Abstract
The Haemophilus ducreyi outer membrane component DsrA (for ducreyi serum resistance A) is necessary for complete resistance to normal human serum (NHS). When DsrA expression in 19 temporally and geographically diverse clinical isolates of H. ducreyi was examined by Western blotting, 5 of the strains expressed a different immunotype of the DsrA protein (DsrAII) than the well-characterized prototypical strain 35000HP (DsrAI). The predicted DsrA proteins expressed by the DsrAII strains were 100% identical to each other but only 48% identical to that of strain 35000HP. In addition to the DsrAII protein, class II strains also expressed variant forms of other outer membrane proteins (OMPs) including NcaA (necessary for collagen adhesion A), DltA (ducreyi lectin A), Hlp (H. ducreyi lipoprotein), major OMP, and/or OmpA2 (for OMP A2) and synthesized a distinct, faster-migrating lipooligosaccharide. Based on these data, strains expressing DsrAI were termed class I, and those expressing DsrAII were termed class II. Expression of dsrAII from strain CIP 542 ATCC in the class I dsrAI mutant FX517 (35000HP background), which does not express a DsrA protein, rendered this strain resistant to 50% NHS. This demonstrates that DsrAII protein is also critical to serum resistance. Taken together, these results indicate that there are two clonal populations of H. ducreyi. The implications of two classes of H. ducreyi strains differing in important antigenic outer membrane components are discussed.
Haemophilus ducreyi is a gram-negative, strict human pathogen and the etiologic agent of the sexually transmitted genital ulcer disease chancroid. Chancroid occurs more commonly in certain areas of Africa, Asia, and Latin America (8, 55, 69) than in the United States (17, 36). Regardless of the socioeconomic conditions where chancroid occurs, underreporting and misdiagnosis make accurate predictions of the prevalence of chancroid difficult (36). The focus on chancroid has intensified because it increases the risk for transmission and acquisition of the human immunodeficiency virus (29, 32, 55). Control of chancroid may result in a decrease in the spread of human immunodeficiency virus.
Chancroid is thought to initiate upon the entry of H. ducreyi into the skin through small abrasions that occur during sexual intercourse (37). Small tender papules form at the site of entry within 2 to 7 days of acquisition. The papules evolve into pustules, that rupture within 2 to 3 days to form soft painful ulcers. The ulcers persist for several weeks to months but may ultimately resolve (37).
The study of chancroid pathogenesis has lead to the identification of a number of H. ducreyi antigens that may be important for the production of disease (4, 11, 12, 18, 19, 25, 31, 33, 35, 39, 45, 50, 54, 65, 67, 71, 72). Thus far, few cell surface determinants, including full-length lipooligosaccharide (LOS), have been shown to be essential for H. ducreyi infection in the human model of chancroid (5, 9, 22, 28, 56). Included among these virulence factors is the protein termed DsrA (for ducreyi serum resistance A), which has been shown to be responsible for serum resistance (19), keratinocyte cell adhesion (14), and binding to the extracellular matrix protein (ECM) vitronectin (14).
DsrA (19) is a member of the Oca (for oligomeric coiled adhesin) family, a group of surface-exposed multifunctional proteins involved in binding to cells and to the ECM and resistance to killing by serum complement (27, 48). This family of proteins includes the Yersinia adhesin YadA (47, 53, 62) found in pathogenic Yersinia species; the ubiquitous surface proteins of Moraxella catarrhalis, UspA1 and UspA2 (1, 2); the NadA proteins of meningococci (16); and the Eib proteins of Escherichia coli (48, 49). Very recently, Cole et al. (15) described a second H. ducreyi Oca family member termed NcaA (for necessary for collagen adhesion A). Hoiczyk et al. (27) proposed that YadA and UspA are capable of forming highly structured oligomers, resulting in so-called lollipop-shaped structures on the cell surface. These structures are thought to be composed of an N-terminal head domain, an intermediate central stalk domain, and a conserved C-terminal anchoring domain. The C-terminal domain of YadA has been shown to be sufficient to grant serum resistance (46), whereas the N-terminal domain confers binding to ECMs such as fibronectin (63) and collagen (CN) (21, 52).
In the course of immunoblot studies with anti-DsrA antibodies, we discovered that a monoclonal antibody (MAb) to 35000HP DsrA failed to bind H. ducreyi strains CIP 542 ATCC and HMC112, even though a polyclonal antiserum made to the 35000HP DsrA antigen was reactive to these strains. We surmised that there could be important antigenic differences between the DsrA proteins from strains CIP 542 ATCC, HMC112, and 35000HP. The objectives of this study were therefore to determine if the DsrA proteins from these strains were different at the nucleotide and amino acid levels, if both types of DsrA possessed the same function in H. ducreyi, and if these strains also expressed different types of other outer membrane determinants.
MATERIALS AND METHODS
Strains and media.
Bacterial strains and plasmids used in this study are listed in Tables 1 and 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αMCR | recA gyrB | Bethesda Research Laboratories |
| BL21(DE3)pLysS | Expression of hexahistidine tag fusion proteins; Cmr | Invitrogen; 61 |
| H. ducreyi | ||
| 35000HP | Wild type, parent, class I | Stanley Spinola, Indiana University |
| CIP 542 ATCC | Wild type, parent, class II | American Type Culture Collection |
| HMC112 | Wild type, parent, class II | P. Totten, University of Washington |
| Plasmids | ||
| pCRT7/CT-TOPO | Expression vector; Ampr | Invitrogen |
| pCRT2.1TOPO/TA | Cloning vector; Ampr | Invitrogen |
| pLSKS | Shuttle vector; replicates in H. ducreyi and E. coli; Smr Cmr | 73 |
| pRSM1791 | Mutagenesis vector; β-Galactosidase+ Ampr | 10 |
| pNC40 | Source of CAT cassette; Ampr Cmr | 64 |
| pUC4K | Source of APT cassette; Kmr Ampr | Pharmacia |
| pUNCH 1250 | dsrAI-pET30a plasmid (35000HP); Ampr | 14 |
| pUNCH 1260 | dsrAI-pLSKS plasmid (35000HP); Cmr Smr | 19 |
| pUNCH 1272 | ncaAI-pCR2.1TOPO/TA, (35000HP) Ampr | This study |
| pUNCH 1274 | ncaAI::CAT-pCR2.1 TOPO/TA plasmid (35000HP); Cmr | This study |
| pUNCH 1275 | ncaAI::CAT-pRSM1791 (35000HP); Cmr | This study |
| pUNCH 1288 | dsrAII-pLSKS plasmid (CIP 542 ATCC); Cmr Smr | This study |
| pUNCH 1291 | dsrAII::CAT-pLSKS plasmid (CIP 542 ATCC); Cmr Smr | This study |
| pUNCH 1292 | dsrAII::CAT-pRS M1791 plasmid (CIP 542 ATCC); Cmr | This study |
| pUNCH 1293 | dsrAI-pCRT7/CT-TOPO plasmid (35000HP); Ampr | This study |
| pUNCH 1294 | dsrAII-pCRT7/CT-TOPO plasmid (CIP 542 ATCC); Ampr | This study |
| pUNCH 1295 | ncaAI-pCRT7/CT-TOPO plasmid (35000HP); Ampr | This study |
| pUNCH 1296 | dsrAII-pLSKS plasmid (CIP 542 ATCC); Cmr Smr | This study |
| pUNCH 1297 | ncaA::KAN-pCR2.1TOPO/TA plasmid; Kmr | This study |
| pUNCH 1298 | ncaA::KAN-pRSM1791 plasmid; Kmr | This study |
CAT, chloramphenicol acetyltransferase; APT, aminoglycoside 3′-phosphotransferase.
TABLE 2.
Panel of H. ducreyi strains used in experiments shown in Fig. 4
| Strain | Other name | Geographic site of isolation | Yr of isolation | Source (reference) |
|---|---|---|---|---|
| 35000HP | Winnipeg | 1975 | S. Spinola (58) | |
| HMC46 | 1023 | Kenya | 1995 | P. Totten |
| HMC48 | M90-02 | Bahamas | 1995 | P. Totten (73) |
| HMC50 | 425 | Jackson, Miss. | 1995 | P. Totten (73) |
| HMC54 | 010-2 | Dominican Republic | 1995 | P. Totten (73) |
| HMC56 | UG030 | Dominican Republic | 1995 | P. Totten (73) |
| HMC60 | HD174 | Florida | 1989 | P. Totten (73) |
| HMC62 | HD301 | Thailand | 1984 | P. Totten (73) |
| HMC64 | HD350 | Kenya | 1984 | P. Totten (73) |
| HMC88 | H1-95 | Seattle | 1995 | P. Totten (73) |
| DMC188 | Bangladesh | J. Boegarts (this study) | ||
| SSMC57 | Bangladesh | J. Boegarts (this study) | ||
| 26V | Atlanta | W. Albritton (18) | ||
| C111 | Nairobi | W. Albritton (18) | ||
| CIP 542 ATCC | Hanoi, Vietnam | 1954 | American Type Culture Collection | |
| DMC64 | Bangladesh | J. Boegarts, this study | ||
| DMC111 | Bangladesh | J. Boegarts, this study | ||
| SSMC71 | Bangladesh | J. Boegarts, this study | ||
| HMC112 | HD342 | CDC | 1984 | P. Totten |
H. ducreyi strains were maintained on chocolate agar plates containing 1× GGC (0.1% glucose, 0.01% glutamine, and 0.026% cysteine) (68) and 5% fetal bovine serum (Sigma, St. Louis, Mo.) at 34.5°C in 5% CO2. E. coli was grown in Luria-Bertani broth or on Luria-Bertani agar plates at 37°C. When appropriate, streptomycin (100 μg/ml), chloramphenicol (1 μg/ml for H. ducreyi and 30 μg/ml for E. coli), kanamycin (30 μg/ml), or ampicillin (100 μg/ml) was incorporated into the media.
Serum susceptibility.
H. ducreyi resistance to 50% normal human serum (NHS) was determined as previously described (13, 19, 33, 41) except that H. ducreyi cultures were grown on chocolate agar plates containing 5% fetal bovine serum. Data are expressed as percent survival in fresh NHS compared to survival in heated NHS (ΔNHS) [(CFU in fresh NHS/CFU in ΔNHS) × 100].
Western blotting.
H. ducreyi total proteins from approximately 0.5 × 107 CFU were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions, transferred to a nitrocellulose membrane, and subjected to Western blotting as previously described (33).
Dot blot.
H. ducreyi cells were suspended in phosphate-buffered saline to an optical density at 600 nm of 0.2 and suctioned onto a nitrocellulose membrane with a dot blot apparatus (Schleicher & Schuell, Keene, N.H.). The membrane was removed from the dot blot apparatus, allowed to dry at room temperature, and blocked for 30 min in 5% skim milk. The membrane was reassembled into the dot blot apparatus, and the following primary antibodies were added to the wells at the given concentrations (in parentheses): anti-recombinant N-terminal DsrAI (anti-rNT-DsrAI) (1:10), anti-rNT-DsrAII (1:200), and anti-outer membrane proteins (anti-OMPs) (1:200) (24). The membrane was incubated at room temperature for 1 h with gentle shaking. Wells were washed four times (high-salt wash buffer of 20 mM Tris and 500 mM NaCl) prior to incubation with alkaline phosphatase-conjugated protein G (1:200) for 45 min with gentle shaking. Finally, the membrane was washed twice, removed from the dot blot apparatus, and washed a third time before addition of the chemiluminescence substrate for development (Lumi-PhosWB; Pierce, Rockford, Ill.).
Analysis of LOS.
Approximately 4 × 108 CFU (optical density at 600 nm, 0.4) were suspended in 200 μl of Laemmli sample buffer and incubated with proteinase K (final concentration, 50 μg/ml) for 1 h at 56°C (26). The preparation was boiled for 1 min and stored at 4°C. Approximately 5 μl of this preparation was subjected to SDS-PAGE and silver stained according to the method of Tsai and Frasch (70).
Cloning, sequencing, and mutagenesis of dsrAII from strain CIP 542 ATCC.
To mutagenize dsrAII from strains CIP 542 ATCC and HMC112, a class II specific mutagenesis plasmid, pUNCH 1292, was constructed. Primers that amplified class I dsrA did not work for class II strains. It was assumed that genes flanking dsrA were conserved between classes, and the genome sequence of strain 35000HP (www.stdgen.lanl.gov) to design primers flanking the dsrA open reading frame (ORF) was used. Using primer 33 located within the upstream manX (HD0768) gene and primer 24 (Fig. 1 and Table 4) at 10 pmol each, we amplified approximately 2.5 kb of DNA from several colonies of strain HMC112 and CIP 542 ATCC. We used Ready to Go PCR beads (Amersham Biosciences, Piscataway, N.J.) according to the manufacturer's instructions. The PCR conditions were a single denaturation at 95°C for 1 min and 30 cycles, each consisting of 1-min denaturation at 95°C, annealing at 52°C for 1 min, and extension at 72°C for 2 min. Following amplification, the product was digested with XbaI and HindIII overnight and ligated to XbaI- and HindIII-digested pLSKS vector to construct pUNCH 1288. Plasmid pUNCH 1288 was digested with StuI and ligated to the BglII- and Klenow-treated chloramphenicol acetyltransferase cassette from plasmid pNC40 to form pUNCH 1291. The mutagenized dsrAII insert was removed by digesting pUNCH 1291 with SmaI and XhoI prior to Klenow treatment. The insert was then ligated to NotI- and Klenow-treated mutagenesis vector pRSM1791 to form pUNCH 1292. pUNCH 1292 was then electroporated into CIP 542 ATCC and HMC112. However, we were unable to recover Cm-resistant putative cointegrates in either strain CIP 542 ATCC or HMC112 by this method on several occasions. Therefore, to study DsrAII function, we cloned and expressed dsrAII in trans in the dsrAI mutant FX517 (19).
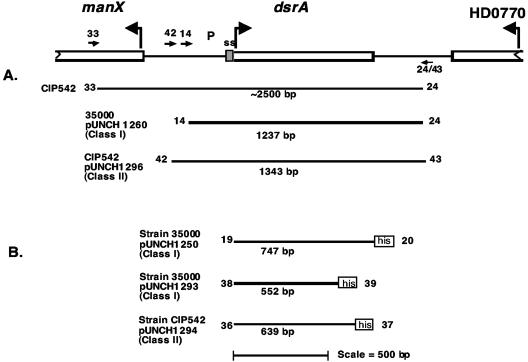
FIG. 1.
Diagram of class I and class II dsrA genes and PCR products. The dsrA ORF is boxed. The partial manX ORF is represented by an open box with a jagged line at the upstream end; the partial HD0770 ORF is represented by an open box with a jagged line at the downstream end. The numbered arrows indicate the direction and position of the dsrA oligonucleotides used for PCR. The proposed signal sequence (ss) is indicated by a shaded box. Curved arrows indicate the polarity of each gene. (A) The PCR products used to construct plasmids pUNCH 1260 and pUNCH 1296 are indicated. (B) The dsrA PCR products are represented by a solid black line followed by a hexahistidine fusion tag depicted by an open box. The PCR-His fusion products were used to produce expression plasmids pUNCH 1250, pUNCH 1293, and pUNCH 1294. Strains and vectors used for PCR are specified to the left of the PCR product. his, hexahistidine, composed of 30 amino acids; P, promoter.
TABLE 4.
List of primers used in this study
| Name | Class | Sequencea | Reference or source |
|---|---|---|---|
| dsrA 14 | I | GACAGCATTCAGTGAATAATGGC | 19 |
| dsrA 19 | I | ATTAATGCAGCAGCCGCCAAAGTTTGCTGG | This study |
| dsrA 20 | I | GCGGCCGCGAATTCATACCCAACAGAACCACC | This study |
| dsrA 24 | I and II | AATGAAGTCCGCACCTTTAACGGC | 19 |
| dsrA 33 | II | CATCGTCGAACGCACACTG | This study |
| dsrA 36 | II | ATGCAGATGCAACCGCAAAATTTTG | This study |
| dsrA 37 | II | CGTGTTTTTAAAGATGTCAGC | This study |
| dsrA 38 | I | ATGCAGCAGCCGCCAAAGTTTGC | This study |
| dsrA 39 | I | TGTATTTTGTTCCATCATACG | This study |
| dsrA 42 | II | ctaggtctagaTGCCTTGCTCTTAATGACG | This study |
| dsrA 43 | II | gcccaagcttTAAAAGCACATAAACAAGCG | This study |
| ncaA 0.01 | I | GAATTATTTTAAGCCAATTTTTTTGC | This study |
| ncaA 0.02 | I | CCACATCCGAAACATATATTAAAGTTAATT | This study |
| ncaA 0.05 | I | TTATTGAAAATTATATACAAAGCCTACACC | This study |
| ncaA 0.08 | I | ATGAAATCAGATACGCTGAGTAAGG | This study |
| ncaA 0.09 | I | ATTTGCAGTAGTTGCTCCGCGTCC | This study |
| ncaA 0.10 | II | GGTTGATTATGTCGAATAATTTG | This study |
| ncaA 0.11 | II | CTAAGCGCGTAAAAATTCGATG | This study |
| DLTA.01 | I | CGCTTGTACAAGCGGGC | 33 |
| DLTA.02 | I | CAGCTTACAAAATGATGGGC | 33 |
| hgbA-F | II | AGCGATTACTCTCTGTATTTTGGGG | This study |
| hgbA-R | II | GCAGATATTGCTGCATCATCGGAG | This study |
| hgbA 2 | I | TTAATGCAGGGCCTGCGTTAAAGC | This study |
| hgbA 3 | I | TTAGCCGTACACACCCAAC | This study |
| hgbA 4 | I | CCTACTTTGCTACTCATTCTGCC | This study |
| hgbA 5 | I | CTAACCCTTCTGGGCTATAC | This study |
| hgbA 8 | I | CGCGCCTGTTTTAGTTTTAACTGG | This study |
| hgbA 9 | I | GCAGTGGTGGGCAAATATC | This study |
| hgbA 10 | I | GCTAGGTAAATACACACGGC | This study |
| hgbA 12 | I | CGTCCTTTGAGTTAAAGTGG | This study |
| hgbA 13 | I | TCAACCTCAAACGCATGTGGTTGGG | This study |
| hgbA 17 | I | GCATCCTTCGTTCTCGATTT | This study |
| hgbA 18 | I | GTACGGCTAACAAATCAAAA | This study |
| hgbA 21 | I | CATCCGTTCTATCCGCCCG | This study |
| 3fl2-3 | I | CCAATTATCTCCTATTGC | Fig. 2 in reference 31 |
| OmpA2-9 | I | GCGATAAATCACTCTTAACCCGAC | Fig. 2 in reference 31 |
Lowercase letters indicate a non-H. ducreyi sequence. Underlined sequences are XbaI and HindIII sites, respectively.
To clone dsrAII, CIP 542 ATCC whole cells were used as a template and subjected to PCR as described above. dsrAII-specific primers, dsrA 42 and dsrA 43 (Table 4), were constructed from the sequence of dsrAII in pUNCH 1288. Following amplification, the approximately 1.3-kb product was restricted with XbaI and HindIII overnight. The dsrAII PCR product was ligated to XbaI- and HindIII-digested pLSKS vector to produce pUNCH 1296. This clone is equivalent to the pUNCH 1260 dsrAI plasmid in the amount of flanking sequences (Fig. 1). With the dsrAII PCR product as template, the dsrAII ORF and promoter region were completely sequenced on both strands with strain-specific primers dsrA 42 and dsrA 43. The ligation product was then electroporated into FX517. Restriction analysis and Western blotting with anti-rNt-DsrAII sera confirmed the transformants (data not shown). Since DsrA class I and class II strains share similar C termini, transformants expressing DsrAII were also recognized by polyclonal antirecombinant full-length DsrAI (rFL-DsrAI) antisera (see below).
Identification of ncaAII.
Primers ncaA 0.08 and ncaA 0.09, which had been used to successfully amplify ncaAI from strain 35000HP, failed to amplify a product from class II H. ducreyi strains. Since ncaAI in strain 35000HP is located between recG, which encodes an ATP-dependent DNA helicase, and proS, which encodes a prolyl-tRNA synthetase (www.stdgen.lanl.gov), we presumed that these flanking genes would be conserved in the dsrA class II strains. Therefore, we designed primers ncaA 0.10 and ncaA 0.11 (Table 4), located 750 bp upstream of the ncaA 0.08 primer site and 978 bp downstream of the ncaA 0.09 primer site, respectively. PCR conditions were the same as described for dsrAII, with the exception of a final magnesium concentration of 2.5 mM, 20 pmol of each primer, and an annealing temperature of 50°C. A 2.2-kb PCR product was amplified from all class II strains and sequenced.
PCR and sequencing of class I and class II hemoglobin receptor A genes (hgbA genes).
To PCR amplify hgbA from H. ducreyi strains, whole cells were used as a template and subjected to PCR as described for dsrAII with the following exceptions: 30 cycles, each consisting of 1-min denaturation at 95°C, annealing at 58°C for 1 min, and extension at 72°C for 3.5 min; and a final cycle of 95°C for 1 min, 58°C for 1 min, and 72°C for 5 min. A 3.3-kb product was predicted with primers hgbA-F and hgbA-R (Table 4), and a product of approximately 3.3 kb was obtained. The hgbA ORF was completely sequenced on both strands with the PCR product as a template and primers hgbA-F and hgbA-R (Table 4).
Cloning of rNt DsrAI, DsrAII, and NcaAI.
To generate class-specific antisera to the mature N-terminal two-thirds of recombinant DsrAI and DsrAII, expression plasmids were constructed by cloning the N-terminal variable regions of the dsrA gene of strains 35000HP and CIP 542 ATCC in a vector containing a C-terminal hexahistidine tag (Fig. 1). Primers dsrA 38 and dsrA 39 were used for strain 35000HP, and dsrA 36 and dsrA 37 were used for CIP 542 ATCC (Fig. 1; Table 4). Conditions for PCR were the same as those described for dsrAII. Products of approximately 0.5 and 0.6 kb were generated for strains 35000HP and CIP 542 ATCC, respectively. Products were ligated to pCRT7/CT-TOPO vector (Invitrogen, Carlsbad, Calif.) and transformed into E. coli DH5α MCR. Plasmid DNA was isolated from the pool of DH5α transformants and electroporated into E. coli BL21(DE3)pLysS. Transformants were screened by Western blotting with anti-rFL-DsrAI (14), following induction with isopropyl-β-d-thiogalactoside (IPTG). Positive clones expressed an approximately 21-kDa protein (expressed by dsrAI from 35000HP) or an approximately 24 kDa protein (expressed by dsrAII from strain CIP 542 ATCC). A single transformant from positive 35000HP and CIP 542 ATCC clones was chosen for each and termed pUNCH 1293 (35000HP) and pUNCH 1294 (CIP 542 ATCC), respectively.
The specific N-terminal sequence encoding NcaAI (Fig. 5) was PCR amplified from strain 35000HP with primers ncaA 0.08 and ncaA 0.09 (Table 4) as described above. The PCR fragment was ligated to the pCRT7/CT-TOPO vector, clones were identified as described above, and a single one was termed pUNCH 1295.
FIG. 5.
Alignment of NcaAI and NcaAII amino acid sequences. The amino acid sequences of the NcaA proteins from 35000HP and CIP 542 ATCC are compared. Boxed, shaded residues indicate identity. The predicted signal peptidase I cleavage site for 35000HP and CIP 542 ATCC is indicated by the inverted triangle. The segment encompassed by the two arrows represents the N-terminal region of recombinant NcaAI from 35000HP that was used for antibody production.
Purification of recombinant protein for immunization purposes.
One-liter cultures of pUNCH 1293 (dsrAI), pUNCH 1294 (dsrAII), and pUNCH 1295 (ncaAI) in BL21(DE3)pLysS were grown in liquid broth with appropriate antibiotics (Table 1). As previously described (14, 44), protein expression was induced from these plasmids with IPTG. The cultures were subsequently treated with rifampin to inhibit bacterial RNA polymerase. Inclusion bodies were isolated and washed extensively until the recombinant protein was >95% pure (data not shown).
Production of specific recombinant DsrAI, DsrAII, NcaAI, and HgbAI antibodies.
Antibodies used in this study are listed in Table 3. Female New Zealand White rabbits (Covance, Denver, Pa.) were immunized a total of four times by administration of 200 μg of rNt-DsrAI, rNt-DsrAII, rNt-NcaAII, or rFL-HgbAI (20) protein every 2 weeks. Freund's complete adjuvant was used for the initial immunization, whereas successive immunizations were in incomplete Freund's adjuvant. Sera were collected prior to the first immunization and 2 weeks following the final immunization.
TABLE 3.
Antibodies used in this study
| Name | Recombinant protein used as immunogen | Reference |
|---|---|---|
| Polyclonal antibodies (rabbit) | ||
| Anti-rFL-DsrAI | Mature full-length DsrA, strain 35000HP (class I), pUNCH 1250a | 14 |
| Anti-rFL-DltAI | Mature full-length DltA, strain 35000HP (class I), pUNCH 1286 | 33 |
| Anti-rFL-HgbAI | Mature full-length HgbA, strain 35000HP (class I), pUNCH 572 | 20 |
| Anti-rFL-HlpI | Mature full-length HlpA, strain 35000HP (class I) | 25 |
| Anti-rNt-DsrAI | N-terminal DsrA, strain 35000HP (class I), pUNCH 1293 | This study |
| Anti-rNt-DsrAII | N-terminal DsrA, strain CIP 542 ATCC (class II), pUNCH 1294 | This study |
| Anti-rNt-NcaAI | N-terminal NcaA, strain 35000HP (class I), pUNCH 1295 | This study |
| Anti-Omp | Outer membrane preparation, strain 35000HP (class I) | 24 |
| MAbs (Mouse) | ||
| 4.79 | Mature full-length DsrA, strain 35000HP (class I); pUNCH 1250 | This study |
| 2C7 | H. influenzae biogroup aegyptius | 34, 57 |
Subsequent to this publication, we found an error in the primer used to construct clone pUNCH 1250 that would result in a single amino acid change, A249G.
Production of MAbs.
MAbs were obtained as described by Patterson et al. (44), except that 5 μg of rFL-DsrAI protein (Table 3) (14) from strain 35000HP was used as the immunogen. The SBA Clonotyping System/AP (Southern Biotech, Birmingham, Ala.) was used to isotype antibodies from established murine hybridomas.
Protein alignments and analysis.
PRETTY BOX and BESTFIT (GCG Computer Group, Wisconsin) were used to generate protein lineups and similarity and identity scores (with a gap weight of 8) (Fig. 3 and 5). Possible signal peptidase I cleavage sites were predicted with LipoP 1.0 (Center for Biological Sequence Analysis, Lyngby, Denmark [www.cbs.dtu.dk/services]) (30).
FIG. 3.
Alignment of DsrAI and DsrAII amino acid sequences. The amino acid sequences of DsrA from 35000HP and DsrA from CIP 542 ATCC are compared. Boxed, shaded residues indicate identity. The first arrow indicates the possible signal peptidase I cleavage site; the second arrow indicates the end of the protein that was used for antibody production. The final 86 amino acids of the C termini are underlined.
Nucleotide sequence accession numbers.
Relevant DNA and deduced amino acid sequences have been submitted to GenBank, and accession numbers are listed in Table 5.
TABLE 5.
GenBank accession numbers for relevant DNA sequences presented in this study
| Strain | Accession no. or reference for:
|
|||
|---|---|---|---|---|
| DsrA | DltA | NcaA | HgbA | |
| 35000HP | 19 | AY371540 | 15 | 18 |
| HMC46 | AY606124 | NSa | NS | AY606118 |
| HMC56 | AY606125 | AY371542 | NS | AY606119 |
| HMC60 | AY606126 | NS | NS | AY606117 |
| C111 | NS | NS | NS | AY603049 |
| CIP 542 ATCC | AY606123 | AY371545 | AY606129 | AY606114 |
| DMC64 | AY606121 | AY371546 | AY612646 | AY606115 |
| DMC111 | AY612644 | AY371547 | AY612645 | AY603046 |
| SSMC71 | AY606120 | AY371548 | AY606128 | AY603048 |
| HMC112 | AY606122 | AY371549 | AY498730 | AY603047 |
NS, not sequenced.
RESULTS
Identification of a novel antigenically variant class of DsrA protein.
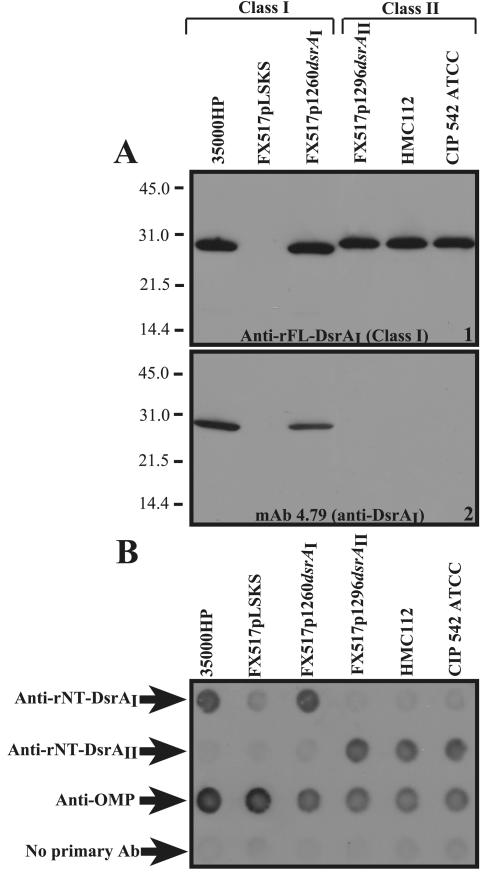
Routine examination of a polyclonal antibody made to the full-length, mature recombinant DsrA protein from 35000HP, rFL-DsrAI, showed that it bound to an approximately 30-kDa protein in strains HMC112 and CIP 542 ATCC in a Western blot (Fig. 2A, panel 1). We therefore hypothesized that the 30-kDa protein found in CIP 542 ATCC and HMC112 was also DsrA. However, the protein in strains CIP 542 ATCC and HMC112 was not recognized by MAb 4.79, also developed to rFL-DsrAI (Fig. 2A, panel 2), and migrated slightly more slowly than the DsrA from strain 35000HP (Fig. 2A, panel 1). This suggested that the DsrA protein from strains CIP 542 ATCC and HMC112 may be antigenicly different than that of strain 35000HP. For the purpose of this communication, we designated the DsrA from 35000HP DsrAI; DsrA from CIP 542 ATCC and HMC112 was designated DsrAII. We chose strain CIP 542 ATCC as the type strain for DsrAII-expressing strains.
FIG. 2.
Identification of antigenically variant DsrA proteins. (A) Two identical SDS-PAGE gels were prepared under reducing conditions with total cellular proteins from the indicated H. ducreyi strains and subjected to Western blotting. Panel 1, DsrA expression is detected using polyclonal anti-rFL-DsrAI made to rDsrAI from strain 35000HP; panel 2, DsrA expression is detected with MAb 4.79 made to rDsrAI from strain 35000HP. (B) H. ducreyi strains were examined for surface expression of class I and II DsrA in a dot blot format.
To determine if the 30-kDa protein in strain CIP 542 ATCC was DsrA, we attempted to construct an isogenic dsrA mutant of CIP 542 ATCC but were unsuccessful (described in Materials and Methods). As an alternative to testing DsrA expression and function in an isogenic mutant, we constructed plasmid pUNCH 1296 (dsrAII from CIP 542 ATCC) and expressed it in host strain FX517 (Table 1), a 35000HP dsrAI isogenic mutant. Like parent strains 35000HP and CIP 542 ATCC, FX517 expressing dsrAI from plasmid pUNCH 1260 or dsrAII from pUNCH 1296 was bound by rFL-DsrAI antibody (Fig. 2A, panel 1), while the MAb 4.79 only bound to FX517 expressing dsrAI (Fig. 2A, panel 2). Neither antibody bound strain FX517 harboring the empty vector pLSKS.
DsrAI was previously shown to be expressed at the surface of H. ducreyi cells (19). To determine if DsrAII was also surface exposed, H. ducreyi strains were examined in a whole-cell dot blot with anti-rDsrA and anti-OMP antibodies. H. ducreyi parent strains 35000HP, CIP 542 ATCC, and HMC112, as well as the dsrAI mutant strain FX517 expressing either class of dsrA or harboring an empty vector, were studied. Anti-rNt-DsrAI, specific for DsrAI, bound whole cells of 35000HP and FX517pUNCH 1260 (dsrAI), while anti-rNt-DsrAII, specific to the DsrAII protein, bound strains CIP 542 ATCC, HMC112, and FX517pUNCH 1296 (dsrAII) (Fig. 2B). Anti-OMP, a positive control antibody to the OMPs of H. ducreyi strain 35000HP (24), bound all strains tested, although it bound class II strains less strongly. Thus, DsrAII is expressed at the surface of H. ducreyi cells.
DNA sequence and deduced amino acid sequence of the H. ducreyi dsrAII locus from strain CIP 542 ATCC.
Since a polyclonal antibody to DsrAI bound to DsrAII from strains CIP 542 ATCC and HMC112 in a Western blot but MAb 4.79 did not, we predicted that there were both antigenically conserved and unique epitopes in the DsrA proteins of the two strain classes. Initial attempts to amplify dsrAII in class II strains using primers 14 and 24, previously used to amplify dsrA from 35000HP and eight other strains of H. ducreyi (19), were unsuccessful (Fig. 1). However, successful amplification occurred using primers 33 (located 975 bp upstream of the 35000HP dsrA start codon) and 24 (277 bp downstream of the dsrA stop codon). An approximately 2.5-kb PCR product was observed from CIP 542 ATCC (Fig. 1). The sequence of the dsrAII locus and 1,621 bp of flanking DNA (Table 5) were obtained from the PCR products. Putative promoter elements similar to the −35 (TTGACA) and −10 (TAGAAT) sites of dsrA from 35000HP were located 74 nucleotides (nt) (TTGACT) and 50 nt (TATAAT) upstream of the start codon, respectively, and separated by 18 nt. The manX (HD0768) gene, encoding a component of a putative mannose-specific uptake system (www.stdgen.lanl.gov), was located upstream of dsrA in strains 35000HP and CIP 542 ATCC (Fig. 1). The G+C content of the dsrAII DNA sequences was 40%, consistent with the AT-rich nature of Haemophilus spp. (3).
The deduced amino acid sequence of DsrAII predicted a protein of 31,646 Da. A possible signal peptidase I cleavage site was predicted to be LSA, unlike TMA in DsrAI from 35000HP (Fig. 3). Cleavage at this site would result in a mature protein of 28,880 Da.
The predicted amino acid sequence of the DsrAII protein was 47.8% identical and 56% similar to the DsrA protein of strain 35000HP. An amino acid lineup of these two DsrA proteins revealed significant variation in the N-terminal two-thirds of the protein (Fig. 3). The DsrAII signal peptide (the first 26 amino acids) from CIP 542 ATCC contained five MK repeats, whereas DsrAI (35000HP) had only one copy of MK. Despite the diversity noted at the N terminus, the C terminus was more highly conserved. The C-terminal sequence of class I and class II DsrAs contained a short motif consisting of alternating hydrophobic residues and ending with a phenylalanine, as is found in the majority of integral outer membrane proteins (OMPs) (60, 66). The final 86 amino acids (Fig. 3) were 88.5% identical and 89.7% similar in DsrAI and DsrAII.
Detection of variable DsrA expression in a panel of H. ducreyi clinical isolates.
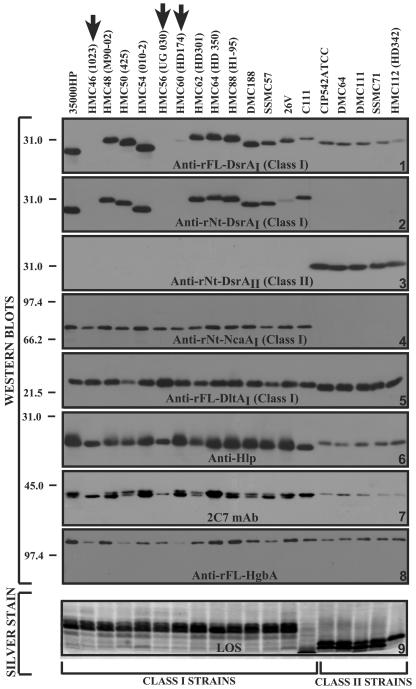
The identification of an antigenically variant DsrA in strain CIP 542 ATCC suggested that other strains of H. ducreyi might also express an alternate form(s) of DsrA. To examine this possibility, a panel of 19 geographically and temporally diverse isolates (Table 2) was chosen for further study. The panel of strains, including 35000HP (DsrAI) and CIP 542 ATCC (DsrAII) controls, was subjected to SDS-PAGE, followed by Western blotting with antibodies listed in Table 3 (Fig. 4). All strains examined reacted with polyclonal anti-rFL-DsrAI (Fig. 4, panel 1), albeit weakly by some, suggesting shared epitopes among the DsrA proteins expressed by these strains. However, when examined with the class-specific sera elicited to the N-terminal two-thirds of each DsrA protein, strains were placed into one of two categories: H. ducreyi strains that expressed a DsrAI protein or a DsrAII protein (Fig. 4, panels 2 and 3).
FIG. 4.
Identification of two classes of OMPs. Nine identically loaded SDS-PAGE gels (panels 1, 3, 6 and 7, 15% PAGE; panel 9, 12.5% PAGE; panels 2 and 5, 12% PAGE; panels 4 and 8, 7.5% PAGE) were prepared under reducing conditions with total cellular proteins or crude LOS from the indicated H. ducreyi strains and subjected to Western blotting, except for gel 9. Crude LOS in gel 9 was prepared as described in Materials and Methods and subjected to silver staining after electrophoresis. The class of each strain is indicated at the bottom of the figure. N-terminal antibodies were made to the N-terminal two-thirds of each recombinant protein (Table 3 and Fig. 1). Arrows indicate strains that reacted weakly with the antisera against DsrAI protein.
Strains HMC46, HMC56, and HMC60 reacted weakly with anti-rFL-DsrAI but did not react with either anti-rNt-DsrAI or anti-rNt-DsrAII sera (Fig. 4, panels 1 to 3). DNA sequencing revealed that the dsrAs from these three strains were identical (Table 5). Each strain contained two single nucleotide differences compared to strain 35000HP, one located between the putative −35 and −10 regions and the other within the −10 region of the promoter (data not shown). A comparison of these dsrAs with eight previously analyzed H. ducreyi isolates (19; data not shown) indicated that the predicted dsrA sequence from strain CHIA was identical to those of HMC46, HMC56, and HMC60 (Table 5). Based on nucleotide sequences, we concluded that HMC46, HMC56, and HMC60 were class I strains despite their weak expression of DsrAI.
H. ducreyi strains DMC64, DMC111, SSMC71, and HMC112 reacted with anti-rFL-DsrAI and anti-rNt-DsrAII, but not with anti-rNt-DsrAI antisera (Fig. 4, panels 1 to 3). Sequence comparison revealed that the dsrA genes from these strains were 100% identical to each other. When these sequences were compared to the dsrA from strain 35000HP, only 48% identity was observed. From these limited studies, we determined that the H. ducreyi strains studied here express either DsrAI or DsrAII proteins, but not both.
Identification of two classes of NcaA, another member of the Oca family from H. ducreyi.
Some bacteria express more than one Oca family member (48); hence, we examined H. ducreyi 35000HP for expression of additional Oca family proteins. A search of the genome sequence of strain 35000HP (http://www.stdgen.lanl.gov/stdgen/bacteria/hduc/) revealed a second member with weak similarity to DsrA (16% identity), termed NcaA (for necessary for collagen attachment A) (HD1920) (15).
To determine the prevalence of NcaA among DsrAI- and DsrAII-expressing strains, the panel of 19 H. ducreyi strains was examined in a Western blot with anti-rNt-NcaAI antiserum (Fig. 4, panel 4). The NcaA antiserum primarily recognized the oligomeric form (approximately 80 kDa) of the NcaA protein from class I but did not recognize either the oligomeric or monomeric (approximately 33-kDa) form of class II strains. Sequence comparison of NcaA from 35000HP (class I) and CIP 542 ATCC (class II) revealed strongly conserved C termini, with little conservation within the N termini (Fig. 5). In addition, a comparison of the NcaA sequence from CIP 542 ATCC with class II strains DMC64, DMC111, SSMC71, and HMC112 showed 100% identity within class II. This finding explained the lack of reactivity to class II NcaA with anti-rNt-NcaAI, since this N-terminal region was the source of immunogen used for antibody production of anti-rNcaAI (Fig. 5). Taken together, these data demonstrate that H. ducreyi strains express two Oca family members, DsrA and NcaA. Among isolates tested thus far, strains expressing DsrAI also express NcaAI, and those expressing DsrAII likewise express NcaAII.
Class I and class II strains express variants of other OMPs.
The detection of two antigenically diverse classes of DsrA and NcaA proteins in a panel of H. ducreyi strains raised the possibility that class II strains may express different versions of other outer membrane antigens. To test this possibility, the panel of 19 H. ducreyi strains was subjected to Western blotting and examined for expression of DltA (33), Hlp (for H. ducreyi lipoprotein) (25), major OMP (MOMP) and OmpA2 (24, 31), and HgbA (18, 59) (Fig. 4, panels 5 to 8). Leduc et al. (33) recently described DltA, a novel lectin required for the full expression of serum resistance phenotype in H. ducreyi strain 35000HP. Anti-rFL-DltA antiserum recognized DltA in all strains tested, although DltAs from class II strains migrated faster than DltAs from class I strains (Fig. 4, panel 5). Comparison of the predicted DltA sequences (Table 5) between five class I and five class II strains revealed that the sequences within each class were identical. However, comparison of the DltA sequences from class II strains with that of strain 35000HP revealed four amino acid differences between the classes: H25N, T40 M, R63H, and E74K (data not shown).
We next examined a Western blot of the 19 strains with an anti-Hlp antibody (25) and MAb 2C7. A putative Hlp protein was expressed in all isolates studied (Fig. 4, panel 6). However, in class II strains, the putative Hlp protein migrated more slowly than that of class I strains. MAb 2C7 was previously shown to recognize the MOMP and OmpA2 homologs as three bands with apparent molecular weights of 37, 39, and 43 kDa on SDS-PAGE gels (57). However, Klesney-Tait et al. (31) showed that depending on the SDS-PAGE system used, the 37- to 39-kDa bands may migrate as a single entity, resulting in the formation of two bands on the gel. Whole-cell preparations of H. ducreyi were subjected to SDS-PAGE with 15% PAGE to separate and visualize the bands in the 37- to 39-kDa range (Fig. 4, panel 7). Class I strains expressed only one or two of the bands in the 37- to 39-kDa range in different intensities, while only a single 2C7-reactive protein was recognized in all class II strains. Furthermore, there was a subtle, consistent mobility difference seen in the single band present in class II strains (Fig. 4, panel 7). PCR of the five class II strains with primers 3f12-3 and OmpA2-9, flanking the tandemly arranged MOMP and OmpA2 genes (31), gave products similar in size to those of 35000HP (data not shown). These data suggest that, although both classes of H. ducreyi appear to contain similar momp and ompA2 gene structures, expression of these proteins is different in class II strains. The slight difference in the mobility of MOMP and/or OmpA2 proteins seen in Western blots (Fig. 4, panel 7) also suggests that there are sequence differences between class I and class II strains.
To examine the expression of the hemoglobin receptor, HgbA, in H. ducreyi class I and class II strains, the panel of 19 strains was subjected to Western blotting with anti-rFL-HgbAI serum (Fig. 4, panel 8). HgbA was recognized in all strains tested. Sequence comparison of HgbA within class I strains revealed 97% similarity, and those within class II strains were 99% similar. Comparisons between class I and class II strains revealed 95.3% similarity (data not shown). These results suggest minimal variability of the HgbA amino acid sequence within or between class I and class II strains.
In addition to the OMPs, LOS was examined in these strains by SDS-PAGE and silver staining. As shown in Fig. 4, panel 9, LOS from class I strains, with the exception of strain C111, exhibited a slower-migrating LOS than that of class II strains. Based on the OMP and LOS expression patterns, these data indicate that at least two classes of H. ducreyi exist among isolates examined thus far.
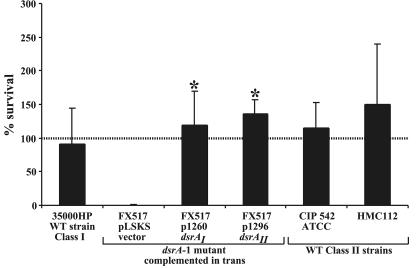
DsrAII mediates serum resistance.
It was previously shown that DsrAI is required for the H. ducreyi serum resistance phenotype (19). We determined that class II strains CIP 542 ATCC and HMC112 were also serum resistant, exhibiting >100% survival in 50% NHS (Fig. 6). In the absence of an isogenic mutant, we tested the ability of cloned dsrAI and dsrAII to confer serum resistance to the isogenic class I dsrA mutant strain FX517 harboring pUNCH 1260 (dsrAI), pUNCH 1296 (dsrAII) and pLSKS (empty vector) (Fig. 6). Expression of either dsrAI from pUNCH 1260 or dsrAII from pUNCH 1296 in host strain FX517 (the dsrAI mutant) conferred serum resistance to FX517, resulting in >100% survival (P = 0.017 and P = 0.001, respectively) (Fig. 6). In contrast, FX517 harboring an empty vector exhibited <5% survival.
FIG. 6.
Bactericidal killing of strains expressing class I and II DsrA. Bactericidal killing of H. ducreyi strains was performed in 50% NHS. The data are compiled from separate experiments done on at least three different days. The asterisk indicates statistically significant differences between the FX517 dsrA strain (35000HP isogenic dsrAI mutant) containing the empty vector pLSKS and FX517 dsrA complemented in trans with vectors expressing either dsrA class I (pUNCH 1260) or dsrA class II (pUNCH 1296) (P = 0.017 and P = 0.001, respectively). WT, wild type.
DISCUSSION
We propose that there are at least two distinct populations, termed classes here, of H. ducreyi strains that express variant forms of the OMPs DsrA, NcaA, DltA, Hlp, MOMP and/or OmpA2, as well as LOS. This conclusion is based upon the following evidence.
Findings. (i) The sequences of two Oca family members, DsrA and NcaA, differ between the two classes of H. ducreyi strains.
Comparison of the N termini of the DsrAI and DsrAII proteins revealed little sequence homology, while the C-terminal domains were nearly identical (Fig. 3). Within the leader sequence at the very N terminus of DsrAII were five MK (ATGAAA) repeats, whereas only one copy of MK existed in DsrAI (Fig. 3). The significance of this pentamer repeat is unclear, although it is conceivable that it may be involved in some form of phase variation resulting in an on/off phenotype in these strains. Transcription of dsrAII is predicted to begin with the first methionine (ATG) of the pentamer. Mutations changing the use to other ATGs would affect the spacing between the ribosome-binding site and the start of the translation, resulting in decreased expression of the protein. Indeed, such a mutation occurred in pUNCH 1288, which expresses DsrAII protein weakly (data not shown). Conversely, mutations may represent in vitro artifacts and additional studies are necessary before any conclusions can be drawn.
Another noticeable difference between the DsrA protein from H. ducreyi classes I and II was the presence of the heptameric repeat sequence NTHNINK in the class I strains (19) that was absent in the class II strains. We previously reported that class I strains contained one, two, or three of these heptameric repeats. The function of these repeats has yet to be determined.
Cole et al. (15) recently described NcaAI, a protein necessary for H. ducreyi binding to the ECM protein type I CN, in strain 35000HP. In this study, we described NcaAII, a variant of NcaAI, composed of a divergent N terminus but a highly conserved C-terminal domain. NcaAI was recently shown to be a virulence factor in the swine model of chancroid (15). Loss of NcaA expression in an isogenic mutant resulted in the decrease of the number of H. ducreyi culture-positive lesions and CFU recovered per lesion on day 7 of infection. Future studies need to be conducted to determine if NcaAII, like NcaAI, binds CN and whether NcaAII is important for virulence in these strains.
(ii) The sequence of the H. ducreyi lectin, DltA, differs between class I and class II strains.
Recently, our laboratory described a novel lipoprotein lectin, termed DltA, found in H. ducreyi strain 35000HP and involved in serum resistance (33). In this study, we determined that all strains tested express DltA and that class II strains possess a variant form of DltA that migrates at a different molecular weight in Western blots than does DltA from class I strains. When DltA was examined at the DNA and the predicted amino acid levels (Table 5), four conserved predicted amino acid differences were observed between the two classes of H. ducreyi strains, likely accounting for the variable mobility seen in Western blots (Fig. 4, panel 5).
(iii) Hlp protein migration differences.
There was a difference in migratory patterns in putative Hlp proteins, since Hlp from class II isolates migrated slower on SDS-PAGE than class I strains (Fig. 4, panel 6). Furthermore, the anti-Hlp antibody reacted weakly to the Hlp in class II strains even though all wells were loaded equally. More studies are needed to determine if differences at the sequence or expression level or in lipidation pattern are responsible for these differences.
(iv) MOMP/OmpA2 migration differences.
The H. ducreyi MOMP (24), comprises two homologs, MOMP and OmpA2. Spinola et al. (57) previously showed that both proteins are recognized by MAb 2C7. In this study, we demonstrated that MAb 2C7 recognized MOMP and/or OmpA2 from class I strains as one or two distinct bands in Western blots while only one, more slowly migrating band was detected in class II strains. Furthermore, MAb 2C7 reacted only weakly to the single band present in class II strains (Fig. 4, panel 7). Future studies may determine which, if either, of the two proteins is not expressed in the class II strains and whether differences exist within the momp and ompA2 gene sequences between strain classes.
(iv) Class I and class II strains express different species of LOS.
LOS may be a possible virulence factor for H. ducreyi and may play a role in pathogenesis of chancroid (12). Our studies have shown that class I strains, with the exception of C111, contain a slower-migrating LOS species typical of strain 35000HP. In contrast, class II LOS species migrate faster than 35000HP (Fig. 4, panel 8). Despite the difference in mobility of the LOS species, the class II strains CIP 542 ATCC and HMC112, expressing faster-migrating LOS, are as resistant to serum killing as 35000HP (Fig. 6). In support of these findings, our laboratory has determined that LOS mutants 35.252 (7) and 35.10 (6) (parent strain 35000HP), expressing severely truncated forms of LOS, resist killing in a bactericidal assay with 50% NHS (unpublished data). During the preparation of the manuscript, Scheffler et al. (51) analyzed the proteome of several strains of H. ducreyi that vary in their LOS structure by two-dimensional gel electrophoresis and mass spectrometry. They concluded that little proteome diversity existed in relation to the LOS of each strain. One exception, strain 33921, showed significant differences in its proteome map and synthesized an unusual LOS with a trisaccharide branch structure. Scheffler et al. (51) concluded that the studies did not preclude the possibility that significant variations exist among the proteomes of the strains tested. The differences seen with strain 33921 may suggest that this strain belongs to the second clonal population of H. ducreyi described here. Studies are under way to determine if strain 33921 is a class II strain.
Implications of two clonal populations of H. ducreyi.
In this report, we presented evidence that two major classes of H. ducreyi strains exist. In the course of this study, we observed that class II strains, CIP 542 ATCC and HMC112, grew more slowly than most class I strains and formed smaller colonies on solid medium (data not shown). These observations raised the possibility that the poor isolation rate of H. ducreyi from primary lesions (38) may in part be due to these differences in growth characteristics. Most importantly, class II strains may be markedly underrepresented in current culture collections compared to their presence in ulcers. Additional information is needed to address this concern.
PCR has been shown to be a more sensitive method of detection than culture (38). Our primer pairs, dsrA 14-dsrA 24, dsrA 42-dsrA 43, ncaA 0.08-ncaA 0.09, and ncaA 0.10-ncaA 0.11 (Table 4), are specific for either class I or class II dsrA or ncaA and may be useful in discriminating between an H. ducreyi class I- or class II-infected lesion. However, these primers have yet to be proven with material from primary lesions.
The discovery of two populations of H. ducreyi raises issues about vaccine development against chancroid. Several OMPs such as DsrA, NcaA, MOMP, hemolysin (43, 67), D15 (65), and HgbA have been suggested as possible vaccine candidates. The hemoglobin-binding OMP HgbA, also termed HupA (59), is a 100-kDa protein essential to the ability of H. ducreyi to utilize both hemoglobin and hemoglobin-haptoglobin as sources of heme (59). HgbA is also required for expression of full virulence in the temperature-dependent rabbit model (59) and the human model of chancroid (5). Patterson et al. (44) reported that HgbA was highly conserved, since three anti-HgbA Mabs bound to all 26 strains tested. The sole exception was MAb 4.23, which bound all strains except HMC112, identified as a class II strain in the present study. Here, we determined that the amino acid sequences of hgbA were strikingly similar between class I and class II strains, considering the degree of variability found in the Oca proteins. The high degree of hgbA sequence conservation between the classes suggests that a protective vaccine containing HgbA from either class I or class II may be efficacious against both classes. However, it is unclear if a monovalent vaccine designed against more variable H. ducreyi targets will be capable of inducing protection against both class I and class II strains.
DsrAI and DsrAII have similar functions.
Elkins et al. (19) previously showed that DsrAI expression was essential for the full serum resistance phenotype in nine strains of H. ducreyi. Serum resistance is an important virulence factor for H. ducreyi, since serum susceptible strains are avirulent (9, 40, 42). In the human infection model of chancroid, Bong et al. (9) demonstrated that inoculation with strains 35000HP and FX517 (the dsrAI mutant) resulted in the formation of papules. However, only subjects who received strain 35000HP developed infections that progressed to the pustular stage of infection. In this report, we demonstrated that both classes of DsrA proteins conferred serum resistance to type strains 35000HP (19) and CIP 542 ATCC in high concentrations of NHS (50%). Based on these observations, it is possible that inoculation with strains expressing DsrAII in the human model of chancroid may result in an infection similar to that observed with strain 35000HP. Studies are needed to confirm this hypothesis. These findings also suggest that the functional domains involved in serum resistance are possibly conserved within the C termini of both DsrAI and DsrAII proteins. Current studies will determine the specific sequences responsible for the serum resistance phenotypes, if DsrAII, like DsrAI, is involved in attachment to keratinocytes and ECM proteins, and whether DsrAII is important for virulence in the animal and human models of chancroid.
In conclusion, the discovery of variant forms of important OMP constituents in strains isolated from Africa and Asia over a period of decades provides strong evidence that two clonal populations of H. ducreyi exist. This data may strongly impact our understanding of the pathogenesis of chancroid, disease diagnosis, treatment, and vaccine development.
Acknowledgments
We thank Annice Roundtree for her outstanding technical support and Patricia Totten, Josef Bogaerts, William Albritton, Robert Munson, and Stanley Spinola for providing H. ducreyi strains, plasmids, and antibodies. Furthermore, we thank Robert S. Munson for providing the sequence of H. ducreyi strain 35000HP, Chris Thomas for help with figures and DNA analysis, and members of the Infectious Diseases Research Laboratory for reviewing the manuscript and providing invaluable suggestions. We especially thank Tom Kawula and Leah Cole for careful review of the manuscript and helpful suggestions. We also thank Walter E. Bollenbacher and Leslie Lerea for their ongoing support and encouragement.
This work was supported by grants AI 31496, R01-AI 53593-02 to C.E. and the Minority Opportunities in Research Division of the National Institute of General Medical Sciences grant GM000678. C.D.W. is currently a fellow in the Seeding Postdoctoral Innovators in Research and Education (SPIRE) program.
Editor: D. L. Burns
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albritton, W. L. 1989. Biology of Haemophilus ducreyi. Microbiol. Rev. 53:377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfa, M. J., and P. DeGagne. 1997. Attachment of Haemophilus ducreyi to human foreskin fibroblasts involves LOS and fibronectin. Microb. Pathog. 22:39-46 [DOI] [PubMed] [Google Scholar]
- 5.Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, B. A., S. R. Lumbley, and E. J. Hansen. 1999. Characterization of a WaaF (RfaF) homolog expressed by Haemophilus ducreyi. Infect. Immun. 67:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer, B. A., M. K. Stevens, and E. J. Hansen. 1998. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect. Immun. 66:4290-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bong, C. T., M. E. Bauer, and S. M. Spinola. 2002. Haemophilus ducreyi: clinical features, epidemiology, and prospects for disease control. Microbes Infect. 4:1141-1148. [DOI] [PubMed] [Google Scholar]
- 9.Bong, C. T., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 69:1488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bozue, J. A., L. Tarantino, and R. S. Munson, Jr. 1998. Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol. Lett. 164:269-273. [DOI] [PubMed] [Google Scholar]
- 11.Brentjens, R., M. Ketterer, M. Apicella, and S. Spinola. 1996. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J. Bacteriol. 178:808-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campagnari, A. A., L. M. Wild, G. E. Griffiths, R. J. Karalus, M. A. Wirth, and S. M. Spinola. 1991. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect. Immun. 59:2601-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbonetti, N., V. Simnad, C. Elkins, and P. F. Sparling. 1990. Construction of isogenic gonococci with variable porin structure: effects on susceptibility to human serum and antibiotics. Mol. Microbiol. 4:1009-1018. [DOI] [PubMed] [Google Scholar]
- 14.Cole, L. E., T. H. Kawula, K. L. Toeffer, and C. Elkins. 2002. The Haemophilus ducreyi serum resistance antigen, DsrA, binds vitronectin and confers attachment to human keratinocytes. Infect. Immun. 70:6158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole, L. E., R. A. Fulcher, K. L. Toffer, P. E. Orndorff, C. Elkins, and T. H. Kawula. Expression of Haemophilus ducreyi outer membrane protein NcaA is required for collagen I binding and virulence in swine model of chancroid, submitted for publication. [DOI] [PMC free article] [PubMed]
- 16.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Arico, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiCarlo, R. P., and D. H. Martin. 1997. The clinical diagnosis of genital ulcer disease in men. Clin. Infect. Dis. 25:292-298. [DOI] [PubMed] [Google Scholar]
- 18.Elkins, C. 1995. Identification and purification of a conserved heme-regulated hemoglobin-binding outer membrane protein from Haemophilus ducreyi. Infect. Immun. 63:1241-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins, C., K. Yi, B. Olsen, C. Thomas, K. Thomas, and S. Morse. 2000. Development of a serological test for Haemophilus ducreyi for seroprevalence studies. J. Clin. Microbiol. 38:1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emödy, L., J. Heesemann, H. Wolf-Watz, M. Skurnik, G. Kapperud, P. O'Toole, and T. Wadström. 1989. Binding to collagen by Yersinia enterocolitica and Yersinia pseudotuberculosis: evidence for yopA-mediated and chromosomally encoded mechanisms. J. Bacteriol. 171:6674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortney, K. R., R. S. Young, M. E. Bauer, B. P. Katz, A. F. Hood, R. S. Munson, Jr., and S. M. Spinola. 2000. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiltke, T., C. Lau, S. Gurunathan, A. Juarez, A. Campagnari, and S. Spinola. 1994. Presented at the 93rd General Meeting of the American Society for Microbiology, Las Vegas, Nev.
- 24.Hiltke, T. J., M. E. Bauer, J. Klesney-Tait, E. J. Hansen, R. S. Munson, Jr., and S. M. Spinola. 1999. Effect of normal and immune sera on Haemophilus ducreyi 35000HP and its isogenic MOMP and LOS mutants. Microb. Pathog. 26:93-102. [DOI] [PubMed] [Google Scholar]
- 25.Hiltke, T. J., A. A. Campagnari, and S. M. Spinola. 1996. Characterization of a novel lipoprotein expressed by Haemophilus ducreyi. Infect. Immun. 64:5047-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janowicz, D. M., K. R. Fortney, B. P. Katz, J. L. Latimer, K. Deng, E. J. Hansen, and S. M. Spinola. 2004. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infect. Immun. 72:4528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jessamine, P. G., and A. R. Ronald. 1990. Chancroid and the role of genital ulcer disease in the spread of human retroviruses. Med. Clin. North Am. 74:1417-1431. [DOI] [PubMed] [Google Scholar]
- 30.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klesney-Tait, J., T. J. Hiltke, I. Maciver, S. M. Spinola, J. D. Radolf, and E. J. Hansen. 1997. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J. Bacteriol. 179:1764-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langley, C. 1996. Update on chancroid: an important cause of genital ulcer disease. AIDS Patient Care STDS 10:221-226. [DOI] [PubMed] [Google Scholar]
- 33.Leduc, I., P. Richards, C. Davis, B. Schilling, and C. Elkins. 2004. A novel lectin, DltA, is required for expression of full serum resistance phenotype in Haemophilus ducreyi. Infect. Immun. 72:3418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesse, A. J., and W. E. Bittner. Unpublished data.
- 35.Lewis, D. A., J. Klesney-Tait, S. R. Lumbley, C. K. Ward, J. L. Latimer, C. A. Ison, and E. J. Hansen. 1999. Identification of the znuA-encoded periplasmic zinc transport protein of Haemophilus ducreyi. Infect. Immun. 67:5060-5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mertz, K. J., D. Trees, W. C. Levine, J. S. Lewis, B. Litchfield, K. S. Pettus, S. A. Morse, Michael E. St. Louis, J. B. Weiss, J. Schwebke, J. Dickes, R. Kee, J. Reynolds, D. Hutcheson, D. Green, I. Dyer, G. A. Richwald, J. Novotny, I. Weisfuse, M. Goldberg, J. A. O'Donnell, and R. Knaup. 1998. Etiology of genital ulcers and prevalence of human immunodeficiency virus coinfection in 10 US cities. J. Infect. Dis. 178:1795-1798. [DOI] [PubMed] [Google Scholar]
- 37.Morse, S. A. 1989. Chancroid and Haemophilus ducreyi. Clin. Microbiol. Rev. 2:137-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morse, S. A., D. L. Trees, Y. Htun, F. Radebe, K. A. Orle, Y. Dangor, C. M. Beck-Sague, S. Schmid, G. Fehler, J. B. Weiss, and R. C. Ballard. 1997. Comparison of clinical diagnosis and standard laboratory and molecular methods for the diagnosis of genital ulcer disease in Lesotho: association with human immunodeficiency virus infection. J. Infect. Dis. 175:583-589. [DOI] [PubMed] [Google Scholar]
- 39.Nika, J. R., J. L. Latimer, C. K. Ward, R. J. Blick, N. J. Wagner, L. D. Cope, G. G. Mahiras, R. S. Munson Jr., and E. J. Hansen. 2002. Haemophilus ducreyi requires the flp gene cluster for microcolony formation in vitro. Infect. Immun. 70:2965-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1987. Relationship between lipopolysaccharide composition and virulence of Haemophilus ducreyi. J. Med. Microbiol. 23:155-162. [DOI] [PubMed] [Google Scholar]
- 41.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1985. Role of lipopolysaccharide and complement in susceptibility of Haemophilus ducreyi to human serum. Infect. Immun. 50:495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1984. Virulence factors of Haemophilus ducreyi. Infect. Immun. 43:607-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer, K. L., and R. S. Munson. 1995. Cloning and characterization of the genes encoding haemolysin of Haemophilus ducreyi. Mol. Microbiol. 18:821-830. [DOI] [PubMed] [Google Scholar]
- 44.Patterson, K., B. Olsen, C. Thomas, D. Norn, M. Tam, and C. Elkins. 2002. Development of a rapid immunodiagnostic test for Haemophilus ducreyi. J. Clin. Microbiol. 40:3694-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purven, M., and T. Lagergard. 1992. Haemophilus ducreyi, a cytotoxin-producing bacterium. Infect. Immun. 60:1156-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosqvist, R., M. Skurnik, and H. Wolf-Watz. 1988. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature 334:522-524. [DOI] [PubMed] [Google Scholar]
- 48.Sandt, C. H., and C. W. Hill. 2000. Four different genes responsible for nonimmune immunoglobulin-binding activities within a single strain of Escherichia coli. Infect. Immun. 68:2205-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandt, C. H., and C. W. Hill. 2001. Nonimmune binding of human immunoglobulin A (IgA) and IgG Fc by distinct sequence segments of the EibF cell surface protein of Escherichia coli. Infect. Immun. 69:7293-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.San Mateo, L. R., M. M. Hobbs, and T. H. Kawula. 1998. Periplasmic copper-zinc superoxide dismutase protects Haemophilus ducreyi from exogenous superoxide. Mol. Microbiol. 27:391-404.I [DOI] [PubMed] [Google Scholar]
- 51.Scheffler, N. K., A. M. Falick, S. C. Hall, W. C. Ray, D. M. Post, R. S. Munson, Jr., and B. W. Gibson. 2003. Proteome of Haemophilus ducreyi by 2-D SDS-PAGE and mass spectrometry: strain variation, virulence, and carbohydrate expression. J. Proteome Res. 2:523-533. [DOI] [PubMed] [Google Scholar]
- 52.Schulze-Koops, H., H. Burkhardt, J. Heesemann, K. von der Mark, and F. Emmrich. 1992. Plasmid-encoded outer membrane protein YadA mediates specific binding of enteropathogenic yersiniae to various types of collagen. Infect. Immun. 60:2153-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 54.Spinola, S., T. Hiltke, K. Fortney, and K. Shanks. 1996. The conserved 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi has homology to PAL. Infect. Immun. 64:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spinola, S. M., M. E. Bauer, and R. S. Munson. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 70:1667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 71:7178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spinola, S. M., G. E. Griffiths, K. L. Shanks, and M. S. Blake. 1993. The major outer membrane protein of Haemophilus ducreyi is a member of the OmpA family of proteins. Infect. Immun. 61:1346-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spinola, S. M., A. Orazi, J. N. Arno, K. Fortney, P. Kotylo, C. Y. Chen, A. A. Campagnari, and A. F. Hood. 1996. Haemophilus ducreyi elicits a cutaneous infiltrate of CD4 cells during experimental human infection. J. Infect. Dis. 173:394-402. [DOI] [PubMed] [Google Scholar]
- 59.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Struyve, M., M. Moons, and J. Tommassen. 1991. Carboxyl-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 61.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 62.Tamm, A., A. M. Tarkkanen, T. K. Korhonen, P. Kuusela, P. Toivanen, and M. Skurnik. 1993. Hydrophobic domains affect the collagen-binding specificity and surface polymerization as well as the virulence potential of the YadA protein of Yersinia enterocolitica. Mol. Microbiol. 10:995-1011. [DOI] [PubMed] [Google Scholar]
- 63.Tertti, R., M. Skurnik, T. Vartio, and P. Kuusela. 1992. Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect. Immun. 60:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas, C. E., N. H. Carbonetti, and P. F. Sparling. 1996. Pseudo-transposition of a Tn5 derivative in Neisseria gonorrhoeae. FEMS Microbiol. Lett. 145:371-376. [DOI] [PubMed] [Google Scholar]
- 65.Thomas, K. L., I. Leduc, B. Olsen, C. E. Thomas, D. W. Cameron, and C. Elkins. 2001. Cloning, overexpression, purification, and immunobiology of an 85-kilodalton outer membrane protein from Haemophilus ducreyi. Infect. Immun. 69:4438-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tommassen, J., M. Struyve, and H. deCock. 1992. Export and assembly of bacterial outer membrane proteins. Antonie Leeuwenhoek 61:81-85. [DOI] [PubMed] [Google Scholar]
- 67.Totten, P. A., D. V. Norn, and W. E. Stamm. 1995. Characterization of the hemolytic activity of Haemophilus ducreyi. Infect. Immun. 63:4409-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Totten, P. A., and W. E. Stamm. 1994. Clear broth and plate media for the culture of Haemophilus ducreyi. J. Clin. Microbiol. 32:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsai, C.-M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 155:115-119. [DOI] [PubMed] [Google Scholar]
- 71.Ward, C. K., S. R. Lumbley, J. L. Latimer, L. D. Cope, and E. J. Hansen. 1998. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J. Bacteriol. 180:6013-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, C. K., J. R. Mock, and E. J. Hansen. 2004. The LspB protein is involved in the secretion of the LspA1 and LspA2 proteins by Haemophilus ducreyi. Infect. Immun. 72:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wood, G. E., S. M. Dutro, and P. A. Totten. 1999. Target cell range of the Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun. 67:3740-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]