Abstract
A proteomic analysis was undertaken to identify cell wall-associated proteins of Streptococcus pyogenes. Seventy-four distinct cell wall-associated proteins were identified, 66 of which were novel. Thirty-three proteins were immunoreactive with pooled S. pyogenes-reactive human antisera. Biotinylation of the GAS cell surface identified 23 cell wall-associated proteins that are surface exposed.
The gram-positive human pathogen Streptococcus pyogenes (group A streptococcus; GAS) is the etiologic agent of numerous suppurative diseases, ranging from mild skin infections, such as pharyngitis, scarlet fever, impetigo, and cellulitis, to severe invasive diseases such as septicemia, streptococcal toxic shock syndrome, and necrotizing fasciitis (8). S. pyogenes expresses a range of multifunctional surface proteins which facilitate adherence to and invasion of host cells, resistance to phagocytosis, and degradation of host proteins (8). Although many surface-exposed and secreted proteins in GAS have been identified and characterized, there has been no systematic analysis to identify the major cell wall-associated proteins.
To identify the major cell wall-associated proteins of GAS, a two-dimensional (2D) sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) proteomic analysis (6) of mutanolysin cell wall extracts (19) was undertaken for GAS strain NS931 (necrotizing fasciitis isolate; serotype M69) (11), NS13 (bacteremia isolate; serotype M53) (11), and S43 (bronchopneumonia isolate; serotype M6) (21). Proteins of interest were excised from 2D Coomassie blue-stained PAGE gels, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) as described by Cordwell et al. (6). Peptide masses were matched by searching the Swiss-Prot and TrEMBL databases at PeptIdent (http://us.expasy.org/tools/peptident.html). Representative 2D PAGE gels from two independent mutanolysin cell wall extractions are shown in Fig. 1A to C. The protein profiles are similar across all strains, with molecular masses ranging from 14.4 to 77.5 kDa and a pI range of 4.4 to 7.9. A total of 155 protein spots (51 for NS931 [Fig. 1A], 33 for NS13 [Fig. 1B], and 71 for S43 [Fig. 1C]), corresponding to 74 unique proteins, were positively identified by MALDI-TOF MS (Table 1). Several proteins were detected as multiple isoforms in one or more strains. These results suggest that some proteins exist in different charge states or may have undergone posttranslational modifications. It remains to be determined whether or not these modifications are physiological or an artifact caused by urea carbamylation, deamidation, or immobilized pH gradient strip overload. With the exception of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (30), enolase (31), manganese-dependent superoxide dismutase (26), collagen-like protein B (38), SpeM (36), FcrA (16), M protein (13), and cysteine protease SpeB precursor (18), all of the proteins identified in this study have not, to our knowledge, previously been reported as cell wall associated in S. pyogenes. Thirty-five of the 74 cell wall-associated proteins have been previously identified in the cellular or extracellular GAS proteomes (2, 4, 22, 27, 37) (Table 2).
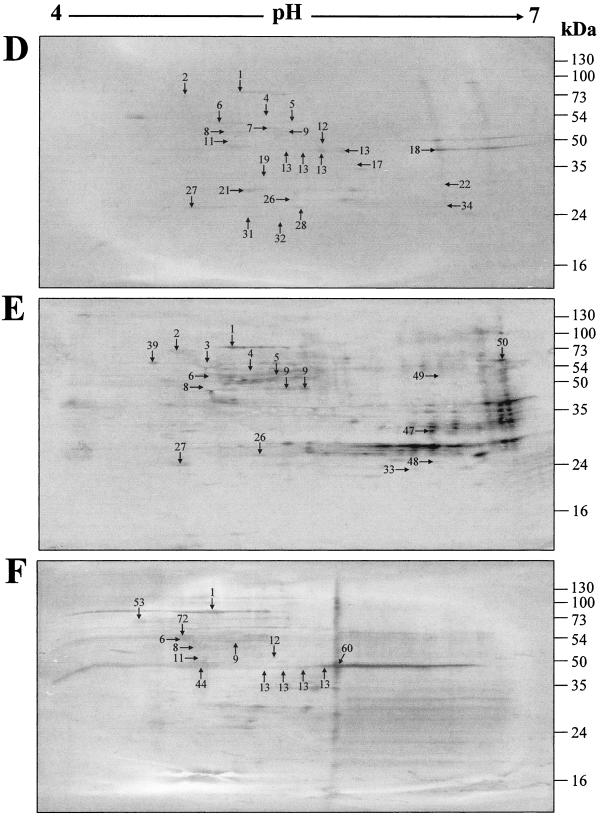
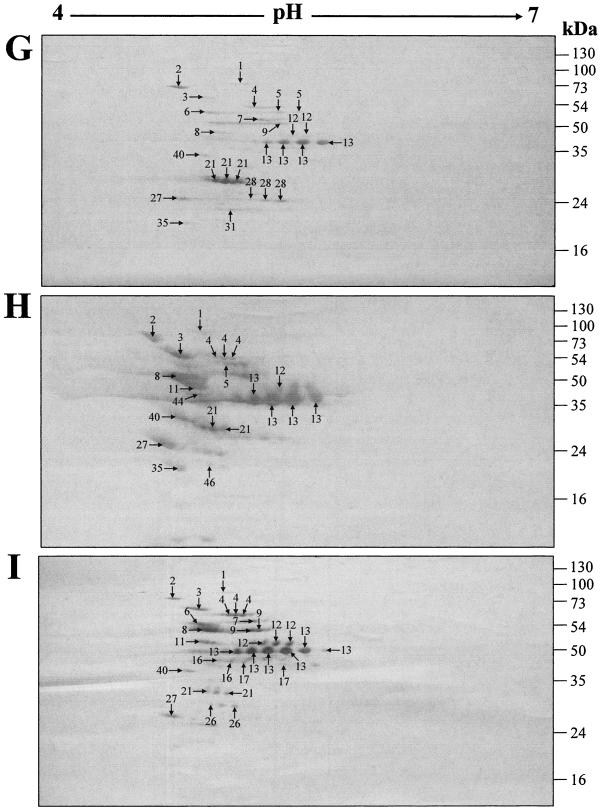
FIG. 1.
Two-dimensional gel electrophoresis profiles of GAS mutanolysin cell wall extracts. The extracts were harvested from GAS strains NS931 (A, D, and G), NS13 (B, E, and H), and S43 (C, F, and I) after growth to late stationary phase (37°C for 16 h) in Todd-Hewitt medium (Difco) supplemented with 1% (wt/vol) yeast extract without shaking. The protein extracts (170 μg) were isoelectric focused over a linear pH gradient of 4 to 7 and resolved with a 12.5% SDS-PAGE gel. (A-C) The gels were stained with colloidal Coomassie blue and destained in 1%anti-human IgG-HRP conjugate (Bio-Rad). Negative-control blots probed only with goat anti-human IgG-HRP conjugate contained no immunoreactive proteins (result not shown). (G-I) The cell surface of each strain was labeled with biotin before the mutanolysin extract was harvested. The proteins were transferred to a PVDF membrane and probed with an SA-HRP conjugate prior to development with diaminobenzidine. Negative-control blots of nonbiotinylated extracts contained no labeled proteins (result not shown). Protein spots identified by peptide mass (vol/vol) acetic acid. (D-F) The proteins were transferred to a PVDF membrane and probed with a 1:100 dilution of pooled human sera from an area of endemicity. Bound antibodies were detected using a goat fingerprinting are denoted by numbered arrows, which correspond to the proteins in Table 1. Molecular mass markers are given in kilodaltons.
TABLE 1.
Major cell wall-associated proteins identified in the mutanolysin extracts of GAS strains NS931, NS13, and S43 by MALDI-TOF peptide mass fingerprinting analysisa
| Function or pathway | Spot | Protein | Accession no.b | Molecular mass (kDa)c | pIc | Peptide matchd | Coverage (%)e | Mutanolysin extract
|
Immunoreactive
|
Biotinylated
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS931 | NS13 | S43 | NS931 | NS13 | S43 | NS931 | NS13 | S43 | ||||||||
| Glycolysis | 4 | Putative pyruvate kinase | Q8K7A3 | 54.5 | 4.96 | 30 | 59.8 | + | + | + | + | + | + | + | + | |
| 5 | Putative NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | Q8K707 | 49.5 | 4.96 | 19 | 56.1 | + | + | + | + | + | + | + | |||
| 11 | Phosphoglycerate kinase | Q8K5W7 | 42.0 | 4.86 | 23 | 70.0 | + | + | + | + | + | + | + | |||
| 17 | 6-Phosphofructokinase | Q8P0S6 | 35.8 | 5.34 | 22 | 50.7 | + | + | + | + | + | |||||
| 21 | Fructose-bisphosphate aldolase | P82486 | 31.1 | 4.87 | 13 | 50.3 | + | + | + | + | + | + | + | |||
| 27 | Triosephosphate isomerase | P82478 | 26.5 | 4.57 | 11 | 63.3 | + | + | + | + | + | + | + | + | ||
| 28 | 2,3-Bisphosphoglycerate-dependent phosphoglycerate mutase | Q8P0C1 | 26.0 | 5.10 | 19 | 77.1 | + | + | + | + | + | |||||
| 69 | l-Lactate dehydrogenase | Q99ZN5 | 35.1 | 5.14 | 9 | 30.7 | + | |||||||||
| Carbohydrate metabolism | 1 | Putative transketolase | Q8K670 | 77.5 | 4.98 | 21 | 42.1 | + | + | + | + | + | + | + | + | + |
| 32 | Putative dTDP-4-keto-6-deoxyglucose-3,5-epimerase | Q9A046 | 22.4 | 5.07 | 10 | 42.1 | + | + | + | |||||||
| 41 | Putative lactoylglutathione lyase | Q9A121 | 14.4 | 5.09 | 4 | 30.4 | + | |||||||||
| 52 | Putative phosphoglucomutase | Q99ZH8 | 63.3 | 4.78 | 10 | 25.7 | + | |||||||||
| 54 | Putative phospho-sugar mutase | Q878L0 | 48.4 | 4.57 | 8 | 17.7 | + | |||||||||
| 60 | Putative dTDP-glucose-4,6-dehydratase | Q8P199 | 38.8 | 5.41 | 4 | 12.4 | + | + | ||||||||
| 61 | Glycerol-3-phosphate dehydrogenase [NAD(P)+] | P58143 | 36.7 | 5.65 | 12 | 45.9 | + | |||||||||
| 67 | Tagatose 1,6-diphosphate aldolase 2 | Q8K5U9 | 36.5 | 4.93 | 15 | 50.5 | + | |||||||||
| Arginine degradation | 12 | Ornithine carbamoyltransferase, catabolic | Q8P052 | 37.8 | 5.19 | 19 | 62.8 | + | + | + | + | + | + | + | + | |
| 40f | Putative carbamate kinase | Q8K6Q9 | 33.2 | 4.71 | 12 | 44.3 | + | + | + | + | + | + | ||||
| Amino acid biosynthesis | 14 | Putative branched-chain-amino-acid aminotransferase | Q8K7U5 | 37.2 | 4.90 | 6 | 21.4 | + | + | |||||||
| 72 | Putative glutamine synthetase | Q8NZG4 | 50.5 | 5.21 | 8 | 19.6 | + | + | ||||||||
| Fatty acid and phospholipid biosynthesis | 47 | Putative malonyl coenzyme A-acyl carrier protein transacylase | Q879J3 | 34.5 | 6.02 | 5 | 19.1 | + | + | |||||||
| Pantothenate (vitamin B5) biosynthesis | 19 | Putative 2-dehydropantoate 2-reductase (ketopantoate reductase) | Q8P1F1 | 33.8 | 4.93 | 6 | 24.8 | + | + | + | ||||||
| Pyridoxine (vitamin B6) biosynthesis | 36 | Putative pyridoxamine-phosphate oxidase | Q8K7X7 | 14.9 | 5.87 | 10 | 68.4 | + | ||||||||
| Nucleoside metabolism | 26 | Putative purine nucleoside phosphorylase | Q878J4 | 28.5 | 4.98 | 12 | 44.4 | + | + | + | + | + | + | |||
| 34 | Uracil phosphoribosyltrans- ferase (UMP pyrophosphorylase) | Q9A194 | 22.8 | 6.30 | 13 | 57.4 | + | + | ||||||||
| 55 | GMP synthase (gluta- mine hydrolyzing) | Q8K7E6 | 57.5 | 4.91 | 7 | 14.8 | + | |||||||||
| 63 | Adenylosuccinate synthetase | Q8P2U1 | 47.4 | 5.29 | 10 | 30.7 | + | |||||||||
| Metabolic enzyme | 16 | Putative phosphotransacetylase | Q878S0 | 35.9 | 5.08 | 13 | 43.8 | + | + | + | + | |||||
| 66 | Putative acetoin reduc- tase | Q8P1U1 | 26.8 | 4.79 | 9 | 44.5 | + | |||||||||
| 68 | Probable manganese-dependent inorganic pyrophosphatase | Q9A1A2 | 33.6 | 4.47 | 7 | 23.8 | + | |||||||||
| Virulence factor | 8 | Enolase (2-phosphoglycerate dehydratase) | P82479 | 47.2 | 4.74 | 28 | 65.4 | + | + | + | + | + | + | + | + | + |
| 9 | Arginine deiminasei | Q8K5F0 | 46.1 | 4.99 | 31 | 70.5 | + | + | + | + | + | + | + | + | ||
| 13 | GAPDH | P50467 | 35.8 | 5.34 | 18 | 61.8 | + | + | + | + | + | + | + | + | ||
| 18f | Cysteine protease SpeB precursor | Q93LQ2 | 37.3 | 7.21 | 7 | 22.4 | + | + | ||||||||
| 23 | Putative C3-degrading proteinase | Q99Y63 | 28.6 | 4.89 | 10 | 43.2 | + | |||||||||
| 31 | Superoxide dismutase (Mn) | Q8P0D4 | 22.5 | 4.87 | 13 | 81.5 | + | + | + | + | + | |||||
| 43f | Pyrogenic exotoxin M (SpeM) (fragment) | Q7WYA3 | 24.1 | 7.87 | 6 | 25.0 | + | |||||||||
| 48fg | FcrA protein precursor | Q54859 | 45.4 | 6.47 | 6 | 15.4 | + | + | ||||||||
| 50f,g | M protein | Q54840 | 61.7 | 6.24 | 4 | 6.5 | + | + | ||||||||
| 73 | M protein (fragment) | Q93LJ0 | 27.2 | 5.22 | 6 | 25.0 | + | |||||||||
| 74 | M protein (fragment) | O86065 | 21.0 | 5.41 | 5 | 21.7 | + | |||||||||
| Protein biosynthesis | 7 | Elongation factor Tu | Q8K872 | 43.8 | 4.91 | 24 | 58.3 | + | + | + | + | + | ||||
| 29 | Peptide deformylase | Q8NZB7 | 22.9 | 5.51 | 11 | 72.5 | + | + | ||||||||
| 33 | Ribosome recycling factor | Q8P274 | 20.5 | 5.68 | 11 | 64.3 | + | + | + | + | ||||||
| 38 | Elongation factor G | P82477 | 76.4 | 4.83 | 13 | 22.9 | + | + | ||||||||
| 44 | Elongation factor Ts | Q8K5L1 | 37.3 | 4.86 | 13 | 45.7 | + | + | + | + | ||||||
| 57 | Probable sigma54 modulation protein (fragments) | P82482 | 18.4 | 4.45 | 4 | 30.6 | + | |||||||||
| 62 | Seryl-tRNA synthetase | Q8K635 | 48.1 | 5.17 | 13 | 38.8 | + | |||||||||
| 64 | Elongation factor P | P82459 | 20.5 | 4.85 | 4 | 29.2 | + | |||||||||
| Protein transport | 22f | Putative ABC trans- porter, substrate- binding protein | Q8P2K8 | 30.6 | 7.69 | 7 | 28.6 | + | + | |||||||
| 39 | Trigger factor | Q879L7 | 47.1 | 4.39 | 11 | 27.6 | + | + | + | |||||||
| 53 | Putative ABC trans- porter, ATP-binding protein | Q99XH2 | 60.7 | 4.77 | 9 | 21.2 | + | + | ||||||||
| 65 | Putative copper homeostasis protein (hypothetical protein) | Q8K8H0 | 22.6 | 4.79 | 4 | 24.4 | + | |||||||||
| Proteolysis and peptidolysis | 6 | Putative dipeptidase | Q8K7L6 | 51.4 | 4.81 | 12 | 33.0 | + | + | + | + | + | + | + | + | |
| 10 | Putative X-His dipeptidase | Q99YT8 | 49.1 | 4.74 | 21 | 46.5 | + | |||||||||
| 30 | Pyrrolidone-carboxylate peptidase | Q8K8C4 | 23.2 | 5.64 | 7 | 38.6 | + | + | ||||||||
| 45 | Putative methionine aminopeptidase | Q8K718 | 31.6 | 4.84 | 5 | 31.5 | + | |||||||||
| Chaperone | 2 | Chaperone protein DnaK | P95831 | 64.8 | 4.62 | 24 | 47.6 | + | + | + | + | + | + | + | + | |
| 3 | 60-kDa chaperonin GroEL | Q8K5M5 | 56.9 | 4.75 | 35 | 69.6 | + | + | + | + | + | + | + | |||
| Stress protein | 35 | Putative alkyl hydroperoxidase | Q99XR7 | 20.5 | 4.65 | 9 | 58.6 | + | + | + | + | + | ||||
| 49 | Putative glutathione reductase | Q8P1H3 | 48.9 | 5.66 | 5 | 11.1 | + | + | ||||||||
| Transcription factor | 58 | Transcription elongation factor GreA | Q9A1C4 | 17.7 | 4.67 | 7 | 61.3 | + | ||||||||
| Adhesin | 59f,g | Collagen-like protein B (fragment) | Q9AGC4 | 46.7 | 4.60 | 4 | 10.8 | + | ||||||||
| Housekeeping | 15 | Putative alcohol dehydrogenase I | Q9A1X7 | 35.4 | 4.89 | 6 | 20.7 | + | ||||||||
| 24 | Putative phosphoprotein phosphatase | Q99YM9 | 27.0 | 4.60 | 19 | 80.9 | + | + | ||||||||
| 25 | Adenylate kinase | Q8K8X1 | 23.7 | 4.83 | 13 | 79.7 | + | + | ||||||||
| Unknown function | 20 | Hypothetical UPF0082 protein SPy0316/SpyM3_0231/SPs16 28/spyM18_0311 | Q9A1E6 | 25.9 | 4.49 | 6 | 29.0 | + | ||||||||
| 37 | Hypothetical phage protein spyM18_0356 | Q8P2H6 | 25.1 | 5.37 | 4 | 13.6 | + | |||||||||
| 42 | Hypothetical phage protein spyM18_1764 | Q8NZS3 | 22.7 | 4.63 | 3 | 20.2 | + | |||||||||
| 46 | Hypothetical protein SPy1262 | Q99ZE5 | 19.9 | 4.93 | 9 | 42.5 | + | + | + | |||||||
| 51 | Hypothetical protein SpyM3_0548 | Q8K7Z2 | 15.4 | 6.05 | 4 | 21.2 | + | |||||||||
| 56 | Conserved hypothetical protein SPs1095 | Q878P1 | 17.5 | 4.65 | 6 | 54.6 | + | |||||||||
| 70 | Conserved protein SpyM18_1567 | Q8P050 | 26.6 | 4.67 | 5 | 28.6 | + | |||||||||
| 71 | Hypothetical protein (phage associated) SPs0647 | Q879B2 | 26.8 | 5.10 | 5 | 21.0 | + | |||||||||
Identified proteins are indicated by a plus sign.
Swiss-Prot or TrEMBL accession number.
Theoretical values obtained from Swiss-Prot or TrEMBL database.
Number of tryptic peptides detected by MALDI-TOF MS that could be matched to the protein.
Percentage of the protein sequence covered by the matched peptides.
Contains a putative secretion signal sequence identified by SignalP3.0 signal peptide prediction server (http://www.cbs.dtu.dk/services/SignalP/).
Contains a C-terminal LPXTG membrane anchor motif identified by Pfam motif search (http://pfam.wustl.edu/hmmsearch.shtml).
TABLE 2.
Cell-wall associated proteins previously characterized as cellular or secreted in GAS
| Functional category | Protein | Positivity for indicated characteristic
|
||
|---|---|---|---|---|
| Cellulara | Secretedb | Cell wallc | ||
| Chaperonin | DnaK | + | + | + |
| GroEL | + | + | + | |
| Plasminogen binding | Enolase | + | + | + |
| Glyceraldehyde-3-phosphate dehydrogenase | + | + | + | |
| Glycolytic pathway | 6-Phosphofructokinase | + | + | + |
| Phosphoglycerate kinase | + | + | + | |
| Fructose-bisphosphate aldolase | + | + | + | |
| Triosephosphate isomerase | + | + | + | |
| Phosphoglycerate mutase | + | + | + | |
| Pyruvate kinase | + | + | + | |
| Virulence factor | M protein | + | + | + |
| SpeB | + | + | ||
| Protein synthesis | Ribosome recycling factor | + | + | + |
| Elongation factor Tu | + | + | + | |
| Elongation factor Ts | + | + | + | |
| Elongation factor G | + | + | + | |
| Elongation factor P | + | + | ||
| Peptide deformylase | + | + | ||
| Urea cycle pathway | Arginine deaminase | + | + | |
| Ornithine carbamoyltransferase | + | + | ||
| Carbamate kinase | + | + | ||
| Cell wall synthesis | dTDP-4-keto-6-deoxyglucose-3,5-epimerase | + | + | |
| Stress protein | Alkyl hydroperoxidase | + | + | |
| Superoxide dismutase (Mn) | + | + | ||
| Nucleotide synthesis | GMP synthase | + | + | |
| Housekeeping | l-Lactate dehydrogenase | + | + | + |
| NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | + | + | + | |
| 2-Dehydropantoate 2-reductase | + | + | + | |
| Transketolase | + | + | + | |
| Manganese-dependent inorganic pyrophosphatase | + | + | + | |
| Dipeptidase | + | + | ||
| Adenylate kinase | + | + | ||
| ABC transporter (ATP-binding protein) | + | + | ||
| Branched-chain-amino-acid aminotransferase | + | + | ||
| Phosphotransacetylase | + | + | ||
Western blot analysis (3) was used to ascertain the immunoreactivities of proteins in cell wall extracts harvested from S. pyogenes NS931, NS13, and S43. Immunoreactive proteins were detected by probing the membranes with pooled human sera obtained from the Menzies School of Health Research, Darwin, Northern Territory, Australia. Serum samples were pooled from 10 school-aged children residing in a remote community in northern Australia, where GAS infections are endemic and up to 70% of children have GAS-associated impetigo (9). To act as a negative control, 2D mutanolysin extract blots for each strain were probed with goat anti-human immunoglobulin G (IgG) horseradish peroxidase (HRP) only prior to development with diaminobenzidine. The extracts were separated in two dimensions over a linear pH range of 4 to 7, transferred to a polyvinylidene difluoride (PVDF) membrane, and probed with the pooled human sera (Fig. 1D to F). Reactive protein spots were identified according to their relative positions (pI and molecular weight) (Fig. 1A to C). Of the 74 cell wall-associated proteins identified in this study, only 33 (45%) were identified as immunoreactive (Table 1) and therefore are presumably expressed during the course of human infection. Multiple immunoreactive proteins situated near the pH 7 end of the NS13 2D immunoblot (Fig. 1E) could not be identified because the protein concentrations in the corresponding region of the Coomassie blue-stained gel (Fig. 1B) were below the detection threshold. Interestingly, several proteins were identified as immunoreactive in only one or two of the GAS strains examined (Table 1). Given the use of pooled sera and the highly conserved nature of these proteins, an immunoreactive protein should presumably be detected in all three strains. However, strain-specific differences affecting protein expression levels, antigenic variation, or the sensitivity of spot detection may account for this discrepancy.
In an attempt to determine which cell wall-associated proteins are surface exposed, the cell surfaces of GAS strains NS931, NS13, and S43 were biotinylated (1) prior to mutanolysin extraction and subsequent 2D Western blot analysis. Biotin-labeled cell surface proteins were detected using a streptavidin-HRP (SA-HRP) conjugate (Sigma). Two-dimensional blots containing nonbiotinylated mutanolysin extract were used as negative controls for all strains (result not shown). Biotinylated spots were identified by MALDI-TOF MS from the corresponding Coomassie blue-stained biotinylated cell wall extract 2D gel. Only 23 (31%) of the identified cell wall extract proteins were biotinylated and therefore surface exposed in at least one strain (Fig. 1G to I) (Table 1).
Gram-positive proteins destined for transport across the cytoplasmic membrane frequently contain a hydrophobic N-terminal signal sequence and a conserved C-terminal membrane anchor motif of Leu-Pro-X-Thr-Gly (LPXTG) (5). Following protein translocation across the cytoplasmic membrane, the signal peptide is proteolytically removed by signal peptidase. Proteolytic cleavage of the LPXTG motif by sortase facilitates the covalent cross-linking of the protein to the cell wall (28). In this study, many GAS cell wall-associated proteins lack apparent secretion signal sequences and the LPXTG membrane anchor sequence (Table 1). Similarly, a number of secreted GAS proteins lack secretion signals (2, 22, 27). The absence of a signal peptide and LPXTG motif suggests that these proteins are either passively released during autolysis or that an alternative secretory pathway may exist for many secreted GAS proteins. Although the mechanism by which these proteins are transported to the cell surface is unknown, internal signal sequences, posttranslational acylation, or an association with a secreted protein may be involved (31). Recently, asymmetric protein secretion of GAS SpeB was shown to occur at distinct cytoplasmic membrane microdomains termed ExPortals (35). The role that this structure plays in the secretion of other GAS proteins is currently unknown. We also note the possibility that cytosolic proteins passively released by autolysis may have adsorbed to the GAS cell surface.
SpeB is an extracellular and surface-associated cysteine protease virulence factor produced by most GAS strains (18) that can efficiently degrade the majority of proteins in the secreted GAS proteome (2). Twenty-five of the cell wall-associated proteins described in this communication have previously been identified in the extracellular proteomes of GAS SpeB mutants (2, 4, 22). The identification of these proteins in the cell walls of SpeB-positive strains suggests that while these proteins are associated with the GAS cell wall during late stationary phase, they are efficiently protected from SpeB-mediated degradation. Future studies may be performed with biochemically or genetically inactivated SpeB to test this hypothesis.
A significant number of traditional cytoplasmic proteins were also identified as cell wall-associated immunogens in this work. Several cytosolic proteins, such as the glycolysis pathway enzymes, have been reported as cell wall associated in GAS or other prokaryotic species. The glycolytic enzyme GAPDH, also referred to as the plasmin receptor protein (Plr), is a well-characterized GAS cell surface protein with plasminogen binding (40) and ADP-ribosylating (29) activities. This multifunctional protein binds fibronectin, lysozyme, myosin, and actin (30) and elicits signal transduction events in human pharyngeal cells (32). Streptococcal enolase is a glycolytic and major plasminogen-binding protein located on the cell surfaces of most GAS strains (12). Streptococcal enolase has been implicated in GAS adherence to and invasion of human pharyngeal cells (33) and is a highly immunogenic autoantigen with a possible role in the initiation of poststreptococcal sequelae (15). Phosphoglycerate kinase is a glycolytic and major outer surface protein of Streptococcus oralis (39) and Streptococcus agalactiae (group B streptococcus) (17).
Consistent with our findings, the normally cytoplasmic chaperonins DnaK and GroEL have been identified as immunoreactive antigens of S. pyogenes (23, 24). Although these chaperones have not previously been characterized as GAS cell wall constituents, homologs of DnaK and GroEL are located in the cell walls of S. agalactiae (17). Elongation factor Tu is localized in the cell walls of S. oralis (39). Other factors involved in protein synthesis, such as ribosome recycling factor and protein translation elongation factors G, Ts, and P, are expressed on the cell surface of S. oralis (39).
The three components of the arginine deiminase pathway, which consists of ornithine carbamoyltransferase, arginine deiminase, and carbamate kinase, were identified as cell wall associated in this study. The enzymes of this system catalyze the breakdown of arginine to ornithine, CO2, and two molecules of ammonia, with the concomitant production of ATP (7). Ornithine carbamoyltransferase is a bona fide cell wall protein of S. agalactiae (17), Streptococcus sanguis (14), and Streptococcus suis (41). GAS arginine deiminase, also known as the streptococcal acid glycoprotein, is thought to play a role in virulence factor expression and GAS internalization into epithelial cells (10, 25).
Although an association between biotinylated proteins and immunoreactivity was established, some biotinylated cell surface proteins were not immunoreactive. To account for this, we suggest that these proteins either are poor immunogens or are expressed at low levels during GAS infection. Conversely, some immunoreactive proteins were not found to be biotinylated, which may indicate the absence of surface-exposed lysine residues for biotinylation. Alternatively, proteins with only a small number of surface-exposed lysine residues may have been below the limit of detection used in this study. For example, the M protein of NS13 exhibited immunoreactivity against the human antiserum (Fig. 1E, spot 50) but was not found to be biotinylated. Cleavage of surface-exposed M protein by SpeB (20, 34) may explain the apparent lack of M protein biotinylation in this study. Lack of M protein immune reactivity in GAS strains NS931 and S43 may suggest that the individuals from an area of endemicity from whom the serum was derived had not been exposed to these GAS M types. M protein fragments were detected in each of these strains (Fig. 1A and C; spots 73 and 74).
Numerous surface-exposed cell wall proteins have been identified as vaccine candidates in GAS (8). However, a safe and efficacious commercial GAS vaccine has yet to be developed. In this study, we have undertaken a systematic proteomic analysis to extend the range of proteins known to associate with the GAS cell wall. In summary, a total of 74 distinct proteins were identified in the cell wall extracts of three GAS strains. Thirty-three of these proteins were immunoreactive against pooled human sera, and 23 were identified as surface exposed. Further characterization of these proteins is required to elucidate their precise role in GAS pathogenesis. Taken together, these data illustrate the usefulness of proteomics in analyzing the cell surface topology of GAS.
Acknowledgments
We thank Tove' Bolken (SIGA Research Laboratories, Oregon) for providing S. pyogenes strain S43 and Jody Wilton for assisting with the 2D gel electrophoresis.
J. N. Cole is the recipient of an Australian postgraduate award. This work was supported by the National Health and Medical Research Council (NHMRC) of Australia.
Editor: V. J. DiRita
REFERENCES
- 1.Altin, J. G., and E. B. Pagler. 1995. A one-step procedure for biotinylation and chemical cross-linking of lymphocyte surface and intracellular membrane-associated molecules. Anal. Biochem. 224:382-389. [DOI] [PubMed] [Google Scholar]
- 2.Aziz, R. K., M. J. Pabst, A. Jeng, R. Kansal, D. E. Low, V. Nizet, and M. Kotb. 2004. Invasive M1T1 group A streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51:123-134. [DOI] [PubMed] [Google Scholar]
- 3.Burnette, W. N. 1981. Western blotting: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 4.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chhatwal, G. S. 2002. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 10:205-208. [DOI] [PubMed] [Google Scholar]
- 6.Cordwell, S. J., D. J. Basseal, B. Bjellqvist, D. C. Shaw, and I. Humphery-Smith. 1997. Characterisation of basic proteins from Spiroplasma melliferum using novel immobilised pH gradients. Electrophoresis 18:1393-1398. [DOI] [PubMed] [Google Scholar]
- 7.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Currie, B. J., and J. R. Carapetis. 2000. Skin infections and infestations in Aboriginal communities in northern Australia. Australas. J. Dermatol. 41:139-143. [DOI] [PubMed] [Google Scholar]
- 10.Degnan, B. A., M. C. Fontaine, A. H. Doebereiner, J. J. Lee, P. Mastroeni, G. Dougan, J. A. Goodacre, and M. A. Kehoe. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delvecchio, A., B. J. Currie, J. D. McArthur, M. J. Walker, and K. S. Sriprakash. 2002. Streptococcus pyogenes prtFII, but not sfbI, sfbII or fbp54, is represented more frequently among invasive-disease isolates of tropical Australia. Epidemiol. Infect. 128:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derbise, A., Y. P. Song, S. Parikh, V. A. Fischetti, and V. Pancholi. 2004. Role of the C-terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect. Immun. 72:94-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischetti, V. A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2:285-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floderus, E., L. E. Linder, and M. L. Sund. 1990. Arginine catabolism by strains of oral streptococci. APMIS 98:1045-1052. [DOI] [PubMed] [Google Scholar]
- 15.Fontan, P. A., V. Pancholi, M. M. Nociari, and V. A. Fischetti. 2000. Antibodies to streptococcal surface enolase react with human alpha-enolase: implications in poststreptococcal sequelae. J. Infect. Dis. 182:1712-1721. [DOI] [PubMed] [Google Scholar]
- 16.Hollingshead, S. K., J. Arnold, T. L. Readdy, and D. E. Bessen. 1994. Molecular evolution of a multigene family in group A streptococci. Mol. Biol. Evol. 11:208-219. [DOI] [PubMed] [Google Scholar]
- 17.Hughes, M. J., J. C. Moore, J. D. Lane, R. Wilson, P. K. Pribul, Z. N. Younes, R. J. Dobson, P. Everest, A. J. Reason, J. M. Redfern, F. M. Greer, T. Paxton, M. Panico, H. R. Morris, R. G. Feldman, and J. D. Santangelo. 2002. Identification of major outer surface proteins of Streptococcus agalactiae. Infect. Immun. 70:1254-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hytonen, J., S. Haataja, D. Gerlach, A. Podbielski, and J. Finne. 2001. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol. Microbiol. 39:512-519. [DOI] [PubMed] [Google Scholar]
- 19.Ji, Y., N. Schnitzler, E. DeMaster, and P. Cleary. 1998. Impact of M49, Mrp, Enn, and C5a peptidase proteins on colonization of the mouse oral mucosa by Streptococcus pyogenes. Infect. Immun. 66:5399-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kansal, R. G., A. McGeer, D. E. Low, A. Norrby-Teglund, and M. Kotb. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun. 68:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lancefield, R. C., and E. W. Todd. 1928. Antigenic differences between matt hemolytic streptococci and their glossy variants. J. Exp. Med. 48:769-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei, B., S. Mackie, S. Lukomski, and J. M. Musser. 2000. Identification and immunogenicity of group A streptococcus culture supernatant proteins. Infect. Immun. 68:6807-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemos, J. A., R. A. Burne, and A. C. Castro. 2000. Molecular cloning, purification and immunological responses of recombinants GroEL and DnaK from Streptococcus pyogenes. FEMS Immunol. Med. Microbiol. 28:121-128. [DOI] [PubMed] [Google Scholar]
- 24.Lemos, J. A., M. Giambiagi-Demarval, and A. C. Castro. 1998. Expression of heat-shock proteins in Streptococcus pyogenes and their immunoreactivity with sera from patients with streptococcal diseases. J. Med. Microbiol. 47:711-715. [DOI] [PubMed] [Google Scholar]
- 25.Marouni, M. J., E. Ziomek, and S. Sela. 2003. Influence of group A streptococcal acid glycoprotein on expression of major virulence factors and internalization by epithelial cells. Microb. Pathog. 35:63-72. [DOI] [PubMed] [Google Scholar]
- 26.McMillan, D. J., M. R. Davies, M. F. Good, and K. S. Sriprakash. 2004. Immune response to superoxide dismutase in group A streptococcal infection. FEMS Immunol. Med. Microbiol. 40:249-256. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura, T., T. Hasegawa, K. Torii, Y. Hasegawa, K. Shimokata, and M. Ohta. 2004. Two-dimensional gel electrophoresis analysis of the abundance of virulent exoproteins of group A streptococcus caused by environmental changes. Arch. Microbiol. 181:74-81. [DOI] [PubMed] [Google Scholar]
- 28.Novick, R. P. 2000. Sortase: the surface protein anchoring transpeptidase and the LPXTG motif. Trends Microbiol. 8:148-151. [DOI] [PubMed] [Google Scholar]
- 29.Pancholi, V., and V. Fischetti. 1993. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. USA 90:8154-8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pancholi, V., and V. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 32.Pancholi, V., and V. A. Fischetti. 1997. Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: signal transduction between streptococci and pharyngeal cells. J. Exp. Med. 186:1633-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pancholi, V., P. Fontan, and H. Jin. 2003. Plasminogen-mediated group A streptococcal adherence to and pericellular invasion of human pharyngeal cells. Microb. Pathog. 35:293-303. [DOI] [PubMed] [Google Scholar]
- 34.Raeder, R., M. Woischnik, A. Podbielski, and M. D. Boyle. 1998. A secreted streptococcal cysteine protease can cleave a surface-expressed M1 protein and alter the immunoglobulin binding properties. Res. Microbiol. 149:539-548. [DOI] [PubMed] [Google Scholar]
- 35.Rosch, J., and M. Caparon. 2004. A microdomain for protein secretion in Gram-positive bacteria. Science 304:1513-1515. [DOI] [PubMed] [Google Scholar]
- 36.Smoot, L. M., J. K. McCormick, J. C. Smoot, N. P. Hoe, I. Strickland, R. L. Cole, K. D. Barbian, C. A. Earhart, D. H. Ohlendorf, L. G. Veasy, H. R. Hill, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Characterization of two novel pyrogenic toxin superantigens made by an acute rheumatic fever clone of Streptococcus pyogenes associated with multiple disease outbreaks. Infect. Immun. 70:7095-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thongboonkerd, V., J. Luengpailin, J. Cao, W. M. Pierce, J. Cai, J. B. Klein, and R. J. Doyle. 2002. Fluoride exposure attenuates expression of Streptococcus pyogenes virulence factors. J. Biol. Chem. 277:16599-16605. [DOI] [PubMed] [Google Scholar]
- 38.Whatmore, A. M. 2001. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology 147:419-429. [DOI] [PubMed] [Google Scholar]
- 39.Wilkins, J. C., D. Beighton, and K. A. Homer. 2003. Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl. Environ. Microbiol. 69:5290-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winram, S. B., and R. Lottenberg. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 142:2311-2320. [DOI] [PubMed] [Google Scholar]
- 41.Winterhoff, N., R. Goethe, P. Gruening, M. Rohde, H. Kalisz, H. E. Smith, and P. Valentin-Weigand. 2002. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the arginine deiminase system of Streptococcus pyogenes. J. Bacteriol. 184:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]