treatment of this type of renal cancer.
Clear cell renal cell carcinoma (ccRCC) is the most common and aggressive subtype of kidney cancer (1). Current treatment includes kidney resection and targeted therapies, but tumors often acquire resistance and combination strategies have proved toxic (2). Better understanding of the mechanisms underlying this malignancy is crucial for the development of new targeted therapeutic approaches that overcome these limitations.
The von Hippel-Lindau (VHL) gene, controlling proteasomal degradation of hypoxia-inducible factors (HIFs) in the presence of oxygen, is lost in approximately 90% of all ccRCC cases (3). Silencing of VHL by loss of chromosome 3p, followed by mutation or methylation of the remaining allele, results in constitutive activation of HIFs, inducing transcription of genes involved in metabolism, angiogenesis, and metastasis (4). Moreover, the MYC oncogene is amplified in 5 to 10% and overexpressed in 20% of ccRCC, associated with aggressiveness (5, 6), In addition, the majority of ccRCC present with alterations in downstream signaling pathways controlled by MYC (5).
Metabolic reprogramming is a major characteristic of ccRCC (7), in part due to activation of partially overlapping transcriptional and metabolic programs controlled by HIF and MYC. Both oncoproteins activate glycolysis and increase glutamine consumption in cancer cells (8, 9). HIF1-α decreases flux of glucose-derived carbons into the tricarboxylic acid (TCA) cycle (10), promoting reductive carboxylation of glutamine, resulting in the synthesis of fatty acids, which can accumulate in lipid droplets (LDs) (11). These are organelles composed of a core rich in triglycerides (TGs) and sterol esters, surrounded by a phospholipid monolayer (12). In recent years, they have emerged as regulators of tumorigenesis, controlling lipid metabolism, signaling and trafficking, as well as redox homeostasis and autophagy, supporting cancer cell survival (13). The presence of LDs can be considered as a biomarker for aggressive disease and a potential target for cancer treatment (14). However, the mechanisms and factors impacting LD accumulation in cancer cells are still poorly understood.
We have previously demonstrated that inhibition of MYCN, one member of the MYC family, induces LD accumulation in neuroblastoma cells via disruption of fatty acid β-oxidation (15). Here, using a combination of lipidomics, metabolic tracing, and xenograft studies, we found that inhibiting MYC in cells with loss of VHL enhanced accumulation of TGs in LDs. Furthermore, impairing glutamine metabolism using BPTES combined with MYC inhibition reduced tumor burden and prevented LD accumulation in vivo. This process is regulated by increased expression of the hypoxia-inducible lipid droplet–associated protein (HILPDA) under MYC control. Moreover, downregulation of HILPDA resulted in reduced ccRCC cell proliferation, LD formation, and tumor burden in vivo, while its overexpression promoted accumulation of lipids. We identified HILPDA as a potential vulnerability in ccRCC since it is expressed in tumoral renal tissue but not in kidney samples from healthy individuals.
Together, these data demonstrate that the metabolic reprogramming induced by HIF and MYC, as manifested by LD formation, confers ccRCC cells with a pathway for regulating lipid trafficking for survival. Identification of HILPDA as a specific marker for ccRCC offers future opportunities to selectively target this renal cancer.
Results
Lipid Droplet Accumulation in ccRCC Is Dependent on VHL and MYC Status.
We previously showed that MYCN inhibition results in LD accumulation in neuroblastoma (15). To examine whether other cancer types respond similarly, we performed a comprehensive screening in around 60 well-established human cancer cell lines from different tumor types (SI Appendix, Table S1). All cell lines were treated with three MYC inhibitors (MYCis), 10058-F4 (F4), 10074-G5 (G5), or JQ1, followed by evaluation of the presence and/or induction of LDs. Many cell lines accumulated LDs upon MYC inhibition independently of cancer type or inhibitor (SI Appendix, Fig. S1A). The most striking difference observed was between the RCC4 subclones: While RCC4 cells lacking VHL (VHL−) accumulated large amounts of lipids, no LDs were detected upon MYC inhibition in RCC4 cells with restored VHL expression (VHL+), indicating that loss of VHL could be a prerequisite for this phenotype (SI Appendix, Fig. S1B and Table S1). Hence, we focused on analyzing the molecular mechanism underlying the effect of MYCis on LD accumulation in ccRCC.
Clear cell renal cell carcinoma cells are characterized by the loss of VHL, a regulator of HIF stability in the presence of oxygen (3). An inverse correlation between VHL and HIF1-α and/or HIF2-α expression was observed in RCC4 VHL+ and VHL− cells (SI Appendix, Fig. S1C). MYC was strongly down-regulated after JQ1 treatment in VHL− cells and with G5 and JQ1 in VHL+ cells. In contrast, no reduction in MYC was observed after treatment with F4 or in VHL− cells incubated with G5. Both F4 and G5 are MYC:MAX dimerization inhibitors and their mechanism of action does not necessarily imply a reduction in MYC levels (16), while the bromodomain and extraterminal domain inhibitor JQ1 reduces MYC transcription as well as expression of additional genes (17). Lipid stainings using Oil-red O were validated with Lipid Droplet–Benzothiadiazole 1 (LD-BTD1) (SI Appendix, Fig. S1D). This fluorophore specifically stains LDs without affecting cell viability (18), or impacting either MYC or HIF1-α expression (SI Appendix, Fig. S1E).
Inhibition of MYC resulted in a reduction in cell numbers. To study this effect, RCC4 cells were incubated with MYCis at different concentrations. RCC4 VHL− cells showed a significantly smaller reduction in proliferation with a correspondingly higher IC50 value than RCC4 VHL+ cells (SI Appendix, Fig. S1F). Importantly, the concentrations chosen (60 μM F4, 40 μM G5, or 5 μM JQ1) were all below their IC50 values, except in RCC4 VHL+ cells, where it was slightly higher for JQ1.
Next, we analyzed two additional ccRCC cell pairs, 786-O and A498 (VHL+ and VHL−), which express HIF2-α but not HIF1-α. Both 786-O and A498 VHL− cells accumulated LDs after MYC inhibition (Fig. 1A and SI Appendix, Fig. S2A). Moreover, cells expressing VHL showed downregulation of HIF2-α, and MYC levels were robustly decreased upon JQ1 exposure in 786-O VHL+ and VHL− cells, as well as after G5 treatment in A498 VHL+ and VHL− cells (Fig. 1B and SI Appendix, Fig. S2B). 786-O VHL− cells also showed a small reduction in proliferation and thus a higher IC50 value than VHL+ cells (SI Appendix, Fig. S2C).
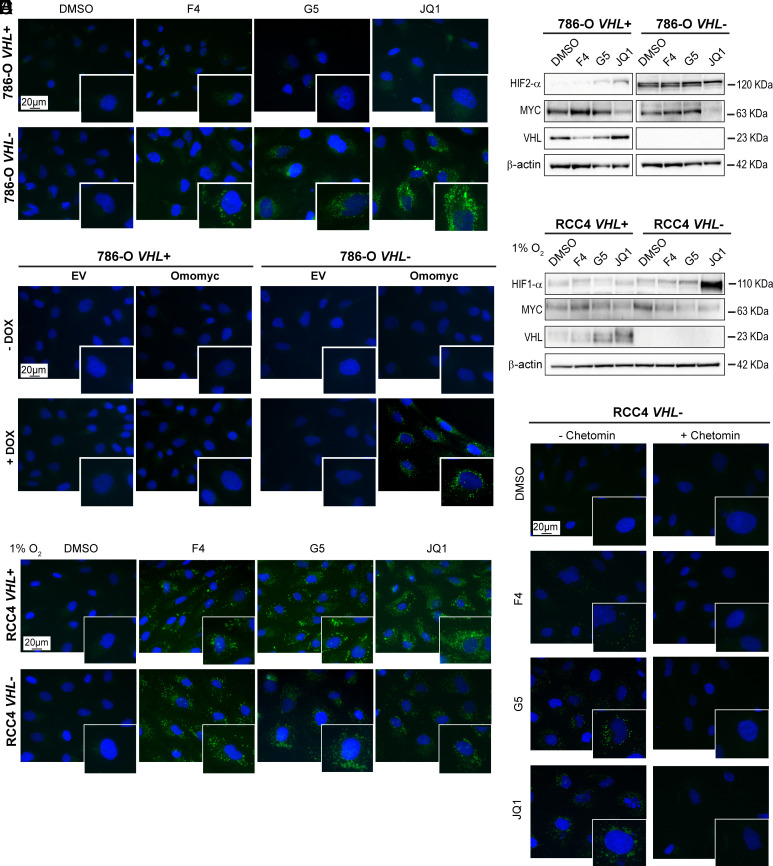
Fig. 1.
HIF stabilization is required for LD accumulation upon MYC inhibition in ccRCC cells. (A) Lipid droplet staining and (B) western blot of the indicated proteins in 786-O VHL+ and VHL− cells after MYC inhibition. (C) Lipid droplet staining in 786-O VHL+ EV, 786-O VHL+ Omomyc, 786-O VHL- EV, or 786-O VHL- Omomyc cells after vehicle (−Omomyc, H2O) or DOX treatment (+Omomyc). (D) Western blot of the indicated proteins in RCC4 VHL+ and VHL− cells after treatment with MYCis in 1% O2 (hypoxia). (E) Lipid droplet staining in RCC4 VHL+ and VHL− cells after treatment with MYCis in 1% O2 (hypoxia). (F) Lipid droplet staining in RCC4 VHL+ and RCC4 VHL− cells after treatment with MYCis in combination with DMSO (−chetomin) or chetomin. For western blots, β-actin was used as loading control. Molecular weight markers are shown to the Right. For fluorescence images: blue, DAPI; green, LD-BTD1 (LDs). (Scale bars: 20 μm.) All results are representative of three independent replicates.
To exclude that the effect on LD accumulation induced by the MYCis was caused by off-target effects, we silenced MYC by shRNA. MYC silencing was accompanied by increased LD formation compared to the empty vector control (shEV) in transduced RCC4 VHL− cells, while no effect was observed in VHL+ cells (SI Appendix, Fig. S2 D and E). We next induced the expression of Omomyc, a dominant negative peptide inhibiting the transcriptional activity of MYC (19). We generated doxycycline (DOX)-inducible 786-O VHL+ and VHL− Omomyc cells, as well as control cells (786-O VHL+ and VHL− EV) carrying RFP for tracing. Induction of Omomyc was confirmed by the presence of RFP in the nuclei, while RFP was in the cytoplasm in EV control cells (SI Appendix, Fig. S2F). Omomyc preferably forms inactive dimers with either MAX or itself, blocking MYC binding sites in promoters of target genes (19). While Omomyc expression did not affect MYC levels (SI Appendix, Fig. S2G), LDs were still accumulated in 786-O VHL- Omomyc cells (Fig. 1C).
Together, our results showed that LDs accumulated following pharmacological or genetic MYC inhibition only in ccRCC cells with VHL loss.
Stabilization of HIF Is essential for Lipid Deposition.
Given that lipid accumulation was only observed in VHL− cells, we asked whether this would occur when HIF expression was induced in VHL+ cells. To this end, cells were cultured at 1% O2 resulting in stabilization of HIFs in RCC4 and 786-O VHL+ cells (Fig. 1D and SI Appendix, Fig. S3A), and correlating with LD formation upon MYC inhibition (Fig. 1E and SI Appendix, Fig. S3B). Similar results were obtained in A498 VHL+ and VHL− cells (SI Appendix, Fig. S3 C and D). Furthermore, inhibition of the interaction of HIFs with their transcriptional coactivators p300 and cAMP response element binding (CREB) binding protein (CBP) using chetomin (20), prevented LD formation when combined with MYCis in RCC4 VHL− cells (Fig. 1F).
Collectively, induction of HIFs in the presence of MYCis led to increased lipid accumulation in both VHL+ and VHL− ccRCC cells, a process prevented by HIF inhibition.
Glutamine Deprivation Prevents Lipid Droplet Formation.
Fatty acids stored in LDs can be obtained from the extracellular milieu or synthesized from glucose or glutamine. To identify the fuel used for LD formation, cells were cultured with reduced concentrations of either of these three nutrients. We first cultured RCC4 and 786-O VHL+ and VHL− cells in either complete, delipidized, or a mixture of both media in different proportions. Viability decreased in all cells as the percentage of delipidized serum increased, with a drastic reduction with 100% delipidized medium (SI Appendix, Fig. S4A). Thus, we incubated cells in a 30:70 mixture of complete and delipidized serum in the presence of MYCis, but decreasing lipid content did not affect LD accumulation in either RCC4 or 786-O VHL− cells (SI Appendix, Fig. S4B).
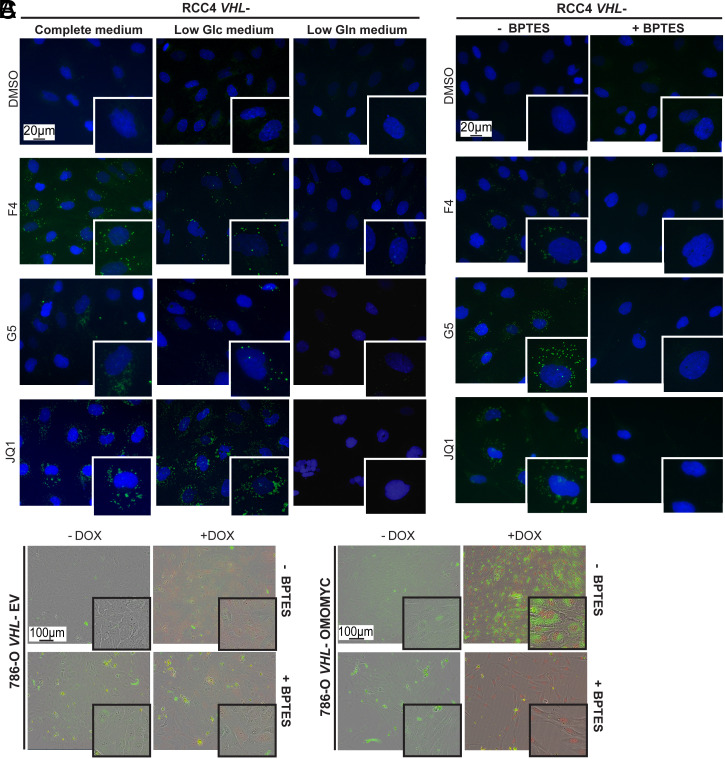
Low glucose medium significantly diminished the viability of RCC4 and 786-O cells, with a higher impact in RCC4 VHL+ cells, resulting in a 50% decrease. In addition, low glutamine medium also significantly reduced viability by 20% in VHL+ and 24% in VHL− RCC4 cells, as well as by 34% in VHL− and 50% in VHL+ 786-O cells (SI Appendix, Fig. S4C). Low glucose conditions did not impact LD formation either during normoxia or hypoxia, while low glutamine prevented LD accumulation in normoxia in RCC4 and 786-O VHL− cells, as well as in RCC4 VHL+ and VHL− cells during hypoxia. This effect was especially evident upon JQ1 incubation (Fig. 2A and SI Appendix, Figs. S4D and S5A).
Fig. 2.
Low glutamine levels or inhibition of glutaminase prevent LD formation. (A) Lipid droplet staining in RCC4 VHL− cells after culture in complete, low glucose (Low Glc) or low glutamine (Low Gln) medium treated with MYCis. (B) Lipid droplet staining in RCC4 VHL− cells after treatment with DMSO or MYCi alone or in combination with BPTES. Blue: DAPI; green: LD-BTD1 (LDs). (C) Bright-field images of live 786-O VHL- EV cells treated with vehicle (-DOX), DOX, or/and BPTES, and 786-O VHL- Omomyc cells treated with vehicle (-DOX), DOX (+Omomyc), or/and BPTES. RFP (red) upon DOX induction. (Scale bars: 20 or 100 μm.) Images are representative of three independent replicates.
Together, these data showed that LD formation relied on glutamine availability, while reduced levels of fatty acids or glucose did not have any significant impact.
Disrupting Glutamine Metabolism Inhibits Lipid Storage.
Our data suggested an important role of glutamine in LD formation, and consequently, we investigated the effect of impairing its metabolism. Glutamine is converted to glutamate by glutaminase (GLS) and is further metabolized to α-ketoglutarate (α-KG), generating citrate via reductive carboxylation. This results in production of cytosolic acetyl-CoA, supporting fatty acid synthesis when HIF is constitutively expressed as well as during hypoxia (21). We thus employed L-DON and BPTES, two well-known GLS inhibitors (22, 23). Formation of LDs was significantly reduced upon combination of MYCis together with either of the two GLS inhibitors in RCC4 VHL− cells (Fig. 2B and SI Appendix, Fig. S5B). Since L-DON also can inhibit other amino acid transporters and transglutaminases (24), we focused on studying the effect of BPTES. The combination of MYCi and BPTES during hypoxia prevented LD accumulation in both RCC4 cell lines (SI Appendix, Fig. S6A). Furthermore, LD formation was robustly induced by Omomyc in 786-O VHL− Omomyc cells but blocked by exposure to BPTES (Fig. 2C).
Treatment with BPTES alone or in combination with JQ1 did not impact proliferation significantly either in 786-O VHL+ EV or Omomyc cells. In contrast, BPTES reduced proliferation in 786-O VHL− EV cells, and the combination with Omomyc further decreased the number of cycling cells (SI Appendix, Fig. S6B).
Since our data suggested that glutamine contributed to fatty acid synthesis, we assessed LD formation upon treatment with inhibitors of two key enzymes in the fatty acid synthesis pathway, TOFA targeting acetyl-CoA carboxylase A (ACACA), and UB006 inhibiting fatty acid synthase (FASN) (25, 26). Neither of these inhibitors induced LD formation alone, while in combination with MYCis, both prevented their appearance, indicating that de novo synthesis of fatty acid was important for LD accumulation (SI Appendix, Fig. S6C).
Altogether, these data demonstrated that glutamine was essential for fatty acid synthesis and survival after MYC inhibition.
MYC Inhibition Manifests in Accumulation of Different Lipid Species in VHL+ Versus VHL− cells.
To investigate whether glutamine-derived carbons were directed to fatty acid synthesis, we next performed stable isotope tracing experiments in RCC4 VHL+ and VHL− cells using U-13C5-glutamine alone or together with U-13C6-glucose following treatment with JQ1. MYC inhibition reduced the contribution of both glucose and glutamine to the cytosolic acetyl-CoA and palmitate pools, indicated by a shift when both substrates were used together. Moreover, glucose was the preferred substrate to produce palmitate in RCC4 VHL+ cells (SI Appendix, Fig. S7A). Treatment with JQ1 slightly reduced the contribution of both glucose and glutamine into acetyl-CoA in RCC4 VHL− cells. However, no alteration in glutamine-derived acetyl-CoA occurred upon MYC inhibition, suggesting that JQ1 reduced the contribution of glucose to the acetyl-CoA pool. Notably, glutamine-derived carbons were used for palmitate synthesis to a far greater extent in VHL− than in VHL+ cells (Fig. 3A).
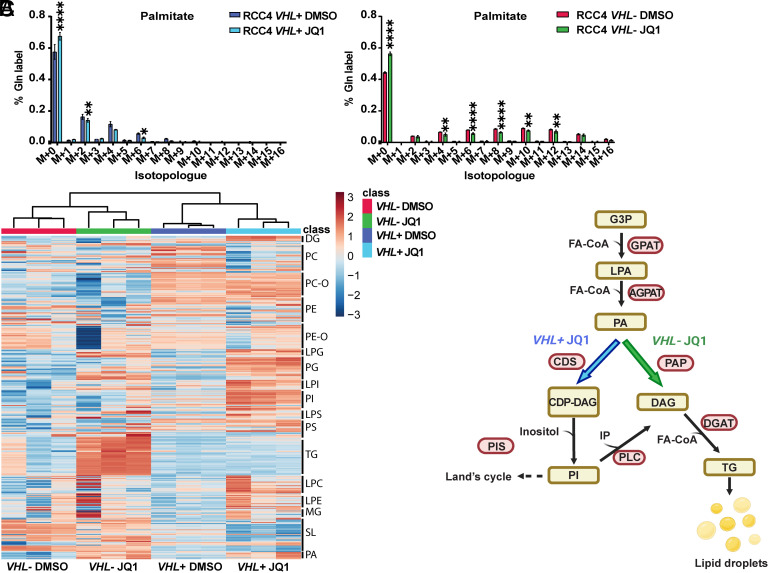
Fig. 3.
Lipidomic analysis reveals different lipid species content in RCC4 VHL+ versus VHL- cells. (A) Mass isotopologue distribution of palmitate using U-13C5-glutamine in RCC4 VHL+ and VHL− cells after treatment with JQ1. Data are presented as mean ± SD. Statistical analysis: two-way ANOVA with *, **, and **** indicating P < 0.05, <0.01, and <0.0001, respectively. (B) Heatmap with different lipid species in RCC4 VHL+ or VHL− cells after DMSO or JQ1 treatment, with red up-regulated and blue down-regulated lipid species. DG/DAG: diacylglycerides, PC: phosphatidylcholine, PC-O: phosphatidylcholine-O, PE: phosphatidylethanolamine, PE-O: phosphatidylethanolamine-O, LPG: lysophosphatidylglycerol, PG: phosphatidylglycerol, LPI: lysophosphatidylinositol, PI: phosphatidylinositol, LPS: lipopolysaccharide, PS: phosphatidylserine, TG: triglycerides, LPC: lysophosphatidylcholine, LPE: lysophosphatidylethanolamine, SL: saccharolipids, PA: phosphatidic acid. (C) Graphical representation of the different lipid species accumulated and the key enzymes in lipid metabolism upon JQ1 treatment in RCC4 VHL+ (purple) and VHL− (green). Image created with Biorender.com. G3P: glyceraldehyde 3-phosphate, FA-CoA: fatty acyl-CoA, LPAT: lysophosphatidyl acyltransferase, LPA: lysophosphatidic acid, A/GPAT: acyl/glycerol-3-phosphate acyltransferase, PAP: phosphatidic acid phosphatase, CDP-DAG: cytidine diphosphate-diacylglycerol, CDS: CDP-DAG synthase, PIS: phosphatidylinositol synthase, IP: inositol phosphate, DGAT: diacylglycerol acyltransferase. Lipidomic experiments were performed in three independent replicates.
Next, we analyzed the effect of MYC on the cellular lipidome and observed that VHL− compared to VHL+ cells exhibited distinct lipid composition (Dataset S1). Triglycerides (TGs) and sterol esters are the most abundant lipids comprising the neutral core of LDs. Indeed, our analysis revealed that TGs were more abundant in RCC4 VHL− cells treated with JQ1 compared to their control as well as to JQ1-treated or control RCC4 VHL+ cells (Fig. 3B and SI Appendix, Fig. S7B). In contrast, RCC4 VHL+ cells incubated with JQ1 resulted in a higher abundance of glycerophospholipids [phosphatidyl inositol (PI), phosphatidylserine (PS), and phosphatidyl-glycerol (PG)], as well as lysophospholipids [lysophosphatidylinositol (LPI), lysophosphatidylserine (LPS), and lysophosphatidylglycerol (LPG)], compared to vehicle-treated or to VHL− cells with any of the treatments (Fig. 3B and SI Appendix, Fig. S7C). Moreover, the levels of phosphatidic acid (PA), an intermediate of glycerolipid synthesis, were increased upon JQ1 exposure compared to controls in both cell lines (Fig. 3B). These data suggested that MYC inhibition increased lipid synthesis in both cell types while only VHL− cells subsequently converted PA into TGs. Instead, VHL+ cells enhanced the synthesis of membrane phosphoglycerides, explaining the differences in LD accumulation (Fig. 3 B and C and SI Appendix, Fig. S7 B and C).
Taken together, we found that RCC4 VHL− cells used glutamine to produce acetyl-CoA for lipid synthesis, while RCC4 VHL+ cells employed glucose. Importantly, MYC inhibition induced TG accumulation only in VHL− cells, explaining the difference in LD abundance between VHL+ and VHL− ccRCC cells.
Combined Inhibition of MYC Signaling and Glutamine Metabolism Reduces Tumor Growth.
To analyze the relevance of combined MYC and GLS inhibition on LD accumulation as well as on tumor growth in vivo, we injected 786-O VHL− Omomyc cells into the right flank of NMRI-Foxn1nu mice. Animals were administered water alone (−Omomyc) or supplemented with 1 mg/mL DOX, (+Omomyc) and injected daily with either vehicle (−BPTES) or with 12.5 mg/mL/kg BPTES (+BPTES) (SI Appendix, Fig. S8A). Compared to control-treated mice (−Omomyc −BPTES), tumor growth was reduced in all three conditions, most evidently in the combination of +Omomyc +BPTES (Fig. 4 A and B and SI Appendix, Fig. S8 B and C). Mouse weight was monitored daily, showing a normal increase throughout the study (SI Appendix, Fig. S8D).
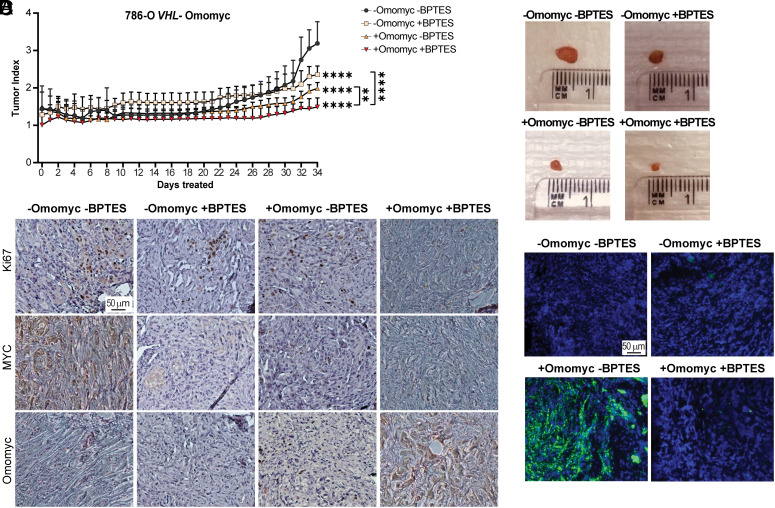
Fig. 4.
Combined inhibition of MYC signaling and glutamine metabolism results in reduced tumor growth. (A) Tumor index (volume each day/initial volume) in mice with tumors from 786-O VHL− Omomyc cells divided into four groups: −Omomyc −BPTES (n = 6), −Omomyc +BPTES (n = 6), +Omomyc −BPTES (n = 8), or +Omomyc +BPTES (n = 7). Data are presented as mean ± SD. Statistical analysis: two-way ANOVA with **, and **** indicating P < 0.01 and <0.0001, respectively. (B) Representative images of tumors at end point. (C) Immunohistochemistry analysis of tumor sections stained with anti-Ki67, anti-c-MYC, or anti-Omomyc. (Scale bar: 50 μm.) (D) Tumor sections stained with LD-BTD1 (green) for LD visualization. Blue: DAPI. (Scale bar: 50 μm.) Results in C and D are representative of at least three tumor sections.
In accordance, vehicle-treated mice expressed higher levels of the proliferation marker Ki67 compared to animals administered with +BPTES or in which +Omomyc expression was induced alone, while it barely was detected in tumors from the +Omomyc +BPTES combination (Fig. 4C and SI Appendix, Fig. S9A). Tumors expressing Omomyc showed reduced MYC levels, although interestingly, BPTES treatment slightly up-regulated its expression. As expected, tumors from PBS or non-treated mice showed high levels of both Ki67 and MYC, and no Omomyc expression (SI Appendix, Fig. S9B). TUNEL staining for apoptotic cells did not reveal any differences in cell death between conditions (SI Appendix, Fig. S9C).
Although LDs were present in tumors from all treatment groups, these were confined to the edges probably due to the presence of fat cells surrounding the tumors, or restricted to the hypoxic inner cores. In contrast, LD accumulation was robust upon MYC inhibition in tumors from the +Omomyc −BPTES group, where glutamine metabolism was intact (Fig. 4D and SI Appendix, Fig. S9D). Since corn oil was used as vehicle, we analyzed whether LD formation was triggered by the added fat. However, tumors from PBS− or non-treated mice showed a similar LD content compared to oil-injected mice, demonstrating that this vehicle did not impact LD accumulation under these conditions (SI Appendix, Fig. S9 E and F).
Collectively, MYC inhibition by Omomyc induced LD formation in VHL-deficient ccRCC cells also in vivo, while concomitant Omomyc expression together with inhibition of glutamine metabolism prevented their formation and further attenuated tumor burden.
Lipid Droplet–associated Protein Is Up-regulated in Cells with VHL Loss.
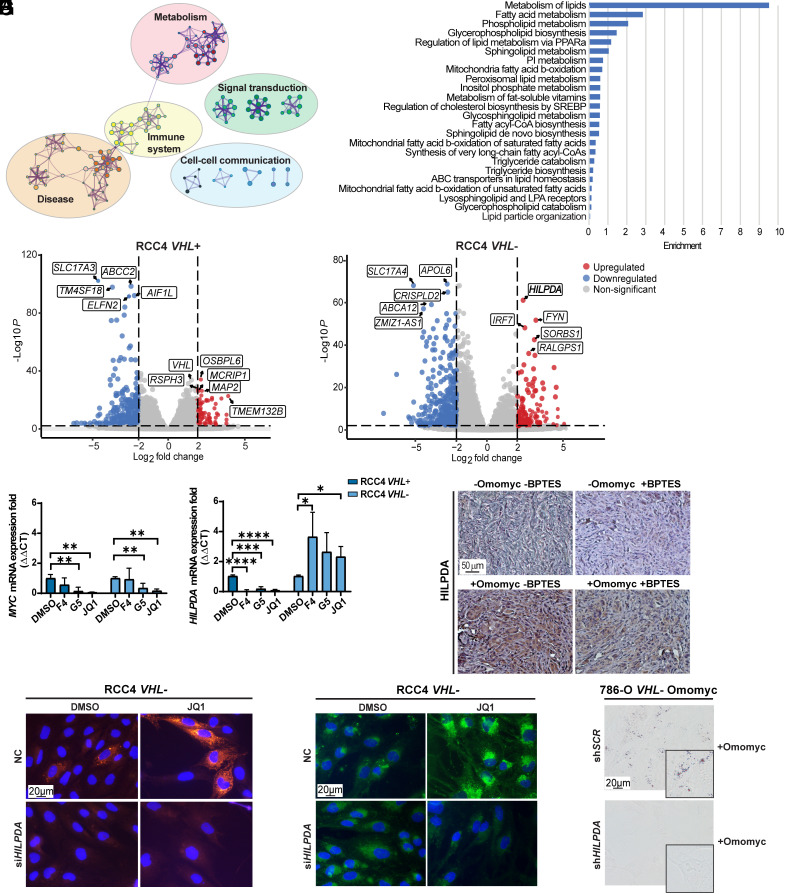
Subsequently, we performed bulk RNA sequencing (RNA-seq) analyses to identify the molecular mechanism resulting in LD accumulation after MYC inhibition in HIF-expressing cells. We detected 515 down-regulated and 308 up-regulated genes comparing JQ1 versus control in RCC4 VHL- cells, while RCC4 VHL+ cells displayed 629 down-regulated and 215 up-regulated genes (for all, log2 fold change < −2 or >2, adjusted P-value < 0.05) in response to JQ1. Using the Reactome database we identified the processes in which these genes are involved (Fig. 5A and SI Appendix, Fig. S10A) and found several related to lipid metabolism (Fig. 5B and SI Appendix, Fig. S10B). For instance, “lipid particle organization” was altered (Fig. 5B), and strikingly, the hypoxia-inducible lipid droplet–associated (HILPDA) gene was one of the most significantly up-regulated genes when inhibiting MYC in RCC4 VHL− cells (log2 fold change 2.31, adjusted P-value 3.09 × 10−62) (Fig. 5C and SI Appendix, Fig. S10C). In contrast, no differences in HILPDA or in other genes participating in lipid accumulation were detected in VHL+ cells. Notably, expression of HILPDA was significantly down-regulated upon MYC inhibition in RCC4 VHL+ cells whereas it remained robustly expressed in VHL− cells (Fig. 5D). Next, we analyzed the presence of HILPDA in the tumors from the in vivo experiment and observed its expression in all samples, while only strongly increased in tumors where MYC was inhibited (+Omomyc −BPTES and +Omomyc +BPTES) (Fig. 5E and SI Appendix, Fig. S10 D and E). Interestingly, HILPDA levels were lower in +Omomyc +BPTES compared to +Omomyc −BPTES tumors, even though MYC was down-regulated in both. Expression of HILPDA mRNA and protein was also reduced in vitro in +Omomyc +BPTES compared to +Omomyc -BPTES conditions (SI Appendix, Fig. S10 F and G), while MYC protein levels were increased in cells upon BPTES treatment, which could explain the lower HILPDA levels (SI Appendix, Fig. S10 G and H).
Fig. 5.
The HILPDA gene is up-regulated in RCC4 VHL− cells after MYC inhibition. (A) Visualization of functional enrichment maps in JQ1− versus DMSO-treated RCC4 VHL− cells (n = 3). The nodes represent significantly enriched genes participating in indicated processes. (B) Bar plot of the most significantly changed lipid metabolic processes in JQ1− versus DMSO-treated RCC4 VHL− cells. (C) Volcano plot (P < 0.05, horizontal dotted line) of the log2 fold change (<−2, >+2, vertical dotted lines) of expression of genes between JQ1− and DMSO-treated RCC4 VHL+ cells (Left plot) and RCC4 VHL− cells (Right plot). The five most down-regulated (blue) and up-regulated (red) genes in each cell line are boxed. Not significantly changed genes are depicted in gray. (D) RT-qPCR of MYC and HILPDA in RCC4 VHL+ and VHL− cells. B2M was used as housekeeping gene. Data are presented as mean ± SD (n = 3). Statistical analysis: one-way ANOVA with *, **, ***, and **** indicating P < 0.05, <0.01, <0.001, and <0.0001, respectively. (E) Immunohistochemistry analysis of tumor sections stained with anti-HILPDA. (Scale bar: 50 μm.) (F) Immunofluorescence staining of HILPDA and (G) lipid droplet staining in RCC4 VHL− negative control (NC) and siHILPDA cells after treatment with DMSO or JQ1. Green: LD-BTD1; red: HILPDA; blue: DAPI. (Scale bar: 20 μm.) (H) Bright-field images of lipid droplet staining with Oil-red O (red) in 786-O VHL− Omomyc shSCR and shHILPDA cells upon Omomyc induction. (Scale bar: 20 μm.) Images in E–H are representative of three independent replicates/sections.
HIF1-α induces expression of HILPDA, inhibiting the release of fatty acids from LDs via inhibition of adipose triglyceride lipase (ATGL), promoting their accumulation during hypoxia (27). We therefore interrogated whether MYC would also regulate HILPDA expression. Using the ENCODE database containing public data from chromatin immunoprecipitation sequencing (ChIP-seq) we found a compelling binding peak for MYC, indicating a direct interaction at the HILPDA promoter (SI Appendix, Fig. S10I). The expression of HILPDA increased following MYC inhibition (Fig. 5 C and D), indicating that while HIF1-α induced HILPDA expression, MYC might instead repress this gene. MYC-mediated repression is frequently achieved via interaction with the MYC-interacting-zinc finger 1 (MIZ-1) protein forming a ternary repressive complex with MAX on initiator (INR) sequences at the promoters of target genes (28). In support, we confirmed MIZ-1, MYC as well as MAX binding to the HILPDA promoter using ENCODE data (SI Appendix, Fig. S10I).
Collectively, our results demonstrated that HILPDA is up-regulated upon combined HIF activation and MYC inhibition correlating with LD accumulation in VHL− cells.
Selective Expression Indicates HILPDA As A New Vulnerability in ccRCC.
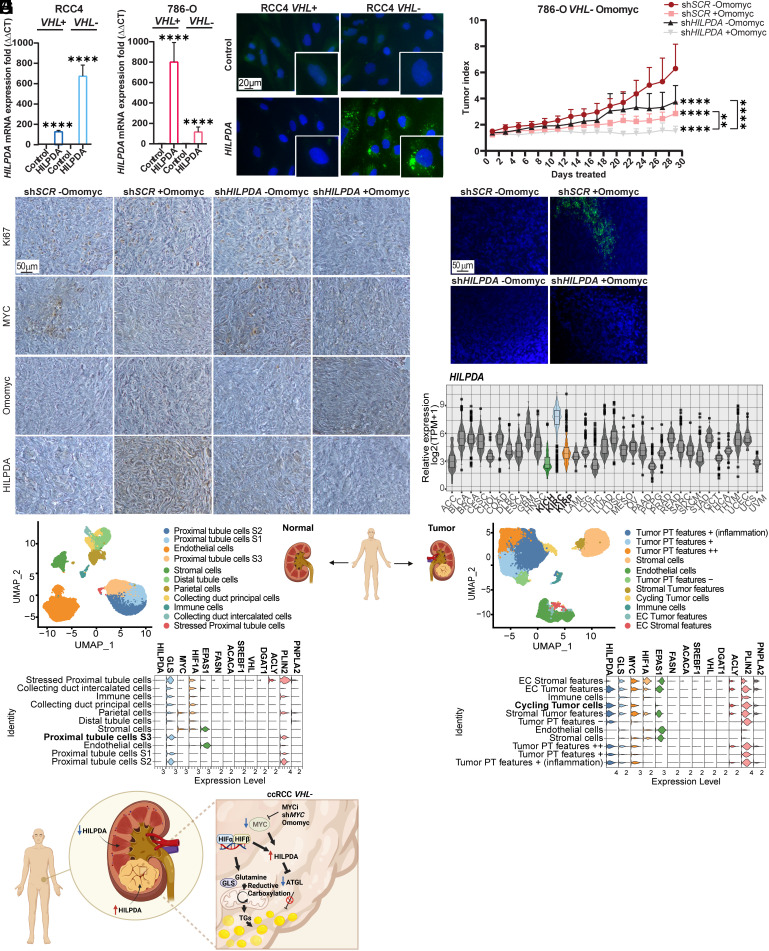
To investigate whether HILPDA expression was required for inducing LD accumulation upon MYC inhibition, we employed siRNA for its downregulation (Fig. 5F and SI Appendix, Fig. S11A), and observed that LD formation was prevented, and proliferation reduced in RCC4, 786-O, and A498 VHL− cells (Fig. 5G and SI Appendix, Fig. S11 B and C). We further generated stable 786-O VHL− Omomyc cells with constitutive HILPDA knockdown (shHILPDA) or control scramble cells (shSCR). The levels of HILPDA were down-regulated by 95% in 786-O VHL− Omomyc shHILPDA cells compared to the corresponding shSCR cells (SI Appendix, Fig. S11D) and correlated as expected with the absence of LD accumulation (Fig. 5H). In contrast, overexpression of HILPDA did not influence proliferation except for a small increase in RCC4 VHL+ cells (Fig. 6A and SI Appendix, Fig. S11E), while promoting LD formation in RCC4 and 786-O VHL− cells (Fig. 6B and SI Appendix, Fig. S11F).
Fig. 6.
Identification of HILPDA as a biomarker for ccRCC. (A) RT-qPCR of HILPDA in RCC4 and 786-O VHL+ and VHL− control or HILPDA overexpressing cells. B2M was used as housekeeping gene. Data are presented as mean ± SD (n = 3). Statistical analysis: t test with **** indicating P < 0.0001. (B) Lipid droplet staining in RCC4 VHL+ and VHL− control and HILPDA overexpressing cells. Green: LD-BTD1, blue: DAPI. (Scale bar: 20 μm.) Images are representative of three independent replicates. (C) Tumor index (volume each day/initial volume) in mice with tumors derived from 786-O VHL− Omomyc shSCR or shHILPDA cells divided into two groups: −Omomyc (n = 8 and n = 7) or +Omomyc (n = 6 for both cell lines). Data are presented as mean ± SD. Statistical analysis: two-way ANOVA with ** and **** indicating P < 0.01 or <0.0001, respectively. (D) Immunohistochemistry analysis of tumor sections stained with anti-Ki67, anti-c-MYC, anti-Omomyc and anti-HILPDA. (Scale bar: 50 μm.) (E) Tumor sections stained with LD-BTD1 (green) for LD visualization. Blue: DAPI. (Scale bar: 50 μm.) Images in D and E are representative of at least three independent tumors per condition. (F) Relative expression of HILPDA across different tumors, including the three renal cell carcinomas: clear cell (KIRC) blue, chromophobe (KICH) green, and papillary (KIRP) orange. (G) Single-cell RNA-seq analysis of normal (Left panel) or tumor (Right panel) renal tissue. The upper graphs represent dimensional reduction plots (UMAP). The lower graphs indicate the levels of different genes of interest in the cell entities found within the samples. (H) Graphical abstract, created with Biorender.com.
To examine the impact of HILPDA downregulation on tumor growth and LD accumulation in vivo, we performed xenograft experiments using 786-O VHL− Omomyc shHILPDA and shSCR cells (Fig. 6C). Mice were administered with vehicle (−Omomyc) or doxycycline (+Omomyc) in the drinking water. Downregulation of HILPDA decreased tumor index (shHILPDA −Omomyc) compared to scramble controls (shSCR -Omomyc). Inhibition of MYC by Omomyc expression robustly reduced tumor burden in control (shSCR +Omomyc) cells while tumor growth was completely prevented by MYC inhibition in shHILPDA (shHILPDA +Omomyc) cells. Analysis of tumor sections revealed higher Ki67 levels in shSCR −Omomyc tumors, and a marked reduction of MYC levels in tumors with Omomyc expression (shSCR +Omomyc and shHILPDA +Omomyc) (Fig. 6D and SI Appendix, Fig. S11G). Notably, HILPDA was highly expressed only in shSCR +Omomyc tumors, thus correlating with LD presence (Fig. 6E and SI Appendix, Fig. S11H).
To investigate the possibility that our results could translate to clinical samples, we used publicly available data from The Cancer Genome Atlas (TCGA) and analyzed the levels of HILPDA and other genes associated with LD accumulation in different adult cancers. Indeed, HILPDA was selectively higher in ccRCC (KIRC) compared to all other tumors including two other RCC subtypes, papillary (KIRP) and chromophobe RCC (KICH) (Fig. 6F and SI Appendix, Table S2). This is in accordance with a previous study demonstrating that RCC cells and patient samples were expressing high HILPDA levels (29). The genes encoding GLS, ATP citrate lyase (ACLY), and perilipin 2 (PLIN2) were also highly expressed in ccRCC. While ACLY regulates fatty acid synthesis, PLIN2 is located in the membrane of LDs. Notably, the genes encoding FASN and ACACA had lower expression in ccRCC than in all other tumor types analyzed, while MYC showed average abundance (SI Appendix, Fig. S12A and Table S2).
Finally, we analyzed single-cell RNA-seq (scRNA-seq) on data from patient-derived normal renal as well as tumor tissue, reported by Obradovic et al. (30). Strikingly, while cycling tumor cells as well as other cells from the tumor stroma revealed high levels of HILPDA, none of the cells from the healthy renal tissue expressed this gene (Fig. 6G). Moreover, MYC levels were only observed in the tumor cells, especially in those with proliferative characteristics. Similar to data from the TCGA database, GLS, ACLY, and PLIN2 were elevated in tumors compared to normal tissue while VHL and genes encoding other enzymes in the fatty acid synthesis pathway were not detected. Finally, EPAS1, encoding HIF2-α was mainly expressed in tumors and no major differences in HIF-1A expression between cells in tumor and normal tissue were observed (Fig. 6G).
Taken together, we have identified HILPDA as a specific biomarker for ccRCC as it is expressed in both cancer as well as in tumor-associated stroma cells but not in normal kidney.
Discussion
Metabolic reprogramming is a major characteristic of ccRCC as manifested by LD accumulation (31). However, this phenotype is not limited to this renal cancer as we and others have demonstrated that several different cancer types accumulate LDs (15, 32, 33). Indeed, LDs are emerging as a new hallmark of cancer (34). Recent studies showed that LDs control the levels of polyunsaturated fatty acids, which could result in lipotoxicity and ferroptosis (35, 36). Furthermore, LD accumulation in colorectal cancer provided resistance to cell death upon treatment with chemotherapeutic drugs (37). This suggests that LDs contribute to cancer cell survival, and inhibiting their formation could be a potential approach to avoid cancer cell dissemination and metastasis. Thus, understanding the mechanism underlying LD accumulation is crucial for identifying new therapeutic targets.
It is well known that LD accumulation occurs during hypoxia (38, 39); however, the key role of MYC in this process has not been elucidated. Notably, we observed that VHL-deficient ccRCC cells induced LD formation upon MYC inhibition using either of four different inhibitors or MYC silencing. We expected that vehicle-treated VHL− cells would also accumulate LDs due to their constitutive HIF expression. However, we found that cells in culture have mostly lost the capacity to accumulate large amounts of LDs, unless MYC is inhibited. Interestingly, G5 treatment not only induced LD accumulation but also reduced the levels of HIF in RCC4 and A498 cells. Further studies are needed to determine whether this was due to off-target effects or by selective modulation of HIF expression.
The use of chemical compounds at high concentrations could lead to nonspecific effects and toxicity. Yet, downregulation of MYC using shMYC lentiviral transduction or inducing Omomyc expression validated that MYC inhibition indeed promoted formation of LDs both in vitro and in vivo. Omomyc was recently in phase I clinical trials for the treatment of different solid tumors, demonstrating tolerability, minimal side effects, and stabilization of disease (40). The Omomyc peptide has previously not been studied in ccRCC nor has its impact on LD accumulation. Together, our results show that this process is governed by the metabolic rewiring induced by the concomitant HIF stabilization and MYC inhibition in ccRCC.
During hypoxia, in situations where HIF is constitutively expressed, or in cells with disrupted mitochondrial respiration, reductive carboxylation of glutamine provides the acetyl-CoA necessary for lipid synthesis (11). We demonstrated that glutamine was essential for fatty acid synthesis and accumulation in LDs after MYC inhibition, as glutamine deprivation or treatment with BPTES both in vitro and in vivo prevented this phenotype. Furthermore, inhibition of fatty acid synthesis also blocked LD accumulation. Hence, preventing the use of glutamine-derived carbons for fatty acid synthesis is a promising strategy for impairing LD accumulation, resulting in disrupted metabolic adaptation and reduced cancer cell survival.
Enzymes regulating the synthesis of diacylglycerol from PA are controlled by the sterol regulatory element binding protein 1 (SREBP1) and previous results indicate that this enzyme can cooperate with MYC as well as HIF1-α in the regulation of lipid synthesis (41, 42). Our isotope tracing experiments demonstrated that VHL+ cells mainly generated cytosolic acetyl-CoA from glucose while glutamine was a major contributor in VHL− cells. Furthermore, while VHL− cells treated with JQ1 were mostly converting PA to TGs, VHL+ cells instead produced high amounts of PI, PG, and LPI. This could explain why these cells have lower amounts of LDs, and why inhibition of GLS or deprivation of glutamine prevented LD accumulation in VHL− cells upon MYC inhibition. The combination of BPTES and Omomyc expression further reduced tumor growth and prevented LD accumulation, with no signs of apoptosis, indicating that the treatment inhibited proliferation, postulating this combination as an attractive approach for preventing LD accumulation induced by MYC inhibition.
We identified HILPDA as one of the most significantly up-regulated genes in VHL- cells exposed to JQ1. MYC, MAX, as well as MIZ-1 have been reported to bind the HILPDA promoter, suggesting that this ternary complex could inhibit its expression. In addition, downregulation of HILPDA prevented LD accumulation and impaired proliferation in vitro and tumor burden in vivo, while its overexpression supported this process, confirming previous results on its crucial role in metabolic rewiring. In accordance with our in vitro findings, enhanced HILPDA expression was indeed observed in tumors upon Omomyc induction, which was attenuated when combined with BPTES. We also observed that BPTES increased MYC which could explain the reduced levels of HILPDA upon treatment with this GLS inhibitor, although further studies will be needed to fully understand this observation. Importantly, in silico analysis of patient data demonstrated that HILPDA is highly expressed in ccRCC compared to other tumor types or healthy renal cells. It has previously been reported that RCC samples have high HILPDA levels (29). Here, we expanded the analysis to different adult cancers, postulating HILPDA as a specific biomarker for ccRCC. In addition, we observed high HILPDA levels only in ccRCC (KIRC), the most aggressive renal cancer type, compared to the other two subtypes, chromophobe (KICH), or papillary (KIRP). Importantly, we found that HILPDA is not only highly expressed in cancer cells but also in the tumor stroma, suggesting that these cells also could accumulate LDs, possibly induced by cancer cells, thereby supporting the tumor and promoting its survival. In fact, heterogeneity in LD accumulation in the same cell population contributes to reduce lipotoxicity and increased survival during periods of nutrient starvation (43–45). This observation paves the way for future studies on the role of LDs in the cross talk between cancer and stroma cells. Moreover, the absence of HILPDA expression in normal renal tissue opens possibilities for targeting ccRCC tumors without disrupting the surrounding healthy tissue.
Collectively, our study identifies the molecular mechanism leading to robust LD accumulation in ccRCC due to enhanced TG content upon simultaneous HIF expression and MYC inhibition involving HILPDA (Fig. 6H). As the role of LDs in cancer progression and survival is gaining more relevance, understanding the process of LD accumulation, as well as the identification of selective targets as HILPDA, could lay the basis for attractive strategies for precision medicine for ccRCC patients.
Materials and Methods
Detailed description of cell lines, mice, inhibitors and all methods including cell culture, hypoxia chamber experiments, cell viability and BrdU proliferation assays, western blot, RT-qPCR, nutrient deprivation experiments, immunofluorescence, siRNA, shRNA knockdown and protein overexpression experiments, lipidomics, stable isotope tracing, in vivo xenograft studies, bulk and single-cell RNA sequencing, immunohistochemistry, ENCODE and TCGA database analysis are provided in SI Appendix.
Statistical Analysis.
All in vitro and in vivo data were analyzed by Student’s t test or one-way or two-way ANOVA test for multiple comparisons with Dunnett’s or Tukey’s corrections using GraphPad Prism (version 8.1.1), as indicated in the Figure legends. All experiments have been performed at least in three independent replicates, and all data are presented as mean ± SD with *, **, ***, and **** indicating P < 0.05, <0.01, <0.001, and <0.0001, respectively.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We are grateful to S. Doloczki (Uppsala University) for synthesis of LD-BTD1, Dr. L. Schlicker (DKFZ, Heidelberg) for assistance with metabolic tracing experiments, Associate Prof. E. Fredlund (DoubleStrand Bioinformatics, Stockholm) for initial bioinformatic analyses, and Dr. J. Liaño-Pons (Karolinska Institutet) for support with the ENCODE database and assistance during the animal experiments together with MSc F. Fahrig. We thank Dr. L. Herrero-Rodriguez (University of Barcelona) for generously sharing UB006, Associate Prof. R. Nilsson (Karolinska Institutet) for metabolic tracers, and Prof. L. Soucek (VHIO, Barcelona) for the pTRIPZ-OMOMYC-RFPshEV construct. We are indebted to many colleagues from Uppsala and Lund University, as well as from Karolinska Institutet for generously sharing tumor cell lines. We acknowledge fruitful discussions with members of the Wilhelm, Schlisio, and Arsenian-Henriksson Laboratories (all Karolinska Institutet). Research funding in the C.D. Lab was supported by the Swedish Research Council (2018-03524), in the V.P. Lab by the Swedish Research Council (2020-01480, 2019-02335), Vinnova (2020-03620), Wallenberg Academy Fellowship (2021.0167), and Karolinska Institutet, in the A.S. Lab by the German Research Foundation (FOR2314, SCHU2670) and the German Cancer Aid (111917), in the H.A. Lab by Swedish Cancer Society (Grant no. 21-1709), Regional ALF Funds (2018-176) and the European Union’s Horizon 2020 research and innovation programme (101017453), and in the M.A.-H. Lab by the Swedish Research Council (2018-02580), the Swedish Cancer Society (19 0510, 22 2266), the Swedish Childhood Cancer Fund (2021-0131), Radiumhemmet Research Funds (19 1073, 22 1031), and Karolinska Institutet. L.S.-A. was supported by the Robert Lundberg Foundation and M.V.R.-P. by Castengren Memorial, Goljes, Karolinska Institutet Funds, as well as by a postdoctoral position from the Swedish Childhood Cancer Fund.
Author contributions
L.S.-A, M.T.S., M.V.R.-P, V.P., H.A., A.S., and M.A.-H. designed research; L.S.-A., E.G.-L., M.T.S., S.S., I.S., M.V.R.-P., V.P., H.A., A.S., and M.A.-H. performed research; C.D. contributed new reagents/analytic tools; L.S.-A., E.G.-L., M.T.S., S.S., I.S., M.V.R.-P., V.P., H.A., A.S., and M.A.-H. analyzed data; and L.S.-A. and M.A.-H. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The RNA-sequencing and corresponding metadata have been deposited in NCBI’s Gene Expression Omnibus (46) and are accessible through GEO Series accession number GSE223069 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223069) (47). This work does not report original code. All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Rini B. I., Campbell S. C., Escudier B., Renal cell carcinoma. Lancet 373, 1119–1132 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Atkins M. B., Tannir N. M., Current and emerging therapies for first-line treatment of metastatic clear cell renal cell carcinoma. Cancer Treat. Rev. 70, 127–137 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Hakimi A. A., Pham C. G., Hsieh J. J., A clear picture of renal cell carcinoma. Nat. Genet. 45, 849–850 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Haase V. H., The VHL tumor suppressor: Master regulator of HIF. Curr. Pharm. Des. 15, 3895–3903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang S.-W., et al. , MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Lett. 273, 35–43 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Shroff E. H., et al. , MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc. Natl. Acad. Sci. U.S.A. 112, 6539–6544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linehan W. M., Srinivasan R., Schmidt L. S., The genetic basis of kidney cancer: A metabolic disease. Nat. Rev. Urol. 7, 277–285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wise D. R., et al. , Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc. Natl. Acad. Sci. U.S.A. 105, 18782–18787 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun R. C., Denko N. C., Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell Metab. 19, 285–292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J., Tchernyshyov I., Semenza G. L., Dang C. V., HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Metallo C. M., et al. , Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thiele C., Spandl J., Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20, 378–385 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Olzmann J. A., Carvalho P., Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruz A. L. S., Barreto E. d. A., Fazolini N. P. B., Viola J. P. B., Bozza P. T., Lipid droplets: Platforms with multiple functions in cancer hallmarks. Cell Death Dis. 11, 105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zirath H., et al. , MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc. Natl. Acad. Sci. U.S.A. 110, 10258–10263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin X., Giap C., Lazo J. S., Prochownik E. V., Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene 22, 6151–6159 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Mertz J. A., et al. , Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U.S.A. 108, 16669–16674 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colas K., et al. , Photophysical characteristics of polarity-sensitive and lipid droplet-specific phenylbenzothiadiazoles. ChemPhotoChem 5, 632–643 (2021). [Google Scholar]
- 19.Demma M. J., et al. , Omomyc reveals new mechanisms to inhibit the MYC oncogene. Mol. Cell. Biol. 39, e00248-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung A. L., et al. , Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell 6, 33–43 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Gameiro P. A., et al. , In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coffey G. L., et al. , 6-Diazo-5-oxo-L-norleucine, a new tumor-inhibitory substance. I. Biologic studies. Antibiot. Chemother. (Northfield) 6, 487–497 (1956). [PubMed] [Google Scholar]
- 23.DeLaBarre B., et al. , Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry 50, 10764–10770 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Lemberg K. M., Vornov J. J., Rais R., Slusher B. S., We’re not “don” yet: Optimal dosing and prodrug delivery of 6-diazo-5-oxo-L-norleucine. Mol. Cancer Ther. 17, 1824–1832 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makowski K., et al. , (-)-UB006: A new fatty acid synthase inhibitor and cytotoxic agent without anorexic side effects. Eur. J. Med. Chem. 131, 207–221 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Harris R. A., McCune S. A., "[41] 5-(Tetradecyloxy)-2-furoic acid" in Lipids Part D (Academic Press, 1981), pp. 552–559. [DOI] [PubMed] [Google Scholar]
- 27.Maier A., et al. , Hypoxia-inducible protein 2 Hig2/Hilpda mediates neutral lipid accumulation in macrophages and contributes to atherosclerosis in apolipoprotein E-deficient mice. FASEB. J. 31, 4971–4984 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Peukert K., et al. , An alternative pathway for gene regulation by Myc. EMBO J. 16, 5672–5686 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Togashi A., et al. , Hypoxia-inducible protein 2 (HIG2), a novel diagnostic marker for renal cell carcinoma and potential target for molecular therapy. Cancer Res. 65, 4817–4826 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Obradovic A., et al. , Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell 184, 2988–3005.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du W., et al. , HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat. Commun. 8, 1769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Q., et al. , Co-administration of 20(S)-protopanaxatriol (g-PPT) and EGFR-TKI overcomes EGFR-TKI resistance by decreasing SCD1 induced lipid accumulation in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 38, 129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotte A. K., et al. , Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat. Commun. 9, 322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petan T., Lipid droplets in cancer. Rev. Physiol. Biochem. Pharmacol. 185, 53–86 (2023). [DOI] [PubMed] [Google Scholar]
- 35.Jarc E., et al. , Lipid droplets induced by secreted phospholipase A2 and unsaturated fatty acids protect breast cancer cells from nutrient and lipotoxic stress. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1863, 247–265 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Listenberger L. L., et al. , Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehdizadeh A., et al. , Common chemotherapeutic agents modulate fatty acid distribution in human hepatocellular carcinoma and colorectal cancer cells. Bioimpacts 7, 31–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du W., et al. , HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat. Commun. 8, 1769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bensaad K., et al. , Fatty acid uptake and lipid storage induced by HIF-1a contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 9, 349–365 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Peptomyc S. L., Phase 1/2 Study to Evaluate Safety, PK and Efficacy of the MYC-Inhibitor OMO-103 in Solid Tumours (MYCure). https://clinicaltrials.gov/ct2/show/results/NCT04808362. Accessed 2 March 2023.
- 41.Gouw A. M., et al. , The MYC oncogene cooperates with sterol-regulated element-binding protein to regulate lipogenesis essential for neoplastic growth. Cell Metab. 30, 556–572.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furuta E., et al. , Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 68, 1003–1011 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Herms A., et al. , Cell-to-cell heterogeneity in lipid droplets suggests a mechanism to reduce lipotoxicity. Curr. Biol. 23, 1489–1496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velázquez A. P., Tatsuta T., Ghillebert R., Drescher I., Graef M., Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J. Cell Biol. 212, 621–631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cabodevilla A. G., et al. , Cell survival during complete nutrient deprivation depends on lipid droplet-fueled β-oxidation of fatty acids. J. Biol. Chem. 288, 27777–27788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar R., Domrachev M., Lash A. E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sainero-Alcolado L., et al. , Data from “Targeting MYC induces lipid droplet accumulation by upregulation of HILPDA in clear cell renal cell carcinoma”. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223069. Deposited 17 January 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
The RNA-sequencing and corresponding metadata have been deposited in NCBI’s Gene Expression Omnibus (46) and are accessible through GEO Series accession number GSE223069 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE223069) (47). This work does not report original code. All other data are included in the manuscript and/or supporting information.