Abstract
Importance
This manuscript will be of interest to most Clinical and Translational Science Awards (CTSA) as they retool for the increasing emphasis on translational science from translational research. This effort is an extension of the EDW4R work that most CTSAs have done to deploy infrastructure and tools for researchers to access clinical data.
Objectives
The Iowa Health Data Resource (IHDR) is a strategic investment made by the University of Iowa to improve access to real-world health data. The goals of IHDR are to improve the speed of translational health research, to boost interdisciplinary collaboration, and to improve literacy about health data. The first objective toward this larger goal was to address gaps in data access, data literacy, lack of computational environments for processing Personal Health Information (PHI) and the lack of processes and expertise for creating transformative datasets.
Methods
A three-pronged approach was taken to address the objective. The approach involves integration of an intercollegiate team of non-informatics faculty and staff, a data enclave for secure patient data analyses, and novel comprehensive datasets.
Results
To date, all five of the health science colleges (dentistry, medicine, nursing, pharmacy, and public health) have had at least one staff and one faculty member complete the two-month experiential learning curriculum. Over the first two years of this project, nine cohorts totaling 36 data liaisons have been trained, including 18 faculty and 18 staff. IHDR data enclave eliminated the need to duplicate computational infrastructure inside the hospital firewall which reduced infrastructure, hardware and human resource costs while leveraging the existing expertise embedded in the university research computing team. The creation of a process to develop and implement transformative datasets has resulted in the creation of seven domain specific datasets to date.
Conclusion
The combination of people, process, and technology facilitates collaboration and interdisciplinary research in a secure environment using curated data sets. While other organizations have implemented individual components to address EDW4R operational demands, the IHDR combines multiple resources into a novel, comprehensive ecosystem IHDR enables scientists to use analysis tools with electronic patient data to accelerate time to science.
Keywords: work force development, informatics, Enterprise Data Warehouse for Research (EDW4R), real-world data, team science, electronic health record (EHR)
Objective
Real-world data plays a critical role in advancing research across the broad spectrum of healthcare. Funded by the NIH, Clinical and Translational Science Award hubs deliver real-world data to researchers using Enterprise Data Warehouses for Research (EDW4R),1,2 ecosystems of technology, services, policies, and collaborations.3–5 At the University of Iowa (UI) Institute for Clinical and Translational Science (ICTS), the EDW4R is used to collect and curate a variety of patient data from diverse sources housed within the health sciences entity. Research using the EDW4R has been limited due to (1) demand for data exceeding workforce capacity, (2) limited data literacy regarding the electronic health record (EHR), (3) lack of an institutionally compliant computational environment, and (4) limited analytic resources such as domain science collaborators.
Originally, the UI EDW4R team was comprised of 3 staff who fulfilled custom data requests to healthcare colleges including medicine, dentistry, nursing, pharmacy, and public health. To expand research access to clinical data, we introduced self-service tools, such as TriNetX.6 This expanded data access accelerated interest in secondary use of EHR data. In addition, requests for data requiring secure computing environments for high-performance computing and machine learning increased, but delivery of the data was delayed due to a lack of adequate environments that met institutional requirements, creating a bottleneck for researchers resulting in delays accessing appropriate computational of up to 2 years. Finally, requests for domain-specific, persistently curated datasets (herein termed transformative datasets) also increased.
Iowa participated in a pilot of the EDW4R maturity model7 by filling out the maturity assessment.8 Analysis of this assessment identified gaps between Iowa’s EDW4R operational practices and EDW4R best practices. Gaps included: EDW4R team size, skillsets, domain scientists, secure, and compliant high-performance computing (HPC) environment; and a standard method for creating and curating domain-specific datasets. In establishing the Iowa Health Data Resource (IHDR), our goals were to address gaps in data access, data literacy, lack of computational environments for processing Personal Health Information (PHI) and the lack of processes and expertise for creating transformative datasets.
Methods
A 3-pronged approach was taken to: (1) build and train a data liaison team, (2) establish a data enclave, and (3) develop a process to create transformative datasets.
Building and training a data liaison team
Research Deans in each health science college identified one faculty member and one staff member to serve as a data liaison and as a member of the Intercollegiate Advisory and Implementation team (IAI). The IHDR funded 5%-25% effort for faculty and staff team members. Prior to starting the program, participants completed training on Health Insurance Portability and Accountability (HIPAA), human subjects research, and information security. The program lasted 8 weeks and included formal training and informal experiential learning, with each team member spending 40-60 h in training (Figure 1).
Figure 1.
Learning timeline.
A curriculum was developed and implemented for the IAI team (Table S3). The formal curriculum covers the following topics: (1) orientation, (2) team science,9–11 (3) self-service data tool training, (4) data sources, infrastructure, data tiers, (5) data request workflows, (6) data governance processes, (7) expanding research questions to national networks, and (8) formal 1 h feedback sessions. Through these training sessions, data liaisons gain a comprehensive understanding of the IHDR’s data sources, infrastructure, and governance processes. They also learn how to effectively use the self-service data tool and navigate the formal request process.
Experiential informal learning includes active participation in data consults, data triage, and request scoping meetings. These working sessions address complex and custom data requests. Consultations consist of data liaisons, analysts, and the research team collaborating to establish the cohort/computable phenotype, the data desired, and the context of the data. Data triage meetings include reviewing requests for datasets, analyzing the information collected during the data consult, and determining whether sufficient information exists to move to the next stage of scoping the request or if additional consultation is needed to finalize request requirements. As part of this work, liaisons also coordinate work with other research cores on campus (eg, biostatistics, imaging, and data analytics). Requirements are reviewed with the data architecture team and at the data request scoping meeting. To measure impact, these metrics were tracked: time from request to data deposit, count of requests supported, complexity of requests supported, and requests supported by data liaisons.
Establishment of data enclave
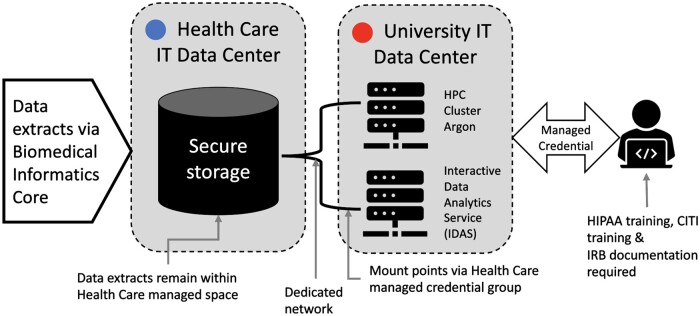
The data enclave is a protected computational environment for complex research analyses of patient data. Created through a unique collaboration between 3 university organizations (University IT, Healthcare IT, and IHDR team), the data enclave established compute-resource-linked storage devices in the hospital data center. All systems involved in this project have been certified as HIPAA compliant; however, healthcare leadership required the additional protection of keeping data on storage systems that are part of the health system. The specific design of the enclave was developed to comply with this internal institutional policy defining the physical location of stored health care data, resulting in requirements that include: (1) datasets extracted from patient data remain in the hospital data center, and (2) access to data is controlled and managed by research dedicated hospital IT employees. The physical design makes use of existing tools, staff, processes, and mirrors existing storage deployments designed to support large volume storage with access to central HPC and analytics environments. There is an additional process component which also leverages existing tools to limit access and provisioning to ensure that data deposited in the enclave has access controls which comply with institutional business practices.
The enclave environment allows processing of data by direct connection to existing computational resources situated outside the hospital firewall. Credentials are issued to IRB-approved research team members to access datasets from computational resources such as the university analytics environment or the central HPC cluster, eliminating any need to download data (Figure 2). Beyond the highly secure systems that are in place, the main protections are the policies, processes, and researcher training to prevent downloading of data from the enclave.
Figure 2.
Data enclave infrastructure.
Through the creation of the data enclave, healthcare leadership has pre-approved placing EHR data and medical imaging extracts into this resource. Other data types (eg, genomic, financial, operational), would generate a data governance review. Data enclave use is managed through biomedical informatics core service requests and processes.
To measure impact, these metrics were tracked: estimated cost savings, number of studies, and number of researchers.
Development of transformative datasets
An efficient process for establishing a set of novel comprehensive, and accessible domain-specific datasets called transformative datasets was developed. Transformative datasets are defined by the following characteristics: (1) Novel: The data does not already exist as a resource, (2) Specific to a domain of science/research: The dataset is tailored to a particular field of study. (3) Contain concepts hard to directly extract from the EHR: The dataset includes information that is not easily obtained from EHRs, (4) Comprehensive as defined by scientist leads: The dataset is extensive and includes all the necessary variables as determined by the leading scientists, (5) Dynamic: The data are regularly updated and new sources are continuously incorporated, (6) Contain data points from EHR(s) and other data sources: The dataset includes information from EHRs as well as other sources such as tumor registries, biospecimens, and patient-reported outcomes, and (7) Contain computationally created variables: The dataset includes variables that are generated through computational methods, such as cancer episodes, mom-baby dyads, gestational age, substance users, and twice-gifted individuals.
The methodology is grounded in Community-Based Decision-Making12 which engages the community through collaborative, cooperative, iterative design of the transformative dataset. The process consists of (1) formal project management practices, (2) interdisciplinary scientist engagement, (3) individual and/or group semi-structured interviews, (4) qualitative analysis and summary of themes from interviews, (5) sharing results of analysis with stakeholder community, and (6) community-based decision-making to develop and finalize the requirements and definitions for the transformative dataset.
Specific transformative datasets were identified and proposed by the research and informatics community to address institutional research strategic goals. Each dataset’s work queue is managed through project management processes and an agile workflow. The first fully operational transformative dataset created was the Intergenerational Health Knowledgebase (IHK),13 built off previous work from EDW4R, the Maternal Child Baby Datamart,13 and UIBioshare14 (the UI bio-sample information management system). The second transformative Neurodevelopmental Registry (NDVR) was founded on the IHK data model. Once launched, the dataset follows change management process to continually curate and further develop the resource. The goal was to reduce the time to creation of the dataset to employing a reproducible methodology. To measure impact, these metrics were tracked: the number of datasets and time to creation.
Results
Building and training data liaison team
To date, all 5 of the health science colleges (dentistry, medicine, nursing, pharmacy, and public health) have had at least one staff and one faculty member complete the 2-month experiential learning curriculum. Over the first 2 years of this project, 9 cohorts totaling 36 data liaisons have been trained, including 18 faculty and 18 staff. Data liaisons have improved their competencies from this experiential learning. They can (1) demonstrate and train faculty and staff to use data self-service tools; (2) collaborate with research teams to understand the research question and identify the appropriate data resources and specific variables; (3) translate the requirements to an actionable data request; (4) increase data literacy through communication of available tools and existing mapped data sources; (5) provide feedback in evaluation of the EDW4R request processes and the experiential learning methods; and (6) facilitate the development and utilization of transformative datasets.
The data liaisons are embedded virtually within the informatics team, expanding the 3 member EDW4R team formally to over 30 part-time members with unique clinical and research domains of expertise (Table S4). Faculty liaisons mentor other faculty and learners in their college and across the health sciences based on expertise, further increasing engagement. Liaisons remain collocated within their college to provide direct support to researchers from their college from the start of the data request through data delivery. Liaisons also support and collaborate across the health sciences on interdisciplinary projects. To maintain community, liaisons participate in working informatics meetings, EDW4R activities, and team-oriented activities with the biomedical informatics core.
Time from data request to data deposit has been reduced by more than 50% from 72 days to 18 days, increasing efficiency, while capacity for complex custom requests has increased by 30%. Data requests facilitated through the data liaisons have resulted in higher-quality requests and reduced the instances of requests for additional or different data after the initial request fulfillment.
Establishment of data enclave
The data enclave eliminated the 2-year bottleneck for access to computational resources appropriate for EHR data, which previously required multiple institutional approvals. Researchers have easily utilized data from the EHR with HPC and analytics tools (Table 1).
Table 1.
Studies utilizing IHDR data enclave since July 2022.
| Study title | TB | # researchers | College(s) | Department/division/unit(s) |
|---|---|---|---|---|
| A data-driven approach to classify safety criteria for ICU patient mobility readiness | 1 | 4 | Nursing, ICTS, | Acute and Critical Care, Design, Biostats & Ethics |
| Functional decline among patients with sepsis | 1 | 4 | Nursing, Engineering | Nursing—Acute and Critical Care, Industrial Engineering, |
| Prediction of substance use outcomes | 1 | 3 | Medicine, Liberal Arts & Science | Pathology, Computer Science, |
| Post-acute neurological sequelae of COVID-19 compared to influenza | 2 | 3 | Graduate, Public Health | Infectious Disease, Biostatistics |
| Advancing symptom science using hidden symptoms and chronic conditions in patients with acute myeloid leukemia | 1 | 4 | Nursing, Business | Acute and Critical Care, Management Sciences, Computer Science, Business Analytics |
| Use of machine learning in the detection of eyelid malignancy using clinical photographs | 2 | 2 | Medicine, Engineering | Ophthalmology & Visual Science, Computer Network |
| Retrospective electronic health record analysis to predict preeclampsia | 1 | 3 | Nursing, Business | Community and Primary Care, Management Sciences, Nursing Research Center |
| Retrospective neonatal research studies involving sickbay | 1 | 2 | Medicine, Engineering | Pediatrics, Engineering Research |
| Machine learning, multimorbidity, & symptom science: mobile app intervention development | 3 | 10 | Nursing, Business, Liberal Arts & Science, Graduate | Community and Primary Care, Management Sciences, Statistics & Actuarial Science, Computer Science, Business Analytics |
| Deep learning in cardiovascular magnetic resonance imaging | 8 | 1 | Medicine | Radiology |
| Automating cohort selection with an attention-based neural network | 3 | 4 | Medicine, Engineering | Internal Medicine, Engineering Research, Electrical-Computer Engineering |
| Natural language processing to measure person-centeredness of care in older persons with palliative care needs | 2 | 1 | Nursing | Nursing |
| Variation in clinical response to observed high blood pressure among patients with no prior diagnosis of hypertension | 1 | 6 | Pharmacy | Health Services Research |
| Pharmacy Practice and Science | ||||
| Exercise vital sign | 1 | 4 | Liberal Arts & Science, Pharmacy | Exercise Science, Health & Human Physiology, Integrative Physiology, Pharmacy Practice and Science, Health Services Research |
| Relation of sarcopenia to poor perioperative outcomes via novel muscular volumetric analysis in emergency surgery | 2 | 1 | Medicine | GI Surgery |
| Exploring automatically extracted “norms” for standard practices in health care | 1 | 2 | Liberal Arts & Science, Graduate | Computer Science |
| Impact of restraints in hospitalized patients with dementia | 1 | 4 | Nursing | Nursing |
Abbreviation: TB = terabytes.
The establishment of the IHDR data enclave eliminated the need to duplicate computational infrastructure inside the hospital firewall which reduced infrastructure, hardware, and human resource costs, while leveraging the existing expertise embedded in the university research computing team. The estimated financial savings include hardware startup cost of $500 000; an ongoing cost of $200 000/year for staffing, and an additional $70 000/year/maintenance and licensing. Furthermore, the toolset and expert staff resources remained unchanged, ensuring that faculty were immediately productive.
Development of transformative datasets
Finally, the creation of a process to develop and implement transformative datasets has resulted in the creation of 7 domain-specific datasets to date (Table 2).
Table 2.
Description of transformative datasets.
| Dataset | Purpose | Sources | Lead | Status | Studies supported |
|---|---|---|---|---|---|
| Intergenerational Health knowledgebase (IHK) | Next version of the of MCKB dataset tracking intergenerational changes | Medical EHR, biospecimen data, patient reported outcomes, community hospital EHRs | Ob/Gyn | Live | 50+ |
| Dental-medical data mart (DMDM) | Cross-referencing medical and dental data | Dental and medical EHRs | Dentistry | Live | 6 |
| Neuro developmental registry (NDVR) | Detection and tracking of mental health | Medical EHR, registry data, patient reported outcomes | Psychiatry | Live | 4 |
| Nursing Care Dataset (NCD) | Extraction of nursing data | Medical EHR, flowsheets, assessments, nursing care documentation | Nursing | Development | |
| Cochlear Implant Knowledgebase | The Iowa Cochlear Implant Clinical Research Center | Medical EHR, auditory assessments and testing, patient reported outcomes | Otolaryngology | Development | |
| Rural Impact on Health (RIoH) | State-wide mapping of health information | Medical and dental EHRs, state hygienic lab data, insurance claims, patient reported outcomes, HIE | UI Health Care | Planning | |
| Imaging EHR dataset (IEHR) | Linking patient data to image data | Medical imaging data and media | Radiology | Planning |
Three are live, 2 are in development, and 2 are in planning. Prior to this methodology, development of these types of datasets took 3-5 years minimum, using this methodology time to dataset creation has been reduced by at least 50%. Combined, the datasets that are live support more than 50 studies.
Discussion
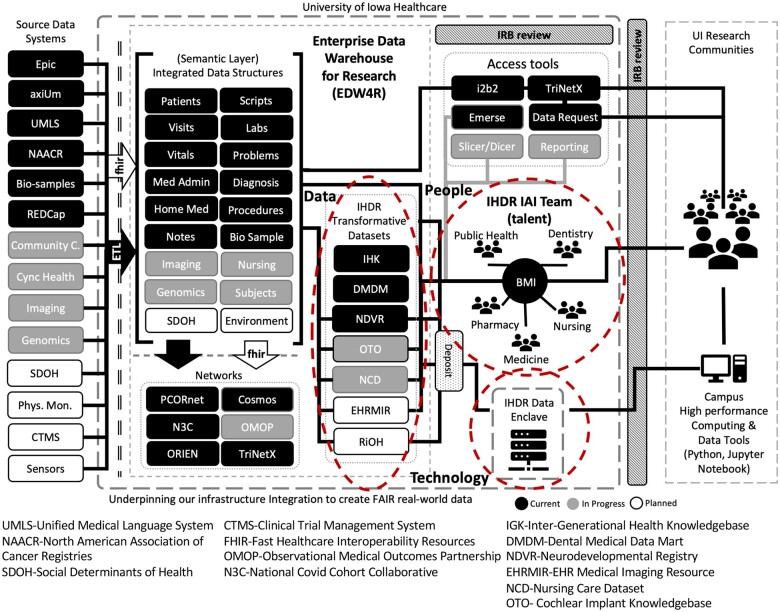
The IHDR was informed by research in EDW4R operations1,2 and the rapidly evolving landscape in research leveraging these data resources. While other organizations have implemented individual components to address EDW4R operational demands, the IHDR combines multiple resources into a novel, comprehensive ecosystem (Figure 3).
Figure 3.
Enterprise Data Warehouse for Research (EDW4R).
IHDR expanded the informatics workforce through reassignment of roles and duties of existing employees. The UI EDW4R workforce has evolved with the addition of scientific and domain expertise (people), leading to a notable improvement in metrics for data delivery services. The IAI team has added non-informatics people in non-informatics units. It is not an increase of IHDR staff; what is particularly valuable is the broadening of data and informatics literacy into clinical and research units. This has created a broader network that informatics, clinical, and research teams can reach out to for support, expertise, and additional training.
IHDR established a secure data enclave via a collaboration between enterprise IT organizations, informatics, and research community stakeholders that resulted in a compliant space currently in use for 17 studies and 58 data researchers. This new resource positions researchers at the institution to engage in research that they were not previously able to conduct. Subsequently, these researchers can both advance the science and be highly competitive for extramural funding.
IHDR empowers scientists using transformative datasets—developed in response to repetitive complex data requests for similar information—to lead new collaborations and expand their area of research. The research community learns about the existence of transformative datasets through the scientific leads and their presentations, the IAI team, and through pairing with the right resource through the data request process. The transformative datasets reduce both the time to access complex domain-specific data for both grant applications and manuscripts and the need to re-validate data being extracted.
Synergistic results of this effort include: (1) offering the IHDR as a data ecosystem for 3 new faculty with data-driven research programs, (2) training health sciences graduate students in using the IHDR dataset, (3) cross-training Assistant Chief Medical Officers in UI Health Care as data liaisons, and (4) training data liaisons from other organizations, enhancing inter-institutional collaboration.
Limitations
There are 3 notable limitations to this work. An institutional pilot grant allowed for funding the program; long-term sustainability approaches will need to be identified or institutional investments made. Due to institutional constraints, cloud-based resources were not considered for the data enclave, an on-premises solution was required. Finally, this work was a single-center study, with an institutional culture of collaboration. It is unknown if this approach can extend elsewhere; however, sustainability is a major issue at all academic medical centers, and the approach of engaging investigators rather than informatics staff alone to deliver data may be of value.
Conclusion
The IHDR has combined people, data, and technology to address addresses multiple institutional strategic priorities: (1) improving research and discovery by boosting the reach and competitiveness of high-impact interdisciplinary health science research linked to a unique, new, integrated data platform; (2) promoting faculty and student success by creating a data-science training program; and (3) expanding research efforts to provide new diagnostic, prognostic, and therapeutic insights. While this effort positions us to meet institutional priorities, it is also closely aligned with goals for CTSAs. This ecosystem will allow us to move the science of using research informatics forward.
Supplementary Material
Acknowledgments
People: James M. Blum, Lee T. Carmen, Tara Carr, Marian F. Carson, Martha L. Carvour, An Gie L. Cech, Betsy A. Chrischilles, Douglas J. Van Daele, Rhonda R. DeCook, Scott J. Egerton, Elaine M. Dettner, Jennie Embree, Sean B. Fain, Steven R. Fleagle, Mike S. Frangi, Carol Geary, Stephanie H. Gilbertson-White, Brian M. Gryzlak, Shannon Hampton, Genevieve R. Johnson, William P. Keating, Aaron K. Kline, Lindsay A. Knake, Brendel Krueger, Anna E. Krupp, Laura Jacobus, Brad D. McDowell, Jason Misurac, Kenneth G. Nepple, Corinne Peek-Asa, Diva Perez, Justin Perez, Kirk T. Phillips, Robert C. Piper, Barbara A. Rakel, Dianne Rohlman, David L. Roman, Greg L. Stewart, Gi-Yung Ryu, Scott D. Stiegelmeyer, Lucas D. Van Tol, Julie Vignato, Joseph F. Wagner, Patricia L. Winokur, Jessica S. Wolf, Xian Jin Xie.
Contributor Information
Heath A Davis, Biomedical Informatics, Institute for Clinical & Translational Science, University of Iowa, Iowa City, IA 52242, United States; Office of Information Technology, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA 52242, United States.
Donna A Santillan, Biomedical Informatics, Institute for Clinical & Translational Science, University of Iowa, Iowa City, IA 52242, United States; Obstetrics and Gynecology, University of Iowa, Iowa City, IA 52242, United States.
Chris E Ortman, Biomedical Informatics, Institute for Clinical & Translational Science, University of Iowa, Iowa City, IA 52242, United States; Office of Information Technology, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA 52242, United States.
Asher A Hoberg, Biomedical Informatics, Institute for Clinical & Translational Science, University of Iowa, Iowa City, IA 52242, United States.
Joseph P Hetrick, Research Services, Information Technology Services, University of Iowa, Iowa City, IA 52245, United States.
Charles W McBrearty, Department of Preventive and Community Dentistry, College of Dentistry, College of Dentistry, University of Iowa, Iowa City, IA 52242, United States.
Erliang Zeng, Department of Preventive and Community Dentistry, College of Dentistry, College of Dentistry, University of Iowa, Iowa City, IA 52242, United States; Division of Biostatistics and Computational Biology, College of Dentistry, University of Iowa, Iowa City, IA 52242, United States.
Mary S Vaughan Sarrazin, Internal Medicine, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA 52242, United States; Center for Access and Delivery Research and Evaluation, Veterans Affairs Medical Center, Iowa City, IA 52246, United States.
Karen Dunn Lopez, Center for Nursing Classification and Clinical Effectiveness, College of Nursing, University of Iowa, Iowa City, IA 52242, United States.
Cole G Chapman, Pharmacy Practice and Science, College of Pharmacy, University of Iowa, Iowa City, IA 52242, United States.
Ryan M Carnahan, Epidemiology, College of Public Health, University of Iowa, Iowa City, IA 52242, United States.
Jacob J Michaelson, Psychiatry, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA 52242, United States.
Boyd M Knosp, Biomedical Informatics, Institute for Clinical & Translational Science, University of Iowa, Iowa City, IA 52242, United States; Office of Information Technology, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, Iowa City, IA 52242, United States.
Author contributions
H.A.D. and B.M.K. conceptualized the overarching vision of the IHDR. H.A.D. and A.A.H. developed and implemented the IHDR IAI curriculum. H.A.D., D.A.S., C.E.O., A.A.H., C.W.M., E.Z., M.S.V.S., K.D.L., C.G.C., R.M.C., J.J.M., and B.M.K. participated in Team Science, IAI experiential learning and provided feedback. H.A.D., B.M.K., D.A.S., J.J.M., C.W.M., E.Z., and K.D.L. provided expertise in developing transformative datasets. H.A.D., B.M.K., J.P.H., and A.A.H. developed the process for the IHDR data enclave. H.A.D. and B.M.K. wrote the manuscript with contributions from D.A.S., K.D.L., and M.S.V.S. J.P.H., A.A.H., C.E.O., and E.Z. provided edits. H.A.D. and B.M.K. revised the manuscript.
Supplementary material
Supplementary material is available at Journal of the American Medical Informatics Association online.
Funding
This work was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health grant number UL1TR002537, the UI Center for Advancing Multimorbidity Science (CAMS), National Institute of Nursing Research, grant number P20NR018081, and the Utility Public-Private Partnership (P3) Award—The Iowa Health Data Resource: Building the Future of Health Informatics at the University of Iowa, https://strategicplan.uiowa.edu/public-private-partnership-p3/p3-program-support-strategic-priorities/p3-proposals-funded-fy-2022.
Conflict of interest
None declared.
Data availability
The data underlying this article will be shared upon reasonable request to the corresponding author.
References
- 1. Campion TR, Craven CK, Dorr DA, et al. Understanding enterprise data warehouses to support clinical and translational research. J Am Med Inform Assoc. 2020;27(9):1352-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knosp BM, Craven CK, Dorr DA, et al. Understanding enterprise data warehouses to support clinical and translational research: enterprise information technology relationships, data governance, workforce, and cloud computing. J Am Med Inform Assoc. 2022;29(4):671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Haendel MA, Chute CG, Bennett TD, et al. ; N3C Consortium. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021;28(3):427-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collins FS, Hudson KL, Briggs JP, et al. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21(4):576-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hernandez AF, Fleurence RL, Rothman RL.. The ADAPTABLE trial and PCORnet: shining light on a new research paradigm. Ann Intern Med. 2015;163(8):635-636. [DOI] [PubMed] [Google Scholar]
- 6. Topaloglu U, Palchuk MB.. Using a federated network of real-world data to optimize clinical trials operations. JCO Clin Cancer Inform. 2018;2:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Knosp BM, Dorr DA, Campion TR.. Maturity in enterprise data warehouses for research operations: analysis of a pilot study. J Clin Transl Sci. 2023;7(1):e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Knosp BM, Campion TH.. EDW4R Maturity Assessment. 2023. Accessed 2023; REDCap-based EDW4R Maturity Assessment]. https://bit.ly/EDW4R_Maturity
- 9. Lowe HJ, Ferris TA, Hernandez PM, Weber SC.. STRIDE—an integrated standards-based translational research informatics platform. AMIA Annu Symp Proc.2009;2009:391-395. [PMC free article] [PubMed] [Google Scholar]
- 10. Hall KL, Vogel AL, Huang GC, et al. The science of team science: a review of the empirical evidence and research gaps on collaboration in science. Am Psychol. 2018;73(4):532-548. [DOI] [PubMed] [Google Scholar]
- 11. Hall KL, Vogel AL, Croyle RT. Strategies for Team Science Success: Handbook of Evidence-Based Principles for Cross-Disciplinary Science and Practical Lessons Learned from Health Researchers. Cham: Springer; 2019. 10.1007/978-3-030-20992-6 [DOI] [Google Scholar]
- 12. Knosp BM. Four ways to manage change in preparation for a major implementation. The Evollution. 2014. Accessed 2023. https://evolllution.com/opinions/ways-manage-change-preparation-major-implementation/
- 13. Santillan MK, Davis H, Crooks M, et al. Development and Utility of a Novel Intergenerational Health Knowledgebase. FASEB J. 2022;36. 10.1096/fasebj.2022.36.S1.R5732 [DOI] [Google Scholar]
- 14. Santillan MK, Leslie KK, Hamilton WS,. et al. Collection of a lifetime: a practical approach to developing a longitudinal collection of women’s healthcare biological samples. Eur J Obstet Gynecol Reprod Biol.2014;179:94-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared upon reasonable request to the corresponding author.