Abstract
Bartonella proteins that elicit an antibody response during an infection are poorly defined; therefore, to characterize antigens recognized by the host, a Bartonella genomic expression library was screened with serum from an infected mouse. This process led to the discovery of a Bartonella vinsonii subsp. arupensis gene encoding a 382-kDa protein, part of a gene family encoding large proteins, each containing multiple regions of repetitive segments. The genes were termed brpA to -C (bartonella repeat protein) and bore significant similarity to genes encoding the BadA adhesin protein and members of the variably expressed outer membrane protein family of proteins from Bartonella henselae and Bartonella quintana, respectively.
Microbes belonging to the genus Bartonella are gram-negative, facultative intracellular, bacillus-shaped bacteria characterized by an ability to invade and parasitize erythrocytes and endothelial cells and are recognized as agents of emerging diseases of humans and animals (4, 11, 21). Important in human disease are Bartonella bacilliformis, the causative agent of Carrion's disease; Bartonella henselae, the causative agent of cat scratch disease and bacillary angiomatosis; and Bartonella quintana, the causative agent of trench fever and also associated with bacillary angiomatosis. Other Bartonella species that have been isolated from humans include B. elizabethae (17), B. vinsonii subsp. arupensis (49), B. vinsonii subsp. berkhoffii (41), and B. washoensis (27).
Bartonella infections are associated with arthropod vector transmission; e.g., B. bacilliformis is transmitted by sand flies, B. henselae by cat fleas, and B. quintana by the human body louse. There is recent evidence of Bartonella association with ticks, fleas, and flies (10, 14, 15, 46). Furthermore, several distinct Bartonella strains have been isolated from various rodent (5, 7, 8, 22, 23, 29) and ruminant (19, 6, 13) species throughout the world. Because of the ubiquitous nature of Bartonella and their association with arthropod vectors, this genus has been implicated as a potential causative agent in diverse disease manifestations seen in humans for which there have been no definitive clinical diagnoses. Our laboratory was interested in exploring whether the prevalence of Bartonella residing in wild-mammal populations may have had an association with unexplained human febrile illnesses in the North American Southwest, as there was some serological evidence suggesting such a link (M. Y. Kosoy et al., Abstr. Am. Soc. Rickettsiol. Bartonella Emerg. Pathog. Group 2001 Joint Conf., abstr. 108, 2001; F. Koster et al., Abstr. Am. Soc. Rickettsiol. Bartonella Emerg. Pathog. Group 2001 Joint Conf., abstr. 133, 2001). Moreover, a Bartonella (B. washoensis) strain isolated from a Nevada patient with myocarditis was shown to be identical to strains cultured from ground squirrels in the same area (27). We wished to further investigate the putative role of rodent-associated Bartonella in human infections and, as an initial step, to identify Bartonella species-specific antigens that elicit antibody responses during infection. The goal was to initiate a primary analysis of immunodominant antigens related to rodent-borne Bartonella and to further develop a more detailed comparative analysis of immunogenic proteins from human Bartonella pathogens to define genus-specific antigens versus species-specific antigens. As a result of these aims, this report describes a novel immunodominant, surface-associated protein from B. vinsonii subsp. arupensis encoded by one of the largest bacterial genes yet identified. Additionally, at least two more related genes are present upstream, forming a multigene family. The genes in this family are characterized by multiple internal repetitive regions that are conserved within and among the individual genes, which we have therefore termed brp (bartonella repeat protein).
Identification of the brp gene complex.
Mice were infected with live Bartonella strains isolated from rodent species to generate antibodies that could be used to clone and characterize genes encoding proteins associated with infection. Strains na19103nm (isolated from Neotoma albigula [wood rat] in New Mexico), Pm15590co and Pm136co (isolated from Peromyscus maniculatus [deer mouse] in Colorado), and Sb944nv (isolated from Spermophilus beecheyi [California ground squirrel] in Nevada) were obtained from Michael Kosoy, Division of Vector-Borne Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins, Colorado. B. henselae, B. quintana, B. elizabethae, B. vinsonii subsp. berkhoffii, B. vinsonii subsp. arupensis, and B. bacilliformis were purchased from the American Type Culture Collection, Manassas, Va. Bartonella strains were cultivated on Trypticase soy agar supplemented with 5% sheep blood. Plates were incubated at 37°C in 5% CO2 until growth was visible, usually between 5 and 10 days. B. bacilliformis was incubated at 26°C under ambient atmosphere. Antiserum was generated from mice infected with Bartonella as follows. Female BALB/cJ mice (The Jackson Laboratory, Bar Harbor, Maine) were inoculated intraperitoneally with live Bartonella strain Pm15590co resuspended in 0.5 ml of phosphate-buffered saline (PBS) following harvesting from a plate culture. A booster injection with live Pm15590co was given 5 weeks after the primary injection, followed by a third booster 6 weeks later with strain Sb944nv. The mice were bled 3 weeks later and provided a source of anti-Bartonella polyclonal antiserum. Polyclonal antibodies against each of strains Sb944nv and Pm136co were generated in a similar fashion. The rodent-associated strains of Bartonella were used in this initial experiment because there was an interest in identifying specific antigens related to these types of Bartonella infections.
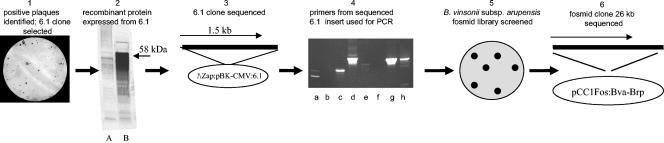
The progression from initially finding an immunoreactive clone from a rodent Bartonella strain genomic library to identifying the brp gene complex in B. vinsonii subsp. arupensis is summarized in Fig. 1. A positive expression clone, termed 6.1, was identified from the Bartonella strain na19203nm genomic library using the mouse antiserum described in the previous paragraph. Clone 6.1 contained a 1.5-kb insert and expressed a recombinant protein product of 58 kDa that reacted in an immunoblot with the same antibody used in the screening process (Fig. 1). DNA sequencing of the 6.1 insert revealed a truncated gene with no start or stop codon at the 5′ or 3′ end, respectively. To extend and obtain the DNA sequence of the 5′ region of the 6.1 clone, probes specific to that area were used for hybridization to the genomic library, which yielded several new clones that were sequenced. It was found that these clones, although identical at the hybridizing ends, were entirely different from one another as the sequences extended. This was the first evidence that this gene was composed of repeat regions and/or was part of a paralogous gene family.
FIG. 1.
Flow chart of steps describing the identification of the initial 6.1 clone through the DNA sequencing of the 26-kb Bva-Brp fosmid clone. (1) Bartonella strain na19103nm genomic lambda library screened with antiserum from a mouse infected with Bartonella strains isolated from Peromyscus and Spermophilus reveals several positive plaques. A positive clone, 6.1, was selected for further characterization. (2) Western blot of the 58-kDa recombinant protein expressed by clone 6.1 reacted with the antiserum used in the library screening in step 1. Lane A, Escherichia coli with expression vector with no insert; lane B, E. coli with the 6.1 cloned insert. (3) The 1.5-kb insert of 6.1 is sequenced. Repeat regions are recognized. (4) PCR primers are synthesized from the 6.1 termini and used for amplification from Bartonella species genomic DNA. A similar-sized amplicon is observed from B. vinsonii subsp. arupensis (lane d). Lanes: a, B. quintana; b, B. henselae; c, B. vinsonii subsp. berkhoffii; d, B. vinsonii subsp. arupensis; e, B. elizabethae; f, B. bacilliformis; g, strain na19103 nm; h, clone 6.1. (5) B. vinsonii subsp. arupensis fosmid library is hybridized with the clone 6.1 probe to identify large inserts harboring analogous region. Positive clones are selected for analysis. (6) A B. vinsonii subsp. arupensis fosmid clone containing a 26-kb insert, Bva-Brp, was sequenced and analyzed.
A PCR primer pair was designed from sequences at each end of clone 6.1, and amplification of homologous regions present in other Bartonella spp. was attempted. Only B. vinsonii subsp. arupensis produced an amplicon of the same size as the rodent Bartonella clone 6.1 (Fig. 1). At this juncture, the decision was made to complete the molecular cloning of this gene using genomic DNA from B. vinsonii subsp. arupensis for four reasons: (i) PCR amplification using B. vinsonii subsp. arupensis DNA and primers generated from clone 6.1 produced an amplicon of the same size, indicating a gene relatedness; (ii) there is a close phylogenetic relationship between B. vinsonii and Bartonella rodent isolates (28, 30); (iii) subspecies of B. vinsonii have been shown to infect humans (41, 49); and (iv) the closely related genomes of B. henselae and B. quintana were being sequenced (and have since been released [2]); therefore, a duplication of effort was avoided.
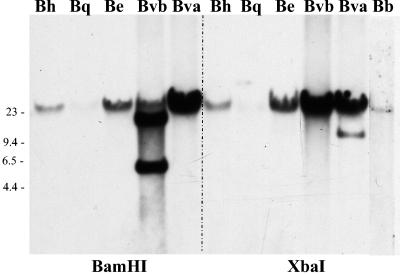
Genomic DNA was purified from Bartonella strains for Southern blot analysis by phenol-chloroform extraction according to standard methods. Two μg of genomic DNA was digested to completion with BamHI or XbaI restriction enzymes (New England Biolabs, Waverly, Mass.) and fractionated on a 0.8% Tris-acetate-EDTA agarose gel, whereby the gel was depurinated, denatured, and neutralized by respective soaking in 0.25 M HCl; 0.5 M NaOH, 1.5 M NaCl; and 0.5 M Tris, pH 7.5, 1.5 M NaCl. Hybridization was performed utilizing the AlkPhos direct labeling reagents (Amersham Biosciences, Little Chalfont, Buckinghamshire, England), and the hybridization probe was the 1.5-kb PCR amplicon amplified from B. vinsonii subsp. arupensis genomic DNA using primers designed from the 6.1 clone of strain na19103nm (Fig. 1). The forward primer, 6.1F, was 5′ ATTGCCATTGGTAAGGAAACAGGC 3′. The reverse primer, 6.1R, was 5′ TTGTAAGGGCACTGACCTCTTCTG 3′. Southern blot analysis revealed orthologs of this gene present in various Bartonella species known to infect humans (Fig. 2). Hybridizing bands were present in B. henselae, B. elizabethae, B. vinsonii subsp. arupensis, and B. vinsonii subsp. berkhoffii, with less prominent hybridizing bands in B. quintana and B. bacilliformis. A single distinct B. vinsonii subsp. arupensis BamHI fragment of approximately 25 kb was observed in the Southern blot; therefore, a fosmid library was constructed with that genomic DNA to ensure a cloned insert large enough to harbor the entire gene plus the flanking regions. A B. vinsonii subsp. arupensis fosmid library was created with the CopyControl fosmid library production kit (Epicentre, Madison, Wis.). Purified genomic DNA was randomly sheared, and fragments were size selected from 25 kb to 40 kb. The sheared DNA was blunt ended and ligated into the fosmid vector, pCC1FOS, and packaged using MaxPlax lambda packaging extracts (Epicentre). Packaged phage particles were allowed to infect E. coli EPI300-T1 (Epicentre), with subsequent colony plating on chloramphenicol selection plates. Three positive fosmid clones were selected by colony hybridization, analyzed by BamHI restriction digestion, and rehybridized with the 1.5-kb probe to ensure the presence of the band seen in the Southern blot. Once confirmed, one of the clones containing a 26-kb insert, Bva-Brp (Fig. 1), was sequenced in its entirety. Purified Bva-Brp fosmid DNA was sequenced by primer walking on both strands to an average fourfold redundancy. Electrophoresis was performed using ABI (PE Biosystems, Foster City, Calif.) Big-Dye 3.1 dye chemistry and an ABI 3730XL automated DNA sequencer. Chromatogram data were analyzed using the program Phred (University of Washington) for base calling and quality assessment. Consensus sequence assembly was performed using the program Phrap, with manual editing being performed using the program Consed. Additional sequencing analyses were performed using SeqManII and MegAlign (DNASTAR, Madison, Wis.) for assembly, DNA and protein alignments, and dot plot matrix analyses of repetitive regions. Protein and DNA GenBank database search and alignments were performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
FIG. 2.
Southern blot of Bartonella spp. genomic DNA. Genomic DNA samples were digested with either BamHI or XbaI as indicated and hybridized to the clone Bva-6.1 probe (for the location within the gene, see Fig. 3A). Bh = B. henselae, Bq = B. quintana, Be = B. elizabethae, Bvb = B. vinsonii subsp. berkhoffii, Bva = B. vinsonii subsp. arupensis, Bb = B. bacilliformis. Molecular weight markers in kb are noted on the left. A 23- to 25-kb weakly hybridizing band for B. quintana is present.
DNA sequence of the B. vinsonii subsp. arupensis brpA gene and flanking regions.
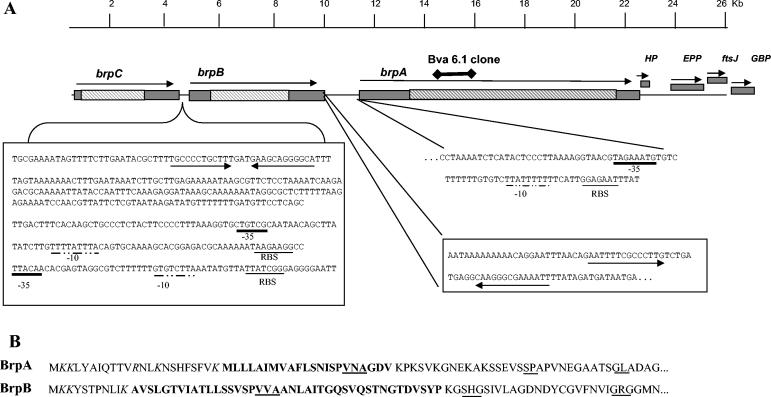
The Bva-Brp insert was 26.4 kb and consisted of the complete gene encoding the immunoreactive protein (brpA), two large open reading frames located upstream of this gene (brpB and brpC), and four smaller genes located downstream (Fig. 3A). The largest gene, brpA, had a sequence of 10,863 bp encoding a protein of 3,619 amino acids with a deduced molecular mass of 382.5 kDa. brpB, upstream of brpA, had a sequence of 5,283 bp encoding a protein of 185 kDa. The Bva-Brp clone contained a truncated brpC gene upstream of brpB but nevertheless indicated a large gene, as the partial version is 4,263 bp long and encodes a truncated protein of 152 kDa. The number of contiguous genes belonging to this family remains to be determined, as the 26-kb fragment was not of sufficient size to contain the entire brpC gene plus others that may be located upstream. The single hybridizing band seen in the Southern blot suggested that there may be few or no more brp-like sequences residing elsewhere in the chromosome, although one cannot rule out the possibility of more brp-like genes or pseudogenes that did not hybridize with the probe.
FIG. 3.
Organization of genes on the 26-kb Bva-Brp clone of B. vinsonii subsp. arupensis. (A) brpA, brpB, and brpC coding sequences are represented by rectangular bars, with the arrows showing the direction of transcription. The hatched bars within the coding sequences denote portions of the genes composed of repetitive sequences. Genes downstream of brpA are abbreviated as HP (hypothetical protein), EPP (exopolyphosphatase), ftsJ (cell division protein), and GBP (GTP-binding protein). The 409-bp intervening sequence between the stop codon of brpC and the start codon of brpB is shown. The bidirectional arrows indicate inverted repeats characteristic of transcriptional termination sites for brpC. Two putative brpB promoter sites based on consensus sequences for the ribosome binding site (RBS) (underlined), the −10 site (underlined with a dot-and-dash line), and the −35 site (heavily underlined line) are denoted. The region immediately downstream of the stop codon for brpB is shown with inverted repeats illustrated with the bidirectional arrows. The immediate upstream noncoding region is shown for brpA with putative RBS, −10, and −35 sites. The location of the original truncated Bva-6.1 clone that generated the recombinant protein used in this study is shown above brpA. A scale in kbp is above the diagram. (B) Signal peptides deduced from the amino acid sequences for brpA and brpB. Charged residues are italicized, the hydrophobic central region is in bold, and putative SPase I cleavage sites are underlined. Cleavage sites predicted by the SignalP program follow the underlined VNA and VVA of BrpA and BrpB, respectively.
Based on the DNA sequence analysis, the genes brpA and brpB appear to be independently transcribed genes and not part of a multicistronic locus or nontranscribed pseudogenes because of the spacing between the coding sequences and the characteristic promoter elements upstream of the coding sequence (Fig. 3A). brpB and brpC show inverted repeats characteristic of transcriptional terminators downstream of the stop codon; however, brpA may be cotranscribed with the hypothetical protein gene immediately downstream as there is only a 2-bp separation between these two coding sequences. The deduced amino acid sequences at the N termini of BrpA and BrpB have properties consistent with signal peptides cleavable by signal peptidase I (SPase I) (Fig. 3B) (38). These peptides begin with an arrangement of charged residues (lysine or arginine) at the N terminus, followed by a central region(s) of hydrophobic amino acids and the SPase I cleavage site (−3 [AGS], −1 [AGS]). BrpA and BrpB are predicted to have cleavable signal peptides by the SignalP program (Technical University of Denmark; http://www.cbs.dtu.dk/services/SignalP/). The presence of putative signal peptides thereby provide evidence that the Brp proteins are exported to the cell surface.
A search of the GenBank protein database with BrpA found the most significant match with portions of a B. henselae protein encoded by a gene termed badA1, submitted as part of the genome sequencing project recently reported by Alsmark et al. (2). The gene product, BadA, has recently been described as a 340-kDa surface protein adhesin of B. henselae that mediates proangiogenic responses in infected cells (40). The Clustal W alignment program of DNASTAR indicated there was a 35.9% amino acid identity between BadA and BrpA. Portions of BrpA also shared significant homology with a recently described B. quintana family of four variably expressed outer membrane proteins (Vomps), A to D (51). The B. quintana Vomps, like BadA, also function as adhesins and mediate autoaggregation. The Vomp gene family encodes proteins of approximately 100 kDa, smaller than the Brp family. Both the B. quintana Vomps and B. henselae BadA contain repeated regions and were shown to be immunogenic, consistent with properties observed with the B. vinsonii subsp. arupensis Brp proteins. The B. henselae badA1 and the B. quintana Vomp gene family members are likely to be orthologous companions of the B. vinsonii subsp. arupensis brp gene family, and it is not surprising that the comparably smaller sized Vomp proteins may be a result of the genome reduction of B. quintana as noted by the genome descriptions of Alsmark et al. (2).
There were four additional genes downstream of brpA, each showing similarity to database proteins by BLAST analysis. With the exception of the gene immediately downstream from brpA, the other genes were conserved and identified from analogous genes present in several other prokaryotic organisms. The gene downstream of brpA encoded a hypothetical protein of 22.3 kDa and has highly conserved orthologous partners in B. henselae and B. quintana, with amino acid identities of 82 and 81%, respectively. Next downstream was a gene encoding a 49-kDa protein described as an exopolyphosphatase, with 79% amino acid identity to the corresponding B. henselae gene. There was no BLAST match to this protein in B. quintana. Following the exopolyphosphatase gene were genes encoding the cell division protein ftsJ (also termed a 23S rRNA methyltransferase) and the GTP-binding protein. Both of these genes were present in B. henselae and B. quintana.
Repeat regions in brp genes.
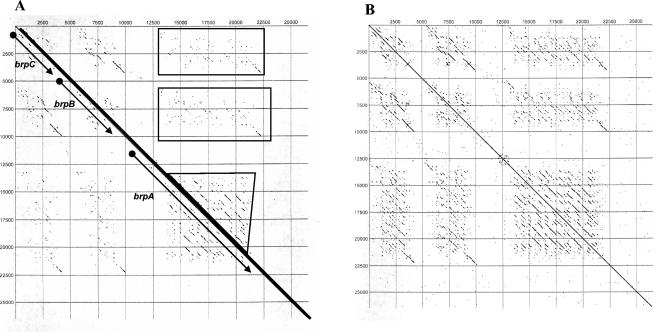
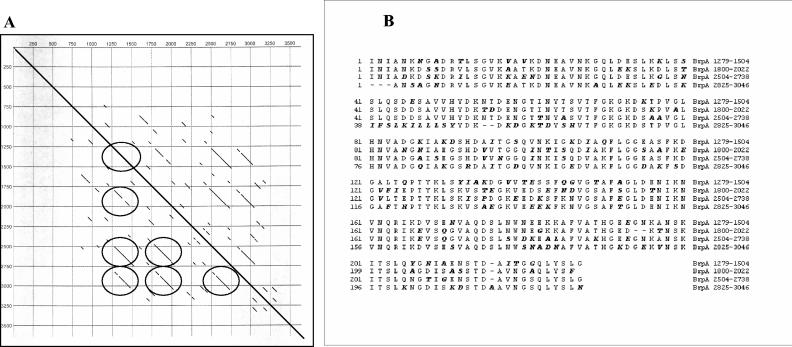
brpA, brpB, and brpC were each found to contain multiple extensive regions of internal repeats sometimes encoding long stretches of repetitive amino acid residues. These regions were not necessarily tandemly oriented but were separated by other repeats in no particular order or arrangement. Additionally, the repeats, although quite similar, were not identical. Figure 4 graphically illustrates the complexity of the repeat regions by dot plot matrix analysis of the brp nucleotide sequences. The dot plot of the entire 26.4-kb insert plotted against itself clearly shows overlapping homologous areas within each gene and also the homology among genes. Dot plot analysis of the amino acid sequence of BrpA reveals the extensive repetitive nature of various lengths of sequence throughout the deduced protein sequence (Fig. 5A). For example, the longest repeats were seen at amino acid positions 1279 to 1504, 1800 to 2022, 2504 to 2738, and 2825 to 3046 (Fig. 5A). These particular repeats range from 221 to 234 amino acids and are aligned in Fig. 5B. The percentages of identity between each repeat ranged from 62.8% (amino acids 2504 to 2738 and 2825 to 3046) to 75.8% (amino acids 1800 to 2022 and 2504 to 2738). Several more smaller repetitive regions are interspersed throughout the BrpA coding region, as illustrated in the dot plot matrix. The apparent randomness of the repetitive arrangements, with some longer than 200 amino acids, are a unique characteristic of these genes. The larger brpA repeats are not tandemly oriented but are interspaced at various lengths, as are the many shorter repetitive regions. The repetitive regions of BrpA are limited to the internal majority of the protein, with the amino and carboxy ends of the protein having few or no repeats. The recent publication describing the genomes of B. henselae and B. quintana reported that these organisms contain repeated or partially repeated gene families, with B. henselae containing the higher proportion, with 78 repeat families of 2 to 14 members (2). The precise number of brp genes in B. vinsonii subsp. arupensis remains to be counted, and comparison of their organization and arrangements to the badA1 and Vomp gene families within Bartonella species remains to be performed.
FIG. 4.
Dot plot matrix analysis at the nucleotide level of the 26-kb Bva-Brp insert plotted against itself. (A) Analysis performed at an 80% homology level. Each line represents an area with a corresponding homologous area with an identity level of at least 80%. The bold diagonal in the center represents the 100% match of the sequence with itself. Areas illustrating the locations of brpA, brpB, and brpC are denoted along the diagonal. The boxed regions of the matrix indicate homologous segments to brpA within brpA, brpB, and brpC. (B) Same as panel A, except the analysis was performed at the 70% identity level, which illustrates the areas of homology within and among the genes more clearly. Numbers on the x and y axes denote the nucleotide positions of the 26-kb Bva-Brp cloned insert.
FIG. 5.
(A) Dot plot analysis at the amino acid level of BrpA plotted against itself at the 70% identity level. The diagonal in the center indicates the 100% match of the sequence with itself. Circles surround the largest repeated regions corresponding to amino acid positions 1279 to 1504, 1800 to 2022, 2504 to 2738, and 2825 to 3046. Numbers on the x and y axes denote the amino acid positions for BrpA. (B) Amino acid alignment of repeats 1279 to 1504, 1800 to 2022, 2504 to 2738, and 2825 to 3046. Residues in bold italics represent divergence from the consensus.
Western blot and immunofluorescence assay analysis.
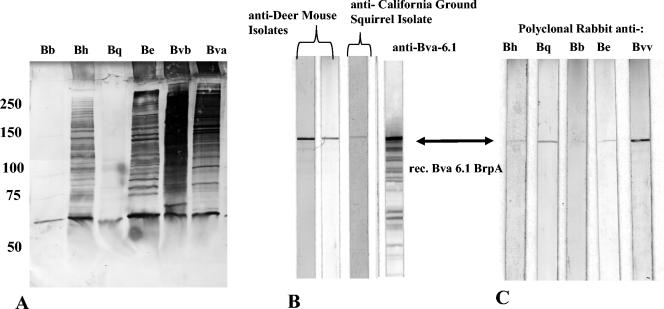
Anti-recombinant Bva-6.1 (the original truncated protein; for the location, see Fig. 3A) was used in immunoblots against whole-cell lysates of various Bartonella species (Fig. 6A). A ladder or smear of several high-molecular-weight immunoreactive bands were apparent in B. henselae, B. elizabethae, B. vinsonii subsp. berkhoffii, and B. vinsonii subsp. arupensis. The reason for the high number of bands is not clear but may reflect polymeric, cleaved, or semidegraded forms of the proteins and/or several epitope-containing repeated regions within the Bartonella protein profiles. A common immunoreactive band of approximately 65 kDa was observed in all Bartonella lysates. B. quintana and B. bacilliformis did not exhibit seroreactivity to the high-molecular-weight bands with this antibody, with the exception of the 65-kDa band. This lack of reactivity is reflected in the Southern blot data which showed little homology to this region of the brpA gene. To determine whether the 65-kDa band may have been an artifactual cross-reactive antigen, the Bartonella cell lysates were blotted against polyclonal mouse preimmune serum and a polyclonal mouse serum against an unrelated protein. No cross-reactive immunoblot bands were observed (data not shown), suggesting that the 65-kDa band was specific to Brp-related proteins and probably not associated with bacterial common antigens, such as heat shock proteins or GroEl. The smaller 65-kDa band that was common among the assayed Bartonella strains may represent a cleaved portion of BrpA or an as-yet-undiscovered smaller version of BrpA that is conserved among species. Even though the antibody was directed against a relatively small segment of the BrpA protein, portions of that particular region are repeated throughout BrpA 18 times, and homologous regions are also present in BrpB and BrpC, according to dot plot analysis at 70% amino acid homology, thereby allowing this antibody to be useful in detecting Brp proteins. Moreover, this region represents a highly conserved immunodominant domain, as it originated from the expression clone found using antiserum from a Bartonella-infected animal.
FIG. 6.
(A) Immunoblot of whole-cell Bartonella lysates with anti-Bva 6.1. Anti-Bva-6.1 was used at a 1:1,000 dilution. Proteins were fractionated on a 7.5% polyacrylamide gel. Molecular size markers (in kilobases) are noted at the left. (B) Recombinant Bva-6.1 protein blotted against antisera from mice infected with rodent Bartonella strains. Sources of antibodies are indicated. Antibodies were used at a 1:500 dilutin. The lower reacting bands in the anti-Bva-6.1 blot are breakdown products of recombinant BrpA. (C) Immunoblot of recombinant Bva-6.1 protein blotted with polyclonal antisera generated against Bartonella sp. whole-cell lysates. Antibodies were used at a 1:500 dilution. Bh = B. henselae, Bq = B. quintana, Bb = B. bacilliformis, Be = B. elizabethae, Bvv = B. vinsonii subsp. vinsonii.
The purified 58-kDa recombinant Bva-6.1 protein was subjected to immunoblot analysis with antiserum from infected animals. Figure 6B demonstrates the seroreactivity of these antibodies against BrpA, indicating that a humoral response is generated during Bartonella infections. Polyclonal antibodies against whole-cell Bartonella spp. lysates reacted with the recombinant Bva-6.1 protein (with the exception of B. bacilliformis antibody), demonstrating a degree of antigenic cross-reactivity among species to BrpA (Fig. 6C). Large-molecular-mass Bartonella immunoreactive bands of >200 kDa have been noted by other researchers in Western blots using antiserum from infected humans and animals, and they may represent orthologous Brp proteins (20, 32, 33). Work remains to be done to identify and characterize Brp-like proteins in other Bartonella species, determine whether infection with specific species or strains elicits an anti-Brp antibody response, and identify the immunodominant regions. There may be enough diversity in orthologous members of the Brp family in Bartonella for species-specific serological reactivity.
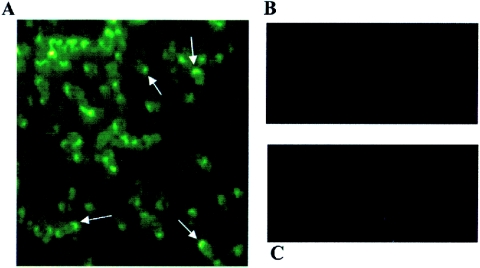
B. vinsonii subsp. arupensis and B. quintana cells were subjected to immunofluorescent staining using anti-Bva-6.1 protein (Fig. 7). B. vinsonii subsp. arupensis and B. quintana cells from a culture plate were separately mixed in 10 μl sterile PBS on a microscope slide and allowed to air dry. The mouse antibody generated against the recombinant Bva-6.1 was incubated with the cells at a 1:50 dilution at 37°C for 1 h. Following five washes in PBS, the cells were incubated with a 1:50 dilution of fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G at 37°C for 1 h. After another series of PBS washes, the slide was air dried and viewed by epifluorescence microscopy. The staining showed strong fluorescent reactivity with B. vinsonii subsp. arupensis cells, providing evidence for an outer surface localization for at least the part of the protein to which the antibody was directed. The antibody reactivity was concentrated in discrete areas of the cell, usually at the ends, indicating a polarized location for BrpA following secretion or export to the outer surface (Fig. 7A). B. quintana cells stained with the same antibody did not demonstrate fluorescent activity (Fig. 7B), reflecting the result seen in the immunoblot. Epitopes in the Bva-6.1 segment may also be present in other Brp proteins; therefore, these could be detected on the surface as well.
FIG. 7.
Immunofluorescent staining of Bartonella cells. (A) B. vinsonii subsp. arupensis stained with anti-Bva 6.1; (B) B. quintana stained with anti-Bva 6.1; (C) B. vinsonii subsp. arupensis stained with preimmunized mouse serum. Arrows in panel A point to the concentrated staining in the polar regions of the cells. Magnification, ×1,000.
Concluding remarks.
During the course of our investigation of Bartonella immunogenic antigens, we discovered orthologous genes encoding large proteins from a rodent Bartonella isolate and from B. vinsonii subsp. arupensis. Taken together, the data describing their immunoreactive properties, probable surface localization, remarkable structure of repeats, and unusually large size reflect characteristics of gene products associated with pathogenic mechanisms of infection. Solid progress has been made in identifying Bartonella-related factors mediating host infection or invasion of host cells. Virulence factors that have been described include the VirB type IV secretion system, which mediates several processes involved in endothelial cell interactions (43, 44); GroEL as a mitogen to enhance the proliferation of endothelial cells (36); the B. bacilliformis invasion-associated locus (ial), which has a direct role in human erythrocyte parasitism (16, 37); flagella involved in host cell adherence (42, 48); deformin associated with cellular invasion (34); the B. quintana hemin binding protein family (12, 35); and the recently described adhesins BadA and Vomp A to D of B. henselae and B. quintana, respectively (40, 51). The function of the Brp family of proteins, other than the elicitation of antibodies following a Bartonella infection, is unknown, although given the homology with BadA and the Vomp proteins, it is not unrealistic to speculate that the Brp proteins may share the same functional properties. In addition, the highly repetitive regions of the brp gene family suggest a gene rearrangement mechanism as a method for possible antigenic variation and immune evasion. Paralogous gene families and individual genes containing repeats have been documented to occur in many bacterial pathogens and have been shown to be essential in mediating infectious processes, such as bacterial adhesion to host cells and gene rearrangements leading to antigenic variation. Examples include the vlsE gene of Borrelia burgdorferi (50), the vsp and vlp families in Borrelia hermsii (24), the ompA gene of Rickettsia rickettsii and other spotted fever group rickettsiae (3), the msp gene family of Anaplasma marginale (1, 18), the Ehrlichia risticii ssa gene (9), Listeria monocytogenes actA (45), the tpr gene family of Treponema pallidum (47), sof and M protein genes of group A streptococci (25, 26, 39), and the alpha-like protein genes of group B streptococci (31).
BrpA is encoded by one of the largest prokaryotic genes thus far identified, and to our knowledge, the brp family is novel in having genes encoding such large proteins with the extensive and complex mosaic of repeated domains. The definition of the numbers of brp gene family members in Bartonella spp., outer membrane protein locations, kinetics of expression, secretion mechanisms, the role of the repeat regions, and the biological function of the Brps are some of the many issues awaiting future investigations.
Nucleotide sequence accession number.
The GenBank accession number for the Bva-Brp 26-kb clone is AY730759.
(Portions of this work were presented at the 18th meeting of the American Society for Rickettsiology, 28 September to 1 October 2003, Cumberland, Maryland, and at the 104th General Meeting of the American Society for Microbiology, 23 to 27 May 2004, New Orleans, Louisiana.)
Acknowledgments
We thank Rendi Bacon, Amber Carpio, Barbara Karl, Ken Gage, and Michael Kosoy for their important contributions to this work. We acknowledge William Nicholson, CDC, for the kind gift of anti-Bartonella antibodies, and we thank Mike Minnick and Jay Carroll for their critical assessment of this work. We also thank the CDC Biotechnology Core Facility for their timely primer synthesis efforts.
Editor: V. J. DiRita
REFERENCES
- 1.Alleman, A. R., G. H. Palmer, T. C. McGuire, T. F. McElwain, L. E. Perryman, and A. F. Barbet. 1997. Anaplasma marginale major surface protein 3 is encoded by a polymorphic, multigene family. Infect. Immun. 65:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, B. E., G. A. McDonald, D. C. Jones, and R. L. Regnery. 1990. A protective protein antigen of Rickettsia rickettsii has tandemly repeated, near-identical sequences. Infect. Immun. 58:2760-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, B. E., and M. A. Neuman. 1997. Bartonella spp. as emerging human pathogens. Clin. Microbiol. Rev. 10:203-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, Y., M. Y. Kosoy, G. O. Maupin, K. R. Tsuchiya, and K. L. Gage. 2002. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am. J. Trop. Med. Hyg. 66:622-627. [DOI] [PubMed] [Google Scholar]
- 6.Bermond, D., H. J. Boulouis, R. Heller, L. G. Van, H. Monteil, B. B. Chomel, A. Sander, C. Dehio, and Y. Piemont. 2002. Bartonella bovis sp. nov. and Bartonella capreoli sp. nov., isolated from European ruminants. Int. J. Syst. Evol. Microbiol. 52:383-390. [DOI] [PubMed] [Google Scholar]
- 7.Bermond, D., R. Heller, F. Barrat, G. Delacour, C. Dehio, A. Alliot, H. Monteil, B. Chomel, H. J. Boulouis, and Y. Piemont. 2000. Bartonella birtlesii sp. nov., isolated from small mammals (Apodemus spp.). Int. J. Syst. Evol. Microbiol. 50:1973-1979. [DOI] [PubMed] [Google Scholar]
- 8.Birtles, R. J., T. G. Harrison, and D. H. Molyneux. 1994. Grahamella in small woodland mammals in the U.K.: isolation, prevalence and host specificity. Ann. Trop. Med. Parasitol. 88:317-327. [DOI] [PubMed] [Google Scholar]
- 9.Biswas, B., R. Vemulapalli, and S. K. Dutta. 1998. Molecular basis for antigenic variation of a protective strain-specific antigen of Ehrlichia risticii. Infect. Immun. 66:3682-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bown, K. J., M. Bennet, and M. Begon. 2004. Flea-borne Bartonella grahamii and Bartonella taylorii in bank voles. Emerg. Infect. Dis. 10:684-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitschwerdt, E. B., and D. L. Kordick. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll, J. A., S. A. Coleman, L. S. Smitherman, and M. F. Minnick. 2000. Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 68:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C. C., B. B. Chomel, R. W. Kasten, R. M. Heller, H. Ueno, K. Yamamoto, V. C. Bleich, B. M. Pierce, B. J. Gonzales, P. K. Swift, W. M. Boyce, S. S. Jang, H. J. Boulouis, Y. Piemont, G. M. Rossolini, M. L. Riccio, G. Cornaglia, L. Pagani, C. Lagatolla, L. Selan, and R. Fontana. 2000. Bartonella spp. isolated from wild and domestic ruminants in North America. Emerg. Infect. Dis. 6:306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, C. C., B. B. Chomel, R. W. Kasten, V. Romano, and N. Tietze. 2001. Molecular evidence of Bartonella spp. in questing adult Ixodes pacificus ticks in California. J. Clin. Microbiol. 39:1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, C. Y., R. W. Kasten, S. M. Paff, B. A. Van Horn, M. Vayssier-Taussat, H. J. Boulouis, and B. B. Chomel. 2004. Bartonella spp. DNA associated with biting flies from California. Emerg. Infect. Dis. 10:1311-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman, S. A., and M. F. Minnick. 2001. Establishing a direct role for the Bartonella bacilliformis invasion-associated locus B (IalB) protein in human erythrocyte parasitism. Infect. Immun. 69:4373-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daly, J. S., M. G. Worthington, D. J. Brenner, C. W. Moss, D. G. Hollis, R. S. Weyant, A. G. Steigerwalt, R. E. Weaver, M. I. Daneshvar, and S. P. O'Connor. 1993. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J. Clin. Microbiol. 31:872-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Fuente, J., J. C. Garcia-Garcia, E. F. Blouin, and K. M. Kocan. 2003. Characterization of the functional domain of major surface protein 1a involved in adhesion of the rickettsia Anaplasma marginale to host cells. Vet. Microbiol. 91:265-283. [DOI] [PubMed] [Google Scholar]
- 19.Dehio, C., C. Lanz, R. Pohl, P. Behrens, D. Bermond, Y. Piemont, K. Pelz, and A. Sander. 2001. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int. J. Syst. Evol. Microbiol. 51:1557-1565. [DOI] [PubMed] [Google Scholar]
- 20.Freeland, R. L., D. T. Scholl, K. R. Rohde, L. J. Shelton, and K. L. O'Reilly. 1999. Identification of Bartonella-specific immunodominant antigens recognized by the feline humoral immune system. Clin. Diagn. Lab. Immunol. 6:558-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greub, G., and D. Raoult. 2002. Bartonella: new explanations for old diseases. J. Med. Microbiol. 51:915-923. [DOI] [PubMed] [Google Scholar]
- 22.Heller, R., M. Kubina, P. Mariet, P. Riegel, G. Delacour, C. Dehio, F. Lamarque, R. Kasten, H. J. Boulouis, H. Monteil, B. Chomel, and Y. Piemont. 1999. Bartonella alsatica sp. nov., a new Bartonella species isolated from the blood of wild rabbits. Int. J. Syst. Bacteriol. 49:283-288. [DOI] [PubMed] [Google Scholar]
- 23.Heller, R., P. Riegel, Y. Hansmann, G. Delacour, D. Bermond, C. Dehio, F. Lamarque, H. Monteil, B. Chomel, and Y. Piemont. 1998. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int. J. Syst. Bacteriol. 48:1333-1339. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch, B. J., A. G. Barbour, B. I. Restrepo, and T. G. Schwan. 1998. Population structure of the relapsing fever spirochete Borrelia hermsii as indicated by polymorphism of two multigene families that encode immunogenic outer surface lipoproteins. Infect. Immun. 66:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus. Repetitive structure and membrane anchor. J. Biol. Chem. 261:1677-1686. [PubMed] [Google Scholar]
- 26.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1987. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol. Gen. Genet. 207:196-203. [DOI] [PubMed] [Google Scholar]
- 27.Kosoy, M., M. Murray, R. D. Gilmore, Jr., Y. Bai, and K. L. Gage. 2003. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 41:645-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosoy, M. Y., R. L. Regnery, O. I. Kosaya, and J. E. Childs. 1999. Experimental infection of cotton rats with three naturally occurring Bartonella species. J. Wildl. Dis. 35:275-284. [DOI] [PubMed] [Google Scholar]
- 29.Kosoy, M. Y., R. L. Regnery, T. Tzianabos, E. L. Marston, D. C. Jones, D. Green, G. O. Maupin, J. G. Olson, and J. E. Childs. 1997. Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am. J. Trop. Med. Hyg. 57:578-588. [DOI] [PubMed] [Google Scholar]
- 30.Kosoy, M. Y., E. K. Saito, D. Green, E. L. Marston, D. C. Jones, and J. E. Childs. 2000. Experimental evidence of host specificity of Bartonella infection in rodents. Comp. Immunol. Microbiol. Infect. Dis. 23:221-238. [DOI] [PubMed] [Google Scholar]
- 31.Lachenauer, C. S., R. Creti, J. L. Michel, and L. C. Madoff. 2000. Mosaicism in the alpha-like protein genes of group B streptococci. Proc. Natl. Acad. Sci. USA 97:9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Litwin, C. M., T. B. Martins, and H. R. Hill. 1997. Immunologic response to Bartonella henselae as determined by enzyme immunoassay and Western blot analysis. Am. J. Clin. Pathol. 108:202-209. [DOI] [PubMed] [Google Scholar]
- 33.McGill, S. L., R. L. Regnery, and K. L. Karem. 1998. Characterization of human immunoglobulin (Ig) isotype and IgG subclass response to Bartonella henselae infection. Infect. Immun. 66:5915-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mernaugh, G., and G. M. Ihler. 1992. Deformation factor: an extracellular protein synthesized by Bartonella bacilliformis that deforms erythrocyte membranes. Infect. Immun. 60:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minnick, M. F., K. N. Sappington, L. S. Smitherman, S. G. Andersson, O. Karlberg, and J. A. Carroll. 2003. Five-member gene family of Bartonella quintana. Infect. Immun. 71:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minnick, M. F., L. S. Smitherman, and D. S. Samuels. 2003. Mitogenic effect of Bartonella bacilliformis on human vascular endothelial cells and involvement of GroEL. Infect. Immun. 71:6933-6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell, S. J., and M. F. Minnick. 1995. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect. Immun. 63:1552-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rakonjac, J. V., J. C. Robbins, and V. A. Fischetti. 1995. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect. Immun. 63:622-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riess, T., S. G. Andersson, A. Lupas, M. Schaller, A. Schafer, P. Kyme, J. Martin, J. H. Walzlein, U. Ehehalt, H. Lindroos, M. Schirle, A. Nordheim, I. B. Autenrieth, and V. A. Kempf. 2004. Bartonella adhesin a mediates a proangiogenic host cell response. J. Exp. Med. 200:1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux, V., S. J. Eykyn, S. Wyllie, and D. Raoult. 2000. Bartonella vinsonii subsp. berkhoffii as an agent of afebrile blood culture-negative endocarditis in a human. J. Clin. Microbiol. 38:1698-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherer, D. C., I. DeBuron-Connors, and M. F. Minnick. 1993. Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of human erythrocytes. Infect. Immun. 61:4962-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmid, M. C., R. Schulein, M. Dehio, G. Denecker, I. Carena, and C. Dehio. 2004. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Mol. Microbiol. 52:81-92. [DOI] [PubMed] [Google Scholar]
- 44.Schulein, R., and C. Dehio. 2002. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 46:1053-1067. [DOI] [PubMed] [Google Scholar]
- 45.Smith, G. A., J. A. Theriot, and D. A. Portnoy. 1996. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin-based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J. Cell Biol. 135:647-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson, H. L., Y. Bai, M. Y. Kosoy, J. A. Montenieri, J. L. Lowell, M. C. Chu, and K. L. Gage. 2003. Detection of novel Bartonella strains and Yersinia pestis in prairie dogs and their fleas (Siphonaptera: Ceratophyllidae and Pulicidae) using multiplex polymerase chain reaction. J. Med. Entomol. 40:329-337. [DOI] [PubMed] [Google Scholar]
- 47.Sun, E. S., B. J. Molini, L. K. Barrett, A. Centurion-Lara, S. A. Lukehart, and W. C. Van Voorhis. 2004. Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity. Microbes Infect. 6:725-737. [DOI] [PubMed] [Google Scholar]
- 48.Walker, T. S., and H. H. Winkler. 1981. Bartonella bacilliformis: colonial types and erythrocyte adherence. Infect. Immun. 31:480-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welch, D. F., K. C. Carroll, E. K. Hofmeister, D. H. Persing, D. A. Robison, A. G. Steigerwalt, and D. J. Brenner. 1999. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J. Clin. Microbiol. 37:2598-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, P., B. B. Chomel, M. K. Schau, J. S. Goo, S. Droz, K. L. Kelminson, S. S. George, N. W. Lerche, and J. E. Koehler. 2004. A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc. Natl. Acad. Sci. USA 101:13630-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]