Abstract
Healthy aquatic ecosystems are essential for human beings. However, anthropogenic activities severely worsen water quality. In this study, using assembling mesocosms, we developed an efficient and easy-to-handle method to monitor the water quality by measuring the electrical conductivity (EC) of water. Our data demonstrate that the growth of two submersed macrophytes, Vallisnerianatans and Vallisneria spinulosa, improves water quality by decreasing EC. Furthermore, using high-throughput DNA sequencing, we analyzed the microbial community abundance and structure in sediment and water columns with or without plant growth. We generated 33,775 amplicon sequence variants from 69 samples of four sediment groups (BkM, CtM, VnR, and VsR) and three water column sample groups (CtW, VnW, and VsW). The results show that the relative abundance of bacteria was higher in the sediment than in the water column. Moreover, the diversity and composition of microbiomes were altered by Vallisneria spp. growth, and the α-diversity of the microbial communities decreased due to submersed macrophytes in both the sediment and water columns. The β-diversity of the microbial communities also varied significantly with or without Vallisneria spp. growth for both the sediment and water columns.
Keywords: Vallisneria natans, Vallisneria spinulosa, water quality, microbial community
Highlights
-
•
Vallisneria spp growth reduced water electrical conductivity.
-
•
Bacterial α-diversity index were significant lower in water columns than sediments.
-
•
Vallisneria spp alert α-diversity index in both sediments and water columns.
-
•
The β-diversity of microbial varied significantly by Vallisneria spp growth.
1. Introduction
Aquatic ecosystems are essential for human society; however, industrial processes and other anthropogenic activities aggravate the pollution of rivers and lakes and severely worsen water quality, further decreasing its self-clarification [[1], [2], [3]]. Phytoremediation through aquatic macrophytes is a green and economic strategy that has proven promising for restoring water quality [4,5]. Aquatic macrophytes are macroscopic flora that can be separated into four groups: (i) emergent macrophytes, (ii) floating-leaved macrophytes, (iii) free-floating macrophytes, and (iv) submerged or submersed macrophytes [6,7]. Submersed macrophytes, as essential maintainers of low trophic levels, are essential in water ecosystems and crucial in nutrient recycling and promoting ecosystem biodiversity and self-clarification, thus reconstructing their health [[8], [9], [10], [11], [12]]. Submersed macrophytes stabilize freshwater ecosystems, promote the removal of pollution from the water environment, and improve water quality by (i) physically adsorbing the pollutants due to their large surface area, (ⅱ) absorbing to support their growth, (ⅲ) absorbing and degrading pollutants, and (ⅳ) providing an adherent surface for the microbial community and producing adherent biofilms. Submersed plants and epiphytic biofilms provide an appropriate microenvironment for microbiomes, thereby removing pollution [[13], [14], [15], [16], [17], [18]].
The physical, chemical, and biological components of aquatic ecosystems exhibit an intimate interdependence [12,[19], [20], [21], [22], [23], [24]]. Submersed macrophytes and interacting microorganisms constitute the main components of the self-purification ability of water bodies. Aquatic plants not only generate oxygen through stem and leaf photosynthesis, creating an oxygen-rich environment in the water, but also recruit the microbial community with strong metabolic ability for organic/inorganic matter by providing carbon sources and other matter to support the microorganisms through secretion from stems, leaves, and roots. Microorganisms provide carbon dioxide and secondary metabolites to host plants and even inhibit the growth of harmful microorganisms, thereby promoting plant growth. Many studies have demonstrated that microbes and submersed macrophytes interact to improve water quality [13,18,[25], [26], [27], [28]]. Microbial diversity affects water quality, and it is also affected by it; thus, microbial diversity can be a bioindicator in aquatic ecosystems [[29], [30], [31], [32]]. Many studies have recently focused on microbiomes in sediments and epiphytic biofilms of urban black odorous ecosystems, as well as other aquatic ecosystems that contain submersed macrophytes [17,[33], [34], [35], [36], [37]]. However, most studies performed in natural water bodies may be affected by many known/unknown factors, and the microbial diversity affected by submersed macrophyte growth under laboratory conditions is rarely reported.
Vallisneria spp. are important submersed macrophytes that are widely used to improve the water quality of freshwater ecosystems and have attracted increasing attention in recent years [11,24,35,[38], [39], [40]]. Vallisneria natans and V. spinulosa are two species of the genus Vallisneria with similar morphologies. V. spinulosa is thought to be endemic to China and is a dominant submersed macrophyte in lakes of the middle-lower reaches of the Yangtze River, often coexisting with V. natans, while V. natans is a cosmopolitan species [41,42].
In this study, we conducted mesocosm experiments to detect the changes in water quality caused by the growth of V. natans and V. spinulosa. Furthermore, using high-throughput DNA sequencing (HTS) technologies (also called next-generation sequencing (NGS) technologies), we analyzed the microbial community abundance and structure in sediments and water columns with or without plant growth. Our data showed that the diversity and composition of the microbiomes were altered by Vallisneria spp. growth.
2. Materials and methods
2.1. Plant materials
Nine plastic tanks (250 L, diameter = 80 cm, and height = 52 cm) were used for this comparative experiment, and each tank was filled with a ∼20-cm layer of soil as sediment and ∼30 cm of water. The soil used in this study was taken from the top 0–10 cm of soil from the Lushan Botanical Garden Poyang Lake Branch Garden, located in Weijia town, Jiujiang city, Jiangxi province, China (latitude 29°40′25″N, longitude 116°5′4″E). V. natans and V. spinulosa were collected from Changhuchi Lake, a shallow sub-lake of Poyang Lake, Jiangxi province. The seedlings were generated from the seeds and winter buds of V. natans and V. spinulosa, respectively, and approximately 10-cm high seedlings were planted in the tanks, with ∼20 plants in each plastic tank. Nine 250-L plastic tanks were divided into three groups as follows: (i) without plants, (ii) planted V. natans (Vn), and (iii) planted V. spinulosa (Vs); each group included three replicates. All tanks were exposed to natural sunlight outside the lab, with little shading. During the experimental period, the light intensity ranged from approximately 5000 to 90,000 LUX. All the tanks were periodically watered with tap water to maintain the initial water level. By the end of the experiment, the final leaf lengths of the plants were approximately 50 and 30 cm for V. natans and V. spinulosa, respectively.
2.2. Electrical conductivity (EC) assays
The EC of the water columns was measured using a portable HQ4200 pH/EC/TDS meter (HACH) according to the manufacturer's instructions. The EC of all nine plastic tanks (three groups) was measured weekly from 167 days (167d) after the plants had grown to 216 days (216d) when the leaves of these plants almost stopped elongating. Each group comprised three biological replicates.
2.3. Experimental Design and samples Collection
For bacterial community assays, 15 soil samples were collected from the soil described in the “plant materials” section (latitude 29°40′25″N, longitude 116°5′4″E) and named BkM (BkM1-BkM15) as the soil control; sediments samples were collected from V. natans (VnR) and V. spinulosa (VsR) root or mud without plant growth controls. Five sediments samples were collected from each plastic tank at different position using an autoclaved 50-mL tube, and marked as VnR (VnR1-VnR15), VsR (VsR1-VsR15) and CtM (CtM1-CtM15), respectively. The soil samples and sediment/mud samples were all treated as sediments in this study. Water column samples (referred to as water samples below) were taken from the three plastic tank groups: those planted with V. natans (Vn), V. spinulosa (Vs), or without plants, marked as VnW (VnW1-VnW3), VsW (VsW1-VsW3), and CtW (CtW1-CtW3), respectively. To prepare the water samples, 500 mL of water was taken from each tank and filtered with gauze and then with sterilized filter paper. Finally, the planktonic microorganisms in the water columns were harvested using an autoclaved vacuum filtration bottle with a 0.22-μm filter membrane (Millipore Filter Membrane, Aquo-system, 0.22 μm/50 mm). All the samples were stored at −80 °C until DNA isolation was performed.
2.4. DNA isolation
For DNA extraction from the sediment samples, 0.2 g (sediments) were transferred to a PowerBead tube and extraction was performed using the MagPure Stool DNA LQ Kit (Magen, Guangzhou, China) following the manufacturer's instructions. For the water samples (planktonic microorganisms), the filter membranes were cut, ground, and then transferred to a PowerBead tube, following the same protocol as for the sediment samples. Isolated DNA was checked for purity using a Nanodrop (Thermo Scientific, USA), and DNA concentrations were measured using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, MA, USA). DNA isolates were stored at −80 °C until sequencing.
2.5. PCR, amplicon library preparation and sequencing
The V3–V4 region of 16S rRNA gene was amplified with 12.5 ng of template DNA using the primers of 341F/805R (F-5′-CCTACGGGNGGCWGCAG-3’; R-5′-GACTACHVGGGTATCTAATCC-3′) in a 30 μL PCR reaction system. The PCR program was performed as follows: denaturation for 3 min at 95 °C; 25 cycles of denaturation at 95 °C for 30 s, at 55 °C for 30 s, and at 72 °C for 15 s; and then final extension at 72 °C for 5 min. The purified PCR production was then quantified using a Qubit 2.0 Fluorometer, and the sequencing libraries were built using the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina (New England Biolabs, MA, USA) following the manufacturer's instructions. The index codes have also been added. The library quality was assessed using a Qubit 2.0 Fluorometer. Finally, the library was sequenced on an Illumina NovaSeq 6000 platform at the Wuhan Benagen Technology Company Limited (Wuhan, China).
2.6. Data processing and Bioinformatic analysis
Cutadapt (v.3.5) software was used to identify and remove primer sequences, and length filtering was performed to obtain clean sequences. To identify feature sequences, that is, amplicon sequence variants (ASVs), DADA2 [43] was used to filter the raw data, splice paired-end reads, and remove chimeric sequences. The QIIME2 software was used to filter low-abundance ASVs (threshold 1) [44], and all samples were flattened with the minimum number of sequences. Subsequently, the bacterial ASVs were assigned to taxonomic groups using the SILVA database (https://www.arb-silva.de/) for species annotation. The α-diversity indices, including Shannon, Simpson, Chao1, and ACE, were calculated using the QIIME2 software. The β-diversity indices, including PCA, PCoA, NMDS and PLS-DA, were calculated using the R packages to investigate the community dissimilarities among the treatments. Furthermore, an analysis of similarities (ANOSIM) was conducted to examine the significance between groups and to determine the significance of the distance between and within groups. Significance was assessed using a one-way analysis of variance (ANOVA), followed by Tukey's post-hoc test. Linear discriminant analysis (LDA) effect size (LEfSe) was used to identify the biomarkers in the community. Most figures were prepared using the ImageGP web tool [45]. Bioinformatics analysis following the EasyAmplicon pipeline showed similar results [46].
3. Results
3.1. The growth of aquatic plants V. natans and V. spinulosa reduced EC of water columns
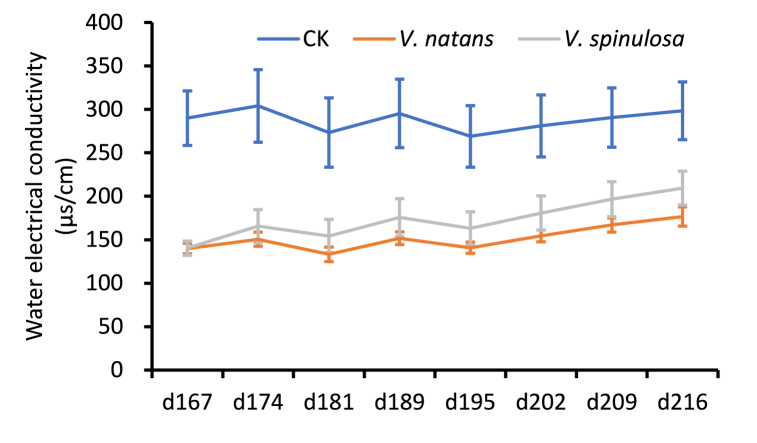
EC is a crucial indicator of several water quality variables. A lower EC indicates better water quality; moreover, EC can be easily measured at a low cost [47,48]. In this study, we used the EC index to reflect the water quality. The EC index of the different plastic tanks was measured every week from 167 to 216 d after the plants were planted. The average EC of the control, which excluded any aquatic plant growth, was approximately 300 μs/cm, whereas the ECs of the groups involving the growth of aquatic plants V. natans and V. spinulosa were approximately 150 μs/cm and from 140.2 to 209.4 μs/cm, respectively (Fig. 1). These data suggest that the growth of the aquatic plants V. natans and V. spinulosa significantly decreased EC, indicating that V. natans and V. spinulosa purified the aquatic ecosystem where they grew. In addition, the data suggest that V. natans has a slightly greater water purification ability than V. spinulosa, with no significant difference as indicated by the t-test (data not shown).
Fig. 1.
Electrical conductivity assays with and without Vallisneria spp. growth. CK: without aquatic plant; V. natans: with Vallisneria natans plants; V. spinulosa: with Vallisneria spinulosa plants.
3.2. The growth of aquatic plants V. natans and V. spinulosa affect the bacterial communities
For the bacterial community assays, 3,391,282 valid sequences were obtained from 69 samples belonging to four sediment groups (BkM, CtM, VnR, and VsR) and three water groups (CtW, VnW, and VsW), generating 33,775 ASVs (Supplementary Tables S1 and S2, and Supplementary Fig. S1). BkM, CtM, VnR, and VsR shared 766 core ASVs, while CtW, VnW, and VsW shared 90 (Supplementary Fig. S2). For the α-diversity of the bacterial community, Shannon index rarefaction curves for sediment and water column samples reached saturation rates, indicating that our data have a sufficient sequence covering most of the bacterial community (Supplement Fig. S3).
The Chao1 index, which represents community richness, ranged from 837 to 3028 and 211to 407 in the sediment and water samples, respectively (Supplementary Table S2, Fig. 2A–C). The Shannon index, representing community diversity, ranged from 8.459 to 10.735 and from 3.707 to 5.569 for the sediment and water samples, respectively (Supplementary Table S2, Fig. 2B–D). The mean α-diversity was much higher in the sediment than that in the water samples, suggesting that richness and diversity are lower in water than in sediment. We found a significant difference (p < 0.01 in the t-test) between the mud control group (CtM) and the groups with the aquatic plants V. natans or V. spinulosa (VnR or VsR) (Fig. 2A and B, and Supplementary Tables S2 and S3). In the two control groups, BkM (soil) and CtM (water), the Chao1 and Shannon indices of CtM were higher than those of BkM, suggesting that water promotes bacterial community richness and diversity. A significant difference (p < 0.01, t-test) in the Shannon index was obtained between the water control group CtW and the water sample VnW, which contained V. natans (Fig. 2D and Supplementary Tables S2 and S4). Together, our data indicate that Vallisneria spp. reduce bacterial community richness and diversity in sediment and water columns.
Fig. 2.
The α-diversity of Chao1 and Shannon indices of the sediment and water bacterial communities in different groups. A–B, sediments; C–D, water columns. BkM: soil samples control; CtM: sediment sampling from tanks without aquatic plant; VnR: sediment sampling from root of V. natans plants; VsR: sediment sampling from root of V. spinulosa plants; CtW: water column sampling from tanks without aquatic plant; VnR: water column sampling from tanks planted V. natans; VsW: water column sampling from tanks planted V. spinulosa.
For β-diversity, unconstrained principal coordinate analyses (PCoAs) based on the Bray–Curtis distance were performed (Fig. 3). Sediments from the plant growth groups (VnR and VsR) were separated into soil and mud control groups (BkM and CtM) across the first principal component (PC1, 29.4 %). VnR, VsR, CtM, and BkM were separated across the second principal component (PC2: 8.42 %) (Adonis: R2 = 0.431, p = 0.001) (Fig. 3A). For water samples, VnW and VsR were separated from the control CtW as the first principal component, which explained 30.93 % of the variation (PC1, 30.93 %), whereas the second principal component explained 20.23 % of the variation (PC2, 20.23 %) (Adonis, R2 = 0.447, p = 0.001) (Fig. 3B). These data suggest that the largest source of variation in microbial communities is introduced by aquatic plant growth, and that different plants (V. natans and V. spinulosa) affect the variation in microbial communities.
Fig. 3.
β-diversity based on the Bray–Curtis distance of bacterial communities of sediment and water columns in different groups. Unconstrained PCoA (for principal coordinates PCo1 and PCo2) with the Bray–Curtis distance showed that the root microbiota of the control groups was separate from that of the groups with V. natans or V. spinulosa. (A) Sediment; (B) Water column.
3.3. Composition of bacterial community
The composition of the bacterial community was analyzed from phylum to family in all 69 samples, generating 58 phyla, 161 classes, 380 orders, 573 families, and 948 genera (Supplementary tables S5–S9). Proteobacteria, Acidobacteriota, Planctomycetota, Verrucomicrobiota, Bacteroidota, and Cyanobacteria were the dominant phyla in the sediment samples with V. natans or V. spinulosa (VnR and VsR, respectively), and all had a relative abundance of more than 5 % (Fig. 4A and Supplementary Table S10). Proteobacteria was the most dominant phylum, with 29 and 35.9 % abundance in VnR and VsR, respectively (Fig. 4A and Supplementary Table S10). In the mud control sample group (CtM), Acidobacteriota, Proteobacteria, Planctomycetota, Chloroflexi, and Verrucomicrobiota were the dominant phyla (CtM; Fig. 4A and Supplementary Table S10). Furthermore, our data showed that the abundance of Proteobacteria, Bacteroidota, and Cyanobacteria increased in the VnR and VsR groups when compared with that in the control sediments BkM and CtM, whereas the phyla Acidobacteriota and Chloroflexi were reduced (Fig. 4A and Supplementary Table S10). At the class level, Gammaproteobacteria and Alphaproteobacteria were the dominant classes in the VnR and VsR groups, and their relative abundance increased nearly two-fold compared with that in the CtM group (Supplementary Table S12).
Fig. 4.
Histograms of the bacterial community at the phylum level. A, sediment samples; B, water column samples.
In water samples with plant growth (VnW and VsW), the top four phyla, Proteobacteria, Bacteroidota, Cyanobacteria, and Actinobacteriota, comprised more than 90 % of the total phyla (Fig. 4B and Supplementary Table S11). Moreover, Bacteroidota and Cyanobacteria increased with plant growth, whereas Actinobacteriota and Verrucomicrobiota decreased (Fig. 4B and Supplementary Table S11).
Network analysis has been widely used to describe the composition of microbial communities and detect associations between microorganisms [[49], [50], [51]]. We performed network analysis with the top 50 genera for each group (Spearman's correlation coefficient r > 0.6, p < 0.05) (Supplementary Fig. S4 and Supplement Tables S14–S28). The clustering coefficient represents the complexity of the network and the strong interactions among microorganisms. For the sediment samples, the VsR group network showed the highest clustering coefficient (0.505), followed by the VnR group. The clustering coefficients for the water column samples were very high (0.959, 1, and 0.959 for CtW, VnW, and VsW, respectively) (Supplementary Fig. S4 and Supplementary Table S14). This indicates that microbial interactions in the water column are stronger than those in the sediment, and that Vallisneria spp. growth strengthens these interactions. The average shortest path length was higher in the VnR and VsR groups than in the CtM group (3.045, 2.876, and 2.709 for VnR, VsR, and CtM, respectively). The number of edges increased in VsR and decreased in VnR. However, in the water column samples, V. natans growth increased the number of edges, and V. spinulosa growth decreased this index (Supplementary Fig. S4 and Supplementary Table S14).
Linear discriminant analysis (LDA) effect size (LEfSe) was performed on genera with relative abundances of more than 0.1 % (LDA thresholds of four and three for sediment and water column samples, respectively). The LEfSe results suggest that in sediment samples, o__Cytophagales, g__Novosphingobium, and g__Arenimonas are biomarkers of the VsR group. In the VnR group, c__Planctomycetes, c__Cyanobacteriia, and f__Chitinophagaceae were identified as biomarkers. The biomarkers in the CtM group were g__Subgroup_7, f__Anaerolineaceae, and o__Myxococcales (Fig. 5A, Supplementary Fig. S5, Supplementary Table S29, and Supplementary Table S31). In the water column samples, g__Lacunisphaera, g__Allorhizobium_Neorhizobium_Pararhizobium_Rhizobium.s__bacterium, and g__Arcicella were biomarkers for the VsW group, and bacteria g__Flavobacterium.s__Flavobacterium_indicum, c__Cyanobacteriia, g__UKL13_1, f__Hyphomicrobiaceae, and g__Hydrogenophaga were biomarkers for the VsW group. The CtW group biomarkers were f__CandidatusMoranbacteria, g__CandidatusPlanktophila, g__Peredibacterstarrii, g__Polynucleobacteracidiphobus, g__Polaromonas, g__MWH_UniP1aquaticgroup, g__FukuN18freshwatergroup, g__SH3_11. s__bacterium_SH3_11, and g__Luteolibacter.s__Verrucomicrobia_bacterium (Fig. 5B, Supplementary Fig. S6, Supplementary Tables S30 and S32).
Fig. 5.
LEfSe analysis of the sediment (A) and water (B) column samples.
4. Discussion
Human society requires pure water for daily life, including direct and indirect consumption during industrial and agricultural processes. However, anthropogenic activities pollute the Earth's aquatic ecosystem, and this pollution worsens daily [[1], [2], [3]]. Using aquatic macrophytes to restore water quality is a green and economical biological method because these aquatic plants can promote biodiversity and the self-clarification of water ecosystems, thus reconstructing their health [4,5,[8], [9], [10], [11]]. Several indices are commonly used to evaluate water quality, including total concentrations of P and N, pH, EC, and turbidity. Many types of pollution influence the EC, and the total ion concentration is correlated with the EC [52]. Moreover, EC measurements are easy to perform and relatively inexpensive [53]. In this study, we found that EC decreased with the growth of the aquatic macrophytes V. natans and V. spinulosa, providing direct evidence that their growth improves water quality (Fig. 1), making them useful for aquatic ecosystem conservation in the future.
We analyzed the sediment and water column microbial community abundance and structure using high-throughput DNA sequencing technologies. Bacterial abundance was much lower in the water columns, as indicated by the α-diversity of the bacterial community (Fig. 2), which was similar to that of previous reports [31,54,55], possibly because the sediments have more nutrition and oxygen than water, thus supporting an increased number and variety of bacteria. The microbial association and interactions were stronger in the water column than in the sediment, as shown by the network analysis data (Supplementary Fig. S4 and Supplementary Tables S14–S28), possibly because the water columns act as one unit, and the microorganisms interact and communicate freely, while in the sediment, microorganism interactions and communication are limited to numerous small units. The growth of V. natans and V. spinulosa reduced the Chao1 and Shannon indices of sediment and water samples (Fig. 2), suggesting that these submersed macrophytes may promote the growth of some microorganisms while inhibiting others, thereby reducing bacterial abundance and diversity. The β-diversity results of the microbial communities in the sediments and water columns significantly differed between the absence and presence of Vallisneria spp. (Fig. 3). In natural wild aquatic ecosystems, submersed macrophytes influence bacterial community composition in their rhizosphere sediments, epiphytic leaves, and surrounding water [56,57].
Microbial diversity is used as a bioindicator in aquatic ecosystems because it is affected by water quality [29,30]. Our data revealed differences in microbial diversity and water quality caused by submersed macrophyte growth. The submersed macrophytes improved water quality and affected microbial diversity, reconstructing the microbial community by recruiting specific microorganisms while inhibiting others; the aquatic plants and the interacting microorganisms improve water quality, mutually promoting each other [31,32,38].
Proteobacteria, Acidobacteriota, Planctomycetota, Verrucomicrobiota, Bacteroidota, and Cyanobacteria were the dominant phyla in the VnR and VsR groups, whereas in the mud control sample group (CtM), Acidobacteriota, Proteobacteria, Planctomycetota, Chloroflexi and Verrucomicrobiota were the dominant phyla (Fig. 4A and Supplementary Table S10). Proteobacteria, Verrucomicrobiota, and Bacteroidota increased due to these Vallisneria spp. aquatic macrophytes, especially for α-proteobacteria and γ-proteobacteria, similar to a previous study [54]. It has been reported that different submersed macrophytes differ in the shapes of their interacting bacterial communities [32,54,55]. Our results showed that V. natans and V. spinulosa, highly related Vallisneria spp., differ in their microbial communities in sediment and water columns. Future research using metagenomics and metaculturomics techniques will provide more solid data to help us use different aquatic plants and their microbial communities to improve and protect aquatic ecosystems.
5. Conclusions
The growth of two submersed macrophytes, V. natans and V. spinulosa, improved the water quality as the EC index decreased. Furthermore, 16S rRNA amplicon analysis was used to analyze the microbial community abundance and structure in the sediment and water columns with or without plant growth. A total of 33,775 ASVs were generated from four sediment groups (BkM, CtM, VnR, and VsR) and three water column sample groups (CtW, VnW, and VsW). The diversity and composition of the microbiomes were altered by Vallisneria spp. growth in the sediment and water columns, and the relative abundance of bacteria was higher in the sediment than that in the water columns. The α-diversity of the microbial communities decreased due to submersed macrophytes in both sediment and water columns, and the β-diversity also varied significantly with or without Vallisneria spp. The relative abundances of Proteobacteria, Bacteroidota, and Cyanobacteria increased, whereas the phyla Acidobacteriota and Chloroflexi were reduced by Vallisneria spp. Gammaproteobacteria and Alphaproteobacteria were the dominant classes in Vallisneria spp. sediment groups. Proteobacteria, Bacteroidota, Cyanobacteria, and Actinobacteriota were the dominant phyla in water columns; Bacteroidota and Cyanobacteria were increased, whereas Actinobacteriota and Verrucomicrobiota were decreased by Vallisneria spp.
CRediT authorship contribution statement
Libing Liao: Writing – review & editing, Writing – original draft, Visualization, Resources, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Deshui Yu: Writing – review & editing, Writing – original draft, Visualization, Project administration, Methodology, Data curation, Conceptualization. Lei Xu: Investigation. Qian Hu: Investigation. Tongjun Liang: Resources. Ludan Chen: Resources, Investigation. Qiuping Zhu: Visualization, Resources, Investigation. Songping Liu: Investigation. Aiwen Zhong: Writing – review & editing, Writing – original draft, Visualization, Supervision, Resources, Project administration, Funding acquisition, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by the National Natural Science Foundation of China (32160075), the Scientific Planning Project of Lushan Botanical Garden, Chinese Academy of Sciences (2020ZWZX03, 2020ZWZX05, 2020ZWZX07 and 2023ZWZX09), the “Double Hundred and Double Thousand” Talent Project of Jiujiang City (jjsbsq2020026), the Talents Program of Jiangxi Province (PR China) (jxsq2020104003), and the Technological Innovation Guidance Project of Jiangxi Province (20212BDH80012).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e25942.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Cao J., et al. A critical review of the appearance of black-odorous waterbodies in China and treatment methods. J Hazard Mater. 2020;385 doi: 10.1016/j.jhazmat.2019.121511. [DOI] [PubMed] [Google Scholar]
- 2.Liang Z., et al. Blackening and odorization of urban rivers: a bio-geochemical process. FEMS Microbiol Ecol. 2018:94. doi: 10.1093/femsec/fix180. [DOI] [PubMed] [Google Scholar]
- 3.Bruno A., et al. It's a long way to the tap: microbiome and DNA-based Omics at the core of Drinking water quality. Int J Environ Res Public Health. 2022;19 doi: 10.3390/ijerph19137940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan A., et al. Phytoremediation: a promising Approach for Revegetation of heavy Metal-polluted Land. Front Plant Sci. 2020;11:359. doi: 10.3389/fpls.2020.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mu X., et al. Impacts of water flow on epiphytic microbes and nutrients removal in constructed wetlands dominated by Vallisneria natans with decreasing temperature. Bioresour Technol. 2020:318. doi: 10.1016/j.biortech.2020.124058. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava J., Gupta A., Chandra H. Managing water quality with aquatic macrophytes. Rev Environ Sci Bio/Technol. 2008;7(3):255–266. doi: 10.1007/s11157-008-9135-x. [DOI] [Google Scholar]
- 7.Chambers P.A., et al. Global diversity of aquatic macrophytes in freshwater. Hydrobiologia. 2008;595:9–26. doi: 10.1007/s10750-007-9154-6. [DOI] [Google Scholar]
- 8.Phillips G., Willby N., Moss B. Submerged macrophyte decline in shallow lakes: what have we learnt in the last forty years? Aquatic Botany. 2016;135:37–45. doi: 10.1016/j.aquabot.2016.04.004. [DOI] [Google Scholar]
- 9.Su H., et al. Morphological traits of submerged macrophytes reveal specific positive feedbacks to water clarity in freshwater ecosystems. Sci Total Environ. 2019;684:578–586. doi: 10.1016/j.scitotenv.2019.05.267. [DOI] [PubMed] [Google Scholar]
- 10.Bakker E.S., et al. Effect of macrophyte community composition and nutrient enrichment on plant biomass and algal blooms. Basic Appl Ecol. 2010;11:432–439. doi: 10.1016/j.baae.2010.06.005. [DOI] [Google Scholar]
- 11.Zhang C., et al. Responses of submerged macrophytes Vallisneria natans and epiphytic biofilm to floating plants Eichhornia crassipes in eutrophic water. Water Environ Res. 2021;93:2237–2249. doi: 10.1002/wer.1596. [DOI] [PubMed] [Google Scholar]
- 12.Wang H., et al. Biodiversity buffers the impact of eutrophication on ecosystem functioning of submerged macrophytes on the Yunnan-Guizhou Plateau, Southwest China. Environ Pollut. 2022;314 doi: 10.1016/j.envpol.2022.120210. [DOI] [PubMed] [Google Scholar]
- 13.Xu X.J., et al. Purification of eutrophic water containing chlorpyrifos by aquatic plants and its effects on planktonic bacteria. Chemosphere. 2018;193:178–188. doi: 10.1016/j.chemosphere.2017.10.171. [DOI] [PubMed] [Google Scholar]
- 14.Davey M.E., O'Toole G A. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wijewardene L., et al. Epiphytic biofilms in freshwater and interactions with macrophytes: Current understanding and future directions. Aquatic Botany. 2022:176. doi: 10.1016/j.aquabot.2021.103467. [DOI] [Google Scholar]
- 16.Mishra V.K., Tripathi B.D. Concurrent removal and accumulation of heavy metals by the three aquatic macrophytes. Bioresour Technol. 2008;99:7091–7097. doi: 10.1016/j.biortech.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Li Q., et al. Response of submerged macrophytes and periphyton biofilm to water flow in eutrophic environment: plant structural, physicochemical and microbial properties. Ecotoxicol Environ Saf. 2020:189. doi: 10.1016/j.ecoenv.2019.109990. [DOI] [PubMed] [Google Scholar]
- 18.Pang S., et al. Characterization of bacterial community in biofilm and sediments of wetlands dominated by aquatic macrophytes. Ecological Engineering. 2016;97:242–250. doi: 10.1016/j.ecoleng.2016.10.011. [DOI] [Google Scholar]
- 19.Wisnoski N.I., et al. Metabolic insight into bacterial community assembly across ecosystem boundaries. Ecology. 2020;101 doi: 10.1002/ecy.2968. [DOI] [PubMed] [Google Scholar]
- 20.Lever M.A., et al. Life under extreme energy limitation: a synthesis of laboratory- and field-based investigations. FEMS Microbiol Rev. 2015;39:688–728. doi: 10.1093/femsre/fuv020. [DOI] [PubMed] [Google Scholar]
- 21.Cydzik-Kwiatkowska A., Milojevic N., Jachimowicz P. The fate of microplastic in sludge management systems. Sci Total Environ. 2022;848 doi: 10.1016/j.scitotenv.2022.157466. [DOI] [PubMed] [Google Scholar]
- 22.Li W., et al. Low-dose biochar added to sediment improves water quality and promotes the growth of submerged macrophytes. Sci Total Environ. 2020;742 doi: 10.1016/j.scitotenv.2020.140602. [DOI] [PubMed] [Google Scholar]
- 23.Ye G., et al. Polystyrene microplastics induce microbial dysbiosis and dysfunction in surrounding seawater. Environ Int. 2021;156 doi: 10.1016/j.envint.2021.106724. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J., et al. Responses of submerged plant Vallisneria natans growth and leaf biofilms to water contaminated with microplastics. Sci Total Environ. 2022:818. doi: 10.1016/j.scitotenv.2021.151750. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava J.K., et al. Plant–microbe interaction in aquatic system and their role in the management of water quality: a review. Appl Water Sci. 2017;7:1079–1090. doi: 10.1007/s13201-016-0415-2. [DOI] [Google Scholar]
- 26.Gan L., et al. Pseudomonas putida inoculation promotes submerged plant Vallisneria natans growth by carbon conversion in a plant–microbe interaction. Mar Freshw Res. 2018;69:851–858. doi: 10.1071/MF17117. [DOI] [Google Scholar]
- 27.Supreeth M. Enhanced remediation of pollutants by microorganisms-plant combination. Int J Environ Sci Technol (Tehran) 2022;19:4587–4598. doi: 10.1007/s13762-021-03354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shahid M.J., et al. Comparing the performance of four macrophytes in bacterial assisted floating treatment wetlands for the removal of trace metals (Fe, Mn, Ni, Pb, and Cr) from polluted river water. Chemosphere. 2020;243 doi: 10.1016/j.chemosphere.2019.125353. [DOI] [PubMed] [Google Scholar]
- 29.Pei Y., et al. Microbial community structure and function indicate the Severity of Chromium Contamination of the Yellow river. Front Microbiol. 2018;9:38. doi: 10.3389/fmicb.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C., et al. How sediment bacterial community shifts along the urban river located in mining city. Environ Sci Pollut Res Int. 2021;28:42300–42312. doi: 10.1007/s11356-020-12031-0. [DOI] [PubMed] [Google Scholar]
- 31.Xian L., et al. Structural Variability and functional Prediction in the epiphytic bacteria Assemblies of Myriophyllum spicatum. Curr Microbiol. 2020;77:3582–3594. doi: 10.1007/s00284-020-02139-4. [DOI] [PubMed] [Google Scholar]
- 32.Sun L., et al. Community structure and function of epiphytic bacteria associated with Myriophyllum spicatum in Baiyangdian lake, China. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.705509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., et al. Inherent bacterial community response to multiple heavy metals in sediment from river-lake systems in the Poyang Lake, China. Ecotoxicol Environ Saf. 2018;165:314–324. doi: 10.1016/j.ecoenv.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., et al. Characterization and co-occurrence of microbial community in epiphytic biofilms and surface sediments of wetlands with submersed macrophytes. Sci Total Environ. 2020:715. doi: 10.1016/j.scitotenv.2020.136950. [DOI] [PubMed] [Google Scholar]
- 35.Yang L., et al. Vallisneria natans decreased CH(4) fluxes in wetlands: interactions among plant physiological status, nutrients and epiphytic bacterial community. Environ Res. 2023;224 doi: 10.1016/j.envres.2023.115547. [DOI] [PubMed] [Google Scholar]
- 36.Guo S., et al. Potamogeton crispus restoration increased the epiphytic microbial diversity and improved water quality in a micro-polluted urban river. Environ Pollut. 2023;326 doi: 10.1016/j.envpol.2023.121485. [DOI] [PubMed] [Google Scholar]
- 37.Manirakiza B., et al. Exploring microbial diversity and ecological function of epiphytic and surface sediment biofilm communities in a shallow tropical lake. Sci Total Environ. 2022;808 doi: 10.1016/j.scitotenv.2021.151821. [DOI] [PubMed] [Google Scholar]
- 38.Yan H., et al. Toward understanding submersed macrophyte Vallisneria natans-microbe partnerships to improve remediation potential for PAH-contaminated sediment. J Hazard Mater. 2022;425 doi: 10.1016/j.jhazmat.2021.127767. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y., et al. Effects of Vallisneria natans on H2S and S2- releases in black-odorous waterbody under additional nitrate: Comprehensive performance and microbial community structure. J Environ Manage. 2022:316. doi: 10.1016/j.jenvman.2022.115226. [DOI] [PubMed] [Google Scholar]
- 40.Hua Z.-l., et al. Removal potential of multiple perfluoroalkyl acids (PFAAs) by submerged macrophytes in aquatic environments: Tolerance of Vallisneria natans and PFAA removal in submerged macrophyte-microbiota systems. J Hazard Mater. 2022:424. doi: 10.1016/j.jhazmat.2021.127695. [DOI] [PubMed] [Google Scholar]
- 41.Wang B., et al. Comparison of the extent of genetic variation of Vallisneria natans and its sympatric congener V. spinulosa in lakes of the middle–lower reaches of the Yangtze River. Aquatic Botany. 2010;92:233–238. doi: 10.1016/j.aquabot.2009.12.006. [DOI] [Google Scholar]
- 42.Chen L., Xu L., Huang H. Genetic diversity and population structure in Vallisneria spinulosa (Hydrocharitaceae) Aquatic Botany. 2007;86:46–52. doi: 10.1016/j.aquabot.2006.09.001. [DOI] [Google Scholar]
- 43.Callahan B.J., et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolyen E., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen T., Liu Y.-X., Huang L. ImageGP: an easy-to-use data visualization web server for scientific researchers. iMeta. 2022;1:e5. doi: 10.1002/imt2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y.-X., et al. EasyAmplicon: an easy-to-use, open-source, reproducible, and community-based pipeline for amplicon data analysis in microbiome research. iMeta. 2023;2:e83. doi: 10.1002/imt2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson M.Y., Brandes D., Kney A.D. Using electronic conductivity and hardness data for rapid assessment of stream water quality. J Environ Manage. 2012;104:152–157. doi: 10.1016/j.jenvman.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 48.Jamei M., et al. Surface water electrical conductivity and bicarbonate ion determination using a smart hybridization of optimal Boruta package with Elman recurrent neural network. Process Saf Environ Prot. 2023;174:115–134. doi: 10.1016/j.psep.2023.03.062. [DOI] [Google Scholar]
- 49.Barberan A., et al. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo B., et al. Microbial co-occurrence network topological properties link with reactor parameters and reveal importance of low-abundance genera. NPJ Biofilms Microbiomes. 2022;8:3. doi: 10.1038/s41522-021-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Vries F.T., et al. Soil bacterial networks are less stable under drought than fungal networks. Nat Commun. 2018;9:3033. doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagahashi E., et al. Characteristics of raw and Acid-Activated Bentonite and its Application for improving electrical conductivity of tap water. Chem Pharm Bull. 2021;69:92–98. doi: 10.1248/cpb.c20-00703. [DOI] [PubMed] [Google Scholar]
- 53.Serrano-Finetti E., et al. Cost-effective autonomous sensor for the long-term monitoring of water electrical conductivity of crop fields. Comput Electron Agric. 2019;165 doi: 10.1016/j.compag.2019.104940. [DOI] [Google Scholar]
- 54.Dai Y., et al. Macrophyte identity shapes water column and sediment bacterial community. Hydrobiologia. 2019;835:71–82. doi: 10.1007/s10750-019-3930-y. [DOI] [Google Scholar]
- 55.Yu W., et al. Community structure and function of epiphytic bacteria attached to three submerged macrophytes. Sci Total Environ. 2022;835 doi: 10.1016/j.scitotenv.2022.155546. [DOI] [PubMed] [Google Scholar]
- 56.Zhao D.Y., et al. Submerged macrophytes modify bacterial community composition in sediments in a large, shallow, freshwater lake. Can J Microbiol. 2013;59:237–244. doi: 10.1139/cjm-2012-0554. [DOI] [PubMed] [Google Scholar]
- 57.Zhu H.Z., et al. Submerged macrophytes recruit unique microbial communities and drive functional zonation in an aquatic system. Appl Microbiol Biotechnol. 2021;105:7517–7528. doi: 10.1007/s00253-021-11565-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.