Abstract
Background
Ischemic stroke (IS) is one of most common causes of disability in adults worldwide. However, there is still a lack of effective and reliable diagnostic markers and therapeutic targets in IS. Furthermore, immune cell dysfunction plays an important role in the pathogenesis of IS. Hence, in-depth research on immune-related targets in progressive IS is urgently needed.
Methods
Expression profile data from patients with IS were downloaded from the Gene Expression Omnibus (GEO) database. Then, differential expression analysis and weighted gene coexpression network analysis (WGCNA) were performed to identify the significant modules and differentially expressed genes (DEGs). Key genes were obtained and used in functional enrichment analyses by overlapping module genes and DEGs. Next, hub candidate genes were identified by utilizing three machine learning algorithms: least absolute shrinkage and selection operator (LASSO), random forest, and support vector machine–recursive feature elimination (SVM-RFE). Subsequently, a diagnostic model was constructed based on the hub genes, and receiver operating characteristic (ROC) curves were constructed to validate the performances of the predictive models and candidate genes. Finally, the immune cell infiltration landscape of IS was explored with the CIBERSORT deconvolution algorithm.
Results
A total of 40 key DEGs were identified based on the intersection of the DEGs and module genes, and we found that these genes were mainly enriched in the regulation of lipolysis in adipocytes, neutrophil extracellular trap formation and complement and coagulation cascades. Based on the results from three advanced machine learning algorithms, we obtained 7 hub candidate genes (ABCA1, ARG1, C5AR1, CKAP4, HMFN0839, SDCBP and TLN1) as diagnostic biomarkers of IS and developed a reliable nomogram with high predictive performance (AUC = 0.987). In addition, immune cell infiltration dysregulation was implicated in IS, and compared with those in the normal group, IS patients had increased fractions of gamma delta T cells, monocytes, M0 macrophages, M2 macrophages and neutrophils and clearly lower percentages of naive B cells, CD8 T cells, CD4+ memory T cells, follicular helper T cells, regulatory T cells (Tregs) and resting dendritic cells. Furthermore, correlation analysis indicated a significant correlation between the hub genes and immune cells in progressive IS.
Conclusion
In conclusion, our study identified 7 hub genes as diagnostic biomarkers and established a reliable model to predict the occurrence of IS. Meanwhile, we explored the immune cell infiltration pattern and investigated the relationship between candidate genes and immune cells in the pathogenesis of IS. Hence, our study provides new insights into the diagnosis and treatment of IS.
Keywords: Ischemic stroke, Identification, Bioinformatics analysis, Hub genes, GEO
Highlights
-
•
The study identified 7 hub candidate genes as diagnostic biomarkers of IS with a high predictive performance (AUC = 0.987).
-
•
We found a significant correlation between hub genes and immune cells in the progression of IS.
-
•
We explored the immune cells infiltration pattern and the relationship between candidate genes and immune cells in the pathogenesis of IS.
1. Introduction
As the second leading cause of death and as the main contributor to the incidence of disability worldwide, stroke is associated with a high socioeconomic burden [[1], [2], [3], [4]]. Notably, ischemic stroke (IS) is the most prevalent type of stroke and accounts for more than 80 % of stroke cases [5]. A sudden occlusion of the brain blood supply may lead to catastrophic damage to brain tissue, including an inflammatory cascade, immune cell infiltration and irreversible neuron death. As the most common acute neurological emergency disease, the early diagnosis of IS largely depends on a CT scan, which is time-consuming and hinders timely treatment. In addition, despite the wide application of drug thrombolysis and interventional thrombolysis, the therapeutic effect remains unsatisfactory due to the narrow therapeutic window and high level of risk. To date, permanent neurological impairment of stroke survivors is still a challenging public health issue worldwide. Therefore, the identification of new biomarkers with high sensitivity and specificity to reduce the time to diagnosis in the early stage is urgently needed to improve the prognosis of stroke patients.
The etiology of ischemic stroke is complex and multifactorial and is associated with hypertension [6,7], diabetes mellitus [8,9], smoking [10,11], atherosclerosis [12,13] and genetic factors [14,15]. The development of ischemic stroke is insidious, and patients do not seek medical help until the attack is acute. However, the molecular mechanism underlying ischemic stroke is not yet known. It is very important to deepen our understanding of ischemic stroke. Recent studies have found that many biomarkers are implicated in the pathogenesis of stroke. For example, Wang et al. found that FGF21 alleviated neuroinflammation following ischemic stroke by modulating the temporal and spatial dynamics of microglia/macrophages [16]. The extensive application and advancement of gene chip technology can help identify dysregulated genes, biological processes and signaling pathways in the pathogenesis of ischemic stroke.
A typical pathological change after IS is the disruption of the blood brain barrier, resulting in the recruitment of different infiltrating immune cells, including T cells, B cells, neutrophils, dendritic cells and macrophages, into the area of edema. On the one hand, immune cells secrete a large number of inflammatory cytokines and promote secondary neuroinflammation. On the other hand, immune cells play a protective role in BBB repair and angiogenesis. For example, regulatory T (Treg) cells promoted microglia-mediated white matter repair after ischemic stroke and could serve as a neuroprotective target for stroke recovery [17]. Zhang et al. found that macrophages play an important role in the phagocytic clearance of dead neurons after ischemic stroke and promote the resolution of inflammation in the brain [18]. Hence, immune cell regulation and the immune microenvironment after stroke are worthy of in-depth study.
In this study, we aimed to investigate novel biomarkers and the immune infiltration landscape in progressive IS based on comprehensive bioinformatic methods and machine learning. We initially downloaded IS-related datasets from the GEO database and performed differential expressed gene (DEG) analysis and WGCNA to identify candidate genes and key modules. Then, machine-learning algorithms were used to filter and identify hub biomarkers of IS, and a diagnostic model was developed based on the biomarkers. Finally, the immune infiltration landscape and correlation between different immune cells and hub genes were investigated, providing promising targets for future research.
2. Materials and methods
2.1. Data processing and differentially expressed gene (DEG) identification
The gene expression profiles in the GSE16561 (including 39 IS and 24 healthy samples) dataset was downloaded and extracted from the public database Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). The gene expression profile was normalized using the normalize Between Arrays function in the limma package [19]. Then, differential analysis between IS patients and healthy controls was performed after using the limma package in R software. An adjusted p value < 0.05 and log2fold change (FC) > 0.65 were set as cutoff thresholds to obtain the differentially expressed genes. Then, the heatmap and volcano plot were generated using the ggplot2 package in R software [20]. The study diagram and data preprocessing were illustrated in Fig. 1.
Fig. 1.
The workflow of the study design.
2.2. WGCNA network construction and module identification
The “WGCNA” package in R was used to analyze the gene expression patterns of multiple samples and screened candidate biomarkers or therapeutic targets [21]. First, the availability of all genes was measured, and then an adjacency matrix was constructed to examine the correlation strength between the nodes. Subsequently, we transformed the adjacency matrix into a topological overlap matrix (TOM) to quantitatively describe the similarity in nodes. Finally, key modules were identified after hierarchical clustering with a minimum size of 50 genes.
2.3. Functional enrichment analyses
Biological function enrichment of Gene Ontology (GO), Disease Ontology (DO), and Kyoto Enrichment of Genes and Genomes (KEGG) analyses were performed with cluster Profiler and the DOSE package in R [22,23]. Gene set enrichment analysis (GSEA) was used to analyze the variations in biological function and pathways between IS and healthy samples in depth. Statistical significance was set at a P value < 0.05.
2.4. Machine learning-based hub gene screening
Three machine learning algorithms, LASSO (least absolute shrinkage and selection operator) regression, SVM-RFE (support vector machine-recursive feature elimination) and random forest, were employed to screen for hub genes in IS. Then, genes selected by machine learning were intersected to obtain candidate hub genes for subsequent research. The expression levels of hub genes were examined in the GSE16561 dataset.
2.5. ROC evaluation and nomogram construction
A nomogram was constructed based on the expression of hub genes to predict the risk of IS development by using the “rms” package in R software. A calibration plot was plotted to evaluate the prediction accuracy of the model. Then, decision curves and clinical impact curves were plotted to determine whether clinical decisions based on the diagnostic model were beneficial to patients. The ROC curve was subsequently established to evaluate the diagnostic value of hub genes and the nomogram, and the area under the curve (AUC) and 95 % CI were calculated.
2.6. Hub gene validation
The GSE58294, GSE58294, and GSE66724 dataset, including 51 control samples and 94 IS samples, were downloaded and used for the validation of the candidate hub genes. A p value < 0.05 was considered statistically significant.
2.7. Assessment of immune cell infiltration
The CIBERSORT algorithm, a deconvolution algorithm, was then utilized to identify the infiltration of 22 different types of immune cells in IS patient tissues via transformation of the normalized gene expression matrix into the infiltrating immune cell composition. A histogram, a heatmap and boxplot diagrams were drawn to visualize the differences in immune cell infiltration between the control and ischemic stroke subjects. Spearman's correlation analysis was performed to with the hub genes and immune infiltrating cells, and lollipop charts were used to analyze the relationships between immune cells and hub genes.
3. Results
3.1. Identification of DEGs of IS
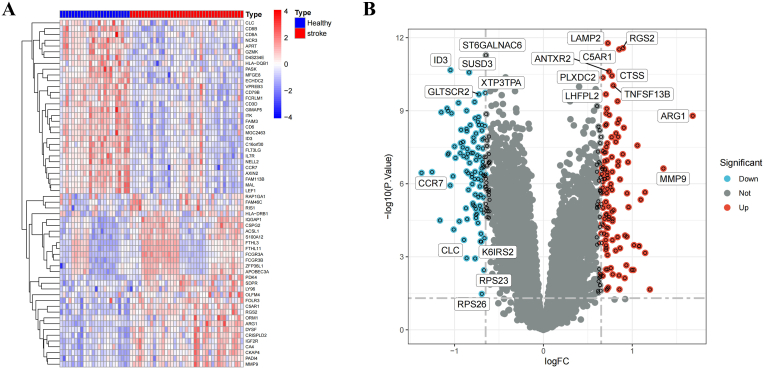
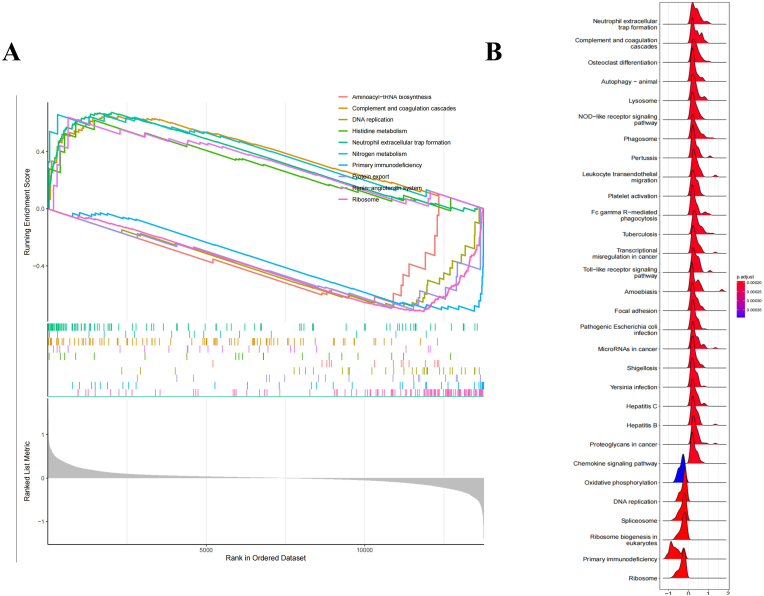
A total of 216 DEGs were identified in the IS patients, including 113 upregulated and 103 downregulated genes compared with the levels in the normal samples. The heatmap and volcano plot of the IS DEGs are shown in Fig. 2A and B, indicating an obvious difference between the IS patients and healthy controls. GSEA of DEGs showed that they were mainly involved in aminoacyl-tRNA biosynthesis, complement and coagulation cascades, DNA replication, histidine metabolism, neutrophil extracellular trap formation, nitrogen metabolism, primary immunodeficiency, protein export, the renin-angiotensin system and ribosomes (Fig. 3A). The GSEA ridge plot is visualized in Fig. 3B.
Fig. 2.
Identification of DEGs between healthy and ischemic stroke patients. (A&B) Heatmap and the Volcano plot of the DEGs.
Fig. 3.
GSEA analysis of DEGs. (A) GSEA plots depicting the enrichment of signal pathways based on the hallmark gene set. (B) Ridge plots of the most enriched 30 pathways of IS patients.
3.2. Significant module gene identification in IS via WGCNA
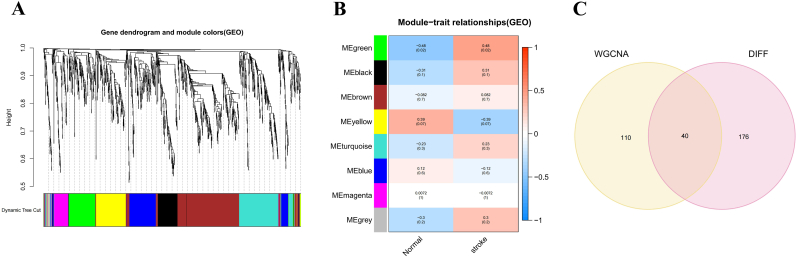
A scale-free coexpression network was established to identify the key modules for IS. As shown in Fig. 4A, a power of β = 9 (scale-free R2 = 0.9) was selected as the soft-thresholding parameter to produce a hierarchical clustering tree. Next, a total of 8 distinct gene modules with different colors were identified through hierarchical clustering, and the number of genes in each module was more than 50. As shown in Fig. 4B, the green module with the highest correlation coefficient was selected as the clinically significant module for further analysis (eigengene value = 0.48 and P = 0.02). After that, an interaction analysis was performed between DEGs and genes in the green module of interest, and a total of 40 important biomarkers were identified for subsequent enrichment analysis (Fig. 4C).
Fig. 4.
Identification of important biomarkers that participate in IS progression through WGCNA. (A) Clustering dendrogram of genes. (B) Module-trait associations where green module has the highest correlation with IS. (C) The Venn plot to identify common biomarkers between DEGs and WGCNA green module genes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Functional enrichment analysis
Functional analysis was performed to learn more about the biological function of the DEGs in the green module. As shown in Fig. 5A, the common DEGs were mainly enriched in the following biological processes (BPs): receptor-mediated endocytosis, negative regulation of lipid localization, negative regulation of MAP kinase activity, regulation of lipid catabolic process and response to peptide. The DEGs were mostly enriched in the following cellular components (CCs): specific granules, tertiary granules, secretory granule membranes, secretory granule lumens and cytoplasmic vesicle lumens. With regard to molecular function, the DEGs primarily participate in hydrolase activity, vinculin binding, monosaccharide binding, low-density lipoprotein particle receptor activity and cargo receptor activity. KEGG enrichment analysis showed that the common DEGs were enriched in the regulation of lipolysis in adipocytes, neutrophil extracellular trap formation, complement and coagulation cascades, shigellosis and amoebiasis (Fig. 5B). The results of the DO analysis revealed that these DEGs were linked to atherosclerosis (Fig. 5C).
Fig. 5.
Functional enrichment results of DEGs. (A) Significant enriched GO terms for DEGs. (BP, biological process; CC, cellular component; MF, molecular function). (B) The significantly enriched KEGG pathways. (C) DO analysis of target genes.
3.4. Diagnostic hub gene identification and validation
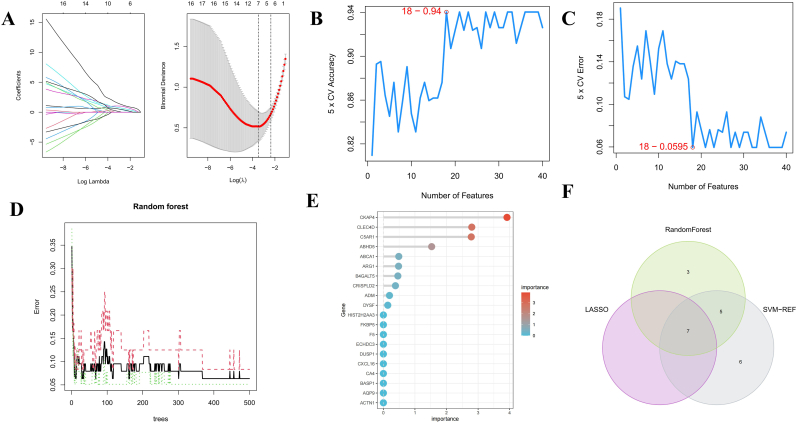
Three different machine learning algorithms were used to screen for reliable candidate hub genes in IS. A total of 7 genes were identified as diagnostic markers for IS by LASSO regression (Fig. 6A). Meanwhile, the top 25 DEGs with the highest accuracy and the lowest error according to SVM-RFE were selected (Fig. 6B&C). The gene importance of the DEGs was calculated (Fig. 5D), and the top 20 most important genes are displayed in Fig. 6E. Finally, after overlapping critical genes using a Venn diagram, 7 common potential genes, namely, ABCA1, ARG1, C5AR1, CKAP4, HMFN0839, SDCBP and TLN1, were selected as the candidate hub genes for IS using the above three algorithms (Fig. 6F).
Fig. 6.
Candidate genes selection by advanced machine learning. (A) Biomarkers screening by Lasso model. (B&C) SVM-RFE algorithm for feature gene selection. (D)The random forest algorithm shows the error in IS and control group. (E) Genes are ranked based on the importance score. (F)Venn diagram showing the feature genes shared by LASSO, SVM-RFE and random forest algorithms.
We further explored the expression levels of hub genes in IS patients and found that all hub genes were significantly upregulated in IS patients compared with the normal controls (Fig. 7A). Correlation analysis revealed that all hub genes were positively correlated and indicated that all genes might play similar biological roles in the pathogenesis of IS (Fig. 7B). Additionally, the analysis of the validation cohort dataset (GSE58294, GSE58294, and GSE66724) indicated that all hub genes were substantially upregulated in IS patients, while there were no statistically significant differences in the expression levels of C5AR1, SDCBP, and TLN1 (Fig. 7C).
Fig. 7.
The expression of DEGs in GSE16561 and external validation in GSE58294, GSE58294, and GSE66724. (A) Boxplot depicting gene expression in ischemic stroke group and healthy controls. (B) Correlation analysis of candidate DEGs. (C) External verification of DEGs in GSE58294, GSE58294, and GSE66724.
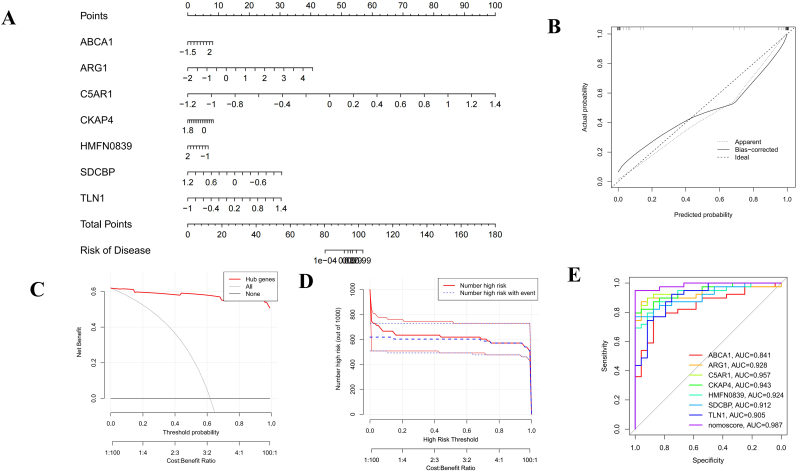
3.5. Nomogram construction and evaluation
A diagnostic nomogram model was established based on 7 candidate hub genes (Fig. 8A). Our calibration curve fit the ideal curve well and indicated reliable predictive ability in terms of stroke development (Fig. 8B). The clinical value of the model was evaluated with DCA, and the curve showed obvious net benefits of the predictive nomogram (Fig. 8C). The clinical impact curve demonstrated the excellent predictive potential of the nomogram (Fig. 8D). ROC curve analysis was used to assess the diagnostic specificity and sensitivity of each gene and the nomogram. The area under the curve (AUC) for each item was as follows: ABCA1 (AUC = 0.841), ARG1 (AUC = 0.928), C5AR1 (AUC = 0.957), CKAP4 (AUC = 0.943), HMFN0839 (AUC = 0.924), SDCBP (AUC = 0.912), and TLN1 (AUC = 0.905). The AUC for the diagnostic model was 0.987, confirming its excellent predictive performance (Fig. 8E).
Fig. 8.
Nomogram construction and predictive value evaluation. (A) Nomogram establishment for the diagnostic model of IS. (B) Calibration curves to evaluate the predictive power of the nomogram. (C) DCA curve to assess the clinical value of the diagnostic model. (D) The clinical impact curve indicating the excellent predictive probability of the diagnostic model. (E) The ROC curves estimating the diagnostic performance of characteristic genes.
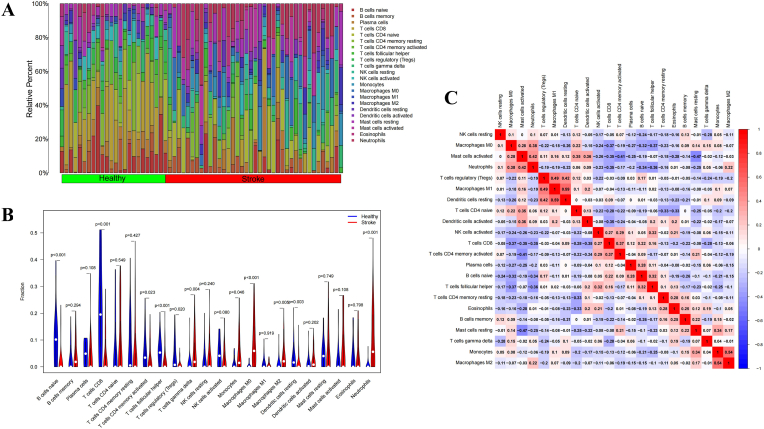
3.6. Immune infiltration analysis
The CIBERSORT algorithm was used to explore immune cell infiltration differences between the IS group and the healthy group. As shown in the histogram and violin diagram (Fig. 9A and B), the immune cell landscape in the IS patients was significantly different from that in the control group. Compared with those in the healthy control group, the IS group had increased fractions of gamma delta T cells, monocytes, M0 macrophages, M2 macrophages and neutrophils and clearly reduced percentages of naive B cells, CD8 T cells, activated memory CD4 T cells, follicular helper T cells, regulatory T cells (Tregs) and resting dendritic cells. Additionally, correlations between immune cells were studied, and we found that multiple immune cell pairs interacted either positively or negatively in the pathogenesis of IS. As shown in Fig. 9C, resting dendritic cells and M1 macrophages showed the most synergistic effect. Meanwhile, resting mast cells and activated mast cells displayed the most competitive effect.
Fig. 9.
Infiltration pattern of immune cells. (A) Heatmap of the 22 immune cell proportions in each sample. (B) Correlation heatmap of all 22 immune cells. (C) Violin plot of all 22 immune cell differentially infiltrated fractions.
We further explored the correlation between the hub genes and immune cells. The results indicated that the hub genes had very close relationships with different immune cells and that those genes might be potential immune-related targets in progressive IS. For example, as indicated in Fig. 10A, ABCA1 displayed significant positive correlations with neutrophils (r = 0.562), T cells gamma delta (r = 0.472), and macrophages M0 (r = 0.343) and significant negative correlations with CD8 T cells (r = −0.290), regulatory T cells (Tregs) (r = −0.336) and resting dendritic cells (r = −0.381). Other hub genes also had complex effects on immune cells, as displayed in Fig. 10B–G.
Fig. 10.
Analysis of the correlation between DEGs expression and the infiltrating immune cells. (A) ABCA1; (B) ARG1; (C) C5AR1; (D) CKAP4; (E) HMFN0839; (F) SDCBP; (G) TLN1.
4. Discussion
Ischemic stroke, as one of the leading threats to human health, has gained increasing attention worldwide. A sudden blockage of cerebral blood supply may result in detrimental neurological deficits and impose substantial social and economic burdens. Despite advances in medicines and recanalization therapy, timely and effective treatments are still lacking. It is estimated that approximately 0.7 million people suffer from acute ischemic stroke in the US annually [24] and 3.94 million in China. With the aging of the population, ischemic stroke has become a major public health problem. Therefore, there is an urgent need to identify new accurate target biomarkers for future experimental studies and the prediction of acute stroke in clinical practice. With the development of gene chip technology, bioinformatic analysis has been widely used to explore different disease target genes and to inform in-depth mechanistic studies.
In the present study, we first obtained 216 DEGs and found that 113 genes were upregulated and 103 genes were downregulated between ischemic stroke patients and control groups. Then, 40 important genes were identified by intersecting DEGs and the WGCNA key module. Subsequent GO enrichment analysis showed that all DEGs were mainly related to the biological processes of receptor-mediated endocytosis, negative regulation of lipid localization, negative regulation of MAP kinase activity, regulation of lipid catabolic processes and response to peptides, while KEGG enrichment analysis showed that DEGs were mostly associated with regulation of lipolysis in adipocytes, neutrophil extracellular trap formation, complement and coagulation cascades. Via the use of several machine learning algorithms, we identified 7 genes (ABCA1, ARG1, C5AR1, CKAP4, HMFN0839, SDCBP and TLN1) most strongly associated with ischemic stroke as candidate hub genes and developed a reliable nomogram model to predict ischemic stroke risk. Finally, we analyzed immune infiltration characteristics and found that immune dysfunction was likely implicated in the pathogenesis of ischemic stroke.
According to previous studies, ATP-binding cassette transporter member A1 (ABCA1) has been identified as a significant cholesterol transporter that contributes to maintaining cholesterol balance in the brain, thereby playing a beneficial role in cholesterol homeostasis [25]. A deficiency of ABCA1 or genetic dysfunction increases the risk of cerebrovascular diseases [26], and studies have suggested that ABCA1 deficiency exacerbates damage to the blood‒brain barrier and white matter after stroke [27,28]. In addition, ABCA1 was found to be upregulated in ruptured atherosclerotic plaques compared with nonruptured plaques as a neuroprotective mechanism via the suppression of inflammatory cytokine secretion [29]. Arginase 1 (Arg1) is an enzyme that is important for tissue repair under pathological conditions via microglia/macrophage phagocytosis. A study has shown that Arg1, as a downstream target of STAT6, can promote microglia/macrophage endocytosis and resolution of inflammation after stroke [30]. C5AR1 (also named CD88) is a potent inflammatory response inducer, and studies indicate that anti-inflammatory therapies targeting C5AR1 inhibition have obvious neuroprotective effects through neuroinflammation and apoptosis alleviation after stroke [[31], [32], [33], [34]]. Unlike the above three genes, there have been few studies on the correlations between CKAP4, HMFN0839, SDCBP, and TLN1 and stroke, and more research is needed.
We further investigated the biological functions of and critical pathways involving hub genes. Interestingly, both GO and KEGG studies indicated that the hub genes are associated with lipid localization, endocytosis and lipolysis, inflammation and coagulation cascades. It is well known that lipid accumulation and invasion into the innermost layer of endothelium of the large and medium-sized arteries contribute to atherosclerotic plaque formation. Based on the pathology, a blood clot caused by coagulation disorders leads to a sudden blockage of the blood supply to the brain tissue. Hence, anticoagulants and lipid-lowering drugs are the most important drugs in the treatment of stroke. For example, ApoE is the most important cholesterol transporter and is strongly implicated in ischemic stroke progression [35,36]. ABCA1 is one of the hub genes we found above, and a recent study further demonstrated that ABCA1 regulates ApoE and high-density lipoprotein (HDL) cholesterol and that the ABCA1/ApoE/HDL signaling pathway is beneficial for ischemic brain areas after stroke by facilitating myelination and oligodendrogenesis [37,38]. Hence, a lipid-lowering strategy targeting ABCA1/ApoE/HDL is promising for the management of ischemic stroke. To date, ischemic stroke is considered a thromboinflammatory disease. In addition to lipid-lowering therapy, anticoagulation and anti-inflammation treatment are important for stroke. Since contact activation and the kallikrein/kinin system (KKS) are procoagulant and proinflammatory response mechanisms, preventing the activation of the KKS pathway protects against stroke. A recent study indicated that as a contact-kinin inhibitor, sylvestin protects brain tissue from ischemic stroke by counteracting intracerebral thrombosis and inflammation and does not increase the risk of bleeding [39]. Overall, further in-depth investigation targeting lipid lowering and thrombo-inflammation relief is expected to improve the prognosis of stroke.
Finally, we studied the immune patterns in stroke. There is growing evidence that immune infiltration and the subsequent inflammatory response play significant roles in stroke development [40,41]. However, activation of the immune response has advantages and disadvantages. On the one hand, immune cells display protective effects by eliminating toxic substances and promoting brain tissue repair. On the other hand, a strong immune response and inflammatory cascade exacerbate tissue injury, while immunodepression leads to subsequent infection, both of which can lead to poor prognosis in stroke. Hence, immunomodulatory therapies are now recognized as the most promising directions in stroke treatment. Meanwhile, the phenotypes of immune cells at different stages of stroke vary and may have dual effects on the stroke outcome. For example, studies have shown that microglia and infiltrated macrophages initially polarize toward a neuroprotective anti-inflammatory phenotype after stroke. However, over time, they gradually transform into a detrimental proinflammatory phenotype [[42], [43], [44]]. Melatonin has been demonstrated as a protective agent against stroke by switching the phenotype of microglia/macrophages to an anti‐inflammatory phenotype via the STAT3 pathway [45]. Similarly, rhFGF21 is considered an anti-inflammatory agent for stroke treatment by modulating the temporal and spatial dynamics of microglia/macrophages in ischemic brain areas16. Therefore, knowledge of the immune cell types, phenotypes and biological functions of immune cells at different stages of stroke may provide a theoretical basis for immune therapy for stroke. The development of bioinformatics, especially the wide application of single-cell sequencing technology, is expected to solve the above problems. Finally, although the disruption of the blood‒brain barrier provides opportunities for drug entry, how to efficiently deliver drugs to ischemic regions is also a question that is worth exploring. Currently, an increasing number of studies have found that immune cell membrane-packaged drugs and immune cell-derived exosomes can efficiently pass through the blood‒brain barrier and reach the ischemic area to promote the recovery of nerve cells [[46], [47], [48]].
In conclusion, although several novel biomarkers were identified beyond those in previous studies, there are still several limitations to be mentioned. First, our study is based on public data, and the results should be verified in vivo and in vitro. Second, the sample size of our study is small, which may result in statistical error. Third, only genetic factors were included when constructing the nomogram. Other clinical information, such as age, sex, BMI, serum lipid levels and smoking status, should be incorporated to improve the accuracy of our nomogram.
5. Conclusion
In summary, we identified 7 hub genes, the key regulatory pathways and immune infiltration characteristics of ischemic stroke based on comprehensive bioinformatic analysis. Furthermore, in our study, we successfully constructed a nomogram that can precisely predict the occurrence of ischemic stroke. Our findings may provide potential targets and valuable perspectives for the future diagnosis and treatment of stroke.
CRediT authorship contribution statement
Shiyu Hu: Data curation, Methodology, Writing – original draft. Jingjing Cai: Methodology. Sizhan Chen: Data curation, Methodology. Yang Wang: Conceptualization, Writing – review & editing. Lijie Ren: Conceptualization, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare there are no competing interests.
Acknowledgements
This study was supported by the Shenzhen Science and Technology Innovation committee (GJHZ20200731095602009), Natural Science Foundation of Guangdong Province (2023A1515010169) and clinical research projects of Shenzhen Second People's Hospital (No. 20203357015).
Contributor Information
Yang Wang, Email: helenwy1978@163.com.
Lijie Ren, Email: 13631605966@126.com.
Data availability
Data will be made available on request.
References
- 1.Feigin V.L., et al. World stroke organization (WSO): global stroke fact sheet 2022. Int. J. Stroke : oficial journal of the International Stroke Society. 2022;17:18–29. doi: 10.1177/17474930211065917. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y.J., et al. China stroke statistics: an update on the 2019 report from the national center for healthcare quality management in neurological diseases, China national clinical research center for neurological diseases, the Chinese stroke association, national center for chronic and non-communicable disease control and prevention, Chinese center for disease control and prevention and institute for global neuroscience and stroke collaborations. Stroke Vascul. Neurol. 2022;7:415–450. doi: 10.1136/svn-2021-001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsao C.W., et al. Heart disease and stroke statistics-2023 update: a report from the American heart association. Circulation. 2023;147:e93–e621. doi: 10.1161/cir.0000000000001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tu W.J., et al. Estimated burden of stroke in China in 2020. JAMA Netw. Open. 2023;6 doi: 10.1001/jamanetworkopen.2023.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamford J., Sandercock P., Dennis M., Burn J., Warlow C. Classification and natural history of clinically identifiable subtypes of cerebral infarction. Lancet (London, England) 1991;337:1521–1526. doi: 10.1016/0140-6736(91)93206-o. [DOI] [PubMed] [Google Scholar]
- 6.C ipolla M.J., Liebeskind D.S., Chan S.L. The importance of comorbidities in ischemic stroke: impact of hypertension on the cerebral circulation. J. Cerebr. Blood Flow Metabol. : oficial journal of the International Society of Cerebral Blood Flow and Metabolism. 2018;38:2129–2149. doi: 10.1177/0271678x18800589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun P., et al. Combined effect of hypertension and hyperuricemia on ischemic stroke in a rural Chinese population. BMC Publ. Health. 2021;21:776. doi: 10.1186/s12889-021-10858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maida C.D., et al. Diabetes and ischemic stroke: an old and new relationship an overview of the close interaction between these diseases. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23042397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Georgakis M.K., et al. Diabetes mellitus, glycemic traits, and cerebrovascular disease: a mendelian randomization study. Neurology. 2021;96:e1732–e1742. doi: 10.1212/wnl.0000000000011555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markidan J., et al. Smoking and risk of ischemic stroke in young men. Stroke. 2018;49:1276–1278. doi: 10.1161/strokeaha.117.018859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkins B.T., Brown R.C., Davis T.P. Smoking and ischemic stroke: a role for nicotine? Trends Pharmacol. Sci. 2002;23:78–82. doi: 10.1016/s0165-6147(02)01893-x. [DOI] [PubMed] [Google Scholar]
- 12.Koton S., et al. Association of ischemic stroke incidence, severity, and recurrence with dementia in the atherosclerosis risk in communities cohort study. JAMA Neurol. 2022;79:271–280. doi: 10.1001/jamaneurol.2021.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau K.K., et al. Low-density lipoprotein cholesterol and risk of recurrent vascular events in Chinese patients with ischemic stroke with and without significant atherosclerosis. J. Am. Heart Assoc. 2021;10 doi: 10.1161/jaha.121.021855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linden A.B., et al. Genetic associations of adult height with risk of cardioembolic and other subtypes of ischemic stroke: a mendelian randomization study in multiple ancestries. PLoS Med. 2022;19 doi: 10.1371/journal.pmed.1003967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traylor M., et al. Genetic basis of lacunar stroke: a pooled analysis of individual patient data and genome-wide association studies. Lancet Neurol. 2021;20:351–361. doi: 10.1016/s1474-4422(21)00031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D., et al. FGF21 alleviates neuroinflammation following ischemic stroke by modulating the temporal and spatial dynamics of microglia/macrophages. J. Neuroinflammation. 2020;17:257. doi: 10.1186/s12974-020-01921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L., et al. Treg cell-derived osteopontin promotes microglia-mediated white matter repair after ischemic stroke. Immunity. 2021;54:1527–1542.e1528. doi: 10.1016/j.immuni.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W., et al. Macrophages reprogram after ischemic stroke and promote efferocytosis and inflammation resolution in the mouse brain. CNS Neurosci. Ther. 2019;25:1329–1342. doi: 10.1111/cns.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie M.E., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter W., Sánchez-Cabo F., Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 21.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G., Wang L.G., Yan G.R., He Q.Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2015;31:608–609. doi: 10.1093/bioinformatics/btu684. [DOI] [PubMed] [Google Scholar]
- 24.Ajoolabady A., et al. Targeting autophagy in ischemic stroke: from molecular mechanisms to clinical therapeutics. Pharmacol. Therapeut. 2021;225 doi: 10.1016/j.pharmthera.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S., et al. Genetic variations in ABCA1/G1 associated with plasma lipid levels and risk of ischemic stroke. Gene. 2022;823 doi: 10.1016/j.gene.2022.146343. [DOI] [PubMed] [Google Scholar]
- 26.Cao X.L., Yin R.X., Huang F., Wu J.Z., Chen W.X. Chromosome 9p21 and ABCA1 genetic variants and their interactions on coronary heart disease and ischemic stroke in a Chinese han population. Int. J. Mol. Sci. 2016;17:586. doi: 10.3390/ijms17040586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui X., et al. Deficiency of brain ATP-binding cassette transporter A- 1 exacerbates blood-brain barrier and white matter damage after stroke. Stroke. 2015;46:827–834. doi: 10.1161/strokeaha.114.007145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ElAli A., Hermann D.M. Liver X receptor activation enhances blood-brain barrier integrity in the ischemic brain and increases the abundance of ATP-binding cassette transporters ABCB1 and ABCC1 on brain capillary cells. Brain Pathol. 2012;22:175–187. doi: 10.1111/j.1750-3639.2011.00517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heo S.H., et al. Differences between the molecular mechanisms underlying ruptured and non-ruptured carotid plaques, and the significance of ABCA1. Journal ofstroke. 2018;20:80–91. doi: 10.5853/jos.2017.02390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai W., et al. STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI insight. 2019;4 doi: 10.1172/jci.insight.131355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., et al. Guhong injection prevents ischemic stroke-induced neuro-inflammation and neuron loss through regulation of C5ar1. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.818245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y., et al. C5aR1 mediates the progression of inflammatory responses in the brain of rats in the early stage after ischemia and reperfusion. ACS Chem. Neurosci. 2021;12:3994–4006. doi: 10.1021/acschemneuro.1c00244. [DOI] [PubMed] [Google Scholar]
- 33.Thundyil J., et al. C5a receptor (CD88) inhibition improves hypothermia-induced neuroprotection in an in vitro ischemic model. NeuroMolecular Med. 2012;14:30–39. doi: 10.1007/s12017-012-8167-0. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y.W., et al. Schisantherin A attenuates ischemia/reperfusion-induced neuronal injury in rats via regulation of TLR4 and C5aR1 signaling pathways. Brain Behav. Immun. 2017;66:244–256. doi: 10.1016/j.bbi.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 35.Pendlebury, S. T., Poole, D., Burgess, A., Duerden, J. & Rothwell, P. M. APOE-ε4 Genotype and Dementia before and after Transient Ischemic Attack and Stroke:. [DOI] [PMC free article] [PubMed]
- 36.Satizabal C.L., et al. APOE and the association of fatty acids with the risk of stroke, coronary heart disease, and mortality. Stroke. 2018;49:2822–2829. doi: 10.1161/strokeaha.118.022132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L., et al. ABCA1/ApoE/HDL signaling pathway facilitates myelination and oligodendrogenesis after stroke. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21124369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui X., et al. ABCA1/ApoE/HDL pathway mediates GW3965-induced neurorestoration after stroke. Stroke. 2017;48:459–467. doi: 10.1161/strokeaha.116.015592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z., et al. Novel contact-kinin inhibitor sylvestin targets thromboinflammation and ameliorates ischemic stroke. Cell. Mol. Life Sci. : CMLS. 2022;79:240. doi: 10.1007/s00018-022-04257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu R., et al. Comprehensive landscape of immune infiltration and aberrant pathway activation in ischemic stroke. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.766724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Y., Yang S., He Q., Zhang D., Chang J. The role of immune cells in post-stroke angiogenesis and neuronal remodeling: the known and the unknown. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.784098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanazawa M., Ninomiya I., Hatakeyama M., Takahashi T., Shimohata T. Microglia and monocytes/macrophages polarization reveal novel therapeutic mechanism against stroke. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang C.T., Wu W.F., Deng Y.H., Ge J.W. Modulators of microglia activation and polarization in ischemic stroke. Mol. Med. Rep. 2020;21:2006–2018. doi: 10.3892/mmr.2020.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wicks E.E., et al. The translational potential of microglia and monocyte-derived macrophages in ischemic stroke. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.897022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z.J., et al. Melatonin protects against ischemic stroke by modulating microglia/macrophage polarization toward anti-inflammatory phenotype through STAT3 pathway. CNS Neurosci. Ther. 2019;25:1353–1362. doi: 10.1111/cns.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li C., et al. Macrophage-disguised manganese dioxide nanoparticles for neuroprotection by reducing oxidative stress and modulating inflammatory microenvironment in acute ischemic stroke. Adv. Sci. 2021;8 doi: 10.1002/advs.202101526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song Y., et al. M2 microglia-derived exosomes protect the mouse brain from ischemia-reperfusion injury via exosomal miR- 124. Theranostics. 2019;9:2910–2923. doi: 10.7150/thno.30879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian T., et al. Targeted delivery of neural progenitor cell-derived extracellular vesicles for anti-inflammation after cerebral ischemia. Theranostics. 2021;11:6507–6521. doi: 10.7150/thno.56367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.