Significance
We demonstrated that CDK9 (Cyclin-dependent kinase 9) phosphorylates LSD1 protein at the C-terminal. The phosphorylated LSD1 protein is then recognized and polyubiquitylated by the E3 ligase RNF20 complex, leading to the accumulation of the LSD1 protein and subsequent epigenetic silencing of the IFN (interferon) response and ERVs (endogenous retrovirus), inducing immune evasion. Our results showed that the CDK9–RNF20–LSD1 axis is vital for gene silencing, inactivation of ERVs, and IFN response, ultimately resulting in immune suppression.
Keywords: LSD1, CDK9, RNF20, epigenetic, immunosuppression
Abstract
Cyclin-dependent kinase 9 (CDK9) plays a critical role in transcription initiation and is essential for maintaining gene silencing at heterochromatic loci. Inhibition of CDK9 increases sensitivity to immunotherapy, but the underlying mechanism remains unclear. We now report that RNF20 stabilizes LSD1 via K29-mediated ubiquitination, which is dependent on CDK9-mediated phosphorylation. This CDK9- and RNF20-dependent LSD1 stabilization is necessary for the demethylation of histone H3K4, then subsequent repression of endogenous retrovirus, and an interferon response, leading to epigenetic immunosuppression. Moreover, we found that loss of RNF20 sensitizes cancer cells to the immune checkpoint inhibitor anti-PD-1 in vivo and that this effect can be rescued by the expression of ectopic LSD1. Our findings are supported by the observation that RNF20 levels correlate with LSD1 levels in human breast cancer specimens. This study sheds light on the role of RNF20 in CDK9-dependent LSD1 stabilization, which is crucial for epigenetic silencing and immunosuppression. Our findings explore the potential importance of targeting the CDK9–RNF20–LSD1 axis in the development of new cancer therapies.
Epigenetic regulations of DNA methylation, histone post-translational modification, and chromatin structure play a critical role in the interactions between tumors and immune cells (1). Recent evidence suggests that tumors commonly hijack various epigenetic mechanisms to evade immune surveillance. Inhibiting these epigenetic regulators normalizes immune function and/or triggers antitumor responses (2). One such epigenetic regulator is LSD1, an enzyme that erases H3K4me1/2 and acts as an inhibitor of anti-tumor immunity and responsiveness to immunotherapy (3, 4). High levels of LSD1 have been identified in various cancers, including leukemia, non-small cell lung, pancreatic, prostate, and breast cancers (5–8). Overexpression of LSD1 has been associated with tumor aggressiveness, metastasis, recurrence, drug resistance, immune suppression, and poor prognosis (9, 10). Recent studies have shown that LSD1 inhibition augments CD8+ T cell infiltration into tumors, reducing tumor burden through enhanced chemokine expression (11) and inducing endogenous retrovirus (ERV)s to activate a type I interferon (IFN) signature, which stimulates anti-tumor T cell immunity (12). Additionally, LSD1 promotes immunosuppressive macrophage polarization in triple-negative breast cancer (13) and modulates T cell exhaustion in cancer (14). Collectively, LSD1 plays a critical role in tumor immunity by regulating not only tumor cells but also intratumoral immune cells.
LSD1 is subject to regulation by various post-translational modifications, including phosphorylation, acetylation, and ubiquitination (15). Our studies and others have shown that LSD1 levels are under tight control by a ubiquitin-proteasome system (UPS) (6, 16–19), with USP28 acting as the LSD1 deubiquitinase and stabilizing the LSD1 protein (18) while FBXO24 functioning as the E3 ligase (19). In addition, USP22 stabilizes LSD1 through GSK3β-mediated phosphorylation (6). Furthermore, lysine methylation and arginine methylation regulate LSD1 stability by inhibiting polyubiquitination modification (20, 21). Despite these findings, the regulation of LSD1 stability remains poorly understood, and the identification of new mechanisms could provide opportunities for the development of LSD1-targeting therapy in breast cancer.
CDK9 (Cyclin-dependent kinase 9), which is the catalytic subunit of P-TEFb, is a transcriptional activator. It promotes the release of RNA polymerase II (RNAP II) from the promoter-proximal pause by phosphorylating negative elongation factors (22), leading to the recruitment of RNA processing factors. CDK9 also controls transcription-coupled chromatin modifications, such as monoubiquitylation of histone H2B (23). In recent studies, it has been observed that CDK9 inhibition can have a significant impact on various biological processes, including transcription, metabolism, DNA damage repair, epigenetics, and the immune response to facilitate an anti-tumor response (24). CDK9 has also been linked to heterochromatin formation and maintenance of epigenetic silencing (25). Treatment with a CDK9 inhibitor results in reactivation of tumor suppressor genes, leading to an increase in immune cells and making the tumor more sensitive to anti-PD1 treatment. Most importantly, treatment with a CDK9 inhibitor leads to higher occupancy of H3K4me2 at the promoter regions of hypermethylated CDK9 targeted genes, particularly for the repetitive elements (25). However, the mechanism remains unclear. Interestingly, integrative chromatin immunoprecipitation (ChIP) analysis data reveal that LSD1 and CDK9 are strongly co-localized at gene promoters and are present along with core RNAP II factors (26). However, the exact mechanism behind this co-localization and the biological function of their relationship remains unclear.
In this study, we found that CDK9 is accountable for the build-up of LSD1 by promoting its binding with and subsequent accumulation through RNF20-mediated K29-linked polyubiquitination. Our results also showed that the CDK9–RNF20–LSD1 axis is vital for gene silencing, inactivation of ERVs and IFN response, ultimately resulting in immune suppression.
Results
CDK9 Stabilizes LSD1 through Ubiquitination.
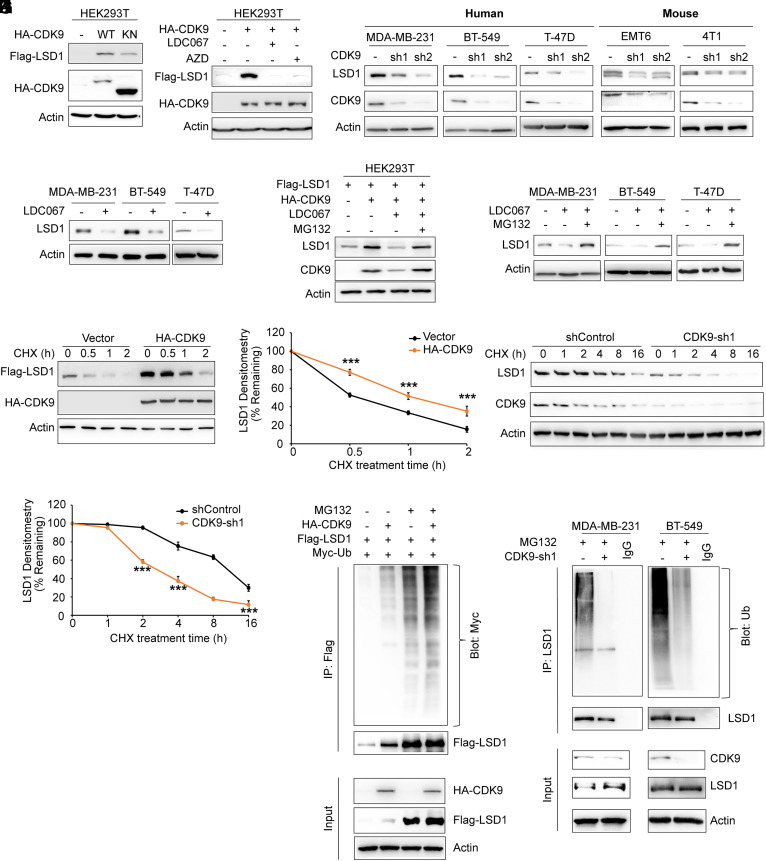
To investigate the relationship between CDK9 and LSD1, which co-localize at gene promoter regions (26), we co-expressed LSD1 with CDK9 in HEK293T cells. Surprisingly, we found that wild-type (WT) CDK9 stabilized LSD1 whereas a kinase negative (KN) mutation, showed no such effect (Fig. 1A). We then treated the cells with two CDK9 highly selective inhibitors, LDC000067 (LDC067) and AZD4573 (AZD), and observed a dramatic reduction in CDK9-induced LSD1 protein level (Fig. 1B). In agreement with these findings, knockdown of endogenous CDK9 resulted in a rapid loss of endogenous LSD1 protein, but had no effect on its mRNA levels, in three human breast cancer cells and two mouse breast cancer cells (Fig. 1C and SI Appendix, Fig. S1). Additionally, treatment with LDC067 reduced endogenous LSD1 protein level (Fig. 1D). Because LSD1 is a liable protein that is readily degraded by proteasome, and because CDK9 stabilizes LSD1 without affecting its mRNA expression, we investigated whether CDK9 blocked LSD1 degradation. First, we treated CDK9 expressing cells with LDC067 alone or with the proteasome inhibitor MG132. Downregulation of LSD1 in LDC067-treated cells was restored by MG132 treatment (Fig. 1E), and similar results were observed for endogenous LSD1 protein level in LDC067-treated cells (Fig. 1F). To further confirm these results, we co-expressed LSD1 with CDK9 or a control vector in HEK293T cells and assessed LSD1 degradation. As expected, LSD1 degraded rapidly in cells transfected with a control vector after treatment with cycloheximide (CHX) to block the new protein synthesis (Fig. 1 G and H). However, its levels were stabilized in the presence of CDK9. Similarly, endogenous LSD1 became unstable and degraded rapidly in CDK9 knockdown cells (Fig. 1 I and J). Since LSD1 is under protein ubiquitination and degradation, we assumed that CDK9 may inhibit LSD1 ubiquitination. To test this, we assessed the effect of CDK9 on the ubiquitination of LSD1. Strikingly, transfection of CDK9 increased LSD1 ubiquitination compared to the control vector (Fig. 1K Lane 2 and lane 1, respectively). As expected, treatment with MG132 enhanced LSD1 ubiquitination. Interestingly, transfection of CDK9 greatly increased LSD1 ubiquitination even in the presence of MG132. Furthermore, knockdown of CDK9 decreased LSD1 ubiquitination in two breast cancer cell lines (Fig. 1L). Taken together, these results suggest that CDK9 stabilizes LSD1 by increasing its ubiquitination.
Fig. 1.
CDK9 stabilizes LSD1. (A) Flag-LSD1 was co-expressed with WT or KN mutant CDK9 in HEK293T cells and protein was detected by western blot. (B) Flag-LSD1 was co-expressed with CDK9 in HEK293T cells treated with CDK9-specific inhibitor LDC000067 (LDC067, 20 µM) or AZD4573 (AZD, 1 µM) for 24 h. Lysates were analyzed by western blot. (C) Protein expression of LSD1 and CDK9 from indicated cell lines was analyzed by western blot. (D) Cells were treated with LDC067 for 24 h. Lysates were analyzed by western blot. (E) Flag-LSD1 was co-expressed with CDK9 in HEK293T cells and then treated with CDK9 specific inhibitor LDC067 with or without MG132 for 6 h. Lysates were analyzed by western blot. (F) Cells were pre-treated with LDC067 for 16 h then treated with or without 10 µM MG132 for 6 h. Lysates were analyzed by western blot. (G) Flag-LSD1 was co-expressed with vector or HA-CDK9 in HEK293T cells. After treatment with CHX for the indicated time intervals, expression of LSD1 and CDK9 was analyzed by western blot using Flag and HA antibodies, respectively. Presented data are representative of three separate experiments. (H) The intensity of LSD1 expression for each time point in (G) was quantified by densitometry and plotted. (I) MDA-MB-231 cells were transfected with control or CDK9 shRNA. After treatment with CHX as indicated above, expression of endogenous LSD1 and CDK9 was analyzed by western blot. Presented data are representative of three separate experiments. (J) Intensity of LSD1 expression for each time point in (J) was quantified by densitometry and plotted. (K) Flag-LSD1 and Myc-ubiquitin were co-expressed with or without CDK9 in HEK293T cells. After treatment with or without 10 μM MG132 for 6 h, Flag-LSD1 was subjected to IP (immunoprecipitated) and the poly-ubiquitination of LSD1 assessed by western blot using Myc antibody. IP LSD1 was blotted using Flag antibody. (L) MDA-MB-231 and BT-549 cells stably transfected with control or CDK9 shRNA were treated with MG132 for 6 h. Extracts were subjected to IP with LSD1 antibody and the poly-ubiquitination of LSD1 assessed by western blot using ubiquitin antibody. Input of LSD1 and CDK9 were analyzed by western blot. ***P < 0.001, compared with controls.
CDK9 Interacts with and Phosphorylates LSD1 at the C-Terminal Domain.
We sought to investigate whether LSD1 interacts with CDK9. When Flag-tagged LSD1 was co-expressed with HA-tagged CDK9 in HEK293T cells, we observed an association between LSD1 with CDK9 and vice versa (Fig. 2A). We also detected an endogenous protein interaction between CDK9 and LSD1 (Fig. 2B), as well as other components of the RNAP II complex, including Cyclin T1. To further explore this interaction, we co-expressed HA-CDK9 and GFP-LSD1 in HEK293 cells. Immunofluorescent staining revealed that CDK9 co-localized and stabilized LSD1 in the nucleus (Fig. 2C). To identify the regions in LSD1 that associate with CDK9, we generated different deletion mutants of LSD1 (16, 18). Our results showed that the amine oxidase-like (AOL) domain of LSD1 is required for its binding to CDK9 (Fig. 2D). Interestingly, the C-terminal domain of LSD1 (residues 727 to 853) is sufficient for its association with CDK9.
Fig. 2.
CDK9 interacts with and phosphorylates LSD1 at C-terminal. (A) Flag-LSD1 was co-expressed with vector or HA-CDK9 in HEK293T cells. LSD1 and CDK9 were IP with Flag or HA antibody, respectively, and the associated CDK9 and LSD1 were analyzed by western blot using either HA or Flag antibody. One-fortieth of the lysate from each sample was subjected to western blot to examine the expression of LSD1 and CDK9 (input lysate). (B) Endogenous CDK9 and LSD1 were captured by IP from MDA-MB-231 and BT-549 cells, and bound endogenous LSD1, CDK9, Cyclin T1, and RNA POL II (Pol II) were examined by western blot. (C) GFP-LSD1 was co-expressed with HA-CDK9 in HEK293 cells. After fixation, the cellular location of GFP-LSD1 (green) and CDK9 (red) was examined by immunofluorescent staining using anti-HA antibody and visualized by fluorescence microscopy (nuclei were stained with DAPI; blue) (Left). (Scale bar, 20 μm.) Arrow indicates the cell without HA-CDK9 expressing. The colocalization intensity was quantified using algorithms (Right). (D) Schematic diagram showing the structure of LSD1 and deletion constructs used (Left). Co-IP of exogenous HA-CDK9 and Flag full length (FL) LSD1 or different deletion mutants (Right). (E) Flag-LSD1 was co-expressed with HA-CDK9 and treated with or without LDC067. Lysates were analyzed by Phos-tag sodium dodecyl-sulfate (SDS)-PAGE and western blot. (F) In vitro kinase assays were performed by incubating purified active CDK9/cyclin T1 with recombinant GST-LSD1 with or without LDC067 treatment in the presence of [γ-32P]ATP. The resultant products were subjected to SDS–polyacrylamide gel electrophoresis and autoradiography. (G) Sequence alignment of LSD1 C-term. (H) Immunoblot of lysates from HEK293T cells transfected with the indicated constructs. (I) Flag-LSD1-WT or Flag-LSD1-5A was co-transfected with or without HA-CDK9 into HEK293T cell. Lysates were analyzed by western blot. (J) In vitro kinase assays were performed by incubating purified active CDK9/cyclin T1 with recombinant WT GST-LSD1 or GST-LSD1-5A in the presence of [γ-32P]ATP. The resultant products were subjected to SDS–polyacrylamide gel electrophoresis and autoradiography. (K) Flag-LSD1-WT or Flag-LSD1-5A was co-transfected with or without HA-CDK9 into HEK293T cells and then treated with MG132 for 6 h. Cell lysates were IP using an anti-Flag antibody and then analyzed by immunoblotting using a pS/T antibody.
To further identify whether CDK9 phosphorylates LSD1, we conducted a Phos-Tag analysis on HEK293T cells that were transiently transfected with CDK9. As shown in Fig. 2E, CDK9 produced the phosphorylation-dependent mobility shift, which was abolished by LDC067 treatment, indicating phosphorylation of LSD1 by CDK9. Next, we performed in vitro kinase assays and found that CDK9 phosphorylated LSD1, which was inhibited by LDC067 treatment (Fig. 2F). To identify the specific phosphorylation sites targeted by CDK9, we co-expressed different mutants of LSD1 with CDK9. Our results showed that CDK9 markedly increased in the C-terminal domain of LSD1, while no significant differences were observed in other mutants (SI Appendix, Fig. S2). Notably, phosphorylation of Ser2 within the repeated YSPTSPS heptapeptide motif of the RNAP II carboxy-terminal domain, and threonine (Thr) within the SPT5 C-terminal repeat are responsible for various aspects of CDK9 function (27). Our analysis showed that the LSD1 C-terminal domain contains an SPS motif and a Thr residue at the LSD1. Furthermore, global proteomics also uncovered that LSD1 is phosphorylated on T810, S817, T841, S849, and S851 (http://www.phosphosite.org). These sites are highly conserved evolutionarily (Fig. 2G). Consistent with this observation, earlier large-scale proteomic studies demonstrated that these five sites are phosphorylated in breast cancer cell lines (28–30). To determine whether these phosphorylation sites are important for LSD1 stability, we generated a series of LSD1 mutants, where these Thr and serine residues were changed to alanine. Our results showed that mutations of all five sites (TS/5A) in the LSD1 protein caused a decrease in stability compared to the LSD1-WT (Fig. 2H). Moreover, the de-phosphomimetic LSD1 was unable to stabilize when co-transfected with CDK9 (Fig. 2I), and we did not detect phosphorylation of the LSD1-5A mutant using the in vitro kinase assay (Fig. 2J). The phosphorylation level of LSD1 was up-regulated by CDK9 in the cells transfected with LSD1-WT, but not in the cells transfected with LSD1-5A (Fig. 2K). These findings indicate that CDK9 interacts with LSD1 and phosphorylates the C-terminal domain.
RNF20 Stabilizes LSD1.
Because phosphorylation by CDK9 increased LSD1 ubiquitination and the CDK9–RNF20–H2Bub axis is associated with H3K4me3 peaks and regulates gene expression (31), we tested whether RNF20 is involved in LSD1 stabilization. We co-expressed LSD1 with RNF20 in HEK293T cells. Strikingly, we observed that RNF20 dramatically increased the protein level of LSD1 (Fig. 3A). The stability of LSD1 was also evaluated by performing RNF20 knockdown experiments in four human breast cancer cells and two mouse cells (EMT6 and 4T1). The results revealed that RNF20 knockdown reduced LSD1 protein level but not LSD1 mRNA levels (Fig. 3 B and C and SI Appendix, Fig. S3A). Additionally, we observed a direct correlation between LSD1 and RNF20 protein level in a panel of breast cancer cell lines (Fig. 3D). Further investigation showed that RNF20 co-localized with GFP-LSD1 in the nuclei of HEK293 cells (Fig. 3E). The effect of RNF20 on LSD1 stability was evaluated and it was found that LSD1 stability was increased in the presence of RNF20 (Fig. 3 F and G). In contrast, downregulation of LSD1 protein levels due to RNF20 knockdown was caused by decreased LSD1 stability (Fig. 3 H and I). These results indicate that RNF20 stabilizes LSD1.
Fig. 3.
RNF20 stabilizes LSD1. (A) Flag-LSD1 was co-expressed with vector or Myc-RNF20 in HEK293T cells and protein was detected by western blot. (B) The protein expression of LSD1 and RNF20 from four human breast cancer cell lines transfected with control or two individual RNF20 shRNAs was analyzed by western blot. (C) The protein expression of LSD1 and RNF20 from two mouse cells (EMT6 and 4T1) transfected with control or two individual RNF20 shRNAs was analyzed by western blot. (D) The protein expression of LSD1 and RNF20 in various breast cancer cell lines was analyzed by western blot. (E) GFP-LSD1 was co-expressed with Myc-RNF20 in HEK293 cells. After fixation, the cellular location of GFP-LSD1 (green) and RNF20 (red) was examined by immunofluorescent staining using anti-Myc antibody and visualized by fluorescence microscopy (nuclei were stained with DAPI; blue) (Left). (Scale bar, 20 μm.) The colocalization intensity was quantified using algorithms (Right). (F) After treatment with CHX for the indicated time intervals, expression of Flag-LSD1 and Myc-RNF20 was analyzed by western blot using Flag and Myc antibodies, respectively. Presented data are representative of three separate experiments. (G) The intensity of LSD1 expression for each time point in (F) was quantified by densitometry and plotted. (H) MDA-MB-231 cells were transfected with control or RNF20 shRNA. After treatment with CHX as indicated above, expression of endogenous LSD1 and RNF20 was analyzed by western blot. Presented data are representative of three separate experiments. (I) The intensity of LSD1 expression for each time point in (H) was quantified by densitometry and plotted. **P < 0.01, ***P < 0.001, compared with controls.
RNF20 Interacts with and Ubiquitinates LSD1 with K29.
To further understand the interaction between RNF20 and LSD1, we co-expressed Flag-LSD1 and Myc-RNF20 in HEK293T cells. Co-IP experiments revealed that RNF20 and LSD1 interacted with each other (Fig. 4A). Endogenous LSD1 and RNF20 were also found to interact in MDA-MB-231 and BT-549 cells (Fig. 4B). To determine the specific region in LSD1 that associates with RNF20, we generated several LSD1 deletion mutants (18). In agreement with association between LSD1 and CDK9, the AOL domain of LSD1 is also necessary for its interaction with RNF20 and the C-terminal domain of LSD1 was sufficient for this interaction (Fig. 4 C and D). This finding was supported by the observation that RNF20 markedly enhanced the expression of the C-terminal domain of LSD1 (SI Appendix, Fig. S3B).
Fig. 4.
RNF20 interacts with and ubiquitinates LSD1 with K29. (A) Flag-LSD1 was co-expressed with vector or Myc-RNF20 in HEK293T cells. LSD1 and RNF20 were IP with Flag or Myc antibody, respectively, and the associated RNF20 and LSD1 were analyzed by western blot using either Myc or Flag antibody. (B) Endogenous RNF20 and LSD1 were captured by IP from MDA-MB-231 and BT-549 cells, and bound endogenous LSD1, CDK9, and RNF20 were examined by western blot. (C) Schematic diagram showing the structure of LSD1 and deletion constructs used. (D) Co-IP of exogenous Myc-RNF20 and Flag full length (FL) LSD1 or different deletion mutants. (E) Flag-LSD1 was co-expressed with Myc-RNF20 and the indicated ubiquitin constructs in HEK293T cells. The cells were treated with MG132 for 6 h. Lysate was collected for Flag-IP, followed by IB analysis with indicated antibodies. (F) HEK293T cells were transfected with Flag-LSD1, Myc-RNF20, and the indicated ubiquitin constructs. After treatment with MG132 for 6 h, extracts were collected for IP with anti-Flag antibody, followed by IB analysis as indicated. (G) HEK293T cells were transfected with the indicated plasmids with or without MG132 treatment. Adjusted extracts were subject to IP with anti-Flag antibody, followed by IB analysis as indicated. (H) MDA-MB-231 and BT-549 cells stably transfected with control or RNF20 shRNA were treated with MG132 for 6 h. Extracts were subjected to IP with LSD1 antibody and the poly-ubiquitination of LSD1 assessed by western blot using ubiquitin antibody. Input of LSD1 and RNF20 were analyzed by western blot.
RNF20 is an E3 ligase, and ubiquitination of LSD1 has been shown to play a critical role in its regulation. Therefore, we examined whether RNF20 could modulate LSD1 ubiquitination. Strikingly, we found that RNF20 remarkably increased the polyubiquitination of LSD1 (SI Appendix, Fig. S3C). To determine the specificity of the ubiquitination linkage, we transfected HEK293T cells with single ubiquitin mutants, each containing only one intact lysine residue at positions 6, 11, 27, 29, 33, 48, or 63, while all other lysine residues were mutated to arginine residues. As shown in Fig. 4E, K29 displayed major polyubiquitinated LSD1 signals. We then used K-to-R ubiquitin mutants, in which only one lysine residue was substituted with an arginine residue, and all other lysine residues were left intact. The Ub-K29R mutant completely abolished ubiquitination compared to Ub-WT and Ub-K29 (Fig. 4F). These results suggest that RNF20 primarily ubiquitinates LSD1 through K29-linked chains. Consistent with this, RNF20 led to an increase of LSD1 polyubiquitination (Fig. 4G, lane 1 and lane 2), especially in the presence of MG132 treatment (Fig. 4G, lane 3 and lane 4). To investigate whether endogenous LSD1 is ubiquitinated by RNF20, we knocked down RNF20 in MDA-MB-231 and BT-549 cells. This resulted in a significant decrease in the polyubiquitination of endogenous LSD1 (Fig. 4H). Collectively, these data demonstrate that RNF20 is a LSD1 ubiquitinase, adding K29-linked polyubiquitination chains to LSD1.
CDK9 Promotes LSD1’s Binding with and Stabilization by RNF20.
We next sought to determine whether the stabilization of LSD1 by RNF20 depends on LSD1 phosphorylation by CDK9. First, we examined whether CDK9 influenced RNF20-mediated LSD1 accumulation, and we found that CDK9 inactivation by LDC067 blocked RNF20-induced LSD1 accumulation (Fig. 5A). In agreement with this, CDK9 increased the interaction between LSD1 and RNF20, while the interaction was completely abolished after LDC067 treatment in HEK293T cells (Fig. 5B). Furthermore, loss of CDK9 in MDA-MB-231 cells weakened the interaction between endogenous RNF20 and LSD1 (Fig. 5C). RNF20 was unable to increase the protein level of LSD1-5A mutation (Fig. 5D). To further confirm this, we measured the half-life of LSD1-5A mutation when co-transfected with RNF20 and found that the stable level of LSD1-5A protein was significantly lower than that of LSD1-WT protein in the presence of RNF20 (Fig. 5 E and F). Additionally, the mutation significantly decreased the interaction between RNF20 and LSD1 in the presence of CDK9 (Fig. 5G). Consequently, the mutation also attenuated LSD1 polyubiquitination by RNF20 (Fig. 5H). In agreement with these results, the CDK9 inhibitor LDC067 reduced polyubiquitylated LSD1 protein bands compared to the control (Fig. 5I). These results demonstrate that CDK9-dependent phosphorylation of LSD1 is critical for LSD1’ binding with and ubiquitination by RNF20.
Fig. 5.
CDK9 promotes LSD1’s binding with and stabilization by RNF20. (A) Flag-LSD1 was co-expressed with Myc-RNF20 in HEK293T cells treated with or without CDK9 specific inhibitor LDC000067 (LDC067, 20 µM) for 24 h. Lysates were analyzed by western blot. (B) HEK293T cells transfected with indicated plasmids with or without LDC067 treatment. Cell lysates were IP using an antibody against Myc and then analyzed by immunoblotting. (C) MDA-MB-231 cells were transfected with control or CDK9 shRNA and then treated with 10 μM MG132 for 6 h. Cell lysates were IP using an antibody against LSD1 (Left) or RNF20 (Right) and then analyzed by immunoblotting. (D) HEK293T cells transfected with indicated plasmids were analyzed by western blot. (E) After treatment with CHX for the indicated time intervals, expression of Flag-LSD1-WT or Flag-LSD1-5A and Myc-RNF20 was analyzed by western blot using Flag and Myc antibodies, respectively. Presented data are representative of three separate experiments. (F) The intensity of LSD1 expression for each time point in (E) was quantified by densitometry and plotted. (G) HEK293T cells were transfected with HA-CDK9, Myc-RNF20, and Flag-LSD1-WT or Flag-LSD1-5A, and then treated with MG132 for 6 h. Cell lysates were IP using an anti-Myc antibody then analyzed by immunoblotting using an anti-Flag antibody. (H) HEK293T cells were transfected with HA-Ub, Myc-RNF20, and Flag-LSD1-WT or Flag-LSD1-5A and then treated with 10 μM MG132 for 6 h. Adjusted cell lysates were IP using an anti-Flag antibody and then analyzed by immunoblotting using an anti-HA antibody. (I) HEK293T cells were transfected with HA-Ub, Myc-RNF20, and Flag-LSD1 and then treated with or without LDC067 and treated with 10 μM MG132 for 6 h. Cell lysates were IP using an anti-Flag antibody and then analyzed by immunoblotting using an anti-HA antibody. ***P < 0.001, compared with controls.
CDK9 and RNF20 Inhibition Activate ERVs and an IFN Response through LSD1.
Both LSD1 and CDK9 inhibition result in genome-wide epigenetic depression, activating ERVs and leading to an IFN response, ultimately resulting in epigenetic immunosensitization (11, 12, 25). To assess whether the loss of RNF20 yields a similar effect, we conducted an RNA-Seq analysis to comprehensively explore how RNF20 regulates ERVs expression and the activation of the IFN response. Our analysis revealed a substantial impact of RNF20 inhibition on gene expression in MDA-MB-231 cells (SI Appendix, Fig. S4A). Gene ontology (GO) enrichment analysis of the differentially expressed genes demonstrated a notable enrichment of up-regulated genes in terms related to type 1 IFN response and antiviral response (Fig. 6A). These findings were further corroborated by GSEA (Fig. 6B). Additionally, we observed an upregulation of repetitive elements and HERVs following RNF20 inhibition (Fig. 6C and SI Appendix, Fig. S4 B and C). Furthermore, H3K4me2 levels of IFN-responsive genes increased with RNF20 knockdown (Fig. 6D and SI Appendix, Fig. S4D). Importantly, many ERVs exhibited elevated H3K4me2 levels upon RNF20 knockdown (Fig. 6D and SI Appendix, Fig. S4 E–G). We validated two ERVs and three IFN signaling pathways, including CCL5, CXCL9, and CXCL10. In line with previous report (12), knockdown of LSD1 increased the expression of these genes (Fig. 6E and SI Appendix, Fig. S5 A and B). Knockdown of CDK9 or RNF20 also resulted in the accumulation of ERVs and IFN response genes, suggesting that CDK9 and RNF20 epigenetically repress the expression of these genes through LSD1 (Fig. 6E and SI Appendix, Fig. S5 A and B). Furthermore, knockdown of CDK9 or RNF20 increased the levels of H3K4me2 in the promoters of CCL5, CXCL9, and CXCL10, which is consistent with the effect of LSD1 deficiency (Fig. 6F). Additionally, the levels of LSD1 in these gene promoters were decreased in response to loss of CDK9 or RNF20 (SI Appendix, Fig. S5C). To confirm that LSD1 phosphorylation is relevant to gene activation, we overexpressed either LSD1-WT or LSD1-5A in LSD1-depleted MDA-MB-231 cells. Similar to previous report (11), depletion of LSD1 up-regulated IFN/antiviral-responsive genes (Fig. 6G and SI Appendix, Fig. S5 D and E). However, overexpression of LSD1-WT, but not LSD1-5A mutant, rescued the expression of these genes. We further tested whether LSD1 is required for gene reactivation by CDK9 inhibition by treating cells with the CDK9 inhibitor LDC067. As previously reported (25), CDK9 inhibition activates ERVs and an IFN response (Fig. 6H). However, this effect was impaired by LSD1-WT but not by the LSD1-5A mutant. Furthermore, overexpression of LSD1-WT, but not by the LSD1-5A mutant, was able to restore the expression of these genes in CDK9-depleted cells (Fig. 6I and SI Appendix, Fig. S5F). To determine the functional impact of RNF20-mediated LSD1 accumulation, we knocked down endogenous RNF20 in both MDA-MB-231 and EMT6 cells. This loss of RNF20 led to an increase in the expression of IFN response genes and ERVs (Fig. 6J and SI Appendix, Fig. S5 G and H). Most importantly, the ectopic expression of LSD1 largely recovered the expression of these genes in both cell lines. In aggregate, these results demonstrate that CDK9 and RNF20-mediated LSD1 stabilization are essential for maintaining the silencing of the IFN response gene and ERVs.
Fig. 6.
CDK9 and RNF20 inhibition activate ERVs and an IFN response through LSD1. (A) A dot map showing top seven immune-related terms in GO analysis of up-regulated genes (log2(FC-KD/Ctrl) > 0.5 and false discovery rate [FDR] < 0.05) in RNF20 knockdown versus control MDA-MB-231 cells. (B) GSEA in RNF20 knockdown versus control MDA-MB-231 cells. Gene list was ranked with signed (from log2FC) likelihood ratio from RNF20 knockdown versus control comparison. (C) Heatmaps for differential expression (FDR < 0.05) of transcripts of ERVs between control and RNF20 knockdown MDA-MB-231 cells. (D) H3K4me2 ChIP-Seq signals at promoter regions of induced IFN-responsive genes or genomic loci of individual ERVs in control and RNF20 knockdown cells. (E) The RT-qPCR analysis of transcript of selected ERVs and IFN response genes in MDA-MB-231 cells transduced with shRNA against scramble, CDK9, RNF20, or LSD1. (F) MDA-MB-231 cells were transfected with sh-Control, sh-CDK9, sh-RNF20, or sh-LSD1 and ChIP assays were performed using an anti-H3K4me2 antibody. The IP DNA was analyzed by real-time PCR using specific primers in the promoter of CCL5, CXCL9, and CXCL10. (G) The RT-qPCR analysis of transcript of selected ERVs and IFN response genes in MDA-MB-231 cells transduced with shRNA against scramble or LSD1, and rescued with either WT-LSD1 or LSD1-5A. (H) The RT-qPCR analysis of transcript of selected ERVs and IFN response genes in (G) treated with LDC067 (20 µM) for 72 h. (I) The RT-qPCR analysis of transcript of selected ERVs and IFN response genes in CDK9-depeleted MDA-MB-231 cells transduced with WT-LSD1 or LSD1-5A. (J) The RT-qPCR analysis of transcript of selected ERVs and IFN response genes in RNF20-depeleted MDA-MB-231 cells transduced with WT-LSD1. *P < 0.05, **P < 0.01, ***P value < 0.001 when control or rescued group was compared with Control shRNA group.
Loss of RNF20 Potentiates In Vivo Response of Breast Tumor Xenografts to Anti-PD-1 Immunotherapy.
Given our in vitro findings that RNF20 regulates cell processes potentially related to tumor immune response, we next investigated whether RNF20 inhibition might trigger anti-tumor immunity in vivo. To explore this possibility, we used mouse syngeneic tumor models by inoculating EMT6 cells into mammary fat pad of BALB/c mice. Deletion of RNF20 in EMT6 cells significantly inhibited tumor growth in vivo, as assessed by both tumor size and weight, in agreement with our in vitro observations (SI Appendix, Fig. S6 A–D). In parallel, immunohistochemical (IHC) analysis revealed that apoptosis (cleaved-Caspase-3) was markedly increased, while cell proliferation (Ki-67) was significantly decreased in RNF20 knockdown EMT6 tumors (SI Appendix, Fig. S6 E and F). Furthermore, the expression of exogenous LSD1 in RNF20-knockdown cells largely rescued the tumor growth (SI Appendix, Fig. S6), consistent with the in vitro function of LSD1.
To further investigate the mechanism connecting RNF20 inhibition to enhanced anti-tumor cell immunity, we assessed the effect of tumor cell-intrinsic RNF20 on immune cell activity in the tumor microenvironment. We observed a significant increase in the numbers of CD45+ immune cells and the percentages of CD8+ T cells in the tumor microenvironment (Fig. 7 A and B). The recruitment of CD8+ T lymphocyte to EMT6 tumors was also increased in RNF20 knockdown tumors (Fig. 7 C and D). Furthermore, we found that RNF20 expression negatively associated with CD8+ T cell infiltration and NK cell infiltration in the TCGA database (Fig. 7E). These findings supported the hypothesis that RNF20 inhibition could sensitize tumors to immune checkpoint inhibitors. To test this, we used the EMT6 mouse model, which is known to express PD-L1 but have poor immunogenicity and be non-responsive to PD-1 blockade. As expected, PD-1 blockade alone has bare effects on EMT6 tumor growth (Fig. 7F). However, PD-1 blockade markedly reduced the growth of RNF20 knockdown tumor. Importantly, the expression of exogenous LSD1 in RNF20-knockdown cells largely recovered the recruitment of CD45+ and CD8+ T cells and restored resistance to PD-1 blockade (Fig. 7). Taken together, these results suggest that RNF20 inhibition triggers cytotoxic CD8+ T cell infiltration, which in turn enhances the in vivo antitumor efficacy of immune checkpoint inhibitors.
Fig. 7.
Loss of RNF20 potentiates in vivo response of breast tumor xenografts to anti-PD-1 immunotherapy. (A) Immune cells (CD45+) in the tumor microenvironment from transplanted EMT6 tumors in immunocompetent mice were analyzed by flow cytometry at day 21 post implantation. (B) Immune cells (CD8+) in the tumor microenvironment as described in (A). (C) IHC analysis of immune infiltration of CD8+ T cells in (A). Representative immunohistochemistry staining of CD8+ T lymphocytes. (Scale bar, 100 µm.) (D) The percentages of CD8+ T lymphocytes in (C) were measured and analyzed by Leica imaging software. (E) Pan-cancer analysis of TCGA dataset for RNF20 expression with indicated gene expression signature. (F) Tumor growth of immunocompetent mice inoculated with EMT6 cells transduced with shControl, RNF20 shRNA, or ectopic expressing LSD1 in RNF20-depleted cells, and treated with anti-PD-1 or isotype control. Arrows indicate time points of anti-PD-1 injection. #, no significance; ***P < 0.001, compared with controls.
RNF20 and LSD1 Are Coordinately Overexpressed in Tumors.
To evaluate the clinical relevance of our finding, we measured RNF20 and LSD1 protein expression patterns in breast cancer patients using IHC on a tumor tissue microarray. We observed a general tendency of positive protein levels between RNF20 and LSD1 in the tumors (Fig. 8A). Quantification of the staining intensity revealed a statistically significant positive correlation among the specimens analyzed (R = 0.77; P < 0.001, Fig. 8B). In all, these results suggest that RNF20 is positively associated with LSD1 in breast tumors.
Fig. 8.
Expression of RNF20 and LSD1 are positively correlated in breast cancer patients. (A) The 227 surgical specimens of breast cancer were immunostained using antibodies against RNF20 and LSD1. Images with consecutive IHC staining of both RNF20 and LSD1 in six cases of breast tumors (Top: three cases of negative staining; Bottom: three cases of positive) are shown. (Scale bar, 100 μm.) (B) Statistical analysis of (A). (C) A proposed model to illustrate CDK9–RNF20–LSD1 axis in epigenetic silencing for ERVs and IFN response suppression.
Discussion
In this study, we have uncovered a unique model in which CDK9 phosphorylates LSD1 protein at the C-terminal. The phosphorylated LSD1 protein is then recognized and polyubiquitylated by the E3 ligase RNF20 complex, leading to accumulation of the LSD1 protein and subsequent epigenetic silencing of the IFN response and ERVs, inducing immune evasion (Fig. 8C). Our mechanistic findings have established that the CDK9–RNF20–LSD1 axis plays a critical role in epigenetic silencing, and targeting this axis presents a potential therapeutic strategy against immune evasion.
CDK9 is known to regulate transcription elongation by forming a complex with Cyclin T1 (24). Recent studies have shown that CDK9 is also required for gene silencing in cancer cells (25). CDK9-mediated BRG1 phosphorylation triggers chromatin remodeling, preventing BRG1 from being recruited to the heterochromatin to move and restructure nucleosomes and mediate gene transcription (25). However, the mechanism by which CDK9 inhibition leads to higher occupancy of H3K4me2 at the promoter regions of hypermethylated CDK9 targeted genes (25) was unknown until our study. We found that CDK9-mediated phosphorylation of LSD1 leads to its stabilization, and CDK9 inhibition induces LSD1 degradation, resulting in higher occupancy of H3K4me2 at the promoter regions. The specific phosphorylation sites we identified are at the C-terminal of LSD1, which is similar to RNAP II. Mutation of these sites impairs interaction between LSD1 and RNF20 and thus facilitates LSD1 degradation. Interestingly, both CDK9 and LSD1 inhibition up-regulate the expression of IFN response gene and ERVs (12, 25). Our results demonstrated that LSD1 is crucial for CDK9-mediated epigenetic silencing (Fig. 6). Additionally, it has been reported that BRG1 is part of the HP1γ–LSD1 complex involved in gene repression (32, 33). It is plausible that BRG1 and LSD1 form the repressing complex to maintain gene silencing. Given that LSD1 overexpression did not fully rescue CDK9-inhibition induced gene expression, it is likely that other factors, such as histone deacetylase (HDAC), may also be involved in CDK9 inhibition-induced gene silencing. It will be important to determine the involvement of other epigenetic enzymes in this process.
Ubiquitination is a process that is regulated by the UPS and involves three enzymes: E1, E2, and E3. Ubiquitin can be attached to lysine (K) residue(s) through any of seven amine groups (K6, K11, K27, K29, K33, K48, and K63), forming polyubiquitin chains with distinct topologies and functional outcomes (34). Previous work suggests that K-6, K-11, and K-48 adopt a compact conformation whereas K-29 and K-63 ubiquitin chains adopt an open conformation (35). The K48 linkage usually leads to protein degradation through the proteasome system, whereas K63 is involved in different biological functions, such as DNA repair and endocytosis. However, little is known about the functions of the other lysine linkage, particularly the K29 linkage. The atypical K29-linked ubiquitination is involved in protein interaction, EV biogenesis, the regulation of a viral infection, protein lysosomal degradation, and protein aggregation (36–38). K29 ubiquitination linkage can constitute a signal leading to protein aggregation. For example, EDD ubiquitinates β-catenin through Lys-29 or lys11-linked ubiquitin chains, leading to enhanced stability of β-catenin (39). HECTD3 also stabilized expression of c-MYC through mediating K29-linked polyubiquitination of c-MYC (40). In addition, K29-linked ubiquitin chains emerge as key contributors to stress response and cell cycle signaling pathways (38). Interestingly. VCP (also known as p97 or Cdc48, an ATPase associated with various activities involved in the processing of ubiquitinated proteins), colocalizes with the K29-linked polyubiquitin signal. This observation suggests a nondegradative role for this modification, distinct from the proteasome (38). These studies underscore the importance of K29-linked ubiquitination in maintaining protein homeostasis through a dual mechanism involving both protein degradation and stabilization. However, a precise understanding of the molecular mechanisms governing when K29-ub leads to protein degradation versus protein stabilization necessitates further investigation. In this study, we found that RNF20 stabilizes LSD1 through K29-linked polyubiquitination (Fig. 4). Acting as a E3 ligases, RNF20 contains a C-terminal RING-finger domain and was first identified as the major E3 ligases responsible for histone H2B lysine (K) 120 monoubiquitylation (H2BK120ub) in mammalian cells (41). In addition to its role in histone ubiquitination, RNF20 has been reported to polyubiquitylate or monoubiquitylate non-histone protein. For example, RNF20 is an E3 ligase for Ebp1, AP-2a, and sterol regulatory element binding protein1c (SREBP1c), leading to their degradation via polyubiquitination (42). In addition, using CRISPR-based pooled screening, RNF20 was identified as an E3 ligase of Foxp3 in regulatory T (Treg) cells and serves as a target for Treg-based immunotherapies for cancer (43). On the other hand, RNF20 stabilizes motor protein Eg5 and a heat shock transcription factor eEF1B δL through monoubiquitylation (44, 45). In the current study, we determined that RNF20 stabilizes another non-histone protein, LSD1, a histone enzyme, through polyubiquitination. Our results were supported by previous study which identified that BRE1A (RNF20) was enriched with LSD1 IP in C4-2B, a prostate cancer cell line using rapid IP mass spectrometry of endogenous proteins (RIME) (46). Furthermore, we found that substrate phosphorylation affects its recognition by RNF20. Specifically, phosphorylation of LSD1 by CDK9 increases its association with RNF20, which subsequently leads to its ubiquitination and stabilization (Fig. 5). This provides a unique mechanism for RNF20-mediated histone remodeling through the regulation of histone modification enzymes.
RNF20 is known to function in transcriptional elongation, DNA damage response, and the maintenance of chromatin differentiation (42). Depletion of RNF20 has been linked to a variety of cellular defects, including genomic instability, impaired tumor suppression, inflammation, and other effects (47). In ovarian cancer, ATAC-Seq and RNA-Seq analyses have identified that RNF20 depletion leads to an open conformation at promoters and alters the immune signaling pathway (48). Consistent with this, previous studies in mice have shown that RNF20 heterozygosity leads to acute and chronic colitis and inflammation-associate colorectal cancer in mice (49). While loss of RNF20 is concomitant with loss of H2BK120ub, the vast majority of genes whose expression increases in RNF20-depleted human cells did not display significant levels of H2BK120ub (50). In addition, H2BK120ub is linked to transcriptional elongation and associated with the transcribed regions of highly expressed genes, whereas RNF20 depletion can both activate and repress distinct genes (51). It was speculated that these two gene cohorts may have different chromatin barriers, indicating the involvement of other epigenetic regulatory proteins and histone modification. Interestingly, it has been reported that RNF20 enzymatically modifies chromatin in the type I IFN response (52). In addition, H3K4me2 levels have been shown to increase and extend in parallel with STAT1 activity in response to IFN treatment (53). Most importantly, knockdown of RNF20 leads to upregulation of IFN response genes through an H3K4me2/3-based mechanism (53). Our study provides potential mechanism underlying this regulation. Specifically, we observed that depletion of RNF20 reduced LSD1 protein level, resulting in a significant increase in H3K4me2 occupancy at the promoter of the IFN response genes (Fig. 6). Moreover, ectopic expression of LSD1 restored the expression levels of these genes. Our study suggests that RNF20 represses IFN response gene by stabilizing LSD1 and promoting its de-methyltransferase activity, thereby reducing H3K4 methylation.
In summary, our findings demonstrate that CDK9 plays a critical role in LSD1 accumulation by phosphorylating LSD1, which enhances its binding with and ubiquitination by RNF20, leading to the repression of ERVs and the IFN response. Moreover, disruption of the CDK9–RNF20–LSD1 axis sensitizes anti-PD-1 treatment, suggesting a potential therapeutic strategy for cancer immunotherapy.
Materials and Methods
The materials used in this study, including cell lines, chemicals, and antibodies, are described in SI Appendix. Detailed descriptions of the study methods, including cell culture and transfection, plasmids and reagents, TMA, IHC, co-IP, WB, RT-qPCR, RNA-Seq, ChIP-Seq, and ChIP-qPCR, in vivo ubiquitination assay, kinase assay, and fluorescence-activated cell sorting, immunofluorescence staining, and in vivo tumorigenesis assay are also provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Drs. Donna Gilbreath for critical reading and editing of this manuscript. This work was supported by grants from NIH (P20GM121327 and CA230758) to Y.W.
Author contributions
B.D. and Y.W. designed research; B.D., X.W., X.S., J.W., X.L., and Y.W. performed research; Z.Y. contributed new reagents/analytic tools; B.D., X.S., Y.Z., J.D., and Y.W. analyzed data; and Y.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. J.-P.I. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All data are available in the main text or the SI Appendix. The accession number for the raw data of ChIP-Seq and RNA-Seq reported in this paper is GSE247308 (54).
Supporting Information
References
- 1.Gomez S., Tabernacki T., Kobyra J., Roberts P., Chiappinelli K. B., Combining epigenetic and immune therapy to overcome cancer resistance. Semin. Cancer Biol. 65, 99–113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao J., Yan Q., Cancer epigenetics, tumor immunity, and immunotherapy. Trends Cancer 6, 580–592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldberg A. D., Allis C. D., Bernstein E., Epigenetics: A landscape takes shape. Cell 128, 635–638 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Shi Y., et al. , Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Schulte J. H., et al. , Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: Implications for therapy. Cancer Res. 69, 2065–2071 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Zhou A., et al. , Nuclear GSK3beta promotes tumorigenesis by phosphorylating KDM1A and inducing its deubiquitylation by USP22. Nat. Cell Biol. 18, 954–966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozono D., et al. , Dynamic epigenetic regulation of glioblastoma tumorigenicity through LSD1 modulation of MYC expression. Proc. Natl. Acad. Sci. U.S.A. 112, E4055–E4064 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi H. J., et al. , UTX inhibits EMT-induced breast CSC properties by epigenetic repression of EMT genes in cooperation with LSD1 and HDAC1. EMBO Rep. 16, 1288–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayami S., et al. , Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer 128, 574–586 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Lim S., et al. , Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31, 512–520 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Qin Y., et al. , Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 38, 390–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng W., et al. , LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan A. H. Y., et al. , Lysine-specific histone demethylase 1A regulates macrophage polarization and checkpoint molecules in the tumor microenvironment of triple-negative breast cancer. Front. Immunol. 10, 1351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., et al. , LSD1 inhibition sustains T cell invigoration with a durable response to PD-1 blockade. Nat. Commun. 12, 6831 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong Z., et al. , OTUD7B deubiquitinates LSD1 to govern its binding partner specificity, homeostasis, and breast cancer metastasis. Adv. Sci. (Weinh) 8, e2004504 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y., et al. , The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 29, 1803–1816 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y. J., et al. , Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Wu Y., et al. , The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 5, 224–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong B., et al. , FBXO24 suppresses breast cancer tumorigenesis by targeting LSD1 for Ubiquitination. Mol. Cancer Res. 21, 1303–1316 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piao L., Suzuki T., Dohmae N., Nakamura Y., Hamamoto R., SUV39H2 methylates and stabilizes LSD1 by inhibiting polyubiquitination in human cancer cells. Oncotarget 6, 16939–16950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., et al. , Arginine methylation-dependent LSD1 stability promotes invasion and metastasis of breast cancer. EMBO Rep. 21, e48597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adelman K., Lis J. T., Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirngruber J., et al. , CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3’-end processing. EMBO Rep. 10, 894–900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ranjan A., et al. , Targeting CDK9 for the treatment of glioblastoma. Cancers 13, 3039 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., et al. , Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell 175, 1244–1258.e26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H. J., et al. , Integrative analysis reveals histone demethylase LSD1 promotes RNA polymerase II pausing. iScience 25, 105049 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamada T., et al. , P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell 21, 227–237 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Van Hoof D., et al. , Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Rigbolt K. T., et al. , System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Hornbeck P. V., et al. , PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W., et al. , RNF40 regulates gene expression in an epigenetic context-dependent manner. Genome Biol. 18, 32 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nacht A. S., et al. , Hormone-induced repression of genes requires BRG1-mediated H1.2 deposition at target promoters. EMBO J. 35, 1822–1843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nacht A. S., Beato M., Vicent G. P., Steroid hormone receptors silence genes by a chromatin-targeted mechanism similar to those used for gene activation. Transcription 8, 15–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochstrasser M., Origin and function of ubiquitin-like proteins. Nature 458, 422–429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristariyanto Y. A., et al. , K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of k29 polyubiquitin. Mol. Cell 58, 83–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nucifora F. C. Jr., et al. , Ubiqutination via K27 and K29 chains signals aggregation and neuronal protection of LRRK2 by WSB1. Nat. Commun. 7, 11792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farooq A. U., et al. , K-29 linked ubiquitination of Arrdc4 regulates its function in extracellular vesicle biogenesis. J. Extracell. Vesicles 11, e12188 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Y., et al. , K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle. Nat. Chem. Biol. 17, 896–905 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hay-Koren A., Caspi M., Zilberberg A., Rosin-Arbesfeld R., The EDD E3 ubiquitin ligase ubiquitinates and up-regulates beta-catenin. Mol. Biol. Cell 22, 399–411 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang G., et al. , HECTD3 promotes gastric cancer progression by mediating the polyubiquitination of c-MYC. Cell Death Discov. 8, 185 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu B., et al. , Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Mol. Cell 20, 601–611 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Fu J., et al. , Epigenetic modification and a role for the E3 ligase RNF40 in cancer development and metastasis. Oncogene 40, 465–474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortez J. T., et al. , CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 582, 416–420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duan Y., et al. , Ubiquitin ligase RNF20/40 facilitates spindle assembly and promotes breast carcinogenesis through stabilizing motor protein Eg5. Nat. Commun. 7, 12648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.In S., Kim Y. I., Lee J. E., Kim J., RNF20/40-mediated eEF1BdeltaL monoubiquitylation stimulates transcription of heat shock-responsive genes. Nucleic Acids Res. 47, 2840–2855 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sehrawat A., et al. , LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc. Natl. Acad. Sci. U.S.A. 115, E4179–E4188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sethi G., Shanmugam M. K., Arfuso F., Kumar A. P., Role of RNF20 in cancer development and progression–A comprehensive review. Biosci. Rep. 38, BSR20171287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hooda J., et al. , Early loss of histone H2B monoubiquitylation alters chromatin accessibility and activates key immune pathways that facilitate progression of ovarian cancer. Cancer Res. 79, 760–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarcic O., et al. , RNF20 links histone H2B ubiquitylation with inflammation and inflammation-associated cancer. Cell Rep. 14, 1462–1476 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung I., et al. , H2B monoubiquitylation is a 5’-enriched active transcription mark and correlates with exon-intron structure in human cells. Genome Res. 22, 1026–1035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shema E., et al. , The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 22, 2664–2676 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fonseca G. J., et al. , Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe 11, 597–606 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Buro L. J., Chipumuro E., Henriksen M. A., Menin and RNF20 recruitment is associated with dynamic histone modifications that regulate signal transducer and activator of transcription 1 (STAT1)-activated transcription of the interferon regulatory factor 1 gene (IRF1). Epigenet. Chromatin 3, 16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong B., et al. , RNF20 contributes to epigenetic immunosuppression through CDK9-dependent LSD1 stabilization. GSE247308. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE247308. Deposited 8 November 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All data are available in the main text or the SI Appendix. The accession number for the raw data of ChIP-Seq and RNA-Seq reported in this paper is GSE247308 (54).