Abstract
Prime editing (PE) technology enables precise alterations in the genetic code of a genome of interest. PE offers great potential for identifying major agronomically important genes in plants and editing them into superior variants, ideally targeting multiple loci simultaneously to realize the collective effects of the edits. Here, we report the development of a modular assembly-based multiplex PE system in rice and demonstrate its efficacy in editing up to four genes in a single transformation experiment. The duplex PE (DPE) system achieved a co-editing efficiency of 46.1% in the T0 generation, converting TFIIAγ5 to xa5 and xa23 to Xa23SW11. The resulting double-mutant lines exhibited robust broad-spectrum resistance against multiple Xanthomonas oryzae pathovar oryzae (Xoo) strains in the T1 generation. In addition, we successfully edited OsEPSPS1 to an herbicide-tolerant variant and OsSWEET11a to a Xoo-resistant allele, achieving a co-editing rate of 57.14%. Furthermore, with the quadruple PE (QPE) system, we edited four genes—two for herbicide tolerance (OsEPSPS1 and OsALS1) and two for Xoo resistance (TFIIAγ5 and OsSWEET11a)—using one construct, with a co-editing efficiency of 43.5% for all four genes in the T0 generation. We performed multiplex PE using five more constructs, including two for triplex PE (TPE) and three for QPE, each targeting a different set of genes. The editing rates were dependent on the activity of pegRNA and/or ngRNA. For instance, optimization of ngRNA increased the PE rates for one of the targets (OsSPL13) from 0% to 30% but did not improve editing at another target (OsGS2). Overall, our modular assembly-based system yielded high PE rates and streamlined the cloning of PE reagents, making it feasible for more labs to utilize PE for their editing experiments. These findings have significant implications for advancing gene editing techniques in plants and may pave the way for future agricultural applications.
Key words: prime editing, rice, bacterial blight, herbicide tolerance, multiplex genome editing
This study reports the development of modularly assembled multiplex prime editors and their capacity to simultaneously target up to four genomic loci at high efficiency in rice. Multiplex prime editing can result in rice lines carrying gene copies that confer broad resistance to bacterial blight and strong tolerance to herbicides.
Introduction
Simultaneously creating multiple genetic variations and breeding them into improved germplasm is the most desirable objective of any crop improvement program. It is necessary to target and modify multiple genes at once to harness the additive benefits of multiple genic combinations in a genotype of interest (Chen et al., 2019). Double-stranded DNA-break-inducing CRISPR systems, which are mainly used to generate insertion/deletion-type knockout mutants, can rarely generate superior alleles, which require precise genomic changes. Prime editing (PE), on the other hand, has broader applications because of its ability to generate DNA insertions and deletions as well as install desired base-pair changes (Anzalone et al., 2019). PE is the latest and most advanced CRISPR-based genome editing technology and has been revolutionizing biology by enabling scientists to search for and replace genomic sequences without the need for a double-stranded DNA break and a donor template carrying the desired sequence changes. In brief, a reverse transcriptase-fused Cas9 nickase (nCas9–RT) targets the genomic region guided by a PE RNA (pegRNA). The pegRNA directs the nCas9–RT/pegRNA complex to the target region; the edit encoded in the 3′ extension is reverse transcribed to the 3′ end of the nicked genomic DNA strand. This leads to the generation of a 3′ flap containing the edited sequence and a 5′ flap of wild-type (WT) sequence surrounding the nicked site. These flaps are resolved via a flap excision and heteroduplex repair system. Excision of the 5′ flap and incorporation of the 3′ flap lead to an editing event (Anzalone et al., 2019). A second guide RNA (nicking guide RNA; ngRNA) is used to nick the opposite strand either upstream or downstream of the original target to render the excision repair in favor of 3′ flap incorporation (Anzalone et al., 2019; Yang et al., 2019). Until recently, PE has suffered from low editing efficiency in plants, but with the new ePPE, PE3max, and PE5max systems, editing efficiencies have been boosted up to 88% for some single-site targets in rice (Anzalone et al., 2019; Li et al., 2022b; Jiang et al., 2022; Gupta et al., 2023b). This high efficiency makes the PE system amenable to multiplexing.
PE can provide an unprecedented opportunity, ideally, to target several heterologous genes or multiple sites of single genes simultaneously and install multiple desired changes in a single transformation event, including knockouts, small insertions/deletions, and specific base pair changes (Yang et al., 2019; Molla et al., 2021). This enables the simultaneous improvement of several agronomically important traits in crop plants. For instance, yield-related genes can be targeted to improve crop productivity, disease-resistance genes can be stacked to provide broader and more durable resistance against numerous diseases, and abiotic stress-related genes can be edited to increase crop resilience in the face of a changing climate (Hassan et al., 2020; Molla et al., 2021; Gupta et al., 2023b; Li et al., 2023). Furthermore, these edits in multiple traits can be combined to engineer new germplasm with improved productivity and enhanced resilience to biotic and abiotic stresses. Moreover, the multiplex PE system can serve as a valuable asset for functional genomics by enabling researchers to change the native gene sequence instead of relying on complementation assays with a transgenic approach. Multiple protein tagging with PE can help us understand complex gene-regulatory networks by tracking protein expression in native conditions (Hua et al., 2022; Kumar et al., 2023).
Rice is an important staple crop in terms of global food security, and it also serves as the model crop for studying cereal genetics and developing genetic tools. It is highly desirable to have a high-efficiency multiplex PE system in rice to facilitate rice crop improvement programs and study the functions of agronomically important genes/traits in an endogenous context (Huang and Puchta, 2021; Xu et al., 2022; Ni et al., 2023). Here, we report the development of a modular PE system amenable to multiplexing pegRNAs together with ngRNAs and editing up to four genomic loci. The modular assembly uses Golden Gate cloning to make individual pegRNA–ngRNA units and subsequently uses Gateway recombination to combine the pegRNA–ngRNA units with the final nCas9–RT vector. We validated the feasibility and efficacy of this system by targeting two, three, or four genes in a single generation, approaches named duplex PE (DPE), triplex PE (TPE), or quadruplex PE (QPE), respectively. In our multiplexing experiments, we achieved high editing efficiencies, with a number of lines carrying simultaneous monoallelic and biallelic edits in the T0 generation. We also obtained lines with both genes edited by DPE, all three genes edited by TPE, and all four genes edited by QPE. In this work, we simultaneously edited genes related to bacterial blight of rice, herbicide tolerance, plant architecture, and grain yield, demonstrating the phenotypic superiority of the edited lines over the unedited lines in terms of bacterial blight resistance and herbicide tolerance.
Results
Modular assembly of multiplex prime editing constructs
The core requirements of PE3 include a nicking Cas9 (H840A) fused to a reverse transcriptase (nCas9–RT), a pegRNA that consists of a single guide RNA with a spacer specifying the target and a reverse transcription template (rtT) encoding information about the edit as well as a prime binding sequence (PBS), and an ngRNA to nick the opposite strand to favor DNA repair toward the edited strand (Figure 1A) (Anzalone et al., 2019). The rtT is located downstream of the pegRNA scaffold followed by the PBS. A nuclease-resistant RNA motif, evopreQ1, is used at the 3′ end to prevent RNA degradation. PBS and evopreQ1 are separated by an 8-bp linker calculated using the webtool pegLIT (Supplemental Protocol). The paired pegRNA–ngRNA format (namely, PE3 or PE3b) is an improvement on the initial PE2 (Anzalone et al., 2019; Yang et al., 2019). Because nCas9–RT is the constant reagent for different PE events, whereas the pegRNA and ngRNA change with every new target, we designed a modular assembly-based PE system for easy cloning of multiple pegRNA–ngRNA units (Figure 1A and 1B). In this modular system, the nCas9–RT is included in a destination binary vector that can accept multiple pegRNA–ngRNA units using the Gateway cloning approach. To construct individual pegRNA–ngRNA units, multiple entry vectors flanked by different Gateway recombination sequences (RS, attL, and attR) were designed and constructed.
Figure 1.
Modular assembly of multiplex prime editing constructs.
(A) Schematic representation of the required components for prime editing.
(B) Binary vector carrying nickase Cas9 fused with reverse transcriptase, the hygromycin-resistance gene, and a Gateway cassette to accept inserts from entry clones.
(C–F) Entry vectors for cloning up to four pegRNA–ngRNA units flanked by variable attachment (att) regions.
(G) Schematic of the Gateway reaction transferring four pegRNA–ngRNA cassettes from entry clones to the destination vector.
(H–K) Resulting final vectors ready for Agrobacterium/bombardment-mediated plant transformation for prime editing of up to four genes.
These Gateway RSs are different combinations of attL and attR sequences that enable the assembly of multiple pegRNA–ngRNA units into a single destination vector. For single pegRNA–ngRNA assembly, attL1–attL2 was used to flank the pegRNA–ngRNA cassette; attL1–attR3 and attL3–attL2 were used for two pegRNA–ngRNA units; attL1–attR5, attL5–attR3, and attL3–attL2 for three pegRNA–ngRNA units; and attL1–attR5, attL5–attL4, attR4–attR3, and attL3–attL2 for four pegRNA–ngRNA units (Figure 1C–1F, Supplemental Sequences 1–11). These entry vectors were designed to insert a spacer sequence for pegRNA at the BsmBI site and rtT-PBS and a spacer sequence for ngRNA at the BsaI site using the Golden Gate procedure or a regular restriction–ligation protocol (Supplemental Protocol). All pegRNA spacer, rtT-PBS, and ngRNA spacer sequences can be derived from respective complementary oligos forming double-stranded fragments with appropriate 5′ overhangs. After cloning of the pegRNA–ngRNA units, up to four units can be mobilized into a single binary vector at the attR1–attR2 site using Gateway cloning (Supplemental Protocol). Using this approach, we successfully assembled one, two, three, and four units and confirmed the cloning via whole-plasmid sequencing (Figure 1G–1K). These constructs remained stable in Agrobacterium and the transfer-DNA transfer of these units was also found to be stable in rice cells. In this approach, expression of each pegRNA–ngRNA unit is driven by an individual promoter, ensuring high and uniform expression of each unit. We used this assembly to design and construct all the pegRNA–ngRNA units used for this study.
Duplex prime editing efficiently generates Xa23SW11 and xa5 co-edited lines in the T0 generation
In our previous study (Gupta et al., 2023b), xa5-edited rice lines exhibited strong broad-spectrum resistance against all Xoo strains dependent on TFIIAγ5 for SWEET gene induction by major transcription activator-like effectors (TALEs). However, Xoo strains carrying the TALE gene pthXo1 can overcome xa5-mediated resistance, as pthXo1 can use both TFIIAγ5 and xa5 efficiently for SWEET11a induction (Huang et al., 2016). Therefore, we decided to utilize the DPE system, with as-yet unknown efficiency, to perform duplex editing of V39E substitution in TFIIAγ5 to generate the xa5 allele and, by inserting the 28-bp-long PthXo1 effector-binding element (EBE) of OsSWEET11a into the promoter of dysfunctional xa23, to generate a functional Xa23SW11 allele variant. The DPE construct encoding TFIIAγ5 to xa5 and xa23 to Xa23SW11 was used for Agrobacterium-mediated rice transformation (Figure 2A and 2B). Twenty-six independent transgenic events were recovered and genotyped for the V39E edit based on the SmlI restriction sequence in TFIIAγ5/xa5 and the PthXo1 EBE knockin based on the BsrGI restriction sequence that arose from successful editing in xa23/Xa23SW11 (Figure 2C and Supplemental Figure 1A and 1B). Of 26 T0 lines, 18 contained the edits for xa5, Xa23SW11, or both, resulting in an editing efficiency of 69.23% (Figure 2C and 2D and Supplemental Figure 1A and 1B; Supplemental Table 3). Of the 18 edited lines, 12 lines were co-edited for both xa5 and Xa23SW11, leading to a co-editing efficiency of 46.1% (Figure 2C and 2D and Supplemental Figure 1A and 1B; Supplemental Table 3). Two of these 12 lines were double biallelic (both alleles of TFIIAγ5 and xa23 edited), 9 were double monoallelic (one allele each of TFIIAγ5 and xa23 edited), and 1 was biallelic for TFIIAγ5 and monoallelic for xa23 (Figure 2C and 2D and Supplemental Figure 1A and 1B; Supplemental Table 3). To validate the accuracy of editing, we deep sequenced the amplicons from several lines of both xa5- and Xa23SW11-edited lines (Supplemental Figure 2A and 2B). For biallelic lines, more than 85% of reads mapped to an edited allele, and for monoallelic lines, >40% of reads out of the total mapped to an edited allele (Supplemental Figure 2A and 2B). We further validated these results by Sanger sequencing of two double-biallelic lines (#2 and #34) (Supplemental Figure 2C and 2D). Sequencing chromatograms also revealed that lines #2 and #34 were biallelic for both xa5 and Xa23SW11 (Supplemental Figure 2C and 2D). These results indicate the feasibility of the highly efficient PE3max system for DPE. This is the first report to generate these two loci in the same genetic background, making it vital to test the activity of both loci, especially Xa23SW11, in the same genetic background. The xa5 allele is known to be less effective at enabling pthXo1-mediated induction of SWEET11a (Huang et al., 2016). Thus, to test the induction of Xa23SW11 in the xa5 background, we infiltrated the PXO99 (pthXo1) and PXO86 (avrXa7) strains into the Xa23SW11/xa5 dual-biallelic, Xa23SW11 monoallelic, and WT lines. Total RNA was extracted 24 h post inoculation, first-strand cDNA was synthesized, and both semi-quantitative RT-PCR and quantitative RT-PCR were performed on all samples with equal amounts of cDNA. The Xa23SW11/xa5 dual-biallelic line (#2) showed the highest induction of the Xa23SW11 gene upon PXO99 infection, followed by the Xa23SW11 monoallelic lines (#19 and #33) (Figure 2E and Supplemental Figure 3). The WT line (#17), PXO86-inoculated edited lines, and uninoculated lines showed no induction of Xa23SW11, suggesting tight regulation and induction due only to presence of PthXo1-EBE (Figure 2E and Supplemental Figure 3). Meanwhile, no difference in SWEET11a gene induction due to PXO99 was observed in any dual-edited line compared with the WT line, again confirming that xa5 has less effect on SWEET11a induction by PthXo1 (Supplemental Figure 3). Furthermore, we challenged the edited and WT lines with PXO86 and PXO99 using the leaf-clipping inoculation method to test resistance in the T0 generation. The dual-biallelic lines were highly resistant to both strains, the Xa23SW11 monoallelic lines were resistant only to PXO99, and the WT lines were susceptible to both strains (Figure 2F and 2G). These results indicate that both engineered loci can work in the same genetic background and could provide strong broad-spectrum resistance against multiple strains.
Figure 2.
Duplex prime editing of Xa23SW11 and xa5 lines.
(A) Gene structures of TFIIAγ5 and xa5. The PE target site (underlined) in the TFIIAγ5 allele and the nick site (underlined) in the edited strand of the xa5 allele are shown.
(B) Gene structures of xa23 and Xa23SW11. The intronless coding sequence (CDS) and untranslated sequences (ND, not determined) are shown.
(C) Counts of monoallelic, biallelic, and WT lines based on PCR-RE-based genotyping of TFIIAγ5/xa5-edited lines with SmlI digestion of relevant PCR amplicons and xa23/Xa23SW11-edited lines with BsrGI digestion of relevant PCR amplicons. Numbers of monoallelic, biallelic, deletion, and WT lines are mentioned in each box.
(D) Summary of genotyping based on PCR-RE. Edited alleles are mentioned in bold.
(E) qRT-PCR of the xa23/Xa23SW11 gene. All samples were normalized against OsActin (housekeeping gene control), and the comparison was made against an unedited line infiltrated with PXO99.
(F and G) Disease phenotypes of edited biallelic lines. Lines #2 and #34 are biallelic for xa5 and Xa23SW11 alleles. Lesion lengths were measured 12 days post inoculation with PXO86 and PXO99 on three to five leaves of individual plants (n = 3–5). Scale bar, 1 cm. Lowercase letters a, b, and c in (E) and letters a and b in (G) represent statistically significant differences among different treatments calculated by Tukey’s test.
xa5/Xa23SW11 dual-edited lines provide broad spectrum resistance to multiple strains in the T1 generation
We grew four T0 lines to test the heritability of xa5/Xa23SW11 dual- and single-edited lines in the T1 generation. All four lines carried the edits to the T1 generation, as confirmed by PCR and restriction enzyme digestion (PCR-RE) and Sanger sequencing. Next, we challenged these T1 lines with multiple Xoo strains carrying different TALEs to test the broad spectrum of resistance. Specifically, PXO86, ME2(pthXo3), ME2(pthXo2B), and ME2(AvrXa7) were selected for testing xa5-mediated resistance, and PXO99 and ME2(pthXo1) were selected for testing Xa23SW11-mediated resistance. The xa5-edited lines were highly resistant to strains PXO86, ME2(pthXo3), ME2(pthXo2B), and ME2(AvrXa7) but susceptible to PXO99 and ME2(pthXo1) (Figure 3A and 3B). By contrast, the Xa23SW11-edited lines were resistant only to PXO99 and ME2(pthXo1) and susceptible to PXO86, ME2(pthXo3), ME2(pthXo2B), and ME2(avrxa7) (Figure 3A and 3B). The duplex-edited lines carrying the xa5/Xa23SW11 genotype were resistant to all the tested strains. These results suggest that the two edits, xa5 and Xa23SW11, can work synergistically to provide resistance against different TALE-carrying strains (Figure 3A and 3B).
Figure 3.
xa5/Xa23SW11 dual-edited lines exhibit broad-spectrum resistance to multiple strains in the T1 generation.
(A and B) Disease phenotypes of edited homozygous T1 lines of xa5/Xa23SW11. The Xoo strains used for inoculation are indicated at the bottom. A numerical key is used to represent the different genotypes in (A), and a color key is used for (B). Lesion lengths were measured 12 days post inoculation on three to five leaves of individual plants (n = 3–5). Scale bar, 1 cm. In the bar graph, letters a, b, and c represent statistically significant differences in lesion lengths of edited and WT lines for all strains calculated by Tukey’s test.
Duplex prime editing efficiently generates EPSPS1 TAP-IVS and SWEET11a EBE-deletion co-edited lines in the T0 generation
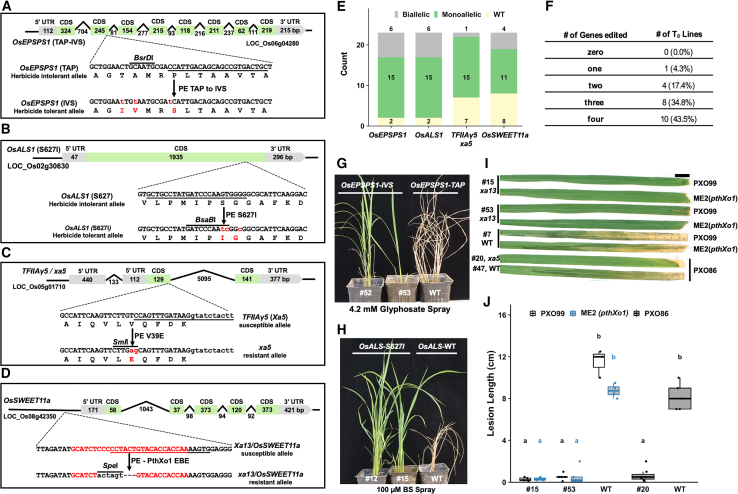
Next, we tested DPE with another set of targets to generate the OsEPSPS1 TAP-IVS triple amino acid substitution and OsSWEET11a EBE-deletion mutation. In OsEPSPS1, the TAP-IVS mutation (T102I, A103V, and P106S) is a naturally occurring triple amino acid substitution linked to strong herbicide tolerance, and OsSWEET11a is a sugar transporter and a susceptibility gene hijacked by Xoo upon infection (Yang et al., 2006; Jiang et al., 2022). Xoo induces the expression of OsSWEET11a via the pthXo1 TALE by binding to the EBE in the OsSWEET11a promoter region. Mutations in the EBE of OsSWEET11a have been shown to render rice resistant (Oliva et al., 2019). Constructs intended for both targets were generated using the modular assembly approach and were used for Agrobacterium-mediated rice transformation (Figure 4A and 4B). Twenty-one callus lines regenerated on medium supplemented with hygromycin, and each callus line produced multiple T0 plants. We considered plants that originated from a single callus line to be a single transformation event, and we performed mutation analysis on a mixture of all the T0 plants from each event. After confirming the presence of a transgene in all lines, we genotyped them for presence of the desired edits using the PCR-RE approach. The BsrDI restriction enzyme was used to detect edits in OsEPSPS1, as the TAP-IVS triple amino acid substitution leads to loss of the BsrDI site (Figure 4A). In OsSWEET11a, we intentionally incorporated a SpeI restriction sequence to disrupt the PthXo1 EBE and facilitate edit detection and genotyping (Figure 4B). As mentioned above, genotyping was performed on a mixture of plantlets that originated from a single callus line, and we considered sites with strong edited bands and weaker WT bands of the PCR amplicons on the agarose gel to be biallelic and sites with strong WT bands and weaker edited bands to be monoallelic. Sites with no band representing an edited site were considered to be WT sites (Figure 4C and Supplemental Figure 4A and 4B; Supplemental Table 4). Of the 21 lines, 16 were edited for either one or both genes, as detected by PCR-RE, reaching an editing frequency of 76.2% (Figure 4C and 4D and Supplemental Figure 4A and 4B; Supplemental Table 4). Of the 16 edited lines, 4 were edited for either OsEPSPS1 or OsSWEET11a, and the remaining 12 were edited for both alleles. Co-editing efficiency of both target genes was thus 57.14% (Figure 4C and 4D and Supplemental Figure 4A and 4B; Supplemental Table 4).
Figure 4.
Duplex prime editing of EPSPS1 for TAP-IVS editing and SWEET11a EBE deletion/knockout.
(A) Gene structures of the OsEPSPS1-TAP allele and OsEPSPS1-IVS allele. The PE target site in the OsEPSPS1-TAP allele is underlined, and the desired amino acid change is shown in red in the OsEPSPS1-IVS allele.
(B) Gene structures of OsSWEET11a/xa13 and xa13. The effector binding element (EBE) in the promoter of susceptible OsSWEET11a is shown in red. The desired edit, including partial EBE deletion, and the SpeI recognition site insertion are shown in the resistant allele xa13.
(C) Counts of monoallelic, biallelic, and WT lines based on genotyping of OsEPSPS1-edited lines with BsrDI digestion of relevant PCR amplicons and OsSWEET11a/xa13-edited lines with SpeI digestion of relevant PCR amplicons.
(D) Summary of genotyping based on PCR-RE. Edited alleles are shown in bold.
(E) Treatment of OsEPSPS1-edited and WT lines with 4.2 mM glyphosate spray. Genotypes are indicated at the top. Picture was taken 10 days after spraying.
(F and G) Disease phenotypes of edited biallelic lines. Lesion lengths were measured 12 days post inoculation with PXO99 and ME2(pthXo1) on three to five leaves of individual plants (n = 3–5). Scale bar, 1 cm. Letters a and b in (G) represent statistically significant differences in lesion lengths of edited versus WT lines for both PXO99 and ME2(pthXo1) calculated by Tukey’s test.
To further validate the approach we used to classify edits as monoallelic or biallelic, we deep sequenced amplicons of both genes from different categories with various intensities of digestion to represent all the possibilities. Deep sequencing of OsEPSPS1 revealed that some lines were purely monoallelic (#1 and #4), with >45% of reads mapping to the edited sequence, whereas one line was partially monoallelic (#5), with >25% of reads mapping to an edited allele (Supplemental Figure 5A). The same line (#5) was mainly edited (>50% reads) in another allele, and only the third amino acid of the TAP-IVS edit was changed (P > S) (Supplemental Figure 5A). For the biallelic lines (#13 and #14), >70% reads mapped to the edited allele. These results confirmed that the callus lines were mixtures, or chimeras, in which some shoots carried biallelic edits and others were monoallelic or WT. Different shoots could carry different edits, as seen in #5, in which partial editing was observed. We Sanger sequenced two lines (#4 and #13) for OsEPSPS1 (Supplemental Figure 5C), and Sanger sequencing also validated the presence of the desired 3-bp substitution. For OsSWEET11a, we deep sequenced the amplicons of several lines and found that all lines carried an undesired deletion ranging from 19 to >40 bp in addition to the desired edit (Supplemental Figure 5B). These deletions may have originated from the ngRNA, which was used to enhance PE efficiency. Thus, further optimization is needed to decrease the frequency of by-product editing. Nevertheless, the OsSWEET11a edits knocked out the EBE from the promoter completely, and we decided to go forward with phenotyping evaluation. First, we screened individual tillers from some monoallelic and biallelic lines to identify the mutant biallelic tillers for phenotype analysis. We sprayed WT lines and lines carrying TAP-IVS edits with 4.2 mM glyphosate. The edited lines survived until maturity and seed set, whereas the WT lines died 5 days after spraying (Figure 4E). Similarly, we inoculated WT lines and lines carrying the OsSWEET11a EBE knockout edit with Xoo strain PXO99 and the ME2 strain carrying the pthXo1 TALE gene. The edited lines were completely resistant to both strains, whereas the WT plants were completely susceptible (Figure 4F and 4G). These results indicate the feasibility of multiplex PE for simultaneous editing of both base substitutions and deletions. The knockouts generated in this case are easier to screen owing to the incorporation of a restriction site with the help of PE. This is an advantage of PE compared with CRISPR–Cas9 for knockout generation. However, further efforts are still needed to reduce the number of by-products generated during editing.
Quadruplex prime editing efficiently edits four genes in the T0 generation
Next, we tested the editing efficiency of four targets using QPE. We selected two genes related to herbicide tolerance (OsEPSPS1 and OsALS1) and two genes associated with bacterial blight resistance (TFIIAγ5 and OsSWEET11a) (Figure 5A–5D). The targets and edits for OsEPSPS1, TFIIAγ5, and OsSWEET11a were the same as those in the DPE constructs. In OsALS1, a single amino acid substitution (S627I) has been shown to confer moderate bispyribac sodium tolerance (Li et al., 2022a). We generated constructs to edit these genes using modular assembly and transformed Kitaake using Agrobacterium. Twenty-three callus events regenerated on medium supplemented with hygromycin, and each callus line produced multiple T0 plants. As in the previous section, plants that originated from a single callus line were considered to represent single transformation events, and mutation analysis was performed on a mixture of all the T0 plants derived from individual callus lines. We first determined whether all the plants carried the nCas9 and the pegRNA units using PCR analysis (primer information in Supplemental Table 1). All plants were found to carry nCas9 and all four pegRNA units, ensuring stable transformation of multiple repeat units into the rice genome. We then genotyped the plants using the PCR-RE approach. OsEPSPS1 had a loss of the BsrDI RE site due to the editing, TFIIAγ5 gained an SmlI RE site with the edit, and OsALS1 had a gain of BsaBI due to editing (Figure 5A–5C). The intentionally incorporated SpeI was used to genotype the OsSWEET11a edits (Figure 5D). We categorized monoallelic, biallelic, and WT lines using the approach described in the last section. PCR-RE on all four genes revealed that at least one site was edited in all 23 lines, making the editing efficiency 100% (Figure 5E and 5F and Supplemental Figure 6; Supplemental Table 5). One line had one site edited, five had two sites edited, eight had three sites edited, and ten lines had all four sites edited (Figure 5E and 5F and Supplemental Figure 6; Supplemental Table 5). Again, to validate this categorization approach and to accurately genotype the edited lines, we deep sequenced several lines for all four genes. For OsEPSPS1, at least 35% of reads mapped to the edited allele in the monoallelic lines (#7, #12, and #37), and at least 70% of reads mapped to the edited allele in the biallelic lines (#21, #28, #53, and #56) (Supplemental Figure 7A). Two lines (#12 and #56) also carried the partially edited allele in which only one amino acid (P > S) of three (TAP > IVS) was edited (Supplemental Figure 7A). For OsALS1, >55% of reads mapped to the edited allele in the monoallelic lines (#22, #37, and #59), and >85% of reads mapped to the edited allele in the biallelic lines (#7, #12, #15, #28, and #58) (Supplemental Figure 7B). In TFIIAγ5, >30% of reads mapped to the edited alleles in the monoallelic lines (#7, #15, #21, #28, and #50), and >75% of reads mapped to the edited allele in the single biallelic line (#20) (Supplemental Figure 7C). In OsSWEET11a, we observed by-product deletions ranging from 12 to more than 40 bp in addition to the desired edits. Some lines (#1 and #59) carried two alleles; one allele was perfectly edited, whereas the other contained the by-product deletion along with the desired edit (Supplemental Figure 7D). We further validated these results using Sanger sequencing of the OsEPSPS1, OsALS1, and TFIIAγ5 genes (Supplemental Figure 8A–8C). All three genes were found to carry the desired edits in Sanger sequencing as well. Sanger sequencing of OsSWEET11a was attempted several times, but a good-quality read was never obtained; therefore, only deep-sequencing data for OsSWEET11a are presented here. Because the deletion in OsSWEET11a was only in the promoter region and did not reach to the coding region, and the other genes had desired edits only, we performed phenotype analysis of the edited lines under respective stresses (Figure 5G–5J). The lines edited for OsEPSPS1 and OsALS1 were sprayed with 4.2 mM glyphosate and 100 μM bispyribac sodium, respectively, and WT lines were sprayed as controls (Figure 5G and 5H). With the glyphosate spray, WT lines began to wilt as soon as 2 days post spray, and the edited lines remained unchanged. After 5 days, the WT lines had wilted completely, but the edited lines remained green, demonstrating that the OsEPSPS1 edits were active in the T0 generation (Figure 5G). The effect of bispyribac sodium spray took longer: WT plants began to show wilting 6 days after spraying and had completely wilted 10 days after spraying. By contrast, the edited plants showed no obvious wilting 10 days after spraying, demonstrating that the ALS1-S627I edit was also active in the T0 lines (Figure 5H). However, the OsALS1 mutant plants did not perform well and remain stunted compared with plants that did not receive any herbicide spray. This might be due to the moderate tolerance against bispyribac conferred by the single amino acid substitution, as opposed to the complete tolerance provided by the W548L and S627I mutations. Lines carrying the OsSWEET11a edits were resistant to Xoo strains PXO99 and ME2(pthXo1), and the line carrying the xa5 edit was resistant to PXO86 (Figure 5I and 5J).
Figure 5.
Quadruplex prime editing of OsEPSPS1, OsALS1, TFIIAγ5, and OsSWEET11a genes.
(A) Gene structures of the OsEPSPS1-TAP allele and the OsEPSPS1-IVS allele. The PE target site (underlined) in the OsEPSPS1-TAP allele and the desired amino acid change (in red) in the OsEPSPS1-IVS allele are shown.
(B) Structure of the OsALS1 gene with herbicide intolerant and tolerant alleles. The PE target site (underlined) in the OsALS1-S627 allele and the desired amino acid change (in red) in the OsALS1-I627 allele are shown.
(C) Gene structures of TFIIAγ5 and xa5. The PE target site (underlined) in the TFIIAγ5 allele and the nick site (in red) in the edited strand of the xa5 allele are shown.
(D) Gene structures of OsSWEET11a and xa13. The effector binding element (EBE) in the promoter of susceptible OsSWEET11a is shown in red. The desired edit, including partial EBE deletion, and the SpeI recognition site insertion are shown in the resistant allele xa13.
(E) Counts of monoallelic, biallelic, and WT lines based on PCR-RE of the T0 lines.
(F) Summary of genotypes based on PCR-RE.
(G) Treatment of OsEPSPS1-edited and WT lines with 4.2 mM glyphosate spray. Genotypes are shown at the top. Picture was taken 10 days after spraying.
(H) Treatment of OsALS1-edited and WT lines with 100 μM bispyribac sodium (BS) spray. Genotypes are shown at the top. Picture was taken 10 days after spraying.
(I and J) Disease phenotypes of edited biallelic lines. Lesion lengths were measured 12 days post inoculation with Xoo strains PXO99 and ME2(pthXo1) for OsSWEET11a/xa13-edited lines and PXO86 for TFIIAγ5-edited lines on three to five leaves of individual plants (n = 3–5). Scale bar, 1 cm. Letters a and b in (J) represent statistically significant differences in lesion lengths of edited versus WT lines for both PXO99 and ME2(pthXo1) for OsSWEET11a/xa13-edited lines and PXO86 for TFIIAγ5-edited lines calculated by Tukey’s test.
To further validate the multiplex PE system, we generated two additional constructs targeting three genes (TPE) and three additional constructs targeting four genes (QPE). In the first TPE construct, we targeted the genes OsEPSPS1 for TAP-IVS mutation, TFIIAγ5 for xa5 (V39E) mutation, and OsSPL14 (SQUAMOSA promoter-binding protein like 14) (Miura et al., 2010) for IPA1 (Ideal plant architecture 1) (Jiao et al., 2010) mutation. We achieved an overall editing efficiency of 70.5% with this construct; 12 out of the 17 T0 lines were edited for at least one gene (Supplemental Table 6), with four edited for one gene, 7 edited for two genes, and 1 edited for all three genes (Supplemental Tables 6 and 7). Sanger sequencing confirmed that line #5 carried the desired edits in all three genes (Supplemental Figure 9A–9C). For the second TPE construct, we targeted OsGS2 (Grain size 2) (Hu et al., 2015) to disrupt the microRNA 396 binding site while keeping the same amino acid sequence, along with TFIIAγ5 for xa5 (V39E) mutation and OsSPL14 for IPA1 mutation. The OsGS2 pegRNA did not lead to any editing (validated with PCR-RE, Sanger sequencing, and deep sequencing). Of the total 25 lines, 8 were edited for TFIIAγ5 to xa5, and 10 were edited for OsSPL14 to IPA1; of these edited lines, 6 were edited for both TFIIAγ5 to xa5 and OsSPL14 to IPA1. No line with edits in all three genes was obtained (Supplemental Tables 8 and 9).
For the first QPE construct, we targeted OsGS2 to disrupt the microRNA 396 binding site, OsSPL13 (SQUAMOSA-promoter binding protein like 13) for GLW7 (Grain length and weight 7) (Si et al., 2016; Gupta et al., 2023a) mutation (originally the GLW7 allele had a 6-bp deletion in the promoter; we replaced the 6 nt with the recognition site for SpeI), TFIIAγ5 for xa5 (V39E) mutation, and OsSPL14 for IPA1 mutation. The pegRNAs targeting genes OsGS2 and OsSPL13 did not lead to any editing in any of the T0 lines, as confirmed via PCR-RE, Sanger sequencing, and deep sequencing. The other two genes were edited at rates of 52.2% for TFIIAγ5/xa5 and 47.8% for OsSPL14/IPA1, and the co-editing rate of the two genes was 21.7% of all lines (Supplemental Tables 10 and 11). Note that the pegRNA–ngRNA used for OsGS2 was the same as that used in the previous TPE segment. We believe that this pegRNA–ngRNA has no activity for OsGS2 editing, similar to OsSPL13 editing. We decided to change the pegRNA–ngRNA of these two genes but, because of PAM (protospacer adjacent motif) restriction, we could change only the ngRNA. A new construct targeting the same four genes (OsGS2, OsSPL13, TFIIAγ5, and OsSPL14) but with a different ngRNA for OsGS2 and OsSPL13 was used for transformation of Kitaake. For OsSPL13, the new ngRNA led to 30% editing frequency based on PCR-RE, but no edits for OsGS2 were recovered (Supplemental Tables 12 and 13). This suggests that the pegRNA of OsGS2 has little to no activity. Overall, of the 20 T0 lines, 8 were edited for OsSPL13, TFIIAγ5, or OsSPL14; 6 were edited for two genes; and 1 was edited for three genes (Supplemental Tables 12 and 13). The OsSPL13 edit was validated using Sanger sequencing (Supplemental Figure 10A). Finally, we replaced the OsGS2 target with the OsEPSPS1 target for another QPE experiment, thus targeting OsEPSPS1, OsSPL13-2, TFIIAγ5, and OsSPL14 in one construct. Of the 18 T0 lines, 3 were unedited, 9 were edited for only one gene, 4 were edited for two genes, 2 were edited for three genes, and none were edited for all four genes (Supplemental Tables 14 and 15). The highest editing frequency was achieved for OsEPSPS1, with 13 lines carrying edits, followed by TFIIAγ5, with 4 lines edited, and OsSPL13 and OsSPL14, each with 3 lines edited (Supplemental Tables 14 and 15).
Discussion
The long-sought goal of biologists and crop breeders is to be able to precisely target and modify genes or genomes in living organisms. PE technology represents a significant advance toward achieving this capability (Jin et al., 2023). Over the past 3 years, remarkable progress has been made in enhancing PE efficiency in plants, elevating it from below 5% to nearly 100% (Jiang et al., 2020; Lin et al., 2020, 2021; Xu et al., 2020, 2021; Molla et al., 2021; Wang et al., 2021; Li et al., 2022a, 2022b; Zong et al., 2022; Gupta et al., 2023b; Jin et al., 2023; Ni et al., 2023; Qiao et al., 2023). This advance has now paved the way for efficient multigene targeting, a breakthrough that we demonstrate in this study. Although the prospect of multiplex PE has been demonstrated in wheat for up to eight genes (Ni et al., 2023) and in rice for up to three genes (Li et al., 2022a), the cloning of PE reagents has remained a daunting task, limiting its use to some labs with expertise in molecular cloning. By developing a modular assembly-based PE system for plants, we successfully targeted up to four genes in a single generation (Figure 1A–1K). The tandem pegRNA–ngRNA cassettes, although rich in repeats, remained stable in both Escherichia coli and Agrobacterium. In addition, the transgenes contained all four units (in the case of QPE) in multiple transformation events. Editing efficiency was found to be dependent on the activity of the pegRNA–ngRNA, and we did not observe any differences in editing rates based on the position of the pegRNA–ngRNA unit in the QPE. All units (except for the OsGS2 pegRNA–ngRNA unit) were found to be active, resulting in mutations in the T0 generation. The introduction of modular assembly not only streamlines the cloning of PE-required reagents for single-gene targeting but also facilitates the targeting of multiple genes, thereby empowering numerous labs to leverage the full potential of PE for their genome-editing experiments. Furthermore, the system can easily be expanded to an even higher number of multiplexed pegRNA–ngRNA units.
In this study, we demonstrated the development and use of a multiplex PE system in rice to target traits related to disease resistance, herbicide tolerance, plant architecture, and grain yield, thereby harnessing the potential of PE to improve multiple agronomic traits in a single editing experiment. A bacterial disease of rice caused by Xanthomonas oryzae is the major threat to global rice production, and it can cause up to 70% yield loss in years of severe infection (Srinivasan and Gnanamanickam, 2005). In our previous study, we successfully employed PE to develop two distinct strategies to impart genetic resistance against bacterial blight of rice. The first strategy involved introduction of the EBE from the OsSWEET14 gene into the promoter of the dysfunctional “Executor” R gene xa23, making it a functional R gene, Xa23SW14, and leading to dominant resistance that effectively protects rice against all Xoo strains carrying pthXo3/avrXa7 TALE genes. The second strategy relied on xa5, which conferred recessive resistance, offering protection against all Asian Xoo strains except those carrying the pthXo1 TALE gene (Gupta et al., 2023b). To build upon these promising outcomes, we further employed DPE to combine the Xa23SW11 (in this case, the EBE from OsSWEET11a corresponding to the pthXo1 TALE gene was incorporated into the promoter of xa23) and xa5 edits in rice (Figures 2A–2G, 3A, and 3B). By creating this novel allelic combination not found in nature, we achieved robust and broad-spectrum resistance against all tested Xoo strains, including PXO99, which harbors the challenging pthXo1 TALE gene (Figures 2A–2G, 3A, and 3B).
We incorporated a third strategy to provide genetic resistance against Asian Xoo strains by combining promoter EBE deletion/knockout of OsSWEET11a with the xa5 edit. In the same construct, we edited two herbicide-related genes, OsEPSPS1 and OsALS1, to their herbicide-tolerant alleles. In this QPE experiment, we achieved a high editing efficiency of 100%; all the lines were edited for at least one gene, and the co-editing efficiency for all four genes was 43.5%. We were able to detect completely biallelic or near biallelic edits (from a mixture of T0 lines originating from a single callus event) for all four genes in the T0 generation. Except for the OsSWEET11a EBE deletion, all genes had the desired edits, whereas OsSWEET11a had undesired deletions along with the desired edits (Figure 5A–5F and Supplemental Figures 7A–7D and 8A–8C; Supplemental Table 5). All edits were found to be active in the T0 generation as tested by challenging the edited plants with Xoo infection or herbicide spray (Figure 5G–5J). These results demonstrate the feasibility of multiplex PE for targeting multiple trait-related genes and testing the activity of new alleles in the T0 generation.
We validated the prospect of multiplex PE with five additional constructs that targeted either three or four genes concurrently. The editing efficiencies of these constructs varied depending upon the target and pegRNA–ngRNA used. Some targets were edited at very high rates, whereas others remained unedited or edited at lower rates. Because of the limitation of the PAM requirement, there is not much flexibility in terms of choosing a pegRNA, and PE rates are thus dependent on the activity of the pegRNA. The success of PE depends largely upon the activity of the pegRNA unit. In this study, we mainly selected pegRNAs that had previously shown activity in rice protoplasts or stable lines (Jiang et al., 2022; Gupta et al., 2023b), except for the pegRNAs of OsSPL13, OsSPL14, and OsGS2. This minimized the effort needed to optimize each pegRNA unit and ensured higher activity of these pegRNA units for testing the multiplexed system. Another component of the PE3 or PE5 system is the ngRNA, which nicks the unedited strand either upstream or downstream of the target region. Flexibility to choose the most active ngRNA near the target site requires optimization for every target. In our case, switching the ngRNA for OsSPL13 targeting increased the editing rate from 0% to 30%, whereas changing the ngRNA for OsGS2 had no effect, and no edits for OsGS2 were obtained with any construct. Perhaps the OsGS2 pegRNA had no or very low activity, and switching the ngRNA did not help in that case, whereas the OsSPL13 pegRNA was active, and switching to an alternative ngRNA (perhaps with better activity than the first ngRNA) complemented the pegRNA to yield 30% editing in T0. This result highlights the need for further optimization of PE for recalcitrant targets and/or development of PAM-flexible or PAM-less Cas9 variants to be used for PE to allow selection of the best pegRNA for the target site.
Our results not only showcase the potential of multiplex PE in rice but also pave the way for more efficient and effective genetic resistance strategies against bacterial blight of rice. This strategy for design and construction of modular pegRNA–ngRNA units is readily applicable to multiplex PE in other crop species. The multiplex approach demonstrated in this study holds immense promise for significantly improving various agronomic traits simultaneously, providing a transformative and sustainable solution for rice production and food security.
Methods
All primers used in this study are listed in Supplemental Table 1, and pegRNAs and ngRNAs are listed in Supplemental Table 2.
Plant materials, bacterial strains, medium, and growth conditions
All editing experiments were performed using the japonica rice variety Kitaake (Oryza sativa spp. japonica). The Xoo strains used in the experiments were from the Yang laboratory’s collection. Rice plants were grown in a greenhouse and growth chambers with a 12-h/30°C light period and a 12-h/28°C dark period and a relative humidity of 60% to 75%. E. coli and Agrobacterium tumefaciens strains were cultivated in Luria-Bertani medium supplemented with appropriate antibiotics at temperatures of 37°C and 28°C, respectively. Xoo was grown on TSA (10 g/l tryptone, 10 g/l sucrose, 1 g/l glutamic acid, 1.5% Difco agar) at a temperature of 28°C. When necessary, the following concentrations of antibiotics were used: 25 μg/ml rifampicin, 50 μg/ml kanamycin, and 100 μg/ml spectinomycin.
Disease assays
The leaf tip-clipping method was used to assess the disease phenotypes of edited rice as described previously (Yang and Bogdanove, 2013). In brief, Xoo glycerol stock stored at −80°C was streaked onto TSA (containing appropriate antibiotics) and grown at 28°C for approximately 3 days. Bacterial cells were then harvested from the plates, suspended in sterile water, washed twice, and resuspended in water. The optical density of the bacterial inoculum was adjusted to 0.5 at 600 nm. To perform the experiment, scissor blades were immersed in the Xoo suspension and used to clip the tips of fully expanded leaves. The resulting lesion lengths were measured either 12 days after inoculation or at specified time points. Each Xoo strain was tested with three to five replicates, each containing multiple leaves.
Data analysis was performed using R software, and the ggplot2 (Villanueva and Chen, 2019) and ggpubr (Kassambara and Kassambara, 2020) packages were used for plotting. The R package rstatix (Kassambara, 2020) was used to perform two-tailed Student’s t-tests, with or without Bonferroni correction for multiple comparisons. Tukey’s post hoc tests were performed using the R package agricolae (de Mendiburu and de Mendiburu, 2019).
Development of the modular prime editing system
To develop the modular PE system, we digested the original PE3max vector with PmeI–AflII to remove the 35S-CmYCLV-AtU6-pegRNA cassette and replaced it with attR1-ccdb-attR2 using a Gibson assembly kit (New England Biolabs), resulting in pG3H-PE3max-attR1R2. The resulting vector served as the destination vector for PE cloning. To construct the entry vectors, the 35S-CmYCLV-AtU6-pegRNA cassette was synthesized as gBlock from Integrated DNA Technologies and cloned into pCR8-attL1-attL2, pCR8-attL1-attR5, pCR8-attL5-attL2, pCR8-attL5-attL4, pCR8-attR4-attL2, pCR8-attR4-attR3, and pCR8-attL3-attL2 vectors between the att regions, resulting in modular pCR8-pegRNA-ngRNA entry vectors. The double-stranded oligonucleotides with proper 4-nt overhangs at each side for the pegRNA spacer were first cloned at the BsmBI site, and, similarly, oligonucleotides corresponding to the extension RNA region and ngRNA were sequentially cloned at the BsaI sites of the respective pCR8-pegRNA vectors. All plasmids were confirmed by whole-plasmid sequencing via Plasmidsaurus. A detailed protocol for the design of pegRNA–ngRNAs and their subsequent cloning entry vectors and destination vector is provided in the Supplemental Protocol.
Rice transformation
Kitaake rice was transformed with the Agrobacterium-based DNA delivery method with slight modifications, following the procedure described by Hiei et al. (1994). In brief, mature seed embryos of Kitaake were used for callus induction in Murashige and Skoog (MS) medium supplemented with 2 mg/l 2,4-dichlorophenoxyacetic acid. Callus cells derived from the scutella were co-cultivated with Agrobacterium strain LBA4404/pVS1-VIR2 carrying the appropriate PE plasmids. The inoculated callus cells were cultured in MS medium supplemented with 2,4-dichlorophenoxyacetic acid (2 mg/l), hygromycin (50 mg/l), and Timentin (200 mg/l) for two rounds of selection (14 days per round) to generate hygromycin-resistant callus lines. The hygromycin-resistant callus lines were then transferred to a regeneration medium (MS supplemented with BAP and NAA) to induce formation of embryogenic shoots. The developed shoots were transferred to a rooting medium (½ MS medium supplemented with 25 mg/l hygromycin) to facilitate root formation, then transferred to soil and cultivated in a greenhouse.
RNA isolation and gene expression analysis
RNA was extracted from the leaves of PE-edited and WT Kitaake lines that had been infiltrated with Xoo inoculum using a needleless syringe. DNase I treatment (Thermo Fisher Scientific) was applied to eliminate any remaining DNA. RNA quality was evaluated using agarose gel electrophoresis, and RNA concentration was measured using a NanoDrop instrument (Thermo Fisher Scientific). First-strand cDNA was synthesized from 1 μg of RNA using the iScript cDNA synthesis kit (Bio-Rad). The resulting cDNA was diluted at 1:20 for use in RT-PCR and RT-qPCR with gene-specific primers. For RT-qPCR, PowerTrack SYBR master mix (Thermo Fisher Scientific) was used. OsActin was used as the housekeeping control gene, and the 2−ΔΔCt method was used to calculate the fold change.
Genotyping of PE callus lines and T0 and T1 plants and deep sequencing analysis
DNA was isolated from T0 and T1 lines using the CTAB method. To detect editing events, primers flanking the target sites were used for PCR amplification of the specific regions, which were then digested with appropriate enzymes. The PCR amplicons from the edited lines were subjected to deep sequencing using the Illumina MiSeq instrument (PE150). In brief, the 150- to 250-bp region flanking the target site was first amplified in the initial PCR round using gene-specific primers extended with sequencing primers. Subsequently, a second nested PCR was performed using dual barcoded Illumina adapters to amplify the gene-specific products from the first round. The resulting PCR products were purified using columns, pooled in equal quantities, and sent for sequencing at the DNA sequencing core facility of the University of Missouri–Columbia and to Azenta–GENEWIZ for deep sequencing. The obtained reads were demultiplexed and trimmed during the sequencing process. For analysis, CRISPResso2 was used with default settings for both NHEJ and PE output (Pinello et al., 2016).
Herbicide treatment
EPSPS1-edited plants were sprayed with 2 ml/l (4.2 mM) commercial glyphosate (Monsanto), and pictures were taken 10 days after treatment. ALS1-edited plants were sprayed with 100 μM bispyribac sodium salt, and pictures were taken 10 days post spraying.
Statistics and data analysis
Data were analyzed and plotted using the R packages ggplot2 (Villanueva and Chen, 2019), ggpubr (Kassambara and Kassambara, 2020), rstatix (Kassambara, 2020), and agricolae (de Mendiburu and de Mendiburu, 2019). Tukey’s test was used for all figures with statistics.
Data and code availability
The plant materials and constructs generated in this study are available upon request.
Funding
The work was partially supported by an NSF award (IOS-2210259 to B.Y.) and a subaward to the University of Missouri from the Heinrich Heine University of Dusseldorf funded by the Bill & Melinda Gates Foundation (OPP1155704). A.G. is partially supported by the Daniel Millikan Award for Outstanding Research in Plant–Microbe Interactions at the University of Missouri.
Author contributions
A.G. and B.Y. designed the research; A.G., B.L., and S.R. performed the research; Q.-J.C., A.G., and B.Y. analyzed the data; and A.G. and B.Y. wrote the paper with revisions from the other authors.
Acknowledgments
No conflict of interest is declared.
Published: October 26, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- de Mendiburu F., de Mendiburu M.F. 2019. Package ‘agricolae’. R Package, version 1. [Google Scholar]
- Gupta A., Hua L., Zhang Z., Yang B., Li W. CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol. J. 2023;21:536–548. doi: 10.1111/pbi.13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A., Liu B., Chen Q.J., Yang B. High-efficiency prime editing enables new strategies for broad-spectrum resistance to bacterial blight of rice. Plant Biotechnol. J. 2023;21:1454–1464. doi: 10.1111/pbi.14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan M.M., Yuan G., Chen J.-G., Tuskan G.A., Yang X. Prime editing technology and its prospects for future applications in plant biology research. Biodes. Res. 2020;2020 doi: 10.34133/2020/9350905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hu J., Wang Y., Fang Y., Zeng L., Xu J., Yu H., Shi Z., Pan J., Zhang D., Kang S., et al. A rare allele of GS2 enhances grain size and grain rield in rice. Mol. Plant. 2015;8:1455–1465. doi: 10.1016/j.molp.2015.07.002. [DOI] [PubMed] [Google Scholar]
- Hua K., Han P., Zhu J.K. Improvement of base editors and prime editors advances precision genome engineering in plants. Plant Physiol. 2022;188:1795–1810. doi: 10.1093/plphys/kiab591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Antony G., Li T., Liu B., Obasa K., Yang B., White F.F. The broadly effective recessive resistance gene xa5 of rice is a virulence effector-dependent quantitative trait for bacterial blight. Plant J. 2016;86:186–194. doi: 10.1111/tpj.13164. [DOI] [PubMed] [Google Scholar]
- Huang T.K., Puchta H. Novel CRISPR/Cas applications in plants: from prime editing to chromosome engineering. Transgenic Res. 2021;30:529–549. doi: 10.1007/s11248-021-00238-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Chai Y., Qiao D., Wang J., Xin C., Sun W., Cao Z., Zhang Y., Zhou Y., Wang X.C., et al. Optimized prime editing efficiently generates glyphosate-resistant rice plants carrying homozygous TAP-IVS mutation in EPSPS. Mol. Plant. 2022;15:1646–1649. doi: 10.1016/j.molp.2022.09.006. [DOI] [PubMed] [Google Scholar]
- Jiang Y.Y., Chai Y.P., Lu M.H., Han X.L., Lin Q., Zhang Y., Zhang Q., Zhou Y., Wang X.C., Gao C., et al. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol. 2020;21:257. doi: 10.1186/s13059-020-02170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010;42:541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- Jin S., Lin Q., Gao Q., Gao C. Optimized prime editing in monocot plants using PlantPegDesigner and engineered plant prime editors (ePPEs) Nat. Protoc. 2023;18:831–853. doi: 10.1038/s41596-022-00773-9. [DOI] [PubMed] [Google Scholar]
- Kassambara A. 2020. rstatix: Pipe-Friendly Framework for Basic Statistical Tests. R package version 0.6. 0. [Google Scholar]
- Kassambara A., Kassambara M.A. 2020. Package ‘ggpubr’. R package version 0.1 6. [Google Scholar]
- Kumar J., Char S.N., Weiss T., Liu H., Liu B., Yang B., Zhang F. Efficient protein tagging and cis-regulatory element engineering via precise and directional oligonucleotide-based targeted insertion in plants. Plant Cell. 2023;35:2722–2735. doi: 10.1093/plcell/koad139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhu Z., Li S., Li J., Yan L., Zhang C., Ma Y., Xia L. Multiplex precision gene editing by a surrogate prime editor in rice. Mol. Plant. 2022;15:1077–1080. doi: 10.1016/j.molp.2022.05.009. [DOI] [PubMed] [Google Scholar]
- Li J., Chen L., Liang J., Xu R., Jiang Y., Li Y., Ding J., Li M., Qin R., Wei P. Development of a highly efficient prime editor 2 system in plants. Genome Biol. 2022;23:161. doi: 10.1186/s13059-022-02730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang C., He Y., Li S., Yan L., Li Y., Zhu Z., Xia L. Plant base editing and prime editing: The current status and future perspectives. J. Integr. Plant Biol. 2023;65:444–467. doi: 10.1111/jipb.13425. [DOI] [PubMed] [Google Scholar]
- Lin Q., Jin S., Zong Y., Yu H., Zhu Z., Liu G., Kou L., Wang Y., Qiu J.L., Li J., et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021;39:923–927. doi: 10.1038/s41587-021-00868-w. [DOI] [PubMed] [Google Scholar]
- Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A.V., Raguram A., Doman J.L., et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020;38:582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010;42:545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- Molla K.A., Sretenovic S., Bansal K.C., Qi Y. Precise plant genome editing using base editors and prime editors. Nat. Plants. 2021;7:1166–1187. doi: 10.1038/s41477-021-00991-1. [DOI] [PubMed] [Google Scholar]
- Ni P., Zhao Y., Zhou X., Liu Z., Huang Z., Ni Z., Sun Q., Zong Y. Efficient and versatile multiplex prime editing in hexaploid wheat. Genome Biol. 2023;24:156. doi: 10.1186/s13059-023-02990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J.C., Perez-Quintero A., Li T., Eom J.S., Li C., Nguyen H., Liu B., et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019;37:1344–1350. doi: 10.1038/s41587-019-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao D., Wang J., Lu M.H., Xin C., Chai Y., Jiang Y., Sun W., Cao Z., Guo S., Wang X.C., et al. Optimized prime editing efficiently generates heritable mutations in maize. J. Integr. Plant Biol. 2023;65:900–906. doi: 10.1111/jipb.13428. [DOI] [PubMed] [Google Scholar]
- Si L., Chen J., Huang X., Gong H., Luo J., Hou Q., Zhou T., Lu T., Zhu J., Shangguan Y., et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016;48:447–456. doi: 10.1038/ng.3518. [DOI] [PubMed] [Google Scholar]
- Srinivasan B., Gnanamanickam S.S. Identification of a new source of resistance in wild rice, Oryza rufipogon to bacterial blight of rice caused by Indian strains of Xanthomonas oryzae pv. oryzae. Curr. Sci. 2005;88:1229–1231. [Google Scholar]
- Villanueva R.A.M., Chen Z.J. Ggplot2: Elegant graphics for data analysis. Measurement: Interdisciplinary research and perspectives. 2019:160–167. 2nd ed.17. [Google Scholar]
- Wang L., Kaya H.B., Zhang N., Rai R., Willmann M.R., Carpenter S.C.D., Read A.C., Martin F., Fei Z., Leach J.E., et al. Spelling changes and fluorescent tagging with prime editing vectors for plants. Front. Genome Ed. 2021;3 doi: 10.3389/fgeed.2021.617553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Li J., Liu X., Shan T., Qin R., Wei P. Development of plant prime-editing systems for precise genome editing. Plant Commun. 2020;1 doi: 10.1016/j.xplc.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Liu X., Li J., Qin R., Wei P. Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice. Nat. Plants. 2021;7:888–892. doi: 10.1038/s41477-021-00942-w. [DOI] [PubMed] [Google Scholar]
- Xu W., Yang Y., Yang B., Krueger C.J., Xiao Q., Zhao S., Zhang L., Kang G., Wang F., Yi H., et al. A design optimized prime editor with expanded scope and capability in plants. Nat. Plants. 2022;8:45–52. doi: 10.1038/s41477-021-01043-4. [DOI] [PubMed] [Google Scholar]
- Yang B., Bogdanove A. Inoculation and virulence assay for bacterial blight and bacterial leaf streak of rice. Methods Mol. Biol. 2013;956:249–255. doi: 10.1007/978-1-62703-194-3_18. [DOI] [PubMed] [Google Scholar]
- Yang B., Sugio A., White F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Yang B., Chen J. One Prime for All Editing. Cell. 2019;179:1448–1450. doi: 10.1016/j.cell.2019.11.030. [DOI] [PubMed] [Google Scholar]
- Zong Y., Liu Y., Xue C., Li B., Li X., Wang Y., Li J., Liu G., Huang X., Cao X., et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 2022;40:1394–1402. doi: 10.1038/s41587-022-01254-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The plant materials and constructs generated in this study are available upon request.