Abstract
During our investigation of saprophytic fungi in Guizhou and Hainan provinces, China, three hyphomycetes were collected from terrestrial and freshwater habitats. Based on morphological characteristics and phylogenetic analyses of combined ITS, LSU, tef1-α, and rpb2 sequence data, two new species are introduced: Distoseptisporahainanensis and D.lanceolatispora. Additionally, one known species, D.tectonae, previously unreported from Edgeworthiachrysantha, is newly reported. Detailed descriptions, illustrations, and a phylogenetic tree to show the two new species and the new host record of Distoseptispora are provided. In addition, a checklist of Distoseptispora species with their locations, lifestyles, habitats, and hosts is provided.
Key words: 2 new taxa, asexual morph, phylogeny, taxonomy
Introduction
Distoseptispora K.D. Hyde, McKenzie & Maharachch. was introduced by Su et al. (2016) with D.fluminicola McKenzie, Hong Y. Su, Z.L. Luo & K.D. Hyde, as the type species. Most Distoseptispora species are reported as saprophytes, typically found on decaying wood in terrestrial and freshwater habitats (Hyde et al. 2016, 2019; Su et al. 2016; Xia et al. 2017; Yang et al. 2018; Crous et al. 2019; Luo et al. 2019). The initial descriptions of Distoseptispora are derived from its asexual morphology (Hyde et al. 2016, 2019, 2020; Su et al. 2016; Yang et al. 2018, 2021; Luo et al. 2019; Sun et al. 2020). The first description of a sexual morph of Distoseptispora was described by Yang et al. (2021). Recently, Konta et al. (2023) identified the second sexual species on dead leaves of Licualaglabra, and provided detailed explanations, enhancing our understanding of Distoseptispora sexual morphology. This sexual morph is characterized by solitary or gregarious, immersed to semi-immersed, subglobose to ellipsoidal, perithecial, dark brown ascomata with a short neck; 8-spored, cylindrical, short pedicellate asci with non-amyloid apical annuli; and fusiform, 0–3-septate, hyaline ascospores with mucilaginous sheaths (Yang et al. 2021; Konta et al. 2023). The asexual morph of Distoseptispora was recently expanded upon by Yang et al. (2021), incorporating macronematous, mononematous, solitary or fasciculate conidiophores, blastic, terminal, percurrent, cylindrical conidiogenous cells; and acrogenous, solitary, obclavate, ellipsoidal, obovoid or fusiform, rostrate or not, euseptate, distoseptate or rarely muriform conidia with or without a septal pore and mucilaginous sheath.
Distoseptispora has been found on various hosts viz. Tectona, Pandanus, bamboo, Clematis, Carex, Dipterocarpus, Licualaglabra, Cocosnucifera, Phragmitesaustralis, Thysanolaenamaxima, Platanusorientalis, and decaying wood and grasses (Shoemaker and White 1985; McKenzie 1995; Hyde et al. 2016, 2019, 2021, 2023; Su et al. 2016; Tibpromma et al. 2018; Crous et al. 2019; Phookamsak et al. 2019; Phukhamsakda et al. 2020, 2022; Sun et al. 2020; Zhai et al. 2022; Afshari et al. 2023; Hu et al. 2023; Konta et al. 2023). Most Distoseptispora species have been described in Asia, mainly in China, Thailand, and Malaysia, and only a few have been described in Europe (Shoemaker and White 1985; McKenzie 1995; Phookamsak et al. 2019; Ma et al. 2022; Zhai et al. 2022; Zhang et al. 2022; Konta et al. 2023). Distoseptispora comprises 74 accepted species in Index Fungorum (2024), but there is an ambiguity in the taxonomic status of D.submersa Z.L. Luo, K.D. Luo et al. (2019) stated that D.submersa is phylogenetically closely related to D.tectonae, and there are only minor size differences in conidiophores and conidia between D.tectonae and D.submersa. Dong et al. (2021) synonymized D.submersa under D.tectonae, thus, Distoseptispora comprises 73 accepted saprobic species, of which 44 were from freshwater habitats, 29 from terrestrial habitats, and five from both terrestrial and freshwater environments (Hyde et al. 2016, 2019; Luo et al. 2019; Monkai et al. 2020; Yang et al. 2021; Ma et al. 2022; Zhang et al. 2022; Afshari et al. 2023; Hu et al. 2023; Konta et al. 2023; Liu et al. 2023).
In this study, three fresh hyphomycetous fungal collections were encountered during a microfungal investigation in Hainan and Guizhou provinces. Based on multi-gene phylogeny and morphological comparison, two new species, Distoseptisporahainanensis and D.lanceolatispora are introduced. In addition, a new host record of D.tectonae from Edgeworthiachrysantha is also reported.
Materials and methods
Sample collection, isolation, and morphological study
Fresh specimens were collected from Hainan and Guizhou provinces in China. Fungal colonies were mounted on a slide with distilled water and were observed and examined using a stereomicroscope (SMZ 745, Nikon, Tokyo, Japan). Micro-morphological characteristics were captured with a Nikon EOS 90D digital camera combined with an ECLIPSE Ni-U compound microscope (Nikon, Tokyo, Japan). The sizes of the fungal structures were measured using the Tarosoft (R) Image Frame Work program (IFW 0.97 version), and the photo plates were processed with Adobe Photoshop CC 2019 (Adobe Systems, San Jose, CA, USA).
Single spore isolations were carried out following the methods described in Senanayake et al. (2020). Germinated conidia were transferred to fresh potato dextrose agar (PDA) plates and incubated at 25–27 °C for four weeks. Culture characteristics, including color, shape, and size, were recorded. Herbarium specimens were deposited in the herbarium of the Guizhou Academy of Agriculture Sciences (GZAAS), Guiyang, China, and the living cultures were deposited at the Guizhou Culture Collection, China (GZCC). Faces of Fungi and Index Fungorum numbers were obtained following the protocols outlined by Jayasiri et al. (2015) and Index Fungorum (2024), respectively.
DNA extraction, PCR amplification, and sequencing
Fresh mycelia were scraped from cultures that were incubated at 25–27 °C for 28 days. Fungal genomic DNA was extracted using the Biospin Fungus Genomic DNA Extraction Kit (BioFlux, Shanghai, China), following the manufacturer’s instructions. Four gene regions: internal transcribed spacer (ITS), large subunit ribosomal DNA (LSU), translation elongation factor 1-alpha (tef1-α), and RNA polymerase II second largest subunit (rpb2) were selected. The primers used in this study for each gene region were as follows: ITS4 and ITS5 for ITS (White et al. 1990), LR0R and LR5 for LSU (Vilgalys and Hester 1990; Cubeta et al. 1991), EF1-983F and EF1-2218R for tef1-α (Rehner and Samuels 1994), and rpb2 with fRPB2-5F and fRPB2-7cR (Liu et al. 1999).
Polymerase chain reaction (PCR) amplifications were carried out in a 50 µL reaction volume containing 44 μL of 1.1 × T3 Super PCR Mix (TsingKe Biotech, Chongqing, China), 2 µL of DNA template, and 2 µL of each forward and reverse primer. The amplification condition for LSU and ITS consisted of initial denaturation at 94 °C for 3 min, followed by 35 cycles of 45 s at 94 °C, 50 s at 56 °C, and 1 min at 72 °C, and a final extension period of 10 min at 72 °C. The amplification condition for the tef1-α gene consisted of initial denaturation at 94 °C for 3 min, followed by 30 cycles of 30 s at 94 °C, 50 s at 56 °C, and 1 min at 72 °C, a final extension period of 10 min at 72 °C. The amplification condition for the rpb2 gene consisted of initial denaturation at 95 °C for 5 min, followed by 35 cycles of 15 s at 95 °C, 50 s at 56 °C, and 1 min at 72 °C, a final extension period of 10 min at 72 °C. The quality of PCR amplification products was examined with 1% agarose electrophoresis gels stained with ethidium bromide, and the PCR products were sent to TsingKe Biotech, Chongqing, China for purification and sequencing.
Phylogenetic analyses
The raw sequences were initially checked with BioEdit v 7.0.5.3 (Hall 1999). Forward and reverse sequences were assembled using SeqMan v. 7.0.0 (DNASTAR, Madison, WI, USA). Sequence data (LSU, ITS, tef1-α, and rpb2) for Distoseptispora were downloaded from GenBank based on the blast results and recent publications (Table 1). Each individual gene dataset was aligned using the online program MAFFT version 7 with the “auto” option (Hall 1999; Katoh and Standley 2013). These alignments were visually inspected and manually improved in BioEdit v 7.0.5.3. Multi-gene alignments were combined by SequenceMatrix (Vaidya et al. 2011). In this study, phylogenetic analyses were performed using maximum likelihood (ML), maximum parsimony (MP), and Bayesian posterior probability (BYPP) methods. The analyses were based on LSU, ITS, tef1-α, and rpb2 combined sequence datasets.
Table 1.
Names, strain numbers, and corresponding GenBank accession numbers of taxa used in this study.
Note: “T” denotes ex-type strain. Newly generated sequences are indicated in black bold. “N/A”: no data available in GenBank.
The phylogenetic analyses were conducted using the CIPRES Science Gateway V. 3.3. “RAxML-HPC v.8 on XSEDE”, “PAUP on XSEDE”, and “MrBayes on XSEDE (3.2.7a)” were utilized for ML, MP, and BYPP methods, respectively (Huelsenbeck and Ronquist 2001; Swofford 2002; Stamatakis et al. 2008; Miller et al. 2010; Ronquist et al. 2012). For the ML analysis, the GTRGAMMA model of nucleotide evolution was employed, and RAxML rapid bootstrapping with 1,000 bootstrap replicates was obtained (Stamatakis et al. 2008).
The MP analysis employed 1,000 random taxa additions to infer trees. Branches of zero length were collapsed, and all multiple parsimonious trees were saved. The maxtrees value was set to 5,000. For trees generated using different optimal criteria, parsimony score values were determined for tree length (TL), consistency index (CI), retention index (RI), and homoplasy index (HI). To assess clade stability, the bootstrap (BT) method was used with 1,000 iterations, each consisting of 100 trials of random stepwise addition of taxa (Hillis and Bull 1993).
The posterior probabilities (PP) were determined based on Bayesian Markov chain Monte Carlo sampling (Huelsenbeck and Ronquist 2001). The best nucleotide substitution model for each data partition was determined using the program MrModeltest 2.2 (Nylander 2004). The GTR + I + G substitution model with gamma rates and Dirichlet base frequencies was selected for all LSU, ITS, tef1-α, and rpb2 sequences. To calculate the posterior probabilities, four simultaneous Markov chains were run for one million generations, with trees sampled every 100th generation, resulting in a total of 10,000 trees. A burn-in parameter of 0.25 was set, indicating that 75% of the trees were remined during the burn-in phase, and the remaining trees were used for calculating the posterior probabilities in the majority rule consensus tree.
FigTree v. 1.4.4. was used for visualizing the phylogenetic trees, and Adobe Illustrator CC 2019v. 23.1.0 was used to edit trees and figure layout.
Phylogenetic analyses results
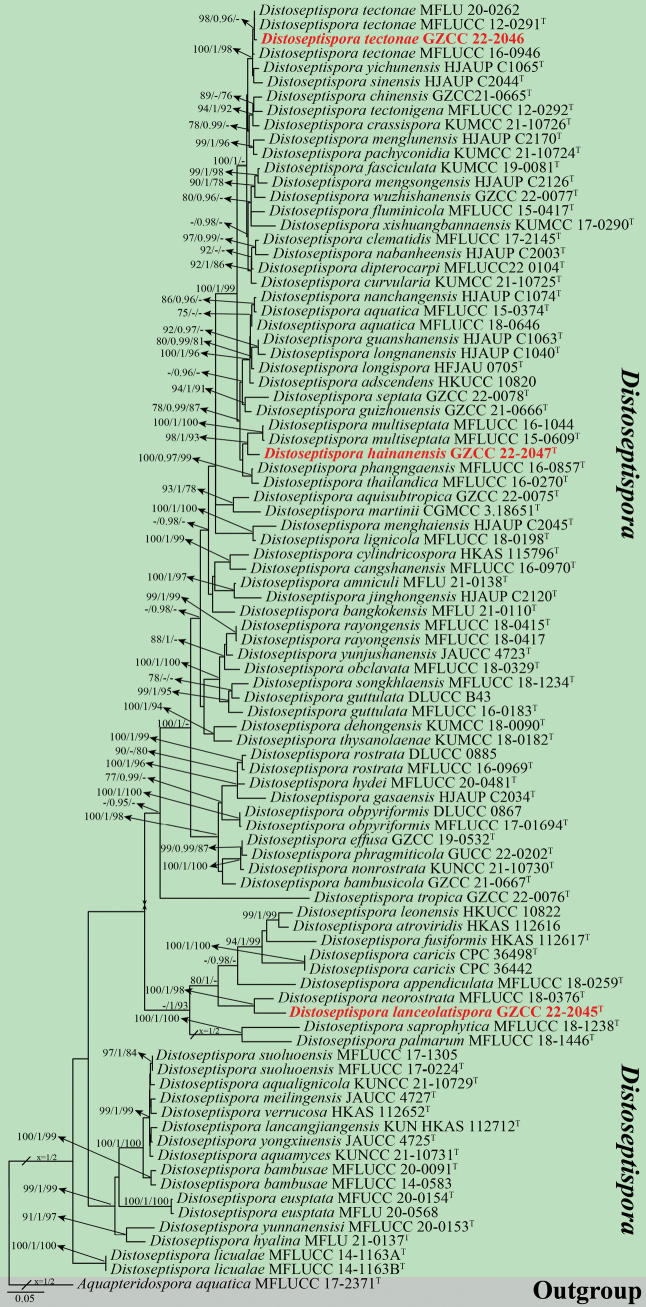
This study utilized a combined multi-gene dataset encompassing ITS, LSU, tef1-α, and rpb2 sequences to assess the phylogenetic relationships among Distoseptispora species. The analyses included a total of 90 taxa, designating Aquapteridosporaaquatica X.D. Yu, W. Dong & H. Zhang (MFLUCC 17-2371) as the outgroup taxon. The combined aligned sequence matrix comprised 3,360 characters, including gaps: LSU (1–840 bp), ITS (841–1406 bp), tef1-α (1407–2321 bp), and rpb2 (2322–3360 bp). The ML, MP, and Bayesian trees analyzed exhibited a high degree of similarity in topology and showed no significant conflicts. The RAxML analysis yielded a best-scoring tree (ln = -31666.963504), which is presented in Fig. 1. The matrix encompassed 1572 distinct alignment patterns, with 27.15% constituted by undetermined characters or gaps. The estimated base frequencies were as follows: A = 0.239306, C = 0.265297, G = 0.281926, T = 0.213472; substitution rates AC = 1.429077, AG = 3.512798, AT = 1.204511, CG = 0.845859, CT = 6.948345, GT = 1.000000; gamma distribution shape parameter α = 0.244431. For the MP analysis, 3360 characters remained unchanged, 330 were variable and parsimoniously uninformative, and 1074 were parsimoniously informative. The most parsimonious tree yielded the following values: TL = 5624, CI = 0.400, RI = 0.738, RC = 0.295, HI = 0.600. For BYPP analysis, Bayesian posterior probabilities from MCMC were evaluated with a final average standard deviation of split frequencies of 0.009754.
Figure 1.
Phylogenetic tree generated from ML analysis based on a combination of LSU, ITS, tef1-a, and rpb2 sequence data. Bootstrap support values of ML and MP equal to or greater than 75%, and PP value equal to or greater than 0.95 are given near the nodes as ML/PP/MP. The tree is rooted with Aquapteridosporaaquatica (MFLUCC 17-2371). Ex-type strains are indicated by the superscript T. The new collections are in bold red text.
In the phylogenetic analyses (Fig. 1), all our newly identified taxa nested within Distoseptispora, affirming their classification within this genus. Distoseptisporahainanensis (GZCC 22-047) formed a sister clade to D.multiseptata strains (MFLUCC 16-1044 and MFLUCC 15-0609) with 98% ML, 1.00 PP, and 93% MP statistical support. Distoseptisporalanceolatispora (GZCC 22-2045) formed a sister clade to D.neorostrata (MFLUCC 18-0376) with 100% ML, 1.00 PP, and 98% MP statistical support. In addition, our new collection GZCC 22-2046 clustered together with three D.tectonae strains (MFLU 20-0262 and MFLUCC 12-0291) with 98% ML and 0.96 PP statistical support, indicating they represent the same species.
Taxonomy
. Distoseptispora hainanensis
X.M. Chen & Y.Z. Lu sp. nov.
1369CFAC-E4DC-5C03-8878-CEA7116DE0F3
Index Fungorum: IF900953
Facesoffungi Number: FoF14663
Figure 2.
Distoseptisporahainanensis (GZAAS 22-2047, holotype) a, b colonies on substrate c–e conidiophores and conidia f–h conidiogenous cells bearing conidia i, j conidiophores k–q conidia r, s colony on PDA (r from front s from reverse). Scale bars: 50 μm (c, d, f–j, l–q); 30 μm (e, k).
Etymology.
The epithet refers to the location “Hainan Province” where the holotype was collected.
Holotype.
GZAAS 22-2047.
Description.
Saprobic on decaying wood in terrestrial habitat. Sexual morph: Undetermined. Asexual morph: Colonies on natural substrate superficial, effuse, dark brown, and hairy. Mycelium mostly immersed, composed of branched, septate, brown to dark brown, smooth hyphae. Conidiophores 70–130 × 5–8.5 μm (x– = 103 × 7 μm, n = 20), macronematous, mononematous, erect, solitary, straight or slightly flexuous, brown to dark brown, paler towards the apex, cylindrical, 4–6-septate, slightly constricted and darkened at septa, unbranched, thick-walled. Conidiogenous cells 6–13 × 3.5–6.5 μm (x– = 10 × 5 μm, n = 20), holoblastic, monoblastic, integrated, terminal, indeterminate, cylindrical, slightly tapering towards the apex, brown, percurrent. Conidia 44–117 μm × 9–18.5 μm (x– = 90 × 14 μm, n = 20), acrogenous, solitary, obclavate or obpyriform, rostrate, truncate at the base, straight or slightly curved, up to 22-distoseptate, slightly constricted at septa, brown, verrucose.
Culture characteristics.
Colonies grown on PDA circular, dense, fluffy, with raised center and lobate edge, pale gray in the center, grayish brown in the outer ring from the front view, dark brown in the center, and blackish brown in the outer ring from the reverse view.
Material examined.
China, Hainan Province, on unidentified decaying wood, 15 May 2021, Xia Tang, HN02 (GZAAS 22-2047, holotype), ex-type living culture, GZCC 22-2047.
Notes.
Morphologically, Distoseptisporahainanensis is similar to D.effusa L.L. Liu & Z.Y. Liu in having macronematous conidiophores, monoblastic conidiogenous cells, and acrogenous, obclavate, rostrate conidia (Yang et al. 2021). However, conidia of D.hainanensis are up to 22-distoseptate, whereas those of D.effusa are only 4–9-distoseptate. In the phylogenetic analyses, D.hainanensis formed a distinct clade sister to D.multiseptata Jiao Yang & K.D. Hyde with 98% ML, 1 PP, and 93% MP statistical support (Fig. 1). Distoseptisporahainanensis differs from D.multiseptata in having brown, longer conidiophores (70–130 μm vs. 23–65 µm) and obclavate or obpyriform, brown, verrucose, smaller conidia (44–117 μm vs. up to 290 µm) (Hyde et al. 2016). Comparing DNA sequence data, D.hainanensis diverges from D.multiseptata (MFLUCC 15-0609) in the ITS by 21/552 bp (3.8% difference), in the LSU by 1/812 bp (0.01% difference), in tef1-α by 33/912 bp (3.6% difference), and no data is available for rpb2 of D.multiseptata (MFLUCC 15-0609) in GenBank. Hence, the novel species, D.hainanensis, is introduced, following the guidelines of Jeewon and Hyde (2016) and Chethana et al. (2021).
. Distoseptispora lanceolatispora
X.M. Chen & Y.Z. Lu sp. nov.
558A79C2-EC0D-5BBD-BFB7-91B2EE3E304A
Index Fungorum: IF900954
Facesoffungi Number: FoF14664
Figure 3.
Distoseptisporalanceolatispora (GZAAS 22-2045, holotype) a, b colonies on substrate c–e conidiophores and conidia f, g conidiogenous cells bearing conidia h–k conidia l germinated conidium m, n colony on PDA (m from front n from reverse). Scale bars: 50 μm (c–g); 30 μm (h–l).
Etymology.
Referring to the lanceolate conidia.
Holotype.
GZAAS 22-2045.
Description.
Saprobic on submerged decaying wood in freshwater habitat. Sexual morph: Undetermined. Asexual morph: Colonies on substrate effuse, gregarious, hairy, pale brown to brown. Mycelium mostly immersed, composed of septate, yellow-brown to brown, smooth hyphae. Conidiophores 120–190 × 4–8 µm (x– = 155 × 6.5 µm, n = 20), macronematous, mononematous, erect, solitary, straight or slightly flexuous, grayish brown to dark brown, slightly tapering towards the apex, cylindrical, 7–8-septate, unbranched, thick-walled, smooth-walled. Conidiogenous cells 15–27 × 3–5.5 µm (x– = 20.5 × 4.5 µm, n = 20), monoblastic, integrated, terminal, cylindrical, slightly tapering towards the apex, pale brown, percurrent. Conidia 31–90 × 9.5–15 µm (x– = 58.5 × 13 µm, n = 20), acrogenous, solitary, fusiform or lanceolate, rostrate, truncate at the base, straight or slightly curved, 5–13-distoseptate, slightly constricted at septa, olivaceous to olivaceous brown, slightly paler at the apex, verrucous, with or without apical, hyalina appendages.
Culture characteristics.
Colonies grown on PDA circular, dense, flat, dry, gray to dark gray, radially striated, and a ring in the middle of the colonies with an entire edge from the front view, dark brown to black with a circular, gray edge from reverse view, not pigmented.
Material examined.
China, Hainan Province, on submerged decaying wood in a freshwater stream, 23 October 2021, Jian Ma, J13 (GZAAS 22-2045, holotype), ex-type living culture, GZCC 22-2045.
Notes.
Distoseptisporalanceolatispora is morphologically similar to D.leonensis (M.B. Ellis) R. Zhu & H. Zhang. However, compared to D.lanceolatispora, D.leonensis has longer conidiophores (120–190 µm vs. 110–130 µm), longer conidiogenous cells (15–27 µm vs. 5–15 µm), and 5–13-distoseptate, fusiform or lanceolate conidia (Zhang et al. 2022). In the phylogenetic analyses (Fig. 1), D.lanceolatispora forms a unique clade adjacent to D.neorostrata D.F. Bao, Z.L. Luo & H.Y. Su with 100% ML, 1 PP, and 98% MP support. Based on a pairwise nucleotide comparison of ITS and LSU sequences, D.lanceolatispora deviates from D.neorostrata by 39/529 bp (6.8%) for ITS and 14/850 bp (1.6%) for LSU, and there is no data available for tef1-α and rpb2 for D.neorostrata (MFLUCC 18-0376) in GenBank. Hence, we introduce the new species, D.lanceolatispora, based on the criteria established by Jeewon and Hyde (2016) and Chethana et al. (2021).
. Distoseptispora tectonae
Doilom & K.D. Hyde, Fungal Diversity 80: 222 (2016)
E502DE4A-6E52-505B-B0FD-A7463654155E
Index Fungorum: IF552223
Facesoffungi number: FoF01877
Figure 4.
Distoseptisporatectonae (GZAAS 22-2046) a, b colonies on substrate c, d conidiophores and conidia e, f conidiophores g–k conidia l germinated conidium m, n colonies on PDA (m from front n from reverse) Scale bars: 50 μm (c, d, g–l); 20 μm (e, f).
Description.
Saprobic on dead twigs of Edgeworthiachrysantha. Sexual morph: Undetermined. Asexual morph: Colonies on natural substrate abundant, superficial, dark brown, hairy. Conidiophores 35–80 μm × 4–7.5 μm (x– = 58 × 5.5 μm, n = 20), macronematous, mononematous, simple, erect to slightly curved, solitary, pale brown to dark brown, cylindrical, 2–4-septate, slightly constricted at the septa, unbranched, thick-walled. Conidiogenous cells 6–10 μm × 3.5–6.5 μm (x– = 8 × 4.5 μm, n = 20), holoblastic, monoblastic, integrated, terminal, cylindrical, slightly tapering towards the apex, brown to reddish brown, percurrent. Conidia 190–255 μm × 9.5–16 μm (x– = 220 μm × 13 μm, n = 20), 5–16 μm (x– = 13 μm, n = 20) wide at the protruding truncate base; 4.5–8 μm (x– = 6.5 μm, n = 20) wide in the tapering part, acrogenous, solitary, obclavate, elongate, rostrate, straight or curved, tapering towards the apex, 9–39-distoseptate, olivaceous-green when young, dark reddish brown at maturity, verrucose.
Culture characteristics.
Conidia germinating on PDA within 24 h, colonies circular, dense, umbonate, spreading, fluffy. The surface is slightly rough with reddish-gray mycelium, colonies somewhat raised in the middle, and with a filiform edge. The reverse side is dark gray with a circular, pale reddish-gray edge, not pigmented.
Material examined.
China, Guizhou Province, Guiyang City, Guiyang Medicinal Botanical Garden, on dead twigs of Edgeworthiachrysantha, 20 August 2022, Xia Tang, JX30 (GZAAS 22-2046), living culture, GZCC 22-2046.
Known host and distribution.
Tectonagrandis (Thailand, Hyde et al. 2016), on dead stems (Thailand, Sun et al. 2020), on dead, submerged, decaying wood of unidentified plants (China & Thailand, Luo et al. 2019; Dong et al. 2021; Zhang et al. 2022), and dead twig and branch of Edgeworthiachrysantha (China, this study).
Notes.
Distoseptisporatectonae was first isolated from a dead twig of Tectonagrandis in Thailand (Hyde et al. 2016). Since then, this species has been identified in various countries on different substrates and hosts (Hyde et al. 2016; Sun et al. 2020; Dong et al. 2021; Zhang et al. 2022). In the phylogenetic tree (Fig. 1), our new isolate forms a close lineage to D.tectonae (GZCC 22-2046) with statistical support of 98% ML and 0.96 PP. Based on pairwise nucleotide comparisons of ITS, LSU, tef1-α, and rpb2, our new isolate diverges from D.tectonae (MFLUCC 12-0291, ex-type) by 6/554 bp (1%) for ITS, 1/852 bp (0.01%) for LSU, 0/980 bp (0%) for tef1-α, and 2/899 bp (0.2%) for rpb2. In addition, the morphological characteristics of our isolate match well with the holotype description of D.tectonae (Hyde et al. 2016). This study reports a new host record of Distoseptisporatectonae on dead twigs of Edgeworthiachrysantha in China.
Discussion
Distoseptispora is one of the sporidesmium-like taxa and is well-known for its asexual morph, which has considerable morphological variations (Su et al. 2016; Yang et al. 2018, 2021). However, the phylogenetic analyses suggest a lack of correlation between phylogenetic relationships and morphological analyses. For instance, species such as D.appendiculata D.F. Bao, Z.L. Luo & H.Y. Su, D.atroviridis J. Yang & K.D. Hyde, D.caricis Crous, D.fusiformis J. Yang & K.D. Hyde, D.lanceolatispora, D.leonensis, D.neorostrata, D.palmarum S.N. Zhang, K.D. Hyde & J.K. Liu, and D.saprophytica W. Dong, H. Zhang & K.D. Hyde cluster together as a subclade in the phylogenetic tree (see Fig. 1). In contrast, morphological analysis reveals significant differences, especially in the characteristics of conidiophores, conidiogenous cells, and conidia (Crous et al. 2019; Hyde et al. 2019; Luo et al. 2019; Dong et al. 2021; Yang et al. 2021; Zhang et al. 2022). This disparity is common within the genus. We recommend adopting a combination approach using molecular and morphological methods for more effective identification within this genus.
Worth noting, among the various species of Distoseptispora, D.martinii (J.L. Crane & Dumont) J.W. Xia & X.G. Zhang stands out due to its unique morphological characteristics, especially its oblate or subglobose conidia, distinguishing it from other species within Distoseptispora (Xia et al. 2017). The species was initially introduced as Acrodictysmartinii J.L. Crane & Dumont by Crane and Dumont (1975) based on morphological characteristics. Then, it underwent several taxonomic revisions based solely on morphology (Baker et al. 2002; Delgado 2009). Later, Xia et al. (2017) reclassified Acrodictysmartinii as D.martinii based on genetic analysis. However, the morphological traits of D.martinii greatly diverge from typical Distoseptispora features (Crane and Dumont 1975; Xia et al. 2017). Therefore, we suggest additional collections and analysis of D.martinii specimens to ensure the reliability of the provided DNA sequence data.
In recent years, Distoseptispora species have been reported worldwide, such as in China, Hungary, Hawaii, Malaysia, and Thailand (Shoemaker and White 1985; McKenzie 1995; Wu and Zhuang 2005; Zhang et al. 2022). Studies on Distoseptispora have been particularly extensive in China and Thailand (Hyde et al. 2016, 2019, 2020; Su et al. 2016; Yang et al. 2018, 2021; Luo et al. 2019; Sun et al. 2020; Hu et al. 2023). To date, 73 species of Distoseptispora have been documented, of which 55 have been recorded in China (including known species, see Table 2). Our collections further highlight the distribution of the genus in China, and we speculate that the country may harbor a greater diversity of the genus. Thus, future studies are needed to validate this hypothesis.
Table 2.
Distoseptispora species and their locations, lifestyles, habitats, hosts, and corresponding references.
| Species | Country | Habitat | Host | References |
|---|---|---|---|---|
| D.adscendens | China; Hungary; Hawaii | Terrestrial | Decaying wood and decaying branches of many woody plant species; Platanusorientalis | Shoemaker et al. (1985); McKenzie et al. (1995); Wu et al. (2005); Zhang et al. (2022) |
| D.amniculi | Thailand | Freshwater | Submerged decaying wood | Yang et al. (2021) |
| D.appendiculata | Thailand | Freshwater | Submerged decaying wood | Luo et al. (2019) |
| D.aqualignicola | China | Freshwater | Submerged decaying wood | Zhang et al. (2022) |
| D.aquamyces | China | Freshwater | Submerged decaying wood | Zhang et al. (2022) |
| D.aquatica | China | Freshwater | Submerged decaying wood | Su et al. (2016); Luo et al. (2019); Li et al. (2021) |
| D.aquisubtropica | China | Freshwater | Submerged decaying wood | Ma et al. (2022) |
| D.atroviridis | China | Freshwater | Submerged decaying wood | Yang et al. (2021) |
| D.bambusae | China | Terrestrial | Decaying bamboo culms | Sun et al. (2020) |
| D.bambusicola | China | Freshwater | Submerged bamboo culms | Jayawardena et al. (2022) |
| D.bangkokensis | Thailand | Freshwater | Submerged decaying wood | Shen et al. (2021) |
| D.cangshanensis | China | Freshwater | Submerged decaying wood | Luo et al. (2018) |
| D.caricis | Thailand | Terrestrial | Leaves of Carex sp. | Crous et al. (2019) |
| D.chinensis | China | Freshwater | Submerged decaying wood | Hyde et al. (2021) |
| D.clematidis | China; Thailand | Freshwater; Terrestrial | Dried stem of Clematissikkimensis; submerged decaying wood | Phukhamsakda et al. (2020); Shen et al. (2021) |
| D.crassispora | China | Freshwater | Submerged decaying wood | Zhang et al. (2022) |
| D.curvularia | China | Freshwater | Submerged decaying wood | Zhang et al. (2022) |
| D.cylindricospora | China | Freshwater | Submerged decaying wood | Phukhamsakda et al. (2022) |
| D.dehongensis | China; Thailand | Freshwater | Submerged decaying wood | Hyde et al. (2019); Zhang et al. (2022) |
| D.dipterocarpi | Thailand | Terrestrial | Woody litter of Dipterocarpus sp. | Afshari et al. (2023) |
| D.effusa | China | Freshwater | Submerged decaying wood | Yang et al. (2021) |
| D.euseptata | China | Freshwater | Submerged decaying wood | Li et al. (2021) |
| D.fasciculata | Thailand | Freshwater | Submerged decaying wood | Dong et al. (2021) |
| D.fluminicola | China | Freshwater | Submerged decaying wood | Su et al. (2016); Luo et al. (2018) |
| D.fusiformis | China | Freshwater | Submerged decaying wood | Yang et al. (2021) |
| D.gasaensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.guanshanensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.guizhouensis | China | Terrestrial | Decaying wood | Hyde et al. (2021) |
| D.guttulata | Thailand | Freshwater | Submerged decaying wood | Yang et al. (2018); Luo et al. (2019) |
| D.hainanensis | China | Terrestrial | Decaying wood | This study |
| D.hyalina | Thailand | Freshwater | Submerged decaying wood | Yang et al. (2021) |
| D.hydei | Thailand | Terrestrial | Decaying bamboo culms | Monkai et al. (2020) |
| D.jinghongensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.lancangjiangensis | China | Freshwater | Submerged decaying wood | Shen et al. (2021) |
| D.lanceolatispora | China | Freshwater | Submerged decaying wood | This study |
| D.leonensis | China; Malaysia | Terrestrial | Decaying culms of grasses or decaying branches | McKenzie et al. (1995); Wu et al. (2005); Zhang et al. (2022) |
| D.licualae | Thailand | Terrestrial | Decaying leaves of Licualaglabra | Konta et al. (2023) |
| D.lignicola | China; Thailand | Freshwater | Submerged decaying wood | Luo et al. (2019); Yang et al. (2021) |
| D.longispora | China | Freshwater | Submerged decaying wood | Song et al. (2020) |
| D.longnanensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.martinii | China | Terrestrial | Decaying branches | Xia et al. (2017) |
| D.meilingensis | China | Freshwater | Decaying bamboo culms | Zhai et al. (2022) |
| D.menghaiensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.menglunensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.mengsongensis | China | Terrestrial | Decaying branches | Liu et al. (2023) |
| D.multiseptata | Thailand | Freshwater | Submerged decaying wood | Hyde et al. (2016); Yang et al. (2018) |
| D.nabanheensis | China | Terrestrial | Decaying branches | Liu et al. (2023) |
| D.nanchangensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.neorostrata | Thailand | Freshwater | Submerged decaying wood | Luo et al. (2019) |
| D.nonrostrata | China | Freshwater | Submerged decaying wood | Zhang et al. (2022) |
| D.obclavata | Thailand | Freshwater | Submerged decaying wood | Luo et al. (2019) |
| D.obpyriformis | China | Freshwater | Submerged decaying wood | Luo et al. (2018) |
| D.pachyconidia | China | Freshwater; Terrestrial | Submerged decaying wood; decaying wood | Ma et al. (2022); Zhang et al. (2022) |
| D.palmarum | Thailand | Terrestrial | Rachis of Cocosnucifera | Hyde et al. (2019) |
| D.phangngaensis | Thailand | Freshwater | Submerged decaying wood | Yang et al. (2018) |
| D.phragmiticola | China | Terrestrial | Decaying Phragmitesaustralis | Hyde et al. (2023) |
| D.rayongensis | Thailand | Freshwater | Submerged decaying wood | Hyde et al. (2020) |
| D.rostrata | China | Freshwater | Submerged decaying wood | Luo et al. (2018) |
| D.saprophytica | Thailand | Freshwater | Submerged decaying wood | Dong et al. (2021) |
| D.septata | China | Freshwater | Submerged decaying wood | Ma et al. (2022) |
| D.sinensis | China | Terrestrial | Decaying branches | Liu et al. (2023) |
| D.songkhlaensis | Thailand | Freshwater | Submerged decaying wood | Dong et al. (2021) |
| D.suoluoensis | China | Freshwater | Submerged decaying wood | Yang et al. (2018) |
| D.tectonae | China; Thailand | Terrestrial; Freshwater | Decaying twig of Tectonagrandis; stems of dead wood; submerged decaying wood; decaying twigs of Edgeworthiachrysantha | Hyde et al. (2016); Luo et al. (2018); Sun et al. (2020); Dong et al. (2021); Li et al. (2021); Zhang et al. (2022); This study |
| D.tectonigena | Thailand | Terrestrial | Decaying twig of Tectonagrandis | Hyde et al. (2016) |
| D.thailandica | Thailand | Terrestrial | Decaying leaves of Pandanus sp. | Tibpromma et al. (2018) |
| D.thysanolaenae | China | Terrestrial; Freshwater | Decaying culms of Thysanolaenamaxima; Submerged decaying wood | Phookamsak et al. (2019); Shen et al. (2021) |
| D.tropica | China | Terrestrial | Decaying wood | Ma et al. (2022) |
| D.verrucosa | China | Freshwater | Submerged decaying wood | Yang et al. (2021) |
| D.wuzhishanensis | China | Freshwater | Submerged decaying wood | Ma et al. (2022) |
| D.xishuangbannaensis | China | Terrestrial; Freshwater | Decaying leaves of Pandanusutilis; submerged decaying wood | Tibpromma et al. (2018); Ma et al. (2022) |
| D.yichunensis | China | Terrestrial | Decaying branches of broadleaf tree | Hu et al. (2023) |
| D.yongxiuensis | China | Freshwater | Decaying bamboo culms | Zhai et al. (2022) |
| D.yunjushanensis | China | Freshwater | Decaying bamboo culms | Zhai et al. (2022) |
| D.yunnanensis | China | Freshwater | Submerged decaying wood | Li et al. (2021) |
Supplementary Material
Acknowledgments
The authors thank Shaun Pennycook, Manaaki Whenua – Landcare Research, New Zealand, for his guidance on the fungal nomenclature and the suggestion on naming the new taxa. The authors also thank the Guizhou Institute of Technology for its support of the experiment. Samantha Chandranath Karunarathna thanks the National Natural Science Foundation of China (Numbers 32260004) and the High-Level Talent Recruitment Plan of Yunnan Province (“High-End Foreign Experts” program) for their support.
Citation
Chen X-M, Tang X, Ma J, Liu N-G, Tibpromma S, Karunarathna SC, Xiao Y-P, Lu Y-Z (2024) Identification of two new species and a new host record of Distoseptispora (Distoseptisporaceae, Distoseptisporales, Sordariomycetes) from terrestrial and freshwater habitats in Southern China. MycoKeys 102: 83–105. https://doi.org/10.3897/mycokeys.102.115452
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This work was funded by the National Natural Science Foundation of China (NSFC 32360011).
Author contributions
Conceptualization - Xue-Mei Chen and Yong-Zhong Lu; data curation - Xue-Mei Chen, Xia Tang, Jian Ma, Ning-Guo Liu; formal analysis - Yuan-Pin Xiao, Xue-Mei Chen, Xia Tang, Jian Ma; funding acquisition - Yong-Zhong Lu; investigation - Saowaluck Tibpromma, Samantha C. Karunarathna, Yuan-Pin Xiao, Yong-Zhong Lu; methodology - Xue-Mei Chen, Yong-Zhong Lu; project administration - Yuan-Pin Xiao, Yong-Zhong Lu; resources - Yong-Zhong Lu, Saowaluck Tibpromma, Samantha C. Karunarathna; software - Xue-Mei Chen; supervision - Yong-Zhong Lu, Saowaluck Tibpromma, Samantha C. Karunarathna; validation - Xue-Mei Chen, Xia Tang, Jian Ma, Ning-Guo Liu; visualization - Saowaluck Tibpromma, Samantha C. Karunarathna; writing original draft - Xue-Mei Chen; writing, review and editing - Xue-Mei Chen, Xia Tang, Jian Ma, Ning-Guo Liu, Saowaluck Tibpromma, Samantha C. Karunarathna, Yuan-Pin Xiao, Yong-Zhong Lu. All authors have read and agreed to the published version of the manuscript.
Author ORCIDs
Xue-Mei Chen https://orcid.org/0009-0004-8631-0735
Xia Tang https://orcid.org/0000-0003-2705-604X
Jian Ma https://orcid.org/0009-0008-1291-640X
Ning-Guo Liu https://orcid.org/0000-0002-9169-2350
Saowaluck Tibpromma https://orcid.org/0000-0002-4706-6547
Samantha C. Karunarathna https://orcid.org/0000-0001-7080-0781
Yuan-Pin Xiao https://orcid.org/0000-0003-1730-3545
Yong-Zhong Lu https://orcid.org/0000-0002-1033-5782
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Afshari N, Gomes de Farias AR, Bhunjun CS, Phukhamsakda C, Hyde KD, Lumyong S. (2023) Distoseptisporadipterocarpi sp. nov. (Distoseptisporaceae), a lignicolous fungus on decaying wood of Dipterocarpus in Thailand. Current Research in Environmental & Applied Mycology 13(1): 68–78. 10.5943/cream/13/1/5 [DOI] [Google Scholar]
- Baker WA, Partridge EC, Morgan-Jones G. (2002) Notes on hyphomycetes LXXXV. Junewangia, a genus in which to classify four Acrodictys species and a new taxon. Mycotaxon 81: 293–319. [Google Scholar]
- Chethana KWT, Manawasinghe IS, Hurdeal VG, Bhunjun CS, Appadoo MA, Gentekaki E, Raspé O, Promputtha I, Hyde KD. (2021) What are fungal species and how to delineate them? Fungal Diversity 109(1): 1–25. 10.1007/s13225-021-00483-9 [DOI]
- Crane JL, Dumont KP. (1975) Hyphomycetes from the West Indies and Venezuela. Canadian Journal of Botany 53(9): 843–851. 10.1139/b75-102 [DOI] [Google Scholar]
- Crous PW, Wingfield MJ, Lombard L, Roets F, Swart WJ, Alvarado P, Carnegie AJ, Moreno G, Luangsaard J, Thangavel R, Alexandrova AV, Baseia IG, Bellanger JM, Bessette AJ, Bessette AR, Peña-Lastra SDL, García D, Gené J, Pham THG, Heykoop M, Malysheva E, Malysheva V, Martín MP, Morozova OV, Noisripoom W, Overton BE, Rea AE, Sewall BJ, Smith ME, Smyth CW, Tasanathai K, Visagie CM, Adamčík S, Alves A, Andrade JP, Aninat MJ, Araújo RVB, Bordallo JJ, Boufleur T, Baroncelli R, Barreto RW, Bolin J, Cabero J, Caboň M, Cafà G, Caffot MLH, Cai L, Carlavilla JR, Chávez R, Castro RRLD, Delgat L, Deschuyteneer D, Dios MM, Domínguez LS, Evans HC, Eyssartier G, Ferreira BW, Figueiredo CN, Liu F, Fournier J, Galli-Terasawa LV, Gil-Durán C, Glienke C, Gonçalves MFM, Gryta H, Guarro J, Himaman W, Hywel-Jones N, Iturrieta-González I, Ivanushkina NE, Jargeat P, Khalid AN, Khan J, Kiran M, Kiss L, Kochkina JA, Kolařík M, Kubátová A, Lodge DJ, Loizides M, Luque D, Manjón JL, Marbach PAS, Massola NS, Mata M, Miller AN, Mongkolsamrit S, Moreau PA, Morte A, Mujic A, Navarro-Ródenas A, Németh MZ, Nóbrega TF, Nováková A, Olariaga I, Ozerskaya SM, Palma MA, Petters-Vandresen DAL, Piontelli E, Popov ES, Rodríguez A, Requejo Ó, Rodrigues ACM, Rong IH, Roux J, Seifert KA, Silva BDB, Sklenář F, Smith JA, Sousa JO, Souza HG, Souza JTD, Švec K, Tanchaud P, Tanney JB, Terasawa F, Thanakitpipattana D, Torres-Garcia D, Vaca I, Vaghefi N, Iperen ALV, Vasilenko OV, Verkbeen A, Yilmaz N, Zamora JC, Zapata M, Jurjević Ž, Groenewald JZ. (2019) Fungal Planet description sheets: 951–1041. Persoonia 43(1): 223–425. 10.3767/persoonia.2019.43.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubeta MA, Echandi E, Abernethy T, Vilgalys R. (1991) Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 81(11): 1395–1400. 10.1094/Phyto-81-1395 [DOI] [Google Scholar]
- Delgado G. (2009) South Florida microfungi: Veramycellabispora, a new palmicolous, anamorphic genus and species, with some new records for the continental USA. Mycotaxon 107(1): 357–373. 10.5248/107.357 [DOI] [Google Scholar]
- Dong W, Hyde KD, Jeewon R, Doilom M, Yu XD, Wang GN, Liu NG, Hu DM, Nalumpang S, Zhang H. (2021) Towards a natural classification of annulatascaceae-like taxa II: Introducing five new genera and eighteen new species from freshwater. Mycosphere 12(1): 1–88. 10.5943/mycosphere/12/1/1 [DOI] [Google Scholar]
- Hall TA. (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. 10.1021/bk-1999-0734.ch008 [DOI] [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42(2): 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Hu YF, Liu JW, Luo XX, Xu ZH, Xia JW, Zhang XG, Castañeda-Ruíz RF, Ma J. (2023) Multi-locus phylogenetic analyses reveal eight novel species of Distoseptispora from southern China. Microbiology Spectrum 11(6): e02468–e23. 10.1128/spectrum.02468-23 [DOI] [PMC free article] [PubMed]
- Huelsenbeck JP, Ronquist FJB. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8): 754–755. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, Silva NID, Goes-Neto AA, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, Santiago ALCMDA, Drechsler-Santos ER, Senanayake IC, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu HX, Yang J, Zeng XY, Zhang H, Zhang JF, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar MA, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev D, Buyck B, Silva GAD, Lima CLFD, Oliveira RJVD, Souza CAFD, Dai YC, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang JC, Karunarathna SC, Kirk PM, Kytovuori I, Lantieri A, Liimatainen K, Liu ZY, Liu XY, Lucking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su HY, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen TC, Xu JC, Zhang ZK, Zhao YC, Zhou JL, Zhu L. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Wanasinghe DN, Lücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo ZL, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Bart B, Randrianjohany E, Hofstetter V, Gibertoni TB, Soares AWS, Plautz Jr HL, Sotão HMP, Xavier WKS, Bezerra JDP, Oliveira TGL, Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang SN, Bao DF, Samarakoon MC, Pem D, Karunarathna A, Lin CG, Yang J, Perera RH, Kumar V, Huang SK, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao YP, Konta S, Niskanen T, Liimatainen K, Dai YC, Ji XH, Tian XM, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu YZ, Jiang HB, Zhang JF, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei DP, Réblová M, Fournier J, Nekvindová J, Barbosa RN, Santos JEF, Oliveira NT, Li GJ, Ertz D, Shang QJ, Phillips AJL, Kuo CH, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng XY, Fryar S, Tkalčec Z, Liang JM, Li GS, Wen TC, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao RL, Zhao Q, Kirk PM, Liu JK, Yan JY, Mortimer PE, Xu JC, Doilom M. (2019) Fungal diversity notes 1036–1150: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Hyde KD, Jeewon R, Chen YJ, Bhunjun CS, Calabon MS, Jiang HB, Lin CG, Norphanphoun C, Sysouphanthong P, Pem D, Tibpromma S, Zhang Q, Doilom M, Jayawardena RS, Liu JK, Maharachchikumbura SSN, Phukhamsakda C, Phookamsak R, Al-Sadi AM, Thongklang N, Wang Y, Gafforov Y, Jones EBG, Lumyong S. (2020) The numbers of fungi: Is the descriptive curve flattening? Fungal Diversity 103: 219–271. 10.1007/s13225-020-00458-2 [DOI]
- Hyde KD, Suwannarach N, Jayawardena RS, Manawasinghe IS, Liao CF, Doilom M, Cai L, Zhao P, Buyck B, Phukhamsakda C, Su WX, Fu YP, Li Y, Zhao RL, He MQ, Li JX, Tibpromma S, Lu L, Tang X, Kang JC, Ren GH, Hofstetter V, Ryoo R, Antonín V, Hurdeal VG, Gentekaki E, Zhang JY, Lu YZ, Senanayake IC, Yu FM, Zhao Q, Bao DF. (2021) Mycosphere notes 325–344–Novel species and records of fungal taxa from around the world. Mycosphere 12: 1101–1156. 10.5943/mycosphere/12/1/14 [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Ma J, Yang HD, Zhang JY, Du TY, Gao Y, Gomes de Farias AR, Gui H, He SC, He YK, Li CJY, Liu XF, Lu L, Su HL, Tang X, Tian XG, Wang SY, Wei DP, Xu RF, Xu RJ, Yang Q, Yang YY, Zhang F, Zhang Q, Bahkali AH, Boonmee S, Chethana KWT, Jayawardena RS, Lu YZ, Karunarathna SC, Tibpromma S, Wang Y, Zhao Q. (2023) Mycosphere notes 387–412–novel species of fungal taxa from around the world. Mycosphere 14(1): 663–744. 10.5943/mycosphere/14/1/8 [DOI] [Google Scholar]
- Index Fungorum (2024) Index Fungorum. https://wwwindexfungorumorg/Names/Namesasp [Accessed 2 January 2024]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai YC, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu JK, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo JM, Ghobad-Nejhad M, Nilsson H, Pang KL, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen TC, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li WJ, Perera RH, Phookamsak R, de Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao RL, Zhao Q, Kang JC, Promputtha I. (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74: 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Jayawardena RS, Hyde KD, Wang S, Sun YR, Suwannarach N, Sysouphanthong P, Abdel-Wahab MA, Abdel-Aziz FA, Abeywickrama PD, Abreu VP, Armand A, Aptroot A, Bao DF, Begerow D, Bellanger JM, Bezerra JDP, Bundhun D, Calabon MS, Cao T, Cantillo T, Carvalho JLVR, Chaiwan N, Chen CC, Courtecuisse R, Cui BK, Damm U, Denchev CM, Denchev TT, Deng CY, Devadatha B, de Silva NI, dos Santos LA, Dubey NK, Dumez S, Ferdinandez HS, Firmino AL, Gafforov Y, Gajanayake AJ, Gomdola D, Gunaseelan S, He SC, Htet ZH, Kaliyaperumal M, Kemler M, Kezo K, Kularathnage ND, Leonardi M, Li JP, Liao CF, Liu S, Loizides M, Luangharn T, Ma J, Madrid H, Mahadevakumar S, Maharachchikumbura SSN, Manamgoda DS, Martín MP, Mekala N, Moreau PA, Mu YH, Pahoua P, Pem D, Pereira OL, Phonrob W, Phukhamsakda C, Raza M, Ren GC, Rinaldi AC, Rossi W, Samarakoon BC, Samarakoon MC, Sarma VV, Senanayake IC, Singh A, Souza MF, Souza-Motta CM, Spielmann AA, Su WX, Tang X, Tian XG, Thambugala KM, Thongklang N, Tennakoon DS, Wannathes N, Wei DP, Welti S, Wijesinghe SN, Yang HD, Yang YH, Yuan HS, Zhang H, Zhang JY, Balasuriya A, Bhunjun CS, Bulgakov TS, Cai L, Camporesi E, Chomnunti P, Deepika YS, Doilom M, Duan WJ, Han SL, Huanraluek N, Jones EBG, Lakshmidevi N, Li Y, Lumyong S, Luo ZL, Khuna S, Kumla J, Manawasinghe IS, Mapook A, Punyaboon W, Tibpromma S, Lu YZ, Yan JY, Wang Y. (2022) Fungal diversity notes 1512–1610: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 117: 1–272. 10.1007/s13225-022-00513-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 7(11): 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) Evolution MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konta S, Tibpromma S, Karunarathna SC, Samarakoon MC, Steven LS, Mapook A, Boonmee S, Senwanna C, Balasuriya A, Eungwanichayapant PD, Hyde KD. (2023) Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 14(1): 107–152. 10.5943/mycosphere/14/1/2 [DOI] [Google Scholar]
- Li WL, Liu ZP, Zhang T, Dissanayake AJ, Luo ZL, Su HY, Liu JK. (2021) Additions to Distoseptispora (Distoseptisporaceae) associated with submerged decaying wood in China. Phytotaxa 520: 75–86. 10.11646/phytotaxa.520.1.5 [DOI] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Liu JW, Hu YF, Luo XX, Xu ZH, Castañeda-Ruíz RF, Xia JW, Zhang XG, Zhang LH, Cui RQ, Ma J. (2023) Morphological and phylogenetic analyses reveal three new species of Distoseptispora (Distoseptisporaceae, Distoseptisporales) from Yunnan, China. Journal of Fungi 9(4): 470. 10.3390/jof9040470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Bhat DJ, Su H, Bao DF, Li WL. (2018) Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9: 444–461. 10.5943/mycosphere/9/3/2 [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Maharachchikumbura SSN, Jeewon R, Bao DF, Bhat DJ, Lin CG, Li WL, Yang J, Liu NG, Lu YZ, Jayawardena RS, Li JF, Su HY. (2019) Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Ma J, Zhang JY, Xiao XJ, Xiao YP, Tang X, Boonmee S, Kang JC, Lu YZ. (2022) Multi-gene phylogenetic analyses revealed five new species and two new records of Distoseptisporales from China. Journal of Fungi 8(11): е1202. 10.3390/jof8111202 [DOI] [PMC free article] [PubMed]
- McKenzie EHC. (1995) Dematiaceous hyphomycetes on Pandanaceae 5 Sporidesmiumsensu lato. Mycotaxon 56: 9–29. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), New Orleans (USA), November 2010, IEEE, 8 pp. 10.1109/GCE.2010.5676129 [DOI] [Google Scholar]
- Monkai J, Boonmee S, Ren G, Wei D, Phookamsak R, Mortimer PE. (2020) Distoseptisporahydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 459: 93–107. 10.11646/phytotaxa.459.2.1 [DOI] [Google Scholar]
- Nylander JAA. (2004) MrModeltest 20 Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University, Uppsala.
- Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBG, Maharachchikumbura SSN, Raspé O, Karunarathna SC, Wanasinghe DN, Hongsanan S, Doilom M, Tennakoon DS, Machado AR, Firmino AL, Ghosh A, Karunarathna A, Mešić A, Dutta AK, Thongbai B, Devadatha B, Norphanphoun C, Senwanna C, Wei DP, Pem D, Ackah FK, Wang GN, Jiang HB, Madrid H, Lee HB, Goonasekara ID, Manawasinghe IS, Kušan I, Cano J, Gené J, Li JF, Das K, Acharya K, Raj KNA, Latha KPD, Chethana KWT, He MQ, Dueñas M, Jadan M, Martín MP, Samarakoon MC, Dayarathne MC, Raza M, Park MS, Telleria MT, Chaiwan N, Matočec N, Silva NI, Pereira OL, Singh PN, Manimohan P, Uniya P, Shang QJ, Bhatt RP, Perera RH, Alvarenga RLM, Nogal-Prata S, Singh SK, Vadthanarat S, Oh SY, Huang SK, Rana S, Konta S, Paloi S, Jayasiri SC, Jeon SJ, Mehmood T, Gibertoni TB, Nguyen TTT, Singh U, Thiyagaraja V, Sarma VV, Dong W, Yu XD, Lu YZ, Lim YW, Chen Y, Tkalčec Z, Zhang ZF, Luo ZL, Daranagama DA, Thambugala KM, Tibpromma S, Camporesi E, Bulgakov TS, Dissanayake AJ, Senanayake IC, Dai DQ, Tang LZ, Khan S, Zhang H, Promputtha I, Cai L, Chomnunti P, Zhao RL, Lumyong S, Boonmee S, Wen TC, Mortimer PE, Xu JC. (2019) Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity 95: 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Phukhamsakda C, McKenzie EHC, Phillips AJL, Jones EBG, Bhat DJ, Marc S, Bhunjun CS, Wanasinghe DN, Thongbai B, Camporesi E, Ertz D, Jayawardena RS, Perera RH, Ekanayake AH, Tibpromma S, Doilom M, Xu JC, Hyde KD. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102: 1–203. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- Phukhamsakda C, Nilsson RH, Bhunjun CS, Farias ARGD, Sun YR, Wijesinghe SN, Raza M, Bao DF, Lu L, Tibpromma S, Dong W, Tennakoon DS, Tian XG, Xiong YR, Karunarathna SC, Cai L, Luo ZL, Wang Y, Manawasinghe IS, Camporesi I, Kirk PM, Promputtha I, Kuo CH, Su HY, Doilom M, Li Y, Fu YP, Hyde KD. (2022) The numbers of fungi: Contributions from traditional taxonomic studies and challenges of metabarcoding. Fungal Diversity 114: 327–386. 10.1007/s13225-022-00502-3 [DOI] [Google Scholar]
- Rehner SA, Samuels GJ. (1994) Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98: 625–634. 10.1016/S0953-7562(09)80409-7 [DOI] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 32: Efficient Bayesian Phylogenetic Inference and model choice across a large model space. Systematic Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Rathnayaka AR, Marasinghe DS, Calabon MS, Gentekaki E, Lee HB, Hurdeal VG, Pem D, Dissanayake LS, Wijesinghe SN, Bundhun D, Goonasekara ID, Abeywickrama PD, Bhunjun CS, Jayawardena RS, Wanasinghe DN, Jeewon R, Bhat DJ, Xiang MM. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Shen HW, Bao DF, Hyde KD, Su HY, Bhat DJ, Luo ZL. (2021) Two novel species and two new records of Distoseptispora from freshwater habitats in China and Thailand. MycoKeys 84: 79–101. 10.3897/mycokeys.84.71905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy BD, Jeewon R, Wu WP, Bhat DJ, Hyde KD. (2006) Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycological Research 110: 916–928. 10.1016/j.mycres.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Shoemaker RA, White GP. (1985) Lasiosphaeriacaesariata with Sporidesmiumhormiscioides and L.triseptata with S.adscendens. Sydowia 38: 278–283. [Google Scholar]
- Song HY, El Sheikha AF, Zhai ZJ, Zhou JP, Chen MH, Huo GH, Huang XG, Hu DM. (2020) Distoseptisporalongispora sp. Nov. from freshwater habitats in China. Mycotaxon 135(3): 513–523. 10.5248/135.513 [DOI] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology 57(5): 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Su HY, Hyde KD, Maharachchikumbura SSN, Ariyawansa HA, Luo ZL, Promputtha I, Tian Q, Lin CG, Shang QJ, Zhao YC, Chai HM, Liu XY, Bahkali AH, Bhat JD, McKenzie EHC, Zhou DQ. (2016) The families Distoseptisporaceae fam. Nov., Kirschsteiniotheliaceae, Sporormiaceae, and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Diversity 80: 375–409. 10.1007/s13225-016-0362-0 [DOI] [Google Scholar]
- Sun Y, Goonasekara ID, Thambugala KM, Jayawardena RS, Wang Y, Hyde KD. (2020) Distoseptisporabambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodiversity Data Journal 8: e53678. 10.3897/BDJ.8.e53678.figure2 [DOI] [PMC free article] [PubMed]
- Swofford DL. (2002) PAUP*: Phylogenetic analysis using parsimony (and other methods), version 40 b10 MA: Sinauer Associates, Sunderland.
- Tibpromma S, Hyde KD, McKenzie EHC, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS. (2018) Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Diversity 100: 1–160. 10.1007/s13225-018-0408-6 [DOI] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SJWT, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: A guide to methods and applications 18(1): 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Wu WP, Zhuang WY. (2005) Sporidesmium, Endophragmiella and related genera from China. Fungal Diversity Research Series 15: 1–351. [Google Scholar]
- Xia JW, Ma YR, Li Z, Zhang XG. (2017) Acrodictys-like wood decay fungi from southern China, with two new families Acrodictyaceae and Junewangiaceae. Scientific Reports 7: e7888. 10.1038/s41598-017-08318-x [DOI] [PMC free article] [PubMed]
- Yang J, Maharachchikumbura SSN, Liu JK, Hyde KD, Jones EBG, Al-Sadi AM, Liu ZY. (2018) Pseudostanjehughesiaaquitropica gen. et sp. nov. and Sporidesmiumsensu lato species from freshwater habitats. Mycological Progress 17: 591–616. 10.1007/s11557-017-1339-4 [DOI] [Google Scholar]
- Yang J, Liu LL, Jones EBG, Li WL, Hyde KD, Liu ZY. (2021) Morphological variety in Distoseptispora and introduction of six novel species. Journal of Fungi 7: e945. 10.3390/jof7110945 [DOI] [PMC free article] [PubMed]
- Zhai ZJ, Yan JQ, Li WW, Gao Y, Hu HJ, Zhou JP, Song HY, Hu DM. (2022) Three novel species of Distoseptispora (Distoseptisporaceae) isolated from bamboo in Jiangxi Province, China. MycoKeys 88: 35–54. 10.3897/mycokeys.88.79346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu R, Qing Y, Yang H, Li CX, Wang GN, Zhang D, Ning P. (2022) Polyphasic identification of Distoseptispora with six new species from freshwater. Journal of Fungi 8: e1063. 10.3390/jof8101063 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.