Abstract
Modern in vitro technologies for preclinical research, including organ-on-a-chip, organoids- and assembloid-based systems, have rapidly emerged as pivotal tools for elucidating disease mechanisms and assessing the efficacy of putative therapeutics. In this context, advanced in vitro models of Parkinson's Disease (PD) offer the potential to accelerate drug discovery by enabling effective platforms that recapitulate both physiological and pathological attributes of the in vivo environment. Although these systems often aim at replicating the PD-associated loss of dopaminergic (DA) neurons, only a few have modelled the degradation of dopaminergic pathways as a way to mimic the disruption of downstream regulation mechanisms that define the characteristic motor symptoms of the disease. To this end, assembloids have been successfully employed to recapitulate neuronal pathways between distinct brain regions. However, the investigation and characterization of these connections through neural tracing and electrophysiological analysis remain a technically challenging and time-consuming process. Here, we present a novel bioengineered platform consisting of surface-grown midbrain and striatal organoids at opposite sides of a self-assembled DA pathway. In particular, dopaminergic neurons and striatal GABAergic neurons spontaneously form DA connections across a microelectrode array (MEA), specifically integrated for the real-time monitoring of electrophysiological development and stimuli response. Calcium imaging data showed spiking synchronicity of the two organoids forming the inter-organoid pathways (IOPs) demonstrating that they are functionally connected. MEA recordings confirm a more robust response to the DA neurotoxin 6-OHDA compared to midbrain organoids alone, thereby validating the potential of this technology to generate highly tractable, easily extractable real-time functional readouts to investigate the dysfunctional dopaminergic network of PD patients.
Graphical abstract
1. Introduction
Parkinson's disease (PD) is a debilitating neurological disorder that affects millions of individuals worldwide [1]. The motor symptoms of PD are primarily caused by the progressive loss of dopamine (DA) input to the striatum via the nigrostriatal pathway, which is responsible for regulating movement and posture [2]. The dopaminergic neurons that project from the substantia nigra to the striatum play a crucial role in this pathway, and their degeneration is a hallmark of PD [3]. Therefore, the development of models that accurately reflect the characteristics of PD has relied significantly on the use of dopaminergic neurons [4]. In this context, organoid-based models of PD are distinctively positioned to integrate the complexity of the native tissue microenvironment within platforms that can be exploited to study the pathophysiology of this neurodegenerative condition while offering functional readouts for drug screening in an individual oriented manner [5]. In recent years, assembloids have garnered significant interest due to their potential in enabling the study of inter-regional connectivity and the formation of pathways within the brain [6]. These models are comprised of two or more organoids that are fused together, creating a 3D structure that better replicates the complexity of neural tissues. The resulting platforms demonstrated high potential for the investigation of developmental diseases, as they enable researchers to investigate the intricate processes involved in the development of the brain and its connections [7,8].
Characterizing the formation and activity of DA pathways in these complex structures poses technical challenges. Traditional imaging methods such as label and viral-based neural tracing are limited in providing the level of resolution necessary to track the complex trajectories of neurites within these 3D structures. Calcium imaging, which is often used to monitor neural activity, is limited by the depth of tissue that can be imaged within an assembloid [9]. Moreover the tools employed to investigate connections within assembloids, such as neurite tracing and calcium imaging, are time- and labor-intensive, requiring individual processing of each sample.
Therefore, given the pressing need for technologies that effectively provide rapid, real-time functional readouts to study events associated to DA pathways in PD, we have created an organoid-based DA neural model. Consisting of two organoids positioned at a distance from each other, it aims to promote the formation of connections between them within an easily accessible region of the platform. To this end, we derived midbrain and striatal organoids from mouse induced pluripotent stem cells (iPSCs), using protocols previously described in the literature [10,11]. Generation of midbrain and striatal organoids were confirmed and comprehensively characterized in these studies. Midbrain and striatal organoids were then matured together in a novel co-culturing surface that involves coating, masking and plasma etching steps that enable designing hydrophilic domains on hydrophobic culture surfaces for spatial control of neurite development. Midbrain and striatal organoids that were co-cultured in this setting, termed dual organoid platforms (DOP), grew neurites towards each other and formed inter-organoid pathways (IOPs) that are predominantly dopaminergic (DA). We were able to record real-time local field potentials (LFPs) from IOPs using microelectrode arrays (MEA) and show that they are more susceptible to DA-selective toxicity of 6-OHDA compared to individual organoids. Our results show that real-time reading approaches can be combined with the high complexity nature of organoid models to potentially generate high-throughput data for accelerating drug discovery.
2. Results
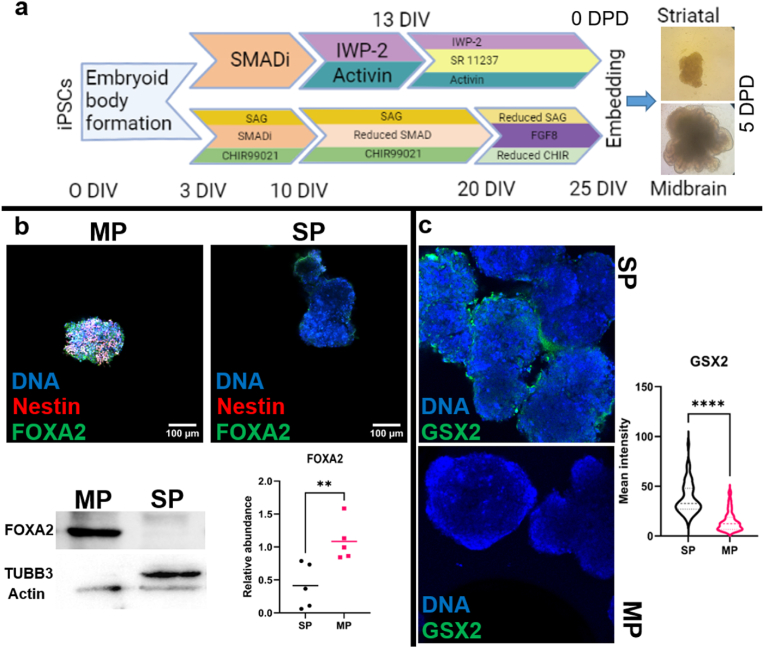
We first optimized the differentiation conditions for iPSCs required to obtain precursor cells resembling both the embryonic floor plate and lateral ganglionic eminence (LGE), two distinctive structures of the native midbrain and striatum, respectively. To this end, midbrain precursor (MP) and striatal precursor (SP) spheroids were derived from iPSCs by sequential treatment with different modulating factors according to the timeline displayed in Fig. 1a. In particular, floor plate progenitors are known to generate midbrain dopaminergic neurons [12] and SHH/WNT signaling activators have been previously used in patterning iPSCs toward floor plate phenotype for midbrain organoids [10]. In order to promote dopaminergic fate in midbrain organoids, FGF-8 was also included in the pre-differentiation stage [13]. Early retraction of SMAD inhibitors in conjunction with WNT inhibition, activin and delayed activation of retinoid-X receptors to obtain LGE progenitors that give rise to striatal projection neurons [14] towards developing striatal organoids was also previously described [11]. Immunofluorescence imaging carried out at the end of pre-differentiation period confirmed the expression of floor plate marker FOXA2 [15] in MP spheroids. Western blot analysis showed 8.6-fold ±2.1 SEM more FOXA2 intensity in MP compared to SP spheroids (Fig. 1b). Similarly, the lateral ganglionic eminence marker, transcription factor GSX2 [16] was expressed by SP spheroids and the fluorescence signal intensity in SP nuclei was 2.6-fold higher (38.0 ± 1.9 SEM vs 14.6 ± 1.2 SEM for SP and MP respectively) compared to MP spheroids. (Fig. 1c). At this stage, the SP spheroids began displaying β-tubulin III expression and a loss of nestin staining, indicating the onset of neuronal differentiation, unlike MP cells which still exhibited nestin (Fig. 1b).
Fig. 1.
Generation of MP/SP spheroids and IOPs. (a) Sequence of reagents used to simultaneously derive SP and MP spheroids from iPSCs. Points of transition during spheroid derivation were denoted with days in vitro (DIV). After Matrigel embedding, the timeline was denoted as days post-differentiation (DPD). Optical microscopy images showing midbrain and striatal organoids at 5 DPD. (b) Immunofluorescence and western blot results given with representative images showing FOXA2 expression in MP spheroids at 25 DIV. FOXA2 western blots were normalized against the total protein in the sample using stain-free gels (Fig. S1) while TUBB3 is shown with actin loading control probed within the same gel. (c) GSX2 immunofluorescence representative images and violin plot showing the mean pixel intensities measured from individual nuclei using 3 different lines.
To prepare a platform capable of supporting the co-maturation of organoids and the subsequent pathway formation, glass and MEA surfaces were first coated with a hydrophobic perfluorooctyl (PFO), and successively oxygen plasma patterning was used to create hydrophilic regions thereby precisely defining the geometry of the Matrigel matrix for organoid culturing (Fig. 2a). Changes in hydrophobicity/hydrophilicity of surfaces after each treatment were monitored by measuring water contact angles (WCA). The initial average WCA of surfaces was determined to be 40.5° ± 2.0° SEM which increased to a 94.0° ± 1.3° SEM after the PFO application (Fig. 2b). Following the patterning process, areas that were exposed to oxygen plasma displayed a mean WCA of 23.3° ± 1.1° SEM, confirming the creation of highly hydrophilic domains within a markedly hydrophobic surface.
Fig. 2.
Surface preparation and formation of dual-organoid platforms. (a) Illustration of surface treatments for generation of IOPs given in cross-section. (b) Water contact angles of hydrophobic and hydrophilic domains given with representative droplets. (c) Top-down view schematics of the mask and resulting geometry of the dual-organoid platform. (d) Immunostaining of an IOP for TUBB3 at 15 DPD.
MP and SP spheroids were placed within hydrophilic domains patterned at a distance of 3 mm from each other and Matrigel was carefully added to cover the spheroids and the inter-organoid hydrophilic channel, thereby creating two separate yet interconnected maturation regions for the organoids, as illustrated in Fig. 2c. In these conditions, midbrain neuroepithelial growth emerged at 3 DPD (days post-differentiation) (Fig. 1a) and neurites started to sprout from the organoids’ body by 7 DPD. β-tubulin III-positive neurites appeared at 14 DPD on both organoids, showing a preferential growth direction oriented along the channel connecting the two organoids (Fig. 2d).
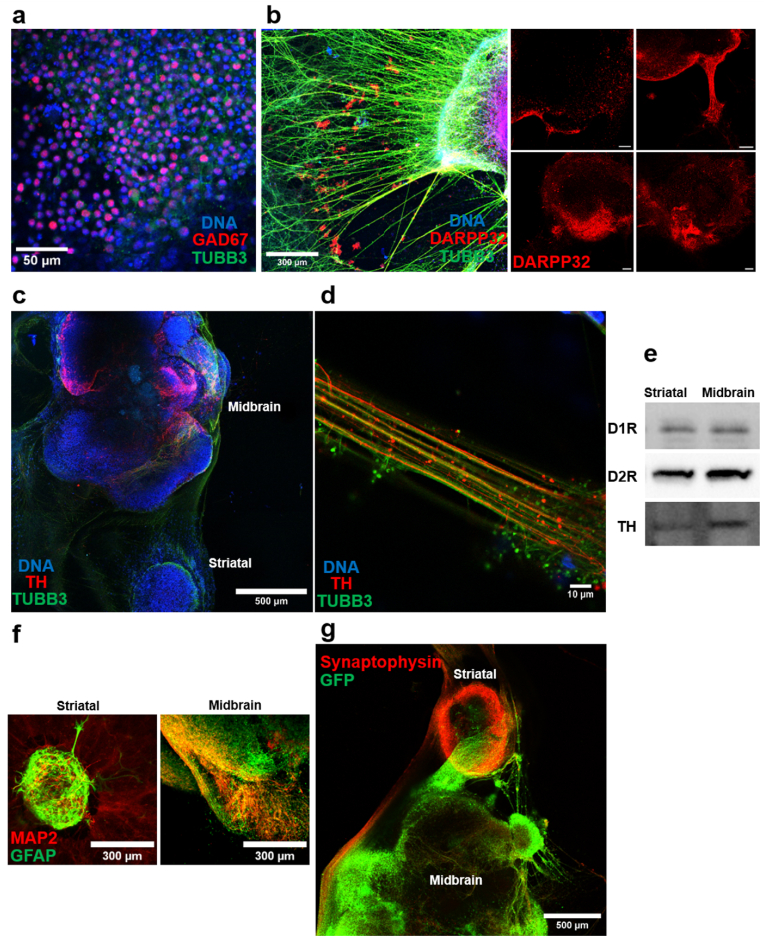
Further characterizations were performed at 30 DPD for sole midbrain organoids or inter-organoid pathways (IOP). At this time point, midbrain organoids reach 1623 ± 93 μm SEM diameter while striatal organoids reach a smaller 737 ± 85 μm SEM diameter. Both organoids retain their spherical shape with 0.81 ± 0.04 SEM and 0.88 ± 0.03 SEM circularity measurements for midbrain and striatal organoids respectively. Striatal organoids in the DOPs presented an outer shell of GABAergic inhibitory neurons consisting of 61.8% ± 1.7% SEM GAD67 expressing cells in addition to abundant expression of DARPP32, a protein associated with synaptic plasticity in the striatum and promoting dopamine receptor D1 expression (Fig. 3a and b). These results validate the striatal identity of the organoids in relation to their neuronal phenotype. Interestingly, DARPP32 expression in striatal organoids was consistently stronger at the side facing the midbrain organoid in DOPs (Fig. 3b). This phenomenon may be ascribed to potential paracrine signaling between the cells of two organoids, leading to the co-regulation of the spatial distribution of cells.
Fig. 3.
Characterization of DOPs at 30 DPD. (a) Immunofluorescence of GAD67 and TUBB3 of striatal organoid. (b) DARPP32 and TUBB3 staining of striatal organoids. Images of replicated DOPs were given in the same panel displaying DARPP32 expression consistently stronger on the edge facing the midbrain organoid. Scale bar 100 μm. (c) Maximum projection image of a single DOP showing both organoids and IOP stained for TH and TUBB3. (d) Close-up image of IOP displaying TH expression. (e) Western blots showing dopamine receptors and TH expression in midbrain and striatal organoids of the same DOP. (f) GFAP and MAP2 staining of midbrain and striatal organoids showing the distribution of neuronal and glial populations. (g) GFP signal observed after VSV-GFP injection into the midbrain organoid given together with synaptophysin staining.
DA neuron regionalization in midbrain organoids was observed by immunofluorescence imaging, as well as fully formed IOPs comprising 57.76% ± 6.4% SEM TH+ neurites (Fig. 3c and d). Fig. 3c also displays symmetrical distribution of DA neurons along the midline axis of the midbrain organoid resembling the symmetric structural organization observed in the brain. D1 and D2 receptors were detected by western blot in both organoids of the DOPs at similar levels. Western blot analyses of tyrosine hydroxylase (TH) in DOPs revealed a 2.80-fold ±0.63 SEM more intense signal for midbrain organoids in comparison to striatal organoids (Fig. 3e). These findings substantiate the phenotypically distinct midbrain and striatal identities of the organoids within the dual organoid platforms. Presence of glial populations in both organoids was also demonstrated by glial fibrillary acidic protein (GFAP) staining shown in Fig. 3f.
Formation of synaptic connections through IOPs was assessed by injecting the midbrain organoids with GFP labeled vesicular stomatitis virus (VSV). This virus was previously shown to enable anterograde trans-synaptic tracing in a wide range of organisms [17]. In Fig. 3g, the progression of neurites from the midbrain organoid to the striatal organoid, labeled with viral green fluorescent protein (GFP), is depicted. The viral GFP is also evident at the central region of the striatal organoid. This observation provides evidence for the existence of continuous synaptic pathways between the organoids. Furthermore, the presence of synapses at the periphery of the striatal organoid is demonstrated through synaptophysin staining, confirming the formation of synapses between the organoids.
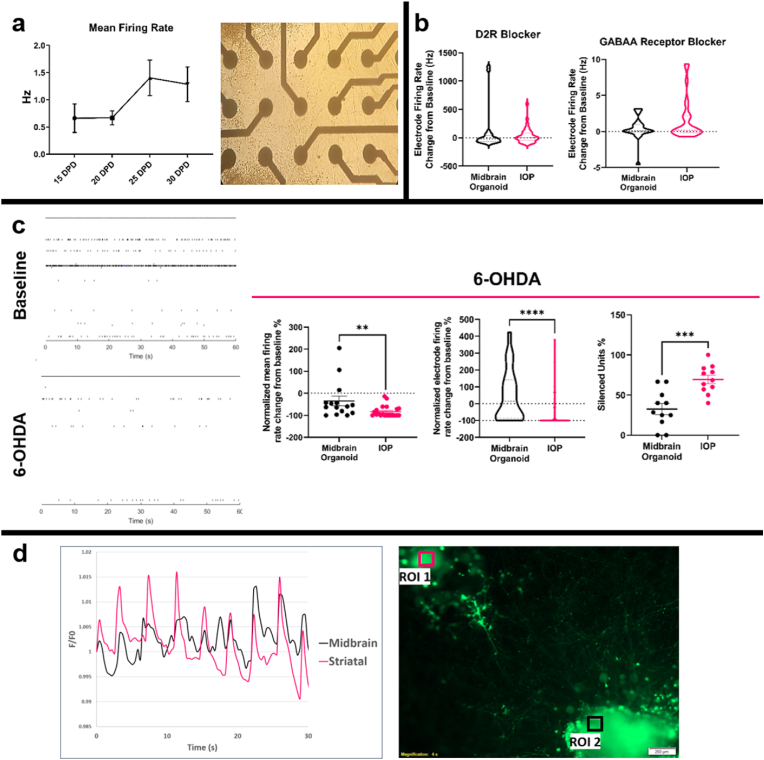
The connectivity of IOPs was tested by monitoring calcium transients in the organoids using Fluo 4-AM calcium imaging. Examination of the spontaneous activities of midbrain and striatal organoids, as depicted in Fig. 4d and supplementary video, revealed temporally synchronized activity. In order to quantify the synchrony between the organoids, correlation coefficients were calculated for each of 20 recorded DOPs. Mean correlation coefficient among them was found to be 0.977 ± 0.007 SEM, substantiating that constructed DOPs consistently result in functional connections and synchronous neural activity.
Fig. 4.
Microelectrode array (MEA) measurements obtained from IOPs and single midbrain organoids. At least 30 samples were recorded for each experiment. (a) IOPs showing the progression of electrical activity maturation in mean firing rate plot given with representative brightfield image of IOP at 25 DPD. (b) Change of IOP activity from baseline 20 min after treatment with D2 dopamine receptor and GABAA receptor blockers displayed in truncated violin plots. (c) 6-OHDA (10 μM) treatment characterization comparing before and 24 h after treatment of IOPs and single midbrain organoids. Representative raster plots were given with firing rate change from baseline at the single electrode and IOP level. Percentage of completely silenced units in each midbrain organoid and IOP were calculated by spike sorting and determining the number of actively firing units. d) Intensity profile of calcium transients over a period of 30 s (85 cycles) and the correlating region of interests (area = 9048 μm2) displayed on Fluo-4, AM loaded organoids, representative from 20 DOPs.
In order to collect local field potential (LFP) measurements from IOPs, DOPs were constructed on MEA surfaces. LFP outputs of IOPs were compared to single midbrain organoids as a control group where a single MP spheroid was placed on the electrode surface of the MEA and allowed to mature in identical conditions to IOPs. The IOPs formed on the electrodes started displaying spontaneous LFPs starting 14 DPD and the electrical activity reached maturity at 25 DPD (Fig. 4a). Transparent MEA surfaces allowed monitoring organoid growth during the experiments and any electrodes overgrown by the organoids were eliminated to enable analyzing signals exclusively derived from the IOP.
In an attempt to evoke activity in IOPs, GABAA receptors and D2 dopamine receptors were blocked by gabazine (5 μM) and raclopride (1 μM) treatments respectively (Fig. 4b). Gabazine treatment induced a mean firing rate increase from 1.6 Hz ± 0.6 SEM to 3.2 Hz ± 1.0 SEM in IOPs. Midbrain organoids alone showed a similar response to gabazine treatment with mean firing rate increasing from 1.4 Hz ± 0.6 SEM to 2.4 Hz ± 0.9 resulting in statistically indistinguishable responses between groups. Blocking D2 dopamine receptors did not significantly change the mean firing rates of either IOPs or midbrain organoids and changes from baseline were statistically similar between these groups. Median change from baseline was −1%±22 SEM for IOPs and −23.3% ± 79.2 SEM for midbrain organoids.
When IOPs and midbrain organoids were treated with 10 μM 6-OHDA, the change in mean firing rate after 24 h was −96.5% ± 5.5 SEM versus −59.9% ± 21.8 SEM respectively (Fig. 4c). The significant response difference between two models is also observable in individual electrode firing rates evident in the truncated violin plot in Fig. 4c. The difference in 6-OHDA responses points to IOPs being a more sensitive model compared to midbrain organoids alone, due to the proximity of DA pathways to the electrode surface. When the signals were analyzed with a valley-seeking, automated spike sorting algorithm, a greater percentage of detected units were completely silenced in IOPs, indicating an electrophysiological DA model that is more sensitive to 6-OHDA.
3. Discussion
High-throughput testing of therapeutic candidates for PD requires disease models that reflect the characteristics of DA neurons as well as spatial distribution of DA connections between the substantia nigra and striatum, a goal severely hindered by the lack of availability of unadulterated DA neuron populations in vitro [18].
In recent years, the development of midbrain organoids has enhanced iPSC-derived models of Parkinson's disease (PD), introducing greater complexity to better simulate the physiological environment [19]. In particular, the latest advancements in assembloids have provided valuable opportunities for investigating the intricate connections between distinct brain regions. These novel platforms allow for the examination of inter-regional connectivity, facilitating a more comprehensive understanding of the complex neural circuitry within the brain. However, these advancements have also presented challenges associated with obtaining electrophysiological output from the 3D volume of the organoids. Notably, the comprehensive characterization of connecting neurites and the assessment of their neuronal activity within assembloid models necessitate the utilization of multiple imaging modalities, which are sequentially applied to each individual assembloid.
Here, we present a novel neural pathway model established between a midbrain and a striatal organoid, primarily composed of dopaminergic neurites. This platform serves as a proof-of-principle, demonstrating its potential for non-destructive, real-time, potentially high-throughput electrophysiological readouts that exhibit heightened sensitivity to 6-OHDA toxicity in comparison to midbrain organoids. As a proof-of-principle, 6-well MEA plates from Axion Biosystems were utilized. This approach allowed data recording from nearly a hundred samples within minutes, showcasing the efficiency of our approach. The potential for conducting even higher throughput experiments is indicated by the availability of microelectrode array (MEA) systems in a 96-well plate format. Such electrophysiological outputs hold promise for assessing the impacts of toxins, therapeutics, and transgenic constructs in a dynamic and time-resolved manner. In our work, striatal organoid differentiation [11] started before Matrigel embedding, as evidenced by a loss of nestin expression, which has not been mentioned elsewhere to our knowledge. Even though the intra-organoid distribution/patterning of DA neurons did not appear to be affected by the co-culture environment, it is noteworthy to observe preferential distribution of DARPP32+ medium spiny neurons closer to the midbrain organoid, possibly due to inter-organoid paracrine signaling. We demonstrate that midbrain and striatal organoids can be matured in a co-culture setting without compromising relevant phenotypes and the spontaneously forming DA pathways can be restricted to predetermined geometries for electrophysiological characterization. Our MEA results show that physical guidance of DA neurites from midbrain organoid creates an electrophysiological model more closely reflecting the DA phenotype compared to sole midbrain organoids. The resulting platform is an invaluable tool for studying DA pathways and the effects of different pharmaceuticals and gene constructs with the limitation that the electrophysiological characteristics of neurons that emerge may differ from the DA neuron subtypes relevant in PD pathophysiology.
In contrast to emerging assembloid models, our DOPs do not feature fused organoids. By enabling real-time electrophysiological recordings, this novel platform provides a means to study and characterize the functional properties of neural connections between two organoids. It allows direct observation of the spatiotemporal patterns of electrical activity occurring between neural organoids without requiring neural tracing and calcium imaging within a 3D assembloid. These real-time readings can provide invaluable insights into the communication and integration of signals across the neural circuitry, mirroring the dynamic nature of the brain in vivo. Applied to different region-specific organoids, the platform offers a powerful tool for studying the development and organization of neural pathways between different brain regions, aiding in understanding normal brain development as well as the pathogenesis of neurodevelopmental disorders. Moreover, the model holds great potential for disease modeling and drug discovery. By utilizing patient-derived cells or genetic modifications, it can allow recreation of specific pathologies or genetic conditions within neural connections. The real-time electrophysiological recordings allow for the assessment of how these pathological factors affect the functionality of the neural connections. This provides a platform for screening potential therapeutic interventions and evaluating their efficacy in a more physiologically relevant context.
In conclusion, the development of this novel in vitro model enabling real-time electrophysiological recordings within interconnected organoids represents a significant advancement in the field. This platform overcomes the limitations of assembloids by providing a means to study and characterize the functional properties of neural connections between two distinct organoids without imaging modalities. It offers new avenues for streamlined disease modeling and drug discovery, allowing the recreation of specific pathologies and the evaluation of therapeutic interventions in a more physiologically relevant context.
4. Materials and methods
4.1. iPSC maintenance and embryoid body formation
Commercially available mouse iPSCs were (Alstem, iPS02 m) propagated in feeder-free conditions on gelatin-coated culture surfaces. Cultures were periodically tested for mycoplasma with Lookout mycoplasma PCR detection kit (Sigma Aldrich, MP0035). iPSC maintenance medium was composed of KnockOut DMEM (Gibco, 10829018) supplemented with 15% knockout serum replacement (Gibco, N10828028), 1% MEM non-essential amino acid solution (Stemcell, 07600), 200 μM L-glutamine (Gibco, 25030), 1% penicillin-streptomycin (Gibco, 15070063), 100 μM 2-mercaptoethanol (Gibco, 31350) and 1000 U/mL leukemia inhibitory factor (LIF).
Embryoid body formation was initiated by detaching iPSCs from culture surfaces using TryplE (Gibco, 12604013) and re-suspending in fresh iPSC maintenance medium without LIF. Cell suspensions were transferred to Aggrewell 800 plates (Stemcell, 34811) treated with anti-adherence rinsing solution (Stemcell, 07010) and embryoid bodies were allowed to form overnight. They were transferred to low-attachment well plates the next day and kept for 2 additional days. The first day of embryoid body formation was regarded as 0 DPD for planning the timeline of subsequent treatments.
4.2. Generation of striatal and midbrain precursor spheroids from embryoid bodies
For neural induction of EBs, N2B27 medium was used which consists of an equal volume mixture of Neurobasal medium (Gibco, 21103049) and DMEM/F12 (Millipore, DF-041-B) supplemented with 1% B27 without vitamin A (Gibco, 12587010), 0.5% N2 (Gibco, 17502048), 1% L-glutamine and 1% penicillin-streptomycin.
In order to generate SP spheroids, embryoid bodies on 3 DIV were transferred to N2B27 medium containing 5 μM TGF-β inhibitor SB525334 (Tocris, 3211) and 250 nM BMP inhibitor dorsomorphin (Tocris, 3093). At 10 DIV, these inhibitors were removed from the medium and 100 nM Wnt inhibitor IWP-2 (Peprotech, 501530240) and 5 ng/mL activin A (Millipore, SRP6057) were added. At 13 DIV, 100 nM retinoid X receptor agonist SR11237 (Millipore, S8951) was added to the medium and cells were continued to be cultured without passaging. Resulting spheroids were used for striatal organoid differentiation starting 25 DIV onwards.
For MP spheroids, 3 DIV embryoid bodies were cultured in N2B27 medium supplemented with 5 μM SB525334, 250 nM dorsomorphin, 500 nM smoothened agonist SAG (Millipore, SML1314), 3 μM Wnt agonist CHIR 99021 (Millipore, SML1046) and 200 μM ascorbic acid (Millipore, A4403). At 10 DIV, SB525334 and dorsomorphin concentrations were reduced to 1 μM and 100 nM, respectively. Spheroids were transferred to pre-differentiation medium at 20 DIV, consisting of N2B27, 500 nM SAG, 700 nM CHIR 99021, 100 ng/mL FGF8 (Invitrogen, RP87811) and 200 μM ascorbic acid. After 25 DIV spheroids were used for midbrain organoid differentiation.
4.3. Fabrication of inter-organoid pathways
Glass bottom tissue culture plates (Greiner, 07000680) and MEA multi-well plates (Axion, M384-tMEA-6B) were prepared for spheroid positioning by treating with Trichloro(1H, 1H, 2H, 2H perfluorooctyl)silane (Millipore, 448931) vapor for 30 min to obtain hydrophobic surfaces. Next, designed and laser cut adhesive masks were applied onto well surfaces and the plates were treated with oxygen plasma for 30 min. After UV sterilization, one of each SP and MP spheroid was placed on each of the hydrophilic spots in the wells using cut pipette tips. Excess medium was aspirated, and the entire hydrophilic surface was filled with 10 μL Matrigel, covering both spheroids and forming a channel between them. This results in initiation of midbrain and striatal organoids, as previously reported in the literature [10,11], in a co-culture setting. Plates were incubated at 37 °C for 10 min and the wells were filled with maturation medium consisting of N2B27, 10 ng/mL BDNF (Stemcell, 78005), 10 ng/mL GDNF (Stemcell, 78058), 5 ng/mL activin A, 200 μM dibutyryl-cAMP (Peprotech, 501529820), 5 μM DAPT (Stemcell, 72082) and 200 μM ascorbic acid. MEA recordings were obtained with Maestro MEA system using a bandpass filter of 10 Hz-2.5 kHz. Spikes were detected with an adaptive thresholding algorithm set to 6 times the standard deviation. For treatments, baseline recordings were obtained and half of the conditioned media were removed from MEA wells and transferred to tubes. 6-OHDA, raclopride and gabazine were added to these tubes in double concentrations and the media were returned to corresponding wells in order to achieve the targeted reagent concentrations. Raclopride and gabazine readings were obtained 20 min after treatment and 6-OHDA treatment readings were taken after 24 h.
4.4. Calcium imaging and trans-synaptic labeling
DOPs were constructed in glass bottom tissue culture plates (Greiner, 07000680) and allowed to mature until 30 DPD. For trans-synaptic labeling, Indiana aero type VSV-GFP construct was grown as previously described [20]. 5 × 107 plaque forming units were diluted in 1 mL maturation medium. Media from DOP wells were completely aspirated and 100 nL of the VSV-GFP solution was injected in the center of midbrain organoids using a Hamilton syringe with 30G needle. DOPs were incubated without medium for 30 min and then washed with PBS before adding maturation medium. 72 h later, the DOPs were fixed and stained with an anti-GFP antibody (Clontech 632376).
For calcium imaging, DOPs at 20–36 DPD were loaded with 5 μM Fluo-4, AM for 60 min at 37 °C in conditioned media. A washout was performed with conditioned media and the DOPs were incubated at 37 °C for 30 min prior to imaging. Calcium transients were recorded over a period of 30 s (85 cycles) with an Olympus IX81 epifluorescence microscope, Ex 494 nm/Em 506 nm. Region of interests with an area of 9048 μm2, were overlaid on each organoid type and intensity profiles were generated using the Olympus cellSens Dimension software.
4.5. Immunofluorescence
DOPs were washed twice with PBS and fixed in 4% formaldehyde for 1 h followed by blocking/permeabilization in IF buffer composed of 5% donkey serum and 0.5% triton X-100 in PBS. Plates were exposed to 3x (3 min-ON/3 min-OFF cycles) of 150-W microwave in a Pelco Biowave tissue processor in order to enhance deep penetration before overnight incubation at 4 °C. Biowave treatments were repeated before every incubation and washing step henceforth. DOPs were incubated in primary antibodies (Table S1) for 48 h at 4 °C and subsequently washed in PBS overnight before being incubated in secondary antibodies for 24 h at 4 °C. After the final overnight wash in PBS, DOPs were optically cleared by covering in Cytovista 3D cell culture clearing reagent and imaged with Leica LSM 880 confocal microscope.
4.6. Statistical analysis
Results are represented as mean ± SEM, individual data point graphs or truncated violin plots showing median, minimum, maximum and quartile values. Normality testing of data distribution was performed with Anderson-Darling test. All experiments were performed in 3 biological replicates to reach the final n values, meaning 3 different sets if precursor spheroid batches were generated from iPSCs. In immunofluorescence experiments, at least 10 samples were stained for DOPs and spheroids. For MEA recordings, 30 DOPs and 30 midbrain organoids were used to monitor local field potentials. Statistical tests were performed using unpaired t-tests (two-tailed) or Mann–Whitney tests for separate organoid constructs using GraphPad Prism version 9.0.0. Paired tests were performed when evaluating western blot results from midbrain and striatal organoid belonging to the same DOP.
CRediT authorship contribution statement
Alp Ozgun: Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. David J. Lomboni: Investigation, Conceptualization. Amy Aylsworth: Writing – review & editing, Visualization, Investigation. Allison Macdonald: Investigation. William A. Staines: Supervision. Marzia Martina: Writing – review & editing, Investigation. Michael G. Schlossmacher: Supervision. Joseph S. Tauskela: Supervision, Investigation. John Woulfe: Supervision. Fabio Variola: Writing – review & editing, Supervision, Project administration, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Fabio Variola and John Woulfe reports financial support was provided by Government of Canada.
Acknowledgements
We acknowledge the support of the Government of Canada's New Frontiers in Research Fund (NFRF), [NFRFE-2019-00039].
The authors acknowledge the Cell Biology and Image Acquisition Core (RRID: SCR_021845) funded by the University of Ottawa, Ottawa, Natural Sciences and engineering Research Council of Canada, and the Canada Foundation for Innovation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2024.100992.
Contributor Information
John Woulfe, Email: jwoulfe@eorla.ca.
Fabio Variola, Email: fabio.variola@uottawa.ca.
Abbreviations
- 6-OHDA
6-Hydroxydopamine
- DA
dopaminergic
- DIV
days in vitro
- DOP
dual-organoid platform
- DPD
days post-differentiation
- IOP
inter-organoid pathway
- MEA
microelectrode array
- SP
striatal precursor
- SEM
standard error of means
- MP
midbrain precursor
- WCA
water contact angle
Appendix A. Supplementary data
The following are the supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Ding C., Wu Y., Chen X., Chen Y., Wu Z., Lin Z., Kang D., Fang W., Chen F. Global, regional, and national burden and attributable risk factors of neurological disorders: the Global Burden of Disease study 1990–2019. Front. Public Heal. 2022 doi: 10.3389/fpubh.2022.952161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weintraub D., Aarsland D., Chaudhuri K.R., Dobkin R.D., Leentjens A.F., Rodriguez-Violante M., Schrag A. The neuropsychiatry of Parkinson's disease: advances and challenges. Lancet Neurol. 2022 doi: 10.1016/S1474-4422(21)00330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kordower J.H., Olanow C.W., Dodiya H.B., Chu Y., Beach T.G., Adler C.H., Halliday G.M., Bartus R.T. Disease duration and the integrity of the nigrostriatal system in Parkinson's disease. Brain. 2013 doi: 10.1093/brain/awt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez-Morales P.L., Liste I. Stem cells as in vitro model of Parkinson's disease. Stem Cells Int. 2012 doi: 10.1155/2012/980941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z., Cong L., Cong X. Patient-derived organoids in precision medicine: drug screening, organoid-on-a-chip and living organoid biobank. Front. Oncol. 2021;11:5625. doi: 10.3389/FONC.2021.762184/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura Y., Li M.Y., Revah O., Yoon S.J., Narazaki G., Pașca S.P. Engineering brain assembloids to interrogate human neural circuits. Nat. Protoc. 2022 doi: 10.1038/s41596-021-00632-z. [DOI] [PubMed] [Google Scholar]
- 7.Panoutsopoulos A.A. Neuroscientist; 2021. Organoids, Assembloids, and Novel Biotechnology: Steps Forward in Developmental and Disease-Related Neuroscience. [DOI] [PubMed] [Google Scholar]
- 8.Reumann D., Krauditsch C., Novatchkova M., Sozzi E., Wong S.N., Zabolocki M., Priouret M., Doleschall B., Ritzau-Reid K.I., Piber M., Morassut I., Fieseler C., Fiorenzano A., Stevens M.M., Zimmer M., Bardy C., Parmar M., Knoblich J.A. In vitro modeling of the human dopaminergic system using spatially arranged ventral midbrain–striatum–cortex assembloids. Nat. Methods. 2023;20:2034–2047. doi: 10.1038/s41592-023-02080-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tasnim K., Liu J. Emerging bioelectronics for brain organoid electrophysiology. J. Mol. Biol. 2022 doi: 10.1016/j.jmb.2021.167165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smits L.M., Reinhardt L., Reinhardt P., Glatza M., Monzel A.S., Stanslowsky N., Rosato-Siri M.D., Zanon A., Antony P.M., Bellmann J., Nicklas S.M., Hemmer K., Qing X., Berger E., Kalmbach N., Ehrlich M., Bolognin S., Hicks A.A., Wegner F., Sterneckert J.L., Schwamborn J.C. Modeling Parkinson's disease in midbrain-like organoids. Npj Park. Dis. 2019;51(5):1–8. doi: 10.1038/s41531-019-0078-4. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura Y., Li M.Y., Birey F., Ikeda K., Revah O., Thete M.V., Park J.Y., Puno A., Lee S.H., Porteus M.H., Pașca S.P. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 2020;38:1421–1430. doi: 10.1038/s41587-020-00763-w. 2020 3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedele S., Collo G., Behr K., Bischofberger J., Müller S., Kunath T., Christensen K., Gündner A.L., Graf M., Jagasia R., Taylor V. Expansion of human midbrain floor plate progenitors from induced pluripotent stem cells increases dopaminergic neuron differentiation potential. Sci. Reports. 2017;71(7):1–11. doi: 10.1038/s41598-017-05633-1. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi J., Liu Y., Liu H., Chen H., Emborg M.E., Zhang S.C. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells. 2012;30:1655–1663. doi: 10.1002/STEM.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenman J., Toresson H., Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J. Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norton W.H., Mangoli M., Lele Z., Pogoda H.M., Diamond B., Mercurio S., Russell C., Teraoka H., Stickney H.L., Rauch G.J., Heisenberg C.P., Houart C., Schilling T.F., Frohnhoefer H.G., Rastegar S., Neumann C.J., Gardiner R.M., Strähle U., Geisler R., Rees M., Talbot W.S., Wilson S.W. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphé neurones and cranial motoneurones. Development. 2005;132:645. doi: 10.1242/DEV.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman H., Waclaw R.R., Pei Z., Nakafuku M., Campbell K. The homeobox gene Gsx2 controls the timing of oligodendroglial fate specification in mouse lateral ganglionic eminence progenitors. Development. 2013;140:2289–2298. doi: 10.1242/DEV.091090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mundell N.A., Beier K.T., Pan Y.A., Lapan S.W., Göz Aytürk D., Berezovskii V.K., Wark A.R., Drokhlyansky E., Bielecki J., Born R.T., Schier A.F., Cepko C.L. Vesicular stomatitis virus enables gene transfer and transsynaptic tracing in a wide range of organisms. J. Comp. Neurol. 2015 doi: 10.1002/cne.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slanzi A., Iannoto G., Rossi B., Zenaro E., Constantin G. In vitro models of neurodegenerative diseases. Front. Cell Dev. Biol. 2020;8:328. doi: 10.3389/FCELL.2020.00328/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marotta N., Kim S., Krainc D. Organoid and pluripotent stem cells in Parkinson's disease modeling: an expert view on their value to drug discovery. Expert Opin. Drug Discov. 2020;15:427. doi: 10.1080/17460441.2020.1703671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diallo J.S., Vähä-Koskela M., Le Boeuf F., Bell J. Methods Mol. Biol. 2012. Propagation, purification, and in vivo testing of oncolytic vesicular stomatitis virus strains. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.