Abstract
We analyzed the segment of DNA which contains the expressed pMGA gene from one strain of Mycoplasma gallisepticum in normal (strain S6) cells and in cells in which pMGA1.1 gene expression had ceased as a consequence of in vitro culture in the presence of pMGA1.1-specific antibodies. Sequence analysis of isolates lacking pMGA1.1 expression revealed that this gene, which is typically expressed, exhibited sequence changes within a region 5′ to its promoter. Specifically, pMGA1.1+ cells contained a (GAA)12 motif upstream of the promoter, whereas in pMGA1.1− cells the corresponding region contained a (GAA)10 motif; when such cells were grown in medium no longer containing pMGA-specific antibodies, pMGA1.1 was reexpressed and the 5′ (GAA)12 motif was restored. Two other genes, pMGA1.9 and pMGA1.2, were also shown to acquire a (GAA)12 motif in clones which expressed these genes. The results imply the evolution by the pMGA genes of M. gallisepticum of a novel transcriptional requirement which facilitates rapid and reversible switches in the pMGA expression pattern.
Previous investigations in this laboratory have shown that the gene for a major surface lipoprotein (pMGA1.1) of Mycoplasma gallisepticum S6 is a member of a multigene family (16, 17). The pMGA gene family in strain S6 contains 33 members comprising a total of 7.7% of the 1,030-kb genome (1). Investigations to date have revealed that three separate field isolates of M. gallisepticum each express single, unique pMGA polypeptides (8). The level of sequence homology between the pMGA1.1 gene and the other pMGA genes of the S6 strain of M. gallisepticum varies widely. Only the pMGA1.2 gene exhibits a notably high level of sequence identity (greater than 95%) to the pMGA1.1 gene, whereas all other known members of the pMGA gene family exhibit much lower overall identity levels. Certain antibodies directed to the pMGA1.1 polypeptide, when included in in vitro growth media, cause a switch within the resultant cell population which results in the loss of pMGA1.1 expression, concomitant with the expression of another pMGA family member, pMGA1.9 (15). The pMGA1.9 gene product has about 42% amino acid identity to pMGA1.1 and, like pMGA1.1, is a plasma membrane protein of M. gallisepticum. In the genome, the pMGA1.1 and pMGA1.9 genes are separated by a third gene, that for pMGA1.7, and all three genes are in the same transcriptional orientation. The surprising attributes of the antibody effect on M. gallisepticum cells were its speed and its reversibility. Specifically, when transferred from the in vitro growth medium onto agar plates containing antibodies, M. gallisepticum cells produced the same number of colonies as control plates containing no antibodies, but in the former case the colonies lacked pMGA1.1 (15). In addition, most or all pMGA1.1− cells obtained by growth in antibody-containing medium were shown to revert to pMGA1.1 expression when transferred to plates lacking antibody. The reexpression of pMGA1.1 occurred within colonies in a sectorial fashion which implied multiple, equivalent reversion events within and between colonies.
The present work was undertaken to investigate the molecular basis of the pMGA1.1-pMGA1.9 transcriptional switch. The findings described here implicate high-frequency alterations in trinucleotide repeat numbers 5′ to the pMGA1.1 and pMGA1.9 genes as the primary cause of the changes in pMGA expression.
The rationale for this study was to establish a number of M. gallisepticum clones, each expressing one or more pMGA genes, and then examine the DNA sequences around the promoter regions of pMGA genes which were either expressed or not expressed in individual clones. Specific PCRs for the amplification of these regions of the pMGA1.1, pMGA1.2, pMGA1.7, and pMGA1.9 genes were developed, the products which they amplified were cloned, and relevant parts of the inserts were then sequenced.
MATERIALS AND METHODS
Antibodies.
The construction and specificities of the monoclonal antibody (MAb), MAb 66, used in this study and the rabbit antiserum directed to purified pMGA1.1 polypeptide were described in previous publications from this laboratory (14, 15). A rabbit antiserum to the pMGA1.9 polypeptide was elicited as follows. The C1 clone (see Results), which expresses pMGA1.9, was grown in liquid culture, and cells were harvested by centrifugation and lysed in the detergent Triton X-114 as previously described (3, 15). The clarified detergent lysate was then partitioned into detergent-rich and detergent-poor fractions (3), and the former fraction was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The resultant gel was subjected to electrophoretic transfer to a nitrocellulose membrane which was stained with Ponceau S, and the zone containing the pMGA1.9 polypeptide (10 to 100 μg) was excised. The nitrocellulose membrane was then physically shredded and sonicated to reduce the particle size. The sample was finally resuspended in phosphate-buffered saline, passed through an 18-gauge needle, and emulsified with an equal volume of Freund’s adjuvant for injection. Two New Zealand White rabbits were injected intramuscularly with antigen, three times at monthly intervals, first in Freund’s complete adjuvant and then in Freund’s incomplete adjuvant. The specificity of the resultant antiserum was verified by Western transfer (see Fig. 1).
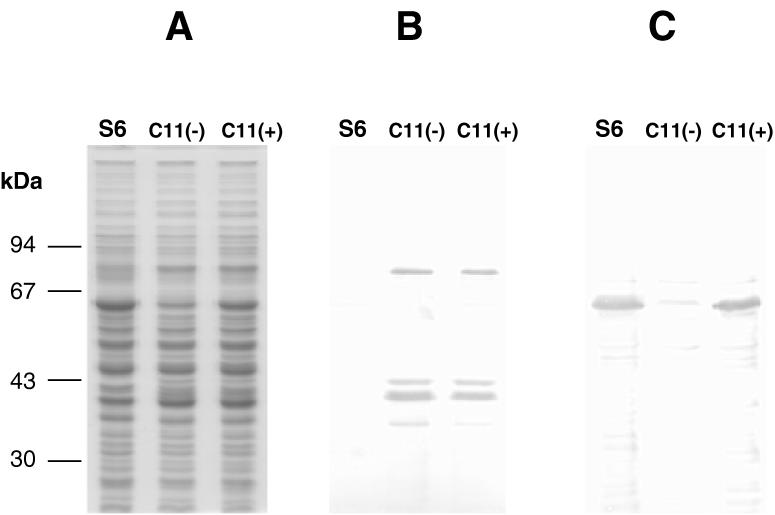
FIG. 1.
SDS-PAGE and Western blot analysis of M. gallisepticum clones C11(−) and C11(+). (A) Coomassie blue staining pattern of protein samples from normal M. gallisepticum cells (S6) and clones C11(−) and C11(+). (B) Replicate gel, transferred to a nitrocellulose membrane and immunostained with rabbit anti-pMGA1.9 serum. (C) Same as for panel B but with rabbit anti-pMGA1.1 used as the primary immunostaining reagent.
Growth of M. gallisepticum cells.
The cells used throughout this study were the S6 strain of M. gallisepticum and have been used in previous studies from this laboratory (8, 15, 16). The composition of the in vitro growth medium used was modified from that of Frey et al. (7) as described by Markham et al. (14). To obtain cell populations in which pMGA1.1 expression was arrested, cells were grown in this medium to which MAb 66 (5 μg ml−1) had been added. Agar culture techniques for mycoplasma cells and methods for lifting colonies onto nitrocellulose discs for immunostaining have been described in previous studies from this laboratory (15).
Colony immunostaining.
Most of the techniques used in this work have previously been described (15). To detect the pMGA1.2 switch variant, nitrocellulose colony lifts were first incubated with MAb 66 hybridoma supernatant (1:100) as previously described, followed by sheep anti-mouse Ig–horseradish peroxidase conjugate (1:2,000) with diaminobenzidine (Sigma) as a chromogenic substrate. The second-round staining procedure consisted of rabbit anti-pMGA1.1 serum (1:1,000) and then swine anti-rabbit Ig–alkaline phosphatase conjugate (1:1,000), followed by Fast Red (Sigma) staining according to the manufacturer’s instructions.
SDS-PAGE and Western blotting.
The SDS-PAGE and Western blotting techniques used have been described in previous publications from this laboratory (8, 14, 16). The concentrations of rabbit anti-pMGA1.1, MAb 66, and horseradish peroxidase-conjugated swine anti-rabbit Ig used in this study were as previously stated (15). The dilution of the rabbit anti-pMGA1.9 used in Western transfer studies was 1:1,000. Molecular weight standards were from Pharmacia.
PCR.
The sequences of the oligonucleotide primers used were based on previously published sequences (GenBank accession no. L28423 and L28424 [16]). They are as follows: 1.1F, 5′-ccgaattcTAGTAATGTTAATTCACAAGG-3′; 1.1R, 5′-atgaattcTTCCACCATTCATACCACCAT-3′; 1.9R, 5′-gcgaattcGCCGCAGTATCCATACCTGG-3′; 1.7F, 5′-AAACCCAACCTGAAACTACTAATGT-3′; 1.9F, 5′-AGTAAAGATGAGACTGGTGCAATCG-3′; 1.2F, 5′-TCCGCTGATTCTAATCCAAC-3′; 1.2R, 5′-CTATCAGCTAGTTTTGTTGCG-3′; 1.2(GAA)F, 5′-ATTTCTGAATTAACTATAAAAATGA-3′; and 1.2(GAA)R, 5′-TAAATCGACTTAATTTTCGCAG-3′. Lowercase nucleotides in these primers denote restriction enzyme cleavage sites appended to facilitate excision from cloning vectors. The locations and orientations of these oligonucleotides relative to the pMGA genes of this study are depicted in Fig. 3 and 6.
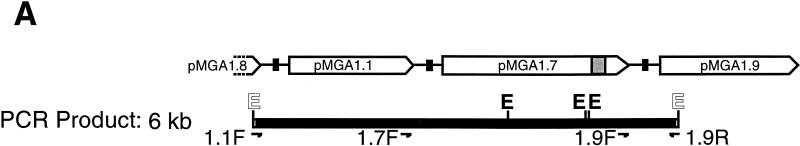
FIG. 3.
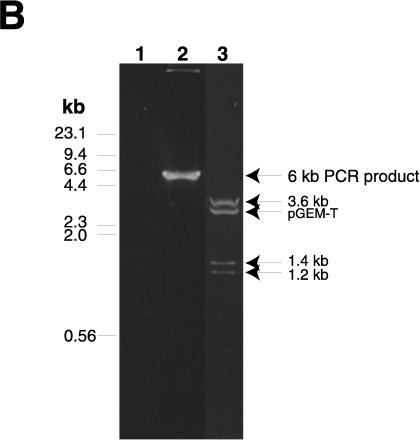
Amplification by PCR of the pMGA1.1-pMGA1.9 locus. (A) Schematic diagram depicting the pMGA1.1, pMGA1.7, and pMGA1.9 genes, along with the positions of EcoRI restriction sites (E) within the segment amplified by the PCR with oligonucleotide primers 1.1F and 1.9R. The oligonucleotide primers, 1.1F, 1.7F and 1.9F, used for the sequence analysis in Fig. 4 to 6 are also depicted. The regions containing the putative promoters of the three pMGA genes shown are indicated as black boxes. The fragment sizes from an EcoRI digest of the approximately 6-kb PCR product expected from the work described by Markham et al. (16) are 3.63, 1.15, 0.04, and 1.34 kb. The EcoRI sites at the 5′ ends of the primers used to amplify the 6-kb fragment are synthetic. The shaded segment within the pMGA1.7 gene contains a sequence without a counterpart in other pMGA genes (15). (B) Agarose gel electrophoresis of PCR products amplified by using oligonucleotides 1.1F and 1.9R as primers and cloned into pGEM-T. Lane 1, no template DNA; lane 2, reaction mixture with strain S6 DNA as the template; lane 3, cloned isolate from the same sample as in lane 2 but digested with EcoRI. Experimentally measured EcoRI fragment sizes are indicated.
FIG. 6.
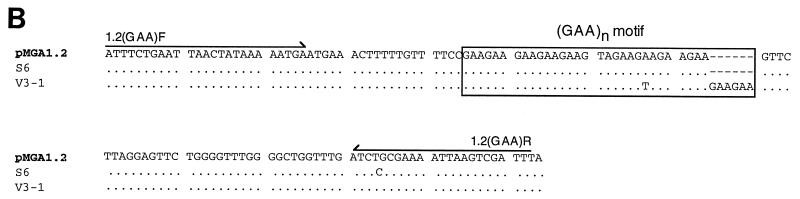
Amplification and analysis of pMGA1.2. (A) Schematic of the gene cluster which contains pMGA1.2 (16, 17). The locations of EcoRI (E), BglII(B), and PstI (P) restriction enzyme cleavage sites are shown. The locations and orientations of the primers used to generate the primary (1,889-bp) and secondary (132-bp) PCR products referred to in the text are shown as arrows. The lengths of PCR products are those expected from the work described by Markham et al. (16). Segments containing putative promoters and flanking GAA repeat motifs are represented as black boxes. (B) The sequences of the secondary PCR products derived from templates from normal (S6) cells and from V3-1 cells are compared to the corresponding segment of the pMGA1.2 gene sequence.
In all but one case, PCRs were performed in a 50-μl volume consisting of 1.25 U of Taq DNA polymerase (Promega), 1× PCR buffer supplied by the manufacturer, 2.5 mM MgCl2, 0.2 mM deoxyribonucleoside triphosphates, 0.4 μM each forward and reverse primer set, and an appropriate amount of DNA template. The amplification reactions with primers 1.1F and 1.9R were performed in 50-μl volumes, and each reaction mixture contained 1.25 U of total DNA polymerase activity (in this case a 40:1 mixture of Taq and Pfu enzymes [Promega]). The buffer was 20 mM Tris–10 mM KCl–10 mM (NH4)2SO4–2 mM MgCl2–0.1% (vol/vol) Triton X-100 (pH 8.8 [HCl]) containing 0.1 mg of bovine serum albumin ml−1. Concentrations of deoxyribonucleoside triphosphates and primers were as described above. Cycling conditions for each primer set (forward/reverse) were as follows: 1.1F/1.9R, one cycle at 94°C for 1 min followed by 30 cycles of 94°C for 10 s, 50°C for 20 s, and 72°C for 6 min; 1.1F/1.1R, one cycle at 94°C for 1 min followed by 30 cycles of 94°C for 10 s, 45°C for 20 s, and 72°C for 30 s; 1.2F/1.2R, one cycle at 94°C for 1 min followed by 30 cycles of 94°C for 10 s, 54°C for 20 s, and 72°C for 2 min; and 1.2(GAA)F/1.2(GAA)R, one cycle at 94°C for 1 min followed by 30 cycles of 94°C for 10 s, 54°C for 20 s, and 72°C for 9 s.
DNA from M. gallisepticum S6 clones was obtained either by washing cells three times in phosphate-buffered saline followed by lysing in water at 95°C for 10 min or by a low-melting-temperature agarose method (1). Fragments amplified by PCR were cloned by insertion into pGEM-T (Promega) and sequenced by using T7 DNA polymerase sequencing mixes (Pharmacia) and either pUC/M13 sequencing primers or pMGA gene-specific primers as indicated in Results.
RNA preparation and Northern blot analysis.
Total RNA was prepared and subjected to Northern blot analysis as previously described (8). The probes for the pMGA1.1 or pMGA1.2 gene were PCR products which encompassed a region encoding 10 amino acids of the leader sequence followed by 45 (pMGA1.1) or 44 (pMGA1.2) amino acids of the mature polypeptides. The probes were labelled and used as previously described (8).
RESULTS
Isolation of clones expressing different pMGA genes.
The parental cell population (pMGA1.1+ pMGA1.9−) was from an isolated clonal colony of M. gallisepticum S6 grown in normal broth medium. The second population (pMGA1.1− pMGA1.9+) was from these cells grown in medium containing MAb 66 as previously described (15). The MAb 66 reagent detects an epitope which is present on the pMGA1.1 polypeptide but is absent from all other known pMGA polypeptides. A sample from the pMGA1.1− population of cells was grown on agar plates without antibody, and one colony, designated C11, which exhibited sectorial reversion to pMGA1.1 expression was chosen for further subcloning. Two derivatives of C11 with relevant pMGA phenotypes were thus isolated. One clone, C11(−), exhibited a stable and uniform pMGA1.1− pMGA1.9+ colony phenotype. The other clone, C11(+), exhibited a rare, uniform pMGA1.1+ pMGA1.9+ colony phenotype in the absence of MAb 66. The pMGA colony phenotypes of these three clones were confirmed by SDS-PAGE followed by Western blotting, and the data for parental S6 cells, C11(+) cells, and C11(−) cells are shown in Fig. 1. The data in Fig. 1C revealed, as expected, that the S6 clone expressed a major 67-kDa polypeptide which bound the anti-pMGA1.1 reagent. The C11(−) clone, however, did not express this species and instead expressed an 82-kDa polypeptide bound by the anti-pMGA1.9 reagent. No cross-reactivity between the pMGA1.1 and pMGA1.9 polypeptides was apparent (Fig. 1B and C). The C11(+) clone expressed both pMGA1.1 and pMGA1.9 polypeptides. Clones C11(+) and C11(−) also expressed lower-molecular-mass species which are probably degradation products of the 82-kDa pMGA1.9 polypeptide (15). By using a protocol similar to that described above to obtain the C11(+) and C11(−) clones, an additional clone, C1, was isolated, which, even in the absence of pMGA antibodies, expressed only the pMGA1.9 polypeptide and which did not revert to the expression of pMGA1.1.
A further clone, V3-1, was obtained as follows. A population of pMGA1.1− cells (grown in MAb 66-containing medium) was inoculated onto agar plates, and nitrocellulose lifts of the resultant colonies were subjected to a double staining procedure designed to detect the expression within colonies of variant pMGA molecules. These variant molecules did not bind MAb 66, which detects pMGA1.1 per se, but bound rabbit antibodies to purified pMGA1.1. One colony (V3) which exhibited sectors which bound the rabbit antibodies but not MAb 66 was identified in this way, and a subclone derived from it, (V3-1) was found to stain uniformly with the rabbit antiserum but not MAb 66. Analysis of the V3-1 clone by SDS-PAGE revealed the expression of an approximately 67-kDa polypeptide which bound rabbit anti-pMGA1.1 antibodies but not MAb 66 (data not shown). The amino-terminal sequence of this polypeptide was TTPTPNPTPNPN. The sequence was identical to the amino terminus of the deduced pMGA1.2 polypeptide sequence and differed at two positions from the corresponding segment of pMGA1.1 (16, 17). To confirm that the V3-1 clone expressed pMGA1.2 rather than another pMGA gene with the same amino-terminal sequence, a Northern blot experiment was conducted (Fig. 2). The RNA species revealed in this experiment were detected by using two PCR probes, derived from the pMGA1.1 and pMGA1.2 genes, respectively; both probes were previously shown to bind highly selectively to their corresponding transcripts (8, 15). A prominent 2.2-kb RNA species which bound the pMGA1.1 probe was evident in S6 cells but was much less apparent in V3-1 cells (Fig. 2). Conversely, a major RNA species of similar size was detected by the pMGA1.2 probe in the V3-1 clone but was much less prominent in S6 cells. These two pMGA probes are known to exhibit minor cross-hybridization with one another under the stringency conditions used but not with other pMGA family members (8, 15). The results in Fig. 2 indicate a switch in expression from pMGA1.1 in S6 cells to pMGA1.2 in V3-1 cells.
FIG. 2.
pMGA gene expression in V3-1 cells. Total RNA samples from parental (S6) and V3-1 cells (3 μg per well) were subjected to Northern blotting with labelled probes as indicated.
Analysis of the region containing the pMGA1.1 gene promoter.
A strategy was devised to facilitate the amplification of the segment of the genome which encoded the pMGA1.1, pMGA1.7, and pMGA1.9 genes. Oligonucleotides were constructed (1.1F and 1.9R) which should amplify the segment indicated in Fig. 3A, namely, the 5′ flanking nucleotides of the pMGA1.1 (GAA)n tract, the coding and 3′ intergenic regions of pMGA1.1, the entire pMGA1.7 coding region and its 3′ intergenic region, and part of the coding region of pMGA1.9. The PCR product size anticipated would be 6.15 kb. The result of gel electrophoresis of one such PCR amplification with DNA template from the parental S6 clone is shown in Fig. 3B along with an EcoRI digest (lanes 2 and 3, respectively). Only one major product of approximately 6 kb was present in this amplification (Fig. 3B), and the mobilities of the EcoRI fragments of this PCR product were highly compatible with size predictions based on the known locations of EcoRI restriction sites within the amplified region (Fig. 3A). Similar 6-kb products were obtained from all of the M. gallisepticum DNA samples in this work. The data of Fig. 4 were obtained by sequencing the region containing the pMGA1.1 promoter with 1.1F as a sequencing primer and several cloned 6-kb DNA isolates as templates. These sequences from five independent 6-kb isolates from the normal (pMGA1.1+) population were all identical to the previously published sequence of the segment 5′ to the pMGA1.1 gene (15). The situation was different within the same region in cells grown in MAb 66-containing medium [designated S6(mAb66) in Fig. 4] and which consequently lacked pMGA1.1 expression (15). Four independent isolates from this source each exhibited a consistent reduction in the number of GAA trinucleotide repeats 5′ to the pMGA1.1 promoter, from (GAA)12 in normal cells to (GAA)10 in S6(mAb66) cells. The only other sequence difference noted between these four isolates and their normal counterparts was a T→C change 5′ to the GAA repeat region in one PCR isolate {S6(mAb66)[10]} (see Fig. 5), which was probably a PCR copying error. The analysis of the same region from the C1 clone (pMGA1.1−) revealed that in three independent isolates the pMGA1.1 GAA repeat length had also decreased from (GAA)12 to (GAA)10. The C11(−) clone (three isolates) contained a (GAA)9 motif, but in contrast, its revertant, C11(+) (three isolates), contained a (GAA)12 sequence. Collectively, the data obtained for the region 5′ to the pMGA1.1 gene and presented in Fig. 4 indicate a link between the transcriptional activity of the pMGA1.1 gene and the presence of a (GAA)12 motif 5′ to its promoter.
FIG. 4.
Sequence analysis of the region 5′ to the pMGA1.1 locus. Each sequence was derived from a single plasmid recombinant obtained by cloning PCR-amplified products from the indicated cloned mycoplasma cell lines. The cellular sources of the DNA templates used to obtain the cloned PCR products were normal cells (S6), cells grown in medium containing mAb66 [S6(mAb66)], and clones C11(+), C11(−), and C1. The numbers in brackets after the cell source indicate individual PCR clones. The sequence denoted pMGA1.1 is from reference 16 and was originally obtained by sequencing part of the M. gallisepticum S6 genome. Only nucleotides which differ from this (top) sequence are shown for the PCR clones, and dots indicate identities. Dashes indicate gaps in the alignment. The locations of the −10 and −35 promoter motifs (8) are indicated by solid lines. The last nucleotide (G) of each sequence represents the start point of transcription (+1). Sequences without asterisks were obtained by using as templates cloned, 6-kb PCR products. PCR clones with asterisks were from a separate experiment in which primers 1.1F and 1.1R (see Materials and Methods) were used to amplify only the region 5′ to pMGA1.1 (504 bp).
FIG. 5.
Relationship between GAA repeat length and pMGA phenotype.
Analysis of the region containing the pMGA1.9 gene promoter.
The regions flanking the putative promoters of the pMGA1.7 and pMGA1.9 genes were next analyzed by using oligonucleotides 1.7F and 1.9R as sequencing primers with the same series of DNA templates as in Fig. 4. The results are summarized in Fig. 5. Apart from single-base differences which occurred outside the pMGA1.9 GAA repeats in 3 of the 15 templates sequenced in this experiment (not shown), the only differences between them occurred within these repeats. Isolates from cells known to express pMGA1.9, namely, S6(mAb66) cells and clones C11(+) and C11(−), contained a (GAA)12 motif 5′ to the promoter. The single exception was found in a C11(+) clone {C11(+)[46]} (Fig. 5), which contained the sequence (GAA)5(GGA)(GAA)6. It seems most likely that the A→G change apparent in this one clone occurred as a consequence of the PCR. In contrast, normal S6 cells which did not express pMGA1.9 contained (GAA)n motifs 5′ to its gene, where, in all cases, n ≠ 12. Heterogeneity was apparent in the pMGA1.9 GAA repeat number even within normal strain S6 cells (which did not express the gene): one isolate contained 20 GAA repeats, and three other isolates each contained 16 repeats.
Analysis of the region containing the pMGA1.7 gene promoter.
In Fig. 5 the trinucleotide enumeration data for the region 5′ to the pMGA1.7 gene are also presented. It is apparent that considerable variation in GAA repeat length occurs both within and between all clones examined. In only 1 case of 13 was a (GAA)12 motif detected. It is not known if the pMGA1.7 gene is expressed under any conditions in the S6 strain of M. gallisepticum.
Expression of pMGA1.2 also requires a (GAA)12 motif.
The V3-1 clone was chosen due to its expression of a pMGA molecule which lacked the MAb 66 epitope which typifies pMGA1.1 and of an alternative molecule with an amino-terminal sequence suggesting its identity as pMGA1.2. Northern blot experiments confirmed V3-1 expression of pMGA1.2 (Fig. 2). To investigate if the pMGA1.2 gene, expressed in the V3-1 clone, had acquired a (GAA)12 motif, the regions which contained the relevant parts of the pMGA1.2 genes from parental cells and from V3-1 cells were amplified and cloned for sequencing. The strategy used to clone the selected region involved two consecutive PCR amplifications and is depicted in Fig. 6A. The first PCR used a pair of primers (1.2F and 1.2R) complementary to sequences common to the pMGA1.1 and pMGA1.2 genes, and the second pair of primers [1.2(GAA)F and 1.2(GAA)R] amplified a segment, nested within the primary PCR product, which amplified only the pMGA1.2 template. Sequence analysis of the cloned, amplified species from normal cells and the V3-1 clone is presented in Fig. 6B. As anticipated, the two-stage PCR had specifically amplified the desired pMGA1.2 segment from both clones. Several nucleotides differ between the pMGA1.1 and pMGA1.2 genes in the sequences flanking the GAA repeats (16, 17), and both sequences in Fig. 6B contain all of these distinctive nucleotides. The S6 sequence in Fig. 6B contained one GTA trinucleotide within the pMGA1.2 GAA motif. This motif within the V3-1 clone was interrupted by two GTA units.
The salient difference between the S6 and V3-1 sequences was in the number of trinucleotide repeats. The V3-1 sequence contained an imperfect (GAA)12 motif, whereas the corresponding segment from normal cells contained an imperfect (GAA)10 motif.
DISCUSSION
This study investigated the molecular basis for pMGA gene switching in M. gallisepticum cells after growth in the presence of antibodies directed to a major surface lipoprotein, pMGA1.1. Previous studies had demonstrated that the presence of certain pMGA1.1-specific antibodies during the growth of M. gallisepticum cells resulted in the growth of colonies which lacked pMGA1.1 (15). The cessation of pMGA1.1 transcription in such cells was found to be accompanied by the expression of another member of the pMGA gene family, pMGA1.9. It was speculated that the switch between expression of pMGA1.1 and pMGA1.9 was due to DNA sequence differences which affected the transcription of these genes (15), and the present work has borne out this proposal.
To facilitate the analysis, an isogenic clonal lineage was derived from the S6 strain of M. gallisepticum. The C11(−) clone was isolated from a population of pMGA1.1− cells on the basis of its failure to produce prominent pMGA1.1+ sectors when plated onto agar medium lacking MAb 66. This clone was shown to exhibit a pMGA1.1− pMGA1.9+ phenotype, and the C11(+) clone (pMGA1.1+ pMGA1.9+) was a pMGA1.1+ revertant of C11(−). The C1 clone was obtained independently of C11(−) but exhibited the same pMGA phenotype. The V3-1 clone was deemed to be interesting because of its ability to attach rabbit polyclonal antibodies directed to pMGA1.1 but not MAb 66, which defines an epitope that is probably exclusive to the pMGA1.1 polypeptide (8, 14, 16). Its expression of pMGA1.2 was demonstrated by amino-terminal sequencing and by Northern blot analysis (Fig. 2).
The data of Fig. 4 and 5 revealed that cells grown in medium containing the pMGA1.1-specific antibody, MAb 66, contained altered sequences 5′ to the promoter sites of both the pMGA1.1 and pMGA1.9 genes. The number of GAA repeats located upstream from the −35 boxes of both pMGA1.1 and pMGA1.9 had altered compared to that in parental S6 cells. Specifically, in parental cells the 5′ region of pMGA1.1 contained 12 GAA repeats, whereas in cells grown with MAb 66 only 10 such repeats were present. In cells grown in medium containing the pMGA1.1-specific antibody, MAb 66, 12 GAA repeats occurred 5′ to pMGA1.9. Cells grown in MAb 66-containing medium are devoid of pMGA1.1 and instead express pMGA1.9 (15). These data strongly suggested a link between the presence of a 5′ (GAA)12 motif and pMGA gene transcription. It is noteworthy in this regard that the pMGA1.1 gene was the only known family member in the S6 strain of M. gallisepticum previously reported to possess this precise motif (14, 17).
The GAA repeat length of the pMGA1.7 gene was also found to vary. Eighteen and 16 GAA repeats were found 5′ to pMGA1.7 from normal (pMGA1.1+) cells. Other sources (pMGA1.1− cells and subclones derived from them) contained from 12 to 20 GAA repeats 5′ to pMGA1.7 (Fig. 5). Variation in the pMGA1.7 motif length was noted even between PCR isolates from the same clone. It seems reasonable, given this observation, to suggest that in M. gallisepticum the GAA repeat sequences which append pMGA genes are intrinsically unstable and are amenable to rapid expansion or contraction events. The sequence data reported in Fig. 4 and summarized in Fig. 5 demonstrated that, in contrast, the segments flanking the GAA repeats were not variable. The few single-base changes which did occur outside the GAA repeats were infrequent and randomly distributed. Whether these differences (Fig. 4 and 5) result from PCR copying errors or from mutations within mycoplasma cells was not investigated.
Collectively, the results of this paper are significant from two viewpoints. First, the apparently obligate association between pMGA gene transcriptional activity and the occurrence of a (GAA)12 motif 5′ to a promoter site suggests the possible existence of a transcriptional activator protein in M. gallisepticum cells which discriminates between the (GAA)12 motif and (GAA)n motifs where n ≠ 12. A functional analogue of the catabolite activator protein of Escherichia coli with strict binding specificity for (GAA)12 would account for the link between (GAA)12 and pMGA transcription. Alternatively, such a hypothetical transcriptional activator may attach to a sequence 5′ to the (GAA)12 motif which would then act to precisely space or orient the transcriptional activator protein and the RNA polymerase molecules on the same DNA template so as to achieve effective initiation of transcription.
The exclusive association and invariable occurrence of (GAA)n motifs with most or all pMGA genes of M. gallisepticum (1) suggest the potential of a single strain of cells to use most or all of its pMGA genes by altering GAA repeat lengths. Most likely, these repeat length changes would be relatively frequent, stochastic events and the fate of individual pMGA switch variants would depend on how well their new pMGA structure supported cell division and/or survival. Studies on short tandem repeat sequences have shown that they are intrinsically prone to frequent alteration in repeat number (11), and slipped-strand mispairing has been invoked to explain their genetic instability. It is easy to see how the instability of GAA repeat numbers demonstrated in this study could rapidly generate variant cells expressing pMGA genes different from that of the original clone and how such variants might selectively prosper. The fact that most of the pMGA genes sequenced to date possess homologous coding sequences, uninterrupted by internal stop codons, suggests a continuing requirement for full-length and functional pMGA polypeptides. This observation implies that gene switches to many, perhaps all, of the pMGA genes occur commonly in M. gallisepticum cells, and studies are now in progress to determine if pMGA switching events occur during infection in this organism’s natural host.
This study provides evidence that the GAA repeat length determines whether a pMGA gene is expressed or not. However, regulation of the pMGA gene family needs to account for several supplementary observations. First, in three independent field isolates of M. gallisepticum, only a single pMGA polypeptide is expressed in cultured cells (8). Second, cultured or stored isolates of S6 indefinitely maintain the expression of pMGA1.1. Third, pMGA expression oscillates highly selectively between only two members of the pMGA gene family, namely, pMGA1.1 and pMGA1.9, depending on the presence or absence of MAb 66 (15). The questions which arise from these observations are as follows. First, what mechanism prevents more than one pMGA gene from being expressed at a time? Second, if most or all pMGA genes in S6 cells are available for expression, why is pMGA1.1 normally favored for expression? Third, as a consequence of selection by certain antibodies to pMGA1.1, why is the pMGA1.9 gene chosen for expression rather than another family member? Finally, it is of interest to ascertain whether switching of pMGA gene expression occurs within the host and what role such events play in the infectious process.
It is significant even in the context of mycoplasma species alone that the mechanism of pMGA gene switching described here is unique. Antigenic variation or phase variation per se is not uncommon among mycoplasmas. In M. hyorhinis, for example, several members of the vlp gene family are variably expressed (20, 21, 24). Transcription of each gene appears to be independently controlled by variation in the number of residues in a polyadenosine tract between the −10 and −35 regions of the promoter. The hsd1 locus of another mycoplasma species, M. pulmonis (5) is an example of a DNA inversion mechanism. One orientation facilitates the expression of restriction-modification functions, and the other orientation prevents expression. Site-specific inversions of DNA elements also account for the rapid phase variation of M. pulmonis V-1 surface antigens (2). M. bovis exhibits high-frequency phenotypic switching of a size-variable lipoprotein (Vsp), and in this species too, switching involves major chromosomal changes, although whether specific inversion, insertion, or recombination events are responsible is not yet known (13). A group of surface antigens in the avian pathogen M. synoviae has recently been described (9, 18). It has been suggested that at least one member of this family may act as an adhesin, the expression of which is phase variable. For M. gallisepticum, the subject of the present work, Levisohn et al. (12) noted that passage of organisms through an avian host resulted in substantial alterations in surface antigen expression. Notably, two of the variable antigens reported were similar in size to pMGA1.1 and pMGA1.9 (67 and 79 kDa, respectively) (12). In the light of these observations, it will be of interest to investigate whether the expression of pMGA1.1 and pMGA1.9 indeed changes in vivo as it does in vitro when grown in medium containing pMGA-specific MAb.
The probable exploitation of the instability of trinucleotide repeat segments within M. gallisepticum contrasts with the deleterious consequences which the same phenomenon can produce in humans, where a number of genetic diseases are due to expansions of these motifs. For example myotonic dystrophy, Huntington’s disease, Kennedy’s disease, and spinocerebellar ataxia type 1 (19, 22, 23) are all due to increases in CAG trinucleotide repeats, which occur in different genes in each case. Fragile X syndrome is due to expansions of the CCG trinucleotide which occur at a locus on the X chromosome. Considerable CCG repeat length heterogeneity occurs even within the somatic cells of single individuals (22). Notably, Fredreich’s ataxia is due to the GAA repeat expansions within the frataxin gene (4).
The pMGA switching mechanism of M. gallisepticum demonstrated in the present work is unique in that it relies upon the genetic instability of tandem GAA repeats. These repeats occur commonly in M. gallisepticum, but only 5′ to pMGA genes (1). They do not occur in the genomes of either M. genitalium or M. pneumoniae, the full sequences of which have recently been published (6, 10). These species are considered to be phylogenetically closely related to M. gallisepticum, and it therefore follows that the exploitation by the latter species of trinucleotide repeat length alterations for antigenic variation may be an independent and relatively recent evolutionary experiment.
ACKNOWLEDGMENTS
This work was supported by project grants from the Australian Research Council (to I.D.W.). M. D. Glew was supported in part by a scholarship from Rural Industries Research and Development Corporation.
We thank Kevin Whithear for his constructive critique of the manuscript.
REFERENCES
- 1.Baseggio N, Glew M D, Markham P F, Whithear K G, Browning G F. Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology. 1996;142:1429–1435. doi: 10.1099/13500872-142-6-1429. [DOI] [PubMed] [Google Scholar]
- 2.Bhugra B, Voelker L L, Zou N X, Yu H L, Dybvig K. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 3.Bordier C. Phase separation of integral membrane proteins in Triton-X114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 4.Campuzano V, Montermini L, Moltò M D, Pianese L, Cossée M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Cañizares J, Koutnikova H, Bidichandani S I, Gellera C, Brice A, Trouillas P, de Michele G, Filla A, De Frutos R, Palau F, Patel P I, di Donato S, Mandel J-L, Cocozza S, Koenig M, Pandolfo M. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 5.Dybvig K, Yu H. Regulation of a restriction and modification system via DNA inversion in Mycoplasma pulmonis. Mol Microbiol. 1994;12:547–560. doi: 10.1111/j.1365-2958.1994.tb01041.x. [DOI] [PubMed] [Google Scholar]
- 6.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 7.Frey M L, Hanson R P, Anderson D P. A medium for the isolation of avian Mycoplasmas. Am J Vet Res. 1968;29:2163–2171. [PubMed] [Google Scholar]
- 8.Glew M D, Markham P F, Browning G F, Walker I D. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum S6. Microbiology. 1995;141:3005–3014. doi: 10.1099/13500872-141-11-3005. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich V A, Ley D H, Markham J F, Whithear K G, Walker I D. Identification of Mycoplasma synoviae immunogenic surface proteins and their potential use as antigens in the enzyme-linked immunosorbent assay. Avian Dis. 1995;39:465–474. [PubMed] [Google Scholar]
- 10.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 12.Levisohn S, Rosengarten R, Yogev D. In vivo variation of Mycoplasma gallisepticum antigen expression in experimentally infected chickens. Vet Microbiol. 1995;45:219–231. doi: 10.1016/0378-1135(95)00039-d. [DOI] [PubMed] [Google Scholar]
- 13.Lysnyansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markham P F, Glew M, Brandon M R, Walker I D, Whithear K G. Characterization of a major hemagglutinin protein from Mycoplasma gallisepticum. Infect Immun. 1992;60:3885–3891. doi: 10.1128/iai.60.9.3885-3891.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markham P F, Glew M D, Browning G F, Whithear K G, Walker I D. The expression of two members of the pMGA gene family of Mycoplasma gallisepticum oscillates and is influenced by pMGA-specific antibodies. Infect Immun. 1998;66:2845–2853. doi: 10.1128/iai.66.6.2845-2853.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markham P F, Glew M D, Sykes J E, Bowden T R, Pollocks T D, Browning G F, Whithear K G, Walker I D. The organisation of the multigene family which encodes the major cell surface protein, pMGA, of Mycoplasma gallisepticum. FEBS Lett. 1994;352:347–352. doi: 10.1016/0014-5793(94)00991-0. [DOI] [PubMed] [Google Scholar]
- 17.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noormohammadi A H, Markham P F, Whithear K G, Walker I D, Gurevich V A, Ley D H, Browning G F. Mycoplasma synoviae has two distinct phase-variable major membrane antigens, one of which is a putative hemagglutinin. Infect Immun. 1997;65:2542–2547. doi: 10.1128/iai.65.7.2542-2547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards R I, Sutherland G R. Dynamic mutations: a new class of mutations causing human disease. Cell. 1992;70:709–712. doi: 10.1016/0092-8674(92)90302-s. [DOI] [PubMed] [Google Scholar]
- 20.Rosengarten R, Wise K S. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science. 1990;247:315–318. doi: 10.1126/science.1688663. [DOI] [PubMed] [Google Scholar]
- 21.Rosengarten R, Wise K S. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct size variant lipoproteins generating high-frequency surface antigenic variation. J Bacteriol. 1991;173:4782–4793. doi: 10.1128/jb.173.15.4782-4793.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross C A, McInnis M G, Margolis R L, Li H-S. Genes with triplet repeats: candidate mediators of neuropsychiatric disorders. Trends Neurobiol. 1993;16:254–260. doi: 10.1016/0166-2236(93)90175-l. [DOI] [PubMed] [Google Scholar]
- 23.Timchenko L T, Caskey C T. Trinucleotide repeat disorders in humans: discussions of mechanisms and medical issues. FASEB J. 1996;10:1589–1597. doi: 10.1096/fasebj.10.14.9002550. [DOI] [PubMed] [Google Scholar]
- 24.Wise K S, Yogev D, Rosengarten R. Antigenic variation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 473–489. [Google Scholar]