Abstract
Background
Cancer growth is significantly influenced by processes such as pyroptosis, apoptosis, and necroptosis that underlie PANoptosis, a proinflammatory programmed cell death. Several studies have examined the long non-coding RNAs (lncRNAs) associated with pancreatic adenocarcinoma (PAAD). However, the predictive value of lncRNAs related to PANoptosis for PAAD has not been established.
Methods
The Clinical Genome Atlas database was used to obtain the transcriptome 、clinical data and the corresponding mutation data of the patients with PAAD in this study. The least absolute shrinkage and selection operator regression analysis was employed to obtain prognosis-related lncRNAs for constructing a risk signature. According to the median risk score of the signature, patients with PAAD were grouped into low- and high-risk groups to further compare the survival prognosis of different risk groups. Time-dependent receiver operating characteristic curves, c-index analysis, nomograms, principal component analysis and univariate Cox and multivariate Cox regression were performed for the internal validation of the signature. In addition, enrichment analysis of different genes was performed using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. Lastly, differences in tumor mutation burden (TMB), immune function, tumor immune dysfunction and rejection (TIDE), and drug response were determined for the two risk groups.
Results
The signature was constructed with six PANoptosis-related lncRNAs (AC067817.2、LINC02004、AC243829.1、AC092171.5、AP005233.2、AC004687.1) that predicted the prognosis of the patients with PAAD. Survival curves showed that patients in the two risk groups had statistically significant differences in prognosis (P < 0.05), and multi-cox regression analysis identified risk score as an independent risk factor for PAAD prognosis, and internal validation of nomograms showed high confidence in the signature. GO and KEGG enrichment analysis showed functional and pathway differences between the high- and low-risk groups. TMB evaluation demonstrated that patients in the high-risk group had a higher frequency of mutations. The TIDE score indicated that the high-risk group had a lower risk of immunotherapy escape and better immunotherapy outcomes. Additionally, the two risk groups revealed significantly different responses to 11 anticancer drugs.
Conclusion
We identified a novel risk signature for PANoptosis-related lncRNAs, which is a standalone prognostic indicator for PAAD. The PANoptosis-related lncRNA risk signature may be relevant for immunotherapy and a therapeutic target for PAAD.
Keywords: Pancreatic adenocarcinoma, PANoptosis, Long non-coding RNAs, Prognostic signature, Immunotherapy
Highlights
-
•
Bioinformatics analysis of the pancreatic adenocarcinomas TCGA database identified 266 lncRNAs associated with PANoptosis.
-
•
Six lncRNAs, including AC067817.2, LINC02004, and AC243829.1, are prognostic-related risk markers for pancreatic adenocarcinomas.
-
•
Six-lncRNAs prognostic signatures have been constructed which can predict the survival and prognosis of patients with pancreatic adenocarcinomas.
1. Introduction
Pancreatic cancer is one of the most common malignant tumors of the gastrointestinal tract and it has the seventh-highest mortality rate globally [1]. Approximately 85% in pancreatic cancers are pancreatic adenocarcinomas (PAADs) [2]. The onset of PAAD is insidious and mostly advanced when it is detected. The treatments for PAAD are usually surgery, chemotherapy, and targeted therapy. But the prognosis of PAAD is poor, almost 80% of patients suffer recurrence after resection [3], and the 5-year survival rate is only 6% [4], with a median survival duration as short as 4–5 months [5]. Therefore, biomarkers for predicting the prognosis and the survival of patients with PAAD are urgently needed.
PANoptosis is a proinflammatory programmed cell death. The processes underlying PANoptosis include pyroptosis, apoptosis, and necroptosis. PANoptosis is regulated by the complex PANoptosome [6,7]. Several previous studies have recently reported that PANoptosis and cancer progression are closely related [[8], [9], [10]]. PANoptosis affects the function of tumor-associated factors and pathways and plays an essential role in immunotherapy [11]. There are several PANoptosis regulators, including TNF-α [12], IFN-γ [13], ADAR1 [14], and ZBP1 [15]. For example, the combined effects of TNF-α and IFN-γ can induce PANoptosis and cancer cell death, which highlights the targeted role of PANoptosis in antitumor therapy [12]. Karki et al. reported that Adar1fl/fl LysMcre mice can inhibit the development of melanomas and colon carcinogenesis [14]. However, tumorigenesis was restored when the Zα2 structural domain of ZBP1 was absent in these mice. Several studies have reported that ADAR1 inhibits PANoptosis by interacting with the Zα2 structural domain of ZBP1, which is related with tumorigenesis [14,15]. However, studies on the regulatory mechanism of PANoptosis in PAAD are lacking. Therefore, identifying the mechanisms underlying PANoptosis that are associated with PAAD is crucial for the development of therapies.
Long non-coding RNAs (lncRNAs) are non-coding RNAs with more than 200 nucleotides in length, and they are involved in many biological functions [16]. LncRNAs have different functions and levels of expression in various tumors, and they are considered promising biomarker candidates and may be targeted for tumor therapy [[17], [18], [19], [20]]. Several PAAD-related lncRNAs influence tumorigenesis, tumor progression, and prognosis [[21], [22], [23]]. Additionally, current research has revealed that the PANoptosis-related lncRNA SNHG7 is associated with colonic adenocarcinoma (COAD). The lncRNA SNHG7 may be crucial for the apoptosis, metastasis, and treatment resistance of COAD cells and is a potential therapeutic target for COAD [24]. The function of lncRNAs associated with PANoptosis in PAAD has not been established. Further research is required to determine how PANoptosis-related lncRNAs affect PAAD treatment and prognosis.
Our study developed a predictive risk model based on PANoptosis-related lncRNAs for predicting the prognosis and immunotherapeutic landscapes of patients with PAAD. The risk signature was predictive of the tumor mutation burden (TMB), tumor immune dysfunction and exclusion score (TIDE), immune-related function, and drug response of patients with PAAD. Our risk signature is a valid prognostic biomarker that can guide the development of immunotherapy.
2. Methods
2.1. Obtain the PAAD-related data
The clinicopathological parameters associated with PAAD were drafted from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/repository). We removed samples with unknown survival status and duration. The mRNA and lncRNA expression data were obtained from the transcriptomic data using Strawberry Perl.
2.2. Collation of PANoptosis gene-associated lncRNAs
We obtained 24 genes (Supplementary Table 1) associated with PANoptosis based on several reports in the literature [[7], [8], [9], [10],[13], [14], [15],[25], [26], [27], [28]]. The “limma” package was utilized to derive expression data related with 24 PANoptosis genes. The data of lncRNAs associated with PANoptosis genes were extracted by co-expression analysis (Pearson correlation coefficient >0.4, P < 0.001). Subsequently, the correlation data of PANoptosis-related genes with lncRNAs were analyzed with the “ggplot2” “ggalluvial” and “dplyr” package and plotted as Sankey plots.
2.3. Derivation of a prognostic risk signature
Patients were randomised into training and testing groups in a 1:1 ratio. The univariate Cox (uni-Cox) regression analysis was employed to screen lncRNAs related to prognosis and a forest plot was established according to the results (P < 0.05) [29]. The least absolute shrinkage and selection operator (LASSO) regression analyses were performed to screen PAAD-related lncRNAs for model building. Then, according to the formula, the risk score was calculated for each PAAD patient: risk score = (coefficient lncRNA1*lncRNA1 expression) + (coefficient lncRNA2*lncRNA2 expression) + … + (coefficient lncRNAn*lncRNAn expression). The coefficients represent coefficients in a multifactorial Cox analysis and the expressions represent lncRNA expression levels [30]. Patients were divided into high- and low-risk groups based on the median risk score of the training group. The correlation of model lncRNAs with PANoptosis genes was analyzed using the “ggplot2”, “tidyverse” and “ggExtra” packages in R language and plotted as a correlation heat map.
2.4. Validation of the prognostic risk signature
The “survival”, “pheatmap” and “survminer” software packages were applied to the training, testing and entire groups for survival analysis, and risk heat map, risk curve, survival status map and survival curve were constructed separately for each group. In addition, the “survival”, “survminer” and “timeROC” packages were employed to construct clinical- and time-dependent receiver operating characteristic (ROC) curves to validate the accuracy and reliability of the signature. Uni-Cox and multivariate Cox regression (multi-Cox) analyses were performed to estimate the capability of the risk model in predicting patient prognosis. Furthermore, “pec”, “survival” and “rms” were utilized to calculation the consistency index (c-index) to evaluate the prognostic signature accuracy. The “limma” and “scatterplot3d” packages were employed to apply principal component analysis (PCA) to the model to visualize the two risk populations' distribution. Additionally, “survival” and “survminer” packages were employed to structure survival curves for patients with early- and late-stage PAAD to determine whether the model was applied to different clinical subgroups of patients.
2.5. Nomogram construction
The “survival”, “regplot” and “rms” packages were utilized to construct Nomograms that predict the prognosis of PAAD patients after 1, 3 and 5 years [31]. And Hosmer-Lemeshow test calibration curve (method = "bootstrap”, B = 1000) was employed to assess the accuracy and reliability of Nomogram.
2.6. Analysis of prognostic risk signature regarding functional and pathways
The “limma” package was applied to identify genes differentially expressed between different risk groups. The “enrichplot”, “clusterProfiler”, “ggplot2”, “GOplot” and “org.Hs.eg.db” were utilized for gene ontology (GO) enrichment analysis to determine the functional enrichment of differential genes [32]. Furthermore, the “enrichplot”, “clusterProfiler”, “ggplot2”, “circlize”, “RColorBrewer”, “dplyr”, “ComplexHeatmap” and “org.Hs.eg.db” packages were employed for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
2.7. Tumor mutation burden
From the TCGA database, the TMB data of the patients with PAAD were downloaded and the TMB data were collated with Strawberry Perl. The “ggpubr” and “limma” packages were utilized to identify TMB differences between the high- and low-risk groups, and to visualize the TMB status of the two risk groups. Subsequently, the optimal threshold of TMB was obtained by R software, and according to the threshold, patients were categorized into high- and low-TMB groups. The “survminer” and “survivor” were applied to build survival curves for the patients in the high- and low-TMB groups, and Kaplan-Meier (KM) survival curves for the patients in the high- and low-TMB groups combined with the high- and low-risk groups. Moreover, “maftools” was utilized to structure waterfall plots of gene mutation rates in different risk groups.
2.8. Immunity and drug sensitivity
R software was employed to analyze the differences in immune-related functions between risk groups. The TIDE algorithm was utilized to predict the efficacy of immunotherapy [33]. *P < 0.05, **P < 0.01, and ***P < 0.001. To identify effective therapeutic agents for PAAD, the R packages “pRRophetic” and “ggpubr” were applied to compute the half-maximal inhibitory concentration (IC50) of each drug in both risk groups [34]. In two risk groups, drugs with different IC50s were represented using box plots (P < 0.001).
3. Results
3.1. Acquisition of PAAD-related data
We retrieved the transcriptome data for four normal and 179 PAAD samples from the TCGA database. Afterwards, lncRNAs were separated from mRNAs in the sample data. By co-expression analysis of lncRNAs and PANoptosis-related genes, we found PANoptosis was significantly associated with 266 lncRNAs (|r| > 0.4, P < 0.001) (Fig. 1A).
Fig. 1.
PANoptosis-related lncRNAs in PAAD. (A) Co-expression network plot of PANoptosis -associated genes and lncRNAs. (B) The 49 PANoptosis-associated lncRNAs extracted by univariate Cox regression analysis. (C-D) Least absolute shrinkage and selection operator coefficient and partial likelihood deviance of the prognostic signature. (E) The correlation between the expression of six lncRNAs and PANoptosis-associated genes. *P < 0.05, **P < 0.01, ***P < 0.001.
3.2. Creation and validation of the PAAD prognostic risk signature
The data from TCGA downloads were randomly classified into training and testing groups, with no significant clinical differences between them (Table 1). As shown in the forest plot (Fig. 1B), the results of the uni-Cox regression analysis indicated that 49 PANoptosis-related lncRNAs were associated with patients' Overall Survival (OS) (P < 0.05). Subsequently, six PANoptosis-associated lncRNAs were selected for signature construction by LASSO regression analysis (Table 2) (Fig. 1C and D). Risk scores were calculated using screened lncRNAs by LASSO regression analysis. Risk score = AC067817.2 × (−1.081) + LINC02004 × (0.561) + AC243829.1 × (−0.803) + AC092171.5 × (−0.986) + AP005233.2 × (0.457) + AC004687.1 × (−0.557). According to the median risk score, patients were further classified into high- and low-risk groups. The relationship between the expression of the six lncRNAs and PANoptosis-associated genes was significant (P < 0.05), in a heat map format (Fig. 1E). In addition, we found that in the training group, the prognosis of the low-risk group was better than the high-risk group, as shown on the expression heat map, risk score distribution, survival time and survival status maps (Fig. 2A–E). According to the expression heat map, we found LINC02004, AP005233.2 were high-risk lncRNAs with increased expression as the risk score increased. And AC067817.2, AC243829.1, AC092171.5 and AC004687.1 were low-risk lncRNAs (Fig. 2A). Survival time decreased with increasing risk score (Fig. 2B and C). KM curves demonstrated (P < 0.001) that the high-risk group had poorer OS and Progression-Free-Survival (PFS) than the low-risk group (Fig. 2D and E).
Table 1.
Comparison of clinicopathological features between the training and testing sets.
| Covariates | Type | Total | Testing set | Training set | p-value |
|---|---|---|---|---|---|
| Age | ≤65 | 94 (52.81%) | 46 (51.69%) | 48 (53.93%) | 0.8807 |
| >65 | 84 (47.19%) | 43 (48.31%) | 41 (46.07%) | ||
| Gender | FEMALE | 80 (44.94%) | 43 (48.31%) | 37 (41.57%) | 0.4512 |
| MALE | 98 (55.06%) | 46 (51.69%) | 52 (58.43%) | ||
| Grade | G1-2 | 126 (70.79%) | 70 (78.65%) | 56 (62.92%) | 0.0739 |
| G3-4 | 50 (28.09%) | 19 (21.35%) | 31 (34.83%) | ||
| unknown | 2 (1.12%) | 0 (0%) | 2 (2.25%) | ||
| Stage | Stage I-II | 168 (94.38%) | 85 (95.51%) | 83 (93.26%) | 0.0897 |
| Stage III-IV | 7 (3.94%) | 3 (3.37%) | 4 (4.49%) | ||
| unknow | 3 (1.69%) | 1 (1.12%) | 2 (2.25%) | ||
| T | T1-2 | 31 (17.41%) | 17 (19.1%) | 14 (15.73%) | 0.2828 |
| T3-4 | 145 (81.47%) | 71 (79.78%) | 74 (83.15%) | ||
| unknow | 2 (1.12%) | 1 (1.12%) | 1 (1.12%) | ||
| M | M0 | 80 (44.94%) | 41 (46.07%) | 39 (43.82%) | 0.678 |
| M1 | 4 (2.25%) | 3 (3.37%) | 1 (1.12%) | ||
| unknow | 94 (52.81%) | 45 (50.56%) | 49 (55.06%) | ||
| N | N0 | 49 (27.53%) | 31 (34.83%) | 18 (20.22%) | 0.0599 |
| N1 | 124 (69.66%) | 57 (64.04%) | 67 (75.28%) | ||
| unknow | 5 (2.81%) | 1 (1.12%) | 4 (4.49%) |
Table 2.
Long non-coding RNA signature models associated with PANoptosis.
| PANoptosis LncSig | Coef | HR | HR (95%CI) | p-value |
|---|---|---|---|---|
| AC067817.2 | −1.081486833 | 0.431 | 0.262–0.708 | <0.001 |
| LINC02004 | 0.561159984 | 1.700 | 1.071–2.697 | 0.024 |
| AC243829.1 | −0.803712507 | 0.394 | 0.169–0.920 | 0.031 |
| AC092171.5 | −0.98609502 | 0.628 | 0.419–0.942 | 0.024 |
| AP005233.2 | 0.457954322 | 1.201 | 1.049–1.375 | 0.008 |
| AC004687.1 | −0.557257657 | 0.566 | 0.386–0.830 | 0.004 |
HR, hazard ratio; CI, confidence interval.
Fig. 2.
Derivation and selection of the PANoptosis-associated lncRNAs signature in the training set. (A) Heat map of expression of the six lncRNAs in the training set. (B) Survival time and status in the training set. (C) Risk score distribution in the training set. (D) Kaplan–Meier curve for PFS in the training set. (E) Kaplan–Meier curve for OS in the training set.
3.3. Internal validation of the PAAD prognostic signature
Subsequently, the model was evaluated with the testing and all sets, and the developed expression heat map, risk score distribution, survival curve, survival status and survival time were consistent with the training set analysis (Fig. 3A–J).
Fig. 3.
Validation of the PANoptosis-associated lncRNAs signature. (A-B) Heat map of expression of the six lncRNAs in the testing, and entire sets. (C-D) Survival time and status in the testing, and entire sets. (E-F) Risk score distribution in the testing, and entire sets. (G-H) Kaplan–Meier curve for PFS in the testing, and entire sets. (I-J) Kaplan–Meier curve for OS in the testing, and entire sets.
Furthermore, we performed uni- and multi-Cox regression analyses to determine whether the signature could be utilized as an independent prognostic factor from other clinical characteristics. According to the results of uni-Cox regression analysis, age, grade and risk score were significantly correlated with the survival time of patients (P < 0.05); the risk score could be an independent prognostic predictor for patients with PAAD based on multi-Cox regression analysis (P < 0.001).The uni- and multi-Cox regression analyses’ hazard ratios (HR) were 1.068 and 1.062, and 95% confidence intervals (CI) of (1.035–1.103) (P < 0.001) and (1.027–1.098) (P < 0.001) (Fig. 4A and B). The ROC curves for the entire dataset at 1, 3, and 5 years were examined (Fig. 4C). The AUCs were 0.772, 0.805, and 0.885, respectively. Clinically relevant ROC curves were developed using the risk, age, gender, grade, and stage as indicators (Fig. 4D), and their AUCs were respectively 0.772, 0.544, 0.563, 0.599, and 0.483; the risk score had the most remarkable AUC and most accurate prediction among these clinical factors.
Fig. 4.
Assessment of the predictive signature. (A) Forest plot for univariate Cox and (B) multivariate Cox regression analysis. (C) ROC curves of 1-, 3-, and 5-year survival for the predictive signature. (D) Comparison of the prediction accuracy of the risk signature with risk, age, gender, grade and stage. (E) Nomogram for predicting the 1-, 3-, and 5-year survival of patients with PAAD. (F) The calibration curves for 1-, 3-, and 5-year OS.
The c-index analysis was further performed to assess the predictive effect of the risk model. We found that the risk score had the highest c-index (Fig. S1A). The PCA results showed that model lncRNAs discriminated significantly the high- and low-risk groups in patients with PAAD (Fig. S1B–D). Subsequently, we found a significant difference in survival time between the two risk groups, when the risk model was applied to patients with early- and late-stage PAAD (P < 0.05) (Figs. S1E and F).
3.4. Construction of nomogram
According to the clinicopathological parameters and risk scores of patients with PAAD, a nomogram was constructed to forecast the prognosis of patients. The corresponding scores for clinicopathological data and risk scores of patients with PAAD were obtained from the nomogram, while the total score was used as a tool to predict prognosis (Fig. 4E). We have also built a calibration curve to further clarify the consistency between expected and actual survival. Fig. 4F shows that the model has a high accuracy in predicting patient survival.
3.5. Enrichment analysis of the PAAD prognostic signature for function and pathways
GO analysis revealed that the biological functions of differentially expressed genes (DEGs) differed significantly between risk groups (Fig. 5A–D). DEGs were mainly enriched in the functions of positive regulation of cell activation, plasma membrane signaling receptor complexes and external side of plasma membrane. It suggests that DEGs may regulate their proliferation by regulating cell function.
Fig. 5.
Gene Ontology Analysis. (A-B) Barplot graph for GO enrichment. (C) Bubble graph for GO enrichment. (D) Circos graph for GO enrichment.
KEGG analysis displayed the DEGs enrichment in the pathway (Fig. 6A–C). The outcomes indicated that DEGs were mainly enriched in cytokine-cytokine receptor interactions, cell adhesion molecules, and hematopoietic cell lineages. The findings may provide guidance in developing targeted clinical therapies for patients with PAAD in different risk populations.
Fig. 6.
Kyoto Encyclopedia of Genes and Genomes Analysis. (A) Barplot graph for KEGG enrichment. (B) Bubble graph for KEGG enrichment. (C) Circos graph for KEGG enrichment.
3.6. TMB analysis
The violin plot illustrates that the TMB differences in patients with PAAD among the different risk groups. The results revealed that TMB was more prevalent in the high-risk group than in the low-risk group (Fig. 7A). According to TMB levels, patients were classified into high- and low-TMB groups.
Fig. 7.
Correlation of risk model with tumor mutation burden in PAAD. (A) Violin plot of TMB status in the high- and low-risk groups (B) Kaplan Meier curve of H-TMB and L-TMB. (C) Kaplan-Meier curve of TMB + Risk. (D) Mutant gene waterfall plot in the high-risk group. (E) Mutant gene waterfall plot in the low-risk group.
The low-TMB group had a better prognosis than the high-TMB group (Fig. 7B). We combined risk scores and TMB in order to explore whether risk model and TMB were correlated with prognosis, and divided into high-risk/high-TMB group, high-risk/low-TMB group, low-risk/high-TMB group, and low-risk/low-TMB group to investigate the prognostic differences between the groups. Subsequently, KM survival curves indicated significant differences in survival between the patient in the different groups (P < 0.001). The low-risk/low-TMB group had the best prognosis, and the high-risk/high-TMB group had the shortest durations of survival (Fig. 7C). Waterfall plot results revealed a higher mutation frequency in the high-risk group (93.67%) than in the low-risk group (69.88%). KRAS, TP53 and SMAD4 had the highest mutation frequencies, in the high-risk group for KRAS (81%), TP53 (65%) and SMAD4 (28%), while in the low-risk group for KRAS (42%), TP53 (48%) and SMAD4 (16%) (Fig. 7D and E).
3.7. Correlation of the PAAD prognostic signature with immunotherapy
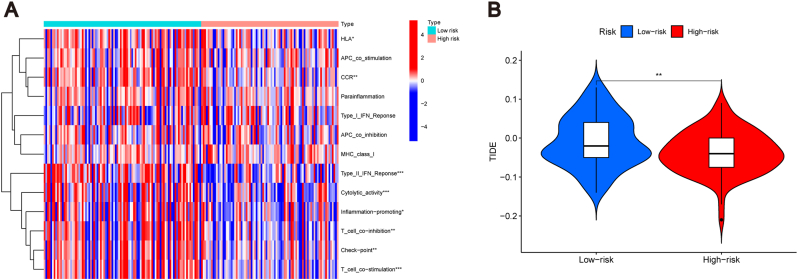
The heat map showed differences between eight immune-related functions (HLA, CCR, Type II IFN response, cytolytic activity, inflammation promotion, T cell co-inhibition, checkpoint, and T cell co-stimulation) in the high- and low-risk groups (P < 0.05). These eight functions were more prominent in the low-risk group (Fig. 8A). It has been found that immune checkpoint inhibitor has a better therapeutic effect in patients with PAAD [35]. TIDE scores revealed patients' response to immunotherapy in both risk groups. The results revealed that low-risk group patients had higher TIDE risk scores than the high-risk group patients (P < 0.01). This indicates that the low-risk group was more probable to avoid immunotherapy, and immunotherapy was more effective in the high-risk group (Fig. 8B).
Fig. 8.
Correlation of risk model with the immunity in PAAD. (A) Heat map of the status of immune-related functions in the high- and low-risk groups. (B) TIDE score of the two groups. *P < 0.05, **P < 0.01, and ***P < 0.001.
3.8. Assessment of drug response
We evaluated the response to some drugs in different risk groups. The two groups showed significantly different drug sensitivities (P < 0.001). Drugs such as all-trans retinoic acid (ATRA), Rac family small GTPases inhibitor EHT 1864 and Phenformin had lower IC50 values in the low-than in the high-risk group (Fig. 9A–K). In the low-risk group, a positive correlation was found between the IC50 value of these drugs and the risk score (Figs. S2A–K). In the high-risk group, the sensitivities of the MEK inhibitors PD-0325901, Gefitinib, Paclitaxel, and Trametinib were higher (Fig. 9A–K), and they increased with increasing risk scores (Figs. S2A–K). This suggested that the PAAD prognostic model helped improve the effectiveness of the drugs for treatment.
Fig. 9.
Investigation of drug sensitivity in risk groups. (A-K) Comparison of IC50 values for different agents in high- and low-risk groups.
4. Discussion
It was reported that the PAAD incidence has steadily increased from 1975 to 2018, with low survival rates [36]. The prognosis remains unfavorable for patients with PAAD even after undergoing radical surgery, which is considered the most efficacious treatment [37]. The immunotherapy has been shown to prolong the overall survival of tumor patients [38]. However, not all patients are well-treated with immunotherapy. Therefore, it is essential to develop a signature that predicts the prognosis and efficacy of immunotherapy in patients with PAAD. A type of inflammatory cell death known as PANoptosis can cause an immunological reaction [39], and recent research has revealed that it is crucial for tumor immunity and growth. For instance, PANoptosis can reduce tumor sizes in immunodeficient mice [13,40]. In addition, the inhibition of PANoptosis promotes tumor progression [41,42]. Thus, PANoptosis has excellent potential for antitumor immunotherapy. However, research on PANoptosis and tumors is still at the primary stage; research on PANoptosis associated with PAAD is lacking. Therefore, it is necessary to develop PANoptosis-related investigations to identify critical prognostic signature in PAAD.
Increasingly, studies have discovered that Panoptosis could be linked to lncRNAs [[43], [44], [45]]. In our study, we identified a total of 266 lncRNAs that exhibited significant associations with PANoptosis. Multiple studies have also shown that lncRNAs can regulate several aspects of tumor development and progression, such as growth, metastasis, genetics, and drug resistance [[46], [47], [48]]. Therefore, PANoptosis-associated lncRNAs hold promising potential as novel prognostic biomarkers and therapeutic targets for neoplastic diseases. According to recent bioinformatics research, the prognosis of patients with PAAD can be predicted by a risk signature linked to lncRNAs [49]. However, the association between PANoptosis-related lncRNAs and the prognosis of PAAD remains to be established.
Our study developed a model for PANoptosis-related lncRNAs that can predict the prognosis of patients with PAAD. To the best of our knowledge, this is the first signature for PANoptosis-related lncRNAs in patients with PAAD. With the model, we identified six PANoptosis-related lncRNAs; two are associated with PAAD [49], and the other four were reported for the first time as risk factors for PAAD. According to the risk model, patients with PAAD were categorized into high- and low-risk groups. The low-risk group demonstrated a more favorable prognosis compared to the high-risk group. Our validation concentration confirmed the predictive stability of the signature. Furthermore, the signature served as an independent prognostic factor for patients with PAAD. The prognostic efficacy of the signature was further validated by the AUC value and ROC curve. The above results collectively suggest that the PANoptosis-related signature we constructed has better validity and stability.
The immune system acts as an irreplaceable role in the anti-tumor fight. Given its success against several tumor types, including liver, kidney, and lung cancers, immunotherapy has become an essential option for PAAD. Currently, ICIs is a hot topic in immunotherapy [[50], [51], [52]].TMB is a predictor of benefit in ICIs, and some studies have shown [[53], [54], [55]] that high-TMB may respond better to ICIs than low-TMB. The TMB levels were significantly higher in the high-risk group compared to the low-risk group in our model, indicating a greater likelihood of favorable response to ICIs treatment for the high-risk group. The findings of a previous study align with this observation, and they also demonstrated a significant correlation between KARS, one of the genes exhibiting the highest mutation rate in PAAD, and an unfavorable prognosis [56]. In our research, KARS demonstrated a higher mutation rate, with 81% in the high-risk group and 42% in the low-risk group. This might contribute to the worse prognosis in the high-risk group. In addition, we utilized TIDE to prognosticate the response to ICIs in both high- and low-risk cohorts, aiming to mitigate adverse events occurrence and thereby enhance the efficacy of immunotherapy. The low-risk group exhibited higher TIDE scores compared to the high-risk group, suggesting a greater susceptibility to immune escape. Conversely, patients with PAAD in the high-risk group demonstrated enhanced responsiveness to ICIs. Additionally, we analyzed the immune functions to explore the association between the predictive model based on the PANoptosis-related lncRNAs and immunity. Immune functions such as type II IFN response, cytolytic activity, and T cell co-stimulation were distinctly different in the risk groups. The prevalence of these functions was higher in the low-risk group.
We also performed a drug screen for the risk model to seek therapeutic drugs that are effective in patients with PAAD. The findings indicated that patients in the two risk categories exhibited distinct responses to 11 pharmaceutical agents. Paclitaxel, as a commonly used chemotherapeutic agent for patients with PAAD, is mainly used to exert anti-tumor efficacy by inducing cell cycle arrest and apoptosis [57]. The IC50 to paclitaxel was higher in the low-risk group in the signature, suggesting that the low-risk group is resistant to paclitaxel, which was more suitable for the high-risk group. ATRA, a natural derivative of retinoic acid, can regulate cell proliferation, apoptosis, and metastasis, and it has been found that the combination of ATRA with chemotherapeutic agents can enhance the tolerability [58,59]. In the low-risk group, patients responded better to ATRA. The purpose of gefitinib is to suppress tumor proliferation and angiogenesis through the inhibition of epidermal growth factor receptor (EGFR) [60]. In tumor, EGFR mutation is an important reason for the sensitivity to gefitinib. It was found that EGFR mutation existed in PAAD, suggesting that gefitinib may be effective in the PAAD therapies [61]. Carneiro et al. reported with the combination of gefitinib and gemcitabine, patients with PAAD responded well and had fewer adverse effects [62]. Trametinib exhibits selective inhibition of MEK1 and MEK2, and a more pronounced response to gefitinib and trametinib in the high-risk group [63].
In conclusion, we identified several biomarkers strongly associated with the survival outcomes of patients with PAAD, and these were used to develop a prognosis model. The signature accurately predicts the prognosis and immune landscapes of patients with PAAD, thereby enhancing treatment effectiveness and preventing drug resistance development. However, this study had some limitations. First, the study was internally validated for the dataset only and lacked external validation. Second, the datasets used for our model were obtained from databases, therefore, the experiments were retrospective. We will continually research to resolve these problems. Further studies should also refine this prognostic model of PAAD to improve accuracy. The findings of this study suggest that a prognostic signature consisting of PANoptosis-related lncRNAs has the potential to accurately predict both the prognosis and immune landscapes of patients with PAAD.
Funding
This research received no external funding.
CRediT authorship contribution statement
Qinying Zhao: Investigation, Resources. Yingquan Ye: Data curation, Investigation. Quan Zhang: Writing – review & editing. Yue Wu: Writing – original draft. Gaoxiang Wang: Writing – original draft. Zhongxuan Gui: Data curation. Mei Zhang: Methodology, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We appreciate the TCGA database for providing their platforms and contributors for uploading their meaningful datasets.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2023.101600.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Further validation of the prediction model. (A) The consistency index analysis curve. Principal component analysis of different groups based on the PANoptosis-related genes (B), PANoptosis-related lncRNAs (C), lncRNAs associated with PANoptosis used to construct prognostic risk models (D). (E-F) Kaplan–Meier survival curves of low- and high-risk groups sorted by TNM stage.
Fig. S2.
Investigation of drug sensitivity in risk groups. (A-K) Linear plot of sensitivity to different drugs with risk scores.
Data availability
Data will be made available on request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA. A Cancer Journal for Clinicians. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Qi B., Liu H., Zhou Q., Ji L., Shi X., Wei Y., Gu Y., Mizushima A., Xia S. An immune-related lncRNA signature for the prognosis of pancreatic adenocarcinoma. Aging. 2021;13:18806–18826. doi: 10.18632/aging.203323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrido-Laguna I., Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 2015;12:319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 4.Li F., Zhang P., Khalaf O.I. The N6-methyladenosine- (m6A-) associated genes act as strong key biomarkers for the prognosis of pancreatic adenocarcinoma. Comput. Math. Methods Med. 2021;2021:1–19. doi: 10.1155/2021/8715823. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.McGuigan A., Kelly P., Turkington R.C., Jones C., Coleman H.G., McCain R.S. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018;24:4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malireddi R.K.S., Gurung P., Kesavardhana S., Samir P., Burton A., Mummareddy H., Vogel P., Pelletier S., Burgula S., Kanneganti T.-D. Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity–independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J. Exp. Med. 2020;217 doi: 10.1084/jem.20191644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christgen S., Zheng M., Kesavardhana S., Karki R., Malireddi R.K.S., Banoth B., Place D.E., Briard B., Sharma B.R., Tuladhar S., Samir P., Burton A., Kanneganti T.-D. Identification of the PANoptosome: a molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10:237. doi: 10.3389/fcimb.2020.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S., Karki R., Wang Y., Nguyen L.N., Kalathur R.C., Kanneganti T.-D. AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature. 2021;597:415–419. doi: 10.1038/s41586-021-03875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng M., Karki R., Vogel P., Kanneganti T.D. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell. 2020;181:674–687 e613. doi: 10.1016/j.cell.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Hong M., Li Y., Chen D., Wu Y., Hu Y. Programmed cell death tunes tumor immunity. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.847345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan H., Pan J., Li P., Gao J. Characterization of PANoptosis patterns predicts survival and immunotherapy response in gastric cancer. Clin. Immunol. 2022;238 doi: 10.1016/j.clim.2022.109019. [DOI] [PubMed] [Google Scholar]
- 12.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., Schreiner P., Neale G., Vogel P., Webby R., Jonsson C.B., Kanneganti T.-D. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e117. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malireddi R.K.S., Karki R., Sundaram B., Kancharana B., Lee S., Samir P., Kanneganti T.-D. Inflammatory cell death, PANoptosis, mediated by cytokines in diverse cancer lineages inhibits tumor growth. ImmunoHorizons. 2021;5:568–580. doi: 10.4049/immunohorizons.2100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karki R., Sundaram B., Sharma B.R., Lee S., Malireddi R.K.S., Nguyen L.N., Christgen S., Zheng M., Wang Y., Samir P., Neale G., Vogel P., Kanneganti T.-D. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malireddi R.K.S., Kesavardhana S., Kanneganti T.-D. ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis) Front. Cell. Infect. Microbiol. 2019;9:406. doi: 10.3389/fcimb.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J., Bai J., Zhang X., Lv Y., Gong Y., Liu L., Zhao H., Yu F., Ping Y., Zhang G., Lan Y., Xiao Y., Li X. A comprehensive overview of lncRNA annotation resources. Briefings Bioinf. 2017;18:236–249. doi: 10.1093/bib/bbw015. [DOI] [PubMed] [Google Scholar]
- 17.Frangoul H., Altshuler D., Cappellini M.D., Chen Y.-S., Domm J., Eustace B.K., Foell J., de la Fuente J., Grupp S., Handgretinger R. CRISPR-Cas9 gene editing for sickle cell disease and β-thalassemia. N. Engl. J. Med. 2021;384:252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 18.Mateo J., Porta N., Bianchini D., McGovern U., Elliott T., Jones R., Syndikus I., Ralph C., Jain S., Varughese M., Parikh O., Crabb S., Robinson A., McLaren D., Birtle A., Tanguay J., Miranda S., Figueiredo I., Seed G., Bertan C., Flohr P., Ebbs B., Rescigno P., Fowler G., Ferreira A., Riisnaes R., Pereira R., Curcean A., Chandler R., Clarke M., Gurel B., Crespo M., Nava Rodrigues D., Sandhu S., Espinasse A., Chatfield P., Tunariu N., Yuan W., Hall E., Carreira S., de Bono J.S. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W., Ye Y., Zhang X., Sun W., Bao L. An angiogenesis-related three-long non-coding ribonucleic acid signature predicts the immune landscape and prognosis in hepatocellular carcinoma. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y., Ye Y., Huang Y., Wu Y., Wang G., Gui Z., Zhang M., Zhang M. Identification and validation of a novel anoikis-related long non-coding RNA signature for pancreatic adenocarcinoma to predict the prognosis and immune response. J. Cancer Res. Clin. Oncol. 2023;149:15069–15083. doi: 10.1007/s00432-023-05285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K., Jutooru I., Chadalapaka G., Johnson G., Frank J., Burghardt R., Kim S., Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ottaviani S., Stebbing J., Frampton A.E., Zagorac S., Krell J., de Giorgio A., Trabulo S.M., Nguyen V.T.M., Magnani L., Feng H., Giovannetti E., Funel N., Gress T.M., Jiao L.R., Lombardo Y., Lemoine N.R., Heeschen C., Castellano L. TGF-β induces miR-100 and miR-125b but blocks let-7a through LIN28B controlling PDAC progression. Nat. Commun. 2018;9:1845. doi: 10.1038/s41467-018-03962-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N., Yang G., Luo L., Ling L., Wang X., Shi L., Lan J., Jia X., Zhang Q., Long Z., Liu J., Hu W., He Z., Liu H., Liu W., Zheng G. lncRNA THAP9-AS1 promotes pancreatic ductal adenocarcinoma growth and leads to a poor clinical outcome via sponging miR-484 and interacting with YAP. Clin. Cancer Res. 2020;26:1736–1748. doi: 10.1158/1078-0432.CCR-19-0674. [DOI] [PubMed] [Google Scholar]
- 24.Huang J., Jiang S., Liang L., He H., Liu Y., Cong L., Jiang Y. Analysis of PANoptosis-related LncRNA-miRNA-mRNA network reveals LncRNA SNHG7 involved in chemo-resistance in colon adenocarcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.888105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karki R., Sharma B.R., Lee E., Banoth B., Malireddi R.K.S., Samir P., Tuladhar S., Mummareddy H., Burton A.R., Vogel P., Kanneganti T.-D. Interferon regulatory factor 1 regulates PANoptosis to prevent colorectal cancer. JCI Insight. 2020:5. doi: 10.1172/jci.insight.136720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samir P., Malireddi R.K.S., Kanneganti T.-D. The PANoptosome: a deadly protein complex driving pyroptosis, apoptosis, and necroptosis (PANoptosis) Front. Cell. Infect. Microbiol. 2020;10:238. doi: 10.3389/fcimb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin J.-F., Hu P.-S., Wang Y.-Y., Tan Y.-T., Yu K., Liao K., Wu Q.-N., Li T., Meng Q., Lin J.-Z., Liu Z.-X., Pu H.-Y., Ju H.-Q., Xu R.-H., Qiu M.-Z. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct. Targeted Ther. 2022;7:54. doi: 10.1038/s41392-022-00889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundaram B., Kanneganti T.-D. Advances in understanding activation and function of the NLRC4 inflammasome. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22031048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Q., Zhang X., Ren X., Hang Z., Yin Y., Wang Z., Chen H., Sun L., Tao J., Han Z., Tan R., Gu M., Ju X. Biological characteristics and predictive model of biopsy-proven acute rejection (BPAR) after kidney transplantation: evidences of multi-omics analysis. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.844709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Zhang W., Wang Y., Wan T., Hu B., Li C., Ge X., Lu S. Construction and validation of a ferroptosis-related lncRNA signature as a novel biomarker for prognosis, immunotherapy and targeted therapy in hepatocellular carcinoma. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.792676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang S., Ren X., Liu S., Lu Z., Xu A., Qin C., Wang Z. Integrated analysis of the prognosis-associated RNA-binding protein genes and candidate drugs in renal papillary cell carcinoma. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.627508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X., Ren X., Zhang T., Zhou X., Chen X., Lu H., Zhou X., Zhang X., Wang S., Qin C. Comprehensive analysis of the association between human non-obstructive azoospermia and plasticisers via single-cell and traditional RNA sequencing methods. Exposure and Health. 2022;14:829–842. [Google Scholar]
- 33.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., Liu J., Freeman G.J., Brown M.A., Wucherpfennig K.W., Liu X.S. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Q., Zhang P., Hu H., Huang H., Pan D., Mao G., Hu B. The DDR-related gene signature with cell cycle checkpoint function predicts prognosis, immune activity, and chemoradiotherapy response in lung adenocarcinoma. Respir. Res. 2022;23:190. doi: 10.1186/s12931-022-02110-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolbeinsson H.M., Chandana S., Wright G.P., Chung M. Pancreatic cancer: a review of current treatment and novel therapies. J. Invest. Surg. 2022:1–11. doi: 10.1080/08941939.2022.2129884. [DOI] [PubMed] [Google Scholar]
- 36.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022, CA. A Cancer Journal for Clinicians. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 37.Mizrahi J.D., Surana R., Valle J.W., Shroff R.T. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 38.Park W., Chawla A., O'Reilly E.M. Pancreatic cancer. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pandian N., Kanneganti T.-D. PANoptosis: a unique innate immune inflammatory cell death modality. J. Immunol. 2022;209:1625–1633. doi: 10.4049/jimmunol.2200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christgen S., Tweedell R.E., Kanneganti T.-D. Programming inflammatory cell death for therapy. Pharmacology & Therapeutics. 2022;232 doi: 10.1016/j.pharmthera.2021.108010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong L., Huang D., Shi Y., Liang Z., Bu H. Regulated cell death in cancer: from pathogenesis to treatment. Chin Med J (Engl) 2022 doi: 10.1097/CM9.0000000000002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan B., Zheng B., Xing C., Liu J. Non-canonical programmed cell death in colon cancer. Cancers. 2022;14 doi: 10.3390/cancers14143309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li R., Zhao M., Sun M., Miao C., Lu J. Construction and validation of a PANoptosis-related lncRNA signature for predicting prognosis and targeted drug response in thyroid cancer. PeerJ. 2023;11 doi: 10.7717/peerj.15884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu W., Qu C., Wang X. Comprehensive analysis of the role of immune-related PANoptosis lncRNA model in renal clear cell carcinoma based on RNA transcriptome and single-cell sequencing. Oncology research. 2023;31:543–567. doi: 10.32604/or.2023.029563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shu J., Yang L., Wei W., Zhang L. Identification of programmed cell death-related gene signature and associated regulatory axis in cerebral ischemia/reperfusion injury. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.934154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q., Chen C., Xu X., Shu C., Cao C., Wang Z., Fu Y., Xu L., Xu K., Xu J., Xia A., Wang B., Xu G., Zou X., Su R., Kang W., Xue Y., Mo R., Sun B., Wang S. APAF1‐Binding long noncoding RNA promotes tumor growth and multidrug resistance in gastric cancer by blocking apoptosome assembly. Adv. Sci. 2022;9 doi: 10.1002/advs.202201889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang B., Thorne R.F., Zhang P., Wu M., Liu L. Vanguard is a glucose deprivation-responsive long non-coding RNA essential for chromatin remodeling-reliant DNA repair. Adv. Sci. 2022;9 doi: 10.1002/advs.202201210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Liu X., Wang Y., Lai S., Wang Z., Yang Y., Liu W., Wang H., Tang B. The m(6)A demethylase ALKBH5-mediated upregulation of DDIT4-AS1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mTOR pathway. Mol. Cancer. 2022;21:174. doi: 10.1186/s12943-022-01647-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye Y., Zhao Q., Wu Y., Wang G., Huang Y., Sun W., Zhang M. Construction of a cancer-associated fibroblasts-related long non-coding RNA signature to predict prognosis and immune landscape in pancreatic adenocarcinoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.989719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Q., Peng X., Li Q., Zhu J., Xue J., Jiang H. Construction and comprehensive analysis of a novel prognostic signature associated with pyroptosis molecular subtypes in patients with pancreatic adenocarcinoma. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1111494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L., Xiao Z., He P., Zou W., Deng Z., Zhang G., Liu R. Molecular subtyping based on TRP family and prognostic assessment for TRP-associated lncRNAs in pancreatic adenocarcinoma. BMC Gastroenterol. 2022;22 doi: 10.1186/s12876-022-02552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J., Yin J., Li W., Wang H., Ni B. Molecular subtypes based on cuproptosis-related genes and tumor microenvironment infiltration characteristics in pancreatic adenocarcinoma. Cancer Cell Int. 2023;23 doi: 10.1186/s12935-022-02836-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garon E.B., Rizvi N.A., Hui R., Leighl N., Balmanoukian A.S., Eder J.P., Patnaik A., Aggarwal C., Gubens M., Horn L., Carcereny E., Ahn M.-J., Felip E., Lee J.-S., Hellmann M.D., Hamid O., Goldman J.W., Soria J.-C., Dolled-Filhart M., Rutledge R.Z., Zhang J., Lunceford J.K., Rangwala R., Lubiniecki G.M., Roach C., Emancipator K., Gandhi L. Pembrolizumab for the treatment of non–small-cell lung cancer. N. Engl. J. Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 54.Moeckel C., Bakhl K., Georgakopoulos-Soares I., Zaravinos A. The efficacy of tumor mutation burden as a biomarker of response to immune checkpoint inhibitors. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms24076710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ke L., Li S., Huang D. The predictive value of tumor mutation burden on survival of gastric cancer patients treated with immune checkpoint inhibitors: a systematic review and meta-analysis. Int. Immunopharm. 2023;124 doi: 10.1016/j.intimp.2023.110986. [DOI] [PubMed] [Google Scholar]
- 56.Yao J., Liang Z., Duan L., G Y., Liu J., An G. Construction of a novel immune response prediction signature to predict the efficacy of immune checkpoint inhibitors in clear cell renal cell carcinoma patients. Heliyon. 2023;9 doi: 10.1016/j.heliyon.2023.e15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duan Y., Du Y., Mu Y., Gu Z., Wang C. Prognostic value, immune signature and molecular mechanisms of the SUMO family in pancreatic adenocarcinoma. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.1096679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kocher H.M., Basu B., Froeling F.E.M., Sarker D., Slater S., Carlin D., deSouza N.M., De Paepe K.N., Goulart M.R., Hughes C., Imrali A., Roberts R., Pawula M., Houghton R., Lawrence C., Yogeswaran Y., Mousa K., Coetzee C., Sasieni P., Prendergast A., Propper D.J. Phase I clinical trial repurposing all-trans retinoic acid as a stromal targeting agent for pancreatic cancer. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-18636-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavudi K., Nuguri S.M., Olverson Z., Dhanabalan A.K., Patnaik S., Kokkanti R.R. Targeting the retinoic acid signaling pathway as a modern precision therapy against cancers. Front. Cell Dev. Biol. 2023;11 doi: 10.3389/fcell.2023.1254612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M.-T., Li B.-Z., Zhang E.-P., Zheng Q. Potential roles of tumor microenvironment in gefitinib-resistant non-small cell lung cancer: a narrative review. Medicine. 2023:102. doi: 10.1097/MD.0000000000035086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwak E.L., Jankowski J., Thayer S.P., Lauwers G.Y., Brannigan B.W., Harris P.L., Okimoto R.A., Haserlat S.M., Driscoll D.R., Ferry D., Muir B., Settleman J., Fuchs C.S., Kulke M.H., Ryan D.P., Clark J.W., Sgroi D.C., Haber D.A., Bell D.W. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin. Cancer Res. : an official journal of the American Association for Cancer Research. 2006;12:4283–4287. doi: 10.1158/1078-0432.CCR-06-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carneiro B.A., Brand R.E., Fine E., Knop R.H., Khandekar J.D., Uhlig W., Locker G.Y. Phase I trial of fixed dose rate infusion gemcitabine with gefitinib in patients with pancreatic carcinoma. Cancer Invest. 2007;25:366–371. doi: 10.1080/15368370701357957. [DOI] [PubMed] [Google Scholar]
- 63.Day H.R., Finn A.P. Serous retinopathy associated with combination MEK and fibroblast growth factor receptor inhibitor. Journal of VitreoRetinal Diseases. 2023;7:352–355. doi: 10.1177/24741264231163393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.