Abstract
Complement attack is a host strategy leading to elimination of pathogens. Yersinia enterocolitica expresses several potential complement resistance factors: the outer membrane proteins YadA and Ail as well as lipopolysaccharide (LPS). To study the contribution of these factors to the survival of Y. enterocolitica serotype O:3 in nonimmune human serum, we constructed 23 mutant strains of Y. enterocolitica O:3 expressing different combinations of YadA, Ail, LPS O antigen, and LPS outer core. Survival of bacteria was analyzed in normal serum (with functional classical, lectin, and alternative complement activation pathways) and EGTA-Mg-treated serum (only alternative pathway functional). Kinetic killing tests revealed that the most potent single-serum resistance factor needed for long-term survival was YadA; Ail was also indispensable, but it provided short-term survival and delayed the bacterial killing. On the contrary, the LPS O antigen and outer core, when in combination with YadA, Ail, or both, had a minor and often negative effect on serum resistance. Bacteria in the exponential phase of growth were more resistant to serum killing than stationary-phase bacteria. After exposing bacteria to EGTA-Mg-treated serum, O antigen could prevent deposition of covalently bound C3b on bacteria at 3 min of incubation, even as a single factor. At later time points (15 and 30 min) it had to be accompanied by YadA, Ail, and outer core. In normal serum, the bacteria were less resistant to C3b deposition. However, no direct correlation between the C3 deposition pattern and bacterial resistance was observed.
The complement system is a key player in defending the host against intruders like microbes. Activation of complement by serum-sensitive microorganisms leads to activation of C3. The cleavage product of C3, C3b, deposits on microbial surface and binds complement component C5. C5b, the cleavage product of C5, subsequently initiates the cascade of interactions that lead to insertion of the membrane attack complex, MAC (C5b-9), into the bacterial membrane. This event results in bacterial lysis and cell death. Many microbes, however, evolved mechanisms to evade this attack.
In general, the serum resistance factors of pathogenic bacteria are surface components. The thick peptidoglycan layer of gram-positive bacteria prevents access of the MAC to the cytoplasmatic membrane, thus protecting bacteria (e.g., Streptococcus pneumoniae) against cell lysis (18). Long O-side chains of gram-negative bacteria can also provide a steric hindrance against MAC formation. Complement resistance of Klebsiella pneumoniae (45) or Coxiella burnetii (47) is mediated especially by the O-antigen chains. Bacterial capsules, on the other hand, not only function as a barrier preventing MAC penetration through the bacterial membrane, but in some cases they also inhibit the alternative pathway activation, e.g., sialilated capsular polysaccharides of type III group B streptococci (22). Finally, some microbes are equipped with membrane proteins conferring complement resistance. For example, TraT protein of Escherichia coli inhibits formation of C5b6 complex (31), and M protein of Streptococcus pyogenes blocks activity of the alternative pathway C3 convertase by binding to the complement regulator, factor H. Microbial structures constantly evolve in response to the host immune system, and the above-given examples illustrate only a few multiple-complement evasion strategies.
Yersinia enterocolitica, a gram-negative pathogen transmitted through contaminated food or water, leads to human and animal enteric infection. In the initial phase of the infection, the pathogen crosses the epithelial barrier (intestinal mucosa) through the M cells of Peyer's patches and infects the underlying tissues. Survival in deeper tissues and serum resistance depends on Yersinia virulence factors encoded by genes located on the Yersinia chromosome, e.g., Ail and lipopolysaccharide (LPS) O-antigen (O-ag) and outer core (OC), and on the 65- to 70-kb Yersinia virulence plasmid pYV, e.g., YadA and Yop proteins. Expression of YadA and Ail occurs exclusively at 37°C; O-side chain is more abundant at room temperature (RT), whereas OC is produced at both RT and 37°C. Due to temperature-dependent gene regulation, only bacteria grown at 37°C are resistant to killing by alternative pathway (AP), classical pathway (CP), or both (CP/AP). C3b, the key protein of complement activation, binds more strongly to pYV-negative or YadA-negative bacteria than to pYV-positive or YadA-positive bacteria grown at 37°C (11, 44). Moreover, pYV-positive strains, which are serum resistant even when opsonized, display much less of the MACs deposited on their surfaces than do the pYV- and YadA-negative strains (30). These results suggest that YadA may be involved in inhibition of complement activation at both the C3b (possibly via factor H binding) and C9 levels.
Another outer membrane (OM) protein conferring resistance on complement killing is Ail. Ail-mediated resistance was prominent not only in YadA- or invasin-negative mutants (7) but also in E. coli carrying the cloned ail gene, which displayed resistance to complement about 100 times as high as that of E. coli lacking Ail (29). No Ail-depending molecular mechanism has been established. Theoretically, Ail could interact with some host factors and interfere with formation of active attack complexes.
In addition to OM proteins, serum resistance of Y. enterocolitica depends on the LPS compounds O-ag and OC. O-ag presumably plays a role in inhibition of the early phase of alternative pathway activation (46). As for OC, our previous studies do not indicate any direct involvement of OC in serum resistance (39). On the contrary, OC-positive, YadA- and Ail-negative strains were efficiently killed by complement. In the absence of YadA, however, OC seemed to potentiate Ail-mediated resistance.
In this report, we analyzed serum resistance of Y. enterocolitica O:3 using a collection of strains expressing different combinations of YadA, Ail, O-ag, and OC. OC and O-ag are linked to different parts of the inner core (39, 40). This particular feature of Y. enterocolitica serotype O:3 LPS made it possible to construct mutants missing OC and still expressing O-ag. The strains were analyzed for serum resistance in a killing assay in normal and EGTA [ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetra-acetic acid]-Mg serum. The results showed that YadA was indispensable for bacterial survival; Ail appeared to delay CP/AP-mediated killing, while O-ag and OC are possibly involved in serum resistance indirectly. Immunoblotting analysis indicated that O-ag plays an important role in blocking the early deposition of C3 on bacterial surfaces. However, no direct correlation between the C3 deposition pattern and bacterial resistance was observed.
MATERIALS AND METHODS
Bacteria, plasmids, phages, and growth conditions.
The bacterial strains, plasmids, and bacteriophages used in this study are listed in Table 1. For bactericidal assay, bacteria were grown to stationary phase overnight in 5 ml of MedECa (0.1 g of MgSO4 · 7H2O liter−1, 2 g of citric acid liter−1, 10 g of K2HPO4 liter−1, 3.5 g of NaNH4HPO4 · 4H2O liter−1, and 1 mg of vitamin B1 liter−1, supplemented with 0.2% glucose, 0.2% Casamino Acids, and 2.5 mM CaCl2) at 22 to 25°C (RT) and 37°C without shaking. To study serum resistance of bacteria at exponential growth phase, overnight bacterial cultures were diluted 1:20 in fresh medium and incubated for 3 h at RT or 37°C without shaking. When appropriate, antibiotics were added to the growth medium at the following concentrations: kanamycin (Km), 100 μg/ml in agar plates and 20 μg/ml in broth; chloramphenicol (Clm), 20 μg/ml; ampicillin (Amp), 50 to 100 μg/ml.
TABLE 1.
Bacteria, plasmids, and phages used in this work
| Bacterial strains, plasmids, and phages | Description | Source or reference |
|---|---|---|
| Y. enterocolitica strains | ||
| 6471/76 (YeO3) | Serotype O:3, patient isolate, wild type | 37 |
| YeO3-Ail | Δail::Km-GenBlock, Kmr | This work |
| YeO3-Ail-R | Spontaneous rough derivative of YeO3-Ail, Kmr | This work |
| YeO3-Ail-OCR | Spontaneous OC mutant derivative of YeO3-Ail-R, Kmr | This work |
| YeO3-trs11 | Δ(wzx-wbcL)::Km-GenBlock, Kmr, derivative of YeO3 | 40 |
| YeO3-trs11-R | Spontaneous rough derivative of YeO3-trs11, Kmr | 40 |
| YeO3-OC | Δ(wzx-wbcQ), derivative of YeO3 | This work |
| YeO3-OCR | Spontaneous rough derivative of YeO3-OC | This work |
| YeO3-Ail-OC | Δail::Km-GenBlock, Kmr, derivative of YeO3-OC | This work |
| YeO3-R2 | Spontaneous rough derivative of YeO3 | 2 |
| YeO3-028 | ΔyadA::Km-GenBlock, Kmr, derivative of YeO3 | This work |
| YeO3-028-R | Spontaneous rough derivative of YeO3-O28, Kmr | This work |
| YeO3-028-OCR | Spontaneous OC mutant derivative of YeO3-028-R, Kmr | This work |
| YeO3-028-OC | Δ(wzx-wbcQ) derivative of YeO3-028, Kmr | This work |
| 6471/76-c (YeO3-c) | Virulence plasmid cured derivative of YeO3 | 37 |
| YeO3-R1 | Spontaneous rough derivative of YeO3-c | 2 |
| YeO3-c-OC | Δ(wzx-wbcQ) derivative of YeO3-c | This work |
| YeO3-c-OCR | Spontaneous rough derivative of YeO3-c-OC | This work |
| YeO3-c-trs8 | Δ(wzx-wbcL)::Km-GenBlock, Kmr, derivative of YeO3-c | 40 |
| YeO3-c-trs8R | Spontaneous rough derivative of YeO3-c-trs8, Kmr | 40 |
| YeO3-c-Ail | Δail::Km-GenBlock, Kmr, derivative of YeO3-c | This work |
| YeO3-c-Ail-OC | Spontaneous OC mutant derivative of YeO3-c-Ail, Kmr | This work |
| YeO3-c-Ail-R | Spontaneous rough derivative of YeO3-c-Ail, Kmr | This work |
| YeO3-c-Ail-OCR | Spontaneous OC mutant derivative of YeO3-c-Ail-R, Kmr | This work |
| E. coli strains | ||
| S17-1λpir | thi pro hsdR−hsdM+recA::RP4-2-Tc::Mu-Km::Tn7, Strr (λpir) | 13 |
| SY327λpir | Δ(lac pro) argE (Am) rif nalA recA56 (λpir) | 24 |
| HBI01/pRK2013 | Triparental conjugation helper strain, Kmr | 14 |
| C600 | thi thr leuB tonA lacY supE | 4 |
| Plasmids | ||
| pAil-8 | A pBR322 clone carrying an 8-kb insert of YeO3-c containing the ail gene; Ampr | This work |
| pAil-8.1 | PvuII deletion derivative of pAil-8; Ampr | This work |
| pAil-8.2 | EcoRV-NruI deletion derivative of pAil-8.1; Ampr | This work |
| pBR322 | Cloning vector; Ampr Tetr | 9 |
| pJM703.1 | Suicide vector, contains R6K origin of replication and RP4 Mob region; must be replicated in (λpir) hosts; Ampr | 24 |
| pRV1 | Suicide vector; derivative of pJM703.1; Clmr | 19 |
| pRV7 | A pBR322 clone carrying a 12.4-kb insert of YeO3-c covering the outer core gene cluster; Ampr | 40 |
| pRV37 | Intermediate to construct OC deletion suicide vector; pBR322 carries a HindIII fragment of pRV7 from which the OC gene cluster is deleted by PCR; Ampr | This work |
| pRV38 | 2-kb HindIII fragment of pRV37 cloned into pRV1 (O:3 OC gene cluster suicide vector); Clmr | This work |
| pRV42 | Suicide vector constructed to inactivate the ail gene; Km-GenBlock inserted between ail flanking fragments and cloned into pRV1; Kmr, Clmr | This work |
| pTM100 | Mobilizable vector, pACYC184-oriT of RK2; Clmr | 23 |
| pUC-4K | Origin of the Km-GenBlock cassette; Ampr, Kmr | Pharmacia |
| pYMS 4505 | yadA gene of YeO3 cloned into BamHI site of pTM100; Clmr | This work |
| pYMS 4506 | ΔyadA::Km-GenBlock derivative of pYMS4505; the internal 1.3-kb NsiI fragment of yadA gene replaced with Km-GenBlock; Clmr Kmr | This work |
| pYMS 4509 | ΔyadA::Km-GenBlock fragment cloned into EcoRI site of pJM703.1; Ampr Kmr | This work |
| pYMS 4515 | Suicide vector to inactivate the yadA gene; the cat gene of pTM100 cloned into the PstI site of pYMS4509; Clmr Kmr | This work |
| Bacteriophages | ||
| φYe03-12 | O-antigen-specific phage of Y. enterocolitica O:3 | 1 |
| φR1-37 | Outer core-specific phage of Y. enterocolitica O:3 | 40 |
Construction of pRV38, a suicide plasmid for deletion of the OC gene cluster.
An intermediate plasmid named pRV37 was constructed based on pRV7 that is a genomic library clone of YeO3-c with a 12.4-kb insert cloned in pBR322 (40). When deleting the OC gene cluster between the hemH and gsk genes in pRV7 by the PCR-based approach (10), pRV7 was used as a template in a PCR with phosphorylated primers trs-47 and gale (Table 2). This resulted in amplification of the approximately 6-kb fragment that contained the complete vector (pBR322) flanked by the hemH and gsk sequences. The 6-kb PCR fragment was gel purified, self ligated, and transformed into E. coli C600. The recovered 6-kb plasmid was named pRV37, and it carried in the pBR322-vector an approximately 2-kb insert where the hemH gene (missing its 5′ end) was immediately followed by the gsk gene (missing its 3′ end). The 2-kb insert was released from pRV37 by HindIII digestion and was gel purified, after which it was ligated with the hindsph adapter (Table 2) into the SphI site of suicide vector pRV1 (Table 1). The ligation mixture was transformed into E. coli SY327λpir, and the recovered 8.6-kb plasmid was named pRV38.
TABLE 2.
Oligonucleotides used in this work
| Name | Sequence (5′-3′) | 5′-3′ position | Accession no. | Purpose |
|---|---|---|---|---|
| ail1 | GACATTACTAGCTAGTTCTC | 496-515 | M29945 | PCR |
| ail2 | GAATCGATACCCTGCACCAAGC | 1021-1000 (2731-2710) | M29945(AJ605740) | Deletion PCR |
| ail4 | CAAGTAAGACGTCAATGGCATACGG | 2592-2616 | AJ605740 | Deletion PCR |
| ail5 | CGTTACTTCCCAGCCCCGGCAA | 1859-1838 | AJ605740 | PCR |
| ail9 | AAGCGCTTGAGGTCAAAGCACAA | 1490-1512 | AJ605740 | PCR |
| ail10 | TTAATGTGTACGCTGCGAGTGAA | 2253-2275 | AJ605740 | PCR |
| trs19 | AGCGTCGACGAAACCTTAAAAAC | 1621-1640 | Z47767 | PCR |
| trs30 | GAGCATTAACAGGAAAATAG | 12935-12916 | Z47767 | PCR |
| trs47 | CTGCAGACAAGCAAAAGGGCCAGGTGAA | 1929-1908 | Z47767 | Deletion PCR, PstI site underlined |
| gale | CCTGG(C/T)GATAT(C/A)GCTGAGTGTTGGGC | 12681-12706 | Z47767 | Deletion PCR |
| hindsph | AGCTCATG | HindIII-SphI adapter | ||
| MS16 | CGCGGATCCATTAGGATTAATACAGGCGCA | 2213-2196 | X13882 | yadA cloning, BamHI site underlined |
| MS17 | CGCGGATCCTGCATGATTATCAGAAAAATA | 478-495 | X13882 | yadA cloning, BamHI site underlined |
| MS20 | CGCGAATTCATTAGGATTAATACAGGCGCA | 2213-2196 | X13882 | yadA::KmGenBlock cloning, EcoRI site underlined |
| MS21 | CGCGAATTCTGCATGATTATCAGAAAAATA | 478-495 | X13882 | yadA::KmGenBlock cloning, EcoRI site underlined |
| MS27 | TCGCTGCAGAATAAATAAATCCTGGTG | 396-375 | X06403 | cat cloning, PstI site underlined |
| MS28 | ATGCTGCAGCAATAGACATAAGCGGC | 3952-3971 | X06403 | cat cloning, PstI site underlined |
Construction of pRV42, a suicide vector for the deletion of the ail gene.
The construction of pRV42 started by isolating the Y. enterocolitica O:3 ail gene carrying clones from the genomic library of YeO3-c (1). The library was screened by colony hybridization using a purified ail-specific 525-bp PCR fragment as a probe. The fragment was obtained using chromosomal DNA of YeO3-c as template and ail1 and ail2 (Table 2) as primers. Four hybridization-positive clones were obtained; one of those carried a plasmid that was named pAil-8. The nucleotide sequence of the ca. 3.9-kb segment over the ail gene and flanking regions in pAil-8 was determined (accession no. AJ605740). Based on the sequence and restriction mapping, a cut-back ligation of PvuII-digested pAil-8 was then performed, and the recovered plasmid was named pAIL-8.1. EcoRV-NruI cut-back ligation was performed with pAil-8.1 to obtain a plasmid named pAIL-8.2. pAIL-8.2 was used as a template for ail deletion-PCR (10) using primers ail4 and ail5 (that are directed from both ends of the ail gene outwards). The resulting ca. 6-kb PCR fragment was gel purified and ligated with the 1.2-kb kanamycin GenBlock fragment obtained from pUC-4K with HincII digestion. The constructed ca. 7-kb plasmid was named pRV40, and it carried the kanamycin GenBlock in place of the deleted ail gene. pRV40 was digested with ScaI and PvuII, and the approximately 5-kb fragment was gel purified and ligated to EcoRV-digested pRV1 to obtain the ail suicide vector pRV42, which was transformed into E. coli S17-1λpir.
Construction of pYMS4515, a suicide vector to eliminate the yadA gene.
The yadA gene of Y. enterocolitica O:3 was first cloned into the BamHI site of pTM100 in a 2-kb PCR fragment amplified using primers MS16 and MS17 (Table 2). The obtained 6.7-kb plasmid, pYMS4505, was digested with NsiI, which released a 1.3-kb NsiI fragment from the yadA gene. The 1.2-kb Km GeneBlock PstI fragment of pUC4-K was ligated with the 5.4-kb fragment of pYMS4505 to obtain plasmid pYMS4506. The Km GenBlock-carrying insert in pYMS4506 was amplified with primers MS21 and MS22 (Table 2), and the fragment was cloned into the EcoRI site of suicide vector pJM703.1 to obtain pYMS4509. Finally, the cat gene of pTM100 was amplified with primers MS27 and MS28 (Table 2), and the obtained 0.9-kb fragment was cloned into the PstI site of pYMS4509, resulting in the final Clmr suicide vector pYMS4515.
Mobilization of suicide vectors by conjugation.
Direct conjugation to Y. enterocolitica O:3 strains was performed to mobilize the suicide plasmids pRV42 (from E. coli S171λpir/pRV42) and pYMS4515 (from E. coli Sm 10λpir/pYMS 4515) to construct ail and yadA mutants, respectively, whereas triparental conjugation was used to mobilize pRV38 (from E. coli SY 327λpir/pRV38) with the help of E. coli HB101/pRK2013 to construct mutants missing the whole OC cluster.
The matings were performed essentially as follows. E. coli strains S171λpir/pRV42 (Clmr, Kmr) and SY 327λpir/pRV38 were grown overnight in 10 ml of Luria-Bertani medium (LB; 10 g of Bacto-tryptone per liter, 10 g of yeast extract per liter, 5 g of NaCl per liter) supplemented with Clm at 37°C without shaking. E. coli HB101/pRK2013 was grown overnight at 37°C with shaking in LB supplemented with Km. Yersinia strains were grown with shaking overnight in 10 ml of LB (supplemented with Km if necessary) at RT or 37°C.
Overnight bacterial cultures were centrifuged (1,500 × g, rpm, 10 min), and pellets were washed (phosphate-buffered saline [PBS]) and resuspended in 1 ml of PBS. Mating mixtures containing 100-μl aliquots of the appropriate bacterial suspensions were mixed on LA plates. After 24 h of incubation at RT, the mating mixtures on plates were suspended into 1 ml of PBS, from which diluted bacterial suspensions were plated out on CIN agar plates (Yersinia-selective agar base; Oxoid) without or supplemented with appropriate antibiotics to select for Yersinia strains. Obtained colonies were screened on appropriate antibiotic plates to select for the successful integration of the suicide vector via homologous recombination into the recipient strain.
Cycloserine enrichment.
Cycloserine was used to enrich clones in which the second recombination event had eliminated the suicide vector and the wild-type gene(s) as follows. A single colony from the above-described matings was grown in LB without any antibiotic for 3 days at RT (cultures were refreshed every day with 200 μl of culture plus 10 ml of LB). Ten milliliters of LB supplemented with 10 μg of Clm/ml was inoculated with 250 μl of the overnight culture (optical density at 600 nm [OD600] ≈ 0.05). The culture was grown until an OD600 of 0.2 was reached. One milliliter of a freshly prepared cycloserine solution (25 mg/ml) was added, and the culture was incubated for an additional 3 h at RT. This treatment would efficiently kill only growing, i.e., Clmr bacteria, while those that had lost the suicide vector after a successful second crossover event would survive. Cells were pelleted, washed once with PBS, and resuspended in 10 ml of PBS, from which 10-fold serial dilutions (up to 10−5) were made and spread on LA plates (100 μl/plate) without any antibiotic to obtain single colonies. To isolate Clms clones, colonies were patched on unsupplemented LA or LA supplemented with Clm.
Isolation of spontaneous OC mutants using φR1-37.
Bacteriophage φR1-37, specific for OC of Y. enterocolitica O:3, was used to select spontaneous OC mutants (i.e., resistant to phage φR1-37). Bacteria grown overnight in LB were spread as a lawn on LA plates supplemented with antibiotic, if necessary. A few drops of φR1-37 were pipetted on the plates after the lawns had dried. After 2 days of incubation, the individual phage-resistant colonies were picked from within the lysis zone and were subjected to the same treatment several times, if necessary. The loss of OC expression was verified by LPS analysis using deoxycholate-polyacrylamide gel electrophoresis (DOC-PAGE; see below).
Selection of OC mutants using enterocoliticin.
Strains of Y. enterocolitica serotypes O:3, O:5,27, and O:9 are killed by enterocoliticin. Enterocoliticin is produced by Y. enterocolitica serotype O:7,8 strain 29930 (43) and consists of phage tail-like particles that are specific for the OC hexasaccharide (42). Enterocoliticin (104 U/ml), kindly provided by E. Strauch (Robert Koch Institute, Berlin, Germany), was used in the same way as described above for φR1-37.
Isolation of spontaneous O-ag mutants using φYeO3-12.
φYeO3-12 infects Y. enterocolitica O:3 strains by using the O-ag as a receptor (1, 25, 26). It was used to select spontaneous rough Y. enterocolitica O:3 mutants. Experiments were carried out in the same way as that described for φR1-37 infection, but strains were incubated at RT to promote better expression of O-ag.
Isolation and analysis of LPS.
LPSs from OC and O-ag mutant candidates were checked by DOC-PAGE. For small-scale LPS isolation, a modified version of the protocol devised by Hitchcock and Brown (17) was used. The bacterial culture grown overnight in 5 ml of LB at RT was diluted to obtain an OD540 of <1. A 1.5-ml aliquot of the culture was then centrifuged in a microcentrifuge (13,000 × g; 3 min), the pellet was resuspended in DOC lysis buffer (2% DOC, 4% 2-mercaptoethanol, 10% glycerol, and 0.002% bromophenol blue in 1 M Tris-HCl buffer, pH 6.8) in a volume adjusted according to the OD540 of the culture (100 μl of DOC lysis buffer/OD540 = 1.0), and the solution was heated at 100°C for 10 min. Two microliters of 20 mg of proteinase K/ml was then added, and lysates were incubated at 55°C for ≥1 h. Samples were stored at −20°C until analyzed by DOC-PAGE as described previously (40).
Extraction of genomic DNA.
Bacteria resuspended in 100 μl of water were incubated for 5 min at 95°C, 100 μl of phenol chloroform/isoamyl alcohol (25:24:1) was added, and samples were vortexed and centrifuged for 5 min at 13,000 × g. A 5-μl aliquot of the upper phase containing the DNA was used as template in PCRs. Alternatively, bacterial DNA was isolated using the cetyltrimethylammonium bromide miniprep protocol (5).
PCR.
PCR mixtures consisted of 1 or 5 μl of genomic DNA template, 1 μl of 10 mM deoxynucleoside triphosphate mixture, 1 U of DynaZyme II DNA polymerase (2 U/μl; FinnZymes), 5 μl of 10× reaction buffer, and 20 to 40 pmol of each primer in a total volume of 50 μl. To confirm the inactivation of the ail gene, PCRs using primers ail2 and ail10 and ail2 and ail9 (Table 2) started with an initial 2-min incubation at 94°C, and then the steps (94°C for 15 s, 52°C for 30 s, 73°C for 3 min) were repeated 30 times, followed by a 5-min extension time at 73°C. The primers for the confirmation of the OC deletion mutants were trs-19 and trs-30 (Table 2). The PCR cycles were the same as those for ail primers, except that the annealing temperature was 55°C and the denaturation time was 30 s.
Immunoblotting and dot blotting to compare YadA, O-ag, and OC expression in mutants.
Bacteria were grown overnight at 37°C in 5 ml of MedECa. Whole-cell lysates were prepared from 5 ml of bacterial cultures (OD600 adjusted to 0.15). The cultures were centrifuged for 15 min (1,500 × g), and pellets were resuspended in 100 μl of Laemmli sample buffer (for O-ag and OC expression verification, samples were digested with proteinase K prior to adding Laemmli sample buffer). The mixtures were heated at 95°C for 10 min before being loaded onto the gel or spotted onto the membrane (dot blotting). For the immunoblotting, aliquots of 10 μl were subjected to electrophoresis using the Hoefer miniVE Vertical Electrophoresis System (Amersham Biosciences) on 8 or 12% resolving polyacrylamide gels (to verify expression of YadA or O-ag and OC, respectively) with 5% stacking gels. The separated samples were transferred to nitrocellulose membranes by semiwet transfer (Blot module; Amersham Biosciences) using a transfer buffer as recommended by the manufacturer. For the dot blot, 1-μl aliquots of serial twofold dilutions were spotted on the membrane. The nonspecific binding sites were blocked by immersing the membranes in a 5% skimmed milk-PBS solution (for >1 h at RT). The membranes were incubated overnight at 4°C with monoclonal antibody (MAb) specific for YadA (MAb 3G12), O-ag (MAb Tom A6), or OC (MAb 2B5), diluted 1:10, 1:25, and 1:10, respectively (27, 38). After four washes (each for 10 min) in PBS, the membranes were incubated with peroxidase-conjugated rabbit anti-mouse immunoglobulins (P0260; dilution 1:2,000; DAKO) for 1 h at RT. Antibody binding was detected by chemiluminescence using the ECL Western blotting detection reagents (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
Outer membrane isolation to determine Ail expression.
Bacteria were grown overnight in 5 ml of MedECa at 37°C without shaking. The bacteria were centrifuged for 15 min (1,500 × g) at 4°C and resuspended into 2 ml of 10 mM Tris-5 mM MgCl2, pH 8.0. Suspensions were sonicated on ice (four times for 15 s with 1-min breaks in between) and centrifuged for 10 min as described above. The supernatants were ultracentrifuged for 1 h (45,000 × g) at 4°C, and the recovered pellets were resuspended in 2 ml of 2% Triton X-100, 10 mM Tris, 5 mM MgCl2 (pH 8.0). Ultracentrifugation was repeated, and the pellets were dissolved in 50 μl of 2× sodium dodecyl sulfate (SDS) sample buffer. The samples were heated at 95°C for 15 min prior analysis by SDS-PAGE (20% separating gel, 5% stacking gel) and silver staining.
Normal human serum.
Normal human serum (NHS) was obtained from healthy human donors who were devoid of anti-Yersinia antibodies. Blood was allowed to clot for 15 min at room temperature and for 60 min at 4°C. Following centrifugation (4°C, 2,500 × g, 30 min), the sera were collected, pooled, and stored at −70°C in aliquots of 0.5 ml. Prior to use, the serum was allowed to thaw on ice. One-third of it was heat inactivated by incubation at 56°C for 30 min (HIS). To block the CP activity, EGTA and MgCl2 were added to another third to final concentrations of 10 and 5 mM, respectively. The last third represented the normal human serum.
Serum-killing assay.
Bacterial cultures were diluted appropriately to obtain ∼500 to 1,000 bacteria in 10 μl. Triplicates of 10 μl of bacterial suspensions were incubated with 20 μl of NHS (final NHS concentration of 66.7%), 20 μl of HIS, or 20 μl of EGTA-Mg serum at 37°C for 30 and 120 min. Before plating on appropriate LA plates, 70 μl of brain heart infusion broth (BHI) was added to each mixture to stop the complement function, and tubes were kept on ice. The serum bactericidal effect was calculated as the survival percentage taking the bacterial counts obtained with bacteria incubated in HIS as 100%. The killing experiment was repeated for each strain at least three times, starting from independent cultures.
Detection of C3 deposition on bacteria by immunoblotting.
Bacteria (2 × 107) grown overnight in MedECa at RT or at 37°C, washed, and resuspended in PBS were added to NHS, EGTA-Mg serum, and HIS, resulting in a reaction mixture volume of 100 μl and a 40% final concentration of serum. Reaction tubes transferred to 37°C were incubated for 3, 15, and 30 min. The reactions were stopped by placing tubes on ice for 1 min. The bacteria were centrifuged at 13,000 × g for 5 min at 4°C; the pellets were washed three times in 500 μl of PBS and were resuspended in 40 μl of SDS-PAGE sample solution. Samples were boiled for 5 min and stored at −20°C before being processed for immunoblotting as described above, using 10% resolving polyacrylamide gels with 5% stacking gels. As a control, in each gel 1.5 μl of NHS diluted 1:100 in PBS was loaded. Monoclonal antibody specific for the α-chain of C3 (MAb755; a kind gift of Ulrich Vogel, University of Würzburg, Germany) diluted 1:200 in blocking solution was used for detection of C3 fragments.
Nucleotide sequence accession number.
The nucleotide sequence of ca. 3.9-kb segment over the ail gene and flanking regions in pAil-8 was determined and deposited in the EMBL/GenBank/DDBJ databases under accession no. AJ605740.
RESULTS
Construction of 23 mutant strains.
To construct strains missing two, three, or four of Yersinia surface factors (YadA, Ail, O-ag, and OC oligosaccharide), we first stemmed from the wild-type (wt) Y. enterocolitica O:3 strain 6471/76 six single-negative direct derivatives: (i) the pYV-cured strain YeO3-c; (ii) the yadA strain YeO3-028 (ΔyadA::Km-GenBlock); (iii) the ail strain YeO3-Ail (Δail::Km-GenBlock); (iv and v) the two types OC mutants, YeO3-OC [Δ(wzx-wbcQ), with the whole OC cluster deleted] and YeO3-trs11 [Δ(wzx-wbcL)::Km-GenBlock], and (vi) the rough strain YeO3-R2 (spontaneous O-ag-negative mutant). The first five served as starting points for further construction steps. Detailed description of the construction strategies of the strains is given in Materials and Methods.
The YadA, O-ag, OC, and Ail expression levels of all the strains were compared. Immunoblotting and dot blotting showed that there were small differences in YadA expression levels between the studied strains (Fig. 1A and data not shown). Compared to the wild type, YeO3-Ail, YeO3-OC, YeO3-Ail-OC, and YeO3-OCR produced slightly more YadA, while YeO3-trs11-R and YeO3-Ail-OCR expressed significantly less protein. When O-ag expression was compared between the strains by immunoblotting, only YeO3-c-trs8 and YeO3-trs11 showed a little fainter O-ag smear than that of the wild type (Fig. 1A and data not shown). A little more O-ag was expressed only by a few strains (for example, YeO3-c; Fig. 1A and data not shown). Most strains expressed amounts of OC similar to those of YeO3, as demonstrated by immunoblotting and dot blotting. There seemed to be marginally more OC in the O-ag-negative strains (Fig. 1A and data not shown). The comparison of Ail expression was performed by analysis of outer membrane proteins (Fig. 1B). These were run on SDS-PAGE and visualized by silver staining. There was, however, a protein comigrating with the Ail band that gave a faint background band in the Ail-negative strains, YeO3-Ail and E. coli carrying pBR322 (Fig. 1B). Based on the band intensities, YeO3-R2, YeO3-c-OC, YeO3-O28-R, YeO3-OCR, and YeO3-trs11 produced slightly more Ail, while YeO3-OC and YeO3-trs11-R produced slightly less Ail, than the wild-type strain.
FIG. 1.
Detection and analysis of YadA, O-ag, OC, and Ail expression in different Y. enterocolitica O:3 strains. (A) Immunoblot analysis using monoclonal antibodies specific for YadA (top), O-ag (middle), and OC (bottom). (B) Silver-stained SDS-PAGE gel showing outer membrane proteins isolated from Ail-expressing and -nonexpressing Y. enterocolitica O:3 and E. coli C600 strains. The arrows point to the migration level of the Ail band. The presence or absence of the factors is indicated below the panels.
CP/AP- and AP-mediated killing of bacteria grown to stationary phase.
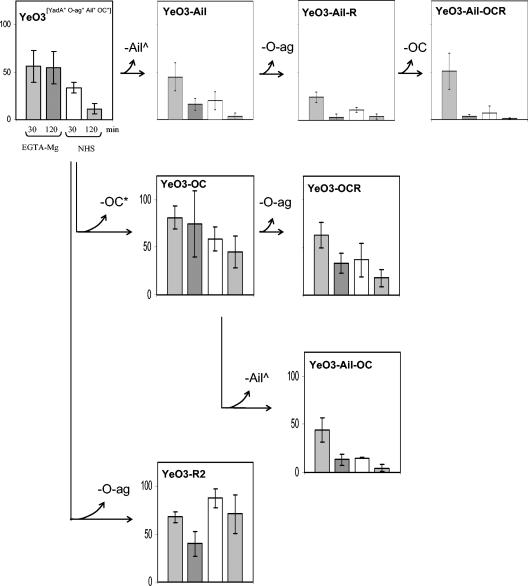
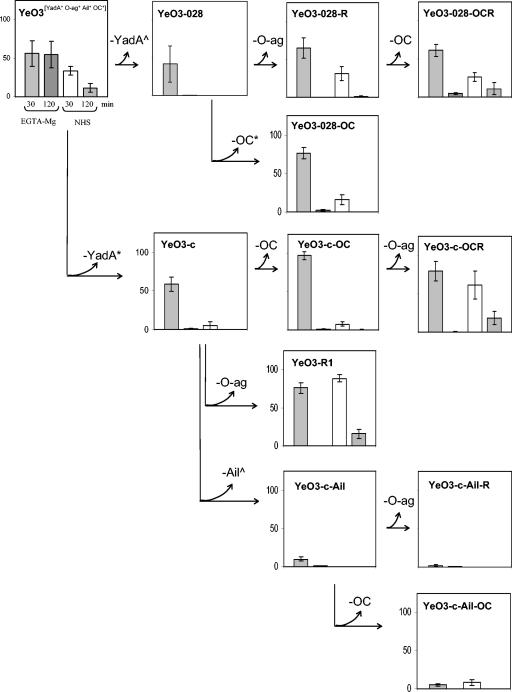
In general, when grown at RT all the studied Y. enterocolitica strains were highly susceptible to AP- and CP/AP-mediated killing, being totally killed already after 30 min of incubation (data not shown). On the contrary, after growing at 37°C the strains expressed various degrees of serum resistance. Several serum resistance phenotypes could be distinguished based on the survival percentages and killing kinetics of the strains (Table 3, Fig. 2 and 3). Below we discuss these results, organized under the construction lines of strains, to demonstrate how stepwise loss of the individual factors affected the bacterial serum resistance.
TABLE 3.
Complement resistance phenotypes and killing kinetics of Y. enterocolitica O:3 wt and mutant strains (grown at 37°C) in 66.7% NHS (based on data presented in Fig. 2 and 3)
| Phenotype (expressed factor)
|
Strain | Phenotype and kinetics at:
|
||||||
|---|---|---|---|---|---|---|---|---|
| 30 min
|
120 min
|
|||||||
| YadA | O-ag | Ail | OC | % Surviving | Killing kineticsa | % Surviving | Resistance phenotypeb | |
| YeO3-c-Ail-OCR | 0.13 ± 0.2 | Rapid | 0 ± 0 | Sensitive | ||||
| O-ag | YeO3-c-Ail-OC | 0.15 ± 0.3 | Rapid | 0.06 ± 0.2 | Sensitive | |||
| Ail | YeO3-c-OCR | 61.08 ± 18.2 | Delayed | 18.21 ± 8.9 | Intermediate | |||
| Ail | YeO3-028-OCR | 26.35 ± 5.5 | Rapid | 10.97 ± 7.8 | Intermediate | |||
| Ail | YeO3-c-trs8-R | 64.04 ± 7.6 | Delayed | 15.69 ± 6.7 | Intermediate | |||
| OC | YeO3-c-Ail-R | 8.1 ± 3.7 | Rapid | 0 | Sensitive | |||
| YadA | YeO3-Ail-OCR | 7.19 ± 1.6 | Rapid | 1.35 ± 0.7 | Intermediate | |||
| O-ag | Ail | YeO3-c-trs8 | 25.08 ± 10.6 | Rapid | 1.2 ± 1.1 | Intermediate | ||
| O-ag | Ail | YeO3-c-OC | 7.61 ± 3 | Rapid | 0.25 ± 0.6 | Sensitive | ||
| O-ag | Ail | YeO3-028-OC | 15.99 ± 6.3 | Rapid | 0.05 ± 0.1 | Sensitive | ||
| O-ag | OC | YeO3-c-Ail | 0 | Rapid | 0.02 ± 0.04 | Sensitive | ||
| Ail | OC | YeO3-028-R | 31.18 ± 9 | Delayed | 0.67 ± 0.6 | Sensitive | ||
| Ail | OC | YeO3-R | 88.93 ± 4.9 | Delayed | 15.94 ± 5.7 | Intermediate | ||
| YadA | O-ag | YeO3-Ail-OC | 15.04 ± 0.6 | Rapid | 4.46 ± 3.7 | Intermediate | ||
| YadA | Ail | YeO3-OCR | 36.84 ± 17.5 | Delayed | 17.73 ± 8.9 | Intermediate | ||
| YadA | Ail | YeO3-trs11-R | 52.75 ± 8.6 | Delayed | 25.78 ± 9.3 | Resistant | ||
| YadA | OC | YeO3-Ail-R | 10.86 ± 2.3 | Rapid | 3.67 ± 2.4 | Intermediate | ||
| O-ag | Ail | OC | YeO3-c | 4.81 ± 5.1 | Rapid | 0 | Sensitive | |
| O-ag | Ail | OC | YeO3-028 | 0.11 ± 0.13 | Rapid | 0.03 ± 0.1 | Sensitive | |
| YadA | O-ag | Ail | YeO3-trs11 | 35.72 ± 18.4 | Delayed | 28.31 ± 15.7 | Resistant | |
| YadA | O-ag | Ail | YeO3-OC | 58.91 ± 12.3 | Delayed | 45.03 ± 16.7 | Resistant | |
| YadA | O-ag | OC | YeO3-Ail | 20.1 ± 9.4 | Rapid | 3.58 ± 2 | Intermediate | |
| YadA | Ail | OC | YeO3-R2 | 87.3 ± 10.1 | Delayed | 70.85 ± 19.7 | Resistant | |
| YadA | O-ag | Ail | OC | YeO3 | 33.67 ± 5.7 | Delayed | 11.49 ± 5.8 | Intermediate |
Rapid, rapidly killed (<30% viable bacteria after 30 min); delayed, delayed type of killing (>30% viable bacteria after 30 min).
Sensitive, <1% viable bacteria after 120 min; intermediate, 1 to 20% viable bacteria after 120 min; resistant, >20% viable bacteria after 120 min.
FIG. 2.
Survival percentages of YadA-positive Y. enterocolitica O:3 strains (grown to stationary phase at 37°C) in normal and EGTA-Mg-treated sera after 30 or 120 min of incubation. Survival in HIS was set to 100%. In each panel, the left-hand columns show the 30- and 120-min survival percentages in EGTA-Mg-serum, and the right-hand columns show the 30- and 120-min survival percentages in NHS. Standard deviations are indicated by bars. The construction lines of the strains are indicated by arrows between the panels. The inactivated factor in each construction step is indicated by the branching curved arrow, and the manner of this inactivation is marked with the following symbols: ^, KmGB insertion; *, knock out (for example, OC*, OC gene cluster knockout; YadA*, pYV-cured strain). No symbol was ascribed to LPS factors (O-ag and OC) inactivated by spontaneous mutations and isolated using bacteriophage or enterocoliticin selection.
FIG. 3.
Survival percentages of YadA-negative Y. enterocolitica O:3 strains grown at 37°C in normal and in EGTA-Mg-treated sera after 30 or 120 min of incubation. See the legend to Fig. 2 for further explanations.
The pYV-positive YadA-positive strains.
In the line YeO3→YeO3-Ail→YeO3-Ail-R→YeO3-Ail-OCR (Fig. 2, top row), all the strains expressed YadA as we deprived the wt strain YeO3 first of Ail and then stepwise of O-ag and OC. Resistance to both sera (resistance to EGTA-Mg serum was more prominent) displayed by the wt strain was significantly weakened by removal of Ail (YeO3-Ail) and then of O-ag (YeO3-Ail-R). Interestingly, final loss of OC (YeO3-Ail-OCR) decreased resistance to NHS even more, but it had no effect on resistance to EGTA-Mg serum.
In the two genetically different but phenotypically identical lines YeO3→YeO3-OC→YeO3-OCR and YeO3→YeO3-trs11→YeO3-trs11-R (Fig. 2 and data not shown), we first deprived the wt strain of OC by complete deletion of the OC gene cluster (YeO3-OC) or by Km-GenBlock insertion (YeO3-trs11). YeO3-OC and YeO3-trs11 displayed comparable and higher resistance to both sera than the wt strain. In both mutant lines, further loss of O-ag (YeO3-OCR and YeO3-trs11-R) caused a decrease in resistance to NHS and EGTA-Mg serum after 2 h of incubation.
In line YeO3→YeO3-OC→YeO3-Ail-OC (Fig. 2), YeO3-OC (see above) was further deprived of Ail, which resulted in decreased resistance to both types of sera.
The last pYV-positive YadA-positive line was YeO3-R2 (Fig. 2, bottom). Compared to the wt strain, YeO3-R2, missing only the O-ag, displayed a significant increase in resistance to NHS. Resistance to EGTA-Mg serum presented by both strains was at about the same level, but surprisingly, YeO3-R2 was more resistant to NHS than to EGTA-Mg serum.
The YadA-negative strains.
Two different YadA-negative strains, YeO3-028 (ΔyadA::Km-GenBlock) and YeO3-c (pYV negative), served as starting points for construction of strains missing Ail, O-ag, and OC. Both YeO3-028 and YeO3-c were very rapidly killed in NHS (Fig. 3). They displayed high-level resistance at 0.5 h in EGTA-Mg serum, but at 2 h they were efficiently eliminated.
In both the pYV-positive, YadA-negative line YeO3→YeO3-028→YeO3-028-R→YeO3-028-OCR and the pYV-negative line YeO3→YeO3-c→YeO3-R1 (Fig. 3), loss of O-ag did not alter the resistance of bacteria to EGTA-Mg serum, but it increased the resistance of bacteria to NHS (see YeO3-028-R and YeO3-R1). However, there were pronounced differences between these two Ail- and OC-positive strains, especially in NHS: YeO3-028-R was more sensitive than YeO3-R1. An explanation for this may be the presence of a fully functional type III secretion system (12) in YeO3-028 and its derivatives but not in YeO3-R1. Type III secretion utilizes a syringe-like structure that extends through the bacterial cell wall. One could speculate that it could impair the OM integrity and make it more susceptible to complement components.
The OC-negative strain YeO3-028-OCR, derived from YeO3-028-R and expressing only Ail, displayed increased resistance to complement, especially at 2 h, in both sera (Fig. 3).
In the pYV-positive YadA-negative line YeO3→YeO3-028→YeO3-028-OC (Fig. 3) as well as the pYV-negative lines YeO3→YeO3-c→YeO3-c-OC→YeO3-c-OCR (Fig. 3) and YeO3→YeO3-c→YeO3-c-trs8→YeO3-c-trs8-R (data not shown), three strains sharing the O-ag Ail-positive phenotype were constructed using different genetic approaches to inactivate or delete the yadA gene and the OC gene cluster (Table 1). Irrespective of their genetic history, serum resistance of the strains was similar (compare YeO3-028-OC, YeO3-c-OC, and YeO3-c-trs8); loss of OC did not dramatically change bacterial resistance to the complement. Of note was the slight increase in resistance of YeO3-c-OC and YeO3-c-trs8 (but not that of YeO3-028-OC) after 0.5 h of incubation in both types of sera. The O-ag-negative derivatives YeO3-c-OCR and YeO3-c-trs8-R (Fig. 3 and data not shown) showed significantly increased resistance, especially to NHS, compared to that of their O-ag-positive precursors. Again, as noted above, the effect of removal of O-ag and exposure of Ail resulted in increased serum resistance.
In the pYV-negative lines YeO3→YeO3-c→YeO3-c-Ail→YeO3-c-Ail-OC and YeO3→YeO3-c→YeO3-c-Ail→YeO3-c-Ail-R (Fig. 3), strains expressing only O-ag and/or OC (YeO3-c-Ail, YeO3-c-Ail-OC, and YeO3-c-Ail-R) were highly susceptible to complement killing, as was YeO3-c-Ail-OCR, derived from YeO3-c-Ail-R and missing all four surface factors (data not shown). These results were identical to those obtained with all the strains grown at RT (see above).
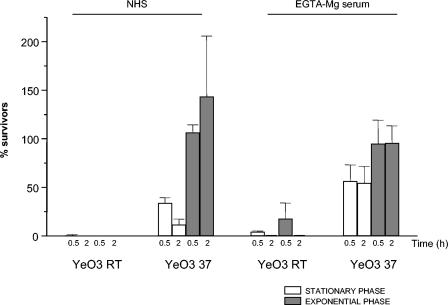
Influence of growth phase on serum resistance.
The growth phase experiments were performed with the wt strain YeO3 and strains expressing only one of the four factors: Ail (YeO3-c-OCR), YadA (YeO3-Ail-OCR), OC (YeO3-c-Ail-R), and O-ag (YeO3-c-OC). The bacteria grown at RT and at 37°C exposed to NHS and EGTA-Mg serum were either in the logarithmic or stationary phase of growth. The results (Fig. 4) clearly showed that wt strain YeO3, when growing exponentially at 37°C (expressing YadA, Ail, O-ag, and OC), was more resistant to NHS and EGTA-Mg serum than when it had reached stationary phase. Interestingly, YeO3 grown at RT (expressing only O-ag and OC) both in the exponential and stationary phase of growth was highly sensitive to NHS and EGTA-Mg serum (Fig. 4). Thus, YadA- and Ail-negative bacteria were rapidly killed irrespective of the growth phase.
FIG. 4.
Effect of growth phase on the survival of the wt strain YeO3 in normal and in EGTA-Mg treated sera. Y. enterocolitica O:3 bacteria, grown at RT and at 37°C to stationary or to exponential phase, were exposed to sera for 0.5 and 2 h.
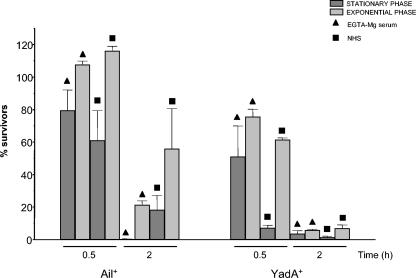
Independent of the phase of growth, strains expressing only OC (YeO3-c-Ail-R grown at 37°C) or O-ag (YeO3-c-OC grown at RT) were highly sensitive to NHS and to EGTA-Mg serum (data not shown). Ail-expressing bacteria (YeO3-c-OCR) at exponential phase were significantly more resistant to serum, especially after 2 h of incubation, than bacteria at the stationary phase of growth (Fig. 5). The same also applied to the YadA-expressing strain YeO3-Ail-OCR (Fig. 5) at 0.5 h of incubation in sera. After 2 h of incubation, differences between YadA-positive bacteria at stationary and exponential phase were very small, and the bacteria were highly sensitive to serum killing.
FIG. 5.
Effect of growth phase on the survival of strains YeO3-c-OCR (Ail positive) and YeO3-Ail-OCR (YadA positive) grown at 37°C and incubated in normal and in EGTA-Mg-treated sera.
Deposition and degradation of complement component C3 on Y. enterocolitica O:3 strains.
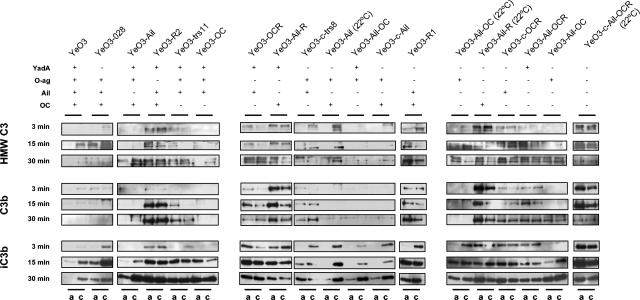
To check C3 deposition and cleavage on Yersinia, we applied the monoclonal antibody, specific for the α-chain of C3 and binding to the C-terminal 40-kDa fragment of iC3b, called MAb755 (48). It detects intact C3, C3b, and iC3b as 118-, 109-, and 40-kDa fragments, respectively (Fig. 6). In addition, C3b molecules covalently bound to Yersinia surfaces are detected as high-molecular-weight (HMW) bands with molecular masses of over 200 kDa. For the deposition experiments, we used Yersinia mutants representing different phenotypes and allowed the bacteria to interact with NHS and EGTA-Mg serum for 3, 15, and 30 min before processing the samples for immunoblotting as described in Materials and Methods. In the interpretation of the C3 deposition data, one needs to remember that some strains were already completely killed at 30 min, thus, significant amounts of C3 originally bound to bacteria could have been washed away with bacterial debris.
FIG. 6.
C3 deposition and degradation on different Y. enterocolitica O:3 mutant bacteria. Bacteria were incubated in EGTA-Mg-treated serum and NHS for 3, 15, and 30 min. After being washed, C3 and its derivatives deposited on bacteria were identified by immunoblotting using monoclonal antibody specific for the α-chain of C3. Shown in frames are the depositions of the C3 derivatives on different types of bacteria at different time points. The bacteria were grown at 37°C unless otherwise indicated. For each bacterial sample, its phenotype with respect to expression of YadA, O-ag, Ail, and OC is given below the strain names. For each bacterial sample, C3 deposition in EGTA-Mg-treated serum (a) and in NHS (c) is indicated at the bottom of the figure.
Deposition in EGTA-Mg-treated serum. (i) HMW C3.
Ten out of 11 O-ag-positive strains used in the experiment did not display HMW C3 binding at 3 min (Fig. 6). Among those strains, there were two single-positive O-ag mutants (YeO3-Ail-OC [at 22°C] and YeO3-Ail-OC), but there were also strains additionally expressing YadA, Ail, or OC in different combinations (Fig. 6). The only O-ag-positive strain that bound some HMW C3 already at 3 min was YeO3-trs11; interestingly, the expression level of O-ag for this strain was lower than that in others (Fig. 1).
Five of the 10 strains still resisted HMW C3 deposition at 15 min or were very efficient in cleaving it (YeO3, YeO3-Ail, YeO3-Ail-OC, YeO3-Ail [at 22°C], and YeO3-c-Ail). Interestingly, three of those strains expressed YadA (YeO3, YeO3-Ail, and YeO3-Ail-OC). The only O-ag-positive Ail-positive strain that did not bind HMW C3 was the wild-type strain expressing all four factors. Ail-positive, O-ag-positive, and YadA- and/or OC-negative strains (YeO3-028, YeO3-trs11, YeO3-OC, and YeO3-c-trs8) were HMW C3 positive at 15 min, as were two single-positive O-ag strains (YeO3-Ail-OC [at 22°C] and YeO3-Ail-OC). The only O-ag-positive YadA-negative strains that did not bind HMW C3 at 15 min were those that expressed OC (YeO3-Ail and YeO3-c-Ail).
Finally, the only strain that did not display HMW C3 on the surface at 30 min was the wild-type strain YeO3.
(ii) C3b.
In general, O-ag-negative strains already deposited C3b on the cell surface by 3 min, while of the 11 O-ag-positive strains, 9 did not (Fig. 6). The O-ag-positive exceptions were YeO3-c-trs8 and YeO3-O28, both being Ail positive and YadA negative. Eight of the above nine O-ag-positive strains did not bind C3b at 15 and 30 min; six of them were Ail negative. Interestingly, the remaining two expressed O-ag and Ail in combination with YadA (YeO3-OC) or OC (YeO3-028). YeO3-trs11, though YadA, O-ag, and Ail positive, had bound some C3b at 15 min and more at 30 min.
(iii) iC3b.
Most of the strains strongly deposited the cleavage product of C3b, iC3b, on the cell surface. The strains that did not show deposition of HMW C3 or C3b also showed no or less deposition of iC3b. After 3 min, from the group of O-ag positive mutants the single-positive strains YeO3-c-Ail-OC and YeO3-Ail-OC (22°C) showed some iC3b deposition that increased with time (Fig. 6). At this time, YeO3-R1 was an exceptional O-ag-negative strain not displaying iC3b. At 15 min, however, only the Ail-negative O-ag-positive strains expressing either YadA or OC (YeO3-Ail [at 22°C], YeO3-Ail-OC, and YeO3-c-Ail) still showed no iC3b deposition; however, some had taken place at 30 min. At 30 min, no iC3b deposition was visible on the wild-type strain (Fig. 6).
Deposition in NHS. (i) HMW C3.
In general, more C3 deposition took place in NHS than in EGTA-Mg-treated serum. Two of the three strains that did not bind HMW C3 at 3 min expressed YadA in combination with Ail (YeO3 and YeO3-OC). YeO3-trs11 was an exception, being the only YadA-, Ail-, O-ag-positive strain that deposited HMW C3. YeO3-c-Ail-OC was the third O-ag-positive strain not depositing HMW C3. The strain with the same phenotype (YeO3-Ail-OC 22°C) showed a little HMW C3 at 3 min. At 15 min, only triple-positive OC mutants (YeO3-trs11 and YeO3-OC) resisted HMW C3 deposition. However, HMW C3 deposition was seen with all the strains at 30 min.
(ii) C3b.
Nine of the 11 O-ag-positive strains showed no C3b deposition at 3 min (Fig. 6). The two O-ag-positive exceptions were the Ail- and OC-expressing strains YeO3 and YeO3-O28. YeO3-c-trs8 started showing some C3b at 15 min. YeO3-trs11 and YeO3-OC, both missing OC, were the only O-ag-, Ail-positive strains that did not bind C3b at 15 min. At 30 min, no C3b binding was observed with the four O-ag-positive, Ail-negative strains missing either YadA, OC, or both (YeO3-Ail [at 22°C], YeO3-Ail-OC, YeO3-c-Ail, and YeO3-Ail-OC [at 22°C]). The other single O-ag-positive strain, YeO3-c-Ail-OC, grown at 37°C and expressing shorter O-ag, bound C3b at 30 min (Fig. 6). No C3b was deposited on YeO3 and O-ag-expressing triple-positive strains (YeO3-028 and YeO3-OC). The exception was again YeO3-trs11, which bound C3b at 30 min.
(iii) iC3b.
In NHS after 3 min of incubation, only the triple-positive, YadA-expressing strains did not display iC3b on the surface (YeO3, YeO3-Ail, and YeO3-OC); again, YeO3-trs11 was an exception, showing some iC3b deposition. At later time points all strains became iC3b positive (Fig. 6).
DISCUSSION
In this study, we have extensively characterized the factors associated with Y. enterocolitica O:3 survival in human serum. Four major conclusions can be reached based on the results of this work.
Firstly, serum resistance in Y. enterocolitica O:3 is multifactorial, and, based on the following observations, it includes YadA, Ail, O-ag, and OC. In stationary-phase bacteria, the most potent single serum-resistant factor turned out to be YadA. YadA-negative strains were efficiently killed by CP/AP and AP, especially if they were additionally deprived of Ail. Ail-positive strains presented the delayed type of killing, being highly resistant at 0.5 h of incubation in EGTA-Mg serum and often efficiently eliminated by 2 h. That was particularly seen with YadA-negative strains in the absence of O-ag, OC, or both, suggesting that these LPS compounds may sterically block Ail in the outer membrane and prevent its interaction with some complement components. Exposed Ail could thus more readily and directly interact with complement components, but the mechanism by which this was achieved is at present unknown. Bliska and Falkow speculated that Ail could subvert the stable formation of active attack complexes by binding a serum factor or complement component (8). Thus, identification of complement components bound by Ail seems to be a prerequisite to revealing the molecular function of this protein.
Secondly, we presume that O-ag may function as a target for some antibodies or lectins, thereby enhancing CP activation. This was based on the observation that YeO3-R2 (O-ag-negative derivative of YeO3) displayed much higher resistance to NHS than the wt strain. Interestingly, we did not observe the same phenomenon, i.e., increased resistance, in the rough derivative of YeO3-Ail (YeO3-AilR; Fig. 2). Also, there was no increase in resistance when O-ag was removed from YeO3-trs11 (YeO3-trs11-R; data not shown) or YeO3-OC (YeO3-OCR; Fig. 2). If anything, in these strains there was a slight decrease in bacterial resistance. These results might suggest that when the three factors YadA, Ail, and OC are expressed, loss of O-ag is beneficial for bacterial survival in NHS. As for EGTA-Mg serum, we found that O-ag may be involved in inhibition of AP activation mostly when it is expressed in conjunction with YadA and/or Ail. Nevertheless, O-ag alone was not potent enough to protect bacteria against AP killing. Our results differ from those obtained earlier by Wachter and Brade (46). They reported that the presence or absence of pYV did not influence the resistance phenotype and that it is O-ag that plays a regulatory role in AP activation (46). These differences may be explained by the fact that sera from different species were used in the studies. The sera differ, for instance, in their hemolytic activity; guinea pig erythrocytes are very susceptible to lysis by human serum. Human erythrocytes are, however, resistant to guinea pig serum action (15). Factor H and factor I from these two species are not compatible, either. Guinea pig factor H and factor I are not able to cleave human C3, but guinea pig C3 is a good substrate for human factor H and factor I. Moreover, the factor I-dependent C3b regulatory system is also species specific (36). It has also been shown that human complement regulatory protein CD59 inhibits the cytolytic activity of C5b-9 if C8 and C9 derive from humans or primates. It cannot, however, recognize C8 and C9 originating from the guinea pig (35).
Thirdly, we observed that wt Y. enterocolitica O:3 bacteria were much more resistant to serum at the exponential than at the stationary phase of growth. No killing of exponentially growing bacteria took place for 2 h, while 50 to 80% of stationary-phase bacteria were killed at the same time (Fig. 4). Furthermore, we noticed that Ail alone was the most potent serum resistance factor in bacteria at exponential phase (Fig. 5).
Fourthly, the C3 deposition experiments gave intriguing results. C3 deposition and degradation patterns did not directly correlate with killing sensitivity or resistance. Generally, we observed that O-ag protected bacteria against C3 deposition. In EGTA-Mg serum among triple-, double-, and single-positive strains, only those expressing O-ag prevented early (3 min) C3 deposition. However, for prolonged protection against C3b deposition, bacteria clearly needed all the other factors. On the other hand, O-ag-negative strains allowed early C3 deposition, and in fact, among these strains, those expressing YadA, Ail, or both seemed to accumulate C3b on their surface.
Binding of the complement regulator, factor H, is exploited by many microbes (3, 6, 16, 19, 20, 32, 41) to block the complement cascade at the C3b level. Factor H not only promotes disassociation of C3 convertase and prevents its assembly, but it also acts as a cofactor for factor I in C3b degradation. Thus, not surprisingly, binding of factor H by Y. enterocolitica O:3 has long been of interest. Affinity blot results revealed YadA as a potential candidate for factor H binding (11, 34). However, further attempts to show direct interaction between YadA and purified factor H have failed (33 and M. Biedzka-Sarek et al., unpublished data). The inconclusive and enigmatic factor H binding still needs thorough investigation.
Our results show that the presence of Ail and O-ag only seems to be insufficient to protect against complement-mediated killing in vitro, but combinations of those factors with YadA strengthen resistance to CP/AP-mediated killing. Thus, the most important serum resistance factor seems to be YadA, which apparently can prevent CP/AP-mediated killing. Relevant to this is the complete sensitivity of the yadA mutant YeO3-028 to CP/AP-mediated killing. According to resistance to CP/AP-mediated killing, the most resistant 37°C-grown Y. enterocolitica strain is YeO3-R2. The most virulent strain is, however, YeO3, while YeO3-R2 is about 50-fold less virulent (2). The explanation for this discrepancy may lie in the important role of O-ag in the resistance of bacteria to host defense mechanisms other than complement, for example, the acidity of the stomach or early colonization of Peyer's patches (39).
MBL (mannose binding lectin) is a key component of the lectin pathway. It binds microbial surface carbohydrates, which results in opsonophagocytosis and/or microbial lysis due to insertion of MAC. In this study, we aimed to compare the extent of Y. enterocolitica O:3 killing by whole human serum (with classical, lectin, and alternative pathways active) and by the alternative pathway only (EGTA-Mg-treated serum). The experimental setup did not allow us to distinguish the activities of the classical and lectin pathways. It has been shown, however, that Y. enterocolitica is able to activate MBL (21). Further research is needed to examine whether the lectin pathway is involved in first-line defense against Y. enterocolitica O:3.
As observed earlier (28), any degree of resistance of Y. enterocolitica O:3 to CP/AP-mediated killing is always dependent on growth at 37°C. One explanation is that bacteria express certain factors at appropriate phases of infection when and where they are most needed. Another explanation is that these in vitro observations may, in fact, be artifacts. The real in vivo situation may be quite different, i.e., the temperature regulation observed in vitro does not take place in vivo. Thus, it seems that Y. enterocolitica O:3, indeed, can express its (virulence) factors in appropriate amounts at appropriate phases of infection. Further work will be needed to clarify the interplay between the various virulence factors in vivo.
Acknowledgments
This work was supported by grants from the Academy of Finland (project numbers 50441, 45820, and 201358).
Editor: J. B. Bliska
REFERENCES
- 1.Al-Hendy, A., P. Toivanen, and M. Skurnik. 1991. Expression cloning of Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb. Pathog. 10:47-59. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hendy, A., P. Toivanen, and M. Skurnik. 1992. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect. Immun. 60:870-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alitalo, A., T. Meri, T. Chen, H. Lankinen, Z. Z. Cheng, T. S. Jokiranta, I. J. Seppala, P. Lahdenne, P. S. Hefty, D. R. Akins, and S. Meri. 2004. Lysine-dependent multipoint binding of the Borrelia burgdorferi virulence factor outer surface protein E to the C terminus of factor H. J. Immunol. 172:6195-6201. [DOI] [PubMed] [Google Scholar]
- 4.Appleyard, R. K. 1954. Segregation of new lysogenic types during growth of doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, O. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 6.Blackmore, T. K., V. A. Fischetti, T. A. Sadlon, H. M. Ward, and D. L. Gordon. 1998. M protein of the group A Streptococcus binds to the seventh short consensus repeat of human complement factor H. Infect. Immun. 66:1427-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bliska, J. B., J. C. Clemens, J. E. Dixon, and S. Falkow. 1992. The Yersinia tyrosine phosphatase-specificity of a bacterial virulence determinant for phosphoproteins in the J774A.1 macrophage. J. Exp. Med. 176:1625-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bliska, J. B., and S. Falkow. 1992. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc. Natl. Acad. Sci. USA 89:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, H. W. Boyer, J. H. Crosa, and S. Falkow. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 10.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404-407. [DOI] [PubMed] [Google Scholar]
- 11.China, B., M. P. Sory, B. T. Nguyen, M. Debruyere, and G. R. Cornelis. 1993. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect. Immun. 61:3129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis, G. R. 2002. The Yersinia Ysc-Yop virulence apparatus. Int. J. Med. Microbiol. 291:455-462. [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 14.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansch, G. M., C. H. Hammer, P. Vanguri, and M. L. Shin. 1981. Homologous species restriction in lysis of erythrocytes by terminal complement proteins. Proc. Natl. Acad. Sci. USA 78:5118-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. Seppala, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 17.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joiner, K., E. Brown, C. Hammer, K. Warren, and M. Frank. 1983. Studies on the mechanism of bacterial resistance to complement-mediated killing. III. C5b-9 deposits stably on rough and type 7 S. pneumoniae without causing bacterial killing. J. Immunol. 130:845-849. [PubMed] [Google Scholar]
- 19.Kotarsky, H., J. Hellwage, E. Johnsson, C. Skerka, H. G. Svensson, G. Lindahl, U. Sjobring, and P. F. Zipfel. 1998. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J. Immunol. 160:3349-3354. [PubMed] [Google Scholar]
- 20.Kraiczy, P., C. Skerka, M. Kirschfink, V. Brade, and P. F. Zipfel. 2001. Immune evasion of Borrelia burgdorferi by acquisition of human complement regulators FHL-1/reconectin and factor H. European J. Immunol. 31:1674-1684. [DOI] [PubMed] [Google Scholar]
- 21.Kuipers, S., P. C. Aerts, and H. van Dijk. 2003. Differential microorganism-induced mannose-binding lectin activation. FEMS Immunol. Med. Microbiol. 36:33-39. [DOI] [PubMed] [Google Scholar]
- 22.Marques, M. B., D. L. Kasper, M. K. Pangburn, and M. R. Wessels. 1992. Prevention of C3 deposition by capsular polysaccharide is a virulence mechanism of type III group B streptococci. Infect. Immun. 60:3986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pajunen, M. I., S. J. Kiljunen, M. E. Söderholm, and M. Skurnik. 2001. Complete genomic sequence of the lytic bacteriophage phiYeO3-12 of Yersinia enterocolitica serotype O:3. J. Bacteriol. 183:1928-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pekkola-Heino, K., M. K. Viljanen, T. H. Ståhlberg, K. Granfors, and A. Toivanen. 1987. Monoclonal antibodies reacting selectively with core and O-polysaccharide of Yersinia enterocolitica O:3 lipopolysaccharide. Acta Pathol. Microbiol. Immunol. Scand. 95:27-34. [DOI] [PubMed] [Google Scholar]
- 28.Perry, R. D., and R. R. Brubaker. 1983. Vwa+ phenotype of Yersinia enterocolitica. Infect. Immun. 40:166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierson, D. E., and S. Falkow. 1993. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect. Immun. 61:1846-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilz, D., T. Vocke, J. Heesemann, and V. Brade. 1992. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect. Immun. 60:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pramoonjago, P., M. Kaneko, T. Kinoshita, E. Ohtsubo, J. Takeda, K. Hong, R. Inagi, and K. Inoue. 1992. Role of TraT protein, an anticomplementary protein produced in Escherichia coli by R100 factor, in serum resistance. J. Immunol. 148:827-836. [PubMed] [Google Scholar]
- 32.Ram, S., A. K. Sharma, S. D. Simpson, S. Gulati, D. P. McQuillen, M. K. Pangburn, and P. A. Rice. 1998. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 187:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roggenkamp, A., K. Ruckdeschel, L. Leitritz, R. Schmitt, and J. Heesemann. 1996. Deletion of amino acids 29 to 81 in adhesion protein YadA of Yersinia enterocolitica serotype O:8 results in selective abrogation of adherence to neutrophils. Infect. Immun. 64:2506-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rollins, S. A., J. Zhao, H. Ninomiya, and P. J. Sims. 1991. Inhibition of homologous complement by CD59 is mediated by a species-selective recognition conferred through binding to C8 within C5b-8 or C9 within C5b-9. J. Immunol. 146:2345-2351. [PubMed] [Google Scholar]
- 36.Seya, T., M. Okada, K. Hazeki, and S. Nagasawa. 1991. Regulatory system of guinea-pig complement C3b: tests for compatibility of guinea-pig factors H and I with human factors. Mol. Immunol. 28:375-382. [DOI] [PubMed] [Google Scholar]
- 37.Skurnik, M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56:355-363. [DOI] [PubMed] [Google Scholar]
- 38.Skurnik, M., Y. El Tahir, M. Saarinen, S. Jalkanen, and P. Toivanen. 1994. YadA mediates specific binding of enteropathogenic Yersinia enterocolitica to human intestinal submucosa. Infect. Immun. 62:1252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skurnik, M., R. Venho, J.-A. Bengoechea, and I. Moriyón. 1999. The lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3 is required for virulence and plays a role in outer membrane integrity. Mol. Microbiol. 31:1443-1462. [DOI] [PubMed] [Google Scholar]
- 40.Skurnik, M., R. Venho, P. Toivanen, and A. Al-Hendy. 1995. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol. Microbiol. 17:575-594. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strauch, E., H. Kaspar, C. Schaudinn, C. Damasko, A. Konietzny, P. Dersch, M. Skurnik, and B. Appel. 2003. Analysis of enterocoliticin, a phage tail-like bacteriocin. Adv. Exp. Med. Biol. 529:249-251. [DOI] [PubMed] [Google Scholar]
- 43.Strauch, E., H. Kaspar, C. Schaudinn, P. Dersch, K. Madela, C. Gewinner, S. Hertwig, J. Wecke, and B. Appel. 2001. Characterization of enterocoliticin, a phage tail-like bacteriocin, and its effect on pathogenic Yersinia enterocolitica strains. Appl. Environ. Microbiol. 67:5634-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tertti, R., E. Eerola, O.-P. Lehtonen, T. Ståhlberg, M. Viander, and A. Toivanen. 1987. Virulence-plasmid is associated with the inhibition of opsonization in Yersinia enterocolitica and Yersinia pseudotuberculosis. Clin. Exp. Immunol. 68:266-274. [PMC free article] [PubMed] [Google Scholar]
- 45.Tomas, J. M., V. J. Benedi, B. Ciurana, and J. Jofre. 1986. Role of capsule and O antigen in resistance of Klebsiella pneumoniae to serum bactericidal activity. Infect. Immun. 54:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wachter, E., and V. Brade. 1989. Influence of surface modulations by enzymes and monoclonal antibodies on alternative complement pathway activation by Yersinia enterocolitica. Infect. Immun. 57:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vishwanath, S., and T. Hackstadt. 1988. Lipopolysaccharide phase variation determines the complement-mediated serum susceptibility of Coxiella burnetii. Infect. Immun. 56:40-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel, U., A. Weinberger, R. Frank, A. Muller, J. Kohl, J. P. Atkinson, and M. Frosch. 1997. Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis. Infect. Immun. 65:4022-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]