Abstract
Root-knot nematodes (RKNs) cause huge agricultural losses every year. They secrete a repertoire of effectors to facilitate parasitism through the induction of plant-derived giant feeding cells, which serve as their sole source of nutrients. However, the mode of action of these effectors and their targeted host proteins remain largely unknown. In this study, we investigated the role of the effector Mi2G02 in Meloidogyne incognita parasitism. Host-derived Mi2G02 RNA interference in Arabidopsis thaliana affected giant cell development, whereas ectopic expression of Mi2G02 promoted root growth and increased plant susceptibility to M. incognita. We used various combinations of approaches to study the specific interactions between Mi2G02 and A. thaliana GT-3a, a trihelix transcription factor. GT-3a knockout in A. thaliana affected feeding-site development, resulting in production of fewer egg masses, whereas GT-3a overexpression in A. thaliana increased susceptibility to M. incognita and also root growth. Moreover, we demonstrated that Mi2G02 plays a role in maintaining GT-3a protein stabilization by inhibiting the 26S proteasome-dependent pathway, leading to suppression of TOZ and RAD23C expression and thus promoting nematode parasitism. This work enhances our understanding of how a pathogen effector manipulates the role and regulation of a transcription factor by interfering with a proteolysis pathway to reprogram gene expression for development of nematode feeding cells.
Key words: Meloidogyne incognita, effector, giant cell, Mi2G02, transcription factor, interaction
Root-knot nematodes establish parasitic relationships with host plants by secreting effectors. This study demonstrates the roles of the effector Mi2G02 and its target GT-3a, a trihelix transcription factor, in the plant nucleus. Mi2G02 maintains GT-3a protein stability by inhibiting the 26S proteasome-dependent pathway, leading to suppression of TOZ and RAD23C expression and thus promoting Meloidogyne incognita parasitism.
Introduction
Root-knot nematodes (RKNs; Meloidogyne spp.) can infect thousands of plant species, causing huge agricultural losses every year (Abad et al., 2008; Jones et al., 2013). RKN juveniles induce the redifferentiation of plant vascular cells to establish feeding structures that support their development into reproductive adult females (Bartlem et al., 2014). The second-stage juveniles (J2s) enter the host in the root elongation area and migrate intercellularly toward the vascular tissues, where they select 5–7 parenchyma cells into which they inject esophageal gland secretions through a syringe-like stylet (Favery et al., 2020). These secretions contain proteinaceous effectors that reprogram the root cells to become giant cells (GCs), hypertrophied multinucleate feeding cells that undergo several rounds of nuclear division without cell division and extensive endoreduplication, with expansion by isotropic growth (Caillaud et al., 2008). The cells surrounding the GCs simultaneously divide to form a typical root knot or gall. The vascular tissues undergo extensive reorganization, and the xylem proliferates (Bartlem et al., 2014; Yamaguchi et al., 2017; Baldacci-Cresp et al., 2020). GCs are the only source of nutrients for RKNs throughout their life cycle; the nematode must therefore maintain this parasitic interaction for several weeks until the female can lay her eggs in a gelatinous matrix on the outside of the root tissues (Favery et al., 2020).
This intricate biotrophic interaction requires the nematode to cope with host defense responses, alter host cell morphology, and hijack the physiology of host cells for its own benefit. The nematode achieves these ends by inducing a deep transcriptional reprogramming of host cells, as demonstrated by a large number of transcriptomic studies (Jammes et al., 2005; Fuller et al., 2007; Portillo et al., 2009; Barcala et al., 2010; Escobar et al., 2011; Cabrera et al., 2014a; Olmo et al., 2017; Yamaguchi et al., 2017; Shukla et al., 2018; Warmerdam et al., 2018; Przybylska and Spychalski, 2021; Sato et al., 2021; Zhu et al., 2022). Genes encoding transcription factors (TFs), which regulate gene expression by binding to appropriate DNA elements and recruiting additional proteins to initiate transcription (Strader et al., 2022), are among the genes known to display differential expression in galls (Cabrera et al., 2014a; Yamaguchi et al., 2017; Przybylska and Spychalski, 2021; Zhu et al., 2022). Many of these plant TFs are known to be key players in the regulation of plant developmental processes and stress responses. They include ETHYLENE-RESPONSIVE FACTORS (ERFs), NO APICAL MERISTEM (NAC), AUXIN RESPONSE FACTORS (ARFs), and LATERAL ORGAN BOUNDARIES DOMAIN (LBD) (Shukla et al., 2018). However, very little is known about the roles of these TFs in GC formation and RKN parasitism. For LBD16, inactivation led to a decrease in infection or even a total absence of feeding-site formation (Cabrera et al., 2014b; Olmo et al., 2017).
RKN effectors are clearly involved in modulating host transcriptional responses. They may act as TFs, as has been shown for the Meloidogyne incognita effector 7H08, which localizes to the plant cell nucleus and functions as a transcriptional activator (Zhang et al., 2015). Other effectors may associate with and dysregulate host TFs. Mi16D10 may be one such effector, as it has been shown to interact with plant SCARECROW-like TFs known to regulate root development (Huang et al., 2006). Another example is provided by MiEFF18, an effector that interacts with the spliceosomal protein SmD1 to trigger alternative splicing events during pre-mRNA maturation in galls, thereby increasing the diversity of host transcripts (Mejias et al., 2021, 2022).
M. incognita secreted protein 2 (Mi-msp2 or Mi2G02) was initially identified as a putative parasitism gene expressed exclusively in the subventral esophageal gland cells of parasitic J2s (Huang et al., 2003). It was subsequently shown to be required for M. incognita parasitism in host-derived RNA interference experiments (Joshi et al., 2019, 2022). Mj2G02, an ortholog from Meloidogyne javanica, has been shown to suppress Gpa2/RBP-1-triggered cell death in Nicotiana benthamiana and jasmonate-mediated plant immune responses (Song et al., 2021). 2G02 proteins are nuclear effectors; they have a ShK toxin (ShKT) domain encoding a potassium-channel inhibitor first identified in a sea anemone (Stichodactyla helianthus) (Tudor et al., 1996). Proteins containing ShKT domains are widely expressed in parasitic and non-parasitic nematodes (Hewitson et al., 2013). There is evidence to suggest that ShKT-like domains act as contact surfaces for protein interactions (Thein et al., 2009) and immune evasion (Chhabra et al., 2014; Niu et al., 2016; Song et al., 2021). However, the plant proteins targeted by Mi2G02 are unknown.
In this study, we show that the Mi2G02 effector interacts with the Arabidopsis thaliana trihelix TF GT-3a and that this interaction is important for nematode parasitism. We also demonstrate that GT-3a functions as a transcription inhibitor, binding to the promoters of TOZ and RAD23C and thereby modulating plant cell development for M. incognita parasitism. We found that Mi2G02 stabilized GT-3a by inhibiting the 26S proteasome-dependent pathway, thereby causing stronger suppression of TOZ and RAD23C expression. Collectively, these results demonstrate the involvement of a M. incognita effector (Mi2G02), a plant TF (GT-3a), and downstream regulatory genes in the formation and development of multinucleate GCs for nematode parasitism.
Results
Mi2G02 is a nematode nuclear effector essential for giant cell development
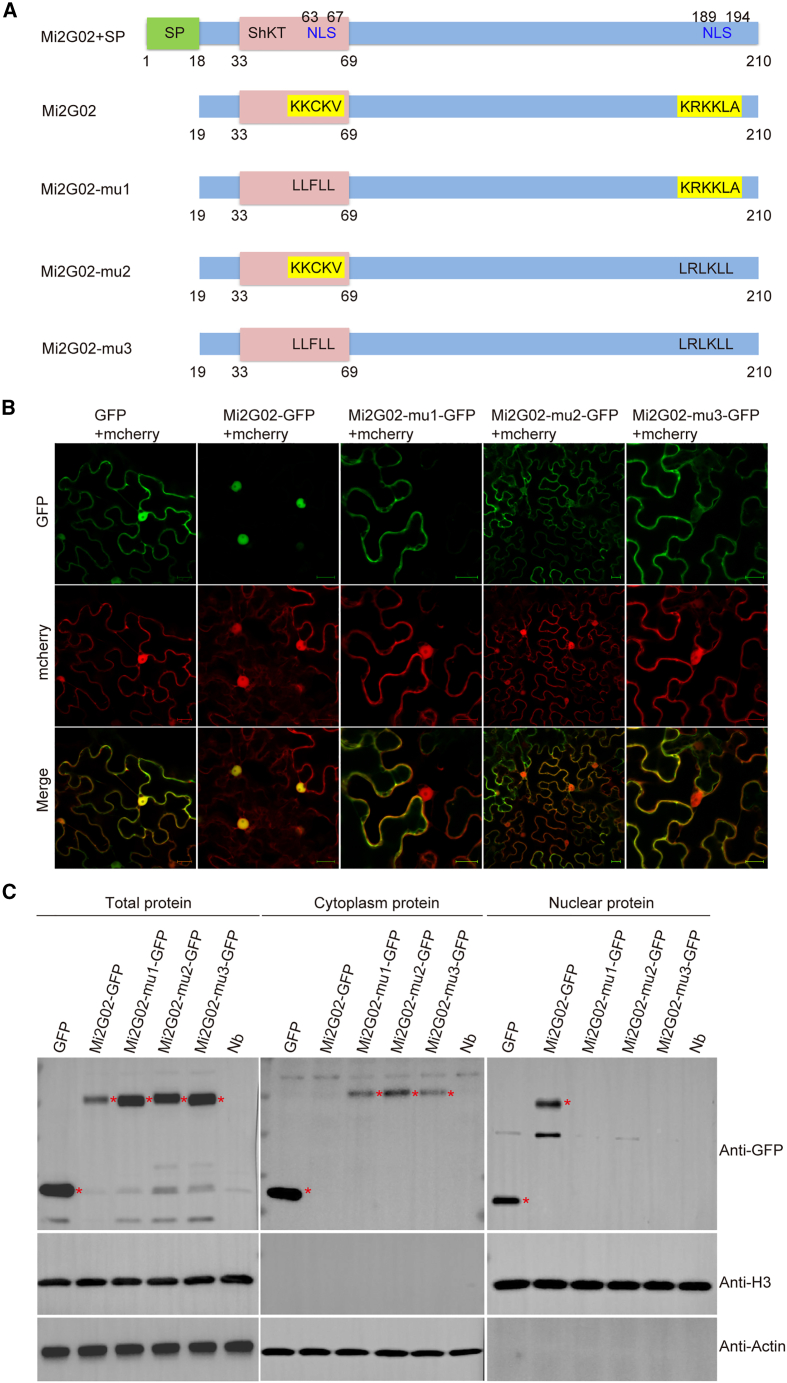
The Mi2G02/MiMSP2 gene encodes a 210-amino-acid (aa) protein with an 18-aa N-terminal signal peptide, a ShKT domain (33–69 aa), and two nuclear localization signals (NLS-1 and NLS-2), one of which is located in the ShKT domain (Figure 1A). Transient expression assays were performed in N. benthamiana leaves to investigate the functionality of the NLSs in planta. Mi2G02-GFP recombinant fusion proteins were detected exclusively in the plant cell nucleus and exclusively in the nucleoplasm but were excluded from the nucleolus (Figure 1B). We mutated the two predicted nuclear localization signals in the effector sequence to generate Mi2G02-mu1 (mutated NLS-1), Mi2G02-mu2 (mutated NLS-2), and Mi2G02-mu3 (mutated NLS-1 and NLS-2) (Figure 1A). Mi2G02-mu1-GFP, Mi2G02-mu2-GFP, and Mi2G02-mu3-GFP were found principally in the cytoplasm (Figure 1B). Immunoblotting of cytoplasmic- and nuclear-fraction proteins extracted from N. benthamiana leaves confirmed the nuclear expression of Mi2G02-GFP but not Mi2G02-mu1-GFP, Mi2G02-mu2-GFP, and Mi2G02-mu3-GFP (Figure 1C). These results demonstrated that both NLSs were required for localization of Mi2G02 to the nucleus.
Figure 1.
Structure and nuclear localization of the M. incognita effector Mi2G02.
(A) Schematic diagram of Mi2G02 and mutant Mi2G02 proteins.
(B) Subcellular localization of Mi2G02 and mutant Mi2G02 in plant cells. Coding sequences were ligated into the ProSuper:GFP vector (C-terminal GFP). Mi2G02 and nuclear localization signal mutants fused with GFP (Mi2G02-GFP, Mi2G02-mu1-GFP, Mi2G02-mu2-GFP, and Mi2G02-mu3-GFP) were co-expressed with mCherry in N. benthamiana leaf cells. Empty vectors were used as controls. The fluorescence signal was detected at 48 h after infiltration. Mi2G02-GFP localized to the nucleus. Mi2G02-mu1-GFP, Mi2G02-mu2-GFP, and Mi2G02-mu3-GFP localized primarily to the plasma membrane and cytoplasm. Images were captured by confocal microscopy (Zeiss LSM 700). GFP, green fluorescent protein. Scale bars, 20 μm.
(C) The relative abundance of Mi2G02-GFP or Mi2G02-mu-GFP in cytoplasmic and nuclear fractions was detected using anti-GFP antibodies after transient expression in N. benthamiana leaf cells. Actin was used as an internal reference for the presence of cytoplasmic proteins, and histone H3 was used as an internal reference for the presence of nuclear proteins.
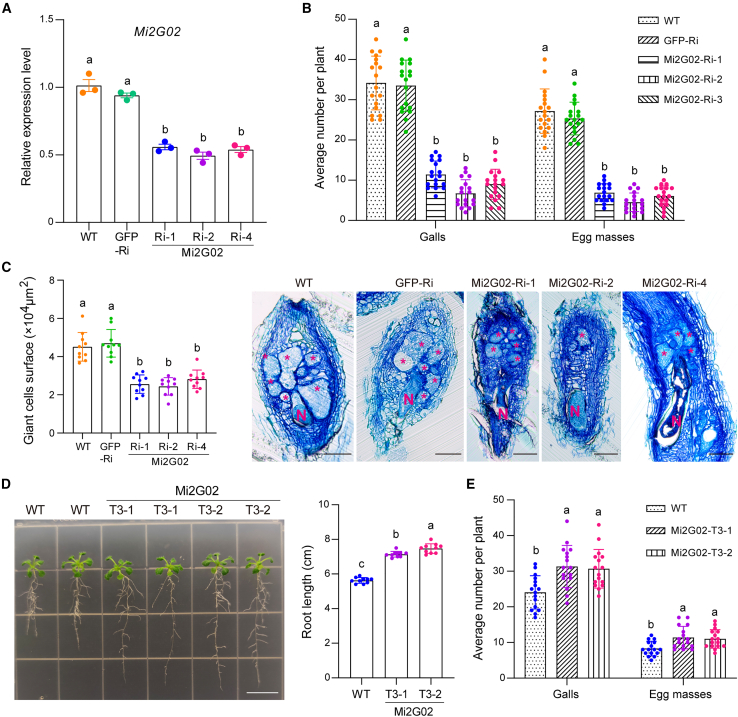
We investigated the role of Mi2G02 in giant cell formation by generating three homozygous RNAi A. thaliana lines expressing the Mi2G02 hairpin double-stranded RNA. Expression of the hairpin construct in these RNAi lines was confirmed by PCR (Supplemental Figure 1A), and the transgenic plants were inoculated with nematodes. Silencing of Mi2G02 by host-derived RNAi was confirmed by quantitative reverse transcription–PCR (qRT–PCR) assays of feeding nematodes recovered from the infested plants (Figure 2A and Supplemental Figure 1B). Consistent with previous findings (Joshi et al., 2019), Mi2G02 silencing decreased the numbers of galls and egg masses by at least 60% in the Mi2G02 RNAi lines relative to the two control lines: wild-type plants and a GFP RNAi line (Figure 2B and Supplemental Figure 1B). We further investigated the role of Mi2G02 in formation of RKN-induced feeding sites by analyzing the morphology of the feeding cells induced in the RNAi lines. All three Mi2G02 RNAi lines showed significantly smaller (30%) GC areas than the controls (Figure 2C). These findings suggest that Mi2G02 plays a role in nematode parasitism, particularly in the development of GCs.
Figure 2.
Host-derived RNA interference and ectopic expression of Mi2G02 in A. thaliana alter plant susceptibility to M. incognita and root development.
(A)Mi2G02 expression level in three homozygous RNAi lines (Mi2G02-Ri-1, Mi2G02-Ri-2, and Mi2G02-Ri-4), a gfp-RNAi line (GFP-Ri), and the wild type (WT) were determined at 10 days post infection (dpi) with M. incognita by qRT–PCR. GAPDH was used as an internal control. The values shown are means ± SE (n = 3). Different letters indicate significant differences (P < 0.05, one-way ANOVA).
(B) Gall numbers and egg mass numbers per plant at 35 dpi. Values are presented as means ± SD (n = 18). Different letters indicate significant differences (P < 0.05, one-way ANOVA). See also Supplemental Figure 1B.
(C) Giant cell areas of M. incognita-induced galls were significantly reduced in the A. thaliana Mi2G02-Ri lines. Gall sections at 14 dpi were stained with toluidine blue. Smaller giant cells were observed in Mi2G02-Ri mature galls at 14 dpi compared with the WT and GFP-Ri controls. Data are the average surface areas ±SD (n = 10) for each line. Different letters indicate significant differences (P < 0.05, one-way ANOVA). Asterisk, giant cell; N, nematode. Scale bars, 100 μm.
(D)A. thaliana phenotypes and relative root lengths of A. thaliana lines ectopically expressing Mi2G02 compared with the wild type (WT). Data represent the average length ± SD (n = 10). Different letters indicate significant differences (P < 0.05, one-way ANOVA). Scale bar, 1 cm. See also Supplemental Figure 2C.
(E) Expression of Mi2G02 in A. thaliana increased susceptibility to M. incognita. Two independent Mi2G02-T3 lines were inoculated with M. incognita pre-J2s. Total numbers of galls and egg masses were counted at 35 dpi. M. incognita inoculated WT A. thaliana inoculated with M. incognita was used as a control. Data are the average number per plant ± SD (n = 16). Different letters indicate significant differences (P < 0.05, one-way ANOVA). See also Supplemental Figure 2D.
We next generated two transgenic A. thaliana lines with ectopic Mi2G02 expression. Semiquantitative RT–PCR (Supplemental Figure 2A) and western blotting (Supplemental Figure 2B) were performed to confirm the expression of Mi2G02 in the transgenic plants. Interestingly, roots of the two independent Mi2G02-expressing transgenic lines were 27% and 33% longer (n = 10) than those of wild-type plants (Figure 2D and Supplemental Figure 3C). In nematode inoculation assays, both transgenic lines were significantly (P < 0.05) more susceptible to M. incognita infection than wild-type plants; the Mi2G02-expressing lines had up to 30% more galls and egg masses than the wild-type plants at 35 days post inoculation (dpi) (Figure 2E and Supplemental Figure 2D). Mi2G02 is therefore an effector essential for M. incognita parasitism and is able to modulate root growth.
Mi2G02 interacts with the trihelix transcription factor GT-3a
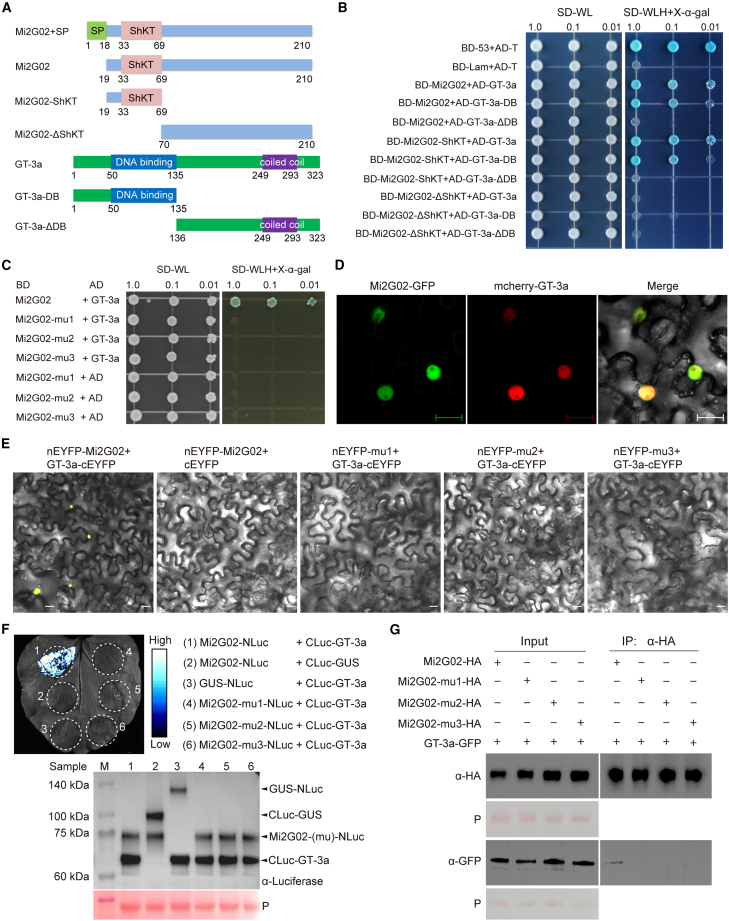
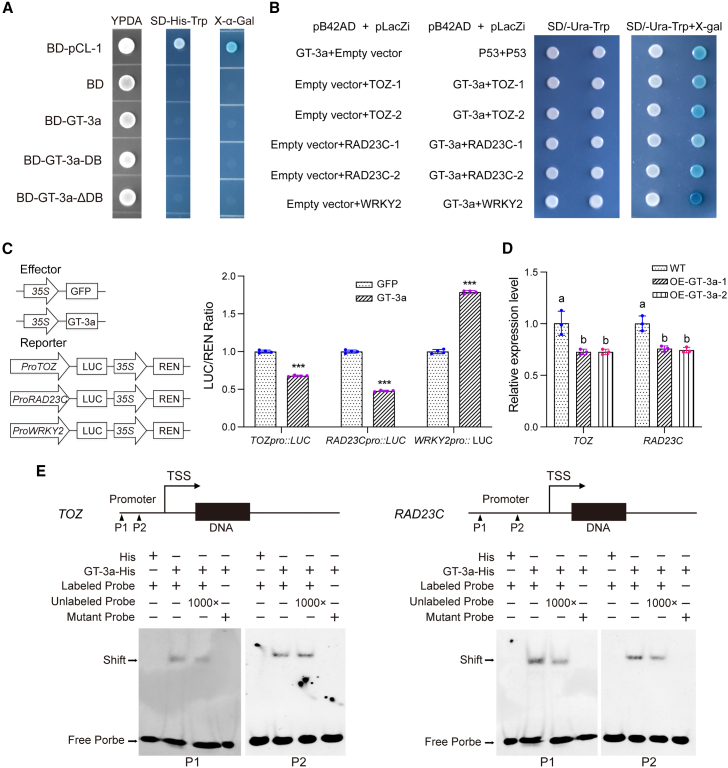
We used a yeast two-hybrid (Y2H) screen to identify A. thaliana proteins targeted by Mi2G02. We used a signal peptide-deficient Mi2G02 as a bait to screen a cDNA library from M. incognita-infected A. thaliana roots. We identified 20 candidate target proteins, including six annotated as predicted nuclear proteins in The Arabidopsis Information Resource (Supplemental Table 1). On the basis of the number of captures and their predicted subcellular distributions and functions, we selected three candidate targets for further study: a trihelix TF (GT-3a, AT5G01380), LATERAL ORGAN BOUNDARIES DOMAIN PROTEIN 41 (LBD41, AT3G020550), and UBIQUITIN EXTENSION PROTEIN 1 (UBQ1, AT3G52990). A pairwise Y2H assay was performed to validate the interactions between Mi2G02 and the full-length GT-3a, LBD41, and UBQ1 proteins. GT-3a was the only protein found to interact with Mi2G02 (Figure 3A and 3B). LBD41 displayed strong autoactivation in yeast, and it was not possible to confirm any interaction between Mi2G02 and UBQ1 (Supplemental Figure 3A).
Figure 3.
The Mi2G02 effector interacts with GT-3a in the nuclei.
(A) Schematic representation of intact and truncated Mi2G02 (with or without the ShKT domain) and GT-3a (with or without the DNA-binding domain) used for yeast two-hybrid assays (Y2Hs).
(B) Pairwise Y2H tests were performed to investigate the interactions between Mi2G02 or the ShKT domain and GT-3a or the DNA-binding (DB) domain. Left column: growth of yeast cells carrying baits in the pGBKT7 vector (BD) and preys in the pGADT7 (AD) on SD/-Trp-Leu (SD-WL) medium, with growth indicating successful transformation of the yeast with both plasmids. Right column: yeast cell growth on selective dropout medium (SD/-Trp-Leu-His, SD-WLH) after addition of 20 mg/ml X-α-gal, with growth indicating protein interaction. Yeast cells containing p53 and SV40 large T antigen were used as a positive control, and yeast cells containing lamin and SV40 large T antigen were used as a negative control.
(C) Pairwise Y2H tests were performed to investigate the interactions between Mi2G02 mutants and GT-3a.
(D) Mi2G02 co-localizes with GT-3a in N. benthamiana nuclei. Mi2G02 fused with GFP at the C terminus (Mi2G02-GFP) and GT-3a fused with mCherry at the N terminus (mCherry-GT-3a) were co-expressed in N. benthamiana leaf cells. The fluorescence signal was detected at 48 h after infiltration. Images were captured by confocal microscopy. Scale bars, 20 μm.
(E) Bimolecular fluorescence complementation experiments demonstrate the interaction between Mi2G02 and GT-3a. N. benthamiana leaves were transformed with different combinations of nEYFP- and cEYFP-fused vectors. Images were obtained 48 h after co-expression. Yellow fluorescent protein (YFP) signals were observed in the nuclei of leaves co-infiltrated with nEYFP-Mi2G02 and GT-3a-cEYFP. Scale bars, 20 μm. See also Supplemental Figure 3C.
(F) Determination of the interaction between Mi2G02 and GT-3a by luciferase complementation assay. A. tumefaciens harboring different combinations of plasmids were infiltrated into indicated regions of N. benthamiana leaves. Luciferase activities were recorded at 2 days post agroinfiltration by spraying 1 mM luciferin solution onto the infiltrated leaves and detecting luciferase activity with a low-light cooled CCD imaging apparatus. Luciferase activity is depicted with false color from low (black) to high (white). The protein levels of Mi2G02-NLuc, Mi2G02-mu1-NLuc, Mi2G02-mu2-NLuc, Mi2G02-mu3-NLuc, CLuc-GT-3a, GUS-NLuc, and CLuc-GUS were determined by western blotting using anti-luciferase antibody. Ponceau S (P) staining provided a loading control.
(G) Mi2G02 associates with GT-3a in a co-IP assay. A. tumefaciens harboring different combinations of plasmids were infiltrated into N. benthamiana leaves. co-IP was performed with BeyoMag Anti-HA magnetic beads, and the eluted protein was detected by western blotting with antibodies against HA and GFP. GFP (green fluorescent protein) and P (Ponceau staining) indicate sample loading.
We investigated the possible involvement of the ShKT domain of Mi2G02 and the DNA-binding domain (DB) of GT-3a in their interaction by generating two truncated versions of Mi2G02 (Mi2G02-ShKT and Mi2G02-ΔShKT) and two truncated versions of GT-3a (GT-3a-DB and GT-3a-ΔDB) (Figure 3A). Subcellular localization results showed that Mi2G02-ShKT-GFP was localized mainly in the cell nucleus (Supplemental Figure 3B). Pairwise Y2H experiments demonstrated that the ShKT domain of Mi2G02 and the DB domain of GT-3a were required for the interaction between these two proteins (Figure 3B). We also investigated whether the NLSs of Mi2G02 were required for its interaction with GT-3a in yeast. Pairwise Y2H experiments with Mi2G02-mu1, Mi2G02-mu2, and Mi2G02-mu3 showed that both NLSs of Mi2G02 were required for interaction with GT-3a (Figure 3C).
Co-expression of Mi2G02-GFP and mCherry-GT-3a in N. benthamiana leaf cells showed that the effector and its target were co-localized in the nucleoplasm of plant cells (Figure 3D). We then investigated the interactions between Mi2G02 and GT-3a in planta by performing bimolecular fluorescence complementation (BiFC) assays. Co-expression of Mi2G02 fused to the N-terminal part of YFP (Mi2G02-nEYFP) and GT-3a fused to the C-terminal part of YFP (GT-3a-cEYFP) in N. benthamiana leaf epidermal cells reconstituted YFP activity in the nucleus, whereas no YFP fluorescence was observed if Mi2G02 with mutated NLSs or an empty vector was used (Figure 3E and Supplemental Figure 3C).
A split luciferase complementation assay (LCA) and a co-immunoprecipitation (co-IP) assay were performed to further verify the interaction between Mi2G02 and GT-3a in planta. A positive luciferase signal was obtained when Mi2G02 was co-expressed with GT-3a in N. benthamiana leaves, as luciferase activity was reconstituted by the interaction between Mi2G02 and GT-3a, but no luciferase signal was observed if Mi2G02 with mutated NLSs or the β-glucuronidase (GUS) control was used (Figure 3F). In the co-IP assay, Mi2G02-HA, the HA empty vector, or GFP-HA was co-expressed with GT-3a-GFP in N. benthamiana leaves. GT-3a-GFP co-precipitated with Mi2G02-HA but not with Mi2G02-mu1-HA, Mi2G02-mu2-HA, or Mi2G02-mu3-HA (Figure 3G). There is therefore a direct interaction between nuclear Mi2G02 and the A. thaliana GT-3a TF in planta.
GT-3a is important for M. incognita parasitism
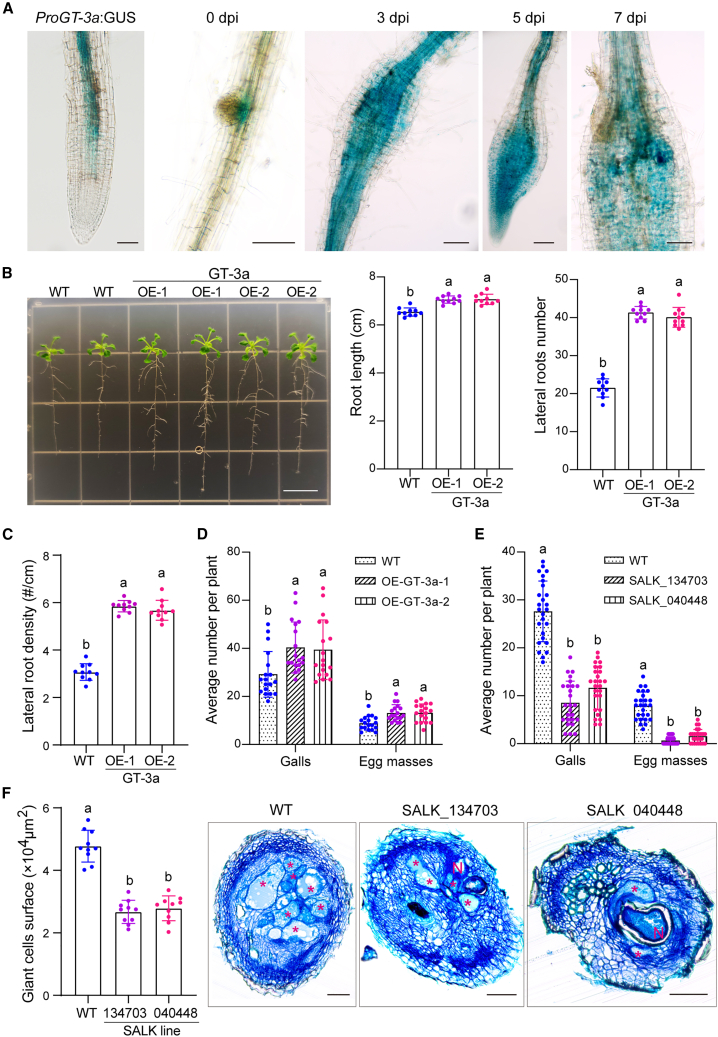
GT-3a has been reported to be predominantly expressed in floral buds and roots, especially at the onset of secondary root development (Ayadi et al., 2004). RNA-sequencing data for M. incognita-infected A. thaliana galls at 3, 5, and 7 dpi and for non-infected roots showed that GT-3a was significantly upregulated by nematode infection at these early time points (Yamaguchi et al., 2017). For analysis of GT-3a expression in galls, we cloned a fragment of the GT-3a promoter (−2023 to 0) and transformed A. thaliana plants with a ProGT-3a:GUS fusion. We then inoculated the transformed plants with M. incognita and performed histochemical assays. We observed a strong GUS signal in uninfected root vascular tissues and lateral root initials and in developing galls at 3, 5, and 7 dpi (Figure 4A). These results suggest that GT-3a plays a role early in nematode feeding-site development.
Figure 4.
GT-3a is involved in M. incognita parasitism and lateral root development.
(A) Activity of the GT-3a promoter was analyzed in uninfected roots and in galls induced by M. incognita in A. thaliana expressing the ProGT-3a:GUS construct. dpi, days post infection. Scale bars, 100 μm.
(B)A. thaliana phenotypes, relative root length, and relative lateral root numbers of A. thaliana lines ectopically expressing Mi2G02 compared with the wild type (WT). Data represent the average length ± SD (n = 10) and the average number ± SD (n = 10). Different letters indicate significant differences (P < 0.05, one-way ANOVA). Scale bar, 1 cm. See also Supplemental Figure 4H.
(C) Lateral root density calculated as the number of emerged lateral roots divided by the total primary root length. Different letters indicate significant differences (P < 0.05, one-way ANOVA).
(D) Overexpression of GT-3a in A. thaliana increased susceptibility to M. incognita. Two independent T3 lines ectopically expressing GT-3a were inoculated with M. incognita pre-J2s. Total numbers of galls and egg masses were counted at 35 dpi. M. incognita inoculated WT A. thaliana inoculated with M. incognita was used as a control. Data are the average number ± SD (n = 18). Different letters indicate significant differences (P < 0.05, one-way ANOVA). See also Supplemental Figure 5A.
(E) The gt-3a T-DNA knockout mutants (SALK_134703 and SALK_040448) were less susceptible to M. incognita compared with the WT, as indicated by mean numbers of galls and egg masses. Data are the average number ± SD (n = 26). Different letters indicate significant differences (P < 0.05, one-way ANOVA). See also Supplemental Figure 5B.
(F) Giant cell areas of M. incognita-induced galls were significantly reduced in the A. thaliana gt-3a T-DNA knockout mutant lines. Gall sections at 21 dpi were stained with toluidine blue. Smaller giant cells were observed in gt-3a T-DNA knockout mutant lines compared with the WT. Data are the average surface area ± SD (n = 10). Different letters indicate significant differences (P < 0.05, one-way ANOVA). Asterisk, giant cell; N, nematode. Scale bars, 100 μm.
We explored the biological functions of GT-3a during gall formation in two gt-3a T-DNA knockout (KO) mutant A. thaliana lines (SALK_134703 and SALK_040448) (Supplemental Figure 4A). We also generated transgenic A. thaliana lines overexpressing a GT-3a-GFP fusion and GFP alone. Homozygous KO mutants were verified by PCR and semiquantitative RT–PCR (Supplemental Figure 4B and 4C). Two independent GT-3a-GFP-overexpressing lines were selected and verified by semiquantitative RT–PCR, western blotting, and observation of GFP fluorescence (Supplemental Figure 5D–5F). No macroscopic root phenotypes were observed in the two gt-3a T-DNA KO lines relative to wild-type Columbia-0 (Supplemental Figure 4G). As observed for Mi2G02, the two independent GT-3a-GFP-overexpressing lines had longer roots (8%; n = 10), more lateral roots (92% and 87%; n = 10), and greater lateral root density than the wild-type plants (Figure 4B and 4C; Supplemental Figure 4H). These lines were then subjected to nematode infection assays. GT-3a-overexpressing plants had larger numbers of galls and egg masses (37% and 48%, n = 18) than control plants (Figure 4D and Supplemental Figure 5A). By contrast, the two gt-3a mutants were significantly less susceptible to M. incognita than control plants, as shown by their smaller numbers of galls (more than 60% fewer, n = 26) and an almost complete absence of egg masses (Figure 4E and Supplemental Figure 5B). The areas covered by GCs were 40% (n = 10) smaller in these KO lines than in control plants (Figure 4F). The GT-3a TF therefore regulates root development and plays an essential role in GC development and M. incognita parasitism.
GT-3a targets and represses TOZ and RAD23C
We investigated the transcriptional activity of GT-3a by fusing the GT-3a coding sequence to the sequence encoding the GAL4 DB domain in the pGBKT7 (BD) vector and using the resulting plasmid to transform the yeast strain AH109. Yeast cells transformed with the positive control pCL-1, which encodes the full-length wild-type GAL4 protein, grew well on SD-Trp-His selection medium and displayed 5-bromo-4-chloro-3-indolyl-α-D-galactopyranoside (X-α-Gal) activity (Figure 5A). By contrast, yeast cells harboring BD-GT-3a or the empty BD plasmid (negative control) were unable to grow on SD-Trp-His selection medium (Figure 5A). These results suggest that GT-3a does not act as a transcriptional activator and probably acts by repressing gene expression.
Figure 5.
Targeting and suppression of TOZ and RAD23C by GT-3a and susceptibility of toz and rad23c knockout mutant lines to M. incognita.
(A) Transcriptional activity of GT-3a in yeast cells. The yeast AH109 strain expressing pCL-1, GT-3a, and GT-3a with or without the DNA-binding domain (GT-3a-DB or GT-3a-ΔDB) grew on yeast peptone dextrose adenine agar (YPDA) or the selective medium SD-His-Trp with or without X-α-gal. pCL-1 encoding full-length GAL4 and the empty vector pGBKT7 (BD) were used as positive and negative controls, respectively.
(B) Yeast one-hybrid experiments showed that GT-3a bound to the promoters of TOZ, RAD23C, and WRKY2. Promoter fragments containing the GTTAC or CACGTG element were cloned into the pLacZi vector, and GT-3a was cloned into the pB42AD vector. The pLacZi vector was then co-transformed with pB42AD-GT-3a into yeast strain EGY48. The yeast transformants were spotted on SD/-Ura-Trp plates with or without 20 mg/ml X-gal. pB42AD-p53 and pLacZi-p53 were used as positive controls.
(C) Luciferase reporter assays of GT-3a-induced suppression of TOZ and RAD23C expression in N. benthamiana. LUC activity was measured by normalizing to the REN signal. Values are means ± SE (n = 4). Asterisks indicate significant differences according to a two-tailed Student’s t-test, ∗∗∗P < 0.001. Similar results were obtained from three independent experiments (biological replicates).
(D) qRT–PCR analysis of TOZ and RAD23C expression in wild-type (WT) A. thaliana and GT-3a-overexpressing A. thaliana lines. UBP22 (AT5G10790) was used as an internal control. Data represent the mean of three independent experiments ± SE (n = 3). Similar results were obtained from three independent experiments (biological replicates). Different letters indicate significant differences (P < 0.05, one-way ANOVA).
(E) EMSAs confirmed that GT-3a-His could bind directly to the promoters of TOZ and RAD23C. Promoter fragments containing the GTTAC element (P1 probe), the CACGTG element (P2 probe), or mutant elements (AAAAA or AAAAAA) were labeled with biotin as probes. 6×His alone served as a negative control. Unlabeled probes were used as competing probes.
We next sought to identify genes whose expression is modulated by GT-3a by searching A. thaliana promoter sequences for the 5′-GTTAC-3′ DNA element, which is known to be specifically targeted by GT-3a (Ayadi et al., 2004), and the 5′-CACGTG-3′ DNA element with the PlantRegMap tool (Tian et al., 2019). We also turned to the ePlant online tools to explore the expression patterns of candidate genes during nematode infection and in developing roots (Waese et al., 2017). We retrieved nine putative GT-3a targets (Supplemental Table 2), which were then further studied with a yeast one-hybrid (Y1H) approach. The Y1H assay revealed that GT-3a bound directly to the promoters of TORMOZ (TOZ; AT5G16750), RADIATION SENSITIVE 23C (RAD23C; AT3G02540), and a WRKY TF (WRKY2; AT5G56270) (Figure 5B and Supplemental Figure 6).
We investigated the ability of GT-3a to repress the expression of TOZ, RAD23C, or WRKY2 in dual-luciferase reporter assays. A construct expressing GT-3a and a reporter construct consisting of the TOZ, RAD23C, or WRKY2 promoter driving transcription of the firefly luciferase (LUC) reporter gene were used for co-infiltration of N. benthamiana leaves. GT-3a decreased the activity of the TOZ and RAD23C promoters, measured as the firefly-to-Renilla (LUC/REN) luciferase ratio, by 30% and 50%, respectively, relative to the GFP control, but it did not decrease WRKY2 promoter activity (Figure 5C). To confirm this result, we used qRT–PCR to quantify TOZ and RAD23c expression in transgenic A. thaliana plants overexpressing GT-3a. Both TOZ and RAD23c appeared to be repressed in the two independent transgenic lines relative to the wild type (Figure 5D).
Finally, we produced a recombinant GT-3a protein, which was used in a gel electrophoretic mobility shift assay (EMSA). GT-3a-His significantly decreased the electrophoretic mobility of TOZ and RAD23C promoter probes containing GTTAC or CACGTG elements but had no effect on the mobility of mutated probes (GTTAC replaced by AAAAA, CACGTG replaced by AAAAAA) (Figure 5E). This result validates the binding of GT-3a to the TOZ and RAD23c promoters and indicates that both the GTTAC and CACGTG elements are important for binding.
These results confirm that GT-3a can bind specifically to the TOZ and RAD23C promoters, downregulating expression of the genes driven by these promoters.
Mi2G02 promotes GT-3a function by stabilizing GT-3a protein level to promote nematode parasitism
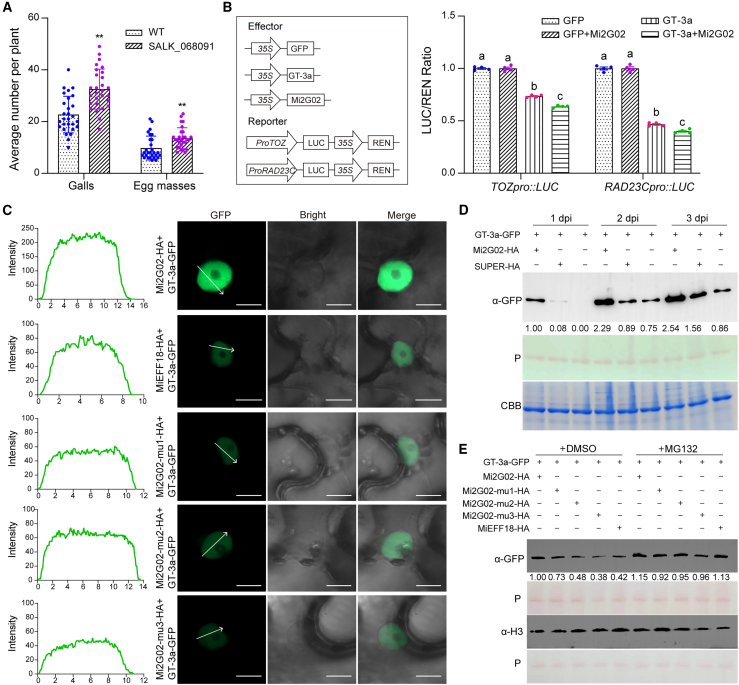
The toz mutant is not viable at postembryonic stages (Griffith et al., 2007) and therefore could not be tested in interaction with the nematode. We investigated the role of RAD23C in the plant response to M. incognita parasitism with a rad23c T-DNA KO mutant A. thaliana line (SALK_068091) obtained from the Arabidopsis Biological Resource Center (Supplemental Figure 7A). Homozygous KO plants were verified by PCR with genomic DNA (Supplemental Figure 7B) and by qRT–PCR with cDNA (Supplemental Figure 7C). No difference in root phenotype was observed between the rad23c T-DNA KO line and the wild type (Supplemental Figure 7D), consistent with previous reports (Farmer et al., 2010). After M. incognita infection, the rad23c KO lines were significantly more susceptible to the nematode than control plants, as shown by the larger numbers of galls (43%, n = 28) and egg masses (39%) (Figure 6A and Supplemental Figure 7E). RAD23C therefore downregulates M. incognita parasitism.
Figure 6.
Mi2G02 stabilizes GT-3a to promote its function in suppression of TOZ and RAD23C expression for nematode parasitism.
(A) The rad23c T-DNA knockout mutant is more susceptible than the wild type (WT) to M. incognita. The rad23c KO mutant (SALK_068091) was inoculated with nematodes, and numbers of galls and egg masses were counted 35 days post inoculation. The data presented are mean numbers per plant ± SD (n = 28). Similar results were obtained in three independent experiments. Asterisks indicate significant differences according to two-tailed Student’s t-tests, ∗∗P < 0.01. See also Supplemental Figure 7E.
(B) Luciferase reporter assays showed that the GT-3a-induced suppression of TOZ and RAD23C expression in N. benthamiana was enhanced by Mi2G02 expression. LUC activity was determined and normalized against the REN signal. The data presented are means of three independent experiments ± SEM (n = 4). Different letters indicate significant differences (P < 0.05, one-way ANOVA).
(C) Mi2G02 stabilizes GT-3a-GFP fluorescence intensity. GT-3a was co-expressed with Mi2G02 in N. benthamiana leaves, and Mi2G02 mutants and MiEFF18 (a nuclear M. incognita effector that does not interact with GT-3a) were used as controls. GT-3a-GFP fluorescence was detected with a confocal microscope (Zeiss LSM700) 48 h after infiltration. Graphs show the fluorescence intensity profiles across the arrows in the GFP images. Scale bar, 10 μm. See also Supplemental Figure 8.
(D) Mi2G02 stabilizes the GT-3a protein, leading to its accumulation. GT-3a was co-expressed with Mi2G02 or GFP in N. benthamiana leaves. The GT-3a protein was detected with an anti-GFP antibody. Band intensity was determined with ImageJ software and is indicated below the bands. Coomassie brilliant blue staining (CBB) and Ponceau staining (P) were used to check protein sample loading.
(E) Mi2G02 inhibits GT-3a degradation via the 26S proteasome pathway in vivo. GT-3a-GFP was co-expressed with Mi2G02-HA and Mi2G02 mutants in N. benthamiana leaves. The 26S proteasome inhibitor MG132 (100 μM) was infiltrated into N. benthamiana leaves 24 h before protein extraction. Band intensity was determined with ImageJ software and is indicated below the bands.
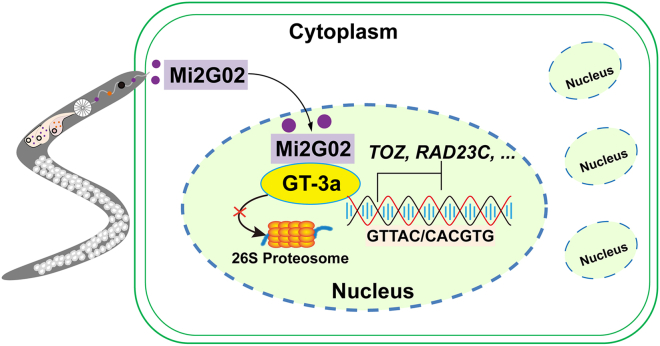
We addressed the potential outcome of Mi2G02 binding to GT-3a more precisely by co-expressing Mi2G02 and GT-3a in N. benthamiana leaves and performing dual-luciferase reporter assays. The previously observed suppression of TOZ and RAD23C expression mediated by GT-3a was significantly enhanced in the presence of Mi2G02 (Figure 6B). We also performed transient expression assays and western blotting to determine whether Mi2G02 affected the amount of GT-3a protein in agroinfiltrated leaves of N. benthamiana. Co-expression of Mi2G02 and GT-3a-GFP in N. benthamiana leaves resulted in a significantly higher GFP fluorescence intensity (500%–660% higher) compared with controls using mutant Mi2G02 or MiEFF18 (Figure 6C and Supplemental Figure 8). Similarly, co-expression of Mi2G02 and GT-3a in N. benthamiana leaves resulted in high levels of GT-3a protein accumulation. No such accumulation was observed with the empty vector, Mi2G02 mutants, or MiEFF18, which were used as negative controls (Figure 6D and 6E). Furthermore, treatment with the proteasome inhibitor MG132 inhibited GT-3a degradation (Figure 6E), suggesting that Mi2G02 stabilizes GT-3a protein levels by inhibiting the 26S proteasome-dependent pathway. Together, these results suggest that the Mi2G02 effector helps to stabilize the GT-3a protein, enabling GT-3a to repress the target genes TOZ and RAD23C and thus promote nematode parasitism (Figure 7).
Figure 7.
A proposed working model illustrating the molecular mechanism of interaction among Mi2G02, GT-3a, TOZ, and RAD23C in nematode parasitism.
In the early stage of M. incognita parasitism, the Mi2G02 effector protein is secreted into the plant cell and translocates to the plant nucleus, where it targets the transcription factor GT-3a and stabilizes its protein level by inhibiting the 26S proteasome pathway, leading to suppression of TOZ and RAD23C expression and thus promoting nematode feeding cell formation and development.
Discussion
Phytopathogen success depends upon secretion of effector proteins that reprogram the host transcriptome to facilitate parasitism. Pathogens have been shown to secrete effectors that can function as TFs or target TFs to manipulate host cell physiology and/or immunity. In plant–nematode interactions, the 10A07 effector from the sugar beet cyst nematode, Heterodera schachtii, is expressed in the nematode dorsal gland cell and targets a plant kinase and the IAA16 TF. There is also evidence to suggest that the 10A07–IAA16 interaction interferes with auxin signaling by modulating the expression of several auxin response factors (Hewezi et al., 2015). Nevertheless, the function of nematode nuclear effectors and the ways in which they manipulate their host targets for feeding-site initiation and development remain largely unknown. In this study, we characterized the function of the nuclear effector protein Mi2G02 and identified its plant target for GC formation, the nuclear trihelix TF GT-3a.
Trihelix TFs (GTs) are unique to plants and have been shown to participate in embryogenesis and subsequent plant growth and development, as well as abiotic stress responses (Kaplan-Levy et al., 2012). A. thaliana AtGT-3b and maize (Zea mays) ZmGT-3b, both members of the GT-1 clade, are induced by pathogens (Park et al., 2004; Zhang et al., 2021). Here, we show that Mi2G02 can interact with the A. thaliana GT-1 clade protein AtGT-3a, resulting in the stabilization of this protein. AtGT-3a is induced during the development of galls induced by M. incognita. Using A. thaliana transgenic plants in which AtGT-3a expression was suppressed or constitutively induced, we demonstrated that AtGT-3a is important for GC development and successful RKN parasitism.
The de novo formation of new organs, such as lateral roots, rhizobium-induced nodules, or nematode-induced galls, from one or a few root cells requires the recruitment of similar developmental programs (Yamaguchi et al., 2017; Soyano et al., 2019; Olmo et al., 2020). Several genes, including ABERRANT LATERAL ROOT FORMATION 4 (ALF4), a RIBULOSE-PHOSPHATE 3-EPIMERASE (RPE), and YUCCA4 (YUC4), have been reported to have functions associated with lateral root initiation and/or development, and their expression is regulated in nematode-induced galls; they have also been shown to be required for normal feeding-site formation and nematode development (Favery et al., 1998; Olmo et al., 2019; Suzuki et al., 2022). These genes include cell-cycle genes and TF genes, such as LBD16 and PUCHI, that play key roles in controlling lateral root initiation and morphogenesis (Torres-Martínez et al., 2019); they are induced following nematode infection and are required for feeding-site development and successful RKN parasitism (Cabrera et al., 2014b ; Suzuki et al., 2021). Similarly, Medicago truncatula LBD16 mutants display nodule initiation defects upon inoculation with Sinorhizobium meliloti (Schiessl et al., 2019). The rewiring of transcriptional networks to alter root-system architecture also involves changes in endogenous levels of growth-related plant hormones and the production of phytohormones or deployment of hormone-mimicking strategies by symbiotic and parasitic microbes (Gheysen and Mitchum, 2019; Eichmann et al., 2021). We show here that expression of Mi2G02 in A. thaliana can promote root growth and GC development and that Mi2G02 acts by stabilizing AtGT-3a, with mutations of the gene encoding this TF also impairing GC formation. AtGT-3a was found to be strongly induced at the onset of lateral root development (this study and Ayadi et al., 2004). AtGT-3a therefore seems to be one of the TFs that regulate both lateral root development and nematode feeding-site neo-organogenesis.
Binding of GT-3a to the promoter of the TOZ and RAD23C genes was confirmed by Y1H and EMSA assays. TOZ is a predicted WD40 repeat protein involved in the regulation of cell-division planes and the expression of patterning genes during embryogenesis. It may therefore be involved in plant embryogenesis and organogenesis, including root development (Griffith et al., 2007), suggesting a possible role in the regulation of root-knot neo-organogenesis. RAD23 probably acts as a shuttle protein, delivering ubiquitinated substrates to the ubiquitin/26S proteasome system for degradation. Roles in plant processes as diverse as the cell cycle, cell morphogenesis, and flower development have been proposed for RAD23 (Farmer et al., 2010; Maclean et al., 2014). Interestingly, a role for RAD23 proteins in plant immunity has been proposed, probably through interactions with stress-associated proteins (SAPs) acting as ubiquitin E3 ligases (Kang et al., 2017; Liu et al., 2019). Consistent with this notion, RAD23 proteins have been shown to be targeted by plant-pathogen effectors, possibly to modulate host protein degradation and suppress host defense responses. A. thaliana RAD23A was identified as a putative target of the Pseudomonas syringae HopM1 effector (Nomura et al., 2006), and the phytoplasma SAP54 effector was shown to interact with both RAD23C and RAD23D (Maclean et al., 2014). Intriguingly, a previous report confirmed that RAD23 proteins were associated with the 26S proteasome and played an essential role in the cell cycle (Farmer et al., 2010), so roles of GT-3a and RAD23C in proteolysis and cell-fate determination are expected. In support of this prediction, transcription activity assays performed in yeast and in planta demonstrated that GT-3a downregulated expression of TOZ and RAD23C, and this suppression was enhanced by co-expression of GT-3a with Mi2G02. Moreover, Mi2G02 stabilized GT-3a protein level by inhibiting the 26S proteasome-dependent pathway. Using rad23c KO A. thaliana lines, we showed that RAD23C inactivation increased susceptibility to RKNs, demonstrating that RAD23C is a key gene for plant–nematode interactions.
It has been suggested that Mj2G02 interferes with both jasmonate signaling and plant immune responses (Song et al., 2021). Mj2G02 expression in planta resulted in the accumulation of jasmonoyl-isoleucine, the endogenous bioactive form of jasmonate (JA), in transgenic A. thaliana, and a dysregulation of the expression of JASMONATE ZIM DOMAIN (JAZ) transcriptional repressors and jasmonate-responsive genes (Song et al., 2021). Our data indicate that Mi2G02 could divert the host plant developmental program to promote formation of the feeding sites important for nematode development and reproduction. RKNs may secrete the 2G02 effector to stabilize GT-3a, maintaining the concentration of this TF at a sufficiently high level to repress the growth regulator genes TOZ and RAD23c, thereby promoting GC development. We hypothesize that, as previously suggested for ZmGT-3a (Zhang et al., 2021), GT-3a acts at the interface between growth and immunity. Microbes can interfere with central regulators of root cell identity and root growth that are also involved in the response to biotic stress (Rich-Griffin et al., 2020; Üstüner et al., 2022). Plant hormones, whose signaling pathways may interact at central hubs, also regulate growth–immunity tradeoffs (Huot et al., 2014; Guo et al., 2018). For instance, crosstalk between signaling pathways of gibberellin (GA)-mediated growth and jasmonic acid (JA)-mediated defense contributes to maintaining the balance between growth and immunity (Huot et al., 2014; Pieterse et al., 2014). DELLA proteins repress growth-related TFs unless they are degraded in the presence of growth-promoting GA. DELLA also binds to JAZs, and DELLA degradation allows JAZs to interact with their cognate TFs, thereby decreasing JA-dependent signaling. Treatment with flg22 suppresses GA-mediated DELLA degradation, leading to an inhibition of root growth dependent on salicylic acid, an antagonist of the JA signaling pathway (Huot et al., 2014; Pieterse et al., 2014). JA is a known growth inhibitor that stabilizes DELLA and has been shown to downregulate the cyclin-dependent kinases CDKA1 and CYCB1 required for cell-cycle progression (Reitz et al., 2015; Qi and Zhang, 2020). Biotic stress may therefore affect cell-cycle regulators, and cell division and hormones influence the underlying regulatory mechanisms.
The plant response to pathogens is highly dependent on the interplay between immunity and development. Regulators of cell identity and TFs may play a crucial role in connecting the developmental and immunity gene networks to reflect response specificity (Rich-Griffin et al., 2020). By modulating GT-3a TF availability in plant cells, the RKN effector Mi2G02 can alter both the root developmental program and JA-dependent signaling pathways to allow GC formation and successful parasitism. In this respect, GT-3a constitutes a novel example of a key regulator recruited by a biotrophic pathogen at the interface between growth and immunity.
Methods
Nematodes and plant materials
M. incognita was reproduced on tomato plants (Solanum lycopersicum var. “Moneymaker”). Egg masses and preparasitic second-stage juveniles (pre-J2s) were collected as described previously (Zhao et al., 2019). A. thaliana seeds were germinated on Murashige and Skoog medium (Coolaber, cat. no. PM1012) at 25°C in a growth chamber, and the seedlings were transplanted into pots of soil at 13 days. The gt-3a T-DNA mutant lines (SALK_014703 and SALK_040448) and the At3g02540 (rad23c) T-DNA mutant line (SALK_068091) were obtained from the Arabidopsis Biological Resource Center (USA). Homozygous plants were verified by PCR and semiquantitative RT–PCR. N. benthamiana and A. thaliana plants were grown in pots and placed in a culture room at 23°C under a 16-h light/8-h dark photoperiod, with fluorescent bulbs used to generate soft white light.
Nematode infection and gall sections
A. thaliana seedlings (1 month after transplant) were inoculated with pre-J2s. For nematode susceptibility assays, A. thaliana roots were inoculated with 200 pre-J2s per plant. The roots were collected, and galls and egg masses were counted under a dissecting microscope (Olympus, Japan) 35 days post inoculation (dpi). For gall collection, roots were inoculated with 500 pre-J2s per plant. Galls were collected at 3, 5, 7, 14, and 21 dpi and fixed as described previously (Gavrilovic et al., 2016). At least 10 galls were fixed for each A. thaliana line. Gall sections were stained with 0.05% toluidine blue and photographed on a Zeiss AxioImager Z2 microscope (Zeiss, Germany). The areas of the GCs were measured with ImageJ software (Schindelin et al., 2012). In brief, the first step was to open the program and draw a line the same length as the image scale, then go to Analyze and Set Scale, enter 100 for known distance and μm for Unit of length, select Global, and click OK. The second step was to go to Analyze and Set Measurements, select Area, and click OK. The third step was to select Freehand selections, select the giant cell area, go to Analyze, select Measure, and click OK.
DNA/RNA isolation and gene amplification
M. incognita RNA was extracted with TRIzol reagent (Invitrogen, USA, cat. no. 10296010) as described previously (Lin et al., 2013; Zhao et al., 2021). Total RNA was extracted from A. thaliana seedlings (10 days after germination) with the RNAprep Pure Plant Kit (TIANGEN, cat. no. DP432) according to the manufacturer’s instructions. The RNA was then used for cDNA synthesis with M-MLV reverse transcriptase (TaKaRa, cat. no. 2641Q) in accordance with the manufacturer’s instructions. DNA was extracted with the Plant Genomic DNA Kit (TIANGEN, cat. no. DP305) according to the manufacturer’s instructions. Gene and promoter sequences were amplified from cDNA or gDNA by PCR with specific primers. All the primers used in this study are provided in Supplemental Table 3 and were synthesized by Tsingke Biotechnology (Beijing, China).
qRT–PCR analysis
RNA was extracted, and cDNA was synthesized for qRT–PCR on the Bio-Rad CFX96 real-time PCR system (Bio-Rad, USA) as follows: 95°C for 5 min and 40 cycles of 95°C for 30 s and 60°C for 30 s. The data were analyzed with the 2−ΔΔCt method (Livak and Schmittgen, 2001). For internal controls, M. incognita GAPDH (Minc12412) or A. thaliana UBP22 (AT5G10790) was used for normalization of qRT–PCR data. qRT–PCR assays were repeated three times.
Plasmid construction and plant transformation
For RNAi experiments in A. thaliana, a 204-nt Mi2G02 fragment was amplified and inserted upstream and downstream of the pSAT5 intron in the forward and reverse orientations (Dafny-Yelin et al., 2007), then inserted into the pSUPER destination vector to construct pSUPER-Mi2G02-RNAi.
A signal-peptide-deficient Mi2G02 sequence and ShKT domain were amplified by PCR and inserted into the Super-GFP vector (C-terminal GFP) to generate Super-Mi2G02-GFP and Super-Mi2G02-ShkT-GFP. The nuclear localization sequences of Mi2G02 were mutated and inserted into the Super-GFP vector to generate Super-Mi2G02-mu1-GFP, Super-Mi2G02-mu2-GFP, and Super-Mi2G02-mu3-GFP. The open reading frame (ORF) of GT-3a was inserted into the pBin-mCherry vector (N-terminal mCherry) to generate pBin-mCherry-GT-3a. Plasmids were checked by sequencing and used to transform Agrobacterium tumefaciens strain GV3101.
For ectopic expression in A. thaliana, the ORFs of Mi2G02 (without signal peptide sequences) and AT5G01380 (GT-3a) were amplified, and the corresponding PCR fragments were inserted into the Super-HA vector (C-terminal HA tag) and the Super-GFP vector (C-terminal GFP tag) to construct Super-Mi2G02-HA and Super-GT-3a-GFP, respectively. The plasmids were verified by sequencing and used to transform A. tumefaciens GV3101, with empty vectors used as a control.
For Y2H screens, Mi2G02 (lacking the signal peptide sequence) was amplified and inserted into the pGBKT7 (BD) vector. For pairwise Y2H verification, the coding sequences of AT5G01380 (GT-3a), AT3G52590, and AT3G02550 were inserted into the pGADT7 (AD) vector. For identification of the key domains for interaction, Mi2G02-ShKT (Mi2G02 retaining the ShKT domain) and Mi2G02-ΔShKT (Mi2G02 without the ShKT domain) were amplified and inserted into BD vectors, and GT-3a-DB (GT-3a retaining the DNA-binding domain) and GT-3a-ΔDB (GT-3a lacking the DNA-binding domain) were inserted into AD vectors. The plasmids were verified by sequencing and used to transform Y2HGold competent yeast cells.
For the Y1H assay, the coding region of GT-3a was amplified and inserted into the pB42AD vector. Fragments of candidate promoters were amplified and inserted into the pLacZi vector.
For the BiFC assay, coding sequences of Mi2G02 (lacking the signal peptide sequence) and GT-3a were inserted into the nE-YFP and cE-YFP vectors (Walter et al., 2004), respectively.
For the LCA, the full-length coding sequence of Mi2G02 (lacking the signal peptide sequence) or GT-3a was inserted into the pCAMBIA1300-nLUC or pCAMBIA1300-cLUC vector. The plasmids were verified by sequencing and used to transform A. tumefaciens GV3101.
For the co-IP assay, the coding regions of Mi2G02 (lacking the signal peptide sequence) or GT-3a were inserted into super1300 vectors with an HA tag and an FLAG tag, respectively, fused to the C-terminal end of the sequence. Plasmids were verified by sequencing and used to transform A. tumefaciens GV3101.
For GUS staining assays, a 2023-bp fragment of the GT-3a promoter was inserted into the pBI101 vector to generate ProGT-3a:GUS. The resulting plasmid was verified by sequencing and used to transform A. tumefaciens GV3101.
For dual-luciferase reporter assays, the coding sequence of GT-3a was amplified and inserted into the pCAMBIA3301 vector to generate pCAMBIA3301-GT-3a. A 623-bp fragment upstream of the start codon of TOZ and a 595-bp fragment upstream of the start codon of RAD23C were amplified and inserted into the pro-LUC-35S-Rluc vector to generate TOZpro-LUC-35S-Rluc and RAD23Cpro-LUC-35S-Rluc, respectively. The plasmids were verified by sequencing and used to transform A. tumefaciens strain GV3101.
A recombinant GT-3a protein was produced by amplifying the coding sequence of GT-3a and inserting it into the pET30a vector (C-terminal His tag) to generate pET30a-GT-3a. The resulting plasmid was verified by sequencing and used to transform Escherichia coli strain BL21 (DE3) cells.
The primers (synthesized by Tsingke Biotechnology) and restriction enzymes (NEB, MA, USA) used for plasmid construction are listed in Supplemental Table 3.
Generation of transgenic A. thaliana plants and nematode infection assays
The sequenced pSUPER-Mi2G02-RNAi, Super1300-Mi2G02-HA, Super1300-GT-3a-GFP, and Super1300-GFP plasmids were used to transform A. tumefaciens strain GV3101. Four-week old A. thaliana plants were transformed with A. tumefaciens by the floral dip method (Clough and Bent, 1998). A. thaliana lines were confirmed to be homozygous for the transgene by quantitative PCR and/or western blotting. Nematode infection assays were performed three times.
Subcellular localization assay
Four-week-old N. benthamiana leaves were infiltrated with A. tumefaciens carrying the appropriate plasmids in infiltration buffer (10 mM MgCl2, 10 mM 2-(N-morpholino)ethanesulfonic acid [MES], and 0.1 mM acetosyringone [AS]) at an OD600 of 0.4. Images were captured with a laser confocal fluorescence microscope (Zeiss LSM 700) 2 dpi at excitation wavelengths of 488 nm for GFP and 561 nm for mCherry.
Yeast two-hybrid and yeast one-hybrid assays
A cDNA library was constructed by extracting RNA from A. thaliana roots infected with M. incognita (1, 3, 5, 10, and 14 dpi) and used to screen for the target proteins of Mi2G02. The Y2H assay was performed according to the Clontech protocol (Clontech, USA). Interactions between Mi2G02 and candidate proteins were assessed in pairwise Y2H assays. Relative BD and AD vectors were used to transform the yeast strain Y2HGold for screening on SD/-Leu-Trp plates. Positive clones were verified and selected for culture on SD/-Leu-Trp-His medium supplemented with 20 mg/ml X-α-Gal (Coolaber, cat. no. SL0940). We investigated whether the nuclear localization sequences of Mi2G02 were required for interaction by inserting a mutated Mi2G02 in the BD vector for pairwise Y2H assays.
The Y1H assay was performed as described previously (Kong et al., 2023). The sequenced pB42AD and pLacZi vectors were integrated into the yeast strain EGY48 grown on SD/-Trp-Ura medium (Coolaber, cat. no. PM2262). Positive transformants were verified and selected for growth on medium containing raffinose (Coolaber, cat. no. SL0990) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal; Coolaber, cat. no. CX11921) for color reactions.
Bimolecular fluorescence complementation assay
BiFC assays were performed as described previously (Zhao et al., 2019). Agrobacterium harboring appropriate pairs of vectors was infiltrated into N. benthamiana leaves for 48 h. YFP fluorescence, indicating protein interaction, was captured with a confocal microscope (Zeiss LSM 700) with excitation at 514 nm and emission at 527 nm.
Luciferase complementation assay
Agrobacterium cultures were resuspended in infiltration buffer, incubated at room temperature for 3 h, and infiltrated into 4-week-old N. benthamiana leaves. Three days after agroinfiltration, 1 mM luciferin (Biovision, cat. no. 7903) was sprayed onto the infiltrated leaves, and luciferase activity was detected with a ChemiScope 6000 low-light cooled CCD imaging apparatus (Clinx Science Instruments, Shanghai, China).
In vivo co-immunoprecipitation assay
Total protein was extracted from 4-week-old N. benthamiana leaves co-expressing Mi2G02 and GT-3a after 48 h of infiltration; co-IP was performed with BeyoMag Anti-HA magnetic beads (Beyotime, cat. no. P2121) as described previously (Zhao et al., 2021). The eluted proteins were checked by western blotting with anti-GFP (ABclonal, cat. no. AE012) and anti-HA (Coolaber, cat. no. AB1105) antibodies, respectively.
Histochemical analysis of GUS activities
Transgenic A. thaliana were generated as described above. The homozygosity of the transgenic lines was confirmed by PCR, and they were inoculated with M. incognita. Root samples were collected at 3, 5, 7, and 14 dpi. More than 10 roots were collected at each sampling time. Histochemical staining for GUS enzyme activity was performed with a GUS staining kit (Coolaber, cat. no. SL7160) in accordance with the manufacturer’s instructions. Images were captured with a stereomicroscope (Zeiss AxioImager Z2).
Dual-luciferase reporter assay
Agrobacterium harboring pCAMBIA3301-GT-3a was infiltrated with TOZpro-LUC-35S-Rluc or RAD23Cpro-LUC-35S-Rluc, with or without Mi2G02, into 4-week-old N. benthamiana leaves. Three days after infiltration, leaf disks with a diameter of 2 cm were harvested and ground in liquid nitrogen. Firefly and Renilla luciferase activities were measured with the Dual-Luciferase Reporter Assay System (Vazyme, cat. no. DL101-01) according to the manufacturer’s instructions using a Synergy SLXFA plate reader (BioTek, USA).
Electrophoretic mobility shift assay
The recombinant GT-3a-His protein was induced with 1 mM isopropyl-β-D-thiogalactoside and purified with the His-tag Protein Purification Kit (Beyotime, cat. no. P2229S) according to the manufacturer’s instructions (Zhao et al., 2021). Biotin-labeled and unlabeled probes for TOZ and RAD23C, containing the GTTAC element or the CACGTG element, were synthesized and purified by Sangon Biotech (Shanghai, China). EMSA was performed with an EMSA chemiluminescence kit (Beyotime, cat. no. GS009). Competition experiments were performed with various amounts of unlabeled oligonucleotides. The mutated competitor was generated by replacing five base pairs or six base pairs in the TOZ and RAD23C binding elements (GTTAC to AAAAA or CACGTG to AAAAAA). EMSA assays were repeated three times.
Transient expression assays in N. benthamiana leaves
Four-week-old N. benthamiana leaves were infiltrated with A. tumefaciens carrying the appropriate plasmids in infiltration buffer (10 mM MgCl2, 10 mM MES, and 0.1 mM AS) at an OD600 of 0.4. MG132 (100 μM) was added 24 h before protein extraction.
Cell fractionation, protein extraction, and western blotting
The cell fractionation assay was performed as described previously (Wang et al., 2018) with some modifications. In brief, N. benthamiana leaves expressing the appropriate proteins (0.5 g) were harvested and ground in liquid nitrogen and mixed with 2 ml/g of lysis buffer (20 mM Tris–HCl [pH 7.5], 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 25% glycerol, 250 mM sucrose, and 5 mM dithiothreitol) supplemented with protease inhibitor cocktail (Beyotime, cat. no. P1045). The homogenate was filtered through a double layer of Miracloth (Millipore, cat. no. 475855). The flow-through was centrifuged at 1500 g for 10 min at 4°C; the supernatant, consisting of the cytoplasmic fraction, was centrifuged at 10 000 g for 10 min at 4°C and then collected as the cytoplasmic fraction. The pellet from the first centrifugation was washed four times with 4 ml of NRBT buffer (20 mM Tris–HCl [pH 7.5], 2.5 mM MgCl2, 25% glycerol, and 0.2% Triton X-100) and then resuspended in 500 μl of NRB2 (20 mM Tris–HCl [pH 7.5], 10 mM MgCl2, 0.25 M sucrose, 0.5% Triton X-100, and 5 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail and carefully overlaid on top of 500 μl of NRB3 (20 mM Tris–HCl, 10 mM MgCl2 [pH 7.5], 1.7 M sucrose, 0.5% Triton X-100, and 5 mM β-mercaptoethanol) supplemented with protease inhibitor cocktail. The suspension was centrifuged at 16 000 g for 45 min at 4°C, and the pellet was collected as the nuclear fraction. 2× SDS loading buffer was then added to the cytoplasmic and nuclear fractions and boiled for 5 min. Actin was detected with an anti-actin antibody (ABclonal, cat. no. AC009) as a cytoplasmic marker. H3 proteins were detected using an anti-H3 antibody (ABclonal, cat. no. A2348) as a nuclear marker. Total proteins were extracted from N. benthamiana leaves or A. thaliana seedlings (10 days after germination) in RIPA lysis buffer (Beyotime, cat. no. P0031K) supplemented with protease inhibitor cocktail. For tag antibodies, we used anti-His (TransGen, cat. no. HT501), anti-GFP (ABclonal, cat. no. AE012), and anti-HA (Coolaber, cat. no. AB1105) antibodies with a goat anti-mouse immunoglobulin G (H + L)–horseradish peroxidase-conjugated secondary antibody (Coolaber, cat. no. AB2102). The protein was detected with the EasySee Western Blot Kit (TransGen, cat. no. DW101). Band intensity was determined with ImageJ software.
Statistical methods
The significance of differences between two groups was assessed using two-tailed t-tests. For multiple comparisons, the significance of differences was assessed by one-way ANOVA followed by Tukey’s test for multiple comparisons. All statistical analyses were performed with GraphPad Prism software version 8.3.0.
Accession numbers
The accession numbers of major genes mentioned in this study are as follows: Mi2G02 (AAQ10016), GAPDH (Minc12412), GT-3a (AT5G01380), LBD41 (AT3G02550), UBQ1 (AT3G52590), TOZ (AT5G16750), RAD23C (AT3G02540), WRKY2 (AT5G56270), UBP22 (AT5G10790), SRP34 (AT1G02840), DVL4 (AT1G13245), SKP2 (AT1G21410), FEI1 (AT1G31420), FEI2 (AT2G35620), and ABS4 (AT1G58340).
Data and code availability
All relevant data are included in the main manuscript and the supplemental information.
Funding
This research was supported by the Youth Innovation Program of the Chinese Academy of Agricultural Sciences (grant no. Y2022QC06), the National Natural Science Foundation of China (grant nos. 32001878, 32172366), the Natural Science Foundation of Beijing (grant no. 6222054), the China Agricultural Research System (CARS-23), and the French Government (National Research Agency, ANR) through “Investments for the Future” LabEx SIGNALIFE (#ANR-11-LABX-0028-01), IDEX UCAJedi (#ANR-15-IDEX-0).
Author contributions
J.Z., B.F., P.A., M.Q., H.J., B.X. and Z.M. conceived the project. J.Z., K.H., B.F., Y.L., J.L., and Y.Y. designed and planned the experiments. J.Z., K.H., R.L., and Y.Q.L. performed the experiments and collected and analyzed the data. J.Z., B.F., P.A., and M.Q. wrote the manuscript.
Acknowledgments
We thank Dr. Panpan Yang (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China) for providing the CP516 vector and the p3301 vector, Dr. Jinzhuo Jian (Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China) for advice concerning gall sections, and Dr. Qian Wei (the Core Facility Platform, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China) for assistance with confocal microscopy. The authors declare no competing interests.
Published: September 22, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Jianlong Zhao, Email: zhaojianlong@caas.cn.
Michaël Quentin, Email: michael.quentin@inrae.fr.
Zhenchuan Mao, Email: maozhenchuan@caas.cn.
Supplemental information
References
- Abad P., Gouzy J., Aury J.M., Castagnone-Sereno P., Danchin E.G.J., Deleury E., Perfus-Barbeoch L., Anthouard V., Artiguenave F., Blok V.C., et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 2008;26:909–915. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- Ayadi M., Delaporte V., Li Y.F., Zhou D.X. Analysis of GT-3a identifies a distinct subgroup of trihelix DNA-binding transcription factors in Arabidopsis. FEBS Lett. 2004;562:147–154. doi: 10.1016/S0014-5793(04)00222-4. [DOI] [PubMed] [Google Scholar]
- Baldacci-Cresp F., Behr M., Kohler A., Badalato N., Morreel K., Goeminne G., Mol A., de Almeida Engler J., Boerjan W., El Jaziri M., Baucher M. Molecular changes concomitant with vascular system development in mature galls induced by root-knot nematodes in the model tree host Populus tremula × P. alba. Int. J. Mol. Sci. 2020;21:406. doi: 10.3390/ijms21020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcala M., García A., Cabrera J., Casson S., Lindsey K., Favery B., García-Casado G., Solano R., Fenoll C., Escobar C. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010;61:698–712. doi: 10.1111/j.1365-313X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
- Bartlem D.G., Jones M.G.K., Hammes U.Z. Vascularization and nutrient delivery at root-knot nematode feeding sites in host roots. J. Exp. Bot. 2014;65:1789–1798. doi: 10.1093/jxb/ert415. [DOI] [PubMed] [Google Scholar]
- Cabrera J., Barcala M., Fenoll C., Escobar C. Transcriptomic signatures of transfer cells in early developing nematode feeding cells of Arabidopsis focused on auxin and ethylene signaling. Front. Plant Sci. 2014;5:107. doi: 10.3389/fpls.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera J., Díaz-Manzano F.E., Sanchez M., Rosso M.N., Melillo T., Goh T., Fukaki H., Cabello S., Hofmann J., Fenoll C., Escobar C. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 2014;203:632–645. doi: 10.1111/nph.12826. [DOI] [PubMed] [Google Scholar]
- Caillaud M.C., Dubreuil G., Quentin M., Perfus-Barbeoch L., Lecomte P., de Almeida Engler J., Abad P., Rosso M.N., Favery B. Root-knot nematodes manipulate plant cell functions during a compatible interaction. J. Plant Physiol. 2008;165:104–113. doi: 10.1016/j.jplph.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Chhabra S., Chang S.C., Nguyen H.M., Huq R., Tanner M.R., Londono L.M., Estrada R., Dhawan V., Chauhan S., Upadhyay S.K., et al. Kv1.3 channel-blocking immunomodulatory peptides from parasitic worms: implications for autoimmune diseases. Faseb. J. 2014;28:3952–3964. doi: 10.1096/fj.14-251967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M., Chung S.M., Frankman E.L., Tzfira T. pSAT RNA interference vectors: A modular series for multiple gene down-regulation in plants. Plant Physiol. 2007;145:1272–1281. doi: 10.1104/pp.107.106062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann R., Richards L., Schäfer P. Hormones as go-betweens in plant microbiome assembly. Plant J. 2021;105:518–541. doi: 10.1111/tpj.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar C., Brown S., Mitchum M.G. In: Genomics and Molecular Genetics of Plant-Nematode Interactions-- Jones J., Gheysen G., Fenoll C.C., editors. Springer Netherlands; 2011. Transcriptomic and Proteomic Analysis of the Plant Response to Nematode Infection; pp. 157–173. [DOI] [Google Scholar]
- Farmer L.M., Book A.J., Lee K.H., Lin Y.L., Fu H., Vierstra R.D. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell. 2010;22:124–142. doi: 10.1105/tpc.109.072660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B., Dubreuil G., Chen M.S., Giron D., Abad P. Gall-inducing parasites: Convergent and conserved strategies of plant manipulation by insects and nematodes. Annu. Rev. Phytopathol. 2020;58:1–22. doi: 10.1146/annurev-phyto-010820-012722. [DOI] [PubMed] [Google Scholar]
- Favery B., Lecomte P., Gil N., Bechtold N., Bouchez D., Dalmasso A., Abad P. RPE, a plant gene involved in early developmental steps of nematode feeding cells. EMBO J. 1998;17:6799–6811. doi: 10.1093/emboj/17.23.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller V.L., Lilley C.J., Atkinson H.J., Urwin P.E. Differential gene expression in Arabidopsis following infection by plant-parasitic nematodes Meloidogyne incognita and Heterodera schachtii. Mol. Plant Pathol. 2007;8:595–609. doi: 10.1111/j.1364-3703.2007.00416.x. [DOI] [PubMed] [Google Scholar]
- Gavrilovic S., Yan Z., Jurkiewicz A.M., Stougaard J., Markmann K. Inoculation insensitive promoters for cell type enriched gene expression in legume roots and nodules. Plant Methods. 2016;12:4. doi: 10.1186/s13007-016-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheysen G., Mitchum M.G. Phytoparasitic nematode control of plant hormone pathways. Plant Physiol. 2019;179:1212–1226. doi: 10.1104/pp.18.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M.E., Mayer U., Capron A., Ngo Q.A., Surendrarao A., McClinton R., Jürgens G., Sundaresan V. The TORMOZ gene encodes a nucleolar protein required for regulated division planes and embryo development in Arabidopsis. Plant Cell. 2007;19:2246–2263. doi: 10.1105/tpc.106.042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Major I.T., Howe G.A. Resolution of growth-defense conflict: mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 2018;44:72–81. doi: 10.1016/j.pbi.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Hewezi T., Juvale P.S., Piya S., Maier T.R., Rambani A., Rice J.H., Mitchum M.G., Davis E.L., Hussey R.S., Baum T.J. The cyst nematode effector protein 10A07 targets and recruits host posttranslational machinery to mediate its nuclear trafficking and to promote parasitism in Arabidopsis. Plant Cell. 2015;27:891–907. doi: 10.1105/tpc.114.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson J.P., Ivens A.C., Harcus Y., Filbey K.J., McSorley H.J., Murray J., Bridgett S., Ashford D., Dowle A.A., Maizels R.M. Secretion of protective antigens by tissue-stage nematode larvae revealed by proteomic analysis and vaccination-induced sterile immunity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Dong R., Allen R., Davis E.L., Baum T.J., Hussey R.S. A root-knot nematode secretory peptide functions as a ligand for a plant transcription factor. Mol. Plant Microbe Interact. 2006;19:463–470. doi: 10.1094/MPMI-19-0463. [DOI] [PubMed] [Google Scholar]
- Huang G., Gao B., Maier T., Allen R., Davis E.L., Baum T.J., Hussey R.S. A profile of putative parasitism genes expressed in the esophageal gland cells of the root-knot nematode Meloidogyne incognita. Mol. Plant Microbe Interact. 2003;16:376–381. doi: 10.1094/MPMI.2003.16.5.376. [DOI] [PubMed] [Google Scholar]
- Huot B., Yao J., Montgomery B.L., He S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jammes F., Lecomte P., de Almeida-Engler J., Bitton F., Martin-Magniette M.L., Renou J.P., Abad P., Favery B. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant J. 2005;44:447–458. doi: 10.1111/j.1365-313X.2005.02532.x. [DOI] [PubMed] [Google Scholar]
- Jones J.T., Haegeman A., Danchin E.G.J., Gaur H.S., Helder J., Jones M.G.K., Kikuchi T., Manzanilla-López R., Palomares-Rius J.E., Wesemael W.M.L., Perry R.N. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013;14:946–961. doi: 10.1111/mpp.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I., Kumar A., Kohli D., Bhattacharya R., Sirohi A., Chaudhury A., Jain P.K. Gall-specific promoter, an alternative to the constitutive CaMV35S promoter, drives host-derived RNA interference targeting Mi-msp2 gene to confer effective nematode resistance. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.1007322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I., Kumar A., Singh A.K., Kohli D., Raman K.V., Sirohi A., Chaudhury A., Jain P.K. Development of nematode resistance in Arabidopsis by HD-RNAi-mediated silencing of the effector gene Mi-msp2. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-53485-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M., Lee S., Abdelmageed H., Reichert A., Lee H.K., Fokar M., Mysore K.S., Allen R.D. Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 2017;40:702–716. doi: 10.1111/pce.12892. [DOI] [PubMed] [Google Scholar]
- Kaplan-Levy R.N., Brewer P.B., Quon T., Smyth D.R. The trihelix family of transcription factors-light, stress and development. Trends Plant Sci. 2012;17:163–171. doi: 10.1016/j.tplants.2011.12.002. [DOI] [PubMed] [Google Scholar]
- Kong D., Li C., Xue W., Wei H., Ding H., Hu G., Zhang X., Zhang G., Zou T., Xian Y., et al. UB2/UB3/TSH4-anchored transcriptional networks regulate early maize inflorescence development in response to simulated shade. Plant Cell. 2023;35:717–737. doi: 10.1093/plcell/koac352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B., Zhuo K., Wu P., Cui R., Zhang L.H., Liao J. A novel effector protein, MJ-NULG1a, targeted to giant cell nuclei plays a role in Meloidogyne javanica parasitism. Mol. Plant Microbe Interact. 2013;26:55–66. doi: 10.1094/MPMI-05-12-0114-FI. [DOI] [PubMed] [Google Scholar]
- Liu S., Yuan X., Wang Y., Wang H., Wang J., Shen Z., Gao Y., Cai J., Li D., Song F. Tomato stress-associated protein 4 contributes positively to immunity against necrotrophic fungus Botrytis cinerea. Mol. Plant Microbe Interact. 2019;32:566–582. doi: 10.1094/MPMI-04-18-0097-R. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- MacLean A.M., Orlovskis Z., Kowitwanich K., Zdziarska A.M., Angenent G.C., Immink R.G.H., Hogenhout S.A. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 2014;12 doi: 10.1371/journal.pbio.1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejias J., Bazin J., Truong N.M., Chen Y., Marteu N., Bouteiller N., Sawa S., Crespi M.D., Vaucheret H., Abad P., et al. The root-knot nematode effector MiEFF18 interacts with the plant core spliceosomal protein SmD1 required for giant cell formation. New Phytol. 2021;229:3408–3423. doi: 10.1111/nph.17089. [DOI] [PubMed] [Google Scholar]
- Mejias J., Chen Y., Bazin J., Truong N.M., Mulet K., Noureddine Y., Jaubert-Possamai S., Ranty-Roby S., Soulé S., Abad P., et al. Silencing the conserved small nuclear ribonucleoprotein SmD1 target gene alters susceptibility to root-knot nematodes in plants. Plant Physiol. 2022;189:1741–1756. doi: 10.1093/plphys/kiac155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Liu P., Liu Q., Chen C., Guo Q., Yin J., Yang G., Jian H. Msp40 effector of root-knot nematode manipulates plant immunity to facilitate parasitism. Sci. Rep. 2016;6 doi: 10.1038/srep19443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Olmo R., Cabrera J., Díaz-Manzano F.E., Ruiz-Ferrer V., Barcala M., Ishida T., García A., Andrés M.F., Ruiz-Lara S., Verdugo I., et al. Root-knot nematodes induce gall formation by recruiting developmental pathways of post-embryonic organogenesis and regeneration to promote transient pluripotency. New Phytol. 2020;227:200–215. doi: 10.1111/nph.16521. [DOI] [PubMed] [Google Scholar]
- Olmo R., Cabrera J., Fenoll C., Escobar C. A role for ALF4 during gall and giant cell development in the biotic interaction between Arabidopsis and Meloidogyne spp. Physiol. Plantarum. 2019;165:17–28. doi: 10.1111/ppl.12734. [DOI] [PubMed] [Google Scholar]
- Olmo R., Cabrera J., Moreno-Risueno M.A., Fukaki H., Fenoll C., Escobar C. Molecular transducers from roots are triggered in Arabidopsis leaves by root-knot nematodes for successful feeding site formation: A conserved post-embryogenic De novo organogenesis program? Front. Plant Sci. 2017;8:875. doi: 10.3389/fpls.2017.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.C., Kim M.L., Kang Y.H., Jeon J.M., Yoo J.H., Kim M.C., Park C.Y., Jeong J.C., Moon B.C., Lee J.H., et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 Box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004;135:2150–2161. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M.J., Pierik R., Van Wees S.C.M. Different shades of JAZ during plant growth and defense. New Phytol. 2014;204:261–264. doi: 10.1111/nph.13029. [DOI] [PubMed] [Google Scholar]
- Portillo M., Lindsey K., Casson S., García-Casado G., Solano R., Fenoll C., Escobar C. Isolation of RNA from laser-capture-microdissected giant cells at early differentiation stages suitable for differential transcriptome analysis. Mol. Plant Pathol. 2009;10:523–535. doi: 10.1111/j.1364-3703.2009.00552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybylska A., Spychalski M. Changes in the expression level of genes encoding transcription factors and cell wall-related proteins during Meloidogyne arenaria infection of maize (Zea mays) Mol. Biol. Rep. 2021;48:6779–6786. doi: 10.1007/s11033-021-06677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Zhang F. Cell cycle regulation in the plant response to stress. Front. Plant Sci. 2020;10:1765. doi: 10.3389/fpls.2019.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz M.U., Gifford M.L., Schäfer P. Hormone activities and the cell cycle machinery in immunity-triggered growth inhibition. J. Exp. Bot. 2015;66:2187–2197. doi: 10.1093/jxb/erv106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich-Griffin C., Eichmann R., Reitz M.U., Hermann S., Woolley-Allen K., Brown P.E., Wiwatdirekkul K., Esteban E., Pasha A., Kogel K.H., et al. Regulation of cell type-specific immunity networks in Arabidopsis roots. Plant Cell. 2020;32:2742–2762. doi: 10.1105/tpc.20.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Uehara T., Holbein J., Sasaki-Sekimoto Y., Gan P., Bino T., Yamaguchi K., Ichihashi Y., Maki N., Shigenobu S., et al. Transcriptomic analysis of resistant and susceptible responses in a new model root-knot nematode infection system using Solanum torvum and Meloidogyne arenaria. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.680151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K., Lilley J.L.S., Lee T., Tamvakis I., Kohlen W., Bailey P.C., Thomas A., Luptak J., Ramakrishnan K., Carpenter M.D., et al. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 2019;29:3657–3668.e5. doi: 10.1016/j.cub.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla N., Yadav R., Kaur P., Rasmussen S., Goel S., Agarwal M., Jagannath A., Gupta R., Kumar A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018;19:615–633. doi: 10.1111/mpp.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Lin B., Huang Q., Sun T., Wang W., Liao J., Zhuo K. The Meloidogyne javanica effector Mj2G02 interferes with jasmonic acid signalling to suppress cell death and promote parasitism in Arabidopsis. Mol. Plant Pathol. 2021;22:1288–1301. doi: 10.1111/mpp.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Shimoda Y., Kawaguchi M., Hayashi M. A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science. 2019;366:1021–1023. doi: 10.1126/science.aax2153. [DOI] [PubMed] [Google Scholar]
- Strader L., Weijers D., Wagner D. Plant transcription factors-being in the right place with the right company. Curr. Opin. Plant Biol. 2022;65 doi: 10.1016/j.pbi.2021.102136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Kanno Y., Abril-Urias P., Seo M., Escobar C., Tsai A.Y.L., Sawa S. Local auxin synthesis mediated by YUCCA4 induced during root-knot nematode infection positively regulates gall growth and nematode development. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.1019427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Yamada M., Higaki T., Aida M., Kubo M., Tsai A.Y.L., Sawa S. PUCHI regulates giant cell morphology during root-knot nematode infection in Arabidopsis thaliana. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.755610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein M.C., Winter A.D., Stepek G., McCormack G., Stapleton G., Johnstone I.L., Page A.P. Combined extracellular matrix cross-linking activity of the peroxidase MLT-7 and the dual oxidase BLI-3 Is critical for post-embryonic viability in Caenorhabditis elegans. J. Biol. Chem. 2009;284:17549–17563. doi: 10.1074/jbc.M900831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Yang D.C., Meng Y.Q., Jin J., Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2019;48:D1104–D1113. doi: 10.1093/nar/gkz1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Martínez H.H., Rodríguez-Alonso G., Shishkova S., Dubrovsky J.G. Lateral root primordium morphogenesis in angiosperms. Front. Plant Sci. 2019;10:206. doi: 10.3389/fpls.2019.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor J.E., Pallaghy P.K., Pennington M.W., Norton R.S. Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone. Nat. Struct. Biol. 1996;3:317–320. doi: 10.1038/nsb0496-317. [DOI] [PubMed] [Google Scholar]
- Üstüner S., Schäfer P., Eichmann R. Development specifies, diversifies and empowers root immunity. EMBO Rep. 2022;23:e55631. doi: 10.15252/embr.202255631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waese J., Fan J., Pasha A., Yu H., Fucile G., Shi R., Cumming M., Kelley L.A., Sternberg M.J., Krishnakumar V., et al. ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell. 2017;29:1806–1821. doi: 10.1105/tpc.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Wang K., He J., Zhao Y., Wu T., Zhou X., Ding Y., Kong L., Wang X., Wang Y., Li J., et al. EAR1 Negatively Regulates ABA Signaling by Enhancing 2C Protein Phosphatase Activity. Plant Cell. 2018;30:815–834. doi: 10.1105/tpc.17.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmerdam S., Sterken M.G., van Schaik C., Oortwijn M.E.P., Sukarta O.C.A., Lozano-Torres J.L., Dicke M., Helder J., Kammenga J.E., Goverse A., et al. Genome-wide association mapping of the architecture of susceptibility to the root-knot nematode Meloidogyne incognita in Arabidopsis thaliana. New Phytol. 2018;218:724–737. doi: 10.1111/nph.15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y.L., Suzuki R., Cabrera J., Nakagami S., Sagara T., Ejima C., Sano R., Aoki Y., Olmo R., Kurata T., et al. Root-knot and cyst nematodes activate procambium-associated genes in Arabidopsis roots. Front. Plant Sci. 2017;8:1195. doi: 10.3389/fpls.2017.01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Davies L.J., Elling A.A. A Meloidogyne incognita effector is imported into the nucleus and exhibits transcriptional activation activity in planta. Mol. Plant Pathol. 2015;16:48–60. doi: 10.1111/mpp.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhong T., E L., Xu M., Dai W., Sun S., Ye J. GT factor ZmGT-3b is associated with regulation of photosynthesis and defense response to Fusarium graminearum infection in maize seedling. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.724133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li L., Liu Q., Liu P., Li S., Yang D., Chen Y., Pagnotta S., Favery B., Abad P., Jian H. A MIF-like effector suppresses plant immunity and facilitates nematode parasitism by interacting with plant annexins. J. Exp. Bot. 2019;70:5943–5958. doi: 10.1093/jxb/erz348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Sun Q., Quentin M., Ling J., Abad P., Zhang X., Li Y., Yang Y., Favery B., Mao Z., Xie B. A Meloidogyne incognita C-type lectin effector targets plant catalases to promote parasitism. New Phytol. 2021;232:2124–2137. doi: 10.1111/nph.17690. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Yuan G., Zhao R., An G., Li W., Si W., Liu J., Sun D. Comparative transcriptome analysis reveals differential gene expression in resistant and susceptible watermelon varieties in response to Meloidogyne incognita. Life. 2022;12:1003. doi: 10.3390/life12071003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the main manuscript and the supplemental information.